Abstract
Background
Levofloxacin prophylaxis is recommended to prevent gram-negative bloodstream infections (BSIs) in patients with prolonged chemotherapy-induced neutropenia. However, increasing fluoroquinolone resistance may decrease the effectiveness of this approach.
Methods
We assessed the prevalence of colonization with fluoroquinolone-resistant Enterobacterales (FQRE) among patients admitted for hematopoietic cell transplantation (HCT) from November 2016 to August 2019 and compared the risk of gram-negative BSI between FQRE-colonized and noncolonized patients. All patients received levofloxacin prophylaxis during neutropenia. Stool samples were collected upon admission for HCT and weekly thereafter until recovery from neutropenia, and underwent selective culture for FQRE. All isolates were identified and underwent antimicrobial susceptibility testing by broth microdilution. FQRE isolates also underwent whole-genome sequencing.
Results
Fifty-four of 234 (23%) patients were colonized with FQRE prior to HCT, including 30 of 119 (25%) allogeneic and 24 of 115 (21%) autologous HCT recipients. Recent antibacterial use was associated with FQRE colonization (P = .048). Ninety-one percent of colonizing FQRE isolates were Escherichia coli and 29% produced extended-spectrum β-lactamases. Seventeen (31%) FQRE-colonized patients developed gram-negative BSI despite levofloxacin prophylaxis, compared to only 2 of 180 (1.1%) patients who were not colonized with FQRE on admission (P < .001). Of the 17 gram-negative BSIs in FQRE-colonized patients, 15 (88%) were caused by FQRE isolates that were genetically identical to the colonizing strain.
Conclusions
Nearly one-third of HCT recipients with pretransplant FQRE colonization developed gram-negative BSI while receiving levofloxacin prophylaxis, and infections were typically caused by their colonizing strains. In contrast, levofloxacin prophylaxis was highly effective in patients not initially colonized with FQRE.
Keywords: levofloxacin prophylaxis, fluoroquinolone resistance, neutropenia, hematopoietic cell transplant recipient
Twenty-three percent of 234 hematopoietic cell transplant recipients were colonized with fluoroquinolone-resistant Enterobacterales (FQRE) upon admission for their transplantation. Thirty-one percent of FQRE-colonized patients developed gram-negative bloodstream infection despite levofloxacin prophylaxis, compared to 1.1% of noncolonized patients.
(See the Editorial Commentary by Pergam and Dadwal on pages 1266–7.)
Patients with hematologic malignancies who receive intensive chemotherapy, including those undergoing hematopoietic cell transplantation (HCT), frequently develop severe neutropenia and gastrointestinal mucositis, placing them at high risk of developing bloodstream infections (BSIs) from gram-negative enteric bacteria (Enterobacterales) [1, 2]. Neutropenic patients often suffer severe consequences from BSIs caused by Enterobacterales, with mortality rates as high as 15%–20% [3–5].
Two randomized trials that were conducted >15 years ago demonstrated that prophylactic levofloxacin during neutropenia decreased the risk of fever and gram-negative BSI among patients with cancer [6, 7]. A subsequent meta-analysis demonstrated that fluoroquinolone prophylaxis during neutropenia decreased all-cause mortality [8]. Based on these data, current guidelines recommend fluoroquinolone prophylaxis for patients with hematologic malignancies and HCT recipients who are treated with intensive chemotherapy [2]. However, fluoroquinolone resistance has recently increased among the most common Enterobacterales that cause BSIs in this population [9]. Moreover, many fluoroquinolone-resistant Enterobacterales (FQRE) also harbor extended-spectrum β-lactamases (ESBLs); thus, breakthrough infections that occur despite fluoroquinolone prophylaxis may be resistant to first-line antimicrobial therapies for fever and neutropenia [10–12]. Finally, adverse effects of fluoroquinolones have become increasingly apparent, including Clostridioides difficile infection, aortic dissection and rupture, dysglycemia, tendinopathy, QT interval prolongation, and mental status changes [13–15]. Thus, fluoroquinolones should only be administered to patients when they are likely to provide clinical benefit to justify these potential adverse effects.
We hypothesized that although fluoroquinolones may decrease the risk of gram-negative BSI in many patients, those who are colonized with FQRE may not benefit from fluoroquinolone prophylaxis. We therefore conducted a prospective cohort study of patients receiving HCT for a hematologic malignancy with the following objectives: (1) to determine the prevalence of FQRE colonization on admission for HCT; (2) to compare the risk of gram-negative BSI in patients with and without pretransplant FQRE colonization; and (3) to compare the genetic relatedness between colonizing and bloodstream FQRE isolates among bacteremic patients.
MATERIALS AND METHODS
Study Population
We conducted this prospective observational study at an 862-bed academic medical center in New York City. We enrolled adult patients (≥18 years of age) who were hospitalized for an autologous or allogeneic HCT between November 2016 and August 2019 and who received levofloxacin prophylaxis. The study was approved by the Institutional Review Board at Weill Cornell Medicine (number 1504016114) and written informed consent was obtained from study participants. Levofloxacin prophylaxis was initiated 1 day before stem cell infusion at a dosage of 500 mg daily and continued until recovery from neutropenia or until fever and neutropenia. Trimethoprim-sulfamethoxazole (TMP-SMX) was administered to allogeneic HCT recipients from 4 to 2 days prior to stem cell infusion. If fever and neutropenia occurred, 2 sets of blood cultures were collected, levofloxacin was discontinued, and an anti-pseudomonal β-lactam agent (most commonly piperacillin-tazobactam) was initiated.
For each participant, we collected data on demographics, underlying malignancy, stem cell source, conditioning regimens, graft-vs-host disease (GVHD) prophylaxis, American Society of Bone Marrow Transplantation (ASBMT) risk category [16], hospitalizations and antibacterial therapies within 90 days prior to transplantation, and history of colonization or infection with resistant enteric bacteria.
We also recorded episodes of fever and neutropenia, defined as a temperature ≥38.0°C with an absolute neutrophil count (ANC) ≤500 cells/µL, and BSIs that occurred during neutropenia. Coagulase-negative staphylococci and other common skin commensals were only considered causes of BSI if isolated from ≥2 separate sets of blood cultures. Any Enterobacterales that were isolated in blood cultures for routine clinical care were stored at –80°C for genotypic analysis.
Specimen Collection and Microbiologic Analysis
We collected a stool specimen upon admission for transplantation and weekly thereafter until recovery from neutropenia (the first day of 2 consecutive days with an ANC ≥500 cells/µL). Stool specimens were collected in FisherBrand Commode Specimen Collection kits (Thermo Fisher Scientific, Waltham, Massachusetts) and stored at 4°C immediately upon collection. A 1-µL loopful of stool was plated onto MacConkey agar that was supplemented with 1 µg/mL of ciprofloxacin within 24 hours of sample collection (Hardy Diagnostics, Santa Maria, California). The agar plate was incubated at 37°C in ambient air for 24 hours. Colonies identified after incubation were subcultured onto BD trypticase soy agar II plates (Becton, Dickinson, and Company, Franklin Lakes, New Jersey) and incubated overnight in ambient air.
The next day, colonies from these plates were identified and underwent antimicrobial susceptibility testing using the MicroScan Walkaway plus System (Panel NM43, Beckman Coulter, Brea, California). If the MicroScan system indicated an identification probability of <95%, organism identification was confirmed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Bruker Daltonics, Billerica, Massachusetts). Interpretive criteria of the Clinical and Laboratory Standards Institute (CLSI) in 2018 were applied [17]. Isolates were considered fluoroquinolone-resistant if resistant to either ciprofloxacin (minimum inhibitory concentration [MIC] ≥4 µg/mL) or levofloxacin (MIC ≥8 µg/mL). Levofloxacin susceptibility testing using Etest strips (bioMérieux, Durham, North Carolina) was also performed on FQRE. Additionally, ceftriaxone- and fluoroquinolone-resistant Escherichia coli, Klebsiella pneumoniae, Klebsiella oxytoca, and Proteus mirabilis isolates underwent phenotypic testing for ESBL production using cefotaxime and ceftazidime disks, with and without clavulanate [17].
When stool samples were not available for analysis, a perianal swab (BD ESwab) was collected and processed instead of the stool sample. Instead of inoculating stool onto the MacConkey agar plate with ciprofloxacin, we inoculated 100 µL of Liquid Amies from the perianal swab onto this agar plate. Subsequent methods to identify FQRE colonization were identical to the analysis of stool samples.
Whole-Genome Sequencing of FQRE
All initial colonizing FQRE isolates and all subsequent bloodstream FQRE isolates underwent whole-genome sequencing (WGS) using Illumina Hiseq platform (Illumina, San Diego, California) with 2 × 150-bp (base pair) paired-end reads. Illumina raw sequencing reads were trimmed using Trimmomatic version 0.39 [18], followed by de novo assembly, using SPAdes version 3.14.1 [19]. The multilocus sequence type (MLST) was determined in silico using MLST (https://github.com/tseemann/mlst), while the acquired resistance genes were determined using AMRFinderPlus 3.9 and ARIBA 2.14.6 [20]. Amino acid substitutions in the quinolone resistance–determining region in gyrA, gyrB, parC, and parE were mined using BLAST. Recombination free core SNP phylogenetic analysis was conducted using Snippy version 4.6 (https://github.com/tseemann/snippy) with default settings.
Statistical Analyses
We first determined the proportion of patients who were colonized with FQRE prior to HCT. Factors associated with pretransplant FQRE colonization were assessed by comparing characteristics of FQRE-colonized patients to those not colonized with FQRE, using Fisher exact or χ 2 for categorical variables and the Wilcoxon rank-sum test for continuous variables. We then compared the cumulative incidence of developing a FQRE BSI, other BSI types, and fever and neutropenia during the transplant admission, as well as 100-day mortality and risk of acute GVHD, between patients with and without pretransplant FQRE colonization using Fisher exact or χ 2 tests. Among patients not initially colonized with FQRE, we assessed the frequency of FQRE acquisition during the transplant admission, including only patients who had a subsequent swab collected within 1 week of neutrophil recovery. Stata version 15.0 software (StataCorp, College Station, Texas) was used for statistical analyses and 2-tailed P values ≤ .05 were considered statistically significant.
RESULTS
Patients and Pretransplant Colonization Status
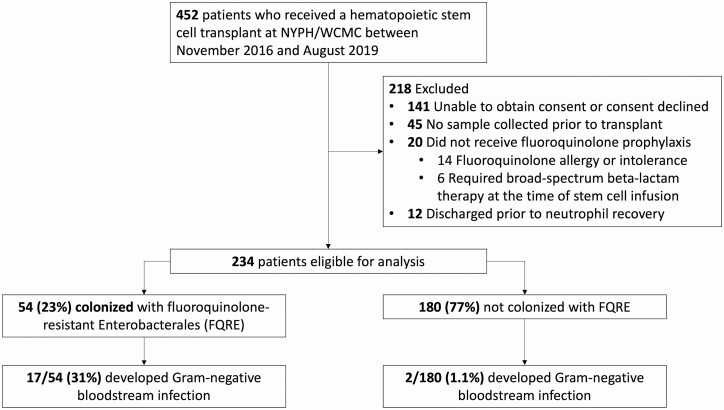
Of 452 patients who received a HCT during the study period, 234 (52%) met criteria to be included in this analysis (Figure 1). The median patient age was 61 years, 44% were women, and 51% received an allogeneic HCT. The most common underlying malignancies were acute myeloid leukemia/myelodysplastic syndrome (31%), multiple myeloma (30%), and non-Hodgkin lymphoma (19%). The median duration of neutropenia was 9 days (interquartile range [IQR], 7–12 days). The first specimen was collected a median of 2 days prior to transplantation (IQR, 1–4 days).
Figure 1.
Flow diagram of patients included in the study, prevalence of fluoroquinolone-resistant Enterobacterales (FQRE) colonization, and risk of gram-negative bloodstream infection in patients colonized and not colonized with FQRE. Abbreviation: NYPH/WCMC, New York–Presbyterian Hospital/Weill Cornell Medical Center.
Fifty-four (23%) patients were colonized with FQRE prior to transplantation (Figure 1), including 21% of autologous and 25% of allogeneic HCT recipients. Pretransplant colonization with FQRE was not associated with age, sex, race, ethnicity, underlying malignancy, ASBMT risk category, prior transplantation, stem cell source, conditioning regimen, recent hospitalizations, or history of positive microbiologic tests for resistant bacteria (Table 1). Seventy-eight percent of FQRE-colonized patients had received an antibacterial agent within the previous 90 days, compared to 63% of patients not colonized with FQRE (P = .048). Prior exposure to TMP-SMX was associated with FQRE colonization, but prior use of β-lactam agents or fluoroquinolones was not. Of the 54 patients colonized with FQRE prior to their transplantation, only 4 (7%) had a prior positive culture for FQRE.
Table 1.
Characteristics of Hematopoietic Cell Transplant Recipients Colonized and Not Colonized With Fluoroquinolone-Resistant Enterobacterales
| Characteristic | FQRE Colonized (n = 54) |
Not Colonized With FQRE (n = 180) | P Value |
|---|---|---|---|
| Demographics | |||
| Age, y, median (IQR) | 61 (50–66) | 61 (52–67) | .85 |
| Female sex | 21 (40) | 81 (45) | .43 |
| Race/ethnicity | |||
| White | 42 (79) | 138 (77) | .87 |
| Black | 9 (17) | 29 (16) | .92 |
| Asian | 3 (6) | 13 (7) | 1.00 |
| Hispanic | 6 (11) | 17 (9) | .72 |
| Underlying malignancy | |||
| AML or MDS | 17 (31) | 55 (31) | .90 |
| ALL | 5 (9) | 16 (9) | 1.00 |
| Multiple myeloma | 13 (24) | 57 (32) | .29 |
| Non-Hodgkin lymphoma | 13 (24) | 32 (18) | .30 |
| Hodgkin lymphoma | 1 (2) | 10 (6) | .46 |
| Other malignancy | 5 (9) | 10 (6) | .35 |
| ASBMT risk category [16] | |||
| Low risk | 28 (52) | 93 (52) | .98 |
| Medium risk | 10 (19) | 27 (15) | .53 |
| High risk | 11 (20) | 46 (26) | .44 |
| NA | 5 (9) | 14 (8) | .78 |
| Prior transplantation | 3 (6) | 14 (8) | .77 |
| Stem cell source | |||
| Autologous | 24 (44) | 91 (51) | .43 |
| Allogeneic | 30 (56) | 89 (49) | .43 |
| Matched related donor | 7 (13) | 20 (11) | .71 |
| Matched unrelated donor | 10 (19) | 31 (17) | .83 |
| Haploidentical-cord blood | 12 (22) | 35 (19) | .66 |
| Other | 1 (2) | 3 (2) | 1.00 |
| Conditioning regimena | |||
| Fludarabine-melphalan-TBIb | 19 (35) | 52 (29) | .38 |
| Melphalan | 10 (19) | 44 (24) | .37 |
| Carmustine-etoposide- cytarabine-melphalan | 10 (19) | 26 (14) | .47 |
| Fludarabine-melphalan | 6 (11) | 19 (11) | .91 |
| Lenalidomide-melphalan | 2 (4) | 11 (6) | .74 |
| Etoposide-TBIc | 2 (4) | 8 (4) | 1.00 |
| Other regimen | 5 (9) | 20 (11) | .70 |
| Use of rituximab in conditioning [33] | 19 (35) | 46 (26) | .17 |
| Myeloablative allogeneic HCT | 5 (9) | 16 (9) | .93 |
| Reduced-intensity conditioning for allogeneic HCT | 25 (46) | 73 (41) | .87 |
| Anti–T-cell therapies for GVHD prophylaxis | |||
| Antithymocyte globulin | 12 (22) | 36 (20) | .72 |
| Alemtuzumab | 16 (30) | 47 (26) | .61 |
| Hospitalization within previous 90 d | 30 (56) | 90 (50) | .47 |
| Antibacterial within previous 90 d | 42 (78) | 114 (63) | .048 |
| β-lactam agent | 24 (44) | 69 (38) | .42 |
| Fluoroquinolone | 19 (35) | 56 (31) | .57 |
| Trimethoprim-sulfamethoxazole | 22 (41) | 43 (24) | .015 |
| Macrolide | 1 (2) | 13 (7) | .20 |
| Intravenous vancomycin | 8 (15) | 16 (9) | .21 |
| History of positive microbiologic tests for specified bacteria | |||
| FQRE | 4 (7) | 8 (4) | .39 |
| Ceftriaxone-resistant Enterobacterales | 2 (4) | 2 (1) | .23 |
| Vancomycin-resistant enterococci | 5 (9) | 6 (3) | .13 |
| Clostridioides difficile | 5 (9) | 9 (5) | .25 |
Data are presented as No. (%) unless otherwise indicated. Bolded P values indicate statistical significance.
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ASBMT, American Society of Bone Marrow Transplantation; FQRE, fluoroquinolone-resistant Enterobacterales; GVHD, graft-vs-host disease; HCT, hematopoietic cell transplantation; IQR, interquartile range; MDS, myelodysplastic syndrome; NA, not applicable; TBI, total body irradiation.
aNo transplant recipients with nonmyeloablative conditioning regimens were enrolled in this study.
bThis regimen used 200–400 cGray of TBI.
cThis regimen used 12 Gray of TBI.
Colonizing FQRE Strains
There were 56 colonizing isolates of FQRE harbored by 54 FQRE-colonized patients, of which 51 (91%) were E. coli (Table 2). Fifty of the 56 (89%) FQRE isolates had levofloxacin MIC values >32 µg/mL. All FQRE isolates were susceptible to piperacillin-tazobactam, carbapenems, and amikacin. Approximately two-thirds of FQRE isolates were susceptible to amoxicillin-clavulanate, aztreonam, ceftriaxone, ceftazidime, and cefepime, and approximately three-quarters were susceptible to gentamicin and tobramycin. Sixteen (29%) FQRE isolates produced an ESBL on phenotypic testing and blaCTX-M was detected in 14 of these isolates.
Table 2.
Characteristics of Colonizing Fluoroquinolone-Resistant Enterobacterales Isolates Prior to Transplant
| Organism Type | No. (% of Total of 56 Isolates) |
|---|---|
| Species and most common MLST | |
| Escherichia coli | 51 (89) |
| ST131 | 23 |
| ST1193 | 9 |
| Klebsiella pneumoniae | 3 (5) |
| Citrobacter freundii | 1 (2) |
| Enterobacter cloacae | 1 (2) |
| Antimicrobial susceptibilities, % susceptible | |
| Amikacin | 100 |
| Amoxicillin-clavulanate | 70 |
| Ampicillin-sulbactam | 34 |
| Ampicillin | 16 |
| Aztreonam | 68 |
| Cefazolin | 34 |
| Cefepime | 70 |
| Cefoxitin | 88 |
| Ceftazidime | 71 |
| Ceftriaxone | 66 |
| Cefuroxime | 59 |
| Ciprofloxacin | 0 |
| Ertapenem | 100 |
| Gentamicin | 79 |
| Levofloxacin | 5 |
| Meropenem | 100 |
| Piperacillin-tazobactam | 100 |
| Tobramycin | 75 |
| Trimethoprim-sulfamethoxazole | 36 |
| Fluoroquinolone resistance determinantsa | |
| Chromosomal mutations | |
| gyrA mutations | 48 (87) |
| Mutation at codon 83 | 48 (87) |
| Mutation at codon 87 | 47 (86) |
| parC mutations | 50 (91) |
| Mutation at codon 80 | 49 (89) |
| Mutation at codon 84 | 21 (38) |
| parE mutations | 41 (75) |
| 5 chromosomal mutations | 20 (36) |
| 4 chromosomal mutations | 22 (40) |
| 3 chromosomal mutations | 5 (9) |
| 2 chromosomal mutations | 1 (2) |
| 1 chromosomal mutations | 2 (4) |
| No chromosomal mutations | 5 (9) |
| Plasmid-mediated resistance determinants | |
| Qnr | 9 (16) |
| aac(6’)-Ib-cr | 4 (7) |
| Oqx b | 2 (4) |
| Notable β-lactamase genesa | |
| ESBL genes: blaCTX-Mc,d | 14 (25) |
| AmpC-encoding genese | 4 (7) |
| Narrow-spectrum β-lactamase genes | 33 (60) |
| bla TEM-1 | 28 (51) |
| blaSHV-1 or blaSHV-11 | 2 (4) |
| bla OXA-1 | 4 (7) |
Abbreviations: ESBL, extended-spectrum β-lactamase; MLST, multilocus sequence type; ST, sequence type.
aThe genetic characterizations are based on 55 isolates that underwent whole-genome sequencing.
bThree additional Klebsiella pneumoniae strains harbored chromosomal oqxA genes, but these genes do not confer fluoroquinolone resistance in K. pneumoniae.
cTwo strains tested positive for ESBL production by phenotypic testing, but no ESBL-encoding genes were identified by sequencing. blaCTX-M was the only type of ESBL gene identified.
dThe most common CTX-M variants were CTX-M-27 (n = 9), CTX-M-15 (n = 3), CTX-M-14 (n = 1), and CTX-M-55 (n = 1).
eThe most common AmpC-encoding genes were blaCMY-2 (n = 2), blaCMY-55 and blaDHA-1 (n = 1), and blaACT-17 (n = 1).
The most common sequence types (STs) among the 50 E. coli that underwent WGS were ST131 (n = 23 [46%]) and ST1193 (n = 9 [18%]; Table 2). In total, 14 different STs were represented by these colonizing E. coli strains. Fifty of the 55 (91%) FQRE isolates that underwent WGS had a chromosomal mutation in at least 1 DNA topoisomerase gene (gyrA, parC, and parE), with 76% of strains having ≥4 different mutations in these genes. Plasmid-mediated fluoroquinolone resistance determinants were less common: 16%, 7%, and 4% of strains had qnr, aac(6′)-Ib-cr, and oqx, respectively.
Risk of Bloodstream Infections and Other Outcomes
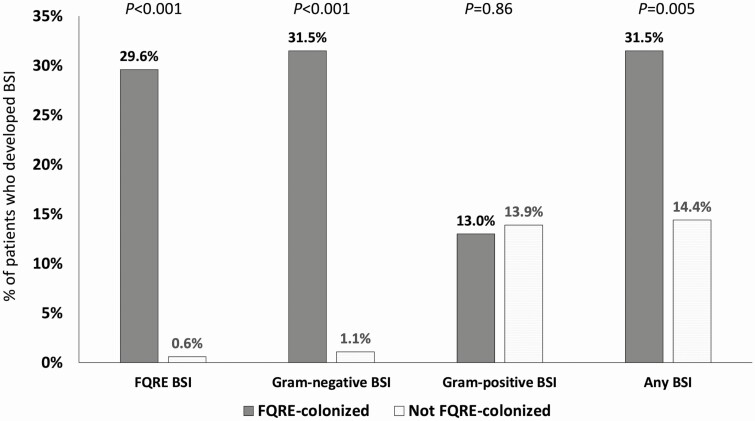
Of 54 patients colonized with FQRE prior to HCT, 17 (31%) developed gram-negative BSIs during their transplant admission, compared to 2 of 180 (1.1%) without pretransplant FQRE colonization (P < .001; Figure 2). All but 1 of the gram-negative BSIs in patients with pretransplant FQRE colonization were caused by FQRE. Unlike gram-negative BSIs, similar proportions of FQRE-colonized (13%) and FQRE-noncolonized (14%) HCT recipients developed gram-positive BSIs. Overall, 31% of FQRE-colonized patients developed any BSI, compared to 14% of patients not colonized with FQRE (P = .005). Fever and neutropenia occurred in 72% of FQRE-colonized patients, compared to 56% of patients not colonized with FQRE (P = .034). There were no differences in 100-day mortality (9% vs 7%; P = .55) or acute GVHD (allogeneic only: 20% vs 27%; P = .45) in patients colonized with FQRE compared to those not colonized with FQRE.
Figure 2.
Proportion of patients who developed fluoroquinolone-resistant Enterobacterales (FQRE) bloodstream infection (BSI), gram-negative BSI, gram-positive BSI, and any BSI, stratified by pretransplant FQRE colonization status. P values represent comparisons between FQRE-colonized and noncolonized patients.
Thirteen of 30 (43%) FQRE-colonized allogeneic HCT recipients developed FQRE BSI, compared to only 3 of 24 (13%) autologous HCT recipients (P = .014; Table 3). Other than type of conditioning regimen (which is linked to transplant type), no other variables were statistically significantly associated with FQRE BSI among FQRE-colonized patients, including demographics, underlying malignancy, ASBMT risk category, GVHD prophylaxis, duration of neutropenia, and FQRE isolate characteristics.
Table 3.
Characteristics of Patients Colonized With Fluoroquinolone-Resistant Enterobacterales (FQRE) Who Did and Did Not Develop FQRE Bloodstream Infection
| Characteristic | FQRE BSI (n = 16) |
No FQRE BSI (n = 38) |
P Value |
|---|---|---|---|
| Demographics | |||
| Age, y, median (IQR) | 61 (50–65) | 61 (55–68) | .53 |
| Female sex | 7 (44) | 14 (37) | .63 |
| Race/ethnicity | |||
| White | 13 (81) | 29 (76) | .69 |
| Black | 3 (19) | 6 (16) | 1.00 |
| Asian | 0 | 3 (8) | .66 |
| Hispanic | 2 (13) | 4 (11) | 1.00 |
| Underlying malignancy | |||
| AML or MDS | 8 (50) | 9 (24) | .057 |
| ALL | 1 (6) | 4 (10) | 1.00 |
| Multiple myeloma | 1 (6) | 12 (32) | .079 |
| Non-Hodgkin lymphoma | 3 (19) | 10 (26) | .73 |
| Hodgkin lymphoma | 0 | 1 (3) | 1.00 |
| Other malignancy | 3 (19) | 2 (5) | .15 |
| ASBMT risk category [16] | |||
| Low risk | 9 (56) | 19 (50) | .68 |
| Medium risk | 1 (6) | 9 (24) | .25 |
| High risk | 4 (25) | 7 (18) | .71 |
| NA | 2 (13) | 3 (8) | .63 |
| Prior transplantation | |||
| Stem cell source | |||
| Autologous | 3 (19) | 21 (55) | .014 |
| Allogeneic | 13 (81) | 17 (45) | .014 |
| Matched related donor | 2 (13) | 5 (13) | 1.00 |
| Matched unrelated donor | 5 (31) | 5 (13) | .14 |
| Haploidentical-cord blood | 6 (38) | 6 (16) | .15 |
| Other | 0 | 1 (3) | 1.00 |
| Conditioning regimen | |||
| Fludarabine-melphalan-TBI | 8 (50) | 11 (29) | .14 |
| Melphalan | 0 | 10 (26) | .024 |
| Carmustine-etoposide-cytarabine-melphalan | 3 (19) | 7 (18) | 1.00 |
| Fludarabine-melphalan | 3 (19) | 3 (8) | .35 |
| Lenalidomide-melphalan | 0 | 2 (5) | 1.00 |
| Etoposide-TBI | 1 (6) | 1 (3) | .51 |
| Other regimen | 1 (6) | 4 (11) | 1.00 |
| Use of rituximab in conditioning | 8 (50) | 11 (29) | .14 |
| Anti–T-cell therapies for GVHD prophylaxis | |||
| Antithymocyte globulin | 5 (31) | 7 (18) | .31 |
| Alemtuzumab | 8 (50) | 8 (21) | .051 |
| Colonizing FQRE characteristics | |||
| Escherichia coli | 15 (94) | 34 (89) | 1.00 |
| ST131 | 8 (50) | 14 (38) | .41 |
| Levofloxacin MIC >32 µg/mL | 14 (88) | 35 (92) | .59 |
| ESBL producer | 4 (25) | 10 (26) | |
| Duration of neutropenia, d | 9 (9–11) | 8 (6–12) | .22 |
Data are presented as No. (%) unless otherwise indicated. Bolded P values indicate statistical significance.
Abbreviations: ALL, acute lymphoblastic leukemia; AML, acute myeloid leukemia; ASBMT, American Society of Bone Marrow Transplantation; BSI, bloodstream infection; ESBL, extended-spectrum β-lactamase; FQRE, fluoroquinolone-resistant Enterobacterales; GVHD, graft-vs-host disease; IQR, interquartile range; MDS, myelodysplastic syndrome; MIC, minimum inhibitory concentration; NA, not applicable; ST, sequence type; TBI, total body irradiation.
Comparison of Colonizing and Bloodstream FQRE Isolates
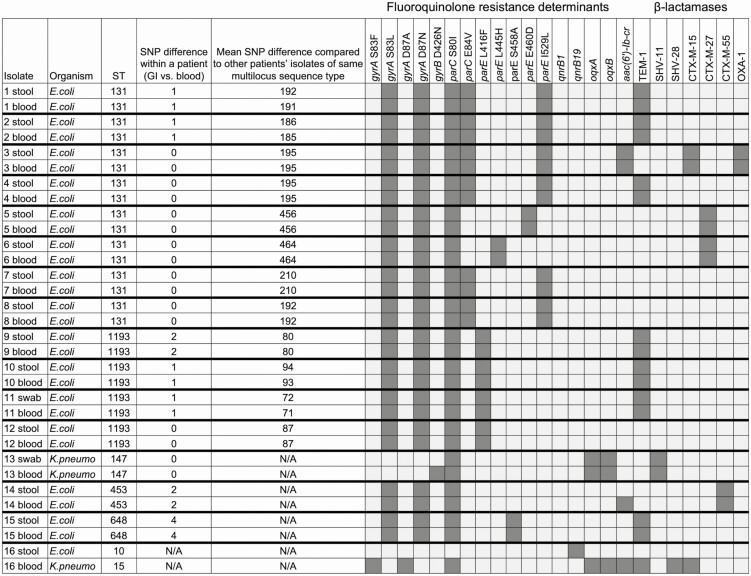
The species of the bloodstream FQRE pathogen was identical to the species of the colonizing FQRE isolate in 15 of 16 (94%) cases where a FQRE-colonized patient developed FQRE BSI. For the 15 pairs of colonizing and bloodstream FQRE isolates that were the same species, all had matching STs (Figure 3). For these pairs, the number of single-nucleotide polymorphism (SNP) differences by WGS was 0–4 bp, suggesting that in each pair the bloodstream isolate and the colonizing FQRE isolate were the same strain. In contrast, the mean number of SNP differences compared to other patients’ colonizing or bloodstream FQRE isolates of the same ST ranged from 71 to 456 bp.
Figure 3.
Comparison of organisms, multilocus sequence types, single-nucleotide polymorphism differences, fluoroquinolone resistance determinants, and β-lactamase genes among stool and bloodstream fluoroquinolone-resistant Enterobacterales (FQRE) isolates among the 16 FQRE-colonized patients who developed FQRE bloodstream infection. Abbreviations: E. coli, Escherichia coli; GI, gastrointestinal; K. pneumo, Klebsiella pneumoniae; NA, not applicable; SNP, single-nucleotide polymorphism; ST, sequence type.
Risk of FQRE Acquisition in Patients Not Initially Colonized With FQRE
Of the 180 patients who were not initially colonized with FQRE upon admission for their transplantation, 170 had subsequent samples (a median of 2 additional samples) collected to determine whether they acquired FQRE during their transplant admission. Eleven of these 170 (6.5%) patients acquired FQRE and this occurred a median of 2 days after their stem cell infusion. The acquired FQRE isolates were E. coli (n = 8), Citrobacter freundii, Enterobacter cloacae, and K. pneumoniae (n = 1 each). Only 1 of the patients who acquired FQRE developed FQRE BSI.
DISCUSSION
Levofloxacin prophylaxis has become a standard of care in patients with prolonged and profound neutropenia, such as patients undergoing HCT, to prevent life-threatening gram-negative BSIs [2]. We found that nearly one-third of patients who were colonized with FQRE prior to their transplantation developed a gram-negative BSI while receiving levofloxacin prophylaxis. Furthermore, the breakthrough gram-negative bloodstream isolates were almost always genetically identical to the colonizing FQRE isolates. In contrast, only 1% of patients who were not colonized with FQRE prior to their transplantation developed gram-negative BSI while receiving levofloxacin prophylaxis. Therefore, we believe that screening for pretransplant FQRE colonization can (1) identify patients who are at high risk of developing breakthrough gram-negative BSI while receiving levofloxacin prophylaxis during neutropenia; (2) predict the likely etiologies of gram-negative BSIs in FQRE-colonized patients who receive levofloxacin prophylaxis; and (3) identify patients who are very unlikely to develop gram-negative BSI while receiving levofloxacin prophylaxis.
We found that 23% of patients admitted for HCT were colonized with FQRE and E. coli was the predominant species. The only other published report of the prevalence of FQRE colonization in patients with hematologic malignancies found that 11 of 68 (16%) patients at a Japanese center were colonized with fluoroquinolone-resistant E. coli [21]. Our data suggest that assessing clinical characteristics will not reliably identify HCT recipients who are colonized with FQRE. Among the many characteristics analyzed, only recent use of antibacterial agents and TMP-SMX in particular were associated with FQRE colonization. Furthermore, we found that relying on prior culture information is unreliable, as only 7% of FQRE-colonized patients had previously had FQRE detected in clinical cultures. Thus, the only way to identify FQRE-colonized patients is to screen for FQRE colonization.
The high rate and absence of risk factors for FQRE colonization suggest that FQRE are prevalent in the community. Indeed, a study of urinary isolates among outpatients in the United States demonstrated that 12% of E. coli isolates from young women and 29% of E. coli isolates from elderly women were fluoroquinolone resistant [22]. A surveillance study of 1831 urinary E. coli isolates from 2017 found that one-quarter were FQ resistant [23]. Furthermore, 13%–16% of men undergoing transrectal prostate biopsies were found to be colonized with fluoroquinolone-resistant E. coli [24, 25]. Thus, we hypothesize that many HCT recipients acquire their colonizing FQRE isolates in the community. In contrast, we found that acquisition of FQRE in the hospital was uncommon. Despite receiving levofloxacin prophylaxis, only 7% of patients who were not initially colonized with FQRE acquired FQRE during their transplant admission.
The colonizing FQRE isolates were typically highly resistant to levofloxacin, with 89% having MIC values >32 µg/mL. These levofloxacin MICs likely exceed the fecal levofloxacin concentrations of 15–94 µg/g observed among patients who receive 500 mg of oral levofloxacin daily [26]. The vast majority of colonizing FQRE isolates had mutations in chromosomal gyrA, parC, and parE genes. In contrast, plasmid-mediated fluoroquinolone resistance determinants were uncommon. This has important implications for designing molecular diagnostic tests to detect colonization with FQRE. Unlike ESBL-producing Enterobacterales or vancomycin-resistant enterococci, where a single gene target may be used to detect the resistant pathogen [27], the detection of the mere presence or absence of a genetic resistance determinant will not suffice to detect FQRE colonization. Instead, a molecular diagnostic assay will need to detect common gyrA, parC, and parE mutations that confer fluoroquinolone resistance among Enterobacterales.
Nearly 30% of colonizing FQRE produced an ESBL and a similar percentage were resistant to cefepime and ceftazidime, agents relied upon for fever and neutropenia [1]. Moreover, many of the gram-negative BSIs were ESBL producers that were resistant to these cephalosporins. This co-resistance between fluoroquinolones and anti-pseudomonal cephalosporins is similar to that observed in a surveillance study of urinary E. coli isolates in the United States, where one-quarter of levofloxacin-resistant isolates were also resistant to ceftazidime [23]. Nearly one-half of fluoroquinolone-resistant E. coli isolates in our study were ST131, a common sequence type that has spread throughout the world and whose isolates are frequently fluoroquinolone resistant and ESBL producers [28, 29]. However, the expansion of FQRE in our cohort was not of clonal origin. There was marked genetic diversity among colonizing FQRE, with 14 different E. coli sequence types represented. Even among ST131 E. coli, strains had hundreds of SNP differences and a variety of resistance determinants.
Patients undergoing HCT are only 1 of multiple types of patients with hematologic malignancies who receive intensive chemotherapy for whom fluoroquinolone prophylaxis is considered. We do not know if our findings will apply to patients with leukemia or lymphoma who receive intensive chemotherapy. Furthermore, our study was conducted at a single center in New York City, and we do not know the degree to which our results are generalizable to other oncology centers. To address these limitations, we are beginning a multicenter observational study of FQRE colonization and risk of gram-negative BSI among HCT recipients and patients with leukemia who receive induction chemotherapy (National Institute of Allergy and Infectious Diseases grant numbers R01 AI151038 and UM1 AI104681). Another limitation is that CLSI recently lowered fluoroquinolone breakpoints for Enterobacterales [30] and thus there may have been FQRE that were not identified in this study because we applied the prior breakpoints. However, surveillance data suggest that only an additional 3%–4% of Enterobacterales isolates would be levofloxacin resistant according to the updated breakpoints [31]. Thus, we do not expect that the prevalence of FQRE colonization would markedly increase using these new breakpoints.
We believe our findings establish a foundation for investigating novel approaches to antibacterial prophylaxis in neutropenic patients, where the choice of prophylactic agent is based on the resistance profile of colonizing enteric bacteria. For the majority of patients who are not colonized with FQRE, our data suggest that fluoroquinolone prophylaxis is highly effective in preventing gram-negative BSI. However, for the substantial proportion of patients colonized with FQRE, our data indicate that other prophylactic approaches are necessary. Potential prophylactic strategies for FQRE-colonized patients that warrant further investigation include oral cefpodoxime if the colonizing isolate is not an ESBL producer and either a carbapenem or omission of antibacterial prophylaxis if the isolate is an ESBL producer [32]. Additional research is needed to evaluate the optimal prophylactic strategies in FQRE-colonized patients with hematologic malignancies who receive intensive chemotherapy.
Notes
Acknowledgments. The authors thank the patients at Weill Cornell Medicine who chose to participate in this study.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (grant numbers R03 AI146612 and K23 AI114994 to M. J. S. and R01 AI090155 to B. N. K.).
Potential conflicts of interest. M. J. S. has received grant support from Allergan, BioFire Diagnostics, and Merck and has received consulting fees from Achaogen and Shionogi. C. B. S. has received grant support from GlaxoSmithKline, ViiV, Abbott, Merck, Gilead, Chimerix, Shire/Takeda, Schering-Plough, Ablynx, Janssen, Ansun Biopharma, and Karyopharm Therapeutics. L. F. W. has received grant support from Accelerate Diagnostics, Hardy Diagnostics, BioFire Diagnostics, and Roche Molecular Systems and has received consulting fees from Roche Molecular Systems, Shionogi, and Talis Biomedical. T. J. W has received grant support from Allergan, The Medicines Company, Merck, Shionogi, and Tetraphase and consulting fees from Allergan, The Medicines Company, Merck, Shionogi, Pfizer, and ContraFect. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Freifeld AG, Bow EJ, Sepkowitz KA, et al. ; Infectious Diseases Society of America. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis 2011; 52:427–31. [DOI] [PubMed] [Google Scholar]
- 2.Taplitz RA, Kennedy EB, Bow EJ, et al. . Antimicrobial prophylaxis for adult patients with cancer-related immunosuppression: ASCO and IDSA clinical practice guideline update. J Clin Oncol 2018; 36:3043–54. [DOI] [PubMed] [Google Scholar]
- 3.Klastersky J, Ameye L, Maertens J, et al. . Bacteraemia in febrile neutropenic cancer patients. Int J Antimicrob Agents 2007; 30(Suppl 1):S51–9. [DOI] [PubMed] [Google Scholar]
- 4.Gudiol C, Bodro M, Simonetti A, et al. . Changing aetiology, clinical features, antimicrobial resistance, and outcomes of bloodstream infection in neutropenic cancer patients. Clin Microbiol Infect 2013; 19:474–9. [DOI] [PubMed] [Google Scholar]
- 5.Girmenia C, Bertaina A, Piciocchi A, et al. ; Gruppo Italiano Trapianto di Midollo Osseo (GITMO) and Associazione Microbiologi Clinici Italiani (AMCLI) . Incidence, risk factors and outcomes of pre-engraftment gram-negative bacteremia after allogeneic and autologous hematopoietic stem cell transplantation: an Italian prospective multicenter survey. Clin Infect Dis 2017; 65:1884–96. [DOI] [PubMed] [Google Scholar]
- 6.Bucaneve G, Micozzi A, Menichetti F, et al. ; Gruppo Italiano Malattie Ematologiche dell’Adulto (GIMEMA) Infection Program . Levofloxacin to prevent bacterial infection in patients with cancer and neutropenia. N Engl J Med 2005; 353:977–87. [DOI] [PubMed] [Google Scholar]
- 7.Cullen M, Steven N, Billingham L, et al. ; Simple Investigation in Neutropenic Individuals of the Frequency of Infection after Chemotherapy +/- Antibiotic in a Number of Tumours (SIGNIFICANT) Trial Group . Antibacterial prophylaxis after chemotherapy for solid tumors and lymphomas. N Engl J Med 2005; 353:988–98. [DOI] [PubMed] [Google Scholar]
- 8.Gafter-Gvili A, Fraser A, Paul M, et al. . Antibiotic prophylaxis for bacterial infections in afebrile neutropenic patients following chemotherapy. Cochrane Database Syst Rev 2012; 1:CDC004386. [DOI] [PubMed] [Google Scholar]
- 9.See I, Freifeld AG, Magill SS. Causative organisms and associated antimicrobial resistance in healthcare-associated, central line-associated bloodstream infections from oncology settings, 2009–2012. Clin Infect Dis 2016; 62:1203–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Satlin MJ, Chavda KD, Baker TM, et al. . Colonization with levofloxacin-resistant extended-spectrum β-lactamase-producing Enterobacteriaceae and risk of bacteremia in hematopoietic stem cell transplant recipients. Clin Infect Dis 2018; 67:1720–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rangaraj G, Granwehr BP, Jiang Y, Hachem R, Raad I. Perils of quinolone exposure in cancer patients: breakthrough bacteremia with multidrug-resistant organisms. Cancer 2010; 116:967–73. [DOI] [PubMed] [Google Scholar]
- 12.Kern WV, Weber S, Dettenkofer M, et al. ; Hospital Infection Surveillance System for Patients with Hematologic/Oncologic Malignancies Study Group (ONKO-KISS). Impact of fluoroquinolone prophylaxis during neutropenia on bloodstream infection: data from a surveillance program in 8755 patients receiving high-dose chemotherapy for hematologic malignancies between 2009 and 2014. J Infect 2018; 77:68–75. [DOI] [PubMed] [Google Scholar]
- 13.McDonald LC, Killgore GE, Thompson A, et al. . An epidemic, toxin gene-variant strain of Clostridium difficile. N Engl J Med 2005; 353:2433–41. [DOI] [PubMed] [Google Scholar]
- 14.Mehlhorn AJ, Brown DA. Safety concerns with fluoroquinolones. Ann Pharmacother 2007; 41:1859–66. [DOI] [PubMed] [Google Scholar]
- 15.US Food and Drug Administration. FDA Drug Safety Communication: FDA updates warnings for oral and injectable fluoroquinolone antibiotics due to disabling side effects. Available at: https://www.fda.gov/drugs/drug-safety-and-availability/fda-drug-safety-communication-fda-updates-warnings-oral-and-injectable-fluoroquinolone-antibiotics. Accessed 3 November 2020.
- 16.American Society for Transplantation and Cellular Therapy. ASBMT RFI 2017—disease classifications corresponding to CIBMTR classifications. Available at: https://www.astct.org/asbmt/practice-resources/rfi-forms. Accessed 6 April 2021.
- 17.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 28th informational supplement. CLSI M100-S28. Wayne, PA: CLSI, 2018. [Google Scholar]
- 18.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bankevich A, Nurk S, Antipov D, et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunt M, Mather AE, Sánchez-Busó L, et al. . ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom 2017; 3:e000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chong Y, Shimoda S, Yakushiji H, et al. . Clinical impact of fluoroquinolone-resistant Escherichia coli in the fecal flora of hematological patients with neutropenia and levofloxacin prophylaxis. PLoS One 2014; 9:e85210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez GV, Babiker A, Master RN, Luu T, Mathur A, Bordon J. Antibiotic resistance among urinary isolates from female outpatients in the United States in 2003 and 2012. Antimicrob Agents Chemother 2016; 60:2680–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Critchley IA, Cotroneo N, Pucci MJ, Mendes R. The burden of antimicrobial resistance among urinary tract isolates of Escherichia coli in the United States in 2017. PLoS One 2019; 14:e0220265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Minamida S, Satoh T, Tabata K, et al. . Prevalence of fluoroquinolone-resistant Escherichia coli before and incidence of acute bacterial prostatitis after prostate biopsy. Urology 2011; 78:1235–9. [DOI] [PubMed] [Google Scholar]
- 25.Liss MA, Johnson JR, Porter SB, et al. . Clinical and microbiological determinants of infection after transrectal prostate biopsy. Clin Infect Dis 2015; 60:979–87. [DOI] [PubMed] [Google Scholar]
- 26.Edlund C, Sjöstedt S, Nord CE. Comparative effects of levofloxacin and ofloxacin on the normal oral and intestinal microflora. Scand J Infect Dis 1997; 29:383–6. [DOI] [PubMed] [Google Scholar]
- 27.Chavda KD, Satlin MJ, Chen L, et al. . Evaluation of a multiplex PCR assay to rapidly detect Enterobacteriaceae with a broad range of β-lactamases directly from perianal swabs. Antimicrob Agents Chemother 2016; 60:6957–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colpan A, Johnston B, Porter S, et al. ; VICTORY (Veterans Influence of Clonal Types on Resistance: Year 2011) Investigators . Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin Infect Dis 2013; 57:1256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee R, Johnson JR. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 2014; 58:4997–5004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: 29th informational supplement. CLSI M100-S29. Wayne, PA: CLSI; 2019. [Google Scholar]
- 31.Chantell C, Humphries RM, Lewis JS II. Clinical and Laboratory Standards Institute rationale document MR02: fluoroquinolone breakpoints for Enterobacteriaceae and Pseudomonas aeruginosa. Available at: https://clsi.org/standards/products/microbiology/companion/mr02. Accessed 27 December 2020.
- 32.Doan VP, Yeh JC, Gulbis AM, Aitken SL, Ariza-Heredia E, Ahmed S. Levofloxacin versus cefpodoxime for antibacterial prophylaxis in allogeneic stem cell transplantation. Biol Blood Marrow Transplant 2019; 25:1637–41. [DOI] [PubMed] [Google Scholar]
- 33.Van Besien K, Bachier-Rodriguez L, Satlin M, et al. . Prophylactic rituximab prevents EBV PTLD in haplo-cord transplant recipients at high risk. Leuk Lymphoma 2019; 60:1693–6. [DOI] [PubMed] [Google Scholar]