Outbreaks of infectious disease often result from exposure to a common source of the etiologic agent. Generally, the etiologic agent causing an outbreak of infection is derived from a single cell whose progeny are genetically identical or closely related to the source organism. In epidemiological terms, the organisms involved in the outbreak are clonally related; that is, they have a common origin. Clonally related organisms are members of the same species that share virulence factors, biochemical traits, and genomic characteristics. However, there is sufficient diversity at the species level that organisms isolated from different sources at different times and in different geographical regions may be differentiated or classified into subtypes or strains.
The process of subtyping is important epidemiologically for recognizing outbreaks of infection, detecting the cross-transmission of nosocomial pathogens, determining the source of the infection, recognizing particularly virulent strains of organisms, and monitoring vaccination programs.
Subtyping or strain classification has been accomplished by a number of different approaches. All of these methods must meet several criteria in order to be broadly useful. All organisms within a species must be typeable by the method used. For some phenotypic methods, such as those based on a reaction with an antibody or the presence of a bacteriophage receptor, the particular characteristic may not be present in all members of the species. Serotypes can be assigned only if a particular serological marker is present. For example, 10 to 15% of Mycobacterium avium strains are untypeable because they fail to react with any available antiserum (78). Similarly, bacteriophage typing has often been applied to the characterization of Staphylococcus aureus, yet S. aureus isolates lacking the bacteriophage receptor cannot become infected with the typing phage (10, 64). In the absence of the marker, the isolates cannot be assigned a phage type.
Any subtyping method must have high differentiation power (3). It must be able to clearly differentiate unrelated strains, such as those that are geographically distinct from the source organism, and at the same time to demonstrate the relationship of all organisms isolated from individuals infected through the same source. As will be discussed in this review, molecular methods can differ widely in their ability to differentiate strains.
A third concern for subtyping methodologies is reproducibility (3). Reproducibility refers to the ability of a technique to yield the same result when a particular strain is repeatedly tested. Reproducibility is especially important for the construction of reliable databases containing all known strains within a species to which unknown organisms can be compared for classification. Variable expression of phenotypic characteristics, such as sporadic expression of virulence genes or antigens, can contribute to problems with reproducibility (3).
Many of the currently used molecular techniques for typing rely on electrophoretic separation of DNA fragments of different molecular lengths. The electrophoretic result is represented by a pattern of bands on a gel. Since these patterns may be extremely complex, the ease with which the patterns are interpreted and related is a factor in evaluating the utility of a particular typing method (3). Along with considerations related to a particular method’s ease of interpretation, its ease of use is also important (3). The technical difficulty, cost, and time to obtain a result must also be evaluated in assessing the utility of a particular typing method.
The shortcomings of phenotypically based typing methods have led to the development of typing methods based on the microbial genotype or DNA sequence, which minimize problems with typeability and reproducibility and, in some cases, enable the establishment of large databases of characterized organisms. In this review, we have compared some of the most current technologies for genotypic typing with regard to the principles of the methods and their reproducibility and discrimination power, ease of use, and cost in order to give the reader some measure of the clinical practicality and utility of the molecular tools available today.
MOLECULAR TYPING METHODS
PFGE of whole chromosomal DNA.
Pulsed-field gel electrophoresis (PFGE) is often considered the “gold standard” of molecular typing methods. For PFGE, bacterial isolates grown either in broth or on solid media are combined with molten agarose and poured into small molds. The results are agarose plugs containing the whole bacteria. The embedded bacteria are then subjected to in situ detergent-enzyme lysis and digestion with an infrequently cutting restriction enzyme. The digested bacterial plugs are then inserted into an agarose gel and subjected to electrophoresis in an apparatus in which the polarity of the current is changed at predetermined intervals. The pulsed field allows clear separation of very-large-molecular-length DNA fragments ranging from 10 to 800 kb (65). The electrophoretic patterns are visualized following staining of the gels with a fluorescent dye such as ethidium bromide. Gel results can be photographed, and the data can be stored by using one of the commercially available digital systems, such as those manufactured by Alpha-Innotech, Bio-Rad, Hitachi, or Molecular Dynamics. Data analysis can be accomplished by using any of a number of commercially available software packages available from Applied Math, Bio-Rad, BioSystematics, Media Cybernetics, or Scanalytics.
Tenover et al. (75) have proposed a system for standardizing the interpretation of PFGE patterns in relation to determining strain relatedness. In their scheme, bacterial isolates yielding the same PFGE pattern are considered the same strain. Bacterial isolates differing by a single genetic event, reflected as a difference of one to three bands, are closely related. Isolates differing by four to six bands, representing two independent genetic changes, are possibly related. Bacterial isolates containing six or more band differences, representative of three or more genetic changes, are considered unrelated. These criteria are applicable to small, local studies in which genetic variability is presumed to be limited.
Numerous studies which define the banding patterns of a variety of bacteria digested with a number of restriction enzymes have been conducted. PFGE has proven to be superior to most other methods for biochemical and molecular typing. It is highly discriminatory and superior to most methods for analysis of Escherichia coli, vancomycin-resistant enterococci, S. aureus, Acinetobacter species, Pseudomonas aeruginosa, and M. avium (4, 5, 7, 30, 46, 52, 57, 63, 64, 73). PFGE was more efficient at differentiating strains of methicillin-resistant S. aureus than other methods tested (58, 63). PFGE was also more discriminatory than repetitive element sequence-based PCR (Rep-PCR) for differentiating strains of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex, vancomycin-resistant enterococci, Neisseria gonorrhoeae, and P. aeruginosa (7, 30, 46, 56).
With the aid of the computerized gel scanning and analysis software, it is possible to create data banks of PFGE patterns for all organisms, enabling the creation of reference databases to which any new strain could be compared for identifying its phylogenetic relationship to other similar strains. This capability is reflected in a recent report to the President on food safety initiatives (22). Two of the recommendations were (i) to enhance surveillance of food-borne outbreaks of disease by equipping FoodNet sites with the equipment to carry out DNA fingerprinting by PFGE in order to identify the source of infectious agents and (ii) to establish an electronic network for rapid fingerprint comparison which would allow sentinel sites to rapidly share data (22).
One of the factors that has limited the use of PFGE is the time involved in completing the analysis. While the procedural steps are straightforward, the time needed to complete the procedure can be 2 to 3 days. This can reduce the laboratory’s ability to analyze large numbers of samples.
Southern blotting and RFLP methods.
Southern blotting has been used for years to detect and locate genomic sequences from a variety of prokaryotic and eukaryotic organisms. For gene detection, whole chromosomal DNA is digested with a restriction enzyme, and the fragments are separated by electrophoresis through an agarose gel. The separated DNA fragments are transferred from the agarose gel to either a nitrocellulose or nylon membrane by Southern blotting (69). The membrane-bound nucleic acid is then hybridized to one or more labeled probes homologous to the gene to be examined. Probes can be labeled with a number of detectable moieties, including enzyme-colorimetric substrates or enzyme-chemiluminescent substrates. This classic method has been adapted to the differentiation of bacterial strains on the basis of the observation that the locations of various restriction enzyme recognition sites within a particular genetic locus of interest can be polymorphic from strain to strain, resulting in gel bands that differ in size between unlike strains. Thus, the name restriction fragment length polymorphism (RFLP) refers to the polymorphic nature of the locations of restriction enzyme sites within defined genetic regions. Only the genomic DNA fragments that hybridize to the probes are visible in RFLP analysis, which simplifies the analysis greatly.
Gene-specific probes have been used to subtype Brucella species (29), Legionella pneumophila (76), and P. aeruginosa (47). Furthermore, ribotyping, a variation of RFLP-Southern blotting in which the probes are derived from the 16S and 23S rRNA genes, has been applied successfully in many studies for differentiating bacterial strains (12, 20, 25, 34, 40, 72). Ribotyping results in only a small number of bands, which simplifies the interpretation. However, this also limits the ability of the technique to distinguish between closely related strains.
Multiple genetic sites can also be targeted in Southern blotting studies to subtype bacteria. Grundmann et al. (30) used a combination of a toxA gene probe with probes for the 16S and 23S rRNA genes to subtype P. aeruginosa isolates. Both RFLP-Southern blotting probe sets yielded information about the evolutionary relationships of the strains examined, yet neither technique was as efficient at discriminating strain differences as PFGE. While RFLP-Southern blotting has been a useful technique for limited studies of strain subtypes, the cumbersome blotting techniques have been largely replaced by PCR-based locus-specific RFLP.
PCR-based locus-specific RFLP.
The PCR has enabled specific genetic loci to be routinely amplified and examined for differences indicative of strain variation and antimicrobial resistance. The specific locus to be examined is amplified with gene-specific primers and subjected to RFLP analysis. The DNA fragments are separated on an agarose or small polyacrylamide gel, and the digestion patterns are visualized following ethidium bromide staining.
Locus-specific RFLP has been applied in a number of situations. Shortridge et al. (67) have used RFLP of the ureC gene to demonstrate the genetic diversity of Helicobacter pylori strains in the United States. The 16S, 23S, and 16S-23S spacer regions have also been used as targets for locus-specific RFLP (30, 83, 93). In this variation of ribotyping, the ribosomal DNA is amplified and subjected to digestion with a restriction enzyme, and the DNA fragments are visualized following separation by gel electrophoresis, alleviating the need for Southern blotting. Ribotyping can be successfully applied for differentiation of bacterial strains that display a high degree of heterogeneity within the rRNA operons.
In a novel application of locus-specific RFLP, Cockerill et al. (16) have identified point mutations in the katG gene of Mycobacterium tuberculosis that correspond to various levels of resistance to isoniazid by examination of RFLP patterns of the amplified gene. However, the discriminatory power of locus-specific PCR is generally not as good as that of other methods, due primarily to the limited region of the genome which can be examined. For example, in a study by Kühn et al. (42), PCR-ribotyping showed poor discriminatory power in comparison to PFGE and biochemical typing methods.
Locus-specific RFLP has found a significant application in epidemiological studies of hepatitis C virus (HCV). HCV can be divided into six major genotypes, numbered 1 through 6. Davidson et al. (17) have developed a simple method for determining the genotype of an HCV isolate based on PCR amplification and RFLP analysis of the 5′ untranslated region of the HCV genome. Digestion with either the combination or single enzymes HaeIII-RsaI, MvaI-HinfI, BstI, and ScrFI allows differentiation of viral isolates into groups 1a, 1b, 2a, 2b, 3a, 3b, 4, 5, and 6. RFLP of the 5′ untranslated region has facilitated studies of the geographical distribution of viral genotypes, the natural history of the disease, and correlation of genotypes with response to interferon.
RAPD assays.
The random amplified polymorphic DNA (RAPD) assay, also referred to as arbitrary primed PCR, was first described by Williams et al. (87) and Welsh and McClelland (86). RAPD assays are based on the use of short random sequence primers, 9 to 10 bases in length, which hybridize with sufficient affinity to chromosomal DNA sequences at low annealing temperatures such that they can be used to initiate amplification of regions of the bacterial genome. If two RAPD primers anneal within a few kilobases of each other in the proper orientation, a PCR product with a molecular length corresponding to the distance between the two primers results. The number and location of these random primer sites vary for different strains of a bacterial species. Thus, following separation of the amplification products by agarose gel electrophoresis, a pattern of bands which, in theory, is characteristic of the particular bacterial strain results (14, 51, 86, 87).
In most cases the sequences of the RAPD primers which generate the best DNA pattern for differentiation must be determined empirically. More recently, a consensus M13 DNA sequencing primer has been used in RAPD fingerprinting assays, which allows for some standardization of the procedure (83, 85).
A number of studies have reported success in using RAPD assays to distinguish bacterial strains among diverse species, including A. baumannii (60), Candida albicans (45, 61), H. pylori (1), Proteus mirabilis (9), Histoplasma capsulatum (37), Haemophilus somnus (53), Leptospira species (59), S. aureus (63), and L. pneumophilia (76, 79).
In comparison to other typing techniques, Vila et al. (83) found that the RAPD assay was more discriminating than RFLP analysis of either the 16S rRNA genes or the 16S-23S rRNA spacer region but less discriminating than Rep-PCR. Furthermore, a number of problems have been reported for RAPD assays that contribute to a lack of reproducibility and standardization. Since the primers are not directed against any particular genetic locus, many of the priming events are the result of imperfect hybridization between the primer and the target site. Thus, the amplification process is extremely sensitive to slight changes in the annealing temperature which can lead to variability in the banding patterns. The use of empirically designed primers, each with its own optimal reaction conditions and reagents, also makes standardization of the technique difficult (6, 51, 86).
Rep-PCR.
Versalovic et al. (80) described a method for fingerprinting bacterial genomes by examining strain-specific patterns obtained from PCR amplification of repetitive DNA elements present within bacterial genomes. Two main sets of repetitive elements are used for typing purposes. The repetitive extragenic palindromic (REP) elements are 38-bp sequences consisting of six degenerate positions and a 5-bp variable loop between each side of a conserved palindromic stem (71). REP sequences have been described for numerous enteric bacteria (28, 33, 66, 71, 92). The palindromic nature of the REP elements and their ability to form stem-loop structures have led to multiple proposed functions for these highly conserved, dispersed elements (27, 54, 92).
The enterobacterial repetitive intergenic consensus (ERIC) sequences are a second set of DNA sequences which have been successfully used for DNA typing. ERIC sequences are 126-bp elements which contain a highly conserved central inverted repeat and are located in extragenic regions of the bacterial genome (33, 66). They have been defined primarily based on sequence data obtained from E. coli and Salmonella typhimurium (33, 66).
While the REP and ERIC sequences are the most commonly used targets for DNA typing, another repetitive element, the BOX sequence, has been used to differentiate Streptococcus pneumoniae (39, 50). BOX elements are located within intergenic regions and can also form stem-loop structures due to their dyad symmetry. They are mosaic repetitive elements composed of various combinations of three subunit sequences referred to as boxA, boxB, and boxC (50). The three subunit sequences have molecular lengths of 59, 45, and 50 nucleotides, respectively (50). The BOX elements have no sequence relationship to either REP or ERIC sequences (50). While initially thought to be unique to S. pneumoniae, BOX elements have now been found in a number of bacterial species (43).
Rep-PCR can be performed with DNA extracted from bacterial colonies or by a modified method using unprocessed whole cells (90). REP or ERIC amplification can be performed with a single primer, a single set of primers, or multiple sets of primers. ERIC patterns are generally less complex than REP patterns, but both give good discrimination at the strain level. Application of both REP and ERIC PCR to samples to be typed increases the discriminatory power over that of either technique used alone.
Rep-PCR is fast becoming the most widely used method of DNA typing. Rep-PCR with primers based on REP and ERIC sequences has been successfully used to differentiate strains of Bartonella (62), Bacillus subtilis (80), Citrobacter diversus (32, 89), Enterobacter aerogenes (24), Rhizobium meliloti (18), methicillin-resistant S. aureus (19), S. pneumoniae (81, 93), A. baumanii (21, 60), Burkholderia cepacia (31), L. pneumophilia (23), H. pylori (43), N. gonorrhoeae (56), and Neisseria meningitidis (91). The technique is easy to perform and can be applied to large or small numbers of isolates. Rep-PCR shows broader species applicability and better discriminatory power than either plasmid profiling or genomic fingerprinting (24). Rep-PCR has considerably better discriminatory power than restriction analysis of the 16S rRNA gene or the 16S-23S spacer region (2, 83). Furthermore, studies which have compared Rep-PCR to other typing methods such as multilocus enzyme electrophoresis (89), biochemical characterizations (15), or ribotyping (2, 68, 83) have shown Rep-PCR to be superior to these methods. Finally, several studies have shown Rep-PCR to have good correlation with PFGE results but, in general, with slightly less discriminatory power (7, 46).
Rep-PCR has been adapted to an automated format in which fluorescently labeled primers are used to create either the REP or ERIC profile and the amplified sequences are separated via a fluorescence-based DNA sequencer (19, 82). This method allows consistent pattern formation and storage of the data in a database as a digitized image. Unknown strains can be compared against the stored database for identification purposes. Thus, like PFGE, Rep-PCR lends itself to standardization, which will enable comparisons of strains isolated in different laboratories.
CFLP.
Cleavase I is a thermostable, engineered endonuclease which consists of the 5′-nuclease domain (amino acids 1 to 281) of Thermus aquaticus DNA polymerase (13). Whereas restriction endonucleases recognize specific DNA sequences as their recognition sites, cleavase I recognizes structures formed by base-paired DNA at elevated temperatures (13). When denatured by heating at 95°C followed by cooling at temperatures ranging between 50 and 60°C, the single strands of DNA go through a self-base-pairing phase during which hairpin structures unique to each strand are formed prior to reannealing with the complementary DNA strand. This process creates a series of stem-loop structures connected by non-base-paired single-stranded regions. Cleavase I recognizes the stem-loop structures and cuts the DNA 5′ to the first paired base at the junction of the single- and double-stranded regions. Theoretically, in any given DNA preparation the denatured single strands represent a mixture of all possible configurations which the strands can assume. For the cleavase reaction, a PCR product labeled at one of its 5′ termini is subjected to partial digestion with the enzyme such that all possible digestion products are represented in the reaction mixture. The resulting mixture is analogous to a dideoxy sequencing reaction in that it contains a collection of randomly cleaved, labeled single-stranded DNA molecules extending from the 5′ labeled end to a terminal cleavage site.
Differences in the structural fingerprints between dissimilar PCR products may appear as a gain or loss of a band, a mobility shift in a band, or a change in the intensity of a particular band. While gains, losses, and shifts of bands are easily recognized, differences in band intensity, which occur frequently, are extremely difficult to detect and can often lead to false results.
The discrimination power of cleavase fragment length polymorphism (CFLP) is dependent on the DNA sequence of the locus examined. For example, while a pattern abundant in cleavage products is obtained from digestion of 16S rDNA, very few cleavage products are generated by digestion of the rpoB gene of M. tuberculosis, precluding identification of many sequence differences in the region critical for drug resistance (13). Furthermore, each DNA strand may digest with different abilities to discriminate sequence changes. For example, whereas differences in the nucleotide sequences of the intergenic regions of Shigella dysenteriae serotypes 1, 2, and 8 are represented in the structural fingerprints of the antisense strand, the fingerprints of the sense strand are indistinguishable. Maddox et al. (48) compared the abilities of CFLP and single-strand conformation polymorphism to detect single point mutations. In that study, single-strand conformation polymorphism detected six of six mutations analyzed, while CFLP could detect only three of the six mutations. Thus, the applicability of CFLP analysis to any given DNA cannot be predicted but must rather be determined empirically. Some DNA molecules give excellent results in terms of differentiation, while others give no results at all due to unknown reasons. CFLP is also size limited; that is, differences in the structural fingerprints of DNA fragments larger than 1 kbp cannot be reliably resolved by electrophoresis.
CFLP has been applied to genotypic differentiation of HCV isolates (49, 70). In both of these studies, genotype analysis by CFLP was successful in differentiating the strains examined. However, while the intralaboratory reproducibilities of the banding patterns for each viral type were consistent, the interlaboratory reproducibilities of the same patterns for the viral subtypes were poor. In the two studies that used CFLP for genotyping HCV, the structural fingerprints of HCV type 1a and 1b isolates in the study by Marshall et al. (49) showed significant differences from those reported by Sreevatsan et al. (70). Both studies presented CFLP fingerprints on a 244-bp fragment derived from the 5′ noncoding region by PCR amplification with the Amplicor HCV kit (Roche Molecular Diagnostics, Branchburg, N.J.). The patterns produced for a given genotype in the study by Marshall et al. (49) were generally consistent, and the patterns were easily interpreted. In contrast, while a consistent, unique fingerprint was generated for each genotype tested in the study by Sreevatsan et al. (70), the fingerprints were considerably more complex and difficult to interpret. Furthermore, the fingerprints for HCV types 1a and 1b differed significantly from the patterns for types 1a and 1b presented by Marshall et al. (49). The implication of this is that while CFLP can achieve acceptable intralaboratory reproducibility, the interlaboratory reproducibility limits its utility. Patterns generated by one laboratory cannot be shared or used as a reference for other laboratories.
The need for empirical determination of the applicability of CFLP to any given genetic system coupled with the difficulty in pattern interpretation and the interlaboratory variation in the CFLP patterns of the same organisms compromise the use of this technology for clinical purposes. CFLP is also considerably more complex to perform than either locus-specific PCR-RFLP, Rep-PCR, or RAPD analysis. With the exception of DNA sequencing, the discriminatory ability of CFLP has not been compared to that of any of the other molecular typing techniques, so its capabilities remain unknown.
AFLP assays.
Amplified fragment length polymorphism (AFLP) is a genome fingerprinting technique based on the selective amplification of a subset of DNA fragments generated by restriction enzyme digestion (84). Originally applied to the characterization of plant genomes, AFLP has been more recently applied to bacterial typing (21, 26, 35, 36, 38). Two variations of AFLP have been described, one with two different restriction enzymes and two primers for amplification and the second with a single primer and restriction enzyme (21, 26, 35, 36, 38). In the most common configuration, bacterial DNA is extracted, purified, and subjected to digestion with two different enzymes, such as EcoRI and MseI. The restriction fragments are then ligated to linkers containing each restriction site and a sequence homologous to a PCR primer binding site. The PCR primers used for amplification contain DNA sequences homologous to the linker and contain one to two selective bases at their 3′ ends. For example, a selective primer directed against an EcoRI site might have the sequence 5′-GAATTCAA-3′, where the first six bases are complementary to the EcoRI site while the two A residues at the 3′ end are selective and allow amplification of only those EcoRI sites with the sequence 3′-CTTAATT-5′. They would not amplify an EcoRI site with the sequence 3′-CTTAATC-5′. Thus, the selective nucleotides allow amplification of only a subset of the genomic restriction fragments. In order to visualize the patterns, one of the PCR primers contains either a radioactive (21, 26, 35, 36) or a fluorescent (38) label, or, alternatively, the gels may be stained with ethidium bromide and the patterns may be examined under UV light (26).
Studies to date have demonstrated that AFLP is reproducible (21, 35, 38) and has good ability to differentiate clonally derived strains (21, 26, 35, 36, 38). The differentiation power of AFLP appears to be greater than that of PCR-based ribotyping; however, it has not yet been compared to Rep-PCR or PFGE (38). The AFLP procedure is more labor-intensive than Rep-PCR, but results are obtained more rapidly than with PFGE. Radioactive formats are impractical for clinical use, and while significant advantages exist with the fluorescent format, the cost of a DNA sequencer may be prohibitive for most laboratories. A commercially available kit can be used in combination with enzymatic detection of the bands; however, this requires that the DNA fragments be transferred to a filter by blotting and adds considerably to the labor costs. Yet, the reproducibility of the banding patterns for a given strain facilitates storing patterns in databases for use in identifying new bacterial strains. This combined with strong discriminatory power may make AFLP attractive to those laboratories performing frequent epidemiological studies such that a DNA sequencer becomes cost-effective.
DNA sequencing.
Ultimately, all molecular genetic methods for distinguishing organism subtypes are based on differences in the DNA sequence. Logically, then, DNA sequencing would appear to be the best approach to differentiating subtypes. However, as discussed below, this is not the case. DNA sequencing was originally performed by using radioactive labels for detection of the reaction products, an approach that is unsuitable for clinical use. Current DNA sequencing protocols employ fluorescent nucleotides to label the DNA. The sequence is then read with an automated instrument.
DNA sequencing generally begins with PCR amplification of a sample DNA directed at genetic regions of interest, followed by sequencing reactions with the PCR products. Modifications of the procedure can be made such that RNA may also be used as the starting material. Instruments that perform automated analysis of DNA sequencing gels are based on real-time detection of fluorescently labeled sequencing reaction products. DNA sequencing instruments most commonly utilize a modification of dideoxynucleotide chain termination chemistry in which the sequencing primer is labeled at the 5′ end with one of four fluorescent dyes. Each of the four fluorescent dyes represents one of the four nucleotides which make up the DNA molecule, and hence four separate annealing and extension reactions must be performed for each DNA sample to be analyzed. Once the DNA sequencing reactions have been completed, the four sets of reaction products are combined, concentrated, and loaded in a single well on a polyacrylamide gel. During electrophoresis, these fluorescently labeled products are excited by an argon laser and automatically detected. The resulting data is stored in digital form for subsequent processing into the final sequence with the aid of specialized software.
There are several considerations that must be evaluated before undertaking the use of DNA sequencing for subtyping. First, for practical purposes in the clinical laboratory, DNA sequencing must be directed at only a very small region of the chromosome of an organism. It is impractical to sequence multiple or large regions of the chromosome of an organism. Thus, in contrast to techniques like PFGE, Rep-PCR, or RAPD analysis, which examine the entire chromosome, DNA sequencing examines only a very small portion of the sites which can potentially vary between bacterial or fungal strains.
Furthermore, the short region of DNA used must meet several criteria before it can be used for strain differentiation. The structure of the region of DNA selected must consist of a variable sequence flanked by highly conserved regions. This enables PCR amplification and typing of all members of a species. The variability within the selected sequence must be sufficient to differentiate different strains of a particular species. Finally, the selected sequence should not be horizontally transmissible to other strains of the species.
Unfortunately, for bacteria and fungi, few sequences meet these criteria. While the 16S rRNA genes have been used to identify new species of organisms, they show limited variability between strains of a bacterial species (11, 44, 55, 88). The intergenic region between the 16S and 23S rRNA genes has also been used with variable results (8). Essentially species-specific genetic loci must be identified and tested empirically for the ability of a locus to provide strain differentiation. The major limitation of DNA sequencing approaches to bacterial and fungal subtyping is the size constraints of the DNA that can be practically sequenced.
In contrast, DNA sequencing is considered the gold standard for viral typing. Nucleic acid sequencing has much greater discriminatory power for determination of HCV genotypes and is the only molecular method by which mutations associated with human immunodeficiency virus (HIV) drug resistance can be reliably determined. The regions used for viral genotyping and detection of drug resistance mutations are in short, well-defined sequences that fulfill the criteria needed for the application of nucleic acid sequencing methodologies.
In applying DNA sequencing to genotypic studies, the laboratory should also be aware of the constraints of the sequencing methodology. One of the most problematic areas involved in DNA sequencing is the preparation of the sequencing gel. Some sequencing instruments either use precast gels or have apparatuses which greatly simplify gel preparation. Sequencing instruments that employ capillary gels, which automate gel preparation and loading, are even more convenient. However, traditional sequencing instruments in which the gel consists of a 40- by 40-cm slab gel are cumbersome to handle and pour.
Care must also be used when interpreting DNA sequencing data. The laboratory should be aware of artifacts that can complicate the interpretation. For example, repeats of a single nucleotide in a sequence can result in band compression artifacts that lead to misreading of the sequence. Both DNA strands should be sequenced and compared in order to minimize incorrect base identification.
In general, DNA sequencing is expensive and requires a high degree of technical competency to perform. Furthermore, automated DNA sequencers can be prohibitively expensive, with some costing in excess of $100,000.
In the future, non-gel-based sequencing methods based on hybridization of an unknown sequence to a DNA array may simplify the task of DNA sequencing for typing purposes as gene-specific chips are made. Kozal et al. (41) have reported the use of high-density DNA arrays to determine the DNA sequences of HIV clade B proteases of 167 HIV isolates taken from 102 patients in order to identify mutations associated with resistance to protease inhibitors. More recently, Troesch et al. (77) have described the use of a high-density DNA array to simultaneously identify mycobacteria at the species level and detect the presence of mutations in the rpoB gene associated with resistance to rifampin. Seventy isolates comprising 27 different species of mycobacteria and 15 rifampin-resistant strains of M. tuberculosis were analyzed. For species determination, the 16S rRNA hypervariable region was amplified by PCR, and the amplicons were hybridized to the array. Twenty-six of 27 species tested were correctly identified. Similarly, mutations associated with rifampin resistance were detected in all 15 rifampin-resistant isolates by hybridization of amplicons obtained by amplification of the rpoB gene to the array. This demonstrates the ability of DNA arrays to simultaneously interrogate more than one genetic locus. The multiplex capabilities of DNA arrays may significantly increase the utility of DNA sequencing-based methods for strain identification, since multiple genes could be rapidly screened for sequence-specific strain differences. The multiplexing capabilities of a DNA array would also enable simultaneous sequencing of both DNA strands, decreasing the amount of material and labor required to complete an analysis and increasing the cost-effectiveness of a chip for typing. While this technology is still some years away from commercialization, DNA arrays show intriguing prospects for the future.
PRACTICAL ASPECTS OF MOLECULAR TYPING METHODS
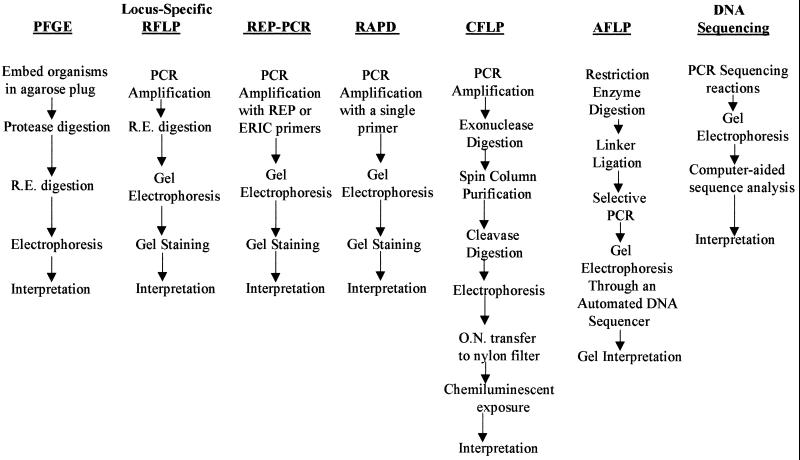
Tenover et al. have identified criteria to aid in the selection and interpretation of molecular typing methods for epidemiological studies (74). Although a particular typing method may have high discriminatory power and good reproducibility, the complexity of the method and interpretation of results as well as the costs involved in setting up and using the method may be beyond the capabilities of the laboratory. The choice of a molecular typing method, therefore, will depend upon the needs, skill level, and resources of the laboratory. As summarized in Fig. 1, methods such as locus-specific PCR, RAPD analysis, and Rep-PCR are similar in their procedures and are generally the easiest to implement. While PFGE appears to be time-consuming, it too is not difficult to implement. In contrast, procedures such as DNA sequencing, which require considerable knowledge about the instrumentation, methods, and potential pitfalls that can lead to uninterpretable results, can be difficult to implement reliably and require a higher skill level. Likewise, CFLP, with its need for DNA purification, careful optimization, and complex interpretation and its questionable reproducibility, can also be difficult to implement in a clinical laboratory.
FIG. 1.
Comparison of the procedural steps involved in various molecular typing methods. R.E., restriction endonuclease; O.N., overnight.
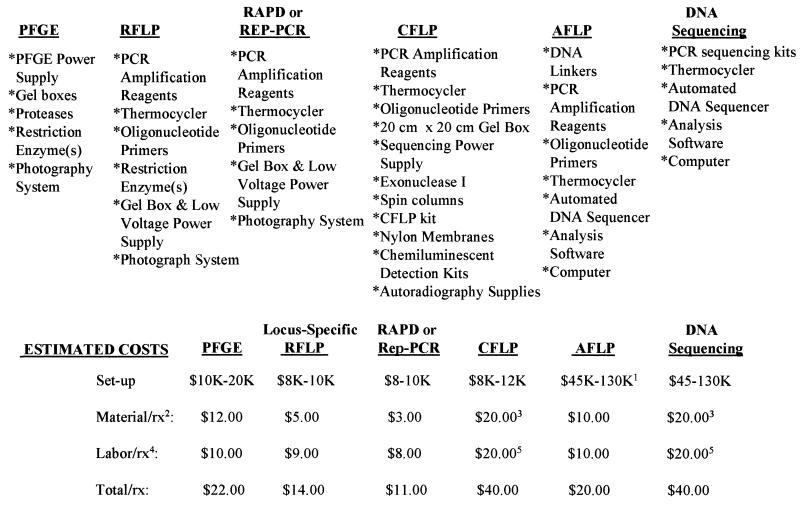
These methods also vary with regard to their equipment needs, setup costs, and cost per test (Fig. 2). For example, while DNA sequencing may ensure that the subtlest of changes can be observed, the cost of an automated sequencer may be prohibitively high. Furthermore, for subtyping bacteria and fungi, the laboratory must weigh the fact that the discriminatory power of techniques such as PFGE and Rep-PCR is sufficiently high to yield excellent subtyping results for a fraction of the setup costs as well as lower costs per test.
FIG. 2.
Equipment, reagents, and costs associated with molecular typing methods. Footnotes: 1, the large range in setup cost corresponds to the various types of automated DNA sequencing available; 2, material costs include enzymes, nucleotides, primers, disposable tubes and pipette tips, chemicals, purification kits, and electrophoresis reagents; 3, the material cost was calculated based on analysis of both DNA strands; 4, labor costs were calculated based on the total hands-on time of a technologist paid at a rate of $20.00/h and analyzing batches of 12 samples per run; 5, labor costs were calculated based on analysis of both DNA strands. rx, reaction.
While the costs per test may be decreased for both DNA sequencing and CFLP if only one DNA strand is examined, this must be weighed against the reliability of the result for strain differentiation. As mentioned above, CFLP can show marked DNA strand-specific differences in discriminatory power. Likewise, DNA sequencing is most accurate when both DNA strands are sequenced.
While the time to obtain a result can be as much as 4 days for PFGE, the labor costs are lower than those for a 2-day result from a technique such as CFLP (Fig. 2). Most of the time required for performance of PFGE is the reaction incubation time, whereas CFLP requires considerably more hands-on manipulation by the technician.
The pertinent characteristics that are important for introduction of a molecular typing method into a clinical laboratory situation are summarized in Table 1. One other criterion, not considered here, is the ability of a method to allow analysis of large numbers of samples. This criterion may not be of importance to some clinical laboratories, while for those routinely involved in large epidemiological studies, high-throughput capability may be crucial. For high throughput, simple techniques with high discriminatory power and low cost, such as Rep-PCR, may be most suitable. However, if storage of genetic profiles for the creation of reference databases of organisms is desirable, then methods such as PFGE or AFLP may be equally attractive for bacterial and fungal subtyping. For viral subtyping, DNA sequencing may be the only method suitable for monitoring genetic change and detecting mutations associated with drug resistance.
TABLE 1.
Summary of the characteristics of the various molecular typing methods
| Methodology | Ease of use | Ease of interpretation | Discrimination powera | Time to result (days) | Intralaboratory reproducibility | Interlaboratory reproducibility | Setup cost | Cost per test |
|---|---|---|---|---|---|---|---|---|
| PFGE | Moderate | Easy | High | 3 | Good | Good | Moderate | Moderate |
| Locus-specific PCR-RFLP | Easy | Easy | Moderate | 1 | Good | Good | Moderate | Low |
| Rep-PCR | Easy | Easy | High | 1 | Good | Moderate | Moderate | Low |
| RAPD | Easy | Easy | High | 1 | Moderate | Poor | Moderate | Low |
| CFLP | Moderate | Moderate | Moderate | 2 | Good | Poor | Moderate | High |
| AFLP | Moderate | Easy | High | 2 | Good | Good | High | Moderate |
| Sequencing | Difficult | Moderate | High | 2 | Good | Good | High | High |
This table is intended to give a relative estimate of discrimination power. For more exacting comparisons, see the references cited in the text.
Modern epidemiologists are fortunate to have a variety of tools which provide good molecular differentiation and which can be tailored to fit the needs of both the laboratory and the clinical study. Several of these molecular methods could enable the creation of large reference libraries of typed organisms to which new outbreak strains can be compared across laboratories in order to monitor changes in microbial populations.
REFERENCES
- 1.Akopyanz N, Bukanov N O, Westblom T U, Kresovich S, Berg D E. DNA diversity among clinical isolates of Helicobacter pylori detected by PCR-based RAPD fingerprinting. Nucleic Acids Res. 1992;20:5137–5142. doi: 10.1093/nar/20.19.5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appuhamy S, Parton R, Coote J G, Gibb H A. Genomic fingerprinting of Haemophilus somnus by a combination of PCR methods. J Clin Microbiol. 1997;35:288–291. doi: 10.1128/jcm.35.1.288-291.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arbeit R D. Laboratory procedures for the epidemiologic analysis of microorganisms. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: ASM Press; 1995. pp. 190–208. [Google Scholar]
- 4.Arbeit R D, Arthur M, Dunn R D, Kim C, Selander R K, Goldstein R. Resolution of recent evolutionary divergence among Escherichia coli from related lineages: the application of pulsed field electrophoresis to molecular epidemiology. J Infect Dis. 1990;161:230–235. doi: 10.1093/infdis/161.2.230. [DOI] [PubMed] [Google Scholar]
- 5.Arbeit R D, Slutsky A, Barber T W, Maslow J N, Niemczyk S, Falkinham J O, O’Connor G T, von Reyn C F. Genetic diversity among strains of Mycobacterium avium causing monoclonal and polyclonal bacteremia in patients with AIDS. J Infect Dis. 1993;167:1384–1390. doi: 10.1093/infdis/167.6.1384. [DOI] [PubMed] [Google Scholar]
- 6.Arbeit R D, Maslow J N, Mulligan M E. Polymerase chain reaction-mediated genotyping in microbial epidemiology. Clin Infect Dis. 1994;18:1018–1019. doi: 10.1093/clinids/18.6.1017. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 7.Barbier N, Saulnier P, Chachaty E, Dumontier S, Andremont A. Random amplified polymorphic DNA typing versus pulsed-field gel electrophoresis for epidemiological typing of vancomycin-resistant enterococci. J Clin Microbiol. 1996;34:106–109. doi: 10.1128/jcm.34.5.1096-1099.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barry T, Colleran G, Glennon M, Dunican L K, Gannon F. The 16S/23S ribosomal spacer region as a target for DNA probes to identify eubacteria. PCR Methods Appl. 1991;1:51–56. doi: 10.1101/gr.1.1.51. [DOI] [PubMed] [Google Scholar]
- 9.Bingen E, Boissinot C, Desjardins P, Cave H, Lambert-Zechovsk N, Denamur E, Blot P, Elion J. Arbitrary primed polymerase chain reaction provides rapid differentiation of Proteus mirabilis isolates from a pediatric hospital. J Clin Microbiol. 1993;31:1055–1059. doi: 10.1128/jcm.31.5.1055-1059.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blumberg H M, Rimland D, Kiehlbauch J A, Terry P M, Wachsmuth I K. Epidemiologic typing of Staphylococcus aureus by DNA restriction fragment length polymorphisms of rRNA genes: elucidation of the clonal nature of a group of bacteriophage-nontypeable ciprofloxacin-resistant methicillin-sensitive S. aureus isolates. J Clin Microbiol. 1992;30:362–369. doi: 10.1128/jcm.30.2.362-369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottger E C. Rapid determination of bacterial ribosomal RNA sequences by direct sequencing of enzymatically amplified DNA. FEMS Microbiol Lett. 1989;53:171–176. doi: 10.1016/0378-1097(89)90386-8. [DOI] [PubMed] [Google Scholar]
- 12.Brenner D J, Mayer L W, Carlone G M, Harrison L H, Bibb W F, Brandileone M C, Sottnek F O, Irino K, Reeves M W, Swenson J M. Biochemical, genetic, and epidemiologic characterization of Haemophilus influenzae biogroup aegyptius (Haemophilus aegyptius) strains associated with Brazillian purpuric fever. J Clin Microbiol. 1988;26:1524–1534. doi: 10.1128/jcm.26.8.1524-1534.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brow M A D, Oldenburg M C, Lyamichev V, Heisler L M, Lyamicheva N, Hall J G, Eagan N J, Olive D M, Smith L M, Fors L, Dahlberg J E. Differentiation of bacterial 16S rRNA genes and intergenic regions and Mycobacterium tuberculosis katG genes by structure-specific endonuclease cleavage. J Clin Microbiol. 1996;34:3129–3137. doi: 10.1128/jcm.34.12.3129-3137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caetano-Anolles G, Bassam B J, Gresshoff P M. DNA amplification fingerprinting using very short arbitrary oligonucleotide primers. Bio/Technology. 1991;9:553–557. doi: 10.1038/nbt0691-553. [DOI] [PubMed] [Google Scholar]
- 15.Clarridge J E, III, Raich T J, Pirwani D, Simon B, Tai L, Rodriguez-Barradas M C, Regnery R, Zollo A, Jones D C, Rambo C. Strategy to detect and identify Bartonella species in routine clinical laboratory yields Bartonella henselae from human immunodeficiency virus-positive patient and unique Bartonella strain from his cat. J Clin Microbiol. 1995;33:2107–2113. doi: 10.1128/jcm.33.8.2107-2113.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cockerill F R, III, Uhl J R, Temesgen Z, Zhang Y, Strockman L, Roberts G D, Williams D L, Kline B C. Rapid identification of a point mutation of the Mycobacterium tuberculosis catalase-peroxidase (katG) gene associated with isoniazid resistance. J Infect Dis. 1995;171:240–245. doi: 10.1093/infdis/171.1.240. [DOI] [PubMed] [Google Scholar]
- 17.Davidson F, Simmonds P, Ferguson J C, Jarvis L M, Dow B C, Follett E A C, Seed C R G, Krusius T, Lin C, Medgyesi G A, Kiyokawa H, Olim G, Duraisamy G, Cuypers T, Saeed A A, Teo D, Conradie J, Kew M C, Lin M, Nuchaprayoon C, Ndimbie O K, Yap P L. Survey of major genotypes and subgenotypes of hepatitis C virus using RFLP of sequences amplified from the 5′ non-coding region. J Gen Virol. 1995;76:1197–1204. doi: 10.1099/0022-1317-76-5-1197. [DOI] [PubMed] [Google Scholar]
- 18.De Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergenic consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. doi: 10.1128/aem.58.7.2180-2187.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DelVecchio V G, Petroziello J M, Gress M J, McClesky F K, Melcher G P, Crouch H K, Lupski J R. Molecular genotyping of methicillin-resistant Staphylococcus aureus via fluorophore-enhanced repetitive-sequence PCR. J Clin Microbiol. 1995;33:2141–2144. doi: 10.1128/jcm.33.8.2141-2144.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dijkshoorn L, Auken H M, Gerner-Smidt P, Kaufmann M E, Ursing J, Pitt T L. Correlation of typing methods for Acinetobacter isolates from hospital outbreaks. J Clin Microbiol. 1993;31:702–705. doi: 10.1128/jcm.31.3.702-705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dijkshoorn L, Aucken H M, Gerner-Smidt P, Janssen P, Kaufman M E, Garaizar J, Ursing J, Pitt T L. Comparison of outbreak and nonoutbreak Acinetobacter baumannii strains by genotypic and phenotypic methods. J Clin Microbiol. 1996;34:1519–1525. doi: 10.1128/jcm.34.6.1519-1525.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Food and Drug Administration; U.S. Department of Agriculture; U.S. Environmental Protection Agency; Centers for Disease Control and Prevention. Food safety from farm to table: a national food safety initiative report to the President, p. 3. Washington, D.C: Food and Drug Administration; 1997. [Google Scholar]
- 23.Georghiou P R, Doggett A M, Kielhofner M A, Stout J E, Watson D A, Lupski J R, Hamill R J. Molecular fingerprinting of Legionella species by repetitive element PCR. J Clin Microbiol. 1994;32:2989–2994. doi: 10.1128/jcm.32.12.2989-2994.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georghiou P R, Hamill R J, Wright C E, Versalovic J, Koeuth T, Watson D A, Lupski J R. Molecular epidemiology of infections due to Enterobacter aerogenes: identification of hospital-associated strains by molecular techniques. Clin Infect Dis. 1995;20:84–94. doi: 10.1093/clinids/20.1.84. [DOI] [PubMed] [Google Scholar]
- 25.Gerner-Smidt P. Ribotyping of Acinetobacter calcoaceticus-Acinetobacter baumannii complex. J Clin Microbiol. 1992;30:2680–2685. doi: 10.1128/jcm.30.10.2680-2685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson J R, Slater E, Xery J, Tompkins D S, Owen R J. Use of an amplified-fragment length polymorphism technique to fingerprint and differentiate isolates of Helicobacter pylori. J Clin Microbiol. 1998;36:2580–2585. doi: 10.1128/jcm.36.9.2580-2585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilson E, Perrin D, Hofnung M. DNA polymerase I and a protein complex bind specifically to E. coli palindromic unit highly repetitive DNA: implications for bacterial chromosome organization. Nucleic Acids Res. 1990;18:3941–3952. doi: 10.1093/nar/18.13.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilson E, Clement J M, Brutlag D, Hofnung D. A family of interspersed extragenic palindromic DNA sequences in E. coli. EMBO J. 1984;3:1417–1421. doi: 10.1002/j.1460-2075.1984.tb01986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grimont F, Verger J M, Cornelis P, Limet J, Lafevre M, Grayon M, Regnault B, Van Brock J, Grimont P A D. Molecular typing of Brucella with cloned DNA probes. Res Microbiol. 1992;143:55–65. doi: 10.1016/0923-2508(92)90034-l. [DOI] [PubMed] [Google Scholar]
- 30.Grundmann H, Schneider C, Hartung D, Daschner F D, Pitt T L. Discriminatory power of three DNA-based typing techniques for Pseudomonas aeruginosa. J Clin Microbiol. 1995;33:528–534. doi: 10.1128/jcm.33.3.528-534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamill R J, Houston E D, Georghiou P R, Wright C E, Koza M A, Cadle R M, Goepfert P A, Lewis D A, Zenon G J, Clarridge J E. An outbreak of Burkholderia (formerly Pseudomonas) cepacia respiratory tract colonization and infection associated with nebulized albuterol therapy. Ann Intern Med. 1995;122:762–766. doi: 10.7326/0003-4819-122-10-199505150-00005. [DOI] [PubMed] [Google Scholar]
- 32.Harvey B S, Koeuth T, Versalovic J, Woods C R, Lupski J R. Vertical transmission of Citrobacter diversus documented by DNA fingerprinting. Infect Control Hosp Epidemiol. 1995;16:564–569. doi: 10.1086/647009. [DOI] [PubMed] [Google Scholar]
- 33.Hulton C S J, Higgins C F, Sharp P M. ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium, and other enterobacteria. Mol Microbiol. 1991;5:825–834. doi: 10.1111/j.1365-2958.1991.tb00755.x. [DOI] [PubMed] [Google Scholar]
- 34.Irino K, Grimont F, Casin I, Grimont P A. rRNA gene restriction patterns of Haemophilus influenzae biogroup aegyptius strains associated with Brazillian purpuric fever. J Clin Microbiol. 1988;26:1535–1538. doi: 10.1128/jcm.26.8.1535-1538.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janssen P, Coopman R, Huys G, Swings J, Bleeker M, Vos P, Zabeau M, Kersters K. Evaluation of the DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology. 1996;142:1881–1893. doi: 10.1099/13500872-142-7-1881. [DOI] [PubMed] [Google Scholar]
- 36.Janssen P, Maquelin K, Coopman R, Tjerberg I, Bouvet P, Kerstens K, Dijkshoorn L. Discrimination of Acinetobacter genomic species by AFLP fingerprinting. Int J Syst Bacteriol. 1997;47:1179–1187. doi: 10.1099/00207713-47-4-1179. [DOI] [PubMed] [Google Scholar]
- 37.Kersulyte D, Woods J P, Kreath E J, Goldman W E, Berg D E. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol. 1992;174:7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koeleman J G M, Stoof J, Biesmans D J, Savelkoul P H M, Vandenbrouke-Grauls C M J E. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J Clin Microbiol. 1998;36:2522–2529. doi: 10.1128/jcm.36.9.2522-2529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koeuth T, Versalovic J, Lupski J R. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res. 1995;5:408–418. doi: 10.1101/gr.5.4.408. [DOI] [PubMed] [Google Scholar]
- 40.Koopman M B H, Kasboher A, Beckmann G, van der Zeijst B A M, Kusters J G. Genetic similarity of intestinal spirochetes from humans and various animal species. J Clin Microbiol. 1993;31:711–716. doi: 10.1128/jcm.31.3.711-716.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kozal M J, Shah N, Shen N, Yang R, Fucini R, Merigan T C, Richman D D, Morris D, Hubbell E, Chee M, Gingeras T R. Extensive polymorphisms observed in HIV-1 clade B protease gene using high-density oligonucleotide arrays. Nat Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 42.Kühn I, Burman L G, Hæggman S, Tullus K, Murray B E. Biochemical fingerprinting compared with ribotyping and pulsed-field gel electrophoresis of DNA for epidemiological typing of enterococci. J Clin Microbiol. 1995;33:2812–2817. doi: 10.1128/jcm.33.11.2812-2817.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kwon D-H, El-Zaatari F A K, Woo J-S, Perng C-L, Graham D Y, Goo M F. Rep-PCR fragments as biomarkers for differentiating gastroduodenal disease-specific Helicobacter pylori. Dig Dis Sci. 1998;43:980–987. doi: 10.1023/a:1018818431828. [DOI] [PubMed] [Google Scholar]
- 44.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analysis. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lehmann P F, Lin D, Lasker B A. Genotypic identification and characterization of species and strains within the genus Candida using random amplified polymorphic DNA. J Clin Microbiol. 1992;30:3249–3254. doi: 10.1128/jcm.30.12.3249-3254.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P Y-F, Wu W-L. Use of different PCR-based DNA fingerprinting techniques and pulsed-field gel electrophoresis to investigate the epidemiology of Acinetobacter calcoacetics-Acinetobacter baumannii complex. Diagn Microbiol Infect Dis. 1997;28:19–28. doi: 10.1016/s0732-8893(97)00080-1. [DOI] [PubMed] [Google Scholar]
- 47.Loutit J S, Tompkins L S. Restriction enzyme and Southern hybridization analyses of Pseudomonas aeruginosa strains from patients with cystic fibrosis. J Clin Microbiol. 1991;29:2897–2900. doi: 10.1128/jcm.29.12.2897-2900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maddox L O, Li P, Bennett A, Descartes M, Thompson J N. Comparison of SSCP analysis and CFLP analysis for mutation detection in the human iduronate 2-sulfatase gene. Biochem Mol Biol Int. 1997;43:1163–1171. doi: 10.1080/15216549700205001. [DOI] [PubMed] [Google Scholar]
- 49.Marshall D J, Heisler L M, Lyamichev V, Murvine C, Olive D M, Ehrlich G D, Neri B P, De Arruda M. Determination of hepatitis C virus genotypes in the United States by cleavase fragment length polymorphism analysis. J Clin Microbiol. 1997;35:3156–3162. doi: 10.1128/jcm.35.12.3156-3162.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martin B, Humbert O, Camara M, Guenzi E, Walker J, Mitchell T, Andrew P, Prudhomme M, Alloing G, Hakenbeck R. A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae. Nucleic Acids Res. 1992;20:3479–3483. doi: 10.1093/nar/20.13.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meunier J R, Grimont P A D. Factors affecting reproducibility of random amplified polymorphic DNA fingerprinting. Res Microbiol. 1993;144:373–379. doi: 10.1016/0923-2508(93)90194-7. [DOI] [PubMed] [Google Scholar]
- 52.Murray B E, Singh K V, Heath J D, Sharma B R, Weinstock G M. Comparison of genomic DNAs of different enterococcus isolates using restriction endonucleases with infrequent recognition sites. J Clin Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myers L E, Silva S V P S, Procunir J D, Little P B. Genomic fingerprinting of Haemophilus somnus isolates using a random-amplified polymorphic DNA assay. J Clin Microbiol. 1993;31:512–517. doi: 10.1128/jcm.31.3.512-517.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newbury S F, Smith N H, Robinson E C, Hiles I D, Higgins C F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987;48:297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- 55.Olsen G J, Woese C R. Ribosomal RNA: a key to phylogeny. FASEB J. 1993;7:113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- 56.Poh C L, Ramachandran V, Tasall J W. Genetic diversity of Neisseria gonorrhoeae IB-2 and IB-6 isolates revealed through whole-cell repetitive element sequence-based PCR. J Clin Microbiol. 1996;34:292–295. doi: 10.1128/jcm.34.2.292-295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Poh C L, Yeo C C, Tay L. Genome fingerprinting by pulsed-field gel electrophoresis and ribotyping to differentiate Pseudomonas aeruginosa serotype 011 strains. Eur J Clin Microbiol Infect Dis. 1992;11:817–822. doi: 10.1007/BF01960881. [DOI] [PubMed] [Google Scholar]
- 58.Prevost G, Jaulhac B, Piedmont Y. DNA fingerprinting by pulsed-field electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992;30:967–973. doi: 10.1128/jcm.30.4.967-973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ralph D, McClelland M, Welsh J, Baranton G, Perolat P. Leptospira species categorized by arbitrarily primed polymerase chain reaction (PCR) and by mapped restriction polymorphisms in PCR-amplified rRNA genes. J Bacteriol. 1993;175:973–981. doi: 10.1128/jb.175.4.973-981.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reboli A C, Houston E D, Monteforte J S, Wood C A, Hamill R J. Discrimination of epidemic and sporadic isolates of Acinetobacter baumannii by repetitive element PCR-mediated DNA fingerprinting. J Clin Microbiol. 1994;32:2635–2640. doi: 10.1128/jcm.32.11.2635-2640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robert F, Lebreton F, Bougnoux M E, Paugam A, Wassermann D, Schlotterer M, Tourte-Schaefer C, Dupouy-Camet J. Use of random amplified polymorphic DNA as a typing method for Candida albicans in epidemiological surveillance of a burn unit. J Clin Microbiol. 1995;33:2366–2371. doi: 10.1128/jcm.33.9.2366-2371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez-Barradas M C, Hamill R J, Houston E D, Georghiou P R, Clarridge J E, Regnery R L, Koehler J E. Genomic fingerprinting of Bartonella species by repetitive element PCR for distinguishing species and isolates. J Clin Microbiol. 1995;33:1089–1093. doi: 10.1128/jcm.33.5.1089-1093.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saulnier P, Bourneix C, Prevost G, Andremont A. Random amplified polymorphic DNA assay is less discriminant than pulsed-field gel electrophoresis for typing strains of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1993;31:982–985. doi: 10.1128/jcm.31.4.982-985.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schlichting C, Branger C, Fournier J-M. Typing of Staphylococcus aureus by pulsed-field gel electrophoresis, zymotyping, capsular typing, and phage typing: resolution of clonal relationships. J Clin Microbiol. 1993;31:227–232. doi: 10.1128/jcm.31.2.227-232.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schwartz D C, Cantor C R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 66.Sharples G J, Lloyd R G. A novel repeated DNA sequence located in the intergenic regions of bacterial chromosomes. Nucleic Acids Res. 1990;18:6503–6508. doi: 10.1093/nar/18.22.6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shortridge V D, Stone G G, Flamm R K, Beyer J, Versalovic J, Graham D W, Tanaka S K. Molecular typing of Helicobacter pylori isolates from a multicenter U.S. clinical trial by ureC restriction fragment length polymorphism. J Clin Microbiol. 1997;35:471–473. doi: 10.1128/jcm.35.2.471-473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Snelling A M, Gerner-Smidt P, Hawkey P M, Heritage J, Parnell P, Porter C, Bodenham A R, Inglis T. Validation of use of whole-cell repetitive extragenic palindromic sequence-based PCR (rep-PCR) for typing strains belonging to the Acinetobacter calcoaceticus-Acinetobacter baumannii complex and application of the method to the investigation of a hospital outbreak. J Clin Microbiol. 1996;34:1193–1202. doi: 10.1128/jcm.34.5.1193-1202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 70.Sreevatsan S, Bookout J B, Ringpis F M, Pottathil M R, Marshall D J, De Arruda M, Murvine C, Fors L, Pottathil R, Barathur R R. Algorithmic approach to high-throughput molecular screening for alpha interferon-resistant genotypes in hepatitis C patients. J Clin Microbiol. 1998;36:1895–1901. doi: 10.1128/jcm.36.7.1895-1901.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stern M J, Ames G F L, Smith N H, Robinson E C, Higgins C F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984;37:1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- 72.Stull T L, LiPuma J J, Edlind T D. A broad-spectrum probe for molecular epidemiology of bacteria: ribosomal RNA. J Infect Dis. 1988;157:280–286. doi: 10.1093/infdis/157.2.280. [DOI] [PubMed] [Google Scholar]
- 73.Tenover F C, Arbeit R D, Archer G, Biddle J, Byrne S, Goering R, Hancock G, Hébert G A, Hill B, Hollis R, Jarvis W R, Kreiswirth B, Eisner W, Maslow J, McDougal L K, Miller J M, Mulligan M, Pfaller M A. Comparison of traditional and molecular methods of typing isolates of Staphylococcus aureus. J Clin Microbiol. 1994;32:407–415. doi: 10.1128/jcm.32.2.407-415.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tenover F C, Arbeit R D, Goering R V. How to select and interpret molecular strain typing methods for epidemiological studies of bacterial infections: a review for healthcare epidemiologists. Molecular Typing Working Group of the Society for Healthcare Epidemiology of America. Infect Control Hosp Epidemiol. 1997;18:426–39. doi: 10.1086/647644. [DOI] [PubMed] [Google Scholar]
- 75.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tram C, Simonet M, Nicolas M-H, Offredo C, Grimont F, Lefevre M, Ageron E, Debure A, Grimont P A D. Molecular typing of nosocomial isolates of Legionella pneumophilia serogroup 3. J Clin Microbiol. 1990;28:242–245. doi: 10.1128/jcm.28.2.242-245.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Troesch A, Nguyen H, Miyada C G, Desvarenne S, Gingeras T R, Kaplan P M, Cros P, Mabilat C. Mycobacterium species identification and rifampin resistance testing with high-density DNA probe arrays. J Clin Microbiol. 1999;37:49–55. doi: 10.1128/jcm.37.1.49-55.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tsang A Y, Denner J C, Brennen P J, McClatchey J K. Clinical and epidemiological importance of typing Mycobacterium avium complex isolates. J Clin Microbiol. 1992;30:479–484. doi: 10.1128/jcm.30.2.479-484.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Van Belkum A, De Jonckheere J, Quint W G V. Typing Legionella pneumophilia strains by polymerase chain reaction-mediated fingerprinting. J Clin Microbiol. 1993;31:2198–2200. doi: 10.1128/jcm.31.8.2198-2200.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Versalovic J, Kapur V, Mason E O, Jr, Shah U, Koeuth T, Lupski J R, Musser J. Penicillin-resistant Streptococcus pneumoniae strains recovered in Houston: identification and molecular characterization of multiple clones. J Infect Dis. 1993;167:850–856. doi: 10.1093/infdis/167.4.850. [DOI] [PubMed] [Google Scholar]
- 82.Versalovic J, Kapur V, Koeuth T, Mazurek G H, Whittam T, Musser J, Lupski J R. DNA fingerprinting of pathogenic bacteria by fluorophore-enhanced repetitive sequence-based polymerase chain reaction. Arch Pathol Lab Med. 1995;119:23–29. [PubMed] [Google Scholar]
- 83.Vila J, Marcos M A, Jimenez de Anta M T. A comparative study of different PCR-based DNA fingerprinting techniques for typing of the Acinetobacter calcoaceticus-A. baumannii complex. J Med Microbiol. 1996;44:482–489. doi: 10.1099/00222615-44-6-482. [DOI] [PubMed] [Google Scholar]
- 84.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Muiper M, Zabeau M. AFLP: a new concept for DNA fingerprinting. Nucleic Acids Res. 1995;21:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Webster C A, Towner K J, Humphreys H, Ehrenstein B, Hartung D, Grundmann H. Comparison of rapid automated laser fluorescence analysis of DNA fingerprints with four other computer-assisted approaches for studying relationships between Acinetobacter baumannii isolates. J Med Microbiol. 1996;44:185–194. doi: 10.1099/00222615-44-3-185. [DOI] [PubMed] [Google Scholar]
- 86.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Williams J G, Kubelik A R, Livak K J, Rafalsky J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Woods C R, Versalovic J, Koeuth T, Lupski J R. Analysis of relationships among isolates of Citrobacter diversus by using DNA fingerprints generated by repetitive sequence-based primers in polymerase chain reaction. J Clin Microbiol. 1992;30:2921–2929. doi: 10.1128/jcm.30.11.2921-2929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Woods C R, Versalovic J, Koeuth T, Lupski J R. Whole-cell repetitive element sequence-based polymerase chain reaction allows rapid assessment of clonal relationships of bacterial isolates. J Clin Microbiol. 1993;31:1927–1931. doi: 10.1128/jcm.31.7.1927-1931.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Woods C R, Koeuth T, Estabrook M M, Lupski J R. Rapid determination of outbreak-related strains of Neisseria meningitidis by repetitive element-based polymerase chain reaction genotyping. J Infect Dis. 1996;174:760–767. doi: 10.1093/infdis/174.4.760. [DOI] [PubMed] [Google Scholar]
- 92.Yang Y, Ames G F L. DNA gyrase binds to the family of prokaryotic repetitive extragenic palindromic sequences. Proc Natl Acad Sci USA. 1988;85:8850–8854. doi: 10.1073/pnas.85.23.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zoe-Jordans J, Paul J, Bates J, Beaumot C, Kimari J, Gilks C. Characterization of Streptococcus pneumoniae from human immunodeficiency virus-seropositive patients with acute recurrent pneumonia. J Infect Dis. 1995;172:983–987. doi: 10.1093/infdis/172.4.983. [DOI] [PubMed] [Google Scholar]