Abstract
The cytoplasmic protein-tyrosine kinase Fes has been implicated in cytokine signal transduction, hematopoiesis, and embryonic development. Previous work from our laboratory has shown that active Fes exists as a large oligomeric complex in vitro. However, when Fes is expressed in mammalian cells, its kinase activity is tightly repressed. The Fes unique N-terminal sequence has two regions with strong homology to coiled-coil-forming domains often found in oligomeric proteins. Here we show that disruption or deletion of the first coiled-coil domain upregulates Fes tyrosine kinase and transforming activities in Rat-2 fibroblasts and enhances Fes differentiation-inducing activity in myeloid leukemia cells. Conversely, expression of a Fes truncation mutant consisting only of the unique N-terminal domain interfered with Rat-2 fibroblast transformation by an activated Fes mutant, suggesting that oligomerization is essential for Fes activation in vivo. Coexpression with the Fes N-terminal region did not affect the transforming activity of v-Src in Rat-2 cells, arguing against a nonspecific suppressive effect. Taken together, these findings suggest a model in which Fes activation may involve coiled-coil-mediated interconversion of monomeric and oligomeric forms of the kinase. Mutation of the first coiled-coil domain may activate Fes by disturbing intramolecular coiled-coil interaction, allowing for oligomerization via the second coiled-coil domain. Deletion of the second coiled-coil domain blocks fibroblast transformation by an activated form of c-Fes, consistent with this model. These results provide the first evidence for regulation of a nonreceptor protein-tyrosine kinase by coiled-coil domains.
The c-fes proto-oncogene is the normal cellular homolog of the transforming oncogenes found in several avian and feline retroviruses (13, 15, 16, 19, 22, 38). The human c-fes locus encodes a cytoplasmic protein-tyrosine kinase (Fes) primarily expressed in hematopoietic cells of the myeloid lineage (7, 28, 40). Distinct from Src and other nonreceptor tyrosine kinases, Fes has a long N-terminal unique region, followed by a central SH2 domain and a C-terminal kinase domain with a total molecular mass of 93 kDa. Previous studies have shown that Fes is activated by numerous hematopoietic growth factors (17, 18, 20, 30), implicating Fes as a downstream component of cytokine receptor signaling. Other work has shown that Fes suppresses the growth and induces the differentiation of the myeloid leukemia cell line, K-562 (46). Taken together, these results strongly suggest that Fes is involved in the regulation of hematopoietic cell proliferation, differentiation and function.
Despite the important roles that Fes plays in hematopoietic development, little is known about the mechanisms that regulate its tyrosine kinase activity. In the absence of activating signals, Fes tyrosine kinase activity is strongly repressed in cells (12). Indeed, high-level overexpression of Fes is required to induce kinase activation in vivo and to release transforming activity in fibroblasts (8). Recent work from our laboratory has shown that Fes autophosphorylation occurs by an intermolecular mechanism, suggesting that oligomerization may be an important initial event in Fes activation (36, 37). We also observed that active Fes elutes from a gel filtration column as a large oligomer and that the unique N-terminal domain is required for oligomerization. Taken together, these data suggest that oligomerization may be required for kinase activation and that suppression of oligomerization may represent a negative regulatory mechanism for Fes in vivo.
Computer analysis of the Fes N-terminal region revealed two strong consensus sequences for the formation of coiled-coil oligomerization domains (36). Coiled-coil domains are comprised of amphipathic α-helices with a characteristic heptad repeat in which the first and fourth residues are hydrophobic and pack against each other to form a hydrophobic core (26). The remainder of the amino acids are often hydrophilic, helping to solvate the oligomeric structure. In this study, we investigated the contribution of the Fes coiled-coil homology domains to the regulation of Fes tyrosine kinase activity and biological function. We observed that disruption or deletion of the more N-terminal coiled-coil domain (CC1) released Fes tyrosine kinase activity in fibroblasts, leading to transformation. In contrast to CC1, deletion of the second coiled-coil domain (CC2) inhibited transformation by an activated form of Fes, suggesting that CC2 may be required for oligomerization. In addition, a truncation mutant of Fes bearing only the unique N-terminal domain strongly suppressed the transforming and tyrosine kinase activity of an activated Fes mutant in fibroblasts, providing further evidence that Fes is active as an oligomer in living cells. These findings are consistent with a mechanism of Fes kinase regulation that involves interconversion of monomeric and oligomeric forms of the protein.
MATERIALS AND METHODS
Fes coiled-coil domain mutants.
The nucleotide sequence encoding the first N-terminal coiled-coil homology domain of Fes (CC1; amino acids 128 to 169) was deleted by using the Gene Editor oligonucleotide-directed mutagenesis system according to the manufacturer’s instructions (Promega). A similar approach was utilized to insert the β-turn motif Leu-Pro-Ala-Gly-Ser between N-terminal amino acids 148 and 149, which form the boundary between the third and fourth heptad repeats of the predicted coiled-coil structure. The coding sequence for the second coiled-coil domain of Fes (CC2; amino acids 310 to 344) was deleted by using a standard PCR-based strategy. The N-terminal myristylation signal sequence of v-Src was added to the N-terminal region of Fes and the coiled-coil domain mutants by using a PCR-based approach. The 5′ end of the Fes cDNA was amplified by using a forward primer encoding a unique HindIII site and the v-Src myristylation signal sequence Met-Gly-Ser-Ser-Lys-Ser-Lys fused to Fes homologous sequences beginning with codon 3 and a reverse primer that maps to the 5′ end of the Fes unique N-terminal domain. The resulting PCR product was digested with HindIII and AccI and swapped with the equivalent restriction fragment in wild-type Fes or the coiled-coil domain mutants to generate the full-length Myr-Fes cDNAs. The coding sequence for the Fes N-terminal region (Fes-NT), the myristylated form of the N-terminal region (Myr-Fes-NT), and related N-terminal constructs lacking the first or second coiled-coil homology domains (ΔCC1-NT, ΔCC2-NT, Myr-ΔCC1-NT, and Myr-ΔCC2-NT) were amplified by PCR from the corresponding full-length Fes cDNAs. All Fes cDNAs used in these studies bear a C-terminal FLAG epitope tag (37).
Expression of Fes proteins in 293T cells.
293T human embryonic kidney cells (35) were maintained in Dulbecco modified Eagle’s medium (DMEM) supplemented with 5% fetal bovine serum (FBS). Cells were transfected with Fes cDNAs in the expression vector pCDNA3 (InVitrogen) by using a modified calcium phosphate method described in detail elsewhere (37). Forty-eight hours later, whole-cell protein extracts were prepared by heating equal numbers of cells directly in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Proteins were resolved by SDS-PAGE, transferred to polyvinylidene difluoride (PVDF) membranes, and probed with antibodies to FLAG (M2; Sigma) to detect Fes protein expression or with antiphosphotyrosine antibodies (PY20; Transduction Laboratories) to detect autophosphorylation.
Production of recombinant retroviruses and infection of Rat-2 fibroblasts.
Wild-type and coiled-coil mutant Fes cDNAs were subcloned into the retroviral vector pSRαMSVtkneo (33). Retroviruses were produced by cotransfecting 293T cells with the pSRα-Fes constructs and an ecotropic packaging vector described elsewhere (2, 24). The retroviral supernatant was collected every 12 h for 3 days, and the supernatants were pooled and stored at −80°C.
Rat-2 cells were maintained in DMEM supplemented with 5% FBS. For viral infection, cells (2.5 × 105) were plated in 60-mm tissue culture dishes and incubated overnight at 37°C. Retroviral supernatants (5 ml) were thawed on ice, supplemented with Polybrene to 4 μg/ml, and added to the cells. After incubation for 4 h at room temperature, the viral supernatants were removed and replaced with 4 ml of fresh medium. The infected cells were then incubated for 48 h at 37°C prior to further analysis. Replicate plates of cells were lysed and analyzed for Fes protein expression as described above for the 293T cells.
Transformation assays.
Rat-2 fibroblast transformation was assessed by using a focus-forming assay. Retrovirally infected Rat-2 cells (2 × 104) were plated in 60-mm tissue culture dishes in the presence of 800 μg of G418 per ml. The cells were incubated at 37°C for 2 weeks, at which time they were Wright-Giemsa stained and observed by light microscopy. For some experiments, stained foci were enumerated from a scanned image by using colony-counting software (Bio-Rad GS-710 Calibrated Imaging Densitometer and Quantity One software).
For coexpression experiments, Rat-2 cells were first infected with recombinant retroviruses carrying the various Fes N-terminal constructs or empty virus as a negative control as described above. At 48 h postinfection, the cells were split and reseeded in 6-well plates at 2 × 105 cells/well. The following day, the cells were reinfected with retroviruses carrying activated forms of Fes or v-Src, incubated for 48 h, and replated at 2 × 104 cells/60-mm culture dish in the presence of G418. Foci were stained and counted 2 weeks later as described above.
In vitro tyrosine kinase assay for Fes.
Populations of Rat-2 cells stably expressing wild-type and mutant forms of Fes were lysed in Fes lysis buffer (50 mM Tris-HCl, pH 7.4; 50 mM NaCl; 1 mM EDTA; 1 mM MgCl2; 0.1% Triton X-100) supplemented with 25 μg of aprotinin per ml, 50 μg of leupeptin per ml, 1 mM phenylmethylsulfonyl fluoride, 20 mM NaF, 1 mM Na3VO4, and 50 μM Na2MoO4. Fes proteins were immunoprecipitated from clarified cell lysates with the M2 anti-FLAG monoclonal antibody resin. The immunoprecipitates were washed with radioimmune precipitation assay buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1 mM EDTA, 1% sodium deoxycholate), followed by kinase assay buffer (50 mM HEPES, pH 7.4; 10 mM MgCl2). The final pellets were incubated with [γ-32P]ATP (10 μCi; Dupont-New England Nuclear) and 2 μg of a glutathione S-transferase (GST) fusion protein containing amino acids 162 to 413 of the human Bcr protein. Previous studies have shown that this protein is strongly phosphorylated by Fes in vitro (23). After incubation for 15 min at 30°C, the reactions were stopped by heating in SDS-PAGE sample buffer. Phosphorylated proteins were resolved on SDS-polyacrylamide gels and visualized by using storage phosphorimaging.
Chemical cross-linking.
Fes N-terminal domain constructs were expressed as FLAG fusion proteins in 293T cells. Cells were lysed by sonication in Fes lysis buffer, and clarified cell lysates were incubated with the bifunctional cross-linking reagent disuccinimidyl suberate (DSS) at a final concentration of 1.0 mM as described previously (36). The reactions were incubated for 5 min at room temperature and stopped by heating in SDS-PAGE sample buffer. Cross-linked Fes proteins were visualized by immunoblotting with anti-FLAG antibodies.
Hematopoietic differentiation assay.
The human erythroleukemia cell line K-562 (25) was obtained from the American Type Culture Collection and grown in RPMI 1640 medium containing 10% FCS. K-562 cells were infected with recombinant Fes retroviruses by using a coculture approach. Cultures of virus-producing 293T cells were initiated by cotransfection with retroviral and packaging plasmids as described above. At 2 days posttransfection, 4 ml of DMEM containing 5% FCS and 3 × 105 K-562 cells were added to the 293T cultures along with 4 μg of polybrene per ml. After incubation for 2 days at 37°C, the K-562 cells were removed from the 293T cell culture by aspiration. Infected K-562 cells were replated on new culture dishes and incubated for an additional 2 days at 37°C, allowing residual 293T cells to readhere. The infected K-562 cells were reharvested, counted by trypan blue exclusion, and plated at 105 cells/60-mm tissue culture dish in 4 ml of RPMI containing 10% FCS and 800 μg of G418 per ml. Four days later, cells were fixed for 20 min in 1% paraformaldehyde and stored at 4°C in phosphate-buffered saline (PBS) prior to staining and flow cytometry. For single-cell analysis of Fes expression, 105 fixed cells were permeabilized with 0.05% saponin in RPMI containing 5% FBS (RPMI-FBS) for 20 min. Cells were resuspended in a minimal volume of RPMI-FBS containing the M2 anti-FLAG monoclonal antibody (20 μg/ml) for 1 h. The cells were washed twice with RPMI-FBS and then incubated with a goat anti-mouse immunoglobulin G-fluorescein isothiocyanate (FITC) conjugate (20 μg/ml in RPMI-FBS; Molecular Probes) for 1 h. The cells were then washed three more times prior to fluorescence-activated cell sorter analysis. Analysis of CD13 and CD33 expression was performed with direct FITC-conjugated antibodies for these myeloid differentiation markers (Southern Biotechnology Associates) (6). Staining was performed as described above for the M2 antibody, except the saponin was omitted. A population of K-562 cells cocultured with 293T cells producing a retrovirus carrying only the neo selection marker served as a negative control in all experiments.
RESULTS
Disruption of the first Fes coiled-coil homology domain (CC1) upregulates Fes kinase activity in vivo.
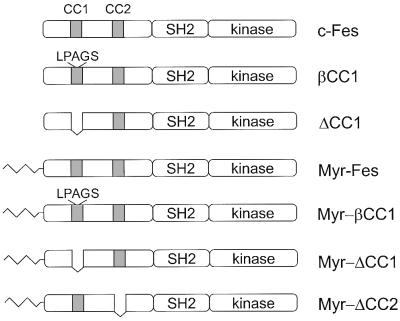
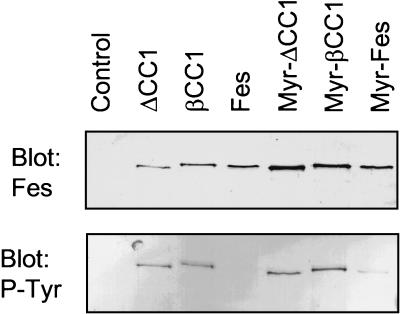
Previous analysis of the Fes unique N-terminal region with the COILS algorithm (27) revealed two regions of strong homology to coiled-coil oligomerization domains (36). We have designated these regions CC1 and CC2, and their relative positions within the Fes structure are illustrated in Fig. 1. To investigate the contribution of CC1 to the regulation of Fes kinase activity, we generated two CC1 domain mutants in which this region was either deleted entirely (ΔCC1) or disrupted by insertion of a β-turn (βCC1; see Fig. 1). The β-turn sequence chosen (Leu-Pro-Ala-Gly-Ser) was previously used to disrupt the coiled-coil oligomerization domain of the human breakpoint cluster region protein (Bcr) (31). Initial characterization of these mutants was conducted after transient expression in the human embryonic kidney cell line, 293T. Whole-cell protein extracts were prepared and analyzed by immunoblotting to determine the expression level and extent of autophosphorylation. As shown in Fig. 2, wild-type Fes was strongly expressed in these cells but displayed no detectable autophosphorylation, a result consistent with the tight negative regulation of Fes kinase activity observed previously in this cell line and in other systems (12, 23). However, disruption or deletion of the CC1 domain led to enhanced autophosphorylation of Fes, a result which is indicative of kinase activation.
FIG. 1.
Fes constructs used in this study. The structure of wild-type human c-Fes is shown at the top, which includes a unique N-terminal region, an SH2 domain, and a C-terminal kinase domain. The locations of the two regions with strong homology to coiled-coil oligomerization domains are indicated as shaded boxes and labeled CC1 and CC2. The CC1 mutant with an insertion of the β-turn sequence Leu-Pro-Ala-Gly-Ser is also shown (βCC1), as well as the CC1 deletion mutant (ΔCC1). Variants of all three proteins with the v-Src myristylation signal added to the N terminus are designated Myr-Fes, Myr-βCC1, and Myr-ΔCC1. Also shown is the structure of the Myr-Fes CC2 deletion mutant (Myr-ΔCC2).
FIG. 2.
Disruption or deletion of the first N-terminal coiled-coil homology domain (CC1) stimulates Fes tyrosine autophosphorylation in 293T cells. Wild-type Fes (Fes), Fes with an N-terminal myristylation signal (Myr-Fes) and the corresponding CC1 mutants (βCC1, ΔCC1, Myr-βCC1, and Myr-ΔCC1) were transiently expressed in human 293T cells as described in Materials and Methods. Whole-cell protein extracts were prepared in SDS-PAGE sample buffer, and Fes expression (top) and autophosphorylation (bottom) were analyzed by immunoblotting. Cells transfected with an empty expression vector were included as a negative control (Control).
Similar experiments were also conducted with a form of Fes bearing the v-Src myristylation signal sequence on its N terminus (Myr-Fes) (11, 24). This modification targets Fes to membranes and releases its transforming activity (see below). As shown in Fig. 2, Myr-Fes exhibits a low level of autophosphorylation, indicating that targeting Fes to membranes partially releases its kinase activity. However, deletion or disruption of the CC1 domain significantly enhanced Myr-Fes autophosphorylation in 293T cells (Fig. 2). This result shows that the effect of CC1 disruption on kinase activity is independent of the subcellular localization of Fes and is consistent with a negative regulatory function for the CC1 domain in the regulation of Fes tyrosine kinase activity in vivo.
Mutagenesis of the CC1 domain enhances Fes transforming and tyrosine kinase activities in Rat-2 fibroblasts.
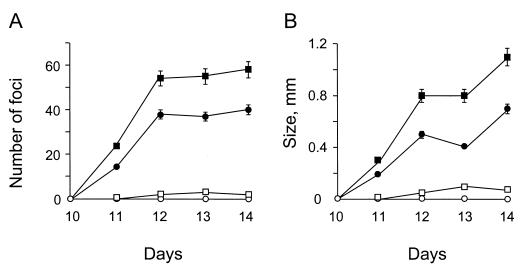
To determine the effect of CC1 mutation on Fes biological activity, we introduced the myristylated versions of the ΔCC1 and βCC1 Fes mutants into Rat-2 fibroblasts by using recombinant retroviruses. The infected cells were plated and observed for the appearance of transformed foci 10 to 14 days after infection, and the results are shown in Fig. 3. Mutation or deletion of CC1 induced the rapid appearance of many transformed foci relative to the Myr-Fes control containing the wild-type CC1 domain (Fig. 3A). In the case of the ΔCC1 mutant, the number of foci observed by the end of the 14 day incubation was more than 50-fold higher than that observed with Myr-Fes under these culture conditions. In addition, foci formed by fibroblasts expressing either of the CC1 mutations attained a much larger size compared to Myr-Fes (Fig. 3B). Control fibroblasts expressing wild-type Fes without the myristylation signal exhibited extremely low focus-forming activity, a result consistent with previous results demonstrating that Fes has tightly regulated tyrosine kinase and transforming activities in Rat-2 cells (12).
FIG. 3.
Deletion or disruption of the first N-terminal coiled-coil domain (CC1) enhances Myr-Fes transforming activity in Rat-2 cells. Wild-type Fes, Fes with an N-terminal myristylation signal (Myr-Fes), and Myr-Fes with insertion (Myr-βCC1) or deletion (Myr-ΔCC1) mutations in the CC1 domain were introduced into Rat-2 fibroblasts by using recombinant retroviruses. Infected cells were selected with G418 for 2 weeks and observed for the appearance of transformed foci. (A) After Wright-Giemsa staining, foci from three independent experiments were counted daily from day 10 to 14, and the average of the results is shown (± the standard deviation [SD]). (B) The average focus size was also determined for each culture under low-power microscopy. Results shown are the average values for three independent experiments ± the SD. Symbols: ○, c-Fes; □, Myr-Fes; ●, Myr-βCC1; ■, Myr-ΔCC1.
We also investigated the transforming activity of the Fes constructs bearing the same CC1 mutations but lacking the N-terminal myristylation signal sequence. However, we were unable to stably express these mutants in the Rat-2 cell background, thus preventing analysis of their transforming activity. Whether CC1 mutation is sufficient for transformation or whether a membrane-targeting or other relocalization signal is also required is not clear at this point.
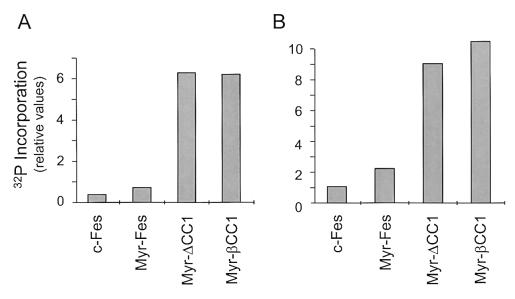
The results shown in Fig. 3 demonstrate that CC1 mutation results in a strong enhancement of Fes focus-forming activity. To determine whether this effect was due to an enhancement of tyrosine kinase activity as observed previously in the transient-transfection system (Fig. 2), Fes kinase activity was measured in an immune complex kinase assay both in terms of autophosphorylation and substrate phosphorylation. Populations of Rat-2 cells expressing Fes, Myr-Fes, Myr-βCC1, and Myr-ΔCC1 were lysed, and the Fes proteins were recovered by immunoprecipitation. After washing, the immunoprecipitates were incubated with [γ-32P]ATP and the substrate protein, GST-Bcr. This recombinant GST fusion protein contains human Bcr amino acids 162-413, which include the major tyrosine phosphorylation sites for this Fes substrate (23, 29). As shown in Fig. 4, both of the CC1 mutants exhibited increased tyrosine kinase activity relative to the wild-type Fes and Myr-Fes controls. This result suggests that the increased transforming activity of the CC1 mutants is due to enhancement of their intrinsic tyrosine kinase activity and is consistent with a role for the CC1 domain in the negative regulation of tyrosine kinase activity in vivo.
FIG. 4.
Deletion or disruption of the first N-terminal coiled-coil domain (CC1) enhances Myr-Fes tyrosine kinase activity in Rat-2 cells. Fes, Myr-Fes, Myr-ΔCC1, or Myr-βCC1 proteins were immunoprecipitated from lysates of Rat-2 fibroblasts and incubated in vitro with [γ-32P]ATP and a recombinant GST-Bcr fusion protein substrate. Phosphorylated GST-Bcr, as well as autophosphorylated Fes proteins, was resolved by SDS-PAGE, and the relative levels of 32P incorporation were determined by storage phosphorimaging. Equivalent levels of Fes proteins were present in the immune complexes, as shown by immunoblot analysis (data not shown). (A) Autophosphorylation. (B) Substrate phosphorylation. Results shown are the average of two separate determinations. The entire experiment was repeated three times with comparable results.
The CC1 domain is not required for oligomerization of the Fes N-terminal region.
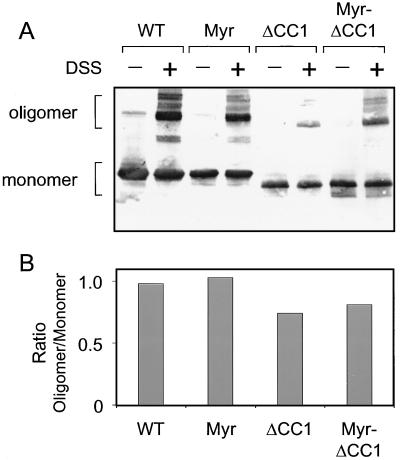
Previous work from our laboratory has shown that the active form of Fes is oligomeric (36). Data shown above indicate that deletion or mutation of the first coiled-coil domain leads to kinase activation and fibroblast transformation. These findings suggest that the Fes CC1 domain is not required for formation of the active oligomer and that CC2 or another portion of the N-terminal domain mediates oligomerization. To test this possibility directly, chemical cross-linking experiments were performed on Fes N-terminal proteins with or without the CC1 deletion. The N-terminal proteins were transiently expressed in 293T cells, and cell lysates were incubated in the presence or absence of the bifunctional cross-linking reagent, DSS. The reactions were analyzed for the presence of cross-linked products by SDS-PAGE and immunoblotting, and the result is shown in Fig. 5A. The relative levels of the monomeric proteins and the major oligomeric cross-linked products were determined by densitometry and are expressed as a ratio in Fig. 5B. All of the N-terminal proteins tested yielded higher-molecular-weight products in the presence of the cross-linking reagent, including those bearing the N-terminal myristyl modification. Thus, the CC1 domain is not required for oligomerization of the N-terminal domain.
FIG. 5.
The CC1 domain is not required for cross-linking of Fes N-terminal proteins. The N-terminal region of wild-type Fes (WT), the myristylated form of the N-terminal region (Myr), the N-terminal region lacking the first coiled-coil homology domain (ΔCC1), and the myristylated version of the N-terminal CC1 deletion mutant (Myr-ΔCC1) were transiently expressed in 293T cells. Clarified cell extracts were incubated in the presence (+) or absence (−) of the bifunctional cross-linking reagent DSS as described in Materials and Methods. (A) Cross-linked products were visualized by immunoblotting. The locations of the monomeric and oligomeric forms of the N-terminal proteins are indicated. (B) The relative levels of the monomeric and major oligomeric forms of the Fes N-terminal proteins were determined by densitometry and then plotted as the ratio shown. Four independent cross-linking experiments produced comparable results.
Transformation of Rat-2 fibroblasts by Myr-Fes is blocked by coexpression with the N-terminal unique domain.
Recent studies from our laboratory have shown that Fes autophosphorylation can be suppressed by a kinase-inactive form of Fes in vitro, suggesting that oligomerization may be an essential part of the activation mechanism in vivo (36). To test this idea, we coexpressed a truncated form of Fes consisting of only the myristylated N-terminal region together with Myr-Fes in Rat-2 fibroblasts and scored the changes in focus-forming activity after 2 weeks. As shown in Fig. 6, coexpression of Myr-Fes with the myristylated N-terminal domain led to a 75% reduction in the number of transformed foci compared to control Rat-2 cells coinfected with Myr-Fes and parental retroviruses. Immunoblots show equivalent expression of Myr-Fes in both cases, indicating that suppression of transforming activity by the N-terminal domain is not due to differences in the expression of Myr-Fes.
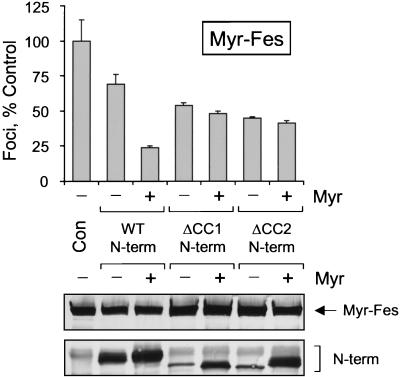
FIG. 6.
Suppression of Myr-Fes transforming activity by coexpression with Fes N-terminal domain proteins. Rat-2 fibroblasts were infected with recombinant retroviruses carrying the wild-type Fes N-terminal sequence (WT N-term), the N-terminal sequence lacking the first coiled-coil homology domain (ΔCC1 N-term), or the N-terminal sequence lacking the second coiled-coil homology domain (ΔCC2 N-term). A parallel series of N-terminal constructs bearing the v-Src myristylation sequence was also tested (indicated as + Myr). Cells infected with a retrovirus carrying only the neo selection marker served as a negative control (Con). Forty-eight hours later, the cells were reinfected with a Myr-Fes retrovirus and selected with G418 for 2 weeks as described in Materials and Methods. Foci were visualized by Wright-Giemsa staining and counted by using a Bio-Rad Model GS-710 Scanning Densitometer and colony-counting software. Foci from three independent cultures were counted and normalized to the negative control average value; the bar graph shows the average normalized value ± the SD. Expression of Myr-Fes and the N-terminal proteins was verified by immunoblotting with the anti-FLAG antibody, which recognizes the FLAG epitope fused to the C terminus of each protein (lower two panels). This entire experiment was performed twice and produced the same pattern of inhibition each time.
The activity of the wild-type Fes N-terminal region (without the myristylation signal) was also tested in this suppression assay. As shown in Fig. 6, removal of the myristyl group reduced the ability of the N-terminal region to induce the suppression of Myr-Fes transforming activity. This difference suggests that targeting of the N-terminal domain to the same compartment as Myr-Fes is important for suppression. In addition, fusion to the N-terminal myristylation signal may disturb intramolecular interaction between the CC1 and CC2 domains, promoting the trans-interaction with Myr-Fes that is required for the dominant-negative effect (see Discussion).
We also tested the suppressive activity of Fes N-terminal proteins with deletions of either CC1 or CC2. Figure 6 shows that both CC1 and CC2 N-terminal protein deletion mutants produced approximately 50% suppression of Myr-Fes focus-forming activity, indicating that the presence of either CC1 or CC2 is sufficient for suppression. In this case, suppression was independent of the presence of the myristylation signal sequence. These results suggest that deletion of one coiled-coil domain may free the remaining coiled-coil to interact with Myr-Fes to produce the dominant-inhibitory effect.
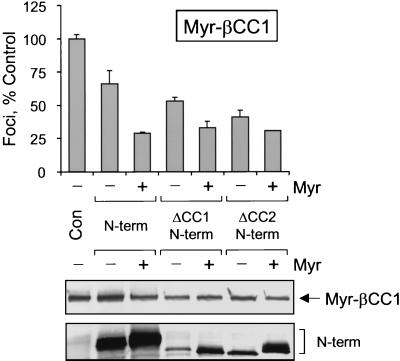
We also used the suppression assay to determine the range of possible coiled-coil interactions. For these experiments, transformation was induced by using the strongly transforming Myr-βCC1-Fes mutant, which has a single functional coiled-coil domain (CC2). As shown in Fig. 7, the wild-type and myristylated forms of the N-terminal domain inhibited Myr-βCC1-Fes transforming activity by approximately 30 and 75%, respectively, as observed for Myr-Fes (Fig. 6). Interestingly, the Fes N-terminal proteins lacking either the CC1 or the CC2 domain also produced strong inhibitory effects. These data suggest that suppression of Myr-βCC1-Fes transforming activity by ΔCC1 N-terminal proteins is mediated by CC2-CC2 interaction, while suppression by ΔCC2 is mediated by CC1-CC2 interaction. Thus, homomeric interactions between CC2 domains, as well as heteromeric interactions between CC1 and CC2, appear to be possible. A very similar pattern of suppression was observed upon coexpression of the same panel of N-terminal proteins with the strongly transforming Myr-ΔCC1 Fes mutant as well (data not shown).
FIG. 7.
Suppression of Myr-βCC1 Fes transforming activity by coexpression with Fes N-terminal domain proteins. Rat-2 fibroblasts were infected with recombinant retroviruses carrying the wild-type Fes N-terminal sequence (N-term), the N-terminal sequence lacking the first coiled-coil homology domain (ΔCC1 N-term), or the N-terminal sequence lacking the second coiled-coil homology domain (ΔCC2 N-term). A parallel series of N-terminal constructs bearing the v-Src myristylation sequence was also tested (indicated as + Myr). Cells infected with a retrovirus carrying only the neo selection marker served as a negative control (Con). Forty-eight hours later, the cells were reinfected with a Myr-βCC1 Fes retrovirus and selected with G418 for 2 weeks as described in Materials and Methods. Foci were visualized by Wright-Giemsa staining and counted by using a Bio-Rad Model GS-710 Scanning Densitometer and colony-counting software. Foci from three independent cultures were counted and normalized to the negative control average value; the bar graph shows the average normalized value ± the SD. Expression of Myr-βCC1 and the N-terminal proteins was verified by immunoblotting with the anti-FLAG antibody, which recognizes the FLAG epitope fused to the C terminus of each protein (lower two panels). This entire experiment was performed twice and produced the same pattern of inhibition each time.
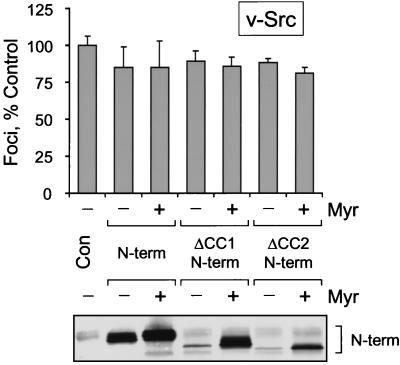
To control for specificity in the N-terminal suppression studies, the Fes N-terminal proteins used for the experiments shown in Fig. 6 and 7 were also coexpressed with v-Src in the focus-forming assay. v-Src tyrosine kinase was chosen because it is dependent upon myristylation for fibroblast transformation (21) but is regulated by mechanisms that do not involve coiled-coil domains. As shown in Fig. 8, none of the c-Fes N-terminal proteins affected v-Src-induced transformation, supporting the conclusion that the suppressive actions of the N-terminal proteins are specific to Fes-mediated transformation.
FIG. 8.
Coexpression with Fes N-terminal domain proteins does not inhibit Rat-2 cell transformation by v-Src. Rat-2 fibroblasts were infected with recombinant retroviruses carrying the wild-type Fes N-terminal sequence (N-term), the N-terminal sequence lacking the first coiled-coil homology domain (ΔCC1 N-term), or the N-terminal sequence lacking the second coiled-coil homology domain (ΔCC2 N-term). A parallel series of N-terminal constructs bearing the v-Src myristylation sequence was also tested (indicated as + Myr). Cells infected with a retrovirus carrying only the neo selection marker served as a negative control (Con). Forty-eight hours later, the cells were reinfected with a v-Src retrovirus and selected with G418 for 2 weeks as described in Materials and Methods. Foci were visualized by Wright-Giemsa staining and counted by using a Bio-Rad Model GS-710 Imaging Densitometer and colony-counting software. Foci from three independent cultures were counted and normalized to the negative control average value; the bar graph shows the average normalized value ± the SD. Expression of the N-terminal proteins was verified by immunoblotting with the anti-FLAG antibody, which recognizes the FLAG epitope fused to the C terminus of each protein (lower panel). This entire experiment was performed twice and produced the same result each time.
Data presented in Fig. 6 to 8 suggest that the ability of Fes N-terminal proteins to suppress Myr-Fes-induced transforming activity involves direct interaction between the proteins, resulting in the formation of inactive mixed oligomers. To test this idea further, we performed kinase assays on Myr-Fes immunoprecipitates from cultures coexpressing the Myr-Fes N-terminal region or matched controls. As shown in Fig. 9, the extent of autophosphorylation of both Myr-Fes and the Myr-ΔCC1 Fes mutant were inhibited by more than 50% in the presence of the N-terminal protein. These results support direct inhibition of kinase activity in vivo as a mechanism for the suppressive action of the N-terminal region and agree with our previous results showing that a purified recombinant N-terminal protein binds directly to Fes and inhibits its kinase activity in vitro (36).
FIG. 9.
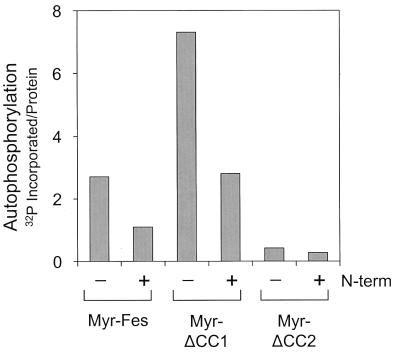
Coexpression with the Fes N-terminal domain inhibits autophosphorylation of Myr-Fes and the Myr-Fes CC1 domain deletion mutant. Rat-2 fibroblasts expressing Myr-Fes, Myr-ΔCC1, and Myr-ΔCC2 in the presence (+) or absence (−) of the myristylated N-terminal domain protein (N-term) were lysed, and the Fes proteins were immunoprecipitated with the anti-FLAG antibody. The immunoprecipitates were washed, incubated with [γ-32P]ATP, and separated by SDS-PAGE. The radiolabeled proteins were transferred to PVDF membranes to allow determination of protein levels by immunoblotting and densitometry. Incorporation of 32P into Fes proteins was determined by storage phosphorimaging, and the resulting values were corrected for protein levels and plotted as shown. This experiment was repeated three times with comparable results.
The Fes CC2 domain is required for Fes transforming and kinase activities.
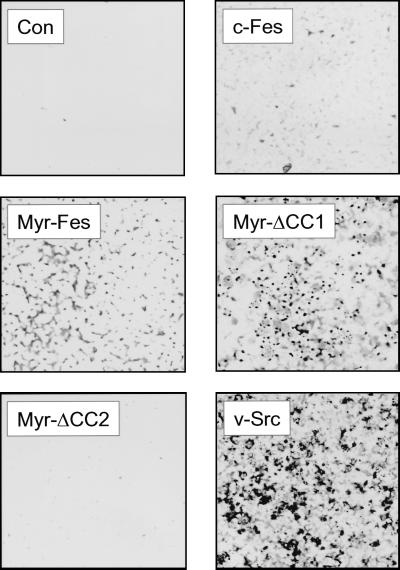
Data presented so far point to the CC1 domain as a negative regulator of Fes kinase activity that is not required for oligomerization and activation and implicate the CC2 domain in oligomer formation. To test this possibility directly, we created a CC2 deletion in the context of Myr-Fes and compared its transforming activity with wild-type Fes, Myr-Fes, and Myr-ΔCC1 Fes, as well as v-Src. As shown in Fig. 10, deletion of the CC2 domain completely inhibited the transforming activity of Myr-Fes. This result is in sharp contrast to the effect of the CC1 mutation, which releases strong transforming activity. Deletion of CC2 also resulted in greatly reduced kinase activity compared to Myr-Fes or Myr-ΔCC1 (Fig. 9). These results show that CC2 is required for both biological and kinase activities and suggest that it may contribute to oligomerization in vivo.
FIG. 10.
Deletion of the second coiled-coil homology domain inhibits the transforming activity of Myr-Fes. Rat-2 fibroblasts were infected with recombinant retroviruses carrying wild-type c-Fes, Myr-Fes, Myr-ΔCC1, Myr-ΔCC2, or v-Src. Cells infected with a retrovirus carrying only the neo selection marker served as a negative control (Con). Infected cells were cultured for 2 weeks in the presence of G418 and were visualized by Wright-Giemsa staining. The experiment was performed in triplicate; representative scanned images of the stained foci are shown.
Fes CC1 domain mutants retain the ability to induce differentiation in K-562 cells.
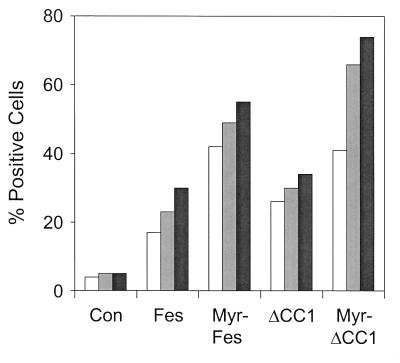
Previous studies have shown that expression of Fes is sufficient to induce differentiation of the myeloid leukemia cell line, K-562 (6, 46). However, studies of Fes-transfected K-562 cell populations suggest that only a fraction of the cells undergo differentiation after transfection with Fes (6). This result suggests that a threshold of Fes tyrosine kinase activity must be reached in order for differentiation to occur. Therefore, we tested the differentiation-inducing activity of the Fes mutants lacking the CC1 domain which demonstrated elevated tyrosine kinase activity and strong transforming potential in fibroblasts. Both the myristylated and nonmyristylated forms of wild-type Fes and the ΔCC1 mutant were tested. Each of these proteins was introduced into K-562 cells by using recombinant retroviruses, and differentiation was assessed as the percentage of cells that express the cell surface myeloid differentiation antigens CD13 and CD33 as determined by flow cytometry. The percentage of cells expressing Fes was also evaluated by this method. As shown in Fig. 11, the ratio of Fes protein to differentiation marker expression was approximately equal for wild-type Fes, Myr-Fes, and the CC1 deletion mutant without the myristylation signal sequence. Interestingly, the myristylated form of Fes with the CC1 deletion exhibited enhanced differentiation-inducing activity, as reflected by the higher ratio of CD13 and CD33 to Fes protein expression. These results show that deletion of the CC1 domain together with a membrane targeting signal leads to robust differentiation signaling in K-562 cells and are consistent with the fibroblast transformation data. Thus, CC1 also appears to function as a negative regulator of Fes biological activity in a physiologically relevant cellular context.
FIG. 11.
Differentiation of K-562 leukemia cells by Fes coiled-coil domain mutants. K-562 myeloid leukemia cells were incubated with 293T cells producing recombinant retroviruses carrying either wild-type Fes, Myr-Fes, ΔCC1, Myr-ΔCC1, or only the G418 resistance marker as a negative control (Con). After 48 h of coculture, K-562 cells were separated from the 293T cells and replated in the presence of G418. Four days later, infected cells were analyzed by flow cytometry for expression of Fes proteins by using the M2 anti-FLAG monoclonal antibody and for cell surface CD13 and CD33 by using direct FITC-conjugated antibodies to these myeloid cell-surface antigens. The percentage of cells positive for Fes (open bars), CD13 (gray bars), and CD33 (solid bars) are shown. Enhancement of differentiation by Myr-ΔCC1 was observed in two independent experiments.
DISCUSSION
Previous studies have established that Fes tyrosine kinase activity is tightly regulated both in physiological sites of expression such as myeloid hematopoietic cells and after overexpression in rodent fibroblasts and human embryonic kidney cells (12, 23, 28). Efficient regulation of Fes kinase activity in these diverse cell types implies that the mechanism of negative regulation may be intrinsic to the structure of the kinase. Data presented in this report show that the coiled-coil domains of Fes may have an important function in the suppression of Fes tyrosine kinase activity in both the fibroblast transformation model and in hematopoietic cells. Mutations in the more N-terminal coiled-coil domain, including deletion, insertion of a β-turn, or even a single point mutation of a conserved leucine residue (data not shown), all released Fes tyrosine kinase and transforming activities in fibroblasts and enhanced differentiation-inducing activity in K-562 cells. These data strongly support an important negative regulatory function for this coiled-coil motif and provide the first example of a coiled-coil domain contributing to the negative regulation of a cellular tyrosine kinase.
Previous data from our laboratory have established that the active form of Fes exists as a large oligomeric complex, at least in vitro (36). In addition, we found that Fes autophosphorylation, which is a critical first step in the activation mechanism, occurs by a trans mechanism reminiscent of growth factor receptor tyrosine kinases (37). Similar results have been reported for the Fes-related tyrosine kinase, v-Fps (42, 43). These previous data suggested that oligomerization is the key to Fes activation in vivo. Thus, suppression of Fes kinase activity may require maintenance of the monomeric state as a mechanism to prevent the trans phosphorylation required for kinase activation. This may be achieved by intramolecular interaction between the first and second coiled-coil domains. Evidence for CC1-CC2 interaction is provided by in vivo suppression experiments in which Fes N-terminal proteins lacking CC2 are shown to suppress transformation by Myr-βCC1 (Fig. 7); this Fes mutant lacks a functional CC1 domain. We have also obtained additional evidence for CC1-CC2 interaction by using the yeast two-hybrid system (data not shown). This model also explains why mutations in the more N-terminal coiled-coil domain release tyrosine kinase activity. Such mutations would prevent CC1-CC2 interaction and promote constitutive oligomerization. The formation of active Fes oligomers appears to require CC2, since deletion of the CC2 domain almost completely inhibited transformation by Myr-Fes and also greatly reduced its kinase activity. Interestingly, mutations in the CC2-homologous region of v-Fps have also been shown to reduce its kinase and transforming activities (34). Determination of the three-dimensional structure of the Fes N-terminal region will provide an important test of this hypothetical model.
Data presented here also provide new evidence that oligomerization is critical to Fes activation in living cells. The Fes N-terminal domain serves as an effective dominant-negative mutant since it was able to suppress Rat-2 cell transformation by Myr-Fes (Fig. 6). This result suggests that the N-terminal domain of Fes is capable of interacting with full-length Myr-Fes molecules, resulting in the formation of nonproductive oligomers that are unable to undergo trans phosphorylation. These results are consistent with earlier work from our laboratory showing that both the Fes N-terminal domain and a full-length kinase-inactive Fes mutant can suppress wild-type Fes autophosphorylation in vitro (36). Importantly, both the isolated N-terminal region and the kinase-dead Fes mutant can form physical complexes with wild-type Fes in vitro (36).
Although our data are consistent with a model in which CC1 may regulate Fes kinase activity via intramolecular interaction with CC2, alternative hypotheses should be considered as well. For example, the CC1 domain or other features of the Fes N-terminal region may interact in trans with inhibitory proteins which suppress Fes oligomerization and activation. A number of trans-inhibitory proteins have been proposed as regulators of c-Abl, another nonreceptor tyrosine kinase with tightly regulated tyrosine kinase and transforming activities (41). In the case of c-Abl, negative regulation seems to require an intact SH3 domain, since mutations in this domain are strongly activating in vivo. Whether or not similar interacting proteins exist that suppress the tyrosine kinase activity of Fes will require further investigation.
Although data presented here provide some of the first evidence demonstrating regulation of a cellular tyrosine kinase by coiled-coil domains in vivo, previous work has shown that coiled coils contribute to the constitutive activation of the Bcr-Abl and TEL-Abl oncoproteins associated with various leukemias (9, 31). In each case, translocations result in the fusion of the c-Abl tyrosine kinase to a coiled-coil oligomerization function provided by Bcr or TEL. In the case of Bcr-Abl, N-terminal Bcr-encoded sequences form a tetramerization motif that is required for constitutive activation of the Abl kinase, as well as for localization to the actin cytoskeleton (31). Interestingly, coexpression of a peptide containing the Bcr oligomerization domain is sufficient to reverse the transformed phenotype of Bcr-Abl-transformed myeloid leukemia cells (14). These data suggest that suppression results from the formation of nonfunctional mixed oligomers and are consistent with our results demonstrating that coexpression with the Fes N-terminal region blocks transformation by activated forms of full-length Fes.
Intramolecular interactions are emerging as a common theme in the regulation of nonreceptor protein-tyrosine kinases. Perhaps the best-studied example involves kinases of the Src family. The X-ray crystal structures of c-Src (44, 45) and the closely related kinase Hck (39) strongly suggest that negative regulation involves two intramolecular interactions. First, tyrosine phosphorylation of the conserved tail tyrosine residue induces intramolecular interaction with the SH2 domain, an interaction long suspected because mutations in either of these regions induce kinase activation (4). A second interaction revealed by the crystal structure involves the SH3 domain and an intramolecular ligand formed by the linker region connecting the SH2 and kinase domains. This intramolecular interaction is also critical for effective negative regulation of Src family kinases, since mutations in the SH3 domain or the linker region also release the tyrosine kinase and transforming activities of c-Src and Hck (3, 5, 10). Activation of Src family kinases can also result from interactions with other proteins that bind to the SH2 or SH3 domain or both, presumably by displacement of these intramolecular interactions (1, 2, 32). These findings with Src-related kinases suggest that association of Fes with other proteins may also lead to activation by disturbing intramolecular, negative-regulatory interactions. For example, previous work from our laboratory has shown that Fes associates with and phosphorylates Bcr, the normal breakpoint-cluster region protein (23, 29). Fes-Bcr interaction is mediated, in part, by the Fes N-terminal region (29). More recently, we observed that coexpression with Bcr induces Fes autophosphorylation (23). Association of Fes with Bcr may disrupt CC1-CC2 interaction, leading to kinase activation. Bcr is also an oligomeric protein (31), suggesting that it is capable of interacting with several molecules of Fes simultaneously. Recruitment of multiple Fes molecules into close proximity may contribute to activation via trans phosphorylation.
ACKNOWLEDGMENT
This work was supported by grant CA58667 from the National Institutes of Health.
REFERENCES
- 1.Alexandropoulos K, Baltimore D. Coordinate activation of c-Src by SH3 and SH2 binding sites on a novel, p130Cas-related protein, Sin. Genes Dev. 1997;10:1341–1355. doi: 10.1101/gad.10.11.1341. [DOI] [PubMed] [Google Scholar]
- 2.Briggs S D, Sharkey M, Stevenson M, Smithgall T E. SH3-mediated Hck tyrosine kinase activation and fibroblast transformation by the Nef protein of HIV-1. J Biol Chem. 1997;272:17899–17902. doi: 10.1074/jbc.272.29.17899. [DOI] [PubMed] [Google Scholar]
- 3.Briggs S D, Smithgall T E. SH2-kinase linker mutations release Hck tyrosine kinase and transforming activities in rat-2 fibroblasts. J Biol Chem. 1999;274:26579–26583. doi: 10.1074/jbc.274.37.26579. [DOI] [PubMed] [Google Scholar]
- 4.Brown M T, Cooper J A. Regulation, substrates, and functions of Src. Biochim Biophys Acta. 1996;1287:121–149. doi: 10.1016/0304-419x(96)00003-0. [DOI] [PubMed] [Google Scholar]
- 5.Erpel T, Superti-Furga G, Courtneidge S A. Mutational analysis of the Src SH3 domain: the same residues of the ligand binding surface are important for intra- and intermolecular interactions. EMBO J. 1995;14:963–975. doi: 10.1002/j.1460-2075.1995.tb07077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang F, Ahmad S, Lei J, Klecker R W, Trepel J B, Smithgall T E, Glazer R I. The effect of mutation of tyrosine 713 in p93c-fes on its catalytic activity and ability to promote myeloid differentiation in K-562 cells. Biochemistry. 1993;32:6995–7001. doi: 10.1021/bi00078a026. [DOI] [PubMed] [Google Scholar]
- 7.Feldman R A, Gabrilove J L, Tam J P, Moore M A S, Hanafusa H. Specific expression of the human cellular fps/fes-encoded protein NCP92 in normal and leukemic myeloid cells. Proc Natl Acad Sci USA. 1985;82:2379–2383. doi: 10.1073/pnas.82.8.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldman R A, Lowy D R, Vass W C, Velu T J. A highly efficient retroviral vector allows detection of the transforming activity of the human c-fps/fes proto-oncogene. J Virol. 1989;63:5469–5474. doi: 10.1128/jvi.63.12.5469-5474.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Golub T R, Goga A, Barker G F, Afar D E H, McLaughlin J, Bohlander S K, Rowley J D, Witte O N, Gilliland D G. Oligomerization of the ABL tyrosine kinase by the Ets protein TEL in human leukemia. Mol Cell Biol. 1996;16:4107–4116. doi: 10.1128/mcb.16.8.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonfloni S, Williams J C, Hattula K, Weijland A, Wierenga R K, Superti-Furga G. The role of the linker between the SH2 domain and catalytic domain in the regulation and function of Src. EMBO J. 1997;16:7261–7271. doi: 10.1093/emboj/16.24.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greer P, Haigh J, Mbamalu G, Khoo W, Bernstein A, Pawson T. The Fps/Fes protein-tyrosine kinase promotes angiogenesis in transgenic mice. Mol Cell Biol. 1994;14:6755–6763. doi: 10.1128/mcb.14.10.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greer P A, Meckling-Hansen K, Pawson T. The human c-fps/fes gene product expressed ectopically in rat fibroblasts is nontransforming and has restrained protein-tyrosine kinase activity. Mol Cell Biol. 1988;8:578–587. doi: 10.1128/mcb.8.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Groffen J, Heisterkamp N, Shibuya M, Hanafusa H, Stephenson J R. Transforming genes of avian (v-fps) and mammalian (v-fes) retroviruses correspond to a common cellular locus. Virology. 1983;125:480–486. doi: 10.1016/0042-6822(83)90219-2. [DOI] [PubMed] [Google Scholar]
- 14.Guo X Y, Cuillerot J M, Wang T, Wu Y, Arlinghaus R, Claxton D, Bachier C, Greenberger J, Colombowala I, Deisseroth A B. Peptide containing the BCR oligomerization domain (AA 1–160) reverses the transformed phenotype of p210bcr-abl positive 32D myeloid leukemia cells. Oncogene. 1998;17:825–833. doi: 10.1038/sj.onc.1201999. [DOI] [PubMed] [Google Scholar]
- 15.Hampe A, Laprevotte I, Galibert F, Fedele L A, Sherr C J. Nucleotide sequences of feline retroviral oncogenes (v-fes) provide evidence for a family of tyrosine-specific protein kinase genes. Cell. 1982;30:775–785. doi: 10.1016/0092-8674(82)90282-3. [DOI] [PubMed] [Google Scholar]
- 16.Hanafusa T, Wang L-H, Anderson S M, Karess R E, Hayward W S, Hanafusa H. Characterization of the transforming gene of Fujinami sarcoma virus. Proc Natl Acad Sci USA. 1980;77:3009–3013. doi: 10.1073/pnas.77.5.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanazono Y, Chiba S, Sasaki K, Mano H, Miyajima A, Arai K, Yazaki Y, Hirai H. c-fps/fes protein-tyrosine kinase is implicated in a signaling pathway triggered by granulocyte-macrophage colony-stimulating factor and interleukin-3. EMBO J. 1993;12:1641–1646. doi: 10.1002/j.1460-2075.1993.tb05809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanazono Y, Chiba S, Sasaki K, Mano H, Yazaki Y, Hirai H. Erythropoietin induces tyrosine phosphorylation and kinase activity of the c-fps/fes proto-oncogene product in human erythropoietin-responsive cells. Blood. 1993;81:3193–3196. [PubMed] [Google Scholar]
- 19.Huang C-C, Hammond C, Bishop J M. Nucleotide sequence of v-fps in the PRC II strain of avian sarcoma virus. J Virol. 1984;50:125–131. doi: 10.1128/jvi.50.1.125-131.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izuhara K, Feldman R A, Greer P, Harada N. Interaction of the c-fes proto-oncogene product with the interleukin-4 receptor. J Biol Chem. 1994;269:18623–18629. [PubMed] [Google Scholar]
- 21.Kamps M P, Buss J E, Sefton B M. Rous sarcoma virus transforming protein lacking myristic acid phosphorylates known polypeptide substrates without inducing transformation. Cell. 1986;45:105–112. doi: 10.1016/0092-8674(86)90542-8. [DOI] [PubMed] [Google Scholar]
- 22.Lee W-H, Bister K, Pawson A, Robins T, Moscovici C, Duesberg P H. Fujinami sarcoma virus: an avian RNA tumor virus with a unique transforming gene. Proc Natl Acad Sci USA. 1980;77:2018–2022. doi: 10.1073/pnas.77.4.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Smithgall T E. Co-expression with Bcr induces activation of the Fes tyrosine kinase and phosphorylation of specific N-terminal Bcr tyrosine residues. J Biol Chem. 1996;271:32930–32936. doi: 10.1074/jbc.271.51.32930. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Smithgall T E. Fibroblast transformation by Fps/Fes tyrosine kinases requires Ras, Rac and Cdc42 and induces extracellular signal-regulated and c-Jun N-terminal kinase activation. J Biol Chem. 1998;273:13828–13834. doi: 10.1074/jbc.273.22.13828. [DOI] [PubMed] [Google Scholar]
- 25.Lozzio B B, Lozzio C B, Bamberger E G, Feliu A S. A multipotential leukemia cell line (K-562) of human origin. Proc Soc Exp Biol Med. 1981;166:546–550. doi: 10.3181/00379727-166-41106. [DOI] [PubMed] [Google Scholar]
- 26.Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- 27.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 28.MacDonald I, Levy J, Pawson T. Expression of the mammalian c-fes protein in hematopoietic cells and identification of a distinct fes-related protein. Mol Cell Biol. 1985;5:2543–2551. doi: 10.1128/mcb.5.10.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maru Y, Peters K L, Afar D E H, Shibuya M, Witte O N, Smithgall T E. Tyrosine phosphorylation of BCR by FPS/FES protein-tyrosine kinases induces association of BCR with GRB-2/SOS. Mol Cell Biol. 1995;15:835–842. doi: 10.1128/mcb.15.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda T, Fukada T, Takahashi-Tezuka M, Okuyama Y, Fujitani Y, Hanazono Y, Hirai H, Hirano T. Activation of Fes tyrosine kinase by gp130, an interleukin-6 family cytokine signal transducer, and their association. J Biol Chem. 1995;270:11037–1109. doi: 10.1074/jbc.270.19.11037. [DOI] [PubMed] [Google Scholar]
- 31.McWhirter J R, Galasso D L, Wang J Y J. A coiled-coil oligomerization domain of bcr is essential for the transforming function of bcr-abl oncoproteins. Mol Cell Biol. 1993;13:7587–7595. doi: 10.1128/mcb.13.12.7587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee C-H, Kuriyan J, Miller W T. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 33.Muller A J, Young J C, Pendergast A M, Pondel M, Landau R N, Littman D R, Witte O N. BCR first exon sequences specifically activate the BCR/ABL tyrosine kinase oncogene of Philadelphia chromosome-positive human leukemias. Mol Cell Biol. 1991;11:1785–1792. doi: 10.1128/mcb.11.4.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park W-Y, Seo J-S. Leucine zipper-like domain regulates the autophosphorylation and the transforming activity of P130gag-fps. Biochem Biophys Res Commun. 1995;211:447–453. doi: 10.1006/bbrc.1995.1834. [DOI] [PubMed] [Google Scholar]
- 35.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read R D, Lionberger J M, Smithgall T E. Oligomerization of the Fes tyrosine kinase: evidence for a coiled-coil domain in the unique N-terminal region. J Biol Chem. 1997;272:18498–18503. doi: 10.1074/jbc.272.29.18498. [DOI] [PubMed] [Google Scholar]
- 37.Rogers J A, Read R D, Li J, Peters K L, Smithgall T E. Autophosphorylation of the Fes tyrosine kinase: evidence for an intermolecular mechanism involving two kinase domain tyrosine residues. J Biol Chem. 1996;271:17519–17525. doi: 10.1074/jbc.271.29.17519. [DOI] [PubMed] [Google Scholar]
- 38.Shibuya M, Hanafusa H. Nucleotide sequence of Fujinami sarcoma virus: evolutionary relationship of its transforming gene with transforming genes of other sarcoma viruses. Cell. 1982;30:787–795. doi: 10.1016/0092-8674(82)90283-5. [DOI] [PubMed] [Google Scholar]
- 39.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 40.Smithgall T E, Yu G, Glazer R I. Identification of the differentiation-associated p93 tyrosine protein kinase of HL-60 leukemia cells as the product of the human c-fes locus and its expression in myelomonocytic cells. J Biol Chem. 1988;263:15050–15055. [PubMed] [Google Scholar]
- 41.Van Etten R A. Cycling, stressed-out and nervous: cellular functions of c-Abl. Trends Cell Biol. 1999;9:179–186. doi: 10.1016/s0962-8924(99)01549-4. [DOI] [PubMed] [Google Scholar]
- 42.Weinmaster G, Zoller M J, Pawson T. A lysine in the ATP-binding site of P130gag-fps is essential for protein-tyrosine kinase activity. EMBO J. 1986;5:69–76. doi: 10.1002/j.1460-2075.1986.tb04179.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinmaster G, Zoller M J, Smith M, Hinze E, Pawson T. Mutagenesis of Fujinami sarcoma virus: evidence that tyrosine phosphorylation of p130gag-fps modulates its biological activity. Cell. 1984;37:559–568. doi: 10.1016/0092-8674(84)90386-6. [DOI] [PubMed] [Google Scholar]
- 44.Williams J C, Weijland A, Gonfloni S, Thompson A, Courtneidge S A, Superti-Furga G, Wierenga R K. The 2.35 Å crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions. J Mol Biol. 1997;274:757–775. doi: 10.1006/jmbi.1997.1426. [DOI] [PubMed] [Google Scholar]
- 45.Xu W, Harrison S C, Eck M J. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 46.Yu G, Smithgall T E, Glazer R I. K562 leukemia cells transfected with the human c-fes gene acquire the ability to undergo myeloid differentiation. J Biol Chem. 1989;264:10276–10281. [PubMed] [Google Scholar]