Abstract
Several Alzheimer’s disease (AD) atrophy subtypes were identified, but their brain network properties are unclear. We analyzed data from two independent datasets, including 166 participants (103 AD/63 controls) from the DZNE-longitudinal cognitive impairment and dementia study and 151 participants (121 AD/30 controls) from the AD neuroimaging initiative cohorts, aiming to identify differences between AD atrophy subtypes in resting-state functional magnetic resonance imaging intra-network connectivity (INC) and global and nodal network properties. Using a data-driven clustering approach, we identified four AD atrophy subtypes with differences in functional connectivity, accompanied by clinical and biomarker alterations, including a medio-temporal-predominant (S-MT), a limbic-predominant (S-L), a diffuse (S-D), and a mild-atrophy (S-MA) subtype. S-MT and S-D showed INC reduction in the default mode, dorsal attention, visual and limbic network, and a pronounced reduction of “global efficiency” and decrease of the “clustering coefficient” in parietal and temporal lobes. Despite severe atrophy in limbic areas, the S-L exhibited only marginal global network but substantial nodal network failure. S-MA, in contrast, showed limited impairment in clinical and cognitive scores but pronounced global network failure. Our results contribute toward a better understanding of heterogeneity in AD with the detection of distinct differences in functional connectivity networks accompanied by CSF biomarker and cognitive differences in AD subtypes.
Keywords: Alzheimer’s disease, brain structure, graph theory, independent component analysis, resting-state connectivity
Introduction
Alzheimer’s disease (AD) shows considerable heterogeneity in central disease characteristics among individual patients, who may differ in their cognitive profiles (Scheltens et al. 2017) and biomarker patterns (Mitelpunkt et al. 2020). Postmortem studies separating groups with distinguishable atrophy patterns and histopathological features (Murray et al. 2011; Janocko et al. 2012) suggest the existence of biologically distinct AD subtypes, supported by evidence from magnetic resonance imaging (MRI), tau positron-emission-tomography (PET) (Whitwell et al. 2018), and clinicopathological research (Whitwell et al. 2012).
An MRI-based classification of subtypes can be achieved by visual atrophy ratings (Persson et al. 2017; Ferreira et al. 2019) or data-driven methods (Noh et al. 2014; Hwang et al. 2016; Zhang et al. 2016; Dong et al. 2017; Park et al. 2017). Most studies, including those in prodromal disease (Ten Kate et al. 2018), subdivide AD atrophy patterns into 1) a typical subtype with accentuated pathology of the hippocampus and association cortex; 2) a limbic predominant subtype with atrophy comprising the limbic system, including the hippocampus; 3) a hippocampal sparing subtype; and 4) a minimal atrophy subtype (Ferreira et al. 2020). These AD subtypes differ in their clinical progression rate, neurocognitive scores, years of education, disease duration, genotype, and cerebrospinal fluid (CSF) biomarker profiles (Ten Kate et al. 2018; Ferreira et al. 2020); further research is warranted to better characterize the underlying pathophysiological differences. To our best knowledge, differences in functional connectivity of resting-state networks (RSNs) between AD subtypes together with neurocognitive and biomarker data have not been explored yet. Furthermore, most previous studies classified patients based on clinical data rather than biomarker information, resulting in heterogeneous datasets. Here, we minimized heterogeneity and potential misdiagnoses by using a biomarker-based classification scheme informed by clinical diagnoses (Jack Jr et al. 2018).
The widespread loss of cortical neuronal connections in AD causes disruptions of brain connectivity (Braskie et al. 2010). Resting-state functional MRI can quantify the degeneration of the cerebral functional architecture and is widely used to investigate intrinsic large-scale neural networks (Biswal et al. 2010). Coherent patterns in spontaneous fluctuations of the blood oxygen level depended (BOLD) signal represent temporarily stable and reproducible intrinsic brain networks, overlapping with individual cognitive and behavioral characteristics (Yeo et al. 2011). The decline in functional connectivity is associated with disease progression and is found typically in AD in the default-mode network (DMN), linked to episodic memory processing (Greicius et al. 2004) and covering hotspots of amyloid-β (Aβ) and tau pathology (Buckner et al. 2005).
Graph theory is a framework used to characterize the behavior of complex brain networks (Bullmore and Sporns 2009). Connectome-based analyses allow measuring network segregation (i.e., “clustering coefficient,” “modularity,” and “transitivity”) and integration (i.e., “global efficiency” and “degree”). Global efficiency, modularity, and transitivity relate to large-scale networks, whereas clustering coefficient and degree characterize network properties at a local level (Watts and Strogatz 1998; Supekar et al. 2008). On a nodal level, highly connected regions, referred to as hub regions, are of primary interest. Regions with a high number of connections can be detected by calculating the degree (Farahani et al. 2019). Previous studies in AD revealed decreased network segregation measures (Supekar et al. 2008) and decreased measures of network integration compared with controls (Sanz-Arigita et al. 2010). In addition, clustering coefficient and modularity are decreased in AD (Brier et al. 2014).
Recently, differences in structural connectivity (Ferreira et al. 2019) and cognitive performance (Ten Kate et al. 2018) between different AD subtypes have been characterized. However, alterations in functional connectivity remain to be explored. Here, we aimed to explore heterogeneity in network properties in the DMN and other RSNs between distinct AD subtypes to investigate how cognitive and AD biomarker differences are associated with these functional network alterations.
Methods and Materials
Data included in this study originate from datasets of two independent study cohorts. The first dataset was obtained from the AD neuroimaging initiative (ADNI) launched in October 2004 (ClinicalTrials.gov IDs: NCT02854033, NCT01231971). The second dataset was obtained from the Deutsches Zentrum für Neurodegenerative Erkrankungen (DZNE)-longitudinal cognitive impairment and dementia study (DELCODE), an observational brain imaging cohort (German Clinical Trials Register: DRKS00007966). Per ADNI and DELCODE protocols, all procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee. Experiments were undertaken with the understanding and written consent of each subject. All local institutional review boards and ethical committees approved the study protocol (Lancichinetti and Fortunato 2012).
Participants
The AD and control groups were defined considering Aβ status and clinical dementia rating (CDR) score. Participants were included based on the availability of T1-weighted structural MRI, resting-state functional MRI, and Aβ status information.
The participants in the ADNI dataset were recruited for the ADNI2, ADNI-go, and ADNI3 convenience cohorts, details about the general ADNI inclusion and exclusion criteria can be found in the ADNI procedures manual available online (https://adni.loni.usc.edu/wp-content/uploads/2008/07/adni2-procedures-manual.pdf). Aβ-positivity in ADNI was defined according to established cut-points as CSF Aβ1–42 concentration < 980 pg/mL (Galasko et al. 2019), or 18F-AV-45 or 18F-Florbetaben Aβ-PET normalized composite score with a cutoff > 1.11 or > 1.08 standardized uptake value ratio, respectively (Landau et al. 2013), resulting in the ADNI dataset of n = 160. After quality assessment and preprocessing of the MRI data, n = 9 participants did not meet the predefined image quality criteria (for details section see MRI Preprocessing) and were excluded from the subsequent analyses, resulting in a final dataset of n = 151 participants (mean age = 75 years, 84 females), including n = 121 Aβ-positive and CDR ≥ 0.5 AD patients (mean age = 75 years, 63 females), and n = 30 Aβ-negative and CDR = 0 controls (mean age = 77 years, 21 females).
In total, 171 participants in the DELCODE dataset met the inclusion criteria. N = 5 participants did not meet the predefined image quality criteria and were excluded from all further analyses, resulting in a final cohort of n = 166 (mean age = 72 years, 93 females). Aβ-positive participants with CSF Aβ1–42 < 496 pg/ml (Jessen et al. 2018) and CDR ≥ 0.5 were defined as AD (n = 103, mean age = 74 years, 57 females), whereas Aβ-negative participants were defined as healthy controls (HC) with CSF Aβ1–42 > 496 pg/ml and CDR = 0 (n = 63, mean age = 69 years, 32 females).
MRI Acquisition
The subjects included in the present study were scanned at various sites with 3T MRI scanners manufactured by GE Healthcare, Philips Medical Systems, or Siemens Healthineers. The Alzheimer’s Disease Neuroimaging Initiative (ADNI) MRI protocol is reported elsewhere (http://adni.loni.usc.edu/methods/mri-tool/mri-acquisition/). DELCODE MRI scanning was performed at nine different DZNE imaging sites on Siemens Healthineers 3T MRI scanners, using synchronized acquisition parameters. We included T1-weighted MPRAGE sequences (repetition time [TR], 2500 ms; echo time [TE], 4.37 ms; flip angle [FA], 7°; and isotropic voxel size, 1 mm) in our analyses. FMRI imaging was performed using the following parameters: DELCODE: 180 volumes; FoV, 224 × 224 × 165 mm; TR, 2580 ms; TE, 30 ms; FA, 80°; isotropic voxel size, 3.5 mm; 7 min 54 s and ADNI: 200 volumes; FoV, 220 × 220 × 160 mm; TR, 3000 ms; TE, 30; FA = 90°; and isotropic voxel size: 3.4 mm; 10 min.
MRI Preprocessing
Every scan was visually inspected by an experienced radiologist for completeness, cuts, subject motion, and other artifacts (e.g., “blurring,” “echoes,” “ghosting”). Following this step, the image was classified as “usable, questionable, and unusable.” We included only images classified as usable in the analysis.
Brain atrophy was analyzed using FreeSurfer version 6 (http://surfer.nmr.mgh.harvard.edu/). All T1-weighted images were processed in the FreeSurfer segmentation recon-all pipeline (Fischl et al. 2002). Segmentations were visually checked for accuracy and corrected if necessary.
Functional connectivity analysis was performed using the CONN-fMRI Functional Connectivity Toolbox (V17, www.nitrc.org/projects/conn) and SPM 12 (www.fil.ion.ucl.ac.uk/spm/). The default preprocessing pipeline for volume-based analyses was used, comprising realignment, slice-time correction, segmentation and structural and functional normalization, ART-based outlier detection, and functional smoothing using a 6-mm kernel (https://web.conn-toolbox.org/fmri-methods/preprocessing-pipeline). Temporal filtering was performed to remove physiological noise. Assessment of motion in both cohorts revealed comparable results (DELCODE: 0.01 ± 0.12 [79.6% match with null hypothesis]; ADNI: 0.02 ± 0.12 [80.1% match with null hypothesis]). After preprocessing, region-of-interest (ROI)-based intrinsic connectivity was obtained with bivariate correlation matrices in cortical and subcortical ROIs, using the multimodal Brainnetome (BN) atlas (Fan et al. 2016), registered to the functional image. Correlation coefficients were Fisher-r-to-z-transformed consecutively.
AD Atrophy Subtype Identification
For subtype classification, individual cortical surfaces obtained from each participant’s T1-weighted MRI using FreeSurfers recon-all were registered to the FreeSurfer standard subject template (fsaverage6) and resampled to 40 962 vertices for each hemisphere to account for intersubject variability of brain shapes and size (Park et al. 2017). Subsequent analyses were performed using in-house MATLAB (The MathWorks, Inc.) scripts in both cohorts.
To obtain an atrophy z-score vector, representing the atrophy pattern of each AD subject, the mean cortical thickness value from every vertex in the AD subjects was subtracted from the cortical thickness values of every vertex in the controls divided by the standard deviation in both hemispheres. Atrophy z-score vectors were consecutively concatenated and a similarity matrix of correlation coefficients between the obtained atrophy z-score vectors of any two AD subjects was calculated.
To identify atrophy subtypes in the AD cohorts based on the correlation of atrophy pattern between any two subjects, an unsupervised cluster detection approach using the Louvain community analysis method implemented in the brain connectivity toolbox was applied (Rubinov and Sporns 2010). This subtyping approach uses the similarity correlation matrix and has previously shown high reproducibility and strong associations with cognitive performance (Park et al. 2017). This unsupervised clustering approach is suggested to be less vulnerable to sampling bias compared with hierarchical clustering approaches. The outcomes in hierarchical clustering tend to cluster based on the overall similarity of the cortical thickness rather than cortical atrophy patterns so that the chosen approach is suggested to be more sensitive to cortical atrophy (Park et al. 2017). In addition, the approach showed excellent reproducibility and the Louvain method was shown to be suitable for high-dimensional data (Blondel et al. 2008).
To determine the ideal cluster number, we tested three-cluster and four-cluster solutions where four-cluster solutions were generally more suitable to subtypes previously found in neuroimaging datasets (Ferreira et al. 2017; Ten Kate et al. 2018; Ferreira et al. 2019), with several studies report four subtypes in a recent review by Ferreira et al. (Ten Kate et al. 2018; Ferreira et al. 2020).
We modified the approach using a consensus community structure approach to obtain stable results through 1000 iterations with a correction of individual-level modular decomposition (Lancichinetti and Fortunato 2012). The level of subtyping can be controlled by the gamma value, a resolution parameter of the Louvain community structure analysis controlling the number of clusters, with a smaller value resulting in a smaller number of subtypes (Blondel et al. 2008). The gamma value was controlled, obtaining subtyping results equivalent to previous imaging and postmortem studies (Murray et al. 2011; Whitwell et al. 2012; Ten Kate et al. 2018).
Dice Overlap
To quantify the overlap of atrophic regions between the two datasets, we compared the regions after setting the threshold level of uncorrected log-p > 1.31 (P < 0.05) on vertex-wise overlay imaging data derived from the statistical comparison with controls. We calculated dice coefficients (DCEs) between atrophy subtypes from both datasets in MATLAB.
Functional Connectivity Analysis
We analyzed functional connectivity characteristics of the atrophy subtypes in seven cortical intrinsic functional connectivity networks (Yeo et al. 2011). Within each network, intra-network connectivity (INC) composite score was calculated by averaging the network ROIs (based on the cortical Brainnetome atlas parcellation) functional connectivity Fisher-r-to-z-transformed correlation values (Brier et al. 2012). The ROIs with the nodes used for the functional network analysis are presented in Supplementary Figure 1 and Supplementary Table 2. To investigate the global and local network properties and differences between the different subtypes in the resting-state brain networks, we performed a graph theory network analysis. An undirected network was constructed from the functional connectivity correlation values with subsequent analysis of graph metrics comparing each subtype using permutation-based analysis of covariance (ANCOVA) statistics with Benjamini and Hochberg false discover rate (FDR) correction to control for multiple comparisons in the GraphVar toolbox (Kruschwitz et al. 2015). The following graph metrics were calculated on a global level: 1) global transitivity (referred to as global clustering coefficient), 2) global efficiency, 3) modularity using the Louvain method, and 4) global strength. On a local level 1) local efficiency, 2) degree, 3) clustering coefficient, and 4) betweenness centrality were investigated (Rubinov and Sporns 2010). The visualization of the global and local network properties was obtained using ggplot2 in R (https://www.r-project.org/) and BrainNetViewer (Xia et al. 2013). We showed our findings on the median threshold.
Clinical Characteristics and CSF Biomarkers
The severity of dementia symptoms was quantified using the CDR sum of the boxes (CDR-SoB) score. The cognitive performance was assessed using established cognitive composite scores for memory (MEM) and executive functions (EXEC) in the DELCODE (Jessen et al. 2018) and ADNI (Crane et al. 2012; Gibbons et al. 2012) datasets. In addition, the Mini-Mental-State Examination (MMSE) score is reported given its high relevance in everyday clinical practice. CSF biomarkers were assessed in both cohorts using established commercially available analysis kits, following standardized procedures (Jessen et al. 2018). The CSF concentrations in the ADNI cohort for Aβ-42, p-tau181 was quantified in aliquoted samples, analyzed using the electrochemiluminescence immunoassay Elecsys on a fully automated Elecsys cobas e 601 instrument (Roche Diagnostics GmbH) using a single lot of each reagent for each of the 3 measured biomarkers. In the DELCODE cohort, V-PLEX Aβ Peptide Panel 1 (6E10) Kit (K15200E) and V-PLEX Human Total Tau Kit (K151LAE; Meso Scale Diagnostics LLC), and Innotest Phospho-Tau (181P; 81 581; Fujirebio Germany GmbH) were used.
Statistical Analysis
Statistical differences between AD atrophy subtype groups and HC in each dataset were tested on cortical z-score maps using two-tailed, two-sample unpaired n = 1000 permutation-based t-tests in FSL-Permutation Analysis of Linear Models (Winkler et al. 2014), applying threshold free cluster enhancement and controlling for family-wise error rate (FWE); additionally, uncorrected contrasts are reported (both P < 0.05).
SPSS (IBM, v25) and R (https://www.r-project.org/) were used for statistical analyses. Subtype group differences in relevant confounding variables (age, gender, APOE genotype, and educational years) were compared with Kruskal–Wallis tests. We detected significant differences in relevant covariates between the subtype groups in the pooled dataset for educational years and gender but not for age or APOE genotype. All consequent subtype group comparisons were therefore adjusted for gender and educational years. All fcMRI analyses were adjusted to account for different imaging acquisition sites using several MRI vendors with harmonized protocols in different cohorts. Functional connectivity scores, neurocognitive scores, and CSF biomarker scores were compared in the entire cohort as well as between subtypes using ANCOVA. Post-hoc pairwise comparisons were Bonferroni corrected as appropriate. Results were considered significant at P < 0.05 (two-tailed). Deviation from normal distribution was assessed by visual inspection of the data distribution and Shapiro–Wilk test. Deviations from the normality distribution were detected for the functional connectivity and CSF biomarker scores. We transformed these variables into normal scores of ranks using the Rankit’s method (Chambers 2018). Cognitive composite scores and CSF biomarkers were z-transformed within each cohort to compare the results independent of the measuring scale.
Comparisons of network properties between the subtypes were performed in the GraphVar Toolbox (Kruschwitz et al. 2015) using nonparametric permutation tests at a range of network thresholds (min = 0.1 to max = 0.4) with a 0.02 interval. Nonparametric analyses were conducted testing against shuffled data with n = 1000 permutations. A median threshold of 0.24 was used for comparisons of network measures. There is currently a no broader consensus on what threshold should be reported in graph-based analyses (Garrison et al. 2015). Our decision to report a median threshold was based on the idea to provide the reader with the most representative number as an overview. A random networks/groups FDR correction for multiple permutation comparisons was used at P < 0.05 (two-tailed) for global and nodal measures at various network densities.
Data Availability Statement
All ADNI data are deposited in a publicly accessible repository and can be accessed at adni.loni.usc.edu. For the DELCODE dataset, anonymized data analyzed in the current study will be made available upon reasonable request from qualified investigators.
Results
Characteristics of the Cohorts
The characteristics of the ADNI and DELCODE cohorts are presented in Table 1. The AD participants in both cohorts demonstrated comparable sociodemographic and neurocognitive measures, except for years of education, with more years in ADNI. In DELCODE, controls were younger and included a lower proportion of female participants compared with ADNI controls.
Table 1.
Characteristics of the two study cohorts (ADNI and DELCODE)
| DELCODE | ADNI | |||||
|---|---|---|---|---|---|---|
| AD (N = 103) | HC (N = 63) | AD (N = 121) | HC (N = 30) | P between AD groups | P between HC groups | |
| Age, mean (SD) | 74 (6) | 69 (5) | 75 (8) | 77 (8) | 0.33a | <0.001a |
| Sex, no. female %, (SD) | 57 (55) | 32 (49) | 63 (52) | 21 (70) | 0.72b | <0.001b |
| Years of education, mean (SD) | 14 (3) | 14 (3) | 16 (2) | 16 (3) | <0.001a | 0.07a |
| MMSE, mean (SD) | 26 (3) | 29 (1) | 25 (4) | 29 (1)d | 0.36a | 0.06a |
| CDR-SoB, mean (SD) | 2.7 (2.2) | 0 | 3.2 (2.6) | 0 | 0.06a | 1a |
| APOE, no. (%) ε4 allele carrier (SD) | 66 (64)d | 9 (14) | 68 (56)c | 6 (21) | 0.39b | 0.64b |
Abbreviation: APOE, apolipoprotein ε4 genotype.
aKruskal–Wallis test.
bChi-squared-test.
cMissing data for n = 4 participants.
dMissing data for n = 1 participant.
Atrophy Pattern in AD Subtypes
In both datasets (DELCODE and ADNI), similar four subtypes were identified, including 1) a medio-temporal predominant subtype (S-MT); 2) a limbic predominant subtype (S-L); 3) a diffuse subtype (S-D); and 4) a mild-atrophy subtype, with relative parahippocampal sparing (S-MA). The differences between the four subtypes within the AD group and compared with the HC are shown in Figure 1A for the ADNI dataset and Figure 1B for the DELCODE dataset. S-MT showed atrophy mainly in the (medial) temporal lobe, while S-L had an atrophy pattern, including the cingulate cortex and parahippocampal brain areas. In contrast, S-D was associated with a diffuse atrophy pattern, including large areas of the neocortex comprising the parietal lobe. The S-MA subtype was characterized by patchy cortical atrophy with a relatively low degree of parahippocampal atrophy. Importantly, cortical atrophy in each of the four subtypes followed a similar pattern in both datasets, with overall more severe atrophy across all subtypes in DELCODE. The spatial overlap of the atrophy subtypes between the two datasets was evaluated using the DCE, showing good overlap for S-MT (DCE = 0.44), S-L (DCE = 0.51), and S-D (DCE = 0.64) and less pronounced overlap for S-MA (DCE = 0.07), most likely explained by the patchy pattern with less atrophy overall.
Figure 1 .
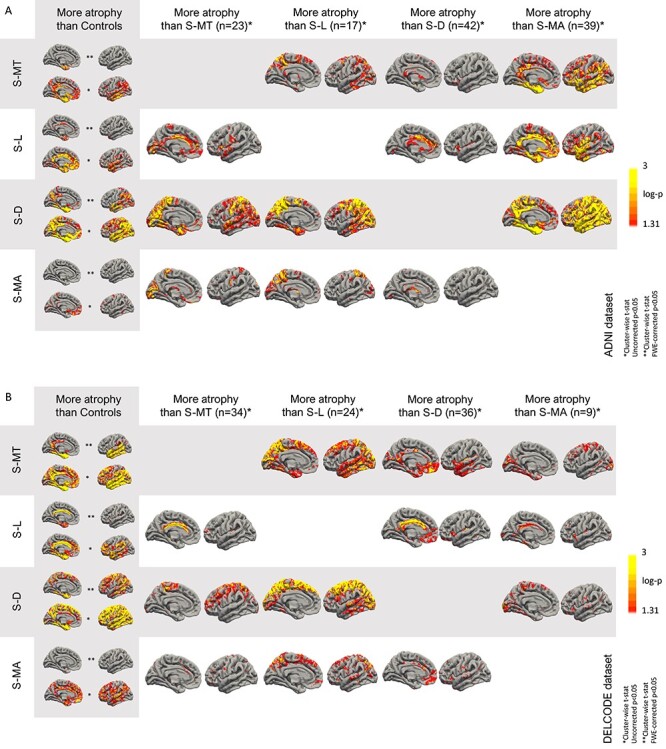
Atrophy regions in Alzheimer’s disease subtypes versus healthy control subjects across atrophy subtypes in the ADNI (A) and DELCODE (B) dataset. *Uncorrected P < 0.05; **FWE-corrected P < 0.05.
Clinical, Cognitive and CSF Biomarker Differences between the Atrophy Subtypes
Similar differences in clinical and cognitive scores and CSF biomarkers between the four subtypes were observed in both datasets. Dementia severity measured by CDR was highest in the S-MT and S-D subgroups with lower scores in S-L and S-MA and HC. Concordantly, cognitive performance measured by the MMSE was lowest in S-MT and S-D with higher scores in S-L and S-MA and HC (Table 2). Since the atrophy subtypes showed good overlap and similar clinical characteristics across DELCODE and ADNI, we pooled the participants in each sub-group across the datasets for all subsequent analyses as shown before (Ten Kate et al. 2018).
Table 2.
Differences in sociodemographic characteristics, APOE genotype, Alzheimer’s disease severity, cognitive performance, and CSF biomarker levels between the atrophy subtypes and HC in the pooled dataset
| HC (n = 93) | S-MT (n = 57) | S-L (n = 41) | S-D (n = 78) | S-MA (n = 48) | P-value (overall) | S-MT versus HC | S-L versus HC | S-D versus HC | S-MA versus HC | S-MT versus S-L | S-MT versus S-D | S-MT versus S-MA | S-L versus S-D | S-L versus S-MA | S-D versus S-MA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Post-hoc comparison P-value | ||||||||||||||||
| Age, mean (SD)a | 72 (7) | 73 (8) | 75 (6) | 75 (6) | 74 (7) | 0.01 | 0.54 | 0.02* | 0.01* | 0.06 | 0.3 | 0.17 | 0.5 | 0.7 | 0.56 | 0.9 |
| Sex, no. female (%)b | 53 (57) | 40 (70.2) | 18 (43.9) | 39 (50) | 23 (47.9) | 0.052 | — | — | — | — | — | — | — | — | — | — |
| Years of education, mean (SD)a | 15 (3) | 14 (3) | 15 (3) | 15 (3) | 16 (3) | <0.001 | 0.01* | 0.45 | 0.47 | 0.01* | 0.5 | 0.2 | <0.001* | 0.6 | 0.002* | 0.002* |
| APOE, no. (%)b ε4 allele carrierb | 15 (16) | 32 (60) | 21 (53) | 50 (64) | 31 (65) | <0.001 | <0.001* | <0.001* | <0.001* | <0.001* | 0.41 | 0.65 | 0.65 | 0.19 | 0.22 | 0.95 |
| CDR-SoB, mean (SD)c | 0 (0) | 3.4 (3) | 2.1 (1.8) | 3.7 (2.5) | 2.0 (1.5) | <0.001 | <0.001* | <0.001* | <0.001* | <0.001* | 0.11 | 0.01* | 0.12 | 0.01* | 0.93 | 0.01* |
| MMSE, mean (SD)c | 29 (1) | 24.8 (4.3) | 27.0 (2.7) | 24.4 (4.6) | 27.5 (2.4) | <0.001 | <0.001* | <0.001* | <0.001* | <0.001* | 0.03* | 0.71 | 0.002* | 0.01* | 0.4 | <0.001* |
| CSF Aβ1–42, z-scoref (SD)c | 0 (1) | −2.2 (0.4) | −2.1 (0.5) | −2.1 (0.4) | −2.0 (0.5) | <0.001 | <0.001* | <0.001* | <0.001* | <0.001* | 0.51 | 0.4 | 0.18 | 0.93 | 0.5 | 0.49 |
| CSF p-tau181, z-scored (SD)c | 0 (1) | 3.3 (3.0) | 1.2 (2.2) | 4.5 (4.0) | 4.4 (5.0) | <0.001 | <0.001* | 0.02* | <0.001* | <0.001* | 0.01* | 0.06 | 0.37 | <0.001* | <0.001* | 0.4 |
| CSF t-tau, z-scored (SD)c | 0 (1) | 2.8 (2.7) | 1.6 (2.4) | 4.5 (4.1) | 4.2 (4.8) | <0.001* | 0.001* | 0.09 | <0.001* | <0.001* | 0.11 | 0.01* | 0.21 | <0.001* | 0.01* | 0.25 |
| Mean Hippocampal volume, z-score (SD)c | 0 (1) | −0.9 (2.0) | 0.2 (2.1) | −0.5 (2.1) | −0.8 (1.3) | 0.001* | 0.03* | 0.32 | 0.12 | 0.001* | 0.01* | 0.47 | 0.24 | 0.02* | <0.001* | 0.047* |
| MEM, z-score (SD)c | 0 (1) | −4.2 (2.3) | −2.8 (1.9) | −3.8 (2.2) | −1.6 (1.7) | <0.001* | <0.001* | <0.001* | <0.001* | <0.001* | <0.001* | 0.16 | <0.001* | 0.01* | 0.3 | <0.001* |
| EXEC, z-score (SD)c | 0 (1) | −2.9 (1.8) | −2.2 (1.5) | −2.8 (1.7) | −1.1 (1.5) | <0.001* | <0.001* | <0.001* | <0.001* | <0.001* | 0.02* | 0.94 | <0.001* | 0.01* | 0.01* | <0.001 |
Notes: aAnalysis of covariance with adjustments for age, sex, sites, APOE genotype and years of education. Post-hoc pairwise comparisons Bonferroni corrected; bChi-squared test; cAnalysis of covariance with adjustments for age, sex, APOE genotype, sites and years of education; dCSF biomarker data in n = 19 participants were missing (n = 8 in S-MT, n = 3 in S-L, n = 6 in S-D, and n = 2 in S-MA). *Significant difference P < 0.05 in post-hoc tests.
Abbreviations: Aβ, amyloid β; p-tau, phosphorylated-tau; t-tau, total-tau; APOE, apolipoprotein ε4 genotype.
ANCOVA test revealed significant differences between the subtypes and HC for MEM (P < 0.001), EXEC (P < 0.001), CSF t-tau (P < 0.001) and p-tau181 (P < 0.001), and Aβ1–42 (P < 0.001). For MEM, post-hoc pairwise comparisons showed lower z-scores in S-MT and S-D compared with S-L and S-MA and HC. A similar pattern was found for EXEC, with lower z-scores in S-MT and S-D compared with S-L and S-MA and HC. CSF t-tau was higher in S-MT and S-D compared with S-L and S-MA and HC; similar differences were also observed for p-tau181 with higher z-scores in S-MT and S-D compared with S-L and S-MA and HC (Fig. 2A). APOE genotype did not differ between the subtypes. In the comparison between subtypes, hippocampal atrophy was most prominent in S-MT and S-D. Differences in participant’s characteristics, cognitive composite, hippocampal volume, and CSF biomarker z-scores between the subtypes are presented in Table 2. Independent analysis results for both cohorts are shown in Supplementary Table 1.
Figure 2 .
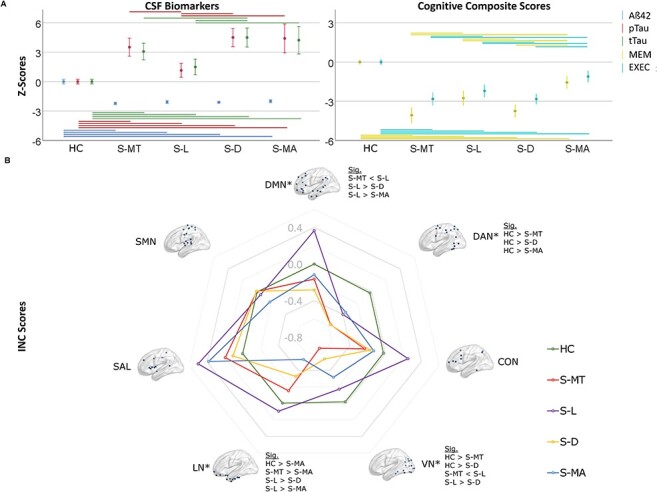
(A) Boxplots of the mean cognitive composite and cerebrospinal fluid biomarker normalized scores ±95% confidence interval (in z-scores). Significant post-hoc comparisons are shown with a line, top: sig. comparisons among the subtypes, bottom: sig. comparisons between HC and subtypes. (B) Spider plot of the estimated mean z-scores of INC in the resting-state networks. Z-scores of the HC are shown for comparison. Abbreviations: Aβ42, amyloid-β42; tTau, total tau; pTau, phosphorylated tau; INC, intrinsic network connectivity; SMN, sensorimotor network; *P (overall) < 0.05; Sig., significantly differing subgroups in post-hoc tests when FDR-corrected two-tailed P (overall) < 0.05.
Intra-network Resting-State Functional Connectivity Differences
Following FDR correction for multiple comparisons assessing the INC in seven resting-state networks, differences between the subtypes and HC in the DMN (P = 0.035), LN (P = 0.035), dorsal attention network (DAN; P = 0.035) and visual network (VN; P = 0.007) but not in the frontoparietal network (CON; P = 0.28), salience network (SAL; P = 0.18)and somatosensory network (SMN; P = 0.89) and were detected using ANCOVA test. Subsequent post-hoc comparisons revealed a higher INC of the DMN in S-L versus S-MT (P = 0.01), S-L versus S-D (P < 0.001) and S-L versus S-MA (P < 0.02), higher INC in the DAN in HC versus S-MT (P = 0.003), HC versus S-D (P = 0.001) and HC versus S-MA (P = 0.04). In the LN, higher INC was revealed in S-MT versus S-MA (P = 0.03), S-L versus S-D (P = 0.03), S-L versus S-MA (P = 0.01) and HC versus S-MA (P = 0.01). In the VN INC was higher in HC versus S-MT (P < 0.001), HC versus S-D (P = 0.001), S-L versus S-MT (P = 0.01) and S-L versus S-D (P = 0.04). Z-score differences between the subtypes and HC in the pooled dataset are presented in Figure 2B and Table 3. INC differences between HC and subtypes for both cohorts independently are shown in Supplementary Table 3 and Supplementary Figure 2.
Table 3.
Adjusted group means of INC scores of the pooled dataset
| RSN | HC | S-MT | S-L | S-D | S-MA | S-MT versus HC | S-L versus HC | S-D versus HC | S-MA versus HC | S-MT versus S-L | S-MT versus S-D | S-MT versus S-MA | S-L versus S-D | S-L versusS-MA | S-D versus S-MA | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean (SE) | P (overall) | P FDR corrected | P post hoc comparisons | ||||||||||||||
| DMN | −0.01 (0.11) | −0.18 (0.14) | 0.38 (0.16) | −0.27 (0.12) | −0.12 (0.15) | 0.02* | 0.035* | 0.36 | 0.054 | 0.11 | 0.57 | 0.01* | 0.58 | 0.78 | <0.001* | 0.02* | 0.40 |
| DAN | 0.01 (0.12) | −0.58 (0.15) | −0.37 (0.17) | −0.58 (0.12)† | −0.42 (0.16) | 0.01* | 0.035* | 0.003* | 0.07 | 0.001* | 0.04* | 0.36 | 1.00 | 0.48 | 0.32 | 0.83 | 0.43 |
| CON | −0.06 (0.12) | −0.24 (0.15) | 0.25 (0.17) | −0.16 (0.12) | −0.09 (0.16) | 0.24 | 0.28 | 0.33 | 0.15 | 0.55 | 0.88 | N.a. | N.a. | N.a. | N.a. | N.a. | N.a. |
| SAL | −0.02 (0.14) | 0.12 (0.18) | 0.53 (0.20) | 0.13 (0.15) | 0.47 (0.19) | 0.13 | 0.18 | 0.57 | 0.03* | 0.51 | 0.05 | N.a. | N.a. | N.a. | N.a. | N.a. | N.a. |
| LN | −0.04 (0.10) | −0.10 (0.13) | 0.07 (0.15) | −0.32 (0.11) | −0.52 (0.14)† | 0.02* | 0.035* | 0.76 | 0.53 | 0.08 | 0.01* | 0.4 | 0.18* | 0.03* | 0.03 | 0.01* | 0.25 |
| VN | 0.02 (0.12) | −0.74 (0.14) | −0.14 (0.16) | −0.55 (0.12) | −0.32 (0.15) | <0.001* | 0.007* | <0.001* | 0.41 | 0.001* | 0.08 | 0.01* | 0.30 | 0.05 | 0.04* | 0.42 | 0.24 |
| SMN | −0.06 (0.10) | −0.02 (0.13) | −0.03 (0.15) | 0.04 (0.11) | −0.13 (0.14) | 0.89 | 0.89 | 0.78 | 0.84 | 0.49 | 0.72 | N.a. | N.a. | N.a. | N.a. | N.a. | N.a. |
Notes: *Two-tailed P < 0.05.
†P < 0.05 in analysis of covariance -with adjustments for age, sex, APOE genotype and years of education—between any atrophy subgroup DELCODE versus ADNI.
AD Subtype Characteristics in Global Network Analysis
In a graph theory analysis of global network properties on whole-brain level, significant differences between the subtypes in global efficiency (P < 0.001), strength (P < 0.001) and transitivity (P < 0.001), but not in modularity (P = 0.68) were revealed. On a network level, DMN but not LN showed significant differences between the subtypes for global efficiency (P < 0.001), strength (P < 0.001) and transitivity (P < 0.006), but not modularity (P = 0.61).
In post-hoc pairwise comparisons on a global level, S-L showed higher global efficiency and transitivity versus S-MT, S-D. Both were lower in S-MT than in S-MA. Moreover, S-L exhibit lower transitivity versus S-MA, and S-MT lower global efficiency than S-D. Global strength was lowest in S-MT and highest in S-L, with S-L significantly higher than S-MT and S-D and S-MA higher than S-MT, but lower than S-L.
Within the DMN, S-L showed the highest global efficiency versus S-MT, S-D and S-MA. Global efficiency was higher in S-MA than in S-MT. Additionally, S-L had higher transitivity in comparison with S-MT and higher transitivity versus S-D and S-MA. Again, global strength was lowest in S-MT and highest in S-L, with S-L significantly higher than S-MT and S-D and S-MA (Table 4 and Fig. 3).
Table 4.
Adjusted group means of graph theory derived global network properties on a whole brain level and within the DMN, the LN, and VN
| Group means | S-MT | S-L | S-D | S-MA | P (overall) | S-MT versus S-L | S-MT versus S-D | S-MT versus S-MA | S-L versus S-D | S-L versus S-MA | S-D versus S-MA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Global network | Efficiency | 0.23 | 0.26 | 0.24 | 0.25 | <0.001* | <0.001* | 0.02* | <0.001* | <0.001* | 0.06 | 0.1 |
| Modularity | 0.4 | 0.4 | 0.4 | 0.4 | 0.68 | — | — | — | — | — | — | |
| Strength | 5266 | 6130 | 5541 | 5745 | <0.001* | <0.001* | 0.05 | 0.002* | 0.001* | 0.03* | 0.18 | |
| Transitivity | 0.21 | 0.25 | 0.22 | 0.23 | <0.001* | <0.001* | 0.11 | 0.04* | <0.001* | 0.01* | 0.46 | |
| Within DMN | Efficiency | 0.3 | 0.34 | 0.31 | 0.32 | <0.001* | <0.001* | 0.17 | 0.02* | 0.001* | 0.03* | 0.21 |
| Modularity | 0.31 | 0.29 | 0.31 | 0.31 | 0.61 | — | — | — | — | — | — | |
| Strength | 199 | 225 | 203 | 207 | <0.001* | <0.001* | 0.41 | 0.24 | <0.001* | 0.006* | 0.56 | |
| Transitivity | 0.46 | 0.52 | 0.47 | 0.47 | 0.006* | 0.004* | 0.84 | 0.74 | 0.001* | 0.001* | 0.86 | |
| Within LN | Efficiency | 0.21 | 0.22 | 0.21 | 0.21 | 0.25 | — | — | — | — | — | — |
| Modularity | 0.37 | 0.36 | 0.38 | 0.37 | 0.30 | — | — | — | — | — | — | |
| Strength | 68 | 73 | 68 | 65 | 0.12 | — | — | — | — | — | — | |
| Transitivity | 0.25 | 0.27 | 0.24 | 0.22 | 0.14 | — | — | — | — | — | — | |
| Within visual network | Efficiency | 0.28 | 0.31 | 0.3 | 0.35 | 0.08 | — | — | — | — | — | — |
| Modularity | 0.33 | 0.34 | 0.33 | 0.3 | 0.28 | — | — | — | — | — | — | |
| Strength | 160 | 175 | 172 | 167 | 0.16 | — | — | — | — | — | — | |
| Transitivity | 0.39 | 0.43 | 0.42 | 0.4 | 0.33 | — | — | — | — | — | — |
Notes: *Two-tailed permutation-based FDR-corrected P < 0.05.
Figure 3 .
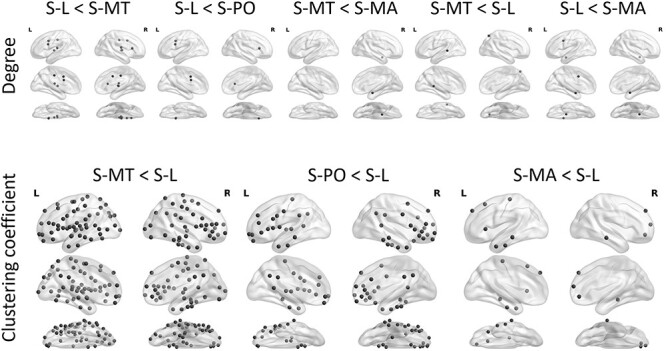
Differences between subtypes in degree and clustering coefficient in Brainnetome atlas derived regions of interest. Permutation based FDR-corrected two-tailed P < 0.05 are shown.
AD Subtype Characteristics in Nodal Network Analysis
Addressing the main research question of this study (i.e., how local changes in network properties of subtypes are related to characteristics of atrophy patterns), we calculated the nodal measures of betweenness centrality, degree, clustering coefficient and local efficiency on whole-brain level (median threshold = 0.24). Differences in degree (a measure of integration and one of the most important measures of network structure) are shown in Figure 3. The S-L subtype showed a reduced degree in the cingulate gyrus versus S-MT, S-D and S-MA. S-MT exhibited a reduced degree in the caudal area of the right parietal and left temporal lobe versus S-L. Clustering coefficient (indicating resilience against random network damage) was reduced in S-MT versus S-L in multiple ROIs comprising the frontal, temporal, parietal, and occipital lobe. A similar pattern, with pronounced changes in lateral temporal and frontal regions, comprising fewer significant ROIs was observed comparing S-D and S-L. S-MA showed reduced clustering coefficient in frontal and temporal regions versus S-L. Significant differences between the subtypes in clustering coefficient are shown in Figure 3.
To compare the differences of nodal measures within RS networks between the subtypes, we selected the nodes belonging to the DMN, LN, and VN, as these networks show significant differences in functional connectivity between the subtypes. Within nodes of the DMN, differences between the subtypes were found for local efficiency, comprising multiple ROIs in the frontal, temporal and parietal lobe as well as cingulate gyrus and precuneus reduced in S-L and S-MA compared to S-MT with a similar pattern in S-L versus S-D. Local efficiency was reduced in the frontal, parietal, and temporal lobe, including the precuneus in S-MA versus S-L. Clustering coefficient was significantly lower in S-MT and S-D versus S-L in the frontal and temporal lobe, the gyrus cinguli and the precuneus. However, clustering coefficient in S-L differs with S-MA in the frontal and temporal lobes. Degree was lower in the posterior temporal lobe in S-MT versus S-L and S-MA. Betweenness centrality was reduced in the cingulate gyrus in S-L versus S-MT. Nodes belonging to the LN showed significant reductions in local efficiency in the frontal and temporal lobe and the fusiform and parahippocampal gyrus. Nodes belonging to the VN showed in S-MT versus S-MA reduced local efficiency but increased local efficiency in S-L versus S-D in several regions. Clustering coefficient was reduced in S-MT versus S-L, comprising mainly parietal and occipital lobes as well as fusiform, parahippocampal, and cingulate gyri; but increased in S-L versus S-D in parahippocampal and cingulate gyri. Results of nodal graph measures on a network level for the DMN, LN, and VN are summarized in Table 5.
Table 5.
Summary of changes in nodal topography in regions of interest (ROIs) associated with the DMN (top) and LN (bottom) at a median threshold of 0.24
| Betweenness centrality | Degree | Clustering coefficient | Local efficiency | |
|---|---|---|---|---|
| DMN | ||||
| S-MT versus S-L | CG ↑ | pSTS ↓ | SFG, MFG, IFG, OrG, STG, MTG, ITG, PCun, CG ↓ | SFG, MFG, IFG, OrG, STG, MTG, ITG, pSTS,IPL, PCun, CG ↓ |
| S-MT versus S-D | — | — | — | |
| S-MT versus S-MA | — | — | — | IFG, OrG, STG, MTG, pSTS, IPL, CG ↑ |
| S-L versus S-D | — | — | SFG, IFG, OrG, MTG, ITG, PCun, CG ↑ | SFG, MFG, IFG, OrG, STG, MTG, ITG, pSTS, IPL, PCun, CG ↑ |
| S-L versus S-MA | — | — | SFG, OrG, MTG, ITG ↑ | SFG, IFG, OrG, MTG, ITG, PCun ↑ |
| S-D versus S-MA | — | — | — | — |
| LN | ||||
| S-MT versus S-L | — | — | MFG, OrG, ITG ↓ | MFG, OrG, STG, ITG, FuG, PhG ↓ |
| S-MT versus S-D | — | — | — | — |
| S-MT versus S-MA | — | — | — | STG, ITG, PhG ↓ |
| S-L versus S-D | — | — | MFG, OrG, ITG ↑ | MFG, OrG, STG, ITG, FuG, PhG ↑ |
| S-L versus S-MA | — | — | — | — |
| S-D versus S-MA | — | — | — | — |
| VN | ||||
| S-MT versus S-L | — | — | FuG, PhG, IPL, CG, LOcC ↓ | — |
| S-MT versus S-D | — | — | — | — |
| S-MT versus S-MA | — | — | — | FuG, PhG, IPL, CG, MVOcC, LOcC ↓ |
| S-L versus S-D | — | — | PhG, CG ↑ | FuG, PhG, IPL, PCun, CG, MVOcC, LOcC ↑ |
| S-L versus S-MA | — | — | — | — |
| S-D versus S-MA | — | — | — | — |
Note: Permutation FDR P < 0.05 (two-tailed).
Abbreviations: S-MT-S-MA, subtype 1–4; SFG, superior frontal gyrus; MFG, medial frontal gyrus; IFG, inferior frontal gyrus; OrG, orbitofrontal gyrus; STG, superior temporal gyrus; MTG, medial temporal gyrus; ITG, inferior temporal gyrus; pSTS, posterior superior temporal sulcus; PCun, precuneus; CG, cingulate gyrus; FuG, fusiform gyrus; PhG, parahippocampal gyrus; IPL, inferior parietal lobule; MVOcC, medial ventral occipital cortex; and LOcC, lateral occipital cortex.
Discussion
Substantial differences between individual AD patients can exist on clinical, cognitive, and biomarker levels. Only recently, the unsupervised classification of atrophy patterns emerged as an approach allowing to distinguish separate AD subtypes with distinct cognitive and biomarker profiles (Ten Kate et al. 2018). However, until now, there was no evidence on brain functional network differences between the subtypes, limiting conclusions about their functional relevance. We addressed this key question by analyzing differences in resting-state functional connectivity networks and graph theory-based brain network measures on a global and nodal level. In addition, we explored biomarker and cognitive differences between atrophy subtypes in two independent datasets from the prospective DELCODE and ADNI cohorts.
The main findings of our study are: 1) in line with previous research, using an unsupervised similarity-based clustering algorithm, we identified four distinct subtypes in two independent datasets exhibiting similar brain atrophy patterns as well as clinical and cognitive characteristics; 2) INC exhibit a heterogeneous alteration pattern for the different subtypes compared among each other and to HC, with distinct INC reductions in S-MT and S-D and a divergent pattern of INC in S-L compared with the other subtypes for most RS networks; 3) the S-MT, S-D, and S-MA subtypes showed reduced global network efficiency compared with the S-L subtype; 4) on a nodal level, network analysis revealed reduced degree and clustering coefficient in regions highly overlapping with the atrophy pattern of the particular subtype; 5) CSF biomarkers were substantially more pathological in all subgroups compared with HC, among subgroups S-L exhibited the lowest tau elevations; and 6) Cognitive scores were reduced in all subgroups compared with HC, with S-MT and S-D revealing pronounced cognitive decline, less prominent in S-L and S-MA.
To the best of our knowledge, this is the first study to assess functional network connectivity changes between different atrophy subtypes in AD. We report differences between subtypes for INC in the DMN, VN, and the LN. Previous research described the DMN as one of the networks most vulnerable to degeneration in AD (Greicius et al. 2004; Buckner et al. 2005). A study comparing multiple imaging biomarkers and intrinsic functional connectivity networks in AD demonstrated substantial overlap between atrophic changes and INC in the anterior LN followed by the DMN (Grothe et al. 2016).
Considering the atrophy pattern, clinical and neurocognitive scores and CSF biomarkers using a two-dimensional framework including typicality and severity, S-MT and S-D can be characterized along the severity dimension, whereas S-L and S-MA appear to be different AD entities along the typicality dimension, exhibiting divergent network features with smaller cognitive differences (Ferreira et al. 2020). The pattern of INC changes between the subtypes accordingly suggests advanced network degeneration within the DMN in S-MT and S-D along the severity dimension. The S-L subtype, exhibits the greatest deviation of the INC pattern across several RSNs, including the DMN. Compared with HC, an increased INC was detected in the DMN. Interestingly, the S-MA subtype exhibits a decrease of INC in the LN and DAN compared with HC, a distinct elevation of tau biomarkers and hippocampal atrophy despite very limited cortical atrophy and significant but modest cognitive changes. Atrophy patterns and functional connectivity changes are related, but as functional connectivity reflects the correlation of BOLD fluctuations between regions not necessarily directly connected by structural tracts, the resulting changes in functional connectivity are not identical to atrophy, emphasizing the need to study both variables to gain a better characterization of the derived subtypes.
All atrophy subtypes show some limbic involvement in the cortical atrophy pattern when compared with HC, but not when compared with each other. A similar atrophy pattern has been found in a recent publication in prodromal and early AD patients, where limbic involvement was also revealed for all subtypes versus HC but not when comparing the subtypes with each other (Ten Kate et al. 2018). Limbic structures are reported to be involved in tau pathology early in the disease (Trzepacz et al. 2013). In PET studies, severe hypometabolism was reported in AD and MCI patients in a network comprising structures of the limbic system, including hippocampus, thalamus and the posterior cingulate cortex (Nestor et al. 2003). These findings emphasize the importance of structures of the limbic systems and the associated limbic network (LN) system. Interestingly, the limbic atrophy subtype, despite pronounced atrophy in the limbic system, shows higher INC in the DMN and LN compared with HC, suggesting that atrophy is not directly correlated with functional connectivity on the network level. In synopsis with the CSF biomarker results, it appears that the moderate changes in cognition in S-L might be mainly associated with changes in INC and point to a significant impact of network disturbances over neurodegeneration traits in this subtype.
Differences between the four subtypes were consistently present on measures of global network properties. Our results suggest that measures of global network integration, most importantly global efficiency, are reduced in the S-MT and S-D subtypes along the severity dimension and strongly associated with cognitive performance, in line with the well-studied disconnection syndrome in AD (Stam et al. 2007). Differences in cognitive performance between the subtypes were previously shown (Liu et al. 2014). In comparison, the S-L, and to a lesser extent the S-MT, subtypes showed less severe disconnectivity on a global network level in conjunction with pronounced network changes on a nodal level (degree), suggesting a more localized underlying network pathology in these subtypes.
Nodal network changes in graph theory analysis showed a noticeable spatial overlap with the characteristic atrophy pattern of the corresponding subtype as measured by degree, expressing the number of links connected to a node as a reflection of the importance of a particular node in the network (Rubinov and Sporns 2010). In contrast, clustering coefficient, a measure of the extent of the local density or cliquishness of a network, was mainly reduced on a nodal level in the S-MT and S-D subtypes, following a typical distribution pattern of neurodegeneration in clinical and prodromal AD (Pereira et al. 2016).
Even though the S-MA subtype exhibits a pattern of sparse atrophy with better cognitive and clinical scores compared with the other subtypes, INC was reduced to a comparable degree as in S-MT and S-D. In addition, on a nodal level clustering coefficient was decreased in frontal and temporal regions, and local efficiency was reduced in areas belonging to the DMN. These changes on a local level might reflect ongoing pathological changes in the absence of clinical or neurocognitive symptoms. Compared with S-L, S-MA demonstrates no difference in Aβ but increased CSF t-tau and p-tau levels. Therefore, patients in the S-MA subgroup may have lower cognitive reserve (Persson et al. 2017) and be more likely to express AD pathology as network disruptions. Previously, the minimal atrophy pattern was shown to be associated with reduced metabolism in the parietal cortex (Shima et al. 2012) and a higher rate of cerebral amyloid angiopathy (Ferreira et al. 2018), causing network disruptions. Consistent with this finding, we observed a locally reduced local efficiency in this area, accompanied by network disruption in the DAN and LN. High vulnerability within nodes of the DMN and other brain network regions in structural graph theory analysis in the S-MA subtype was previously demonstrated (Ferreira et al. 2019).
Compared with some previous studies, this study did not show differences between the atrophy subtypes and HC in the DMN. Numerically, S-L showed increased INC, and in contrast to the other subtypes, decreased INC compared with HC. It is possible that the results of the comparison in DMN functional connectivity between controls and AD patients might vary depending on the proportion of S-L subtype in a particular cohort. Furthermore, the individuals represent a spectrum of AD patients, including MCI and AD participants. A recent meta-analysis shows that depending on the clinical disease stage, hyper- or hypoconnectivity can occur in the DMN (Badhwar et al. 2017). The question of how differences in neuropathology among subtypes affect INC changes depending on the AD stage should be addressed in further research.
There are potential limitations of our study. First, pathological data to verify the clinical diagnoses were not available; however, we selectively included participants in the AD groups of both datasets following a biomarker-based diagnostic scheme including only Aβ-positive individuals with CDR ≥ 0.5, according to the ATN classification system following the recommendations of the NIA-AA Research Framework (Jack Jr et al. 2018), minimizing heterogeneity and potential misdiagnosis. Second, the dice overlap in the minimal atrophy subtype was comparatively low; this can be partly explained by the low number of vertices with reduced thickness in participants belonging to this group with a high probability of unequal distribution, although the clinical and neurocognitive scores showed high similarity. Third, the atrophy similarity clustering method could detect clinical AD stages rather than distinct subtypes. Indeed, this is likely the case for the S-MA and the S-D subtypes, however, the S-L and the S-MA subtypes exhibit aberrant network properties and CSF biomarker concentrations and are most likely entities along the typicality and not the severity dimension. Future investigations should consider tau PET as an additional imaging parameter to gain important information about spatial associations between tau distribution and network degeneration particularly in the S-MA subtype. Finally, the controls in the DELCODE dataset were younger compared with the AD patients, which may have resulted in more severe atrophy measures in this cohort; however, it is unlikely that this difference affected the resulting atrophy patterns and therefore the main results of the study.
In conclusion, we demonstrate a robust detection of different AD subtypes using a similarity-based clustering approach. These subtypes show distinct differences in functional connectivity networks and network properties on the local and global levels, accompanied by CSF biomarker and cognitive differences. Our study contributes toward a better understanding of heterogeneity in AD, with important ramifications for a more individualized approach to diagnosis and treatment. A better characterization of the heterogeneity of functional connectivity changes in AD subtypes lays the foundation for advanced neuromodulatory noninvasive brain stimulation, pharmacological treatment or tailored cognitive interventions aimed at modifying functional connectivity networks. Known differences in patterns of network degeneration may lead to a better-informed, individualized treatment strategy. Follow-up studies should address the longitudinal consequences of the identified heterogeneity.
Supplementary Material
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Funding
Neuroimaging Initiative (ADNI): Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI; National Institutes of Health Grant U01 AG024904). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott; Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Amorfix Life Sciences Ltd; AstraZeneca; Bayer HealthCare; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by the National Institutes of Health (NIH) grants P30 AG010129 and K01 AG030514. The sponsors did not have any role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
DZNE-Longitudinal Cognitive Impairment and Dementia Study (DELCODE): Study funded by the German Center for Neurodegenerative Diseases (Deutsches Zentrum für Neurodegenerative Erkrankungen, DZNE) reference number BN01. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Notes
Conflict of Interest: Boris-Stephan Rauchmann, Ersin Ersoezlue, Sophia Stöcklein, Daniel Keeser, Frederic Brosseron, Katharina Buerger, Peter Dechent, Laura Dobisch, Birgit Ertl-Wagner, Klaus Fliessbach, John Dylan Haynes, Michael T. Heneka, Enise I. Incesoy, Daniel Janowitz, Ingo Kilimann, Christoph Laske, Coraline D. Metzger, Matthias H. Munk, Oliver Peters, Alfredo Ramirez, Sandra Roeske, Nina Roy, Klaus Scheffler, Anja Schneider, Annika Spottke, Eike Jakob Spruth, Stefan Teipel, Maike Tscheuschler, Ruth Vukovich, Michael Wagner, Jens Wiltfang, Renat Yakupov, Emrah Duezel, and Robert Perneczky report no disclosures.
Dr Jessen received fees for consultation from Eli Lilly, Novartis, Roche, BioGene, MSD, Piramal, Janssen, Lundbeck. Dr Priller received fees for consultation, lectures, patents from Neurimmune, Axon, Desitin, Epomedics.
References
- Badhwar A, Tam A, Dansereau C, Orban P, Hoffstaedter F, Bellec P. 2017. Resting-state network dysfunction in Alzheimer's disease: a systematic review and meta-analysis. Alzheimers Dement (Amst). 8:73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo XN, Gohel S, Kelly C, Smith SM, Beckmann CF, Adelstein JS, Buckner RL, Colcombe S, et al. 2010. Toward discovery science of human brain function. Proc Natl Acad Sci U S A. 107:4734–4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. 2008. Fast unfolding of communities in large networks. J Stat Mech. 2008:P10008. [Google Scholar]
- Braskie MN, Klunder AD, Hayashi KM, Protas H, Kepe V, Miller KJ, Huang SC, Barrio JR, Ercoli LM, Siddarth P, et al. 2010. Plaque and tangle imaging and cognition in normal aging and Alzheimer's disease. Neurobiol Aging. 31:1669–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier MR, Thomas JB, Fagan AM, Hassenstab J, Holtzman DM, Benzinger TL, Morris JC, Ances BM. 2014. Functional connectivity and graph theory in preclinical Alzheimer's disease. Neurobiol Aging. 35:757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier MR, Thomas JB, Snyder AZ, Benzinger TL, Zhang D, Raichle ME, Holtzman DM, Morris JC, Ances BM. 2012. Loss of intranetwork and internetwork resting state functional connections with Alzheimer's disease progression. J Neurosci. 32:8890–8899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, et al. 2005. Molecular, structural, and functional characterization of Alzheimer's disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 25:7709–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 10:186–198. [DOI] [PubMed] [Google Scholar]
- Chambers JM. 2018. Graphical methods for data analysis. London, UK: CRC Press. [Google Scholar]
- Crane PK, Carle A, Gibbons LE, Insel P, Mackin RS, Gross A, Jones RN, Mukherjee S, Curtis SM, Harvey D, et al. 2012. Development and assessment of a composite score for memory in the Alzheimer's disease neuroimaging initiative (ADNI). Brain Imaging Behav. 6:502–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong A, Toledo JB, Honnorat N, Doshi J, Varol E, Sotiras A, Wolk D, Trojanowski JQ, Davatzikos C, Alzheimer's Disease Neuroimaging I . 2017. Heterogeneity of neuroanatomical patterns in prodromal Alzheimer's disease: links to cognition, progression and biomarkers. Brain. 140:735–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Li H, Zhuo J, Zhang Y, Wang J, Chen L, Yang Z, Chu C, Xie S, Laird AR, et al. 2016. The Human Brainnetome Atlas: a new brain atlas based on connectional architecture. Cereb Cortex. 26:3508–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farahani FV, Karwowski W, Lighthall NR. 2019. Application of graph theory for identifying connectivity patterns in human brain networks: a systematic review. Front Neurosci. 13:585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D, Nordberg A, Westman E. 2020. Biological subtypes of Alzheimer disease: a systematic review and meta-analysis. Neurology. 94:436–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D, Pereira JB, Volpe G, Westman E. 2019. Subtypes of Alzheimer's disease display distinct network abnormalities extending beyond their pattern of brain atrophy. Front Neurol. 10:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira D, Shams S, Cavallin L, Viitanen M, Martola J, Granberg T, Shams M, Aspelin P, Kristoffersen-Wiberg M, Nordberg A, et al. 2018. The contribution of small vessel disease to subtypes of Alzheimer's disease: a study on cerebrospinal fluid and imaging biomarkers. Neurobiol Aging. 70:18–29. [DOI] [PubMed] [Google Scholar]
- Ferreira D, Verhagen C, Hernández-Cabrera JA, Cavallin L, Guo CJ, Ekman U, Muehlboeck JS, Simmons A, Barroso J, Wahlund LO, et al. 2017. Distinct subtypes of Alzheimer's disease based on patterns of brain atrophy: longitudinal trajectories and clinical applications. Sci Rep. 7:46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, et al. 2002. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 33:341–355. [DOI] [PubMed] [Google Scholar]
- Galasko D, Xiao M, Xu D, Smirnov D, Salmon DP, Dewit N, Vanbrabant J, Jacobs D, Vanderstichele H, Vanmechelen E, et al. 2019. Synaptic biomarkers in CSF aid in diagnosis, correlate with cognition and predict progression in MCI and Alzheimer's disease. Alzheimers Dement (N Y). 5:871–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison KA, Scheinost D, Finn ES, Shen X, Constable RT. 2015. The (in)stability of functional brain network measures across thresholds. Neuroimage. 118:651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons LE, Carle AC, Mackin RS, Harvey D, Mukherjee S, Insel P, Curtis SM, Mungas D, Crane PK, Alzheimer's Disease Neuroimaging I . 2012. A composite score for executive functioning, validated in Alzheimer's disease neuroimaging initiative (ADNI) participants with baseline mild cognitive impairment. Brain Imaging Behav. 6:517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V. 2004. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A. 101:4637–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grothe MJ, Teipel SJ, Alzheimer's Disease Neuroimaging I . 2016. Spatial patterns of atrophy, hypometabolism, and amyloid deposition in Alzheimer's disease correspond to dissociable functional brain networks. Hum Brain Mapp. 37:35–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J, Kim CM, Jeon S, Lee JM, Hong YJ, Roh JH, Lee JH, Koh JY, Na DL, Alzheimer's Disease Neuroimaging I . 2016. Prediction of Alzheimer's disease pathophysiology based on cortical thickness patterns. Alzheimers Dement (Amst). 2:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR Jr, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, Holtzman DM, Jagust W, Jessen F, Karlawish J, et al. 2018. NIA-AA research framework: toward a biological definition of Alzheimer's disease. Alzheimers Dement. 14:535–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janocko NJ, Brodersen KA, Soto-Ortolaza AI, Ross OA, Liesinger AM, Duara R, Graff-Radford NR, Dickson DW, Murray ME. 2012. Neuropathologically defined subtypes of Alzheimer's disease differ significantly from neurofibrillary tangle-predominant dementia. Acta Neuropathol. 124:681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen F, Spottke A, Boecker H, Brosseron F, Buerger K, Catak C, Fliessbach K, Franke C, Fuentes M, Heneka MT, et al. 2018. Design and first baseline data of the DZNE multicenter observational study on predementia Alzheimer's disease (DELCODE). Alzheimers Res Ther. 10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruschwitz JD, List D, Waller L, Rubinov M, Walter H. 2015. GraphVar: a user-friendly toolbox for comprehensive graph analyses of functional brain connectivity. J Neurosci Methods. 245:107–115. [DOI] [PubMed] [Google Scholar]
- Lancichinetti A, Fortunato S. 2012. Consensus clustering in complex networks. Sci Rep. 2:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau SM, Breault C, Joshi AD, Pontecorvo M, Mathis CA, Jagust WJ, Mintun MA, Alzheimer's Disease Neuroimaging I . 2013. Amyloid-beta imaging with Pittsburgh compound B and florbetapir: comparing radiotracers and quantification methods. J Nucl Med. 54:70–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yu C, Zhang X, Liu J, Duan Y, Alexander-Bloch AF, Liu B, Jiang T, Bullmore E. 2014. Impaired long distance functional connectivity and weighted network architecture in Alzheimer's disease. Cereb Cortex. 24:1422–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelpunkt A, Galili T, Kozlovski T, Bregman N, Shachar N, Markus-Kalish M, Benjamini Y. 2020. Novel Alzheimer's disease subtypes identified using a data and knowledge driven strategy. Sci Rep. 10:1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray ME, Graff-Radford NR, Ross OA, Petersen RC, Duara R, Dickson DW. 2011. Neuropathologically defined subtypes of Alzheimer's disease with distinct clinical characteristics: a retrospective study. Lancet Neurol. 10:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestor PJ, Fryer TD, Smielewski P, Hodges JR. 2003. Limbic hypometabolism in Alzheimer's disease and mild cognitive impairment. Ann Neurol. 54:343–351. [DOI] [PubMed] [Google Scholar]
- Noh Y, Jeon S, Lee JM, Seo SW, Kim GH, Cho H, Ye BS, Yoon CW, Kim HJ, Chin J, et al. 2014. Anatomical heterogeneity of Alzheimer disease: based on cortical thickness on MRIs. Neurology. 83:1936–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JY, Na HK, Kim S, Kim H, Kim HJ, Seo SW, Na DL, Han CE, Seong JK, Alzheimer's Disease Neuroimaging I . 2017. Robust identification of Alzheimer's disease subtypes based on cortical atrophy patterns. Sci Rep. 7:43270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira JB, Mijalkov M, Kakaei E, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen H, Spenger C, Lovestone S, et al. 2016. Disrupted network topology in patients with stable and progressive mild cognitive impairment and Alzheimer's disease. Cereb Cortex. 26:3476–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson K, Eldholm RS, Barca ML, Cavallin L, Ferreira D, Knapskog AB, Selbaek G, Braekhus A, Saltvedt I, Westman E, et al. 2017. MRI-assessed atrophy subtypes in Alzheimer's disease and the cognitive reserve hypothesis. PLoS One. 12:e0186595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Sporns O. 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage. 52:1059–1069. [DOI] [PubMed] [Google Scholar]
- Sanz-Arigita EJ, Schoonheim MM, Damoiseaux JS, Rombouts SA, Maris E, Barkhof F, Scheltens P, Stam CJ. 2010. Loss of 'small-world' networks in Alzheimer's disease: graph analysis of FMRI resting-state functional connectivity. PLoS One. 5:e13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheltens NME, Tijms BM, Koene T, Barkhof F, Teunissen CE, Wolfsgruber S, Wagner M, Kornhuber J, Peters O, Cohn-Sheehy BI, et al. 2017. Cognitive subtypes of probable Alzheimer's disease robustly identified in four cohorts. Alzheimers Dement. 13:1226–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima K, Matsunari I, Samuraki M, Chen WP, Yanase D, Noguchi-Shinohara M, Takeda N, Ono K, Yoshita M, Miyazaki Y, et al. 2012. Posterior cingulate atrophy and metabolic decline in early stage Alzheimer's disease. Neurobiol Aging. 33:2006–2017. [DOI] [PubMed] [Google Scholar]
- Stam CJ, Jones BF, Nolte G, Breakspear M, Scheltens P. 2007. Small-world networks and functional connectivity in Alzheimer's disease. Cereb Cortex. 17:92–99. [DOI] [PubMed] [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD. 2008. Network analysis of intrinsic functional brain connectivity in Alzheimer's disease. PLoS Comput Biol. 4:e1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Kate M, Dicks E, Visser PJ, van der Flier WM, Teunissen CE, Barkhof F, Scheltens P, Tijms BM, Alzheimer's Disease Neuroimaging I . 2018. Atrophy subtypes in prodromal Alzheimer's disease are associated with cognitive decline. Brain. 141:3443–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzepacz PT, Yu P, Bhamidipati PK, Willis B, Forrester T, Tabas L, Schwarz AJ, Saykin AJ, Alzheimer's Disease Neuroimaging I . 2013. Frontolimbic atrophy is associated with agitation and aggression in mild cognitive impairment and Alzheimer's disease. Alzheimers Dement. 9:S95–S104 e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. 1998. Collective dynamics of 'small-world' networks. Nature. 393:440–442. [DOI] [PubMed] [Google Scholar]
- Whitwell JL, Dickson DW, Murray ME, Weigand SD, Tosakulwong N, Senjem ML, Knopman DS, Boeve BF, Parisi JE, Petersen RC, et al. 2012. Neuroimaging correlates of pathologically defined subtypes of Alzheimer's disease: a case-control study. Lancet Neurol. 11:868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitwell JL, Graff-Radford J, Tosakulwong N, Weigand SD, Machulda M, Senjem ML, Schwarz CG, Spychalla AJ, Jones DT, Drubach DA, et al. 2018. [(18) F]AV-1451 clustering of entorhinal and cortical uptake in Alzheimer's disease. Ann Neurol. 83:248–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler AM, Ridgway GR, Webster MA, Smith SM, Nichols TE. 2014. Permutation inference for the general linear model. Neuroimage. 92:381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Wang J, He Y. 2013. BrainNet viewer: a network visualization tool for human brain connectomics. PLoS One. 8:e68910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, Roffman JL, Smoller JW, Zollei L, Polimeni JR, et al. 2011. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 106:1125–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Mormino EC, Sun N, Sperling RA, Sabuncu MR, Yeo BT, Alzheimer's Disease Neuroimaging I . 2016. Bayesian model reveals latent atrophy factors with dissociable cognitive trajectories in Alzheimer's disease. Proc Natl Acad Sci U S A. 113:E6535–E6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All ADNI data are deposited in a publicly accessible repository and can be accessed at adni.loni.usc.edu. For the DELCODE dataset, anonymized data analyzed in the current study will be made available upon reasonable request from qualified investigators.


