Abstract
The SARS-CoV-2 virus like many other viruses has transformed in a continual manner to give rise to new variants by means of mutations commonly through substitutions and indels. These mutations in some cases can give the virus a survival advantage making the mutants dangerous. In general, laboratory investigation must be carried to determine whether the new variants have any characteristics that can make them more lethal and contagious. Therefore, complex and time-consuming analyses are required in order to delve deeper into the exact impact of a particular mutation. The time required for these analyses makes it difficult to understand the variants of concern and thereby limiting the preventive action that can be taken against them spreading rapidly. In this analysis, we have deployed a statistical technique Shannon Entropy, to identify positions in the spike protein of SARS Cov-2 viral sequence which are most susceptible to mutations. Subsequently, we also use machine learning based clustering techniques to cluster known dangerous mutations based on similarities in properties. This work utilizes embeddings generated using language modeling, the ProtBERT model, to identify mutations of a similar nature and to pick out regions of interest based on proneness to change. Our entropy-based analysis successfully predicted the fifteen hotspot regions, among which we were able to validate ten known variants of interest, in six hotspot regions. As the situation of SARS-COV-2 virus rapidly evolves we believe that the remaining nine mutational hotspots may contain variants that can emerge in the future. We believe that this may be promising in helping the research community to devise therapeutics based on probable new mutation zones in the viral sequence and resemblance in properties of various mutations.
Keywords: SARS-CoV-2, Mutations, Clustering, Shannon entropy
Contributions of the work:
-
1.
The paper proposes a computational methodology to identify potential mutational hotspots in spike protein of SARS-CoV-2. The high throughput methodology can also identify some of the dangerous mutations emerging in the distant future
-
2.
Understand and identify the similarities and patterns among the different type of mutations using clustering analysis. Such an analysis may possibly help biologists to better understand the relationships between SARS-CoV-2 mutations.
1. Introduction
The SARS-CoV-2 virus has rapidly evolved by continually mutating, affecting more than 180 million people across the globe. Ever since the genome sequence of SARS-CoV-2 became available, mutations at several sites in the genome have been identified raising concerns regarding enhanced transmissibility of the virus [1]. The mutating nature of the virus has inspired global efforts from research community to actively track and understand the emergence of variants of concern[[2], [3], [4]]. One of the first mutation that rapidly spread throughout the world, mutation D614G, was first reported in April 2020 [5]. This mutation has now been classified under several lineages and is found to be a factor in increased transmission of the virus [[6], [7], [8], [9]]. The discovery of this mutation was followed by identification of a series of mutations in the virus belonging to the B. 1.1.7 lineage, which was first found in the Southeast of England [10]. The mutations, namely A222V, S477 N, N501, H69, N439K, Y453F,11S98F, D80Y, A626S, V1122L, have been noted as variants of interest in many studies [[10], [11], [12], [13]] and are the focus of this work as well. These variants were selected because they were marked as the Variant Under Investigation SARS-CoV-2 VUI 202012/01 (Variant Under Investigation, the year 2020, month 12, variant 01) by different studies done in the United Kingdom [14]. The mutation, A222V belongs to the B.1.177 lineage and has been noted to have a dominating presence in European countries[15,16]. N439K and Y453F have been found to have a higher binding affinity to the hACE2 receptor and are noted to reduce the neutralizing potential of antibodies specific to SARS-CoV-2 [[17], [18], [19]]. N439K often co-occurs with 69–70 deletion in the spike protein, the effect of this combined double mutation is being investigated by researchers (COVID-19 Genomics UK consortium, 2021; [20]. N501Y is the causative factor in the increased infectiousness of the disease [21]. The numerous effects of such mutations on the increased transmissibility and lethality of SARS-CoV-2, make it imperative to study these mutations and understand their effects[22].
To tackle the COVID -19 pandemic, efforts from the researchers have involved exploring traditional paradigm of in-vitro experimentation and data analysis-based methodologies like machine learning. Data driven modelling techniques, with their ability to analyze large amounts of data, build a functional mapping between the input parameters and output. This paper explores the use of data-driven methodologies to understand the mutations in the SARS-CoV-2 spike proteins. To understand and identify the mutation hotspots we have examined the sequence entropy and its correlation with experimentally identified variants of concern.
Tomaszewski et al., defined mutational entropy as a measure of molecular heterogeneity of the SARS-CoV-2 proteome which is estimated from the positional variance in these sequences [7]. In our work, we measure the positional variance in the sequence of the SARS-CoV-2 spike proteins by calculating Shannon Entropy. In case of proteins, Shannon entropy is shown to have a strong correlation with protein structural entropy [23], and can provide insights into the compositional stability of the proteins. The Shannon entropy is also directly proportional to the inverse packing density of proteins [24], and the packing density is further related to increased mutagenesis. Moreover, higher local flexibility regions have an increased value of entropy and are prone to mutations [21]. Our study explores these relationships of Shannon entropy to estimate the mutational hotspots in the SARS-CoV-2 spike protein. Higher value of entropy at a position in the sequence is indicative of increased randomness at that site whereas low value of entropy at a certain site is indicative of an increased stability and decreased randomness at the said location.
Apart from identifying the hotspots of interest, we also analyze the similarity of these mutations by employing a k-means clustering algorithm. To generate the embedding for the clustering algorithm we leverage the protein sequence data by using language modeling approaches. Through transfer learning, some of the highly successful models in the Natural Language Processing (NLP) domain have been applied to protein sequence to generate meaningful representations that can be used in tasks like structure prediction [25]. We used the Prot-BERT language modeling to represent these spike protein sequences in the form of semantic rich embeddings [26]. The Prot-BERT model has been trained on 80 billion amino acids, representing wide variety of protein sequences. The embeddings generated via the Prot-BERT model can be used for different downstream tasks. In our work, we use embeddings to determine the similarities between mutations using unsupervised machine learning techniques. This analysis will help in understanding the relationships between the mutations and assist the research community to tackle the virus.
1.1. Related work
Machine learning models have been used in many ways to study and understand the different aspects of COVID-19 pandemic. These models have been previously used for forecasting the COVID-19 cases [[27], [28], [29]], propose the potential antibodies [30], understand the possible evolutions of the virus [31], understand the economic and social effects of social distancing [32,33], understand the efficiency of lockdowns [34], study the transmission and spread of the virus [35,36]. Data driven models have also been used to analyze the SARS-CoV-2 mutations. In their paper [37], use techniques topological like persistent homology to understand the SARS-CoV-2 mutations and uncover some underlying patterns. In another study [38], develop the Informative Subtype Markers (ISM) to visualize and analyze the spread of different mutated SARS-CoV-2 sequences.
2. Methods
2.1. Data
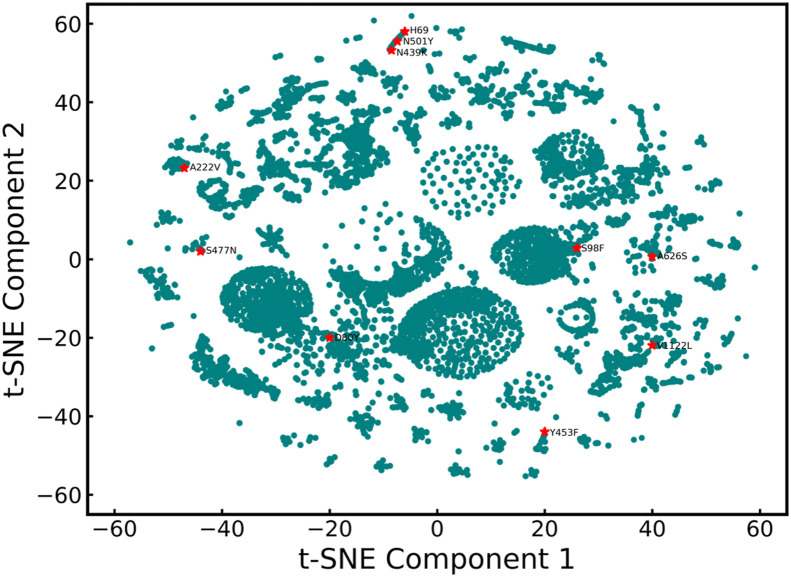
To understand the effect of the mutations we focus only on the spike protein of the virus sequence. We select the spike protein region because it is the major component of the SARS-CoV-2 virus that is responsible for eliciting host immune responses of neutralizing antibodies. It is the presence of this spike protein on the antigen that allows it to interact and penetrate the host cells. Therefore, more attention to spike protein has been given in the analysis of the mutations of the SARS-CoV-2 virus. To this end, we collect the spike protein data from the GISAID server to analyze the effect of the mutations on the spike protein on its transmissibility. We downloaded three hundred eleven thousand two hundred and fifty-six spike protein sequences from the GISAID server (https://www.gisaid.org/) on January 3, 2020 [11,39]. The comprehensive dataset had sequences related to the SARS-CoV-1 virus too, therefore the first stage of preprocessing involved the elimination of sequences that were not from 2020. This resulted in a dataset comprising three hundred ten thousand five hundred and ten sequences. Most of these sequences are comprised of 1273 amino acids, with maximum length being 1278 amino acids. To ensure uniformity in our calculation of the positional entropy, the ones with length less than 1278 were made up to length 1278 by appending the relevant number of ‘X's to the end of the gene sequence for the entropy analysis. The original spike protein sequence found in Wuhan is referenced from Zhao et al. [1] and the mutations in all the collected sequences in the data are analyzed with respect to this sequence. There was a large presence of repeated spike protein sequences found in different countries, so we decided to curate the data further and create data with only the unique sequences as featurizing the same sequence twice using Prot-BERT would have been redundant. We found fifty-three thousand eight hundred and ninety-eight belonging to prime variants of interest that are unique sequences of the spike protein. Subsequently, this dataset was used to generate embedding via the ProtBERT Model. These embeddings were further used to carry out unsupervised machine learning analysis. To understand the spread of the data and visualize it, we generated the plot using t-SNE [40] shown in Fig. 1 .
Fig. 1.
t-SNE plot capturing the distribution of the data collected from the GISAID server. Some of the variants of concern like N439K, N501Y are clustered near each other. From the t-SNE, we can easily infer that the SARS-CoV-2 mutations have unique characteristics.
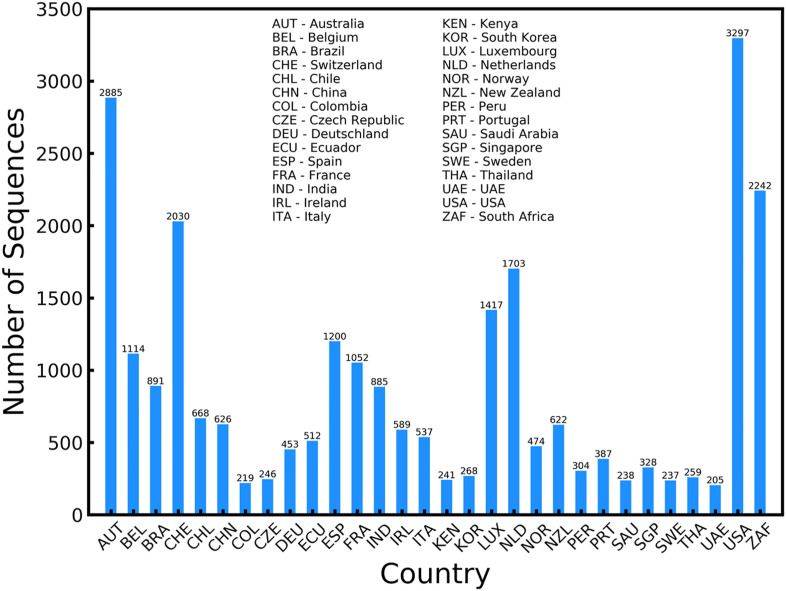
Further, we also analyze the geographical locations and the general distribution of the countries that were a part of the dataset we found that United Kingdom and Denmark contributed to over 50% of the mutation sequences in the dataset with 140458 mutation sequences from United Kingdom and 20346 from Denmark. These two countries have proactively studied the different mutations and made the data available for public use via the GISAID server. To analyze the mutation sequence data from other countries, a distribution of the dataset comprising of countries with more than 200 but less than 5000 mutation sequences is shown in Fig. 2 .
Fig. 2.
Plot showing the distribution of the sequences in the data. Apart from United Kingdom and Denmark, the other countries actively tracking the variants of concern include USA, Australia, South Africa, and Switzerland.
2.2. Positional entropy calculations
The positional entropy is a measure of the randomness at the given position in the sequence [41]. To calculate the positional entropy for our dataset we use Shannon Entropy formulation stated in Equation (1) [42]:
| (1) |
Where L is a list of all possible amino acids in all the sequences is the probability of finding the kth amino acid at that position.
We use equation (1) to find the positional entropy for all the positions in the SARS-CoV-2 spike protein sequence. Using the dataset obtained from the GISAID server, we first pre-process the data using Biopython[43] to extract the sequences from the FASTA file downloaded from the server. We found that the length of the spike protein sequence varied from 1270 to 1278, the distribution of the sequence lengths is shown in Fig. S1. We also observed that the positions that contain ambiguous sites or unidentified amino acid in the spike protein sequence have been denoted with character “X” in the dataset. These positions with character “X” are handled by a masking operation that calculates the entropy without considering them [38]. We proceed by calculating the positional entropy values using equation (1) and all the values for the positional entropy are stored in an array.
To identify the regions of high entropy that can possibly be associated with harmful mutations, we use a running mean (window length = 15, step size = 1), here the first positional index of the window gets assigned the value of the running mean. In the running mean calculation, we don't consider the first 60 and last 60 amino acids in the sequences because of the sequencing uncertainty. After calculating the running mean (window length = 15, step size = 1) for positional entropy, we stored it in another array. The array containing all the running means is then sorted and top 100 entropy values in the sequence are selected. Subsequently, we define the hotspots in the sequence as having ≥2 consecutive high entropy positions among the top-100 positional entropy values. For example: 210 and 211 both belong to the top 100 positional entropy values, and hence region 210–224 has been identified as a hotspot. To ensure both the positions (210 & 211) are included, we select the lowest index (210) as the start position of the hotspot and next 15 positions (included in the running mean) are considered as the hotspot (210–224). Additional details about the distribution of sequence lengths (Fig. S1) in the data and the starting positions of running mean windows for the top 100 positional entropy values are provided in the supplementary information (Table S1).
2.3. Prot-BERT model
The Prot-BERT trained on the UniRef100 dataset was used to generate sequence embeddings [26]. The Prot-BERT model has 30 layers, 16 attention heads, and embedding hidden size 1024. The Prot-BERT model was chosen because the embeddings generated have been used for different downstream tasks successfully increasing our confidence in using the same. We generate the embedding for the spike proteins of the mutated sequences using the pre-trained model on the hugging face api [44]. The Hugging face interface allows the users to easily use the pre-trained models on various Natural Language Processing (NLP) tasks. The curated data containing the unique sequences of spike protein were entered in the pre-trained Prot-BERT model and an embedding of size 1024 for every sequence. These embeddings are then used to study similarities and understand distributions between the mutations via K-Means clustering.
2.4. K means
Clustering is an unsupervised learning technique used to group a collection of unlabeled data sharing similarities. Each cluster comprises data sharing common traits which are distinct from members of other clusters, thereby resulting in clusters with high internal homogeneity and high external heterogeneity [45]. Clustering can be broadly classified into two categories, hierarchical and non-hierarchical clustering.
The k-means clustering technique used in this study is a non-hierarchical clustering approach. This technique involves defining the number of clusters ‘k’. Each cluster is represented by a central location defined as the centroid, where k is the cluster number and j are the number of attributes. The algorithm allocates each data point to the nearest cluster by minimizing the distance from centroid. It starts off by randomly assigning centroids and thereafter continues as an iterative process to optimize the centroid locations depending on the points assigned to that cluster. This process continues until there is no further change in the centroid values or until the maximum number of iterations is reached [46].
Clustering is one of the most important data mining techniques to group unlabeled data based on common traits. In this work, we used K means clustering to group the different mutations based on similarities in properties. The embeddings generated using the ProtBert model were used as features for the clustering model.
To perform k-means clustering we use the scikit-learn library, that builds k-means model under the hood after entering the model parameters [47,48]. The number of clusters chosen for this task was 10, based on the number of different mutation types being 10 and also because we got the highest silhouette score of 0.7228 [49] when using 10 clusters. We also implemented the MST-kNN clustering technique but the algorithm did not perform very well, it had a very low silhouette score of −0.7638 and hence was not used for any further clustering analysis. We use the silhouette scores metric as it is a measure of how well an algorithm can differentiate between different clusters in the data. The score varies from −1 to +1 and high silhouette score indicates that the datapoints have been clustered appropriately, with similar datapoints clustered together and dissimilar datapoints clustered differently. Other parameters for k-means such as the maximum number of iterations was chosen to be 1000 and the total number of initializations was chosen as 50 after multiple trials with other values in order to stabilize the cluster formation.
3. Results
3.1. Positional entropy
The advantage of analyzing the entropy lies in the fact that sequential entropy is correlated to molecular motility is an important factor for the mutation [7,23,24]. Furthermore, studies have found a significant relationship between these high entropy hotspot regions of the viral sequence and enhanced virulence in the mutations associated with these regions, which have had a crucial role in the evolution of this disease. Hence, these sites are regions of interest in vaccine development and medicine formulation[38]. We calculated the positional entropy for all positions of the spike protein genomic sequence and have estimated the mutational hotspot regions in these viral sequences. Table 1 highlights some of these regions of interest we have identified which correspond to some of the most dominant mutations that have been noted in various countries. From this analysis, we have noted that the regions of interest have successfully captured the D614G mutation, which is one of the most dominant mutation and is found to enhance the replication of SARS-CoV-2 in the lung cells [50]. The regions of interest also captured the following mutations - A222V, N439K, Y453F, S477 N, N501, D614G and V1122L [12].
Table 1.
Hotspots found by analyzing the positional entropy. To determine a hotspot region a running mean (window length = 15, step size = 1) is calculated and top 100 value are selected. We found six such regions of interest in our analysis in which ten mutations of interest emerged.
| Hotspots | Mutation |
|---|---|
| 211–225 | A222V |
| 439–453 | N439K, L452R, Y453F |
| 473–487 | S477 N, T478K, E484K |
| 487–501 | N501Y |
| 602–616 | D614G |
| 1121–1135 | V1122L |
Apart from the above mutations, the following other mutations have also been correctly identified in our hotspots - E484K, T478K, and L452R. It has been shown that for the mutation, E484K along with the some mutations from B.1.1.7 lineage requires increased amounts of antibody serum to prevent infection [51] making it especially dangerous. Interestingly, our methodology is capable of capturing some of the potentially harmful mutations that may emerge in the future. For example: Our model that uses sequence data before 2020 identifies one of the hotspot regions from 439 to 453. A mutation of significance, L452R which was first identified by the California Dept of Public Health on 17th Jan 2021 [52] and was later found to be dominant mutation in the months of April and May 2021 worldwide. Similarly, another mutation E484K belonging to the B.1.25 family was recognized as variant of concern was recognized in South Africa in April 2021 [53]. This mutation lies in the region 473–487 which includes another mutation of significance S477 N [16,54]. This emergence of variants of concern from hotspot regions identified by our methodology demonstrates the accurate prediction of Shannon entropy based analysis.
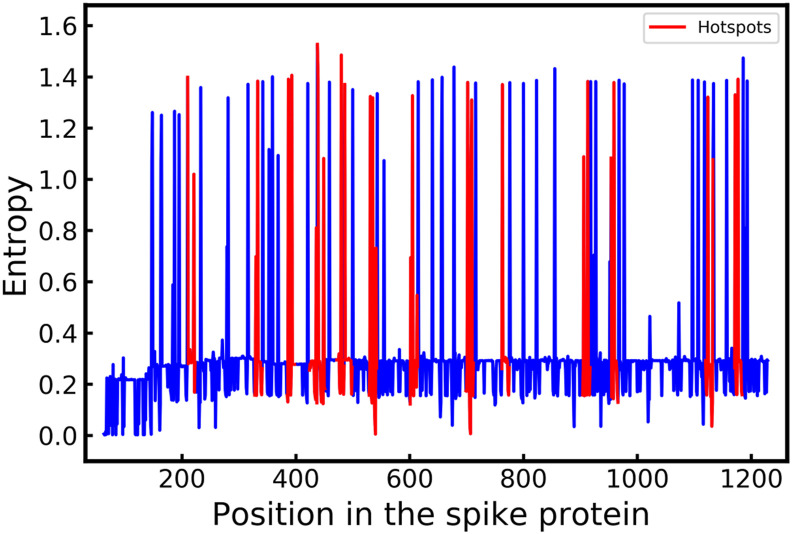
To further illustrate the positional entropy hotspots, we have plotted the positional entropy for the entire sequence of the spike protein of SARS-CoV-2 in Fig. 3 . Based on our analysis, we found nine other hotspot regions including 329–343, 386–400, 425–439, 530–544, 700–714, 763–777, 905–919, 955–968, 1172–1186. Based on validation analysis presented in Table 1 it is likely that the new mutation of concern may emerge in these hotspot regions.
Fig. 3.
Variation of entropy and the position in the spike protein. Hotspots with higher likelihood of mutagenesis and high entropy have been marked in red in the plot. The red regions(hotspots) have the maximum mean entropy over a window of length 15. The blue regions in the plot indicate the regions of relatively lower mean entropy over the window of length 15. According to positional entropy analysis the dangerous spike protein mutations are more likely to emerge from the hotspots (red regions).
To structurally understand the mutations further, we also identified the regions where the dangerous mutations belong in the structure of the spike protein. The analysis was based on study done by Huang et al., where they identify the different regions in the spike protein based on the positions in the sequence [55]. It must be noted that there are seven possible dangerous mutations in the receptor binding domain of the spike protein, these mutations are possibly more lethal because of their location on the binding interface. The locations of these mutations on the spike protein have been presented in Table 2 .
Table 2.
Location of the mutations in the spike protein of the SARS-CoV-2, we have 3 regions of the spike protein where mutations can be located.
| Spike Protein Region | Mutation |
|---|---|
| N – Terminal domain | A222V |
| Receptor-Binding Domain | N439K, L452R, Y453F, S477 N, T478K, E484K, N501Y |
| Heptapeptide repeat sequence | V1122L |
We also validate the mutations in Table 1 by using– EV mutation[56] methodology that determines the favorability of a mutation by calculating the prediction epistatic score. The data for mutation effect using EV mutation for SARS-CoV-2 is available on the server created by Ref. [57], we used the data from this server to analyze the epistatic mutation effect predict for mutations presented in Table 1. The novel aspect of the EV mutation method is its ability to take into account epistasis by taking into consideration the interactions between all pairs of amino acids residues in the neighborhood to quantify the mutational effects. A higher value of the prediction score using EV mutation indicates a highly favorable mutation. The analysis using EV mutation has been presented in Table 3 .
Table 3.
Analysis of the SARS-CoV-2 mutations using EV mutation, the prediction epistatic score is an indicator of whether a mutation is fit or not fit. The higher score indicates that the mutation indicates that the mutation is a better fit. The third column indicates the rank among all the possible mutations at the site. The possible values for rank range from 1 to 19 as there are 20 amino acids and a single amino acids can mutate into 19 other amino acids. The rank depends on the EV mutation score, highest score will get rank-1 that indicates the mutation is highly favorable and lowest score gets rank-19 indicates that mutation is not favorable according to EV mutation calculations.
| Mutation | Prediction epistatic score | Rank among all mutation possibilities |
|---|---|---|
| A222V | 0.5465 | 1 |
| N439K | −3.8605 | 10 |
| L452R | −6.1483 | 15 |
| Y453F | −6.5665 | 7 |
| T478K | 0.4154 | 1 |
| D614G | −4.7144 | 2 |
| V1122L | −6.9294 | 9 |
Among the ten different mutations in Table 1, Table 3 presents the EV mutation score for seven different mutations. The data for S477 N, E484K and N501Y is unavailable on the server (Nathan Rollins*, Kelly Brock*, Joshua Rollins* et al., 2020), and hence is not presented in Table 3. We observe that A222V and T478K are highly favorable mutations as they have the highest possible prediction epistatic score among all mutations for the wild-type residue (A for site 222 and T for site 478). The D614G mutations is also highly favorable, and mutations Y453F, V1122L and N439K may be considered as moderately favorable. On the other hand, the mutation L452R may not be as favorable based on prediction epistatic score. The EV mutation scores validate most mutations identified in the hotspots from our methodology in Table 1, further indicating the calculating the positional entropy of the sequence can be a useful metric for identifying future mutation hotspots.
The positional entropy formulation developed in this work used the data from the year 2020 and yet was able to identify some of the mutations that emerge later in April and May 2021 such as E484K and L452R validating our methodology further. We believe that our method may potentially be used to identify the dangerous mutations in advance and aid in the fight against the pandemic.
3.2. Clustering with K-means
The clustering analysis was done on the embeddings generated from the Prot BERT model. The embeddings for all the sequences are a 2D array of shape (sequence length, 1024) where 1024 is the hidden dimension of the model. Subsequently, we applied mean pooling to the sequence length dimension of the embeddings and generate a vector of dimension 1024 for each sequence. This 1024-dimensional vector is used for k-means clustering analysis.
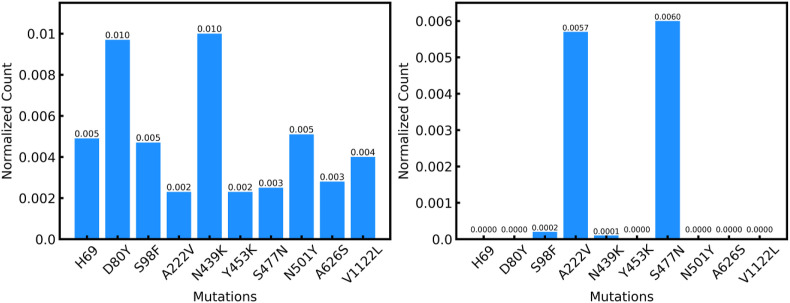
The cluster centers resulting from k-means clustering correspond to the different mutation types, thereby verifying our assumption that the different cluster types get grouped separately. We find that 7 out of 10 different mutations are identified as cluster centers with a few repeats. On analyzing the spike protein sequences that form the clusters and the sequence representative of the cluster center, we find that in most cases most of the sequences are identified to be of the same type as the cluster center whereas in most other cases the mutation type of the cluster center is amongst the top 3 mutation types present in the cluster, the other two types of possibly similar characteristics (Table 4 ). For example, from the plots (Fig. 4 ) show the clusters of S477 N and N439K have a majority of S477 N and N439K components. Furthermore, A222V has the second highest count in the cluster representing S477 N (Fig. 4) indicating similarities between them. D80Y is one of the majorities in the N439K cluster, thereby implying similarity in characteristics. In a study done by Ref. [58], it was found that A222V and S477 N are both stabilizing mutations thereby validating our findings that these two mutations may have some similar characteristics. This similarity analysis between the mutations is significant because when designing therapeutics that can counter new mutations understanding characteristics of mutations computationally can save a lot of experimental time and accelerate the therapeutic development process.
Table 4.
Clusters where the top 3 dominant mutations in the cluster concur with the cluster center mutation. The top-3 dominant mutations are most likely to be similar in characteristics to the mutation in cluster.
| Cluster Centers | Dominant Mutations in the Cluster |
|---|---|
| S477 N | S477N, A222V, S98F, N439K |
| N439K | N439K, D80Y, N501Y, H69-70 |
| N501Y | D80Y, N439K, N501Y, H69 |
| A222V | V1122L, A222V, N501Y, S477 N |
Fig. 4.
A.) Clustering analysis for N439K mutation on the spike protein of SARS-CoV-2. After analyzing the cluster with cluster center as N439K we can conclude that the D80Y may have similar characteristics to that of N439K. B.) Clustering analysis for S477 N mutation on the spike protein of SARS-CoV-2. The majority of sequences in this cluster belong to the mutation S477 N and the next highest number is that of A222V suggesting similarity between them.
4. Conclusion
In this study, we developed a methodology to determine the hotspots for mutations in spike protein sequences of SARS-CoV-2. This study can enable us to know variants of interests beforehand so that therapeutics can be developed for them. We found fifteeen regions of interest in the sequence of the spike protein that may be the potential hotspots for novel mutations in SARS-CoV-2. Six of these hotspots contain ten mutations which have already been flagged as possibly more transmissible by the previous research. Interestingly, some of the new emerging variants from India and South Africa which have been marked dangerous in April 2021 and May 2021 were identified by our methodology even though we use the sequence data on the GISAID server before December 2020. Identifying hotspots beforehand may have implications in the development of therapeutics and be aware of the potential threats posed by the mutations in the virus. We also use the unsupervised learning-based clustering technique k-means to find the similarities between the variants of interests that have previously been found to be dangerous. The encode the protein sequences we use the Prot-BERT model and use features generated by it, for the k-means analysis. Clustering the mutation variants based on similarity reduces redundancy of time and resources, similar treatment techniques can be implemented for mutations that fall into the same cluster. One of the results of our analysis was the similarity between the S477 N and the A222V mutations, it implies that these mutations share common traits and occurrences and may be subjected to similar treatment strategies.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Prakarsh Yadav and Parisa Mollaei for their useful inputs and comments on the paper. This work is supported by the start-up fund provided by CMU Mechanical Engineering and support from Center for Machine Learning and Health (CMLH).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104915.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alam I., Radovanovic A., Incitti R., Kamau A.A., Alarawi M., Azhar E.I., Gojobori T. CovMT: an interactive SARS-CoV-2 mutation tracker, with a focus on critical variants. Lancet Infect. Dis. 2021;21:602. doi: 10.1016/S1473-3099(21)00078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen A.T., Altschuler K., Zhan S.H., Chan Y.A., Deverman B.E. COVID-19 CG enables SARS-CoV-2 mutation and lineage tracking by locations and dates of interest. eLife. 2021;10 doi: 10.7554/eLife.63409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xing Y., Li X., Gao X., Dong Q. MicroGMT: a mutation tracker for SARS-CoV-2 and other microbial genome sequences. Front. Microbiol. 2020;11:1502. doi: 10.3389/fmicb.2020.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C., Angyal A., Brown R.L., Carrilero L., Green L.R., Groves D.C., Johnson K.J., Keeley A.J., Lindsey B.B., Parsons P.J., Raza M., Rowland-Jones S., Smith N., Tucker R.M., Wang D., Wyles M.D. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827. doi: 10.1016/j.cell.2020.06.043. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laha S., Chakraborty J., Das S., Manna S.K., Biswas S., Chatterjee R. Characterizations of SARS-CoV-2 mutational profile, spike protein stability and viral transmission. Infect. Genet. Evol. 2020;85:104445. doi: 10.1016/j.meegid.2020.104445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomaszewski T., DeVries R.S., Dong M., Bhatia G., Norsworthy M.D., Zheng X., Caetano-Anollés G. New pathways of mutational change in SARS-CoV-2 proteomes involve regions of intrinsic disorder important for virus replication and release. Evol. Bioinf. Online. 2020;16 doi: 10.1177/1176934320965149. 117693432096514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Volz E., Hill V., McCrone John T., Price A., Jorgensen D., O'Toole Á., Southgate J., Johnson Robert, Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R., Koshy C., Wise E., Cortes Nick, Lynch J., Kidd S., Mori M., Fairley D.J., Curran T., McKenna J.P., Adams H., Fraser C., Golubchik T., Bonsall D., Moore Catrin, Caddy S.L., Khokhar F.A., Wantoch M., Reynolds N., Warne B., Maksimovic J., Spellman K., McCluggage K., John M., Beer R., Afifi S., Morgan S., Marchbank A., Price A., Kitchen C., Gulliver H., Merrick I., Southgate J., Guest M., Munn R., Workman T., Connor T.R., Fuller W., Bresner C., Snell L.B., Charalampous T., Nebbia G., Batra R., Edgeworth J., Robson S.C., Beckett A., Loveson K.F., Aanensen D.M., Underwood A.P., Yeats C.A., Abudahab K., Taylor B.E.W., Menegazzo M., Clark G., Smith W., Khakh M., Fleming V.M., Lister M.M., Howson-Wells H.C., Berry Louise, Boswell T., Joseph A., Willingham I., Bird P., Helmer T., Fallon K., Holmes C., Tang J., Raviprakash V., Campbell S., Sheriff N., Loose M.W., Holmes N., Moore Christopher, Carlile M., Wright V., Sang F., Debebe J., Coll F., Signell A.W., Betancor G., Wilson H.D., Feltwell T., Houldcroft C.J., Eldirdiri S., Kenyon A., Davis T., Pybus O., du Plessis L., Zarebski A., Raghwani J., Kraemer M., Francois S., Attwood S., Vasylyeva T., Torok M.E., Hamilton W.L., Goodfellow I.G., Hall G., Jahun A.S., Chaudhry Y., Hosmillo M., Pinckert M.L., Georgana I., Yakovleva A., Meredith L.W., Moses S., Lowe H., Ryan F., Fisher C.L., Awan A.R., Boyes J., Breuer J., Harris K.A., Brown J.R., Shah D., Atkinson L., Lee J.C.D., Alcolea-Medina A., Moore N., Cortes Nicholas, Williams R., Chapman M.R., Levett L.J., Heaney J., Smith D.L., Bashton M., Young G.R., Allan J., Loh J., Randell P.A., Cox A., Madona P., Holmes A., Bolt F., Price J., Mookerjee S., Rowan A., Taylor G.P., Ragonnet-Cronin M., Nascimento F.F., Jorgensen D., Siveroni I., Johnson Rob, Boyd O., Geidelberg L., Volz E.M., Brunker K., Smollett K.L., Loman N.J., Quick J., McMurray C., Stockton J., Nicholls S., Rowe W., Poplawski R., Martinez-Nunez R.T., Mason J., Robinson T.I., O'Toole E., Watts J., Breen C., Cowell A., Ludden C., Sluga G., Machin N.W., Ahmad S.S.Y., George R.P., Halstead F., Sivaprakasam V., Thomson E.C., Shepherd J.G., Asamaphan P., Niebel M.O., Li K.K., Shah R.N., Jesudason N.G., Parr Y.A., Tong L., Broos A., Mair D., Nichols J., Carmichael S.N., Nomikou K., Aranday-Cortes E., Johnson N., Starinskij I., da Silva Filipe A., Robertson D.L., Orton R.J., Hughes J., Vattipally S., Singer J.B., Hale A.D., Macfarlane-Smith L.R., Harper K.L., Taha Y., Payne B.A.I., Burton-Fanning S., Waugh S., Collins J., Eltringham G., Templeton K.E., McHugh M.P., Dewar R., Wastenge E., Dervisevic S., Stanley R., Prakash R., Stuart C., Elumogo N., Sethi D.K., Meader E.J., Coupland L.J., Potter W., Graham C., Barton E., Padgett D., Scott G., Swindells E., Greenaway J., Nelson A., Yew W.C., Resende Silva P.C., Andersson M., Shaw R., Peto T., Justice A., Eyre D., Crooke D., Hoosdally S., Sloan T.J., Duckworth N., Walsh S., Chauhan A.J., Glaysher S., Bicknell K., Wyllie S., Butcher E., Elliott S., Lloyd A., Impey R., Levene N., Monaghan L., Bradley D.T., Allara E., Pearson C., Muir P., Vipond I.B., Hopes R., Pymont H.M., Hutchings S., Curran M.D., Parmar S., Lackenby A., Mbisa T., Platt S., Miah S., Bibby D., Manso C., Hubb J., Chand M., Dabrera G., Ramsay M., Bradshaw D., Thornton A., Myers R., Schaefer U., Groves N., Gallagher E., Lee D., Williams D., Ellaby N., Harrison I., Hartman H., Manesis N., Patel V., Bishop C., Chalker V., Osman H., Bosworth A., Robinson E., Holden M.T.G., Shaaban S., Birchley A., Adams A., Davies A., Gaskin A., Plimmer A., Gatica-Wilcox B., McKerr C., Moore Catherine, Williams C., Heyburn D., De Lacy E., Hilvers E., Downing F., Shankar G., Jones H., Asad H., Coombes J., Watkins J., Evans J.M., Fina L., Gifford L., Gilbert L., Graham L., Perry M., Morgan M., Bull M., Cronin M., Pacchiarini N., Craine N., Jones R., Howe R., Corden S., Rey S., Kumziene-Summerhayes S., Taylor S., Cottrell S., Jones S., Edwards S., O'Grady J., Page A.J., Wain J., Webber M.A., Mather A.E., Baker D.J., Rudder S., Yasir M., Thomson N.M., Aydin A., Tedim A.P., Kay G.L., Trotter A.J., Gilroy R.A.J., Alikhan N.-F., de Oliveira Martins L., Le-Viet T., Meadows L., Kolyva A., Diaz M., Bell A., Gutierrez A.V., Charles I.G., Adriaenssens E.M., Kingsley R.A., Casey A., Simpson D.A., Molnar Z., Thompson T., Acheson E., Masoli J.A.H., Knight B.A., Hattersley A., Ellard S., Auckland C., Mahungu T.W., Irish-Tavares D., Haque T., Bourgeois Y., Scarlett G.P., Partridge D.G., Raza M., Evans C., Johnson K., Liggett S., Baker P., Essex S., Lyons R.A., Caller L.G., Castellano S., Williams R.J., Kristiansen M., Roy S., Williams C.A., Dyal P.L., Tutill H.J., Panchbhaya Y.N., Forrest L.M., Niola P., Findlay J., Brooks T.T., Gavriil A., Mestek-Boukhibar L., Weeks S., Pandey S., Berry Lisa, Jones K., Richter A., Beggs A., Smith C.P., Bucca G., Hesketh A.R., Harrison E.M., Peacock S.J., Palmer Sophie, Churcher C.M., Bellis K.L., Girgis S.T., Naydenova P., Blane B., Sridhar S., Ruis C., Forrest S., Cormie C., Gill H.K., Dias J., Higginson E.E., Maes M., Young J., Kermack L.M., Hadjirin N.F., Aggarwal D., Griffith L., Swingler T., Davidson R.K., Rambaut A., Williams T., Balcazar C.E., Gallagher M.D., O'Toole Á., Rooke S., Jackson B., Colquhoun R., Ashworth J., Hill V., McCrone J.T., Scher E., Yu X., Williamson K.A., Stanton T.D., Michell S.L., Bewshea C.M., Temperton B., Michelsen M.L., Warwick-Dugdale J., Manley R., Farbos A., Harrison J.W., Sambles C.M., Studholme D.J., Jeffries A.R., Darby A.C., Hiscox J.A., Paterson S., Iturriza-Gomara M., Jackson K.A., Lucaci A.O., Vamos E.E., Hughes M., Rainbow L., Eccles R., Nelson C., Whitehead M., Turtle L., Haldenby S.T., Gregory R., Gemmell M., Kwiatkowski D., de Silva T.I., Smith N., Angyal A., Lindsey B.B., Groves D.C., Green L.R., Wang D., Freeman T.M., Parker M.D., Keeley A.J., Parsons P.J., Tucker R.M., Brown R., Wyles M., Constantinidou C., Unnikrishnan M., Ott S., Cheng J.K.J., Bridgewater H.E., Frost L.R., Taylor-Joyce G., Stark R., Baxter L., Alam M.T., Brown P.E., McClure P.C., Chappell J.G., Tsoleridis T., Ball J., Gramatopoulos D., Buck D., Todd J.A., Green A., Trebes A., MacIntyre-Cockett G., de Cesare M., Langford C., Alderton A., Amato R., Goncalves S., Jackson D.K., Johnston I., Sillitoe J., Palmer Steve, Lawniczak M., Berriman M., Danesh J., Livett R., Shirley L., Farr B., Quail M., Thurston S., Park N., Betteridge E., Weldon D., Goodwin S., Nelson R., Beaver C., Letchford L., Jackson D.A., Foulser L., McMinn L., Prestwood L., Kay S., Kane L., Dorman M.J., Martincorena I., Puethe C., Keatley J.-P., Tonkin-Hill G., Smith C., Jamrozy D., Beale M.A., Patel M., Ariani C., Spencer-Chapman M., Drury E., Lo S., Rajatileka S., Scott C., James K., Buddenborg S.K., Berger D.J., Patel G., Garcia-Casado M.V., Dibling T., McGuigan S., Rogers H.A., Hunter A.D., Souster E., Neaverson A.S. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184:64–75. doi: 10.1016/j.cell.2020.11.020. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity (preprint) Microbiology. 2020 doi: 10.1101/2020.06.12.148726. [DOI] [Google Scholar]
- 10.Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O'Toole Á., Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V., Boyd O., Loman N.J., McCrone J.T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D.K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D.P., The Covid-19 Genomics UK (COG-UK) consortium. Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N.M. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data (preprint) Infectious Diseases (except HIV/AIDS) 2021 doi: 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- 11.Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID's innovative contribution to global health: data, Disease and Diplomacy. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayashi T., Yaegashi N., Konishi I. Effect of RBD mutations in spike glycoprotein of SARS-CoV-2 on neutralizing IgG affinity (preprint). Infectious Diseases (except HIV/AIDS) 2021. [DOI] [PubMed]
- 14.Galloway SE, Paul P, MacCannell DR, et al., n.d. Emergence of SARS-CoV-2 B.1.1.7 Lineage — United States, December 29, 2020–January 12, 2021, MMWR Morb Mortal Wkly Rep 2021. [DOI] [PMC free article] [PubMed]
- 15.Covid-19 Genomics UK consortium . 2021. COG-UK Report on SARS-CoV-2 Spike Mutations of Interest in the UK 15th January 2021. [Google Scholar]
- 16.Hodcroft E.B., Zuber M., Nadeau S., Vaughan T.G., Crawford K.H.D., Althaus C.L., Reichmuth M.L., Bowen J.E., Walls A.C., Corti D., Bloom J.D., Veesler D., Mateo D., Hernando A., Comas I., González Candelas F., SeqCOVID-SPAIN consortium, Stadler T., Neher R.A. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020 (preprint) Epidemiology. 2020 doi: 10.1101/2020.10.25.20219063. [DOI] [PubMed] [Google Scholar]
- 17.Bayarri-Olmos R., Rosbjerg A., Johnsen L.B., Helgstrand C., Bak-Thomsen T., Garred P., Skjoedt M.-O. The SARS-CoV-2 Y453F mink variant displays a pronounced increase in ACE-2 affinity but does not challenge antibody neutralization. J. Biol. Chem. 2021;296:100536. doi: 10.1016/j.jbc.2021.100536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., Lytras S., Czudnochowski N., Shah R., Meury M., Jesudason N., De Marco A., Li K., Bassi J., O'Toole A., Pinto D., Colquhoun R.M., Culap K., Jackson B., Zatta F., Rambaut A., Jaconi S., Sreenu V.B., Nix J., Zhang I., Jarrett R.F., Glass W.G., Beltramello M., Nomikou K., Pizzuto M., Tong L., Cameroni E., Croll T.I., Johnson N., Di Iulio J., Wickenhagen A., Ceschi A., Harbison A.M., Mair D., Ferrari P., Smollett K., Sallusto F., Carmichael S., Garzoni C., Nichols J., Galli M., Hughes J., Riva A., Ho A., Schiuma M., Semple M.G., Openshaw P.J.M., Fadda E., Baillie J.K., Chodera J.D., Rihn S.J., Lycett S.J., Virgin H.W., Telenti A., Corti D., Robertson D.L., Snell G. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187. doi: 10.1016/j.cell.2021.01.037. e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meng B., Kemp S.A., Papa G., Datir R., Ferreira I.A.T.M., Marelli S., Harvey W.T., Lytras S., Mohamed A., Gallo G., Thakur N., Collier D.A., Mlcochova P., Duncan L.M., Carabelli A.M., Kenyon J.C., Lever A.M., De Marco A., Saliba C., Culap K., Cameroni E., Matheson N.J., Piccoli L., Corti D., James L.C., Robertson D.L., Bailey D., Gupta R.K. Recurrent emergence of SARS-CoV-2 spike deletion H69/V70 and its role in the variant of concern lineage B.1.1.7. Cell Rep. 2021:109292. doi: 10.1016/j.celrep.2021.109292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M.-A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Alcantara L.C.J., Pond S.L.K., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa (preprint) Epidemiology. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 22.Callaway E. The coronavirus is mutating — does it matter? Nature. 2020;585:174–177. doi: 10.1038/d41586-020-02544-6. [DOI] [PubMed] [Google Scholar]
- 23.Koehl P., Levitt M. Sequence variations within protein families are linearly related to structural variations. J. Mol. Biol. 2002;323:551–562. doi: 10.1016/S0022-2836(02)00971-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao H., Yeh W., Chiang D., Jernigan R.L., Lustig B. Protein sequence entropy is closely related to packing density and hydrophobicity. Protein Eng. Des. Sel. 2005;18:59–64. doi: 10.1093/protein/gzi009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao R., Bhattacharya N., Thomas N., Duan Y., Chen X., Canny J., Abbeel P., Song Y.S. 2019. Evaluating Protein Transfer Learning with TAPE. [PMC free article] [PubMed] [Google Scholar]
- 26.Elnaggar A., Heinzinger M., Dallago C., Rehawi G., Wang Y., Jones L., Gibbs T., Feher T., Angerer C., Steinegger M., Bhowmik D., Rost B. ProtTrans: towards cracking the language of life's code through self-supervised learning (preprint) Bioinformatics. 2020 doi: 10.1101/2020.07.12.199554. [DOI] [PubMed] [Google Scholar]
- 27.ArunKumar K.E., Kalaga D.V., Kumar C.M.S., Kawaji M., Brenza T.M. Forecasting of COVID-19 using deep layer recurrent neural networks (RNNs) with gated recurrent units (GRUs) and long short-term memory (LSTM) cells. Chaos, Solit. Fractals. 2021;146:110861. doi: 10.1016/j.chaos.2021.110861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chimmula V.K.R., Zhang L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos, Solit. Fractals. 2020;135:109864. doi: 10.1016/j.chaos.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarkar K., Khajanchi S., Nieto J.J. Modeling and forecasting the COVID-19 pandemic in India. Chaos, Solit. Fractals. 2020;139:110049. doi: 10.1016/j.chaos.2020.110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magar R., Yadav P., Barati Farimani A. Potential neutralizing antibodies discovered for novel corona virus using machine learning. Sci. Rep. 2021;11:5261. doi: 10.1038/s41598-021-84637-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y., Yadav P., Magar R., Farimani A.B. bioRxiv; 2020. Bio-informed Protein Sequence Generation for Multi-Class Virus Mutation Prediction. [DOI] [Google Scholar]
- 32.Memon Z., Qureshi S., Memon B.R. Assessing the role of quarantine and isolation as control strategies for COVID-19 outbreak: a case study. Chaos, Solit. Fractals. 2021;144:110655. doi: 10.1016/j.chaos.2021.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva P.C.L., Batista P.V.C., Lima H.S., Alves M.A., Guimarães F.G., Silva R.C.P. COVID-ABS: an agent-based model of COVID-19 epidemic to simulate health and economic effects of social distancing interventions. Chaos, Solit. Fractals. 2020;139:110088. doi: 10.1016/j.chaos.2020.110088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharov K.S. Creating and applying SIR modified compartmental model for calculation of COVID-19 lockdown efficiency. Chaos, Solit. Fractals. 2020;141:110295. doi: 10.1016/j.chaos.2020.110295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cooper I., Mondal A., Antonopoulos C.G. A SIR model assumption for the spread of COVID-19 in different communities. Chaos, Solit. Fractals. 2020;139:110057. doi: 10.1016/j.chaos.2020.110057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ndaïrou F., Area I., Nieto J.J., Torres D.F.M. Mathematical modeling of COVID-19 transmission dynamics with a case study of Wuhan. Chaos, Solit. Fractals. 2020;135:109846. doi: 10.1016/j.chaos.2020.109846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang R., Chen J., Gao K., Hozumi Y., Yin C., Wei G.-W. Analysis of SARS-CoV-2 mutations in the United States suggests presence of four substrains and novel variants. Commun. Biol. 2021;4:1–14. doi: 10.1038/s42003-021-01754-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao Z., Sokhansanj B.A., Malhotra C., Zheng K., Rosen G.L. Genetic grouping of SARS-CoV-2 coronavirus sequences using informative subtype markers for pandemic spread visualization. PLoS Comput. Biol. 2020;16 doi: 10.1371/journal.pcbi.1008269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 2017;22 doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laurens van der Maaten, Hinton Geoffrey. Visualizing Data using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- 41.Crooks G.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 43.Cock P.J.A., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M.J.L. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolf T., Debut L., Sanh V., Chaumond J., Delangue C., Moi A., Cistac P., Rault T., Louf R., Funtowicz M., Davison J., Shleifer S., von Platen P., Ma C., Jernite Y., Plu J., Xu C., Scao T.L., Gugger S., Drame M., Lhoest Q., Rush A.M. 2020. HuggingFace's Transformers: State-Of-The-Art Natural Language Processing. ArXiv191003771 Cs. [Google Scholar]
- 45.Bustamam A., Tasman H., Yuniarti N., Frisca, Mursidah I. Presented at the INTERNATIONAL SYMPOSIUM on CURRENT PROGRESS IN MATHEMATICS and SCIENCES 2016 (ISCPMS 2016): Proceedings of the 2nd International Symposium on Current Progress in Mathematics and Sciences 2016, Depok, Jawa Barat, Indonesia. 2017. Application of k-means clustering algorithm in grouping the DNA sequences of hepatitis B virus (HBV) [DOI] [Google Scholar]
- 46.Mannor S., Jin X., Han J., Jin X., Han J., Jin X., Han J., Zhang X. In: Encyclopedia of Machine Learning. Sammut C., Webb G.I., editors. Springer US; Boston, MA: 2011. K-means clustering; pp. 563–564. [DOI] [Google Scholar]
- 47.Buitinck L., Louppe G., Blondel M., Pedregosa F., Mueller A., Grisel O., Niculae V., Prettenhofer P., Gramfort A., Grobler J., Layton R., VanderPlas J., Joly A., Holt B., Varoquaux G. ECML PKDD Workshop: Languages for Data Mining and Machine Learning. 2013. API design for machine learning software: experiences from the scikit-learn project; pp. 108–122. [Google Scholar]
- 48.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V., Vanderplas J., Passos A., Cournapeau D., Brucher M., Perrot M., Duchesnay É. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 49.Rousseeuw P.J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 1987;20:53–65. doi: 10.1016/0377-0427(87)90125-7. [DOI] [Google Scholar]
- 50.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.-Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.The CITIID-NIHR BioResource Covid-19 Collaboration, The Covid-19 Genomics UK (COG-UK) Consortium. Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C., Bianchi S., Tortorici M.A., Bowen J., Culap K., Jaconi S., Cameroni E., Snell G., Pizzuto M.S., Pellanda A.F., Garzoni C., Riva A., Elmer A., Kingston N., Graves B., McCoy L.E., Smith K.G.C., Bradley J.R., Temperton N., Ceron-Gutierrez L., Barcenas-Morales G., Harvey W., Virgin H.W., Lanzavecchia A., Piccoli L., Doffinger R., Wills M., Veesler D., Corti D., Gupta R.K. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in southern California. J. Am. Med. Assoc. 2021;325:1324. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wise J. Covid-19: the E484K mutation and the risks it poses. BMJ. 2021;n359 doi: 10.1136/bmj.n359. [DOI] [PubMed] [Google Scholar]
- 54.Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., Alford B., Buchser W.J., Ellebedy A.H., Fremont D.H., Diamond M.S., Whelan S.P.J. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488. doi: 10.1016/j.chom.2021.01.014. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Y., Yang C., Xu X., Xu W., Liu S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hopf T.A., Ingraham J.B., Poelwijk F.J., Schärfe C.P.I., Springer M., Sander C., Marks D.S. Mutation effects predicted from sequence co-variation. Nat. Biotechnol. 2017;35:128–135. doi: 10.1038/nbt.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nathan Rollins, Kelly Brock, Joshua Rollins, et al. 2020. SARS-CoV-2 proteins [WWW document]https://marks.hms.harvard.edu/sars-cov-2/Spike 9.13.2021. [Google Scholar]
- 58.Jacob J.J., Vasudevan K., Pragasam A.K., Gunasekaran K., Veeraraghavan B., Mutreja A. Evolutionary tracking of SARS-CoV-2 genetic variants highlights an intricate balance of stabilizing and destabilizing mutations (preprint) Genomics. 2020 doi: 10.1101/2020.12.22.423920. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.