Abstract
PDK1 (phosphoinositide-dependent kinase 1) is a mammalian growth factor-regulated serine/threonine kinase. Using a genetic selection based on a mutant form of the yeast MAP kinase kinase Ste7, we isolated a gene, PKH2, encoding a structurally and functionally conserved yeast homolog of PDK1. Yeast cells lacking both PKH2 and PKH1, encoding another PDK1 homolog, were nonviable, indicating that Pkh1 and Pkh2 share an essential function. A temperature-sensitive mutant, pkh1D398G pkh2, was phenotypically similar to mutants defective in the Pkc1–mitogen-activated protein kinase (MAPK) pathway. Genetic epistasis analyses, the phosphorylation of Pkc1 by Pkh2 in vitro, and reduced Pkc1 activity in the pkh1D398G pkh2 mutant indicate that Pkh functions upstream of Pkc1. The Pkh2 phosphorylation site in Pkc1 (Thr-983) is part of a conserved PDK1 target motif and essential for Pkc1 function. Thus, the yeast PDK1 homologs activate Pkc1 and the Pkc1-effector MAPK pathway.
Mammalian PDK1 was first identified as a kinase that activates protein kinase B (PKB) (2, 39). PKB is a growth factor-regulated serine/threonine kinase that contains a pleckstrin homology (PH) domain at its amino-terminal end. Binding of phosphoinositide 3-OH kinase products to the PH domain results in translocation of PKB to the plasma membrane, where it is activated by phosphorylation. In response to growth factor treatment of cells, PKB becomes phosphorylated at two major sites, Thr-308 in the kinase domain and Ser-473 in the C-terminal tail (1). PDK1 phosphorylates Thr-308 in PKB, resulting in its activation. This PDK1-PKB cascade mediates the physiological effects of insulin and of several growth factors and stimuli. Furthermore, PKB plays a role in mediating the protective effects of survival factors against apoptosis (11, 20, 21). PDK1 has been shown to phosphorylate and activate a number of other protein kinases such as p70S6K (3, 34) and PKC isoforms (24).
Extracellular molecules that regulate cell proliferation and differentiation in eukaryotes exert their effects through pathways that transmit signals from the cell surface through the cytoplasm to the nucleus. Some of these signals are transmitted by protein kinase cascades that involve mitogen-activated protein kinases (MAPKs). MAPKs are activated by phosphorylation on both tyrosine and threonine residues, catalyzed by a family of dual-specificity MAPK kinases (MAPKKs). MAPKKs are in turn phosphorylated and activated by MAPKK kinases (MAPKKKs). This sequential activation of MAPKKK, MAPKK, and MAPK constitutes the MAPK module (16, 22, 35).
Elements of MAPK activation pathways have been conserved across evolution, as evidenced by the existence of MAPK pathways in organisms ranging from yeast to mammals. In Saccharomyces cerevisiae, several MAPK cascade modules that mediate distinct responses to different extracellular or cell autonomous signals have been identified (4, 13, 15, 27). For example, the mating pheromone response pathway is activated by peptide pheromones and prepares cells for mating (23). The Pkc1-activated pathway, which includes the Mpk1 MAPK, responds to heat stress and hypotonic shock, and it controls both cell wall synthesis and organization of the actin cytoskeleton (14, 27). The high-osmolarity glycerol response pathway is activated by hypertonic stress (33).
The yeast MAPK cascade involved in mating comprises Ste11 (MAPKKK), Ste7 (MAPKK), and FUS3/KSS1 (MAPKs) (23). Ste11 is activated by a mechanism that probably involves Ste20 (a PAK-related protein kinase)-dependent phosphorylation. Phosphorylation activates Ste11, which consequently phosphorylates and activates Ste7. Ste11 phosphorylation of Ste7 appears to involve phosphorylation at two residues (Ser-359 and Thr-363) that are located in a region analogous to the phosphorylation lip of MAPKs. Once activated, Ste7 then activates Fus3 and Kss1 by phosphorylation at Thr and Tyr residues in the signature TEY motif within the respective Fus3 and Kss1 MAPK phosphorylation lips. Fus3 and Kss1 function redundantly in the transmission of pheromone-induced signals and induce the transcriptional activation of genes required for the process of mating.
We have recently identified a hyperactive mutant of Ste7, Ste7S368P, that is generated by mutation of serine to proline at position 368 (17, 45). Ste7S368P stimulates mating-specific transcriptional responses in the absence of mating pheromone. Although activity of Ste7S368P is higher than that of the wild-type enzyme in the absence of pheromone stimulation, its activity is still dependent on the presence of the upstream kinase Ste11. Pheromone stimulation causes a further increase in Ste7S368P activity to an amount equivalent to that of the fully activated Ste7 (45). These results suggest that Ste7S368P requires phosphorylation for full catalytic activity but that some fraction of Ste7S368P is present in the modified and active form even without pheromone stimulation. The proline substitution has one other interesting characteristic; it transforms Ste7S368P into a relatively nonspecific substrate for a variety of heterologous upstream MAPKKKs. For example, an activated form of mammalian Raf (RafΔN), but not normal c-Raf-1, can activate Ste7S368P but not wild-type Ste7 (17). This relaxed specificity allowed us to identify other MAPKKKs as activators of Ste7S368P. The genetic potential of this system has already been exploited in the identification of TAK1, a novel member of the MAPKKK family from mammalian cells, as a kinase that activates Ste7S368P in a manner homologous to Ste11 (44).
In this study, we have taken advantage of this genetic system to identify yeast genes that activate Ste7S368P. We isolated two yeast genes that were capable of activating Ste7S368P in a ste11Δ background when expressed from multicopy plasmids. The first gene was BCK1, which encodes the MAPKKK of the Pkc1-activated MAPK pathway (10, 25). The second gene, PKH2, encodes a protein kinase most similar to mammalian PDK1. We present evidence suggesting that yeast PDK1 homologs, Pkh1 and Pkh2, activate Pkc1 and the Pkc1-effector MAPK pathway. Furthermore, we isolated a multicopy suppressor of a pkh1D398G pkh2 mutant. This gene, PKH3, encodes a third PDK1 homolog.
MATERIALS AND METHODS
Strains and general methods.
Escherichia coli DH5α was used for DNA manipulation; E. coli BL21 was used as the host for expression of heterologous proteins. Standard procedures were followed for yeast manipulations (18). The media used in this study included rich medium, synthetic complete medium (SC), synthetic minimal medium (SD), and sporulation medium. SC lacking amino acids or other nutrients (e.g., SC-Leu lacks leucine) were used to score auxotrophies and to select transformants. Recombinant DNA procedures were carried out as described by Sambrook et al. (37).
Plasmids.
Plasmid YCpG22-PKH1 expresses PKH1 under the control of the GAL1 promoter. The N-terminal portion of the PKH1 coding sequence was amplified by PCR using a 5′ primer (5′-CTCGGATCCATGGGAAATAGGTCTTTGACA-3′, incorporating a BamHI site) and a 3′ primer (5′-ATCTCGTGCTGCATACCCGGGTCATCATTTGCCAGG-3′). A 190-bp BamHI-SpeI fragment generated by PCR and a 2.1-kb SpeI-XhoI fragment containing the C-terminal portion of PKH1 were inserted into the BamHI-SalI gap of YCpG22 harboring the GAL1 promoter to generate YCpG22-PKH1 (GAL1p-PKH1 TRP1). Plasmids pKT10-GAL-HA2-PKH2 and pKT10-GAL-HA2-PKH2(KN) express influenza virus hemagglutinin epitope (HA)-tagged Pkh2 and Pkh2K208R, respectively, under the control of GAL1 promoter. The N-terminal portion of the PKH2 coding sequence was amplified by PCR using a 5′ primer (5′-CTCGGATCCATGTATTTTGATAAGGATAATTCC-3′, incorporating a BamHI site), and a 3′ primer (5′-CGATAGTCTGTAAAACTGTCATTTAAA-3′). A 206-bp BamHI-StuI fragment generated by PCR and a 3-kb StuI-SalI fragment containing the C-terminal portion of Pkh2 and Pkh2K208R were inserted into the BamHI-SalI gap of pKT10-GAL-HA2, harboring the GAL1 promoter and two copies of an HA tag, to generate pKT10-GAL-HA2-PKH2 and pKT10-GAL-HA2-PKH2(KN), respectively. Plasmid pKT10-PDK1 expresses human PDK1 under the control of the TDH3 promoter. The 1.6-kb EcoRI fragment containing the PDK1 cDNA (kindly provided by D. Stokoe and F. McCormick) was inserted into the EcoRI gap of pKT10 harboring the TDH3 promoter, generating pKT10-PDK1. Plasmid pYS116 is a YEp-based URA3 plasmid harboring the lacZ reporter gene containing LexA DNA binding sites in its promoter (31). Plasmid pYW71 is a YEp-based TRP1 plasmid expressing LexA-Rlm1ΔN (amino acids 222 to 676) fusion protein from the ADH1 promoter (43). Plasmids pDL293 and pDL295 (kindly provided by D. Levin) express HA-tagged Pkc1 and Pkc1K853R, respectively, under the control of the GAL1 promoter. Plasmids pE722 and pE738 are YCp-based URA3 plasmids expressing wild-type Pkc1 and Pkc1T983A, respectively. The pkc1T983A mutant allele was generated by site-directed mutagenesis with PCR, and the mutation was confirmed by DNA sequencing.
Construction of yeast strains containing deletion alleles of PKH1 or PKH2.
The pkh1Δ::URA3 and pkh2Δ::URA3 disruption alleles were constructed by the one-step gene replacement method (36). After appropriate conversion of restriction sites, the 1.3-kb SpeI-BglII fragment of PKH1 and the 0.85-kb StuI-SnaBI fragment of PKH2 were replaced with the 1.1-kb BamHI fragment of URA3. The DNAs containing the entire pkh1Δ::URA3 and pkh2Δ::URA3 constructions were used to transform a diploid strain 15Du by selection for Ura+. Restriction mapping and Southern analysis of genomic DNAs from the resulting transformants were conducted to confirm that transplacement had occurred at each locus. The pkh1Δ::LEU2 and pkh2Δ::LEU2 strains were obtained by replacing URA3 with LEU2 in the pkh1Δ::URA3 and pkh2Δ::URA3 strains, respectively, with plasmid pUL2.
Isolation of pkh1 temperature-sensitive mutants.
Temperature-sensitive alleles of pkh1 were created by the PCR mutagenesis method (29). A region of PKH1 (−543 to +2,636) was amplified under mutagenic PCR conditions (50 mM KCl, 10 mM Tris-HCl [pH 8.0]), 2 mM MgCl2, 2 mM each dATP, dCTP, dTTP, and dGTP]. After amplification, PCR products were digested and gel purified. The mutagenized PCR products were cotransformed into a strain of pkh1Δ::LEU2 pkh2Δ::LEU2 carrying YCpG33-PKH1 (GAL1p-PKH1 URA3) with gapped plasmids that contain homology to both ends (−543 to −7 and +2,328 to 2,636) of the mutagenized PCR product. The transformants were plated on YPGlu plates and incubated at 25°C. Then the colonies were replica stamped to YPGlu plates at 37°C for selecting temperature-sensitive (ts) colonies. Plasmids were recovered, rescued in E. coli, and reintroduced into yeast to confirm the phenotypes.
Assessment of cell lysis.
Qualitative assessment of cell lysis in colonies was done by an alkaline phosphatase assay (6). Cells were spotted onto YPGlu plates, incubated at 25°C, and then shifted to 37°C. The plates were then overlaid with an alkaline phosphatase assay solution containing 0.05 M glycine hydrochloride (pH 9.5), 1% agar, and 10 mM chromogenic substrate 5-bromo-4-chloro-3-indolylphosphate. Colonies which contained lysed cells stained blue, whereas intact colonies remained white.
β-Galactosidase assays.
β-Galactosidase assays were performed as described previously (18).
Fluorescence microscopy.
Cells were grown to early logarithmic phase, fixed in formaldehyde, stained with tetramethyl rhodamine isocyanate (TRITC)-phalloidin to visualize the actin cytoskeleton, and observed by fluorescence and Nomarski microscopy as described previously (5, 38).
Expression of Pkc1 in bacteria.
To express Pkc1 in E. coli, 1.3-kb ScaI-SphI fragments (787 to 1151 amino acids) of Pkc1K853R and Pkc1K853R, T983A were inserted into the SmaI-SalI gap of pGEX-5T-2 (Pharmacia Biotech) to produce pGEX-PKC1-K853R and pGEX-PKC1-K853R,T983A, respectively. A pkc1K853R mutant allele was kindly provided by D. Levin, and a pkc1K853R, T983A mutant was generated by site-directed mutagenesis with PCR. Glutathione S-transferase (GST)–Pkc1 fusion proteins were purified as previously described (43).
Preparation of yeast extracts and immunoprecipitations.
Yeast cells were grown to an optical density (at 600 nm) of 0.5 to 1.0 and treated with 2.5% galactose for 1 h to induce the GAL1 promoter. After treatment, yeast cultures were quickly chilled, and cells were collected by rapid centrifugation. The cell pellet was washed twice with Tris-buffered saline (TBS; 20 mM Tris-HCl [pH 7.5], 150 mM NaCl) and then suspended in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5]), 100 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 5 mM NaF, 1 mM sodium pyrophosphate, 1 mM dithiothreitol, 1 mg of leupeptin per ml, 1 mg of pepstatin A per ml, 0.5% aprotinin, 1 mM phenylmethylsulfonyl fluoride). An equal volume of glass beads (0.4- to 0.6-mm diameter) was added to this suspension, and cells were broken by vigorous vortexing for 5 min at 4°C. The beads and cell debris were removed by centrifugation at 10,000 × g at 2°C, and the supernatant was further clarified by centrifugation at 100,000 × g at 2°C. Cell extracts (100 mg of protein) were incubated at 4°C for 2 h with 20 μl of protein A-Sepharose beads (Sigma) containing covalently coupled mouse monoclonal HA.11 anti-HA immunogloblin (Berkeley Antibody). Immune complexes were washed three times with lysis buffer and once with kinase buffer (100 mM Tris-HCl [pH 7.5], 50 mM MgCl2). Protein concentrations of cell extracts were measured with Bio-Rad protein determination reagent.
In vitro phosphorylation assays.
Immunoprecipitated HA-Pkh2 or HA-Pkh2K208R was suspended in 20 μl of kinase buffer with 70 μg of GST-Pkc1ΔN fusion proteins. The reaction was initiated by the addition of ATP to a final concentration of 0.1 mM along with 10 μCi of [γ-32P]ATP. After incubation for 30 min at 30°C, the reaction was terminated by the addition of sodium dodecyl sulfate (SDS) sample buffer, and samples were boiled and subjected to SDS-polyacrylamide gel electrophoresis (PAGE) on 10% acrylamide gels. After electrophoresis, gels were stained with Coomassie brilliant blue R250 and washed in 45% isopropanol–10% acetic acid. Dried gels were subjected to autoradiography. Pkc1 kinase assays were performed as described previously (41). A synthetic peptide corresponding to the sequence surrounding Ser-939 of Bck1, a phosphorylation site for Pkc1, was used as the substrate in the Pkc1 kinase assays (41).
Immunoblots.
Immunoprecipitated complexes were subjected to SDS-PAGE on 7.5% acrylamide gels followed by electroblotting onto Immunobilon-P membranes (Millipore Corporation). Blots were blocked by incubation for 1 h at room temperature in TBS-M (TBS with 5% nonfat dry milk). Blots were then incubated with monoclonal antibody HA.11 diluted 1:1,000 in TBS-M for 20 h at 4°C. After three washes with TBS-M, blots were incubated for 2 h with peroxide-linked secondary antibody (Calbiochem) diluted 1:2,500 in TBS-M. After four final washes with TBS-M, blots were developed using an enhanced chemiluminescence detection kit (Amersham).
RESULTS
Overexpression of BCK1 or PKH2 enhances Ste7S368P function.
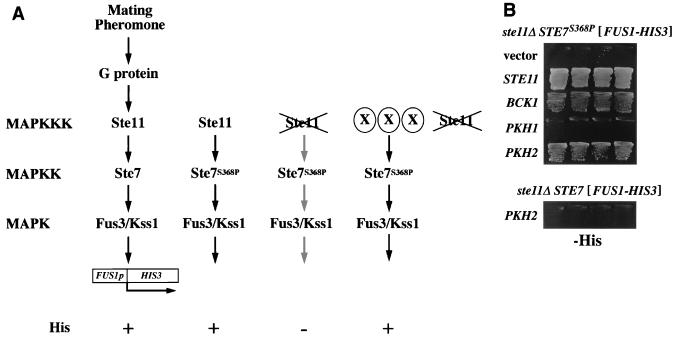
The MAPK pathway mediating mating pheromone signal transduction is regulated by a cascade of protein kinases consisting of Ste11, Ste7, and Fus3/Kss1 (23). A terminal target of this pathway is Ste12, a DNA-binding protein that recognizes the promoter regions of mating pheromone-inducible genes such as FUS1 (12). Using a his3Δ yeast strain containing a FUS1p-HIS3 fusion (40), one can readily assay activity of the mating pheromone signaling pathway by growth on media lacking histidine (His phenotype) (Fig. 1A). We previously isolated a strain containing a hyperactive mutation of STE7, STE7S368P (17, 45). The Ste7S368P enzyme has higher kinase activity than wild type in the absence of pheromone stimulation but still requires modification by Ste11 for its activity. Therefore, ste11Δ FUS1p-HIS3 cells containing STE7S368P exhibited a His− phenotype (Fig. 1). From a multicopy (YEp13) yeast genomic library we isolated two different clones that suppressed the His− phenotype (Fig. 1B). One contained the BCK1 gene, which encodes the MAPKKK of the Pkc1-activated Mpk1 MAPK pathway (10, 25); the other contained the PKH2 gene, which encodes a PDK1 homolog (Fig. 2). A multicopy plasmid carrying the PKH2 gene (YEp-PKH2) failed to support FUS1p-HIS3 expression in a ste11Δ STE7+ background (Fig. 1B). This indicates that overexpression of PKH2 can enhance the function of the Ste7S368P variant but not of wild-type Ste7.
FIG. 1.
Isolation of Ste7S368P activators. (A) Model for the yeast pheromone-stimulated MAPK pathway. The pheromone-stimulated MAPK pathway induces transcription of mating-specific genes such as FUS1. The FUS1p-HIS3 reporter gene consists of the FUS1 upstream activation sequence joined to the HIS3 open reading frame. This reporter allows signal activity in a his3Δ FUS1p-HIS3 strain to be monitored by its ability to grow on medium lacking exogenous histidine (His phenotype). A his3Δ ste11Δ FUS1p-HIS3 STE7S368P strain has a His− phenotype because the activity of Ste7S368P is still dependent on the presence of the upstream Ste11 MAPKKK (17). Presence of Ste7S368P activators (shown as X) in this strain should confer a His+ phenotype. (B) Suppression of ste11Δ STE7S368P by BCK1 or PKH2. Strain SY1984-P (his3Δ ste11Δ FUS1p-HIS3 STE7P368; top) (45) or SY1984 (his3Δ ste11Δ FUS1p-HIS3 STE7; bottom) (40) was transformed with the indicated plasmids, and each transformant was streaked onto SC-His plates and incubated at 30°C. Plasmids were YEplac195 (vector), YCp-STE11 (STE11), YEp-BCK1 (BCK1), YEp-PKH1 (PKH1), and YEp-PKH2 (PKH2).
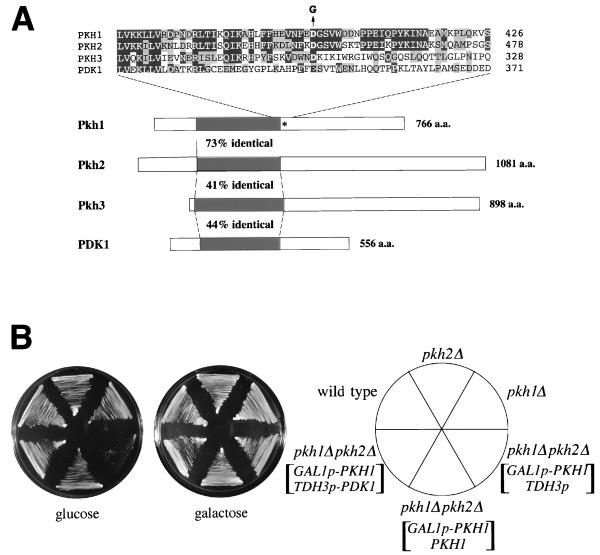
FIG. 2.
Identification of yeast PDK1 homologs. (A) Schematic diagrams of the structures of PDK1, Pkh1, Pkh2, and Pkh3. Kinase domains are indicated by black boxes. The mutated residue in pkh1D398G is indicated by an asterisk. The amino acid (a.a.) residues in the region of the mutation site are aligned above the diagram. The Asp-to-Gly change in the pkh1D398G mutation is indicated as a G above the Pkh1 sequence. Amino acids which are identical or conserved are indicated by black or gray boxes, respectively. (B) Effect of the pkh1Δ and pkh2Δ mutations on cell growth. Yeast strains carrying the indicated plasmids were streaked onto YPGlu (glucose) medium and YPGal (galactose) medium and incubated at 30°C. Yeast strains were 15Dau (wild type), INA25-3B (pkh1Δ::URA3), INA28-1B (pkh2Δ::URA3), and INA38-6A (pkh1Δ::URA3 pkh2Δ::URA3). Plasmids were YCpG22-PKH1 (GAL1p-PKH1), pKT10 (TDH3p), YCplac22-PKH1 (PKH1), and pKT10-PDK1 (TDH3p-PDK1).
PKH1 and PKH2 encode protein kinases homologous to mammalian PDK1.
The PKH2 gene encodes a 1,081-residue protein that shows most similarity to PDK1 from mammals (2, 39) (Fig. 2A). A GenBank database search revealed that PKH2 shares homology with the S. cerevisiae gene YDR490c, also designated PKH1, which encodes a protein of 766 amino acids. The predicted protein kinase domains of these proteins share 73% amino acid identity (Fig. 2A), suggesting the possibility that Pkh1 and Pkh2 proteins functionally overlap. However, in contrast to PKH2, multicopy plasmids containing the PKH1 gene failed to support FUS1p-HIS3 expression in the ste11Δ STE7S368P strain (Fig. 1B).
To examine the phenotypic defect associated with loss of PKH1 and/or PKH2 function, we constructed yeast strains containing deletion alleles of these genes (pkh1Δ, pkh2Δ, and pkh1Δ pkh2Δ) as described in Materials and Methods. Whereas the individual pkhΔ mutants were viable and grew normally at any temperature, the pkh1Δ pkh2Δ double mutant was not viable (Fig. 2B). To characterize the phenotype of the pkh1Δ pkh2Δ mutant cells, we constructed this strain carrying a plasmid expressing PKH1 under the control of the GAL1 promoter, YCpG22-PKH1, allowing PKH1 to be induced or repressed on galactose- or glucose-containing medium, respectively. Haploid pkh1Δ pkh2Δ cells containing YCpG22-PKH1 grew normally on galactose-containing medium but failed to grow on glucose-containing medium (Fig. 2B). Microscopic examination of these nongrowing cells revealed a high frequency of nonrefractile ghosts, suggesting that cell lysis had occurred. However, pkh1Δ pkh2Δ mutants containing YCpG22-PKH1 were not rescued on glucose-containing medium supplemented with 1.2 M sorbitol (data not shown).
To test whether the structural similarity of mammalian PDK1 to Pkh1 and Pkh2 has any functional significance, we investigated whether expression of mammalian PDK1 would suppress the defect associated with loss of both PKH1 and PKH2. We constructed a plasmid, pKT10-PDK1, that constitutively expresses PDK1 under the control of the yeast TDH3 promoter (TDH3p-PDK1). This plasmid was transformed into the pkh1Δ pkh2Δ strain carrying YCpG22-PKH1 (GAL1p-PKH1). On glucose-containing medium, where Pkh1 expression is repressed, expression of PDK1 allowed growth of pkh1Δ pkh2Δ cells (Fig. 2B). Similar results showing complementation of pkh1Δ pkh2Δ by mammalian PDK1 were obtained by Casamayor et al. while this study was in progress (7).
Isolation of a ts pkh1 mutation.
To isolate a ts mutant of PKH1, DNA encoding the entire PKH1 gene was randomly mutagenized by PCR. The resulting amplification products were then introduced along with the TRP1 marker into a pkh1Δ pkh2Δ strain which was maintained by the presence of YCpG33-PKH1 (GAL1p-PKH1 URA3). Cells transformed with a functional PKH1 gene were selected by growth on glucose-containing medium at 25°C. The colonies were then replica plated onto glucose-containing medium and cultured at 37°C. Colonies that grew normally at 25°C but failed to grow at 37°C were subjected to further analysis. From a total of 5,000 transformants, 6 ts colonies were isolated, and their plasmid DNAs were retransformed into the parent strain. Among six candidate plasmids that conferred a ts growth defect, we characterized one pkh1 allele (Fig. 3). This allele was found to harbor a single-base-pair mutation that resulted in an aspartate-to-glycine change at position 398 (Fig. 2A). Alignment of the Pkh1, Pkh2, and PDK1 protein sequences revealed that an acidic residue (Asp in Pkh1 and Pkh2 or Glu in PDK1) corresponding to position 398 in Pkh1 is conserved among these proteins (Fig. 2A). The ts phenotype was recessive, indicating that the phenotype was caused by a loss of PKH1 function at 37°C. The mutant allele was named pkh1D398G. In strains wild type for PKH2, the pkh1D398G allele caused no growth defect at 37°C (data not shown).
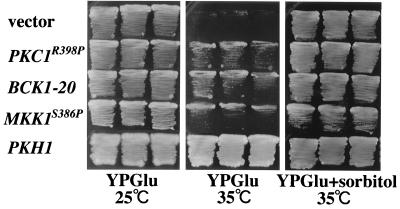
FIG. 3.
A ts pkh1 mutation. Transformants of the yeast strain INA106-3B (pkh1D398G pkh2Δ::LEU2) carrying the indicated plasmids were streaked onto YPGlu medium and YPGlu medium supplemented with 1.2 M sorbitol and incubated at 25 or 35°C. Each patch represents an independent transformant. Plasmids were YCplac22 (vector), pRS316-PKC1(R398P) (PKC1R398P), pRS314-BCK1-20 (BCK1-20), YCplac22-MKK1(S386P) (MKK1S386P), and YCplac22-PKH1 (PKH1).
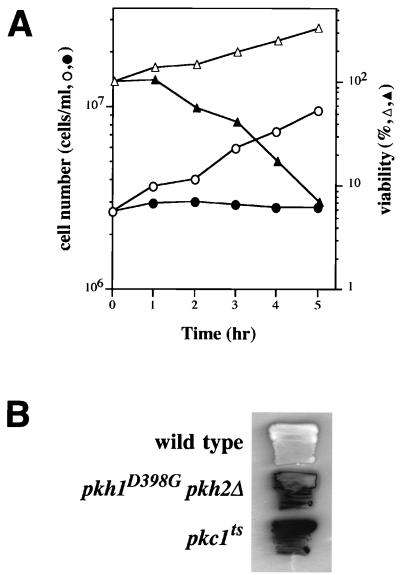
When liquid cultures of exponentially growing pkh1D398G pkh2Δ cells were transferred from the permissive to the restrictive temperature, growth and viability rapidly decreased (Fig. 4A), and cell lysis and the accumulation of cell ghosts and debris were observed. As a further indication of cell lysis, the pkh1D398G pkh2Δ mutant turned blue when cultured at the restrictive temperature on agar plates containing 5-bromo-4-chloro-3-indolylphosphate (Fig. 4B). Also, pkh1D398G pkh2Δ cells survived at 35°C in medium containing 1.2 M sorbitol (Fig. 3). This is characteristic of yeast mutants defective in cell wall integrity; i.e., lysis occurs in normal medium but not in high-osmolarity medium.
FIG. 4.
Growth phenotypes of pkh1D398G pkh2Δ mutants. (A) Viability of pkh1D398G pkh2Δ mutants. INA106-3B cells (pkh1D398G pkh2Δ::LEU2) were grown in YPGlu medium at 25°C (open symbols) and shifted to 35°C (closed symbols). At the times indicated, cell number and viability were assayed. Cell number was determined by phase-contrast light microscopy with a hemacytometer. Percent viability was determined by comparing the colony number obtained after incubation for 48 h in YPGlu with that expected from the number of cells observed in the plated sample. (B) Cell lysis of pkh1D398G pkh2Δ::LEU2 mutants. Yeast strains were patched onto YPGlu medium, incubated at 25°C for 1 day, and then shifted to 37°C for 1 day. The patches were assayed in situ for release of alkaline phosphatase as an indication of cell lysis. Yeast strains were 15Dau (wild type), INA106-3B (pkh1D398G pkh2Δ::LEU2), and SYT11-12A (pkc1ts stt1-1).
Pkh1 and Pkh2 function in the Pkc1-MAPK pathway.
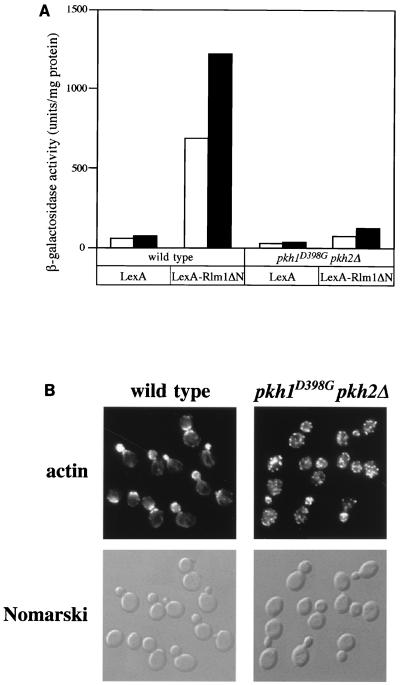
The cell lysis defect displayed by pkh1D398G pkh2Δ mutants was similar to that seen with mutants defective in the Pkc1-activated MAPK pathway, suggesting that Pkh1 and Pkh2 may function in this pathway. The Pkc1 pathway activates a MAPK cascade that consists of sequentially activated kinases: Bck1 (MAPKKK), the redundant Mkk1 and Mkk2 (MAPKKs), and Mpk1 (MAPK) (27). To test how loss of PKH1 and PKH2 affects the Pkc1-Mpk1 pathway, we examined the ability of pkh1D398G pkh2Δ mutants to activate a nuclear target of Mpk1, the Rlm1 transcription factor (42, 43). The extent of Mpk1 pathway activation can be monitored using a LexA-Rlm1ΔN fusion protein, which is phosphorylated by Mpk1, and a LexA-lacZ reporter gene, which is transcriptionally activated by LexA-Rlm1ΔN (43). Wild-type and pkh1D398G pkh2Δ cells expressing LexA-Rlm1ΔN were transformed with the LexA-lacZ plasmid, and transformants were tested for β-galactosidase activity (Fig. 5A). In contrast to the wild-type strain, LexA-Rlm1ΔN was unable to activate transcription of the reporter gene in a pkh1D398G pkh2Δ mutant. These results suggest that both Pkh1 and Pkh2 signal through the MAPK Mpk1.
FIG. 5.
Effect of Pkh on the Pkc1-MAPK pathway. (A) Effect of the pkh1D398G pkh2Δ mutation on Rlm1 transcriptional activity. Cells carrying the LexA-lacZ reporter gene and pBTM116 (LexA) or pYW71 (LexA-Rlm1ΔN) were grown at 23°C (open bars) and then shifted to 37°C for 30 min (solid bars) and assayed for β-galactosidase activity. The units shown are the average of two or three experiments. Yeast strains were 15Dau (wild type) and INA106-3B (pkh1D398G pkh2Δ). (B) Effect of the pkh1D398G pkh2Δ mutation on actin organization. Cells were grown at 24°C, shifted to 35°C for 2.5 h, fixed, stained with TRITC-phalloidin, and observed by fluorescence (top) and Nomarski microscopy (bottom). Yeast strains were 15Dau (wild type) and INA106-3B (pkh1D398G pkh2Δ). A field of mutant cells lacking lysed ghost cells was chosen.
The Pkc1-MAPK pathway is also required for organization of the actin cytoskeleton (14, 28). To investigate further whether Pkh1 or Pkh2 plays a role in the Pkc1-MAPK pathway, wild-type and pkh1D398G pkh2Δ mutant cells were grown at 24°C, shifted to 35°C, and processed for visualization of the actin cytoskeleton (Fig. 5B). Whereas wild-type cells displayed the normal cell cycle-dependent polarized distribution of actin, pkh1D398G pkh2Δ cells were growth arrested at 35°C and exhibited random distribution of actin. Cells lacking only PKH2 (PKH1+ pkh2Δ) displayed a normal distribution of the actin cytoskeleton (data not shown). These results indicate that Pkh1 and Pkh2 are redundantly required for polarization of the actin cytoskeleton and further support a role for Pkh1 and Pkh2 in the Pkc1-MAPK pathway.
To determine the genetic relationship between the PKH genes and the Pkc1-MAPK pathway, we performed epistasis analyses. Mutant alleles encoding constitutively active Bck1 (BCK1-20) and Mkk1 (MKK1S386P) have been shown to suppress the phenotype of a pkc1 deletion (25, 45). Expression of BCK1-20 or MKK1S386P partially suppressed the temperature sensitivity of the pkh1D398G pkh2Δ mutant (Fig. 3). Furthermore, a constitutively active mutant of PKC1, PKC1R398P (30), also partially suppressed the growth defect of the pkh1D398G pkh2Δ mutant (Fig. 3). In contrast, overexpression of PKH1 or PKH2 failed to suppress the phenotype of a ts pkc1 allele (stt1-1) (data not shown). These results suggest that Pkh1 and Pkh2 function upstream of Pkc1 in the Pkc1-MAPK pathway.
Pkh phosphorylates and activates Pkc1.
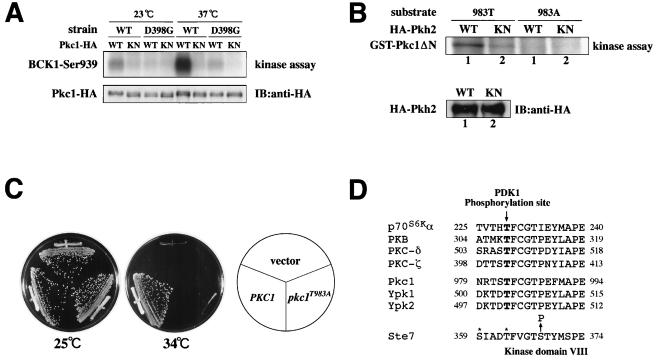
The above results raise the possibility that Pkh proteins directly phosphorylate and activate Pkc1. To test this possibility, we assayed Pkc1 kinase activity in wild-type and pkh1D398G pkh2Δ mutant strains. Wild-type and mutant strains were transformed with a plasmid encoding Pkc1 tagged at its COOH terminus with HA (Pkc1-HA) and expressed under the control of the GAL1 promoter (41). Transformants were grown at 23°C, treated with galactose for 1 h to induce the GAL1 promoter, and then shifted to 37°C for 1 h. Pkc1-HA was immunoprecipitated from yeast extracts, and its protein kinase activity was measured by using a synthetic Bck1 peptide as a substrate (41). Compared to the wild-type strain, Pkc1 activity was significantly reduced in pkh1D398G pkh2Δ mutants even at the permissive temperature (Fig. 6A). Furthermore, the heat induction of Pkc1 kinase activity was severely compromised in the pkh1D398G pkh2Δ mutant relative to the wild type (Fig. 6A). These results indicate that Pkh proteins are required for Pkc1 activation, further supporting the results of the previous epistasis analyses.
FIG. 6.
Effect of Pkh on Pkc1. (A) Pkc1 kinase activity. Yeast strains 15Dau (wild type [WT]) and INA106-3B (pkh1D398G pkh2Δ::LEU2 [D398G]) were transformed with pDL293 [GAL1p-PKC1-HA] or pDL295 [GAL1p-PKC1(K853R)-HA]. Transformants were grown at 23°C, treated with galactose for 1 h to induce the GAL1 promoter, and then shifted to 37°C for 1 h. Pkc1-HA (wild type [WT]) or Pkc1K853R-HA (KN) was immunoprecipitated from cell extracts. In vitro protein kinase assays were conducted with a synthetic peptide substrate corresponding to the sequence surrounding Ser-939 of Bck1, a phosphorylation site for Pkc1 (top). A parallel set of immune complexes was subjected to immunoblotting (IB) for detection of Pkc1-HA or Pkc1K853R-HA (bottom). (B) In vitro phosphorylation of Pkc1 by Pkh2. Yeast strain 15Dau (wild type) was transformed with pKT10-GAL-HA2-PKH2 (GAL1p-HA-PKH2) or pKT10-GAL-HA2-PKH2(KN) [GAL1p-HA-PKH2(K208R)]. Transformants were grown at 30°C and treated with galactose for 1 h to induce the GAL1 promoter. HA-Pkh2 (WT) or HA-Pkh2K208R (KN) was immunoprecipitated from cell extracts. In vitro protein kinase assays were conducted with GST-Pkc1ΔN(787-1151, K853R) (983T) and GST-Pkc1ΔN(787-1151, K853R, T983A) (983A) as substrates (top). A parallel set of immune complexes was subjected to immunodetection of HA-Pkh2 or HA-Pkh2K208R (KN) (bottom). (C) Mutation of Pkc1 phosphorylation site. Transformants of the yeast strain SYT11-12A (pkc1ts stt1-1) carrying the indicated plasmids were streaked onto YPGlu medium and incubated at 25 or 34°C. Plasmids were YCplac33 (vector), pE722 (PKC1), and pE738 (pkc1T983A). (D) Alignment of amino acid sequences of the protein kinase subdomains VII and VIII. The amino acid sequences surrounding the residues equivalent to Thr-308 of PKB are highly conserved in these protein kinases. The positions of sites in Ste7 that are phosphorylated by Ste11 are indicated by asterisks, and the position of the substituted Ser residue in Ste7S368P is indicated by an arrow.
To test whether Pkc1 is directly phosphorylated by Pkh in vitro, we developed an immune complex assay to measure Pkh2 protein kinase activity. HA-tagged wild-type Pkh2 (HA-Pkh2) was expressed under the control of the inducible GAL1 promoter. Expression of HA-Pkh2 was able to complement the growth defect of pkh1Δ pkh2Δ mutants, indicating that HA-Pkh2 is functional (data not shown). As a substrate, we used GST-Pkc1ΔN(787-1151; K853R), containing the inactivating K853R mutation, to eliminate background Pkc1 autophosphorylation. The epitope-tagged Pkh2 proteins were immunoprecipitated with an anti-HA antibody and incubated with [γ-32P]ATP and GST-Pkc1ΔN(787-1151; K853R). We found that Pkc1 was phosphorylated by immune complexes from cells expressing HA-Pkh2 (Fig. 6B). To rule out the possibility that this activity is due to another kinase that is tightly associated with HA-Pkh2 during the immunoprecipitation procedure, we also tested an HA-tagged kinase-inactive mutant (HA-Pkh2K208R) containing a lysine-to-arginine change in the kinase domain. Immunocomplexes from cells expressing HA-Pkh2K208R exhibited much lower kinase activity toward Pkc1, although the HA-Pkh2K208R protein was expressed at levels comparable to those for wild-type Pkh2 (Fig. 6B).
PDK1 activates PKB by phosphorylating a Thr residue in a conserved sequence motif located within the activation loop of its catalytic domain situated between conserved protein kinase subdomains VII and VIII (2, 39). This motif is also conserved in Pkc1 and in mammalian PKCs (Fig. 6D). PDK1 has been shown to phosphorylate sites within the activation loops of PKCξ and PKCδ (23). To test whether the conserved Thr-983 in the activation loop of Pkc1 is indeed the site of Pkh2 phosphorylation, we generated the GST fusion protein Pkc1ΔN(787-1151; K853R; T983A), in which the Thr residue at 983 was replaced with Ala. We found that this mutant protein was not phosphorylated by Pkh2 (Fig. 6B), indicating that Thr-983 is the site of Pkh2 phosphorylation.
We next asked whether Thr-983 is important for Pkc1 function. We examined the ability of Pkc1T983A containing a mutation of Thr-983 to Ala to complement the growth defect of pkc1ts (stt1-1) mutants. Whereas wild-type Pkc1 complemented the pkc1ts defect, the T983A mutant did not (Fig. 6C), indicating that Thr-983 is required for Pkc1 function.
Isolation of PKH3 as a multicopy suppressor of pkh1D398G pkh2.
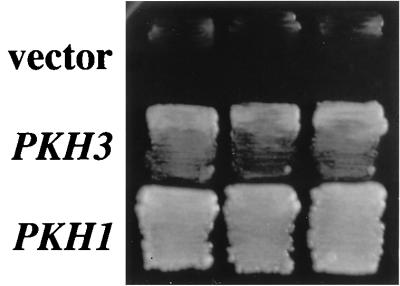
To identify additional components of the Pkh pathway, we isolated multicopy suppressors of the growth defect of a pkh1D398G pkh2 mutant. A yeast genomic library cloned into the multicopy vector YEp13 was transformed into a pkh1Δ pkh2Δ strain that carried plasmid YCplac22-PKH1D398G (pkh1D398G TRP1). A total of 3,500 Leu+ transformants were obtained and subsequently screened for the ability to grow at the restrictive temperature. Six transformants capable of forming colonies at 37°C were isolated (Fig. 7). The plasmids recovered from these yeast transformants were of two classes, based on restriction digest patterns. Sequence analysis revealed that three of these six plasmids contained the PKH2 gene itself. The DNA sequences of the three remaining clones was determined, and all were found to contain a gene designated YDR466w by the Yeast Genome Project. The YDR466w gene encodes a protein of 898 amino acids and contains a protein kinase domain. The predicted protein has 40% sequence identity to Pkh1 and Pkh2 throughout its protein kinase domains (Fig. 2A). We therefore named this gene PKH3. Interestingly, PKH3 restored growth only when coexpressed with pkh1D398G, even though pkh1D398G appears inactive at 37°C. This was shown by the observation that when pkh1Δ pkh2Δ strains carrying both YCplac22-PKH1D398G and YEp-PKH3 were cultured at 37°C, the YCplac22-PKH1D398G plasmid did not segregate away. Furthermore, pkh1Δ pkh2Δ cells transformed with both YEp-PKH3 and YCpG22-PKH1 (GAL1p-PKH1 TRP1) failed to grow in glucose-containing medium (data not shown). Thus, overexpression of PKH3 does not compensate for disruption of both PKH1 and PKH2, and the ts protein Pkh1D398G has some activity at the restrictive temperature that is required together with PKH3 for viability.
FIG. 7.
Isolation of the PKH3 gene. Transformants of the yeast strain INA106-3B (pkh1D398G pkh2Δ::LEU2) carrying the indicated plasmids were streaked onto YPGlu medium and incubated at 35°C. Each patch represents an independent transformant. Plasmids were YEp13 (vector), YEp-PKH3 (PKH3), and YCplac111-PKH1 (PKH1).
We constructed strains containing a deletion allele of PKH3 (pkh3Δ). These strains grew normally at any temperature. Furthermore, pkh1Δ pkh3Δ and pkh2Δ pkh3Δ double-mutant cells also grew normally at any temperature (data not shown), indicating that unlike Pkh1 and Pkh2, Pkh3 is dispensable for growth.
DISCUSSION
We show that S. cerevisiae has two PDK1 homologs, Pkh1 and Pkh2. Single pkh1Δ and pkh2Δ mutants are viable, but the pkh1Δ pkh2Δ double mutant is nonviable, indicating that Pkh1 and Pkh2 share a role that is essential for cell growth. Expression of mammalian PDK1 suppresses the lethality of pkh1Δ pkh2Δ cells, demonstrating that Pkh1 and Pkh2 are functionally similar to PDK1. As many signal transduction pathways and mechanisms that regulate cell growth and proliferation are conserved between mammalian and yeast cells, it is not unexpected that PDK1 homologs are present in yeast. PDK1 activates PKB, PKCs and p70S6K by phosphorylating the Thr residue in a conserved sequence motif located within the activation loops of their catalytic domains (Fig. 6D). This conserved sequence motif is also found in the activation loop of the S. cerevisiae Pkc1 protein, raising the possibility that Pkc1 is a physiological substrate for Pkh1 and Pkh2.
In mammalian cells, PDK1 has been shown to phosphorylate PKC isoforms (24). However, the biological role of this phosphorylation in mammalian cells is not understood. Here we provide evidence indicating that Pkh1 and Pkh2 function in the Pkc1-MAPK pathway. First, temperature-sensitive pkh1ts pkh2Δ mutants display phenotypes similar to those of mutants defective in the Pkc1-MAPK pathway, notably osmoremedial cell lysis and loss of actin cytoskeleton polarity. Second, pkh1ts pkh2Δ mutants are defective in activation of the transcription factor Rlm1, whose activity is dependent on the Pkc1-MAPK pathway. Third, an activating mutation in PKC1, BCK1, or MKK1 partially suppresses the growth defect of a pkh1ts pkh2Δ mutant. Fourth, Pkc1 activity is decreased in a pkh1ts pkh2Δ mutant. Finally, the Pkh2 protein phosphorylates Pkc1 in vitro at Thr-983; this residue is part of the conserved PDK1 target motif in the Pkc1 activation loop and is essential for Pkc1 function. Thus, Pkh1 and Pkh2 are in the Pkc1-MAPK pathway as activators of Pkc1.
Many protein kinases require phosphorylation within their activation loops to be fully activated. Phosphorylation within the activation loop is also important for protein kinases stringently regulated by allosteric effectors. This is exemplified by PKC, where activation of the Ca2+/diacylglycerol-dependent isotypes PKCα and PKCβ absolutely requires phosphorylation of respective activation loops (8, 32). PDK1 regulates multiple protein kinases, including PKB, p70S6K, and PKC isoforms (2, 3, 24, 34, 39). The specificity of PDK1 action on its downstream protein kinase targets could be determined by target-specific regulators. In S. cerevisiae, the GTP-bound form of Rho1 functions as an activator of Pkc1 (19, 30), raising the possibility that this Rho1-dependent activation of Pkc1 is controlled through Pkh1/Pkh2-dependent phosphorylation of the Pkc1 activation loop.
Several observations suggest that Pkc1 is not the only target of the Pkh kinases. pkh1ts pkh2Δ mutants resemble pkc1 mutants in that they also exhibit defects in cell integrity resulting from aberrant cell wall construction. However, whereas cell lysis caused by loss of PKC1 function is suppressed by osmotic stabilizing agents (26), the growth defect in pkh1Δ pkh2Δ mutants was not. Based on the observation that mammalian PDK1 activates PKB (2, 39), it is possible that the yeast PKB-like protein kinases, Ypk1 and Ypk2, which play an essential role in yeast cell growth (9), are also targets of Pkh. Consistent with this possibility, the sequence surrounding the site in PKB phosphorylated by PDK1 is also conserved in Ypk1 and Ypk2 (Fig. 6D). Furthermore, Casamayor et al. have recently shown that Pkh1 activates Ypk1 in vitro by phosphorylating the Thr-504 residue that corresponds to the site of PDK1 phosphorylation (7). Thus, Pkh1 and Pkh2 are likely to play a role in activating at least two types of protein kinases that are essential for cell growth, Pkc1 and Ypk1/Ypk2.
In this study, we isolated six different ts pkh1 mutants and divided them into two classes. The growth defect of the first class, typified by pkh1D398G, was rescued by osmotic stabilization, whereas the growth defect of the second class (unpublished data) was not. This suggests that the second class of mutants may be defective in activation of Ypk1/Ypk2. We thus attempted to rescue this class of mutants by overexpression of Ypk1 or Ypk2, but growth was not restored even in the presence of osmotic stabilizing agents (unpublished data). We suspect that activation of Ypk1 and Ypk2 may absolutely require phosphorylation by Pkh1 and Pkh2. If true, this hypothesis suggests a genetic approach to identify constitutive mutations in YPK1 or YPK2, i.e., by isolating mutations that suppress the growth defect of a pkh1ts pkh2Δ strain. This work may elucidate the different modes by which mammalian PDK1 and PKB are regulated.
Overexpression of the PKH2 gene, but not the PKH1 gene, was shown to activate Ste7S368P, which suggests that Pkh2 basal kinase activity is higher than that of Pkh1. Consistent with this possibility, we could detect Pkh2 but not Pkh1 kinase activity when Pkc1 was used as a substrate (unpublished data). However, the pkh2Δ single mutant grows normally, indicating that endogenous Pkh1 activity is sufficient for growth. It is therefore likely that the activity of Pkh1 is tightly regulated. The fact that PDK1 contains a PH domain and can bind lipid vesicles containing phosphatidylinositol 3,4,5-trisphosphate or phosphatidylinositol 3,4-bisphosphate (39) may suggest that 3-phosphorylated lipids can activate PDK1 in some way. In contrast, Pkh1 and Pkh2 lack any obvious PH domain. It will be interesting to identify upstream activators of Pkh1. These analyses should further our understanding of the signal(s) that activates the Pkh-Pkc1 pathway.
The STE7S368P mutant, which activates mating response genes in the absence of pheromone, can be upregulated by overexpression of PKH2, whereas the wild-type STE7 cannot. This suggests that Pkh2 phosphorylates and activates the Ste7S368P variant but not wild-type Ste7. The Ste7 protein contains a threonine residue in its activation loop, Thr-363, which is known to be the site of Ste11 phosphorylation. This residue is also analogous to a conserved Thr residue in the phosphorylation site of PDK1 (Fig. 6D). The residue in Ste7 that is mutated in Ste7S368P, i.e., Ser-368, lies within the activation loop between subdomains VII and VIII in proximity to the Thr-363 residue. Interestingly, all PDK1 target protein kinases except p70S6K have a proline residue at the site corresponding to Ser-368 of Ste7, which may be important for their interaction with PDK1, Pkh1, and/or Pkh2. Thus, the serine-to-proline change in Ste7S368P may convert Ste7 to a substrate of Pkh2, perhaps by altering the conformation of the activation loop in a way that makes the Thr-363 residue accessible to Pkh2.
We also identified a novel protein kinase, Pkh3, that functions as a multicopy suppressor of a pkh1ts pkh2Δ mutant and which exhibits homology to PDK1, Pkh1, and Pkh2. Growth of the pkh3Δ single mutant is normal and indistinguishable from that of wild-type cells. Also, pkh1Δ pkh3Δ and pkh2Δ pkh3Δ double mutants display no apparent phenotype. These results suggest that Pkh3 is able to phosphorylate the same essential target substrates as Pkh1 and Pkh2 when overexpressed but that Pkh1 and Pkh2 play a more important role in regulating cell growth under physiological conditions.
ACKNOWLEDGMENTS
We thank D. Levin, F. McCormick, M. Shirayama, D. Stokoe, Y. Takai, and K. Tanaka for materials; E. Nishida and H. Shibuya for helpful discussions; and M. Lamphier and R. Ruggieri for critical reading of the manuscript.
This work was supported by the Boehringer Ingelheim Fonds (T.S.), the Swiss National Science Foundation (M.N.H.), and special grants for CREST, Advanced Research on Cancer from the Ministry of Education, Culture and Science of Japan, and HFSP (K.M.).
REFERENCES
- 1.Alessi D R, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings B A. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- 2.Alessi D R, James S R, Downes C P, Holmes A B, Gaffney P R, Reese C B, Cohen P. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase B α. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 3.Alessi D R, Kozlowski M T, Weng Q P, Morrice N, Avruch J. 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 4.Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benedetti H, Raths S, Crausaz F, Riezman H. The END3 gene encodes a protein that is required for the internalization step of endocytosis and for actin cytoskeleton organization in yeast. Mol Biol Cell. 1994;5:1023–1037. doi: 10.1091/mbc.5.9.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabib E, Duran A. Simple and sensitive procedure for screening yeast mutants that lyse at nonpermissive temperatures. J Bacteriol. 1975;124:1604–1606. doi: 10.1128/jb.124.3.1604-1606.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casamayor A, Torrance P D, Kobayashi T, Thorner J, Alessi D R. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol. 1999;9:186–197. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- 8.Cazaubon S, Bornancin F, Parker P J. Threonine-497 is a critical site for permissive activation of protein kinase C α. Biochem J. 1994;301:443–448. doi: 10.1042/bj3010443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P, Lee K S, Levin D E. A pair of putative protein kinase genes (YPK1 and YPK2) is required for cell growth in Saccharomyces cerevisiae. Mol Gen Genet. 1993;236:443–447. doi: 10.1007/BF00277146. [DOI] [PubMed] [Google Scholar]
- 10.Costigan C, Gehrung S, Snyder M. A synthetic lethal screen identifies SLK1, a novel protein kinase homolog implicated in yeast cell morphogenesis and cell growth. Mol Cell Biol. 1992;12:1162–1178. doi: 10.1128/mcb.12.3.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudek H, Datta S R, Franke T F, Birnbaum M J, Yao R, Cooper G M, Segal R A, Kaplan D R, Greenberg M E. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science. 1997;275:661–665. doi: 10.1126/science.275.5300.661. [DOI] [PubMed] [Google Scholar]
- 12.Elion E A, Satterberg B, Kranz J E. FUS3 phosphorylates multiple components of the mating signal transduction cascade: evidence for STE12 and FAR1. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustin M C, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Helliwell S B, Schmidt A, Ohya Y, Hall M N. The Rho1 effector Pkc1, but not Bni1, mediates signalling from Tor2 to the actin cytoskeleton. Curr Biol. 1998;8:1211–1214. doi: 10.1016/s0960-9822(07)00511-8. [DOI] [PubMed] [Google Scholar]
- 15.Herskowitz I. MAP kinase pathways in yeast: for mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- 16.Ip Y T, Davis R J. Signal transduction by the c-Jun N-terminal kinase (JNK)—from inflammation to development. Curr Opin Cell Biol. 1998;10:205–219. doi: 10.1016/s0955-0674(98)80143-9. [DOI] [PubMed] [Google Scholar]
- 17.Irie K, Gotoh Y, Yashar B M, Errede B, Nishida E, Matsumoto K. Stimulatory effects of yeast and mammalian 14-3-3 proteins on the Raf protein kinase. Science. 1994;265:1716–1719. doi: 10.1126/science.8085159. [DOI] [PubMed] [Google Scholar]
- 18.Kaiser C, Michaelis S, Mitchell A. Method in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 19.Kamada Y, Qadota H, Python C P, Anraku Y, Ohya Y, Levin D E. Activation of yeast protein kinase C by Rho1 GTPase. J Biol Chem. 1996;271:9193–9196. doi: 10.1074/jbc.271.16.9193. [DOI] [PubMed] [Google Scholar]
- 20.Kauffmann Z A, Rodriguez V P, Ulrich E, Gilbert C, Coffer P, Downward J, Evan G. Suppression of c-Myc-induced apoptosis by Ras signalling through PI(3)K and PKB. Nature. 1997;385:544–548. doi: 10.1038/385544a0. [DOI] [PubMed] [Google Scholar]
- 21.Kennedy S G, Wagner A J, Conzen S D, Jordan J, Bellacosa A, Tsichlis P N, Hay N. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- 22.Kyriakis J M, Avruch J. Protein kinase cascades activated by stress and inflammatory cytokines. Bioessays. 1996;18:567–577. doi: 10.1002/bies.950180708. [DOI] [PubMed] [Google Scholar]
- 23.Leberer E, Thomas D Y, Whiteway M. Pheromone signalling and polarized morphogenesis in yeast. Curr Opin Genet Dev. 1997;7:59–66. doi: 10.1016/s0959-437x(97)80110-4. [DOI] [PubMed] [Google Scholar]
- 24.Le Good J A, Ziegler W H, Parekh D B, Alessi D R, Cohen P, Parker P J. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–2045. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 25.Lee K S, Levin D E. Dominant mutations in a gene encoding a putative protein kinase (BCK1) bypass the requirement for a Saccharomyces cerevisiae protein kinase C homolog. Mol Cell Biol. 1992;12:172–182. doi: 10.1128/mcb.12.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levin D E, Bartlett-Heubusch E. Mutants in the S. cerevisiae PKC1 gene display a cell cycle-specific osmotic stability defect. J Cell Biol. 1992;116:1221–1229. doi: 10.1083/jcb.116.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levin D E, Errede B. The proliferation of MAP kinase signaling pathways in yeast. Curr Opin Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 28.Mazzoni C, Zarov P, Rambourg A, Mann C. The SLT2 (MPK1) MAP kinase homolog is involved in polarized cell growth in Saccharomyces cerevisiae. J Cell Biol. 1993;123:1821–1833. doi: 10.1083/jcb.123.6.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 30.Nonaka H, Tanaka K, Hirano H, Fujiwara T, Kohno H, Umikawa M, Mino A, Takai Y. A downstream target of RHO1 small GTP-binding protein is PKC1, a homolog of protein kinase C, which leads to activation of the MAP kinase cascade in Saccharomyces cerevisiae. EMBO J. 1995;14:5931–5938. doi: 10.1002/j.1460-2075.1995.tb00281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogawa N, Saitoh H, Miura K, Magbanua J P, Bun-ya M, Harashima S, Oshima Y. Structure and distribution of specific cis-elements for transcriptional regulation of PHO84 in Saccharomyces cerevisiae. Mol Gen Genet. 1995;249:406–416. doi: 10.1007/BF00287102. [DOI] [PubMed] [Google Scholar]
- 32.Orr J W, Newton A C. Requirement for negative charge on “activation loop” of protein kinase C. J Biol Chem. 1994;269:27715–27718. [PubMed] [Google Scholar]
- 33.Posas F, Takekawa M, Saito H. Signal transduction by MAP kinase cascades in budding yeast. Curr Opin Microbiol. 1998;1:175–182. doi: 10.1016/s1369-5274(98)80008-8. [DOI] [PubMed] [Google Scholar]
- 34.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 35.Robinson M J, Cobb M H. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–186. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 36.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schmidt A, Kunz J, Hall M N. TOR2 is required for organization of the actin cytoskeleton in yeast. Proc Natl Acad Sci USA. 1996;93:13780–13785. doi: 10.1073/pnas.93.24.13780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stephens L, Anderson K, Stokoe D, Erdjument B H, Painter G F, Holmes A B, Gaffney P R, Reese C B, McCormick F, Tempst P, Coadwell J, Hawkins P T. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science. 1998;279:710–714. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 40.Stevenson B J, Rhodes N, Errede B, Sprague G J. Constitutive mutants of the protein kinase STE11 activate the yeast pheromone response pathway in the absence of the G protein. Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe M, Chen C Y, Levin D E. Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J Biol Chem. 1994;269:16829–16836. [PubMed] [Google Scholar]
- 42.Watanabe Y, Irie K, Matsumoto K. Yeast RLM1 encodes a serum response factor-like protein that may function downstream of the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1995;15:5740–5749. doi: 10.1128/mcb.15.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe Y, Takaesu G, Hagiwara M, Irie K, Matsumoto K. Characterization of a serum response factor-like protein in Saccharomyces cerevisiae, Rlm1, which has transcriptional activity regulated by the Mpk1 (Slt2) mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:2615–2623. doi: 10.1128/mcb.17.5.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi K, Shirakabe K, Shibuya H, Irie K, Oishi I, Ueno N, Taniguchi T, Nishida E, Matsumoto K. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science. 1995;270:2008–2011. doi: 10.1126/science.270.5244.2008. [DOI] [PubMed] [Google Scholar]
- 45.Yashar B, Irie K, Printen J A, Stevenson B J, Sprague G J, Matsumoto K, Errede B. Yeast MEK-dependent signal transduction: response thresholds and parameters affecting fidelity. Mol Cell Biol. 1995;15:6545–6553. doi: 10.1128/mcb.15.12.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]