Abstract
Cortical interneurons (GABAergic cells) arise during embryogenesis primarily from the medial and caudal ganglionic eminences (MGE and CGE, respectively) with a small population generated from the preoptic area (POA). Progenitors from the lateral ganglionic eminence (LGE) are thought to only generate GABAergic medium spiny neurons that populate the striatum and project to the globus pallidus. Here, we report evidence that neuronal precursors that express the LGE-specific transcription factor Islet1 (Isl1) can give rise to a small population of cortical interneurons. Lineage tracing and homozygous deletion of Nkx2.1 in Isl1 fate-mapped mice showed that neighboring MGE/POA-specific Nkx2.1 cells and LGE-specific Isl1 cells make both common and distinct lineal contributions towards cortical interneuron fate. Although the majority of cells had overlapping transcriptional domains between Nkx2.1 and Isl1, a population of Isl1-only derived cells also contributed to the adult cerebral cortex. The data indicate that Isl1-derived cells may originate from both the LGE and the adjacent LGE/MGE boundary regions to generate diverse neuronal progeny. Thus, a small population of neocortical interneurons appear to originate from Isl-1-positive precursors.
Keywords: genetic fate mapping, lateral ganglionic eminence, Islet1, interneuron, whole-cell physiology
Introduction
GABAergic interneurons account for ~25–30% of neurons in the mammalian cortex (Gabbott and Somogyi 1986; DeFelipe and Fariñas 1992; Beaulieu 1993; Tamamaki et al. 2003) and play a major role in many neurodevelopmental, epileptic, and psychiatric disorders (Lewis 2012; Galanopoulou 2013; Southwell et al. 2014). Cortical interneurons originate from defined germinal niches in the ventral forebrain and migrate to the cortex, possessing a wide range of laminar positions, morphologies, and physiological properties (Xu 2004; Wonders and Anderson 2006; Gelman et al. 2011; Rudy et al. 2011).
The ventral germinal niches lie within the ganglionic eminence (GE), which is traditionally subdivided into lateral, medial, and caudal eminences (LGE, MGE, and CGE, respectively) based upon anatomical location during development. Expression of specific transcription factors (TF) overlap within the physical structures (Flames et al. 2007) and the domains give rise to various GABAergic cell types. The MGE and CGE are the major source of neocortical interneurons (Stenman et al. 2003; Fogarty et al. 2007; Miyoshi et al. 2007; Xu et al. 2008). Nkx2.1+ progenitors in the MGE produce the majority of parvalbumin+ (PV+) and somatostatin+ (SST+) interneurons (Miyoshi et al. 2007; Xu et al. 2008), and CoupTF-II+ progenitors in the CGE generate calretinin/VIP/5-HT3AR+ neurons (Nery et al. 2002; Lee et al. 2010; Miyoshi et al. 2010). The LGE generates olfactory bulb (OB) interneurons derived from Er81+ neuronal precursor cells (Wichterle et al. 2001; Stenman et al. 2003; Kohwi et al. 2007) and striatal medium spiny neurons (MSNs) derived from Islet1+ cells (Deacon et al. 1994; Olsson et al. 1998; Wichterle et al. 2001; Ehrman et al. 2013; Lu et al. 2014).
A number of studies have attempted to clarify the full extent of cortical interneuron heterogeneity including the contribution of smaller, less characterized progenitor pools. A combination of dye labeling and, more recently, fate-mapping studies have demonstrated cortical interneuron contributions from Nkx2.1, Nkx6.2, CoupTF-II, and Prox1 expressing progenitor and precursor pools as the primary sources of cortical interneurons (Rudy et al. 2011; Miyoshi et al. 2015). Other studies of SP8+ migratory neuroblasts suggest the possibility that LGE-derived cortical interneurons may also be present in the cerebral cortex (de Carlos et al. 1996; Tamamaki et al. 1997; Pleasure et al. 2000; Anderson et al. 2001; Ma et al. 2012) since ER81, SP8, and ISL1 subdomains co-exist within the LGE and the LGE/CGE, and at the LGE/MGE boundary.
To determine whether or not other TF subdomains present within the developing basal ganglia produce a fraction of cortical interneurons, we used Isl1 and dual linage (Isl1 and Nkx2.1) fate-mapping to determine if Isl1 precursors contribute to the cortical interneuron population. Islet1 (Isl1) is a LIM-homeodomain TF expressed in the largest domain of the LGE proper (Karlsson et al. 1990; Pfaff et al. 1996; Cai et al. 2003; Elshatory et al. 2007) and in cells adjacent to the medial (MGE) and caudal (CGE) progenitor pools. In the mature CNS, ISL1 is primarily expressed by striatal MSNs and spinal cord motor neurons (Pfaff et al. 1996; Sun et al. 2008). In the present study a combination of genetic fate-mapping, live cell imaging, and electrophysiological analysis was used to demonstrate that a subset of Isl1-derived cells are present in the cerebral cortex and differentiate into mature fast-spiking GABAergic interneurons. Dual lineage fate-mapping of Isl1 and Nkx2.1-derived cells showed that loss of Nkx2.1 TF domains in the MGE and POA did not eliminate cortical Isl1-derived cells from the cortex, indicating that a subpopulation of Isl1 + precursors contribute to cortical development, which arise from a respecified SP8+ domain, most likely nested within the LGE boundaries.
Materials and Methods
Animals
Isl1cre/+ knock-in mice were used to fate map the complete lineage of the Islet1-derived population. Dr Catherine Lee-May (Children’s Hospital of Philadelphia, Philadelphia, PA) generously provided this mouse line following approval by Dr Sylvia Evans, in whose lab the mice were generated (Yang et al. 2006). BAC-Nkx2.1-Cre mice (Xu et al. 2008) were kindly provided by Dr Stewart Anderson (Children’s Hospital of Philadelphia, Philadelphia, PA). Nkx2.1GFP/+ mice (Longmire et al. 2012) were kindly provided by Dr Darrell Kotton (Boston University School of Medicine, Boston, MA). Floxed B6; 129S6-Gt (Rosa)26Sortm14(CAG-tdTomato)Hze/J reporter mice (Madisen et al. 2010) were purchased from Jackson Laboratory. Male Isl1cre/+ or BAC-Nkx2.1-Cre mice were crossed to female B6; 129S6-Gt (Rosa)26Sortm14(CAG-tdTomato)Hze/J reporter mice to generate the desired genetic recombination events. Male Nkx2.1GFP/+ mice were crossed to female Nkx2.1GFP/+ mice to generate Nkx2.1GFP/GFP, referred to as Nkx2.1 HD. Male Isl1cre/+:129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J mice were crossed to female Nkx2.1GFP/+ mice to generate Isl1cre/+:tdTomato; Nkx2.1GFP/+ knock-in mice. Male Nkx2.1GFP/+ were crossed to female B6; 129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J reporter mice to generate Nkx2.1GFP/+; 129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J progeny. Male Isl1cre/+; Nkx2.1GFP/+ mice were mated to female Nkx2.1GFP/+; 129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J to generate the desired genetic recombination events. All mice were maintained on a C57BL/6J background and the F1 generation was used in all experiments. Fate-labeled offspring were confirmed through visual detection of tdTomato+, GFP+ and genotyping by polymerase chain reaction (PCR). The authors declare that they have complied with all relevant ethical animal use regulations and that all animal studies were approved by the Children’s Hospital of Philadelphia Institutional Animal Care and Use Committee.
PCR-Based Genotyping
DNA was extracted from toe clips obtained from mice at the time of harvest. Isl1cre/+ knock-in mice were identified using the following primers: Islet1cre primer 1 (5′ ACT ATT TCG CAC CTA GCC ACA GCA 3′), Islet1cre primer 2 (5′ TCC CTG AAC ATG TCC ATC AGG TTC 3′), and Islet1cre primer 3 (5′ CCA AGC CCC ACA AGG TTA AA 3′) resulting in 300 bp for Cre and 150 bp for wildtype. B6; 129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J transgenic mice were identified using the following primers: wildtype forward (5′ AAG GGA GCT GCA GTG GAG TA 3′), wildtype reverse (5′ CCG AAA ATC TGT GGG AAG TC 3′), mutant forward (5′ GGC ATT AAA GCA GCG TAT CC 3′), and mutant reverse (5′ CTG TTC CTG TAC GGC ATG G 3′) resulting in a 297 bp wildtype product, a 196 bp mutant product and a combination for heterozygotes. BAC-Nkx2.1-Cre mice were identified using the following primers: Nkx2.1 intron 1 (5′ CCA CAG GCA CCC CAC AAA AAT G 3′) and (5′ GCC TGG CGA TCC CTG AAC AT 3′) in Cre, resulting in a 666 bp amplicon. Nkx2.1GFP/+ mice were identified using the following primers: transgene forward (5′-AAAGTAGCGAGGCTTCGCCTTCC) and transgene reverse (5′ GGA CTG GAT GCC GCA CGT CAC 3′).
Birth-Dating Experiments Using EdU Detection
For the staging of embryos, the morning of vaginal plug detection was designated embryonic day (E) 0.5, and the day of birth was considered postnatal day (P) 0. Pregnant dams were pulsed by a single injection at E12.5, E13.5, E14.5, or E15.5 via intraperitoneal injection of 50 ug/mg 5-ethynyl-2′-deoxyurdine (EdU) to label dividing cells. TdTomato+ animals were sacrificed and transcardially perfused at P21. For in-vivo LGE progenitor transplantation, pregnant dams were single pulsed at E14.0 90-min prior live extraction and transplantation. EdU label was detected using Click-iT EdU Alexa Fluor 647 Imaging Kit (Life Technologies C10458).
Slice Culture Model
Embryonic organotypic slice culture protocol was adapted from Polleux and Ghosh (2002). Isl1cre/+: tdTomato+/+ embryos were extracted via caesarian section at E14.5 and maintained on ice-cold 1X HBSS. TdTomato positive and negative mouse brains were embedded in 2.75% low melt agarose (IBI Scientific) and sectioned on a vibrating microtome (Leica VT-1000S, Leica Instruments) at 250 μm in the presence of oxygenated artificial cerebrospinal fluid (aCSF). Live brain sections were collected in ice-cold culture medium (DMEM +10% horse serum +5% FBS + 1% Pen/Strep +1% Glutamax) to preserve slice health while sections containing either the LGE or POA were chosen. TdTomato+ sections were matched to tdTomato− brain sections at similar positions along the rostral–caudal axis. TdTomato+ LGE or POA regions were microdissected using 350-μm sample corer and the explant was transplanted onto tdTomato− slices into either the LGE, MGE, or POA, depending upon experimental condition. To ensure that transplanted microdissected tissue was embedded into the slice, a small incision was made at the predetermined locations to accept the transplanted tissue. The transplanted brain section was transferred on to a poly-d-lysine and laminin coated cell culture insert (Millipore) within glass-bottom imaging chambers in culture medium. Lastly, 5% rat collagen gel (in 1X PBS) was placed over the full brain section to maintain tissue composition, stability, and adherence of microdissected tissue to host brain slice. Slices settled for 5-h before imaging chambers were moved from 37 °C incubator to an incubated Olympus FV10i scanning confocal microscope (Olympus Scientific Solutions) for live cell imaging. The transplanted brain hemisphere was imaged by selecting overlapping regions of interest. Confocal stacks were acquired with phase contrast and TRITC fluorescence to image tdTomato+ cells and the background slice using a 10X objective. The slices were imaged every hour for ~60 h. Movie z-projection reconstruction was performed offline using Olympus imaging software (Olympus Scientific Solutions) and National Institutes of Health ImageJ program. To conduct Isl1cre/+:tdTomato+ and Nkx2.1GFP/+ heterozygous experiments, the same procedure was followed as listed above, with the exception of exclusively harvesting LGE and transplanting onto littermate WT slices at the matched site. Confocal imaging (Leica SP8) was conducted at 5 h and subsequently at 43 h. Total GFP (green), tdTomato (red), and yellow (GFP + tdTomato) cells were quantified within 10 ROIs (250 × 250 pixel) within neocortex at the 43-h time point. Values within slice were determined and subsequently averaged across brain.
In Vivo Cell Transplantation
B6;129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J reporter females impregnated by Isl1cre/+ males were used in this experiment. Pregnant dams were EdU pulsed, as described above, 2-h prior to sacrifice. TdTomato+ embryos were extracted and microdissected as described in the slice culture protocol. Microdissection was performed to isolate the ventricular (VZ) and subventricular zone (SVZ) of the LGE using morphological landmarks, which differentiated germinal zones from non-germinal, mantle zone. Isolated tissue from multiple tdTomato+ embryos was gathered on ice-cold medium (Hibernate-E + 2% B27 + 1% Glutamax). Microdissected tissue was transferred to trypsin +0.05% EDTA solution for ~10 min at 37 °C. Tissue was dissociated and an equal proportion of trypsin inhibitor was added to suspension. Cells were centrifuged at 800 RPM for 10 min and resuspended in medium (NBM + 2% B27+ 1% Y-27632 dihydrochloride) and quantified. Cell suspension containing 50 000 cells/μL was transplanted into somatosensory cortex of wild type C57/BL6 neonatal mice between the ages of P1–P3. Briefly, pups were anesthetized using ice for ~5 min. Mice were kept on a Styrofoam mold and secured in place with pinned parafilm cover. Pulled and beveled glass pipettes attached to Drummond “Nanoinject II” (Cat. 3-000-204) were used to transplant ~150 000 cells (3 μL) bilaterally into somatosensory cortex of neonates. Pups were allowed to recover on a heating pad for 10–15 min and returned to mother. Transplanted pups were allowed to survive until P15, at which time they were sacrificed and tissue was processed using the Click-iT EdU Alexa Fluor 647 Imaging Kit.
Tissue Processing and Immunohistochemistry
Embryonic time points were harvested by caesarean section and immersion fixed for 1–3 h at 4 °C in 4% paraformaldehyde (PFA). Postnatal time points were fixed with 4% PFA using transcardial perfusion and subsequently postfixed overnight at 4 °C. All brains were washed in 1X phosphate-buffered saline (PBS) before tissue sectioning. Brains were cryoprotected in 30% sucrose in 1X PBS and sectioned at 25 μm on a cryostat (Leica CM3050-S, Leica Instruments) or embedded in 2% agarose and sectioned at 50 μm on a vibrating microtome (Leica VT-1000S, Leica Instruments). Primary antibodies were used at the following concentrations: rabbit anti-calbindin (1:200; Swant), goat anti-calretinin (1:2000; Chemicon), goat anti-CHAT (1:200; Chemicon), mouse antihuman COUP-TFII (1:1000; Perseus Proteomics & 1:500; Abcam), mouse anti-CRE (1:800; Chemicon), rabbit anti-CRE (1:1000; Novagen), rabbit anti-CTIP2 (1:500; Abcam), goat anti-DCX (1:200; Santa Cruz Biotechnology), rabbit anti-dsRed (1:1000; Clonetech), rat anti-dsRed (1:800; Chromotek), rabbit anti-ER81 (1:1000; gift from T. Jessell Lab), mouse anti-GAD67 (1:200; Millipore), chicken anti-GFP (1:1000; Invitrogen), mouse anti-GFP (1:1000; Millipore), mouse anti-ISL1 (1:250 [40.2D6] & 1:200 [39.3F7]; Developmental Studies Hybridoma Bank), rabbit anti-ISL1 (1:200; Abcam), rabbit anti-Ki67 (1:100; Chemicon), rabbit anti-NPY (1:2000; Abcam), mouse anti-parvalbumin (1:1000; Chemicon), rabbit anti-somatostatin (1:250; Millipore), rat anti-somatostatin (1:150; Chemicon), goat anti-SP8 (1:500; Santa Cruz Biotechnology), mouse anti-TH (1:200; Millipore), rabbit anti-TTF1 (1:1000; Abcam), and rabbit anti-VIP (1:400; Immunostar). The following Alexa Fluor secondary antibodies were used to detect immunofluorescence: goat anti-rabbit 488, goat anti-rabbit 568, goat anti-rabbit 647, goat anti-mouse 488, goat anti-mouse 647, donkey anti-chicken 488, donkey anti-goat 488, donkey anti-goat 647, donkey anti-rabbit 488, donkey anti-rabbit 647, donkey anti-rat 568, and donkey anti-mouse 647.
Imaging and Analysis
Fluorescence microscopy images were acquired using an epifluorescence Leica DM6000B upright microscope with a high-resolution monochrome CCD (Leica Instruments) or Olympus FV1000 confocal microscope (Olympus Scientific Solutions), Leica SP8 confocal microscope or Olympus Fluoview FV10i live imaging confocal. Images were acquired at 512 × 512 or 1024 × 1024 pixel dimensions and were processed offline for contrast and brightness in Adobe Photoshop CS6 Extended (Adobe Systems Inc.). GFP+ or TdTomato+ cells were co-stained with appropriate markers to determine immunohistochemical identity of labeled cells. Only labeled cells with the morphology of neurons were quantified in statistical analyses. TdTomato+ cells with small soma and processes of astrocytes were omitted from analysis. Cells were counted using ImageJ and the average per section was calculated. All data are expressed as mean SEM and statistical analyses were performed using GraphPad Prism.
Analysis of organotypic slice culture explants was performed using ImageJ (NIH). Z-sections of confocal stack were resliced to include only ROIs with tdTomato+ cells after which maximum projection images were reassembled. Single z-section of phase image with optimal neuroanatomy was used to generate merged tdTomato+ and phase image per time point. Concatenated tdTomato and phase images were generated for each ROI for the duration of imaging experiment. ROIs were stitched together using pairwise stitching plug-in (Preibisch et al. 2009). Movies were generated and isolated time points were generated for figure production.
Electrophysiology
P15-21 mice were deeply anesthetized with isoflurane, decapitated, and the brains were quickly removed and immersed in ice-cold pre-oxygenated aCSF solution with sucrose containing in (mM): sucrose 234, KCl 2.5, MgSO4 10, NaH2PO4 1.25, CaCl2, 0.5, NaHCO3 26, and glucose 11. Transverse brain slices (350 μm) were acutely prepared using a vibrating microtome (Leica VT-1000S, Leica Instruments). Slices containing the cerebral cortex were transferred into an incubation chamber containing aCSF (in mM): NaCl 128, KCl 2.5, CaCl2 2.5, MgSO4, 2, NaHCO3 24, NaH2PO4 1.4, and d-glucose 13 at 37 °C for 45 min. Slices were further incubated at room temperature for at least 1 h in aCSF until recording. For recordings, the slices were transferred into a submerged recording chamber and continuously suffused with aCSF (~2 mL/min) at 31–32 °C. All solutions were continuously bubbled with carbogen gas (95% CO2, 5% O2).
Whole-cell voltage recordings were made from tdTomato+ cortical interneurons visually identified with fluorescence and infrared differential interference contrast video microscopy using an Olympus BX51WI microscope (Olympus Scientific Solutions), equipped with a 40X water-immersion objective and a Rolera-XR digital camera (Q-Imaging). Patch pipettes (4–6 ΜΩ) were pulled from thick borosilicate glass (WPI) and filled with (in mM) K-gluconate 135, KCl 5, NaCl 2, HEPES 10, EGTA 4, MgATP 4, and Na3GTP 0.3 (pH ≈ 7.2; 285 mosmol) for current-clamp and voltage-clamp recordings. Recordings were made using a Multi-Clamp 700B amplifier (Molecular Devices) and data were filtered at 10 kHz and sampled at 20 kHz with a Digidata 1440 interface (Molecular Devices). Liquid junction potential was corrected prior to GigaOhm seal formation, whereas access resistance and capacitance were compensated immediately after going whole-cell. Recordings were accepted if series resistance was < 25 MΩ, action potentials were overshooting beyond +10 mV, electrode resistance changed by <10% during the experiment, and resting membrane potential (RMP) remained more negative than −40 mV. Synaptic recordings of spontaneous and miniature excitatory/inhibitory currents were sampled for 5 min on each cell in voltage clamp mode following acquisition of intrinsic membrane data. For synaptic recordings, the holding potential was set at −40 mV for simultaneous recordings of excitatory (inward) and inhibitory (outward) spontaneous currents (Zhou et al. 2009). All recordings were performed at 31–32 °C.
Membrane properties of Islet1-derived tdTomato+ neurons were characterized in response to hyperpolarizing and depolarizing rectangular current pulses. Current steps were applied for 500 ms in decreasing 10- to 20-pA steps at 0.5 Hz. Membrane data were analyzed with ClampFit 10 (Molecular Devices) and Minianalysis (Synaptosoft). The following parameters were measured: RMP, membrane resistance, membrane time constant, spike threshold potential, action potential amplitude, duration of the action potential at its half amplitude, after-hyperpolarization (AHP) amplitude, and firing frequency. All AP measures were taken from the first AP of the first sweep that reached the spike threshold (Rheobase). Input resistance (Rin) was measured from the slope of a linear regression fit to the voltage–current relation in a hyperpolarizing range relative to the RMP. The membrane time constant was determined by single-exponential fitting to the average voltage responses activated by hyperpolarizing current steps (10–20 pA). Isl1-derived interneurons were classified electrophysiologically based on the nomenclature previously described for interneurons (Miyoshi et al. 2007, 2010; Ascoli et al. 2008; Lee et al. 2010). Excitatory and inhibitory synaptic currents were analyzed for amplitude and frequency using MiniAnalysis. Peak events were first detected automatically using an amplitude threshold of 2 times the average RMS noise and rechecked by visual inspection to remove false events. Frequencies of spontaneous and miniature EPSCs and IPSC events were determined over 5 min for each cell.
In Situ Hybridization
Digoxigenin-labeled RNA probe for Nkx5.1 (kindly provided by D. Epstein, University of Pennsylvania, Philadelphia, PA) was generated from a pBluescript plasmid with a DNA template insert. The plasmid was linearized with restriction enzyme and cleaned with phenol-chloroform. The RNA probe was then labeled and amplified with T3 polymerase in a 1× mixture of digoxigenin-labeled dNTPs, RNasin, DTT, and enzyme buffer. The DNA template was digested with DNaseI before the probe was concentrated and resuspended in RNase-free water for in-situ hybridization (ISH) staining. Twenty-five-micrometer coronal sections were cut on a cryostat (Leica CM3050-S, Leica Instruments, Nussloch, Germany) and stained as described previously (Simonet et al. 2015).
Results
ISL-Expressing Cells Represent a Distinct Subset of LGE Precursors
Immunostaining for ISL1 at E12.5 showed 2 separate populations, one confined to the mantle zone of LGE, with another extending into developing hypothalamus (see Supplementary Fig. 1a). Subsequently, ISL1 expression expands ventrally into the septal commissure and POA (see Supplementary Fig. 1b,c). In sharp contrast, analysis of ISH data from Allen Developing Mouse Brain Atlas of adult mouse brains sections shows a dramatic reduction in Isl1 mRNA in basal ganglia with a few positive cells in the subthalamus (Henry and Hohmann 2012). This indicated that Isl1 expression is globally downregulated in postnatal forebrain structures and permanent fate-mapping was required to track Isl1-derived cells in situ.
To achieve this, Isl1cre/+ knock-in mice (Liang et al. 2011) were crossed with 129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J reporter mice (Fig. 1a). Cre/Lox driven recombination resulted in labeling of all Isl1-derived cell lineages throughout the developing embryo. Even after downregulation of CRE recombinase, floxed CAG-tdTomato expression was visualized in known precursor pools and progeny (Fig. 1). The experiments showed no indication of leaky CRE expression (Fig. 1b, sagittal section) beyond established cell populations (see Supplementary Fig. 2) in the developing embryo, consistent with an earlier report (Liang et al. 2011).
Figure 1 .
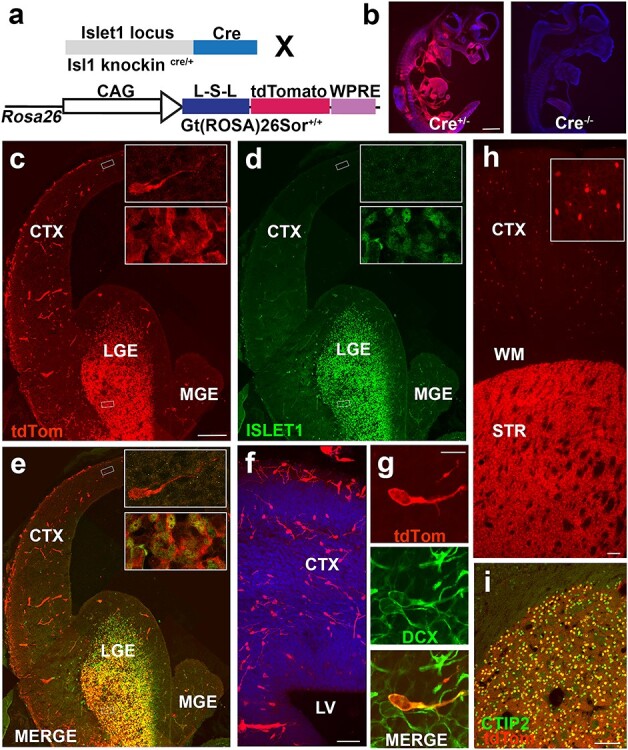
A subset of Isl1-derived tdTomato+ neurons tangentially migrate into developing neocortex. (a) Schematic of cross to identify lineage of Isl1 precursor cells. (b) TdTomato+ expression at E11.5 in Is1cre/+; 129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J and Isl1+/+ and 129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J mice indicate no tdTomato expression was observed in the absence of the Cre allele. (c–e) E14.5 mouse brain section indicating areas of (c) Isl1-derived tdTomato+ cells (d) ISL1, and co-expression (e) between the 2 populations in developing LGE. Insets depict example of bipolar migratory neuron in neocortex (upper inset) and cells in developing striatum (lower inset) corresponding with boxed regions in low mag. (f) Migrating tdTomato+ neurons crossing corticostriatal junction. Leading apical processes indicate direction of migratory stream from GE into neocortex. (g) TdTomato+ cell is co-positive for DCX confirming migratory immature neuron identity. (h) Low magnification image of tdTomato+ coronal section at P21 showing cortical and striatal labeled cells. Inset shows high magnification image of tdTomato+ neurons in cerebral cortex. (i) All tdTomato+ cells in striatum are CTIP2+ indicating a medium-sized spiny striatal projection neuron identity. CTX—cerebral cortex; LGE—lateral ganglionic eminence; MGE—medial ganglionic eminence; LV—lateral ventricle; WM—white matter; STR—striatum. Scale bars represent 1000 μm (b); 200 μm (c–e); 50 μm (f); 10 μm (g) ; 100 μm (h) and 100 μm (i).
Isl1-Derived Progeny Migrate throughout the Brain, Including Cerebral Cortex
To determine the fate of Isl1-derived cells in forebrain development, Isl1cre/+; tdTomato mice were analyzed at mid-neurogenesis. At E14.5, tdTomato+ cells were seen throughout the outer SVZ and mantle zone of the LGE (Fig. 1c–e) with robust overlap of ISL1 protein within the LGE (Fig. 1e, lower inset). Some tdTomato+ expression was also seen in blood vessels (Sun et al. 2007) but they were morphologically distinguishable from precursor cells. In addition, scattered bipolar cells, which were tdTomato+ but ISL1−, were observed throughout the neocortex (Fig. 1e, upper inset). These tdTomato+ cells appeared to migrate tangentially from basal ganglia to developing neocortex (Fig. 1f) and expressed DCX—a marker of immature migratory neuronal fate (Fig. 1g).
At P21, some Isl1cre/+:tdTomato+ cells were present in the cerebral cortex with dense expression in the striatum (Fig. 1h). Striatal cells were found to be CTIP+ (Fig. 1i)—a marker consistent with GABAergic MSN fates (Ehrman et al. 2013).
ISL1+ Cells Remain Largely Separate from Tangentially Migrating Nkx2.1-Derived Progeny during Subpallial Neurogenesis
The fate-mapping results demonstrated that Isl1-derived progeny migrate from subpallium to diverse forebrain structures, including the neocortex. This result may indicate either that some ISL1-expressing cells develop into cortical interneurons or that ectopic expression of ISL1 (through Cre-mediated recombination) may persist within MGE-derived neuroblasts, since the migrating MGE precursor cells must tangentially migrate past ISL1+ corridor cells within the LGE (López-Bendito et al. 2006; Simpson et al. 2009). To directly test the latter possibility, BAC-Nkx2.1-Cre transgenic mice (Xu et al. 2008) were crossed to 129S6-Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J reporter mice (Fig. 2a) to permanently fate-map Nkx2.1-derived progeny.
Figure 4 .
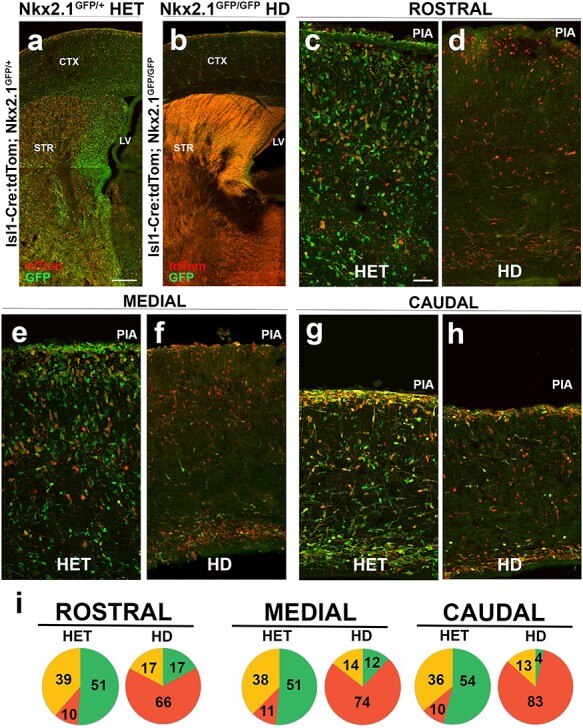
Loss of Nkx2.1 is dispensable for cortical interneuron identity of Isl1-derived precursors. (a) Isl1cre/+:tdTomato; Nkx2.1GFP/+ heterozygous and (b) Isl1cre/+:tdTomato; Nkx2.1GFP/GFP HD mice were sacrificed at E18.5 for comparison. Heterozygous mice showed tdTomato+ only (red), GFP+ only (green), and combined expressing (yellow) cells. GFP+ and tdTomato+ subcortical populations were mostly segregated in heterozygous mice. In contrast, HD mice showed a marked absence of GFP+ cortical cells with a shift to substantial overlap amongst GFP+/tdTomato+ (yellow) cells in basal ganglia. (c–h) E18.5 rostral (c,d), medial (e,f), and caudal (g,h) coronal sections. (i) Pie graphs show relative contribution of GFP+ only, tdTomato+ only and yellow cells within neocortex across conditions. For quantification, refer to text. One-way ANOVA P < 0.01 [rostral], P < 0.002 [medial], and P < 0.05 [caudal] was conducted across sample. N = 6 for pie graphs. Scale bars represent 200 μm (a,b) and 50 μm (c–h).
Since tangential migration from MGE and CGE occurs between E12.5 and E16.5 (Miyoshi and Fishell 2011), BAC-Nkx2.1-Cre:tdTomato+ brains were analyzed at E12.5, E13.5, and E14.5 during neural development. Labeled sections were stained with antibodies against ISL1, NKX2.1, and dsRed. At E12.5, a distinct tangential migratory stream of tdTomato+ cells began to emerge from NKX2.1 immmunopositive progenitors (blue) of the MGE (Fig. 2b and c). Most tdTomato+ cells were NKX2.1 negative, reflecting either a sharp downregulation in Nkx2.1 or short temporal dynamics of CRE expression in MGE progenitors. Nkx2.1-Cre: tdTomato+ neuroblasts migrated past ISL1+ nuclei passing through the LGE without evidence of co-labeling. At E13.5 and E14.5, a larger tdTomato-labeled tangential migratory stream was visible in the GE leading into the neocortex (Fig. 2d, inset and Fig. 2e, inset). The vast majority of tdTomato+ neurons were observed adjacent to densely populated ISL1+ nuclei (green) within the LGE, with only rare examples of tdTomato+/NKX2.1+ and/or ISL1 co-positive cells (Fig. 2d,e). Co-expression of NKX2.1+/ISL+ cells, as demonstrated here, has been previously reported during globus pallidus production (Flandin et al. 2010). NKX2.1 expression extends into the VZ of the hypothalamus/POA (Sussel et al. 1999), a structure known to generate heterogeneous subsets of cortical interneurons (Gelman et al. 2011).
To determine whether or not NKX2.1+ (blue) progenitors overlapped with ISL1+ nuclei (green) in the POA at E14.5 (Fig. 2g), ISL1 expression in the hypothalamic regions during early neurogenesis was assessed (see Supplementary Fig. 1a). The data showed little overlap in expression, which was consistent with NKX2.1 expression in germinal zones and ISL1 expression in the mantle zone. Thus, Isl1-derived progeny appeared to segregate into at least 2 subsets: a small minority that showed overlap with NKX2.1 and the vast majority that remained independent of NKX2.1. While NKX2.1 is associated with striatal, septal and globus pallidus fates (Magno et al. 2009), ISL1 expression in the subpallium is associated with cholinergic and striatal MSN fates (Fragkouli et al. 2009).
Loss of NKX2.1 Is Dispensable for Production of Isl1-Derived Cortical Neurons
To resolve whether or not Isl1-derived cortical cells share a common lineage with Nkx2.1, we performed complementary fate-labeling in Nkx2.1GFP/+ reporter mice (Longmire et al. 2012) and Is1cre/+ knock-in mice. In Nkx2.1GFP/+ reporter mice, eGFP is targeted to one allele of endogenous Nkx2.1 locus by homologous recombination, thereby replacing one copy of Nkx2.1 to generate a functional heterozygous knock-in mouse line with GFP labeling in forebrain structures of Nkx2.1-derived progeny. The fidelity of reporter gene expression was tested in Nkx2.1GFP/+ mouse embryos at W18.5 (n = 5), a time point when tangential migration is largely complete (Miyoshi and Fishell 2011). Medial hemi-sections of Nkx2.1GFP/+ heterozygous (HET) mice showed robust GFP expression spanning the cerebral hemisphere (Fig. 3a) with densely populated GFP+ cells found in the neocortex (Fig. 3a, upper boxed inset) and widely distributed in basal ganglia. The morphologies of the positive cortical cells were consistent with immature neurons. Subpallial NKX2.1 IHC showed scattered co-staining with GFP+ cells indicating normal brain development (Fig 3c,d,e). Quantification of co-labeled neurons within the striatum of Nkx2.1GFP/+ HET animals (n = 4) showed a combination of GFP+/NKX2.1 immuno-positive (42.5% ± 6.3) co-labeled cells, 40.1 ± 5.3 Nkx2.1GFP/+ only labeled striatal neurons, and 17.2% ± 3.4 NKX2.1 only immuno-positive cells within the striatum. Therefore, the majorities of labeled striatal cells either co-expressed GFP and NKX2.1 protein, which is consistent with heterozygous expression of NKX2.1 within animals, or were found to be GFP expressing only, suggesting NKX2.1 expression may have begun to be downregulated within some postmitotic neurons as cells differentiated. The remaining NKX2.1 only cells were the smallest component of the population and likely show MGE-derived cells that had yet to express GFP, had incomplete fate labeling, or had dim expression not detectable by low magnification visualization.
Figure 3 .
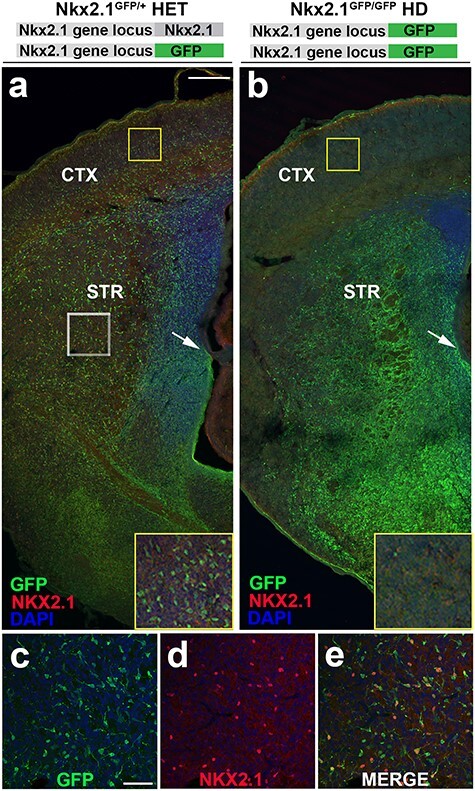
Residual GFP reporter retained in Nkx2.1 heterozygous and HD mice serves to fate map Nkx2.1-derived cells. (a,b) Low magnification E18.5 hemi-section of Nkx2.1GFP/+ heterozygous and Nkx2.1GFP/GFP HD mice. (a) Nkx2.1GFP/+ HET brain slice co-stained with NKX2.1 antibody and DAPI shows widely distributed precursor cells across neocortex and basal ganglia. Yellow boxed inset shows high magnification of GFP+ cells occupying neocortex with immature neuronal morphology. White boxed region shown in (c–e) reveal numerous GFP+ neuroblasts are immmunopositive for NKX2.1, consistent with normal NKX2.1 expression in heterozygous animals. In contrast, (b) Nkx2.1GFP/GFP HD mice reveal near complete absence of GFP+ cells from neocortex (yellow boxed inset) with concomitant increase in overall GFP expression in developing basal ganglia. Immunohistochemistry shows almost complete loss of NKX2.1 expression consistent with global knockout resulting from GFP insertion into both Nkx2.1 gene loci. HET—heterozygous and HD—homozygous deletion. Scale bars represent 200 μm (a,b) and 50 μm (c–e).
Nkx2.1GFP/+ HET mice were crossed to each other to produce offspring with 2 copies of GFP inserted into the Nkx2.1 locus; they thus lacked functional Nkx2.1 transcription to produce a homozygous deletion (HD) of Nkx2.1 (Fig. 3b; n = 3). These Nkx2.1GFP/GFP HD animals showed bright GFP positivity in the basal ganglia with subcortical cells that were nearly indistinguishable from one another (Fig. 3b), indicating a marked abundance compared with HETs. The size of the MGE appeared to be greatly diminished, as reflected by the loss of a visible boundary between LGE and MGE (arrow in 3b). Further, cortical GFP+ cells were greatly reduced in numbers, with only scattered cells present (box in Fig. 3b). Finally, IHC for NKX2.1 concurrently showed a near complete loss of NKX2.1 protein expression in the Nkx2.1GFP/GFP HD mice, consistent with a global knockout line. Quantification of cells within the striatum was not feasible due to increased cell density and lack of discernable GFP+ soma, which had been clearly visible in the HET condition.
Isl1cre/+;tdTomato mice were crossed with Nkx2.1GFP/+ HET mice to produce Isl1cre/+;tdTomato; Nkx2.1GFP/+ amongst the mixed offspring. Interneuron migration and expression of ISL1 was assessed at E13.5 in Isl1cre/+; tdTomato; Nkx2.1GFP/+ HET mice to evaluate neurogenesis and simultaneous tangential migration of Isl1cre/+; tdTomato and Nkx2.1GFP/+ subsets across the rostral–caudal axis (see Supplementary Fig. 3a,d,g). This aspect of brain development was found normal in the sections examined. Additionally, we assessed precursor heterogeneity at the LGE/MGE border regions since the LGE & MGE clades fuse together (see Supplementary Fig. 3j–l), therefore mixed precursor identities may exist and could account for co-labeling visualized in Fig. 2d,e. Our data indicate that 4 subpopulations of precursors express ISL1 protein at the LGE/MGE border: exclusively ISL1+ nuclei (blue); ISL1+/Nkx2.1GFP/+ (cyan); ISL1+/Isl1cre/+; tdTomato (magenta); and ISL1+/Isl1cre/+;tdTomato; Nkx2.1GFP/+ (gray). Quantification within single z-sections were conducted across rostral (n = 5), medial (n = 4), and caudal (n = 5) regions at E13.5. Blue-labeled cells were 31.8% ± 4.7 (rostral), 31.3% ± 1.7 (medial), and 34.0% ± 2.7 (caudal). Cyan-labeled cells were 3.0% ± 0.9 (rostral), 2.2% ± 0.5 (medial), and 1.1% ± 0.3 (caudal). Magenta-labeled cells were 53.5% ± 6.3 (rostral), 58.2% ± 2.1 (medial), and 57.2% ± 2.9 (caudal). Gray-labeled cells were 11.6% ± 1.7 (rostral), 8.3% ± 1.6 (medial), and 7.7% ± 0.4 (caudal). Overall, the data indicate that the majority of the cells (~56%) were ISL1+/Isl1cre/+; tdTomato, consistent with the data in Figure 1. ISL1+ only cells represented ~1/3 of the total labeled precursor pool suggesting that some cells had yet to proceed through Isl1-driven Cre/loxP recombination. Mixed lineage Nkx2.1/Isl1 cells represented only ~ 9% of the total co-labeled ISL1+ precursor pool suggesting that mixed lineages are relatively uncommon at the LGE/MGE border at E13.5. These populations may be larger at earlier stages of subpallial development amongst more limited subsets of progenitor pools. Approximately 2% of ISL1 immuno-positivity was present exclusively within Nkx2.1GFP/+ cells. These precursors may have not yet gone through Cre/loxP recombination and thus may be destined to become mixed lineage Nkx2.1/Isl1 precursors at subsequent time points not captured in our analysis.
Figure 2 .
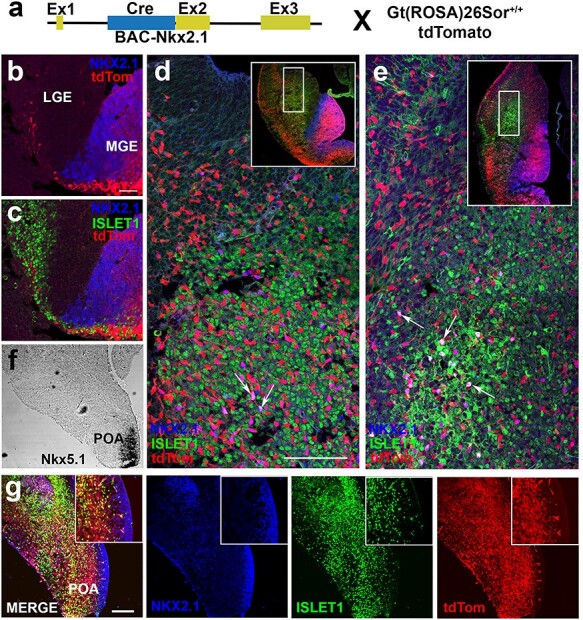
NKX2.1+ cells remain largely distinct from ISL1+ precursors in basal ganglia and preoptic area during neural development. (a) Schematic of genetic cross to identify complete lineage of Nkx2.1 cells following downregulation of NKX2.1 protein. BAC-Nkx2.1-Cre male mice crossed with female Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J reporter mice. (b–e) Triple IHC of LGE for NKX2.1 (blue), ISL1 (green), and BAC-Nkx2.1-Cre:tdTomato (red). (b,c) Early tangential migration at E12.5 illustrates beginning of Nkx2.1-derived migratory stream through ISL1+ corridor in developing LGE. Sharp downregulation of NKX2.1 seen in tdTomato+ cells. (d) At 13.5, few NKX2.1+/tdTomato+ (purple) cells visualized in LGE identified by arrows with majority tdTomato+ only. Inset in (d) shows low magnification of tangential migratory stream. (e) At E14.5, most tdTomato+ cells migrate through LGE towards cortex with few scattered triple-labeled cells (NKX2.1+/ISL1+/BAC-Nkx2.1-Cre:tdTomato+) in LGE. Inset in (e) shows low magnification image with many Nkx2.1-derived cells inhabiting cortex. (f) ISH of coronal hemi-section at E14.5 of Isl1cre/+; tdTomato mice demarcates the area of Nkx5.1 expression, a marker for postmitotic cells derived from the POA but not the MGE. (g) IHC for NKX2.1, ISL1, and tdTomato on adjacent section from (f) demonstrates that ISL1+ and tdTomato+ cells are distinct from NKX2.1-labeled nuclei in the POA. Scale bars represent 50 μm (b,c); 100 μm (d,e) and 100 μm in low magnification images (g).
To determine if loss of Nkx2.1 perturbed fate-specification of neocortical neurons within the Isl1 lineage, Isl1cre/+; tdTomato; Nkx2.1GFP/+ mice were subsequently crossed to each other to yield quadruple transgenic mice in which Isl1 and Nkx2.1 dual lineage fate-mapping occurred in both the Nkx2.1 HET (Fig. 4a) and Nkx2.1 HD (Fig. 4b) mouse lines. The mice were analyzed at E18.5 because HD mice are perinatal lethal due to lack of lung development (Costa et al. 2001). Nkx2.1GFP/+ HET only mice (Fig. 3) and dual lineage fate-mapped HET mice (see Supplementary Fig. 3) showed consistency of GFP labeling across the basal ganglia and neocortex (Fig. 4a,c,e,g), with labeled cells spanning all neocortical laminae and possessing neuronal morphologies (green cells in Fig. 4a,c,e,g). As occurred with the Isl1cre/+: tdTomato mice (Fig. 1), the tdTomato+ cells of the Isl1 lineage were also present (red cells in Fig. 4a,c,e,g) and were highly enriched in the striatum and basal ganglia (Fig. 4a). The majority of GFP+ and tdTomato+ cells appeared to be adjacent to each other in subcortical structures. In contrast, in the cerebral cortex, there were 3 fluorescently labeled populations of cells: GFP+ only (green); tdTomato+ only (red); and GFP+/tdTomato+ (yellow cells) (Fig. 4c,e,g).
The variability of fluorescence across animals and litters was analyzed in multiple Isl1cre/+:tdTomato; Nkx2.1GFP/+ HET mice (n = 12). Although a range of low and high tdTomato expression was observed (see Supplementary Fig. 4), there was no significant difference in the ratios of labeled cells. Thus, all of the HET samples were pooled for comparison to Isl1cre/+:tdTomato; Nkx2.1GFP/GFP HD mice.
In the basal ganglia of HD mice, GFP+ and tdTomato+ populations were indistinguishable and showed dense overlapping reporter gene expression, with no discerning morphological features separating the Nkx2.1 and Isl1 lineages (Fig. 4b; yellow cells). However, in the neocortex, GFP+ fluorescent cells were greatly diminished as compared with the HET condition, leaving predominantly tdTomato+ cells. Analysis across the complete rostral–caudal axis of HD mouse brains showed GFP+ neurons were found to be consistently absent in cerebral cortex regardless of cortical position (Fig. 4d,f,h).
The phenotypic differences within the cerebral cortex of labeled cells were quantified for both conditions in matched rostral, medial, and caudal brain regions (Fig. 4c–h). Three neuronal cell populations were detected and quantified: GFP+ only cells (green); tdTomato+ only cells (red); and GFP+/tdTomato+ (yellow) cells. The GFP+ only population was the largest subset and the tdTomato+ population was the smallest. The fluorescently labeled neurons spanned all cortical layers and the developing white matter. However, in HD conditions, very few GFP+ only cells were seen (Fig. 4d,f,g) and the GFP+ only cells were mostly co-positive for tdTomato (yellow). The differences were determined by counting GFP+ only, tdTomato+ only, and GFP+/tdTomato+ cortical cells at rostral, medial, and caudal regions spanning layer II to layer VI (excluding layer I and white matter; Fig. 4i). In the HET condition, primarily GFP+ only cells were detected at all levels (~50% with 51.2% ± 7.4 at rostral, 51.5% ± 5.5 at medial, and 53.8% ± 6.2 at caudal regions), with a small contribution of tdTomato+ only cells (~10% with 10.1% ± 1.7 at rostral, 10.9% ± 1.5 at medial, and 10.0% ± 1.4 at caudal regions) and the remainder being yellow cells (~40% with 38.6% ± 5.9 at rostral, 37.5% ± 4.6 at medial, and 36.0% ± 5.1 at caudal regions). In the HD condition, the GFP+ only cells were fewer in all regions (~11% with 17.1% ± 7.5 at rostral, 11.8% ± 4.0 at medial, and 3.5% ± 2.1 at caudal regions), the tdTomato+ only cells increased (~74% with 65.5% ± 4.8 at rostral, 73.6% ± 3.4 at medial, and 83.2% ± 4.3 at caudal regions), and the yellow cells were reduced (~15% with 17.3% ± 5.2 at rostral, 14.2% ± 3.2 at medial, and 13.2% ± 4.9 at caudal regions) across all sections (Fig. 4i; n = 6 animals with 4 slices per animal in each condition). Although a relative shift in the proportion of fluorescently labeled cortical cells towards the tdTomato+ only cell fate was visualized, the total numbers of tdTomato+ remained consistent thus this did not represent an absolute increase in the numbers of tdTomato+ cells within the HD condition.
Isl1-Derived Cortical Neurons Re-Specify to SP8+ Cells in Nkx2.1 HD Mouse Neocortex
To further determine the identity for Isl1-derived cells in neocortex, the TFs CoupTF-II and Sp8 were assessed in Nkx2.1 HET and HD mice. CoupTF-II and Sp8 are expressed in the CGE and LGE/CGE, respectively. They border the ISL1+ precursor population of the LGE and the LGE/MGE boundary, and are known to generate cortical interneuron subtypes (Miyoshi et al. 2010; Ma et al. 2012; Alfano et al. 2014). IHC analysis of either COUPTF-II or SP8 expression at E14.5 in Isl1cre/+:tdTomato coronal brain sections showed minimal co-expression of COUPTF-II+/tdTomato+ (9.6% ± 1.1, n = 4) or SP8+/tdTomato+ cells (9.0% ± 1.0, n = 3) in the cerebral cortex (data not shown) or the GE (see Supplementary Fig. 5e), thus these cells appear to inhabit distinct precursor subdomains under normal developmental conditions.
At E18.5, a comparison of COUPTF-II and SP8 co-expression in Nkx2.1 HET (Fig. 5b) and HD (Fig. 5e) mice showed distribution of both TFs across the neocortex and basal ganglia. Although Nkx2.1 HETs (n = 7) generally showed both COUPTF-II and SP8 positive nuclei across cortical layers, Nkx2.1 HD brains (n = 6) had a reduction in overall labeling of COUPTF-II, whereas numbers of SP8+ nuclei were preserved. Quantifications of COUPTF-II only (red), SP8 only (cyan), and co-labeled nuclei (pink) in layers II–VI were conducted (Fig. 5j) under both conditions: 48.1% ± 0.5 (HET) versus 80.0% ± 0 E (HD) for SP8 only; 28.2% ± 2.0 D (HET) versus 4.6% ± 6.0 E (HD) for COUPTF-II only; and 23.7% ± 0.7 (HET) versus 15.4% ± 1.2 (HD) for co-labeled nuclei. The quantification showed a relative shift in TF type, as well as an absolute increase in SP8+ nuclei and decrease in COUPTF-II+ nuclei in the HD mice compared with the HETs.
Figure 5 .
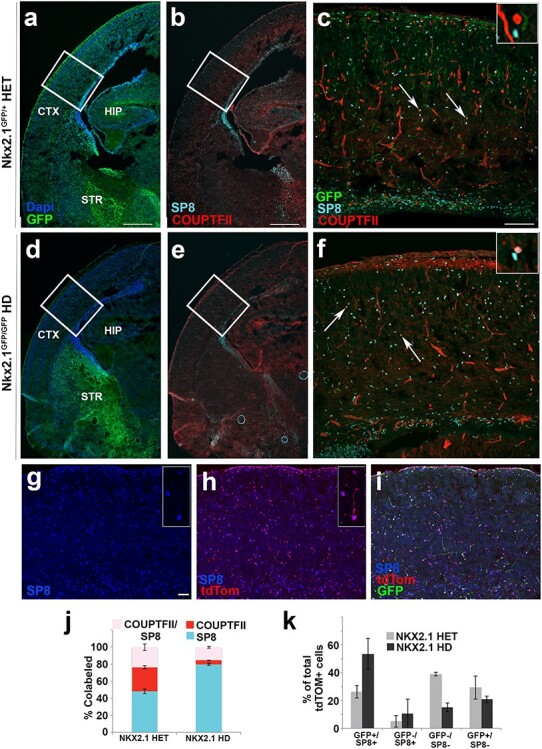
Isl1-derived tdTomato+ cortical cells express SP8 positivity in cerebral cortex. Low magnification coronal sections of (a) Nkx2.1GFP/+ heterozygous and (d) Nkx2.1GFP/GFP HD mice sacrificed at E18.5 show presence and absence of GFP+ cortical cells in (a) and (d), respectively. Co-staining of GFP+ tissue with TFs, SP8 (cyan), and COUPTF-II (red; b,e). High magnification images of boxed regions depicted in (a,b) are shown in (c); (d,e) are shown in (f). Arrows point to co-labeled SP8+/COUPTF+ (pink) cells that differentiate from SP8 only (cyan) and COUPTF-II only (red) (see inset). Confocal images (g–i) of Isl1cre/+:tdTomato; Nkx2.1GFP/GFP (HD) mice at E18.5 showed that many of tdTomato+ cells were re-specified to SP8+ cell fates in cerebral cortex following HD of NKX2.1. Stacked bar graph (j) shows co-expression of SP8 and COUPTF-II in Nkx2.1 HET & HD conditions. HET animals contain 48.1% ± 2.5 (SP8 only), 28.2% ± 1.8 (COUPTF-II only), and 23.7% ± 3.8 (co-labeled) cells, whereas HD animals contain 80.0% ± 1.3 (SP8 only), 4.6% ± 0.3 (COUPTF-II only), and 15.4% ± 1.2 (co-labeled) cells in cortical layers II–VI. N = 7 (HET) & N = 6 (HD), (2-way ANOVA, P < 0.0001). (k) Bar graph shows quantification of fluorescent cells present in cortical layers II–VI for HET and HD conditions whereby tallied cells are quantified relative to total number of tdTomato+ cells revealing 26.2% ± 2.2 (GFP+/SP8+ cells) in HET condition contrasted by 53.6% ± 6.3 (GFP+/SP8+ cells) in HD condition indicating trend. N = 4 (HET) & N = 3 (HD) for bar graphs. Scale bars represent 500 μm (a,b,d,e); 100 μm (c,f) and 50 μm (g–i).
Similarly, Isl1cre/+:tdTomato; Nkx2.1GFP/GFP HD mice also had an abundance of SP8+ nuclei across cortical laminae (Fig. 5g). Some cells co-labeled with scattered tdTomato+/GFP+ immature neurons (Fig. 5h). The relative contributions of labeled cells were quantified for the total tdTomato+ population across cortical layers II–VI. There was a trend towards increased contribution of GFP+/SP8+ expression of total tdTomato+ cortical cells in HD mice (26.2% ± 2.2) compared with HET animals (53.6 ± 6.3%), suggesting a relative expansion of Isl1-derived tdTomato+ cells towards SP8 positivity in the absence of functional NKX2.1 protein expression. Although the possibility cannot be completely excluded that Nkx2.1 HET mice showed some effect on baseline SP8 expression as a result of haplo-insufficiency, the relative independence with COUPTF-II positivity suggests that Isl1-derived cells expand from a SP8+ population residing within the LGE and not a shared COUPTF-II co-positive subset of the CGE.
Isl1 Fate-Mapped Neurons Bifurcate into Neocortical and Striatal Cell Fates
To determine which subset of Isl1 neuroblasts tangentially migrated into the neocortex, an organotypic slice explant model was used in combination with live imaging (Fig. 6). E14.5 Isl1cre/+:tdTomato+ embryos were microdissected at the location of the POA (Fig. 6a) or LGE (Fig. 6d,g) and transferred to unlabeled littermate brain slices. As a control, homotopic transplants of tdTomato+ cells were isolated from POA and inserted into the wildtype POA structure (data not shown). The labeled cells remained near the transplant site and extended axonal processes into surrounding basal ganglia. Similarly, heterotopic transplants of tdTomato+ POA cells placed at the LGE showed local migration into basal ganglia but never crossed the cortico-striatal junction (Fig. 6b). A small number of tdTomato+ LGE cells placed at the POA migrated into the neocortex, but the majority remained in the subpallium (Fig. 6e). In contrast, homotopic transplantation of tdTomato+ LGE cells into wildtype LGE demonstrated robust migration into the surrounding host tissue, with most labeled cells dispersing locally within basal ganglia (Fig. 6g). Notably, a subset of these tdTomato+ neuroblasts (~50–100 cells per slice) crossed the corticostriatal junction into developing cortical plate and intermediate zone (Fig. 6h). These results indicate that Isl1-derived neuroblasts that are isolated from LGE, but not from hypothalamic structures, are capable of responding to local cues and migrating into neocortical destinations within a slice transplant model.
Figure 6 .
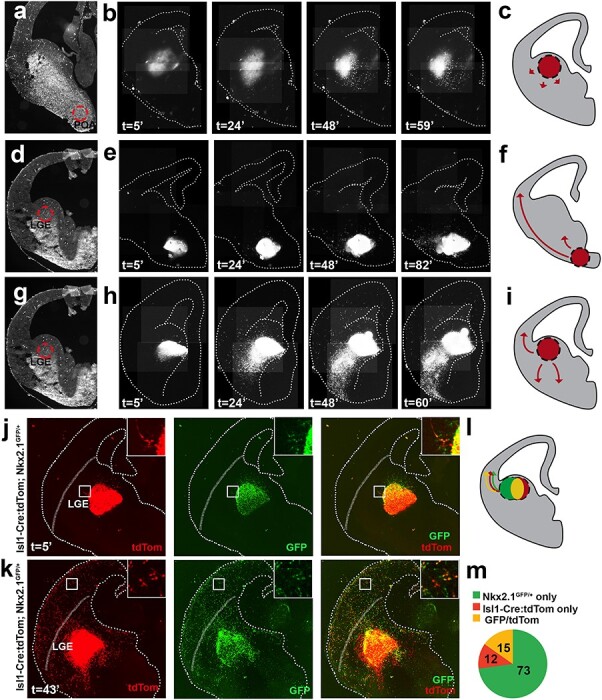
Isl1-derived cells from LGE tangentially migrate to cerebral cortex. (a) Representative acute organotypic section of E14.5 Isl1cre/+; Gt(Rosa)26Sortm14(CAG-tdTomato)Hze/J microdissection of POA at demarcated region and transplanted into LGE of Cre- littermates. (b) Time lapse imaging shows bolus of tdTomato+ tissue. Labeled cells migrate locally and disperse within subpallium but do not cross cortical boundary. (c) Schematic summarizing (b). (d) Example of brain section microdissected at LGE and transplanted into POA. (e) Time lapse shows tdTomato+ tissue bolus migrates proximal to transplanted site. Few scattered LGE cells are found in cortex after 82 h. (f) Schematic summarizing (e). (g) Example of brain section microdissected at LGE and transplanted into LGE. (h) Time lapse shows homotopic transplantation results in dispersion of labeled LGE cells by 24 h. Between 24 and 60 h, the vast majority of neuroblasts are dispersed within subpallium. Labeled neocortical cells are identified emerging for transplant site and tangentially migrating into cortical plate. By 60 h, many cells are detected in cortex. (i) Schematic summarizing (h). (j) Acute organotypic section of E14.5 WT mouse brain slice transplanted with fluorescently labeled Isl1cre/+:tdTomato+; Nkx2.1GFP/+ heterozygous LGE explant placed within LGE. Labeled cells are located at or near explant. (k) After 43 h, the transplanted slice was re-imaged and shows robust migration and dispersion away from LGE explant within basal ganglia and neocortex. GFP, tdTomato and co-labeled (GFP/tdTomato) precursors are dispersed across developing neocortex. The dotted line traces the slice edge, while the blurred line demarcates the corticostriatal boundary (j,k). Insets (j,k) show high magnification boxed region of labeled cells. (l) Schematic summarizing (j,k). (m) Pie graph shows 3 precursor populations present within neocortex: GFP+ only cells (72.8% ± 7.4), GFP/tdTomato+ yellow cells (15.2% ± 6.4) and tdTomato+ only cells (11.9% ± 3.0). N = 4 slice replicates (a); n = 3 slice replicates (d); n = 6 slice replicates (g); n = 6 slices replicates (j,k).
Next, we further sought to clarify whether the cortical contribution of tdTomato+ cells (see Fig. 6h) were from exclusive tangential migration of Isl1-derived precursors or from mixed LGE/MGE precursor subsets as identified by Isl1cre/+:tdTomato+ and Nkx2.1GFP/+ co-expression (yellow cells). The data in Figure 4i indicated that almost 40% of Isl1-derived precursors shared a common lineage with MGE-derived cells. Thus, to determine the contribution of each subpopulation, we conducted a similar LGE homotopic slice transplant experiment, with the exception of using Isl1-Cre:tdTomato; Nkx2.1GFP/+ HET mice harvested from E14.5 mouse embryos (Fig. 6j). LGE explants were microdissected distal to MGE to reduce the likelihood of contamination from neighboring MGE precursors.
Following transplantation and waiting 43-h later, the transplanted slice was imaged and analyzed for fluorescently labeled precursors present in neocortex. The results showed that a combination of 3 fate-labeled identities were present within the extracted explant from the LGE (Fig. 6j). The exclusive Isl1-derived cells were labeled with tdTomato+ only, exclusively Nkx2.1-derived neuroblasts were GFP+ only and co-labeled yellow cells (of Isl1/Nkx2.1 mixed origin) were identified in neocortex (Fig. 6k). Quantification (n = 6) showed the majority of labeled cortical precursors were GFP+ (72.8% ± 7.4), followed by yellow cells (15.2% ± 6.4) and tdTomato+ cells (11.9% ± 3.0). The GFP only population most likely originated from tangentially migrating MGE precursors that were captured as they migrated past LGE into cortex. Most relevant, a combination of tdTomato only and co-labeled (yellow) cells were distributed throughout developing neocortex. Given that the mantle region of LGE-proper was targeted for microdissection, these cohorts were most consistent with LGE and LGE/MGE bordering precursor pools, respectively. Consistent with earlier fixed time point analysis, the tdTomato+ cells were ~10–12% of the total tangentially migrating precursor pool. This test of Nkx2.1-GFP; Isl1-Cre/tdTomato heterozygous animals directly addresses the question of the fate of co-labeled and Isl1-derived precursors following birth within subpallial structures. The data support that these 3 cell types are lineally separate from conception, and that they differentiate from separate precursor pools present within the LGE and LGE/MGE boundaries. Subsequently, at least a small subset of co-labeled (yellow) and tdTomato+ only cells are biased towards neocortical fates.
Neocortical Isl1 Fate-Mapped Cells Are Born in the LGE Germinal Zone
A time course for the production of Isl1-derived cortical cells was determined by pulse-labeling with EdU in Isl1cre/+; tdTomato+ pregnant dams. EdU+/tdTomato+ was injected into dams at E12.5, E13.5, E14.5, and E15.5 and the pups were analyzed on postnatal day 21. In mice that had been labeled at E12.5, the cells were present in the postnatal mice in layer VI, whereas those labeled at E15.5 had cells present in layers II/III, which is consistent with known cortical patterning (Gardette et al. 1982). The mice labeled at E13.5 had the greatest scattering of co-labeled cells (60% ± 1.1) across multiple cortical layers, which suggested that the majority of cortical Isl1cre/+:tdTomato+ were generated at this time point (Fig. 7a). Subsequently, this birth-dating range was used to determine a time point for isolation of LGE cells from the developing mouse brain.
Figure 7 .
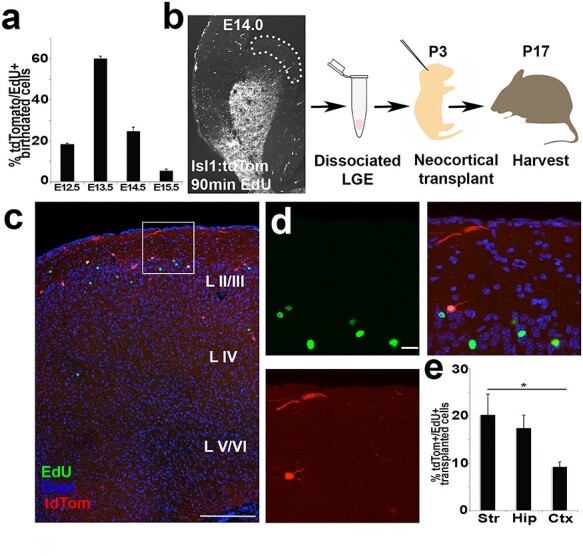
Isl1-derived precursors are born within germinal zones of the LGE. (a) Bar graph of % tdTomato+/EdU+ birth-dating following E12.5 EdU pulse (18.4% ± 0.5), E13.5 EdU pulse (60.1% ± 1.1), E14.5 EdU pulse (24.7% ± 2.0), and E15.5 EdU pulse (5.5% ± 1.6). n = 3–4 brains per condition in Isl1cre/+:tdTomato animals analyzed at P21. (b) Experimental design for birth-dating Isl1-derived cells using B6 neonatal mice transplantation model. Note, microdissected LGE cells are tdTomato−, except for labeled blood vessels within proliferative zone. (c) Cross section of neocortex in B6 tdTomato− mouse brain transplanted 2 weeks earlier with LGE progenitor cells. EdU+ (green), tdTomato+ (red), or tdTomato+/EdU+ (yellow) cells dispersed with the majority of labeled neurons located in superficial layers. (d) High magnification image of (b) shows EdU+/tdTomato+ cells with EdU+ neighboring cells. (e) Bar graph of tdTomato+/EdU+ of total transplanted cells shows 20.2% ± 4.4 striatal cells, 17.4% ± 2.7 hippocampal cells and 9.2% ± 1.1 cortical cells, P < 0.05, Student’s t-test. n = 4 brains. Scale bars represent 200 μm (c) and 20 μm (d).
To test whether Isl1-derived cells were specifically generated within LGE germinal zones, birth-dated LGE germinal zone progenitors were transplanted into neonatal mouse brains (Fig. 7b). The areas with shared boundaries between LGE and MGE were avoided and caudal sections were excluded to avoid inclusion of CGE-derived cells. The Isl1cre/+; tdTomato+ pregnant dams were labeled with EdU at E14.0 and sacrificed 90-min later. The LGE germinal zones (VZ/SVZ) of tdTomato+ embryos were micro-dissected using a dissecting microscope with adjustable fluorescence, allowing the extracted tissue to be clearly identified as tdTomato− and EdU+. The acutely dissociated cells were then microinjected into the cerebral cortex of wildtype neonates at P3 and analyzed at P17. Cells expressing EdU+/tdTomato+ (yellow), EdU+/tdTomato− (green), and tdTomato+/EdU− (red) were quantified to determine the percentage of tdTomato+ cells that were isolated and birth-dated to the VZ/SVZ of LGE at E14.0 (Fig. 7c,d, n = 4). The donor-derived cells were found primarily in superficial layers of the cortex (Fig. 7c). Many EdU+/tdTomato− (green) cells were present in the neocortex and were most likely derived from other LGE lineages (i.e., Er81 or Sp8 lineages). Additionally, tdTomato+/EdU− (red) cells were assumed to be non-S-phase or postmitotic at the time of birth-dating. EdU+/tdTomato+ (yellow) cells were present in forebrain structures including the neocortex, striatum and hippocampus. These labeled cells possessed short dendrites with localized axonal arbors, reminiscent of cortical interneurons in the cortex and hippocampus, and reminiscent of MSNs in the striatum. The double positive cells were 9.0% ± 1.1 of the neocortex compared with 20.2% ± 4.4 of cells in the striatum and 17.4% ± 3.2 in the hippocampus (Fig. 7e). Since isolated cells were tdTomato−, we concluded that some cells must have first incorporated EdU, undergone recombination, and then differentiated following transplant into host mice brains. Taken together, the birth-dating analysis and transplantation studies indicate that at least a small subset of Isl1-derived cells isolated from LGE are capable of differentiating into interneurons in the cerebral cortex.
Fate-Bias towards Fast-Spiking Cortical Interneurons within Isl1 Lineage
The majority of cortical Isl1cre/+:tdTomato+ neurons were immunoreactive for GAD67 at P21 (68.6% ± 3.6) across layers II–VI in the somatosensory cortex. The remaining labeled cells either overlapped with the mature astrocyte marker S100β or lacked immunoreactivity to either marker (not shown). The tdTomato+ cells were ~12% ± 0.6 of the total parvalbumin+ (PV) cells in the neocortex (Fig. 8g), which represented 29.1% ± 2.7 of the entire tdTomato+ population. Of the tdTomato+ cells, the largest group was PV-positive (32.3% ± 2.4), with others being somatostatin+ (SST; 21.5% ± 1.5), calbindin+ (CB; 15% ± 1.2), calretinin+ (CR; 12.3% ± 1.6), or neuropeptide-Y+ (NPY; 6.3% ± 1.4; Fig. 8h).
Figure 8 .
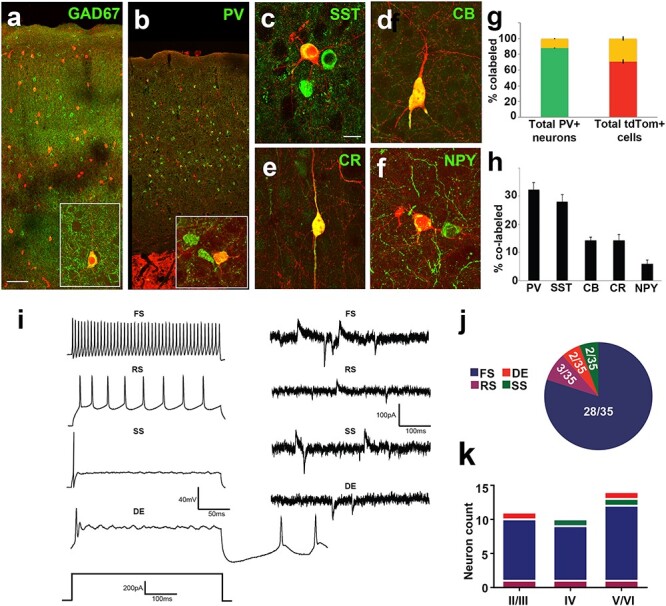
Neurochemical and functional analysis of Isl1-derived neurons shows fate bias towards fast-spiking cortical interneurons. (a) TdTomato+ neurons span all cortical layers and are largely co-positive with GAD67. (b inset, c–f) High magnification of cortical interneuron subtypes (b) parvalbumin, (c) somatostatin, (d) calbindin, (e) calretinin and (f) neuropeptide-Y co-labeled with tdTomato+ in cerebral cortex. (g) Left bar graph of co-positivity of total parvalbumin+ cells shows 12.1% ± 0.7 co-labeled (PV+/tdTomato+, yellow) cells and 87.9% ± 0.7 (PV only, green) cells. Right bar graph of co-positivity of total tdTomato+ cells shows 29.1% ± 2.7 (PV+/tdTomato+, yellow) cells and 70.9% ± 2.7 (tdTomato+ only, red) cells. (h) Bar graph shows % co-label of cortical tdTomato+ cells with all analyzed neurochemical markers shown in (b–f). (i) Electrophysiological traces of examples interneuron subtypes. Population data of recorded cells indicates the majority are fast spiking. Isl1-derived neurons exhibit 4 different electrophysiological properties. Current-clamp (left) and synaptic (right) traces for fast spiking (FS), regular spiking (RS), delayed excitation (DE), and single spiking (SS) interneurons. (j) Pie chart displaying the contribution of each electrophysiological property noted for Isl1-derived neurons. (k) Histogram summarizing the cortical distribution of each electrophysiological subtype of recorded Isl1-derived neurons. Scale bars represent 100 μm (a,b) and 20 μm (c–f).
The intrinsic electrophysiological properties of the cells were evaluated in the barrel cortex using whole-cell current clamp recordings in acute slices from Isl1cre/+:tdTomato+ mice (P15-P21). All of the recorded fate-labeled neurons displayed active and passive membrane properties consistent with functionally mature inhibitory neurons (see Supplementary Fig. 6) (Ascoli et al. 2008; Zhou et al. 2009; Miyoshi et al. 2010). Consistent with the IHC data, tdTomato+ interneurons showed 4 distinct firing patterns: 1) fast spiking (FS); 2) regular spiking (RS); 3) delayed excitation (DE); and 4) single spiking (SS) cells (Fig. 8i). The FS subset was most abundant (80%, n = 28 of 35 cells; Fig. 8j) and fired at high rates in response to larger depolarization steps (data not shown) with brief action potentials and more pronounced AHP amplitudes (Fig. 8i and see Supplementary Fig. 6a). These FS cells exhibited the major hallmarks of cortical basket cells including brief action potentials (<1 ms), high-frequency discharge (>80 Hz), and very low adaption ratio (Rudy and McBain 2001). The RS subtype (8.6%, n = 3 of 35 cells) fired at relatively low rates with typical wide spike waveforms and modest AHP (Fig. 8i and see Supplementary 6a). The DE and SS subpopulations each represented 5.7% of the recorded cells (Fig. 8j, n = 2 of 35 cells) and were characterized by a single spike waveform during depolarization, but the DE cells often fired post-depolarization spikes (Fig. 8i), as previously described in non-cortical tissues (Zhao and Wu 1997; Jiang et al. 1999). The results from laminar analysis suggested that firing characteristics of the Isl1-derived interneurons were independent of laminar distribution (Fig. 8k). To determine whether or not the fate-mapped cortical interneurons formed functional synapses, the inhibitory, and excitatory synaptic inputs onto these cells were analyzed using the whole-cell patch clamp approach in voltage clamp mode. Simultaneous spontaneous EPSCs and IPSCs were recorded in the same cell at a holding potential at −40 mV (Zhou et al. 2009) and excitatory and inhibitory inputs were observed in all 4 types of Isl1-derived interneurons (see Supplementary Fig. 7). The synaptic events were variable for both EPSCs and IPSCs, ranging from 0.05 to 1.93 Hz in the 4 cell types. Similarly, mean amplitude, rise time and decay kinetics were also broad for the examined GABAergic subtypes as reflected in cumulative distribution and mean histograms. These results are consistent with functional integration of Isl1-derived interneurons into cortical circuits.
Discussion
The major progenitor pools responsible for generating cortical interneurons are primarily derived from MGE and CGE progenitors, but do not account for all of the inhibitory neurons present in the neocortex. What remains unresolved is if smaller, less well-defined precursor cells, contribute to mature cortical interneurons. The data presented in this study uses a preponderance of evidence to demonstrate that a small population of tangentially migrating cells, originating from Isl1-derived precursors, diversify into both striatal and neocortical neuron types (Figs.1 and 6), primarily of the fast-spiking subtype (Fig. 8j). Although most Isl1-derived and Nkx2.1-derived cells share a common lineage in the production of cortical interneuron subtypes at LGE/MGE boundaries, our data indicate that each subdomain contains distinct expression patterns that contribute towards mature neocortical function.
Contribution to Neocortical GABAergic Interneurons
Early studies using fluorescent lipophilic dyes suggested the origins of cortical interneurons were in the MGE and LGE (de Carlos et al. 1996; Anderson et al. 1997, 2001). The inability to label smaller precursor pools that concurrently migrated during neurogenesis with lipophilic dyes made it difficult to distinguish stationary corridor cells (Wichterle et al. 2003) from tangentially migrating MGE, POA, CGE, or LGE-derived populations. Cells isolated from the LGE, MGE, or CGE and transplanted homotopically and homochronically into their respective structures, showed that only MGE and CGE neuroblasts migrated into the neocortex (Wichterle et al. 2001; Nery et al. 2002), whereas LGE cells appeared to differentiate into ER81+ OB, ISL1+ striatal medium spiny, and nucleus accumbens neuron subtypes (Wichterle et al. 2001). Although some PV+ cells appeared to migrate into the neocortex following LGE to LGE grafts, the experiments were not definitive (Wichterle et al. 2001). More recent data established that LGE or LGE/CGE border populations defined by SP8 expression (Ma et al. 2012) contribute to neocortical interneuron fates. Thus, fate-mapping the complete Isl1-derived progeny allows for discriminating amongst progeny that target the basal ganglia versus those cells that migrate towards the neocortex. SP8+ cells or Isl1-derived cells can be followed to track the LGE origin from LGE/CGE or LGE/MGE bordering cells, respectively.
Distinct Nkx2.1 and Isl1 Cortical Interneuron Lineages
The goal to fate-map all TF domains simultaneously (Flames et al. 2007; Gelman et al. 2011) has been difficult due the lack of fully compatible reporter mice necessary for multigene lineage tracing. A study that fate mapped MGE and CGE interneuron subtypes and performed single cell gene expression analysis found ~65 TF shared by these 2 populations (Taniguchi et al. 2011), but Isl1 was not significantly expressed in MGE or CGE cells (Paul et al. 2017). Thus, Isl1 expression may generally be excluded from cells originating in the MGE and CGE, and can therefore function as a readout of precursor fates from LGE. In contrast, Sp8 was highly expressed in the CGE cohort, suggesting a difference in overlap across Sp8 and Isl1 within the CGE and MGE regions, respectively. CoupTF-II was highly expressed In the CGE, as expected. Thus, our finding of an increase in SP8 in the HD mice supports the notion that Isl1 is not expressed in MGE. Further, quantification of LGE/MGE border cells with ISL1 immuno-positivity suggests that a dynamic expansion of progenitor pools occurs during brain development. Although Isl1/Nkx2.1 mixed lineages represented only ~ 9% of quantified precursor cells in the subpallium at E13.5, our data indicate ~ 38% of cortical interneurons share both Nkx2.1 & Isl1 mixed lineages at E18.5 in cerebral cortex. This may be the result of earlier subpallial VZ/SVZ progenitor zone divisions that differentially propagate during earlier stages of neurogenesis where adjacent LGE/MGE clades are even less segregated. Although our data supports the concept of Isl1 LGE precursors migrating into the cerebral cortex, whether or not ISL1 protein is necessary and sufficient for the specification of such cells, or merely functions as a transitional label for cells in the LGE mantle zone as they migrate into the cerebral cortex remains to be determined. Future studies using more refined single cell RNA-seq approaches may be able to further subdivide neighboring precursors with respect to their lineal outputs. This would also be able to determine if precursor-identity remains static or changes over the course of neurogenesis within GE based upon transcriptional priming of precursor identity via presence of ISL1 protein. This remains an outstanding question for future analysis of subpallial progenitor diversity.
Our study uses both single (Fig. 2) and dual lineage fate-mapping (Fig. 4) strategies to demonstrate that an Isl1+ population exists within the Nkx2.1 precursor pool and are associated with cortical interneuron fates. However, a small Isl1-derived subset also appears to exist that is Nkx2.1 independent, which is the most consistent with a LGE origin (Fig. 1). The primary line of evidence that supports this conclusion is the data from crossing Nkx2.1GFP/+ or Nkx2.1GFP/GFP with Isl1cre/+:tdTomato mice. A subpopulation of tdTomato+ cells shared a common lineage with Nkx2.1-derived GFP+ cells in heterozygotes (Fig. 4i), but a distinct population of Isl1cre/+:tdTomato+ only cell was also present (Fig. 4i). Complete removal of Nkx2.1 using homozygous Nkx2.1GFP/GFP mice, showed persistent expression of tdTomato+ only cells within the fate-labeled population (Fig. 4i) despite the substantial loss of Nkx2.1+ only cells and significant reduction in Nkx2.1/Isl1 co-positive cells.
The results indicate that Nkx2.1 and Isl1 have both shared (mostly likely on the LGE/MGE border) and independent domains. The domains themselves appear to be continuous along the border regions and thus absolute separation is not possible within these fused structures using the technologies employed. The preponderance of evidence presented here demonstrates the major finding that single lineage Isl1+ only (Nkx2.1- independent) cells exist within normal cortical development and separately contribute to the total cortical interneuron precursor population.
In the Nkx2.1 null mice, some Nkx2.1GFP/GFP labeled precursors may be sensitive to gene silencing and re-specify cell fates towards an Isl1-derived lineage (Fig. 4a,b), which is supported by the near complete overlap between Isl1 and Nkx2.1 lineages in the basal ganglia (Fig. 4b). We cannot exclude the possibility that Nkx2.1GFP/GFP was lost in some remaining cortical cells through turnover of the GFP reporter gene. Although a conditional system could be considered, Flp versus Cre recombination efficiencies can also differ in recombination efficiency and may contribute to differences in visualized phenotypes. However, findings shown here are consistent with work showing that Nkx2.1 (MGE) cells acquire a LGE/CGE phenotype following loss of Nkx2.1 in a temporally specific manner (Butt et al. 2008). Additionally, overlap is known to exist across some Isl1 and Nkx2.1 cells in striatum where Isl1 is required for the production of cholinergic cells derived from an MGE lineage (Cho et al. 2014).
Although the data presented strongly support both shared and separate populations, there were a few GFP+ only and yellow cells following Nkx2.1 deletion (Fig. 4i). This may be due to incomplete deletion of Nkx2.1 in all target interneurons after genetic crossing of Nkx2.1 heterozygotes with some cells able to tangentially migrate into the neocortex. Since the NKX2.1 antibody robustly detected NKX2.1+ nuclei in WT and HET conditions, detection of cells did not appear to be a confounding variable. This suggests that some Nkx2.1- and Isl1-derived cells (GFP+/tdTomato+ cells) represent a common lineage between Isl1 and Nkx2.1 TF domains in vLGE/dMGE border populations, respectively. These results suggest that fixed TF domains are not precisely nested within distinct boundaries of LGE, MGE, CGE, and POA since Nkx2.1 only, Isl1 only, and co-expressing populations were present, which is consistent with a shift in interpretation away from strictly anatomically restricted precursor pools (Bandler et al. 2017).
Further evaluation of Nkx2.1- and Isl1-derived cells was conducted using TFs for neighboring subdomains of the LGE/CGE border populations via expression of Sp8 and CoupTF-II. These genes are known to be highly expressed in the GE and within postmitotic neurons of the cerebral cortex (Fig. 5). The results indicate a shift in normal distribution across Sp8, CoupTF-II and overlapping subtypes in Nkx2.1 HETs predominantly towards SP8+ cells, with a concurrent reduction of COUPTF-II expression in Nkx2.1 HD mice. This re-specification resulted in near exclusive co-labeling of Isl1-derived cells (tdTomato+) with SP8+. This suggests that SP8+/tdTomato+ cells most likely arise from an expanded LGE and are unlikely to be of CGE origin. However, the possibility of overlap with other smaller CGE subdomains such as PROX1 could not be ruled out since this was not tested. The Nkx2.1 HD results in this study should be interpreted with some caution since transcriptional loss of the master regulating gene Nkx2.1 induces significant perturbations in subpallial patterning and expansion of neighboring TF subdomains. Although this does not occur under normal circumstances, it was included in this study to better understand the shared Nkx2.1/Isl1 domains with respect to cortical interneuron fate-specification.
Quantifying Total Cortical Interneuron Numbers
A small proportion of the total PV+ cells in the primary somatosensory cortex were Isl1cre/+:tdTomato+/PV+ (~12%; Fig. 8g). This extends prior fate-mapping studies that show Nkx2.1-derived lineages produce the vast majority of fast-spiking GABAergic cells in cerebral cortex. Compared with other recent studies of LGE/CGE progenitors, which found most labeled cells were of REELIN+ upper layer fates and very few (>2%) PV+ neurons (Torigoe et al. 2016), the data here found Isl1cre/+:tdTomato+ cells present across cortical laminae and the majority differentiates as PV+ neurons. Together, the data indicate that while a majority of cortical interneurons are of Nkx2.1 fates and some have both Nkx2.1/Isl1 lineage markers, a small yet distinct population of Isl1-derived fast-spiking neurons is also present. Our estimate of the contributions of these smaller precursor pools is up to 6% fast-spiking cells originating from Isl1/Nkx2.1 shared transcriptional domains and an additional 2% fast-spiking cells unique to the Isl1 TF domain.
Mapping Interneuron Birth Location to Cortical Network Function
The mapping of interneuron origins may yield insights into their contribution into functional microcircuits. Somewhat surprisingly, the results indicate that Isl1 and Nkx2.1 generate similar mature neocortical interneuron subtypes through both shared and distinct lineages. Determining the differences between Isl1-derived and Nkx2.1-derived PV+ cells would presumably aid in understanding the precise function of these neuronal subsets in the mature cortex. The finding that a subset of Isl1-positive precursor cells located in the LGE proper or at the LGE/MGE boundary contributes to cortical interneuron fate specification expands understanding of the origin of cortical interneurons in rodents to all of the eminence populations, even if these contributions are small.
Authors’ Contributions
FS, EDM and JHW designed the overall experimental directions and specific analyses, and wrote and edited the manuscript. FS, ALT and ML performed mouse experiments, DJ performed electrophysiological experiments, and all analyzed data and aided in manuscript preparation.
Funding
US National Institutes of Health (R01-NS088667 to J.H.W., R01-NS082761 to E.D.M.); NIH training (grant T32-NS007180 partially supported to F.S.); Epilepsy Foundation Post-Doctoral fellowship (partially supported to D.J.).
Notes
We thank: SA Anderson, O Marin, G Fishell, ES Tucker, JJ LoTurco and MV Fuccillo for scientific input, discussions and critique; E Azim, T Clarke, A Polesky for technical assistance; SM Evans and C Lee-May for Isl1cre/+ mice, SA Anderson for BAC-Nkx2.1-Cre mice, and D Kotton for Nkx2.1GFP/+ mice; SA Anderson for antibodies; and all members of the Marsh and Wolfe labs for support, discussions and scientific input. Conflict of Interest: None declared.
Data Availability
The authors declare that most of the data supporting the findings of this study are available within the paper and its supplementary information. Additional data or information about the study are available on request from the corresponding authors (JHW, EDM).
Supplementary Material
Contributor Information
Faez Siddiqi, Children's Hospital of Philadelphia Research Institute, Philadelphia, PA 19104, USA.
Alexandria L Trakimas, Children's Hospital of Philadelphia Research Institute, Philadelphia, PA 19104, USA; Departments of Neurology and Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Donald J Joseph, Children's Hospital of Philadelphia Research Institute, Philadelphia, PA 19104, USA; Division of Child Neurology, Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA.
Margaret L Lippincott, Children's Hospital of Philadelphia Research Institute, Philadelphia, PA 19104, USA.
Eric D Marsh, Children's Hospital of Philadelphia Research Institute, Philadelphia, PA 19104, USA; Division of Child Neurology, Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA; Departments of Neurology and Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
John H Wolfe, Children's Hospital of Philadelphia Research Institute, Philadelphia, PA 19104, USA; Division of Child Neurology, Children's Hospital of Philadelphia, Philadelphia, PA 19104, USA; Departments of Neurology and Pediatrics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA; W.F. Goodman Center for Comparative Medical Genetics, School of Veterinary Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
References
- Alfano C, Magrinelli E, Harb K, Studer M. 2014. The nuclear receptors COUP-TF: a long-lasting experience in forebrain assembly. Cell Mol Life Sci. 71:43–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. 1997. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 278:474–476. [DOI] [PubMed] [Google Scholar]
- Anderson SA, Marín O, Horn C, Jennings K, Rubenstein JL. 2001. Distinct cortical migrations from the medial and lateral ganglionic eminences. Development. 128:353–363. [DOI] [PubMed] [Google Scholar]
- Ascoli GA, Alonso-Nanclares L, Anderson SA, Barrionuevo G, Benavides-Piccione R, Burkhalter A, Buzsáki G, Cauli B, DeFelipe J, Fairén A, et al. 2008. Petilla terminology: nomenclature of features of GABAergic interneurons of the cerebral cortex. Nat Rev Neurosci. 9:557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandler RC, Mayer C, Fishell G. 2017. Cortical interneuron specification: the juncture of genes, time and geometry. Curr Opin Neurobiol. 42:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu C. 1993. Numerical data on neocortical neurons in adult rat, with special reference to the GABA population. Brain Res. 609:284–292. [DOI] [PubMed] [Google Scholar]
- Butt SJB, Sousa VH, Fuccillo MV, Hjerling-Leffler J, Miyoshi G, Kimura S, Fishell G. 2008. The requirement of Nkx2-1 in the temporal specification of cortical interneuron subtypes. Neuron. 59:722–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai C-LL, Liang X, Shi Y, Chu P-HH, Pfaff SL, Chen J, Evans S. 2003. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 5:877–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H-H, Cargnin F, Kim Y, Lee B, Kwon R-J, Nam H, Shen R, Barnes AP, Lee JW, Lee S, et al. 2014. Isl1 directly controls a cholinergic neuronal identity in the developing forebrain and spinal cord by forming cell type-specific complexes. PLoS Genet. 10:e1004280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RH, Kalinichenko VV, Lim L. 2001. Transcription factors in mouse lung development and function. Am J Physiol Cell Mol Physiol. 280:L823–L838. [DOI] [PubMed] [Google Scholar]
- de Carlos JA, López-Mascaraque L, Valverde F. 1996. Dynamics of cell migration from the lateral ganglionic eminence in the rat. J Neurosci. 16:6146–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon TW, Pakzaban P, Isacson O. 1994. The lateral ganglionic eminence is the origin of cells committed to striatal phenotypes: neural transplantation and developmental evidence. Brain Res. 668:211–219. [DOI] [PubMed] [Google Scholar]
- DeFelipe J, Fariñas I. 1992. The pyramidal neuron of the cerebral cortex: morphological and chemical characteristics of the synaptic inputs. Prog Neurobiol. 39:563–607. [DOI] [PubMed] [Google Scholar]
- Ehrman LA, Mu X, Waclaw RR, Yoshida Y, Vorhees CV, Klein WH, Campbell K. 2013. The LIM homeobox gene Isl1 is required for the correct development of the striatonigral pathway in the mouse. Proc Natl Acad Sci. 110:E4026–E4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y, Everhart D, Deng M, Xie X, Barlow RB, Gan L. 2007. Islet-1 controls the differentiation of retinal bipolar and cholinergic amacrine cells. J Neurosci. 27:12707–12720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flames N, Pla R, Gelman DM, Rubenstein JLR, Puelles L, Marin O. 2007. Delineation of multiple subpallial progenitor domains by the combinatorial expression of transcriptional codes. J Neurosci. 27:9682–9695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flandin P, Kimura S, Rubenstein JLR. 2010. The progenitor zone of the ventral medial ganglionic eminence requires Nkx2-1 to generate most of the globus pallidus but few neocortical interneurons. J Neurosci. 30:2812–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogarty M, Grist M, Gelman D, Marín O, Pachnis V, Kessaris N. 2007. Spatial genetic patterning of the embryonic neuroepithelium generates GABAergic interneuron diversity in the adult cortex. J Neurosci. 27:10935–10946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkouli A, van Wijk NV, Lopes R, Kessaris N, Pachnis V. 2009. LIM homeodomain transcription factor-dependent specification of bipotential MGE progenitors into cholinergic and GABAergic striatal interneurons. Development. 136:3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbott PL, Somogyi P. 1986. Quantitative distribution of GABA-immunoreactive neurons in the visual cortex (area 17) of the cat. Exp brain Res. 61:323–331. [DOI] [PubMed] [Google Scholar]
- Galanopoulou AS. 2013. Basic mechanisms of catastrophic epilepsy—overview from animal models. Brain Dev. 35:748–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardette R, Courtois M, Bisconte JC. 1982. Prenatal development of mouse central nervous structures: time of neuron origin and gradients of neuronal production. A radioautographic study. J Hirnforsch. 23:415–431. [PubMed] [Google Scholar]
- Gelman D, Griveau A, Dehorter N, Teissier A, Varela C, Pla R, Pierani A, Marin O. 2011. A wide diversity of cortical GABAergic interneurons derives from the embryonic preoptic area. J Neurosci. 31:16570–16580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry AM, Hohmann JG. 2012. High-resolution gene expression atlases for adult and developing mouse brain and spinal cord. Mamm Genome. 23:539–549. [DOI] [PubMed] [Google Scholar]
- Jiang MC, Liu L, Gebhart GF. 1999. Cellular properties of lateral spinal nucleus neurons in the rat L6-S1 spinal cord. J Neurophysiol. 81:3078–3086. [DOI] [PubMed] [Google Scholar]
- Karlsson O, Thor S, Norberg T, Ohlsson H, Edlund T. 1990. Insulin gene enhancer binding protein Isl-1 is a member of a novel class of proteins containing both a homeo-and a domain. Nature. 344:879–882. [DOI] [PubMed] [Google Scholar]
- Kohwi M, Petryniak MA, Long JE, Ekker M, Obata K, Yanagawa Y, Rubenstein JLR, Alvarez-Buylla A. 2007. A subpopulation of olfactory bulb GABAergic interneurons is derived from Emx1- and Dlx5/6-expressing progenitors. J Neurosci. 27:6878–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Hjerling-Leffler J, Zagha E, Fishell G, Rudy B. 2010. The largest group of superficial neocortical GABAergic interneurons expresses ionotropic serotonin receptors. J Neurosci. 30:16796–16808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DA. 2012. Cortical circuit dysfunction and cognitive deficits in schizophrenia--implications for preemptive interventions. Eur J Neurosci. 35:1871–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Song M-R, Xu Z, Lanuza GM, Liu Y, Zhuang T, Chen Y, Pfaff SL, Evans SM, Sun Y. 2011. Isl1 is required for multiple aspects of motor neuron development. Mol Cell Neurosci. 47:215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longmire TA, Ikonomou L, Hawkins F, Christodoulou C, Cao Y, Jean JC, Kwok LW, Mou H, Rajagopal J, Shen SS, et al. 2012. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell. 10:398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bendito G, Cautinat A, Sánchez JA, Bielle F, Flames N, Garratt AN, Talmage DA, Role LW, Charnay P, Marín O, et al. 2006. Tangential neuronal migration controls axon guidance: a role for neuregulin-1 in thalamocortical axon navigation. Cell. 125:127–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K-M, Evans SM, Hirano S, Liu F-C. 2014. Dual role for Islet-1 in promoting striatonigral and repressing striatopallidal genetic programs to specify striatonigral cell identity. Proc Natl Acad Sci U S A. 111:E168–E177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Zhang Q, Cai Y, You Y, Rubenstein JLR, Yang Z. 2012. A subpopulation of dorsal lateral/caudal ganglionic eminence-derived neocortical interneurons expresses the transcription factor Sp8. Cereb Cortex. 22:2120–2130. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magno L, Catanzariti V, Nitsch R, Krude H, Naumann T. 2009. Ongoing expression of Nkx2.1 in the postnatal mouse forebrain: potential for understanding NKX2.1 haploinsufficiency in humans? Brain Res. 1304:164–186. [DOI] [PubMed] [Google Scholar]
- Miyoshi G, Butt SJB, Takebayashi H, Fishell G. 2007. Physiologically distinct temporal cohorts of cortical interneurons arise from telencephalic Olig 2-expressing precursors. J Neurosci. 27:7786–7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Fishell G. 2011. GABAergic interneuron lineages selectively sort into specific cortical layers during early postnatal development. Cereb Cortex. 21:845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJB, Battiste J, Johnson JE, Machold RP, Fishell G. 2010. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. J Neurosci. 30:1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Young A, Petros T, Karayannis T, McKenzie Chang M, Lavado A, Iwano T, Nakajima M, Taniguchi H, Huang ZJ, et al. 2015. Prox 1 regulates the subtype-specific development of caudal ganglionic eminence-derived GABAergic cortical interneurons. J Neurosci. 35:12869–12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nery S, Fishell G, Corbin JG. 2002. The caudal ganglionic eminence is a source of distinct cortical and subcortical cell populations. Nat Neurosci. 5:1279–1287. [DOI] [PubMed] [Google Scholar]
- Olsson M, Björklund A, Campbell K. 1998. Early specification of striatal projection neurons and interneuronal subtypes in the lateral and medial ganglionic eminence. Neuroscience. 84:867–876. [DOI] [PubMed] [Google Scholar]
- Paul A, Crow M, Raudales R, He M, Gillis J, Huang ZJ. 2017. Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell. 171, e20:522–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaff SL, Mendelsohn M, Stewart CL, Edlund T, Jessell TM. 1996. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron–dependent step in interneuron differentiation. Cell. 84:309–320. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. 2000. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 28:727–740. [DOI] [PubMed] [Google Scholar]
- Polleux F, Ghosh A. 2002. The slice overlay assay: a versatile tool to study the influence of extracellular signals on neuronal development. Sci STKE. 2002:pl9. [DOI] [PubMed] [Google Scholar]
- Preibisch S, Saalfeld S, Tomancak P. 2009. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics. 25:1463–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, Fishell G, Lee S, Hjerling-Leffler J. 2011. Three groups of interneurons account for nearly 100% of neocortical GABAergic neurons. Dev Neurobiol. 71:45–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudy B, McBain CJ. 2001. Kv3 channels: voltage-gated K+ channels designed for high-frequency repetitive firing. Trends Neurosci. 24:517–526. [DOI] [PubMed] [Google Scholar]
- Simonet JC, Sunnen CN, Wu J, Golden JA, Marsh ED. 2015. Conditional loss of Arx from the developing dorsal telencephalon results in behavioral phenotypes resembling mild human ARX mutations. Cereb Cortex. 25:2939–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson TI, Pratt T, Mason JO, Price DJ. 2009. Normal ventral telencephalic expression of Pax 6 is required for normal development of thalamocortical axons in embryonic mice. Neural Dev. 4:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southwell DG, Nicholas CR, Basbaum AI, Stryker MP, Kriegstein AR, Rubenstein JL, Alvarez-Buylla A. 2014. Interneurons from embryonic development to cell-based therapy. Science. 344:1240622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenman J, Toresson H, Campbell K. 2003. Identification of two distinct progenitor populations in the lateral ganglionic eminence: implications for striatal and olfactory bulb neurogenesis. J Neurosci. 23:167–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Dykes IM, Liang X, Eng SR, Evans SM, Turner EE. 2008. A central role for Islet 1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci. 11:1283–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liang X, Najafi N, Cass M, Lin L, Cai C-L, Chen J, Evans SM. 2007. Islet 1 is expressed in distinct cardiovascular lineages, including pacemaker and coronary vascular cells. Dev Biol. 304:286–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sussel L, Marin O, Kimura S, Rubenstein JL. 1999. Loss of Nkx2.1 homeobox gene function results in a ventral to dorsal molecular respecification within the basal telencephalon: evidence for a transformation of the pallidum into the striatum. Development. 126:3359–3370. [DOI] [PubMed] [Google Scholar]
- Tamamaki N, Fujimori KE, Takauji R. 1997. Origin and route of tangentially migrating neurons in the developing neocortical intermediate zone. J Neurosci. 17:8313–8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamamaki N, Yanagawa Y, Tomioka R, Miyazaki J-I, Obata K, Kaneko T. 2003. Green fluorescent protein expression and colocalization with calretinin, parvalbumin, and somatostatin in the GAD67-GFP knock-in mouse. J Comp Neurol. 467:60–79. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsani D, Fu Y, Lu J, Lin Y, et al. 2011. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 71:995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torigoe M, Yamauchi K, Kimura T, Uemura Y, Murakami F. 2016. Evidence that the laminar fate of LGE/CGE-derived neocortical interneurons is dependent on their progenitor domains. J Neurosci. 36:2044–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Alvarez-Dolado M, Erskine L, Alvarez-Buylla A. 2003. Permissive corridor and diffusible gradients direct medial ganglionic eminence cell migration to the neocortex. Proc Natl Acad Sci U S A. 100:727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Turnbull DH, Nery S, Fishell G, Alvarez-Buylla A. 2001. In utero fate mapping reveals distinct migratory pathways and fates of neurons born in the mammalian basal forebrain. Development. 128:3759–3771. [DOI] [PubMed] [Google Scholar]
- Wonders CP, Anderson SA. 2006. The origin and specification of cortical interneurons. Nat Rev Neurosci. 7:687–696. [DOI] [PubMed] [Google Scholar]
- Xu Q. 2004. Origins of cortical interneuron subtypes. J Neurosci. 24:2612–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Tam M, Anderson SA. 2008. Fate mapping Nkx2.1-lineage cells in the mouse telencephalon. J Comp Neurol. 506:16–29. [DOI] [PubMed] [Google Scholar]
- Yang L, Cai C-L, Lin L, Qyang Y, Chung C, Monteiro RM, Mummery CL, Fishman GI, Cogen A, Evans S. 2006. Isl1Cre reveals a common Bmp pathway in heart and limb development. Development. 133:1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ML, Wu CF. 1997. Alterations in frequency coding and activity dependence of excitability in cultured neurons of Drosophila memory mutants. J Neurosci. 17:2187–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou F-W, Chen H-X, Roper SN. 2009. Balance of inhibitory and excitatory synaptic activity is altered in fast-spiking interneurons in experimental cortical dysplasia. J Neurophysiol. 102:2514–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that most of the data supporting the findings of this study are available within the paper and its supplementary information. Additional data or information about the study are available on request from the corresponding authors (JHW, EDM).


