Abstract
Background
Nongonococcal urethritis (NGU) is a common syndrome with no known etiology in ≤50% of cases. We estimated associations between urethral bacteria and NGU in men who have sex with men (MSM) and men who have sex with women (MSW).
Methods
Urine was collected from NGU cases (129 MSM, 121 MSW) and controls (70 MSM, 114 MSW) attending a Seattle STD clinic. Cases had ≥5 polymorphonuclear leukocytes on Gram stain plus symptoms or discharge; controls had <5 PMNs, no symptoms, no discharge. NGU was considered idiopathic when Neisseria gonorrhoeae, Chlamydia trachomatis, Mycoplasma genitalium, Trichomonas vaginalis, adenovirus, and herpes simplex virus were absent. The urethral microbiota was characterized using 16S rRNA gene sequencing. Compositional lasso analysis was conducted to identify associations between bacterial taxa and NGU and to select bacteria for targeted qPCR.
Results
Among NGU cases, 45.2% were idiopathic. Based on compositional lasso analysis, we selected Haemophilus influenzae (HI) and Mycoplasma penetrans (MP) for targeted qPCR. Compared with 182 men without NGU, the 249 men with NGU were more likely to have HI (14% vs 2%) and MP (21% vs 1%) (both P ≤ .001). In stratified analyses, detection of HI was associated with NGU among MSM (12% vs 3%, P = .036) and MSW (17% vs 1%, P < .001), but MP was associated with NGU only among MSM (13% vs 1%, P = .004). Associations were stronger in men with idiopathic NGU.
Conclusions
HI and MP are potential causes of male urethritis. MP was more often detected among MSM than MSW with urethritis.
Keywords: nongonococcal urethritis, urethral microbiota, men who have sex with men, men who have sex with women, urine microbiome
This case-control study of the urethral microbiota identified associations between Haemophilus influenzae and Mycoplasma penetrans with nongonococcal urethritis (NGU). M. penetrans was associated with NGU in men who have sex with men and not men who have sex with women.
Nongonococcal urethritis (NGU) is a male genital tract syndrome characterized by urethral symptoms including dysuria and pruritis, urethral discharge, elevated polymorphonuclear leukocytes (PMNs) in the urethra and the absence of Neisseria gonorrhoeae [1]. Nongonococcal urethritis complications are rare, but occasionally include epididymitis, prostatitis, and reactive arthritis [2, 3]. Organisms associated with NGU are typically sexually transmitted, posing a risk for partners, particularly women among whom transmission may result in sequelae such as pelvic inflammatory disease and infertility [4]. The most common cause of NGU is Chlamydia trachomatis (CT), accounting for 20% to 40% of NGU cases [5–8]. Mycoplasma genitalium (MG) is also consistently associated with 10% to 30% of NGU cases [5, 9–11]. Less common causes include Trichomonas vaginalis (TV), herpes simplex virus (HSV), and adenovirus [1, 5, 7, 10, 11]. In up to 50% of NGU cases there is no known etiology, making treatment and management of this syndrome challenging. Emerging evidence suggests differences in etiology among men who have sex with men (MSM) and men who have sex with women (MSW) [5, 7, 8].
Among studies investigating whether bacteria other than CT and MG are associated with NGU, several have evaluated the role of Ureaplasma urealyticum (UU), but results have been inconsistent [1, 7, 12–14]. The hypothesis that bacterial communities, rather than individual bacteria, may contribute to idiopathic NGU has been examined but there was no consistent separation of bacterial communities between cases and controls in 1 study [13]. We conducted a case-control study to investigate whether bacteria other than CT and MG are associated with NGU among MSM and MSW.
METHODS
Study Design and Study Population
Participants who were 16 years or older, assigned male sex at birth, attending the Public Health–Seattle & King County STD (sexually transmitted disease) Clinic, and previously recruited into a cross-sectional study between August 2014 and April 2018 [15] formed our case-control study population. The Institutional Review Boards at the University of Washington and Fred Hutchinson Cancer Research Center approved the study. All men provided written informed consent. Men had exclusively male or exclusively female sex partners in the past year. Two transgender women who had sex only with men also enrolled. Men reporting both male and female partners in the past year, known contact to a partner with urogenital Neisseria gonorrhoeae (NG), no sex in the past 60 days, antibiotic use in the past 30 days, or with NG by Gram stain or nucleic acid amplification testing (NAAT) were excluded. Men with severe symptoms characteristic of adenovirus and HSV were not explicitly excluded, but rarely enrolled. All participants completed a computer-assisted self-interview, underwent a standard genital examination, and provided 30–50 mL of first-void urine and urethral swab specimens. Urethral exudates were Gram-stained to quantitate PMNs and examined for the presence of gram-negative intracellular diplococci indicative of NG. Cases of NGU had urethral symptoms or visible discharge and 5 or more PMNs per high-power field (HPF). Controls had no urethral symptoms, no discharge and less than 5 PMNs/HPF. Urine specimens were tested for NG, CT, and MG using Aptima assays with analyte-specific reagents that are for research use only (Hologic, San Diego, CA), while TV, adenovirus, and HSV were measured using quantitative polymerase chain reaction (qPCR) [16, 17]. Nongonococcal urethritis in the absence of CT, MG, TV, adenovirus, and HSV was considered idiopathic.
DNA Extraction and Quantification
DNA was extracted from urine samples stored at 4oC for 1–3 days prior to processing in the laboratory [18] using the QIAamp BiOstic Bacteremia Kit (Qiagen, Hilden, Germany). The Tris-EDTA (-ethylenediaminetetraacetic acid) buffer for DNA elution was filtered twice to minimize contamination. Sham extraction negative controls were included to monitor for potential contamination during processing of urine pellets. DNA from mock communities (positive controls) with known bacterial composition was extracted. PCR inhibition was monitored using an internal amplification control assay [19], and samples were considered inhibited if delayed by 2.0 or more cycles. Bacterial DNA concentrations were measured using a TaqMan-based qPCR assay targeting the V3–V4 region of the 16S ribosomal RNA (rRNA) gene [20].
Broad-range Polymerase Chain Reaction and Sequencing
Broad-range PCR amplification of the V3–V4 region of the 16S rRNA gene was performed on samples and positive and negative controls. Amplicons were sequenced on the Illumina MiSeq instrument (San Diego, CA). Sequence reads were demultiplexed and the DADA2 package [21] was used for processing reads, resulting in a list of unique sequence variants (SVs). Taxonomy was assigned to unique SVs based on location on a phylogenetic tree (Supplementary Materials).
Quantitative Polymerase Chain Reaction
Species-specific qPCR assays were developed to measure DNA concentrations of Mycoplasma penetrans (MP), Haemophilus influenzae (HI), TV, and UU (Supplementary Materials). Assays targeted the 16S rRNA genes of MP and HI, the urease accessory protein G gene of UU (adapted from [22]), and the 18S rRNA gene of TV.
Statistical Analyses
Patient characteristics were compared using Fisher’s exact tests for categorical variables and Wilcoxon rank-sum test for continuous variables. Samples yielding more than 1000 sequence reads were included in all subsequent analyses. ɑ-Diversity was calculated using the Shannon Diversity Index and compared between cases and controls using Wilcoxon rank-sum tests. A sequential PCR approach (broad-range PCR→qPCR) was used to identify associations between bacterial taxa and NGU. First, compositional lasso analysis [23] of bacterial taxa was conducted to identify associations between bacteria and NGU among MSW and MSM separately. We replaced zeros in the sequence count data with 0.5 and re-calculated relative abundances [23]. ß-Coefficients estimating change in probability of NGU per log2 change in relative abundance were calculated using the CVS R package (http://www.math.pku.edu.cn/teachers/linw/software.html). Bacterial taxa with nonzero ß-coefficients were considered to be associated with NGU; compositional lasso does not yield P values. Additional analyses to examine associations between bacterial taxa and NGU were conducted to confirm taxa identified in the compositional lasso approach (Supplementary Table 1). Odds ratios (ORs) from exact logistic regression analyses and Wilcoxon rank-sum tests were used to compare relative abundances. Multiple comparisons were accounted for using the Benjamini-Hochberg false discovery rate. Taxa positively associated with NGU were selected for measurement of concentrations using targeted qPCR to validate the associations noted. Fisher’s exact test was used for binary variables (detected, not detected). Concentrations of potential pathogens were evaluated for their associations with NGU and idiopathic NGU among participants in whom the bacterium was detected using Wilcoxon rank-sum tests. All analyses used R version 3.5.1 (R Foundation for Statistical Computing) and Stata version 15 (StataCorp).
RESULTS
Participant Characteristics
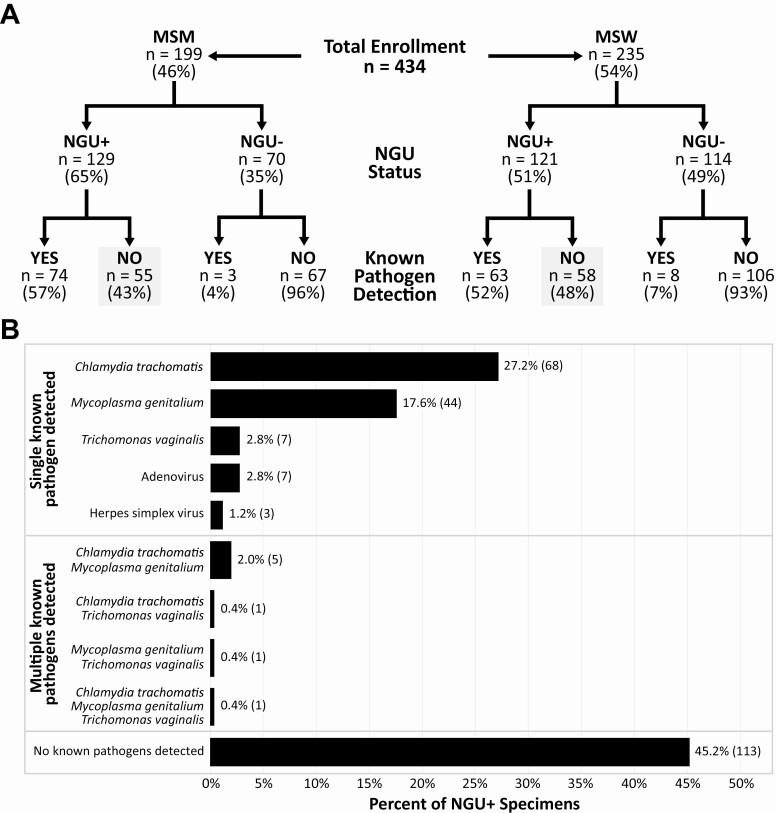
We identified 434 eligible men including 199 MSM (46%) and 235 MSW (54%), comprising 250 cases of NGU and 184 controls (Figure 1, Table 1, Supplementary Table 2). Detailed characteristics have been previously reported [15]. Among cases of NGU, 129 (52%) were infected with a single known pathogen, 3% had 2 pathogens detected, and less than 1% had 3 pathogens detected using NAAT or qPCR (Figure 1). Of NGU cases who were colonized with a single pathogen, the majority were infected with either CT (52.7%) or MG (34.1%) while the prevalence of HSV, adenovirus, or TV was low. In this study, 45.2% of men were classified as having idiopathic NGU with negative testing for CT, MG, TV, adenovirus, and HSV (Figure 1).
Figure 1.
Participants enrolled in the case-control study. A, Of 434 men, 46% were MSM and 54% were MSW. Urethral samples were tested for known causes of NGU including Chlamydia trachomatis, Mycoplasma genitalium, Trichomonas vaginalis, adenovirus, and herpes simplex virus. B, Of the 250 men with NGU, 45.2% did not have a known pathogen detected, hence were classified as having idiopathic NGU. Numbers in parentheses in the bar chart indicate number of men in each group. Abbreviations: MSM, men who have sex with men; MSW, men who have sex with women; NGU, nongonococcal urethritis.
Table 1.
Characteristics of 434 Study Participants With and Without Nongonococcal Urethritis Stratified by Sex of Sex Partners
| MSMa | MSW | |||||
|---|---|---|---|---|---|---|
| Characteristicsb | No NGU (n = 70) | NGU (n = 129) | P c | No NGU (n = 114) | NGU (n = 121) | P c |
| Demographic | ||||||
| Gender, n (%) | 1.000 | |||||
| Transgender | 1 (1) | 1 (1) | 0 (0) | 0 (0) | ||
| Cisgender | 69 (99) | 128 (99) | 114 (100) | 121 (100) | ||
| Age, median (IQR), y | 30.5 (26–39) | 30 (27–39) | .895d | 32 (27–41) | 33 (26–36) | .673d |
| Race, n (%) | .567 | .006* | ||||
| White | 49 (70) | 82 (63) | 73 (64) | 54 (44) | ||
| Black | 3 (4) | 10 (8) | 21 (18) | 42 (35) | ||
| Other/multiple/unknown | 18 (26) | 37 (29) | 20 (18) | 25 (21) | ||
| Hispanic or Latino ethnicity, n (%) | .810 | .821 | ||||
| Yes | 15 (21) | 24 (19) | 11 (10) | 9 (8) | ||
| No | 53 (76) | 102 (79) | 4 (3) | 107 (88) | ||
| Unknown | 2 (3) | 3 (2) | 99 (87) | 5 (4) | ||
| Education completed, n (%) | .025* | .045* | ||||
| ≤High school or GED | 31 (44) | 38 (30) | 40 (36) | 58 (48) | ||
| Some college | 8 (12) | 33 (26) | 22 (20) | 27 (23) | ||
| College graduate | 31 (44) | 57 (44) | 50 (44) | 35 (29) | ||
| Behavioral | ||||||
| Number of sex partners past 2 m, median (IQR) | 3 (2–5) | 3 (2–5.5) | .821d | 1 (1–2) | 2 (1–3) | .014* d |
| New sex partner in past 2 m, n (%) | 51 (75) | 106 (83) | .185 | 74 (65) | 88 (74) | .198 |
| Behaviors at last sexual episode,e n (%) | ||||||
| Vaginal sex | … | … | 107 (95) | 116 (96) | .763 | |
| Insertive oral sex | 47 (69) | 97 (76) | .315 | 62 (55) | 63 (52) | .695 |
| Insertive anal sex | 25 (37) | 90 (70) | <.001* | 3 (3) | 1 (2) | .675 |
| Always uses condoms with last sex partner | 20 (29) | 21 (17) | .065 | 31 (27) | 18 (16) | .036* |
| History, n (%) | ||||||
| Reason for visitf | ||||||
| Urethral symptoms | 0 (0) | 94 (73) | <.001* | 0 (0) | 98 (81) | <.001* |
| Other symptoms | 5 (7) | 1 (1) | .021* | 26 (23) | 0 (0) | <.001* |
| STD testing | 58 (83) | 87 (67) | .020* | 85 (75) | 87 (73) | .768 |
| History of previous STD | ||||||
| History of NGU | 5 (7) | 39 (30) | <.001* | 7 (6) | 20 (17) | .014* |
| History of chlamydial infection | 33 (49) | 76 (59) | .174 | 21 (19) | 46 (38) | .001* |
| History of gonococcal infection | 33 (49) | 72 (56) | .296 | 10 (9) | 25 (21) | .016* |
| Known HIV-positive | 2 (3) | 13 (10) | .091 | 0 (0) | 0 (0) | |
| HIV pre-exposure prophylaxis useg | 13 (19) | 18 (16) | .546 | 0 (0) | 0 (0) | |
| Presentation, n (%) | ||||||
| Urethral symptoms | 0 (0) | 115 (89) | 0 (0) | 110 (91) | ||
| Visible urethral discharge | 0 (0) | 117 (91) | 0 (0) | 107 (88) | ||
| PMNs/HPF on a urethral Gram stain | ||||||
| 0–4 | 70 (100) | … | 114 (100) | … | ||
| 5–9 | … | 25 (19) | … | 31 (26) | ||
| ≥10 | … | 104 (81) | … | 90 (74) | ||
| Urethral pathogen detection, n (%) | ||||||
| Chlamydia trachomatis (NAAT) | 0 (0) | 41 (32) | <.001* | 0 (0) | 34 (28) | <.001* |
| Mycoplasma genitalium (NAAT) | 3 (4) | 29 (22) | .001* | 3 (3) | 22 (18) | <.001* |
| Herpes simplex virus (qPCR) | 0 (0) | 1 (1) | .000 | 1 (1) | 2 (2) | .000 |
| Adenovirus (qPCR) | 0 (0) | 5 (4) | .164 | 1 (1) | 2 (2) | .000 |
| Trichomonas vaginalis (qPCR) | 0 (0) | 1 (1) | .000 | 4 (4) | 9 (7) | .256 |
Abbreviations: GED, General Education Diploma; HIV, human immunodeficiency virus; HPF, high-power field; IQR, interquartile range; MSM, men who have sex with men only; MSW, men who have sex with women only; NAAT, nucleic acid amplification test; NGU, nongonococcal urethritis; PMN, polymorphonuclear leukocyte; qPCR, quantitative polymerase chain reaction; STD, sexually transmitted disease.
aIncludes 2 transgender women who have sex with men only.
bn = 4 missing education completed, n = 14 missing number sex partners past 2 months, n = 7 missing new sex partner past 2 months, n = 1 missing vaginal sex at last sex, n = 4 missing insertive oral sex at last sex, n = 4 missing insertive anal sex at last sex, n = 13 missing recent condom use with last sex partner, n = 1 missing reason for visit is STD testing, n = 4 missing history of NGU, n = 4 missing history of chlamydial infection, n = 3 missing history of gonococcal infection. Participants missing herpes simplex virus (n = 18), adenovirus (n = 18), and Trichomonas vaginalis (n = 3) test results were assumed to be negative given the low prevalence of these pathogens.
cFisher’s exact test unless otherwise specified. *Significance at the ɑ = 0.05 level.
dWilcoxon rank-sum test.
eNot mutually exclusive categories.
fMultiple reasons for visit possible. Other symptoms include genital lesions/rash, nongenital rash, anorectal symptoms, testicular symptoms, oral/pharyngeal symptoms, and other symptoms.
gAmong participants who were not known to be HIV-positive.
Urethral Bacterial Communities in Men With and Without Nongonococcal Urethritis
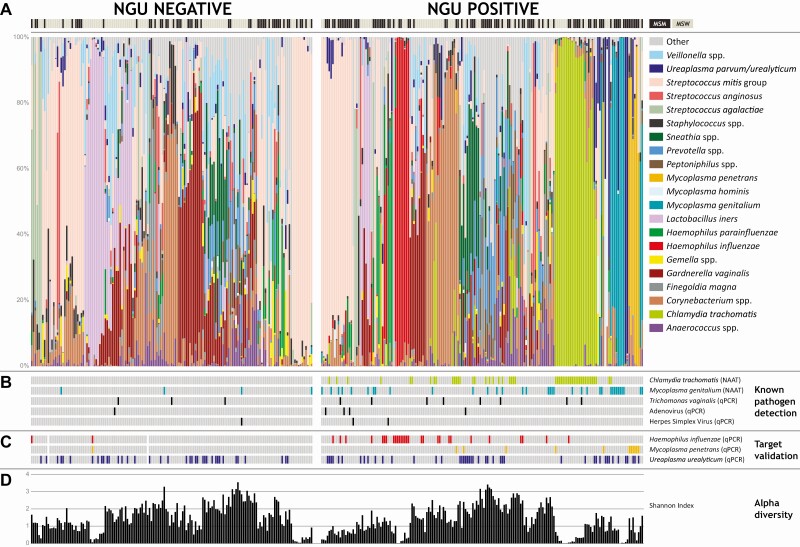
Of 434 urine samples collected, 330 samples (76%) contained sufficient bacterial DNA for 16S rRNA gene sequencing; 328 yielded sequence data. Samples from NGU cases were less likely to yield useful sequence data (70% vs 84%, P = .001). Overall, 13.35 million sequence reads were generated with a mean of 29 423 reads and a median of 40 711 reads per sample. Most (96.2%) reads were classified to the species level. ɑ-Diversity among men with NGU was lower compared with men without NGU (1.21 vs 1.53, P = .005). When stratified by sex of sex partner, diversity was lower among both MSM and MSW with NGU than those without NGU, but the latter was not statistically significant (1.09 vs 1.31, P = .050, and 1.39 vs1.66, P = .085, respectively).
Relative abundance data (Supplementary Table 3) obtained from 328 men were evaluated to determine associations of bacteria with NGU. A dominant taxon was defined as having more than 50% relative abundance within a sample. Haemophilus influenzae– and MP-dominant communities were noted in men with NGU, in addition to CT- and MG-dominant communities (Figure 2). Two men with NGU had Ureaplasma-dominant communities. Among men with NGU in whom we detected CT sequence reads, 40% were dominated by this bacterium and 35% of men with MG reads were dominated by this bacterium. While other taxa were dominant (eg, Corynebacterium spp. and Streptococcus mitis group), no other bacterial communities were consistently different between cases and controls. On stratifying by sex of sex partner, both MSM and MSW had HI-dominant communities, while only MSM had MP-dominant communities in men with NGU (Supplementary Figures 1 and 2). Lactobacillus iners–dominant communities were noted mostly among MSW, with only 1 in MSM. Compositional lasso analysis showed that higher relative abundances of HI (ß = 0.00844) and MP (ß = 0.00112) were positively associated with NGU in MSM, while HI was positively associated with NGU in MSW (ß = 0.00949) (Table 2). As expected, higher relative abundances of CT and MG were associated with NGU among MSM and MSW (Table 2). The compositional lasso model also identified bacterial species that were inversely associated with NGU in MSM and MSW, although the taxa were different among MSM and MSW (Table 2). Notably, L. iners was inversely associated with NGU among MSW (ß = −0.00690) but not MSM. Atopobium vaginae (MSW), Streptococcus mitis (MSW), and Veilonella atypica (MSM) were also associated with the absence of NGU, among other bacteria (Table 2). Haemophilus influenzae and MP were selected for measurements of their concentrations using qPCR to validate their associations with NGU. Although Ureaplasma parvum/urealyticum was not associated with NGU in analyses of relative abundances of bacterial taxa, UU was selected for targeted qPCR given previous associations with NGU in some studies [12, 14, 24–26].
Figure 2.
A, Urethral bacterial communities among MSM and MSW with and without NGU. Relative abundances of 21 of the most abundant bacteria are shown either at the species or genus level for visualization. All other taxa have been placed in the “Other” category. For all statistical analyses, sequences were placed at the most specific level (for complete taxon table, see Supplementary Table 3). Men with NGU had bacterial communities dominated by Chlamydia trachomatis and Mycoplasma genitalium. Other dominant taxa that were noted among men with NGU included Haemophilus influenzae and Mycoplasma penetrans. B, Known pathogen detection by NAAT or qPCR including C. trachomatis, M. genitalium, Trichomonas vaginalis, adenovirus, or herpes simplex virus. C, Target validation of bacteria that were selected in this study for quantitative assessments including H. influenzae, M. penetrans and Ureaplasma urealyticum. D, ɑ-Diversity as measured by the Shannon Diversity Index. Abbreviations: MSM, men who have sex with men; MSW, men who have sex with women; NGU, nongonococcal urethritis; NAAT, nucleic acid amplification testing; qPCR, quantitative polymerase chain reaction.
Table 2.
Association Between Specific Bacterial Taxa and Nongonococcal Urethritis Among 328 Men With Sequence Data
| Taxon | No NGU, n (%) | NGU, n (%) | ß Comp. Lassoa |
|---|---|---|---|
| MSM,b total N | 52 | 93 | |
| Chlamydia trachomatis | 0 (0) | 27 (29) | 0.01859 |
| Haemophilus influenzae | 1 (2) | 16 (17) | 0.00844 |
| Mycoplasma genitalium | 1 (2) | 16 (17) | 0.00926 |
| Mycoplasma penetrans | 0 (0) | 8 (9) | 0.00112 |
| Actinobaculum massiliense | 2 (4) | 0 (0) | −0.00578 |
| Caulobacter | 2 (4) | 0 (0) | −0.00196 |
| Gemella haemolysans | 2 (4) | 0 (0) | −0.00886 |
| Veillonella atypica | 18 (35) | 6 (6) | −0.02102 |
| MSW, total N | 101 | 82 | |
| Chlamydia trachomatis | 3 (3) | 25 (30) | 0.01900 |
| Haemophilus influenzae | 7 (7) | 16 (20) | 0.00949 |
| Mycoplasma genitalium | 0 (0) | 9 (11) | 0.00467 |
| Actinomyces ihumii | 16 (16) | 1 (1) | −0.00004 |
| Atopobium vaginae | 33 (33) | 10 (12) | −0.00546 |
| Corynebacterium pyruviciproducens | 36 (36) | 9 (11) | −0.01680 |
| Lactobacillus iners | 50 (50) | 22 (27) | −0.00690 |
| Streptococcus mitis group | 72 (71) | 45 (55) | −0.00393 |
Abbreviations: Comp., compositional; MSM, men who have sex with men only; MSW, men who have sex with women only; NGU, nongonococcal urethritis.
aThe ß-coefficient (ß) is an estimate of the change in probability of NGU per log2 change in relative abundance. Compositional lasso does not yield P values for each ß. Only taxa with positive or negative ß-coefficients are included in this table. All other taxa had a value of ß = 0, hence are not shown.
bIncludes 2 transgender women who have sex with men only.
Association of Presence and Concentrations of Bacterial Targets and Nongonococcal Urethritis
The presence and concentrations of MP, HI, and UU were measured using targeted qPCR in 431 of 434 men with sufficient remaining DNA. The presence of MP (OR, 8.3; 95% CI 1.98–73.60) and HI (OR, 9.8; 95% CI 2.99–50.22) were significantly associated with NGU, including idiopathic NGU (ORs, 14.9 [95% CI 3.41–135.15] and 20.24 [95% CI 5.97–105.57], respectively), as were higher concentrations (P < .03 for both). Neither the presence nor concentrations of UU were associated with NGU (Table 3). In analyses stratified by sex of sex partner, the presence of HI was significantly associated with NGU in MSM (P = .036) and in MSW (P < .001), but higher concentrations of HI were only associated with NGU (P = .025) including idiopathic NGU (P = .03) among MSM (Table 4). The presence, but not concentrations, of MP was associated with NGU and idiopathic NGU among MSM (P < .01 for both), but this was not observed among MSW. Most cases with HI or MP had monoinfections (12% and 6.8%, respectively); HI was detected in 2 cases with CT and 2 cases with TV, while MP was detected in 4 cases with CT.
Table 3.
Association Between Concentrations of Potential Pathogens and Nongonococcal Urethritis Among 431 Men With qPCR Data
| Any NGU | Idiopathic NGUa | |||||
|---|---|---|---|---|---|---|
| No NGU (n = 182) | NGU (n = 249) | P b | No NGU (n = 171) | Idiopathic NGU (n = 113) | P b | |
| Mycoplasma penetrans | ||||||
| Detected, n (%) | 2 (1) | 21 (8) | .001* | 2 (1) | 17 (15) | <.001* |
| Log10 concentration, median (IQR),c copies/mL | 0.93 (0.62–1.25) | 3.80 (3.09–4.50) | .029* | 0.93 (0.62–1.25) | 4.11 (3.54–4.50) | .034* |
| Haemophilus influenzae | ||||||
| Detected, n (%) | 3 (2) | 35 (14) | <.001* | 3 (2) | 30 (27) | <.001* |
| Log10 concentration, median (IQR),c copies/mL | 0.62 (0.40–0.91) | 3.45 (2.44–4.12) | .005* | 0.62 (0.40–0.91) | 3.47 (2.44–4.14) | .005* |
| Ureaplasma urealyticum | ||||||
| Detected, n (%) | 39 (21) | 56 (22) | .815 | 37 (22) | 28 (25) | .566 |
| Log10 concentration, median (IQR),c copies/mL | 2.96 (2.23–3.75) | 3.09 (2.08–4.12) | .496 | 2.82 (2.23–3.60) | 3.30 (2.08–4.18) | .266 |
n = 3 missing Mycoplasma penetrans, Haemophilus influenzae, Ureaplasma urealyticum, and Trichomonas vaginalis test results. n = 18 missing herpes simplex virus and adenovirus test results.
Abbreviations: IQR, interquartile range; NGU, nongonococcal urethritis; qPCR, quantitative polymerase chain reaction.
aRestricted to participants who tested negative or had missing data for Chlamydia trachomatis and Mycoplasma genitalium by nucleic acid amplification test and for adenovirus, herpes simplex virus, and Trichomonas vaginalis by qPCR.
bFisher’s exact test (binary variables), Wilcoxon rank-sum test (continuous variables). *Significance at the ɑ = 0.05 level.
cAmong participants with the bacterium detected.
Table 4.
Association Between Concentrations of Potential Pathogens and Nongonococcal Urethritis Among 431 MSM and MSW With qPCR Data
| Any NGU | Idiopathic NGUa | |||||
|---|---|---|---|---|---|---|
| No NGU | NGU | P b | No NGU | Idiopathic NGU | P b | |
| MSM,c N | 70 | 129 | 67 | 55 | ||
| Mycoplasma penetrans | ||||||
| Detected, n (%) | 1 (1) | 17 (13) | .004* | 1 (1) | 14 (25) | <.001* |
| Log10 concentration, median (IQR),d copies/mL | 1.25 (1.25-1.25) | 4.20 (3.64–4.53) | .101 | 1.25 (1.25-1.25) | 4.25 (3.65–4.53) | .105 |
| Haemophilus influenzae | ||||||
| Detected, n (%) | 2 (3) | 15 (12) | .036* | 2 (3) | 11 (20) | .003* |
| Log10 concentration, median (IQR),d copies/mL | 0.65 (0.40–0.91) | 3.67 (2.77–4.18) | .025* | 0.65 (0.40–0.91) | 3.89 (3.10–4.49) | .030* |
| Ureaplasma urealyticum | ||||||
| Detected, n (%) | 11 (16) | 28 (22) | .354 | 11 (16) | 13 (24) | .365 |
| Log10 concentration, median (IQR),d copies/mL | 2.45 (1.36–2.97) | 3.02 (2.35–4.13) | .105 | 2.45 (1.36–2.97) | 3.05 (2.12–4.16) | .235 |
| MSW,e N | 112 | 120 | 104 | 58 | ||
| Mycoplasma penetrans | ||||||
| Detected, n (%) | 1 (1) | 4 (3) | .371 | 1 (1) | 3 (5) | .131 |
| Log10 concentration, median (IQR),d copies/mL | 0.62 (0.62-0.62) | 1.46 (1.20–2.30) | .157 | 0.62 (0.62-0.62) | 1.34 (1.06–1.57) | .180 |
| Haemophilus influenzae | ||||||
| Detected, n (%) | 1 (1) | 20 (17) | <.001* | 1 (1) | 19 (33) | <.001* |
| Log10 concentration, median (IQR),d copies/mL | 0.62 (0.62-0.62) | 3.06 (2.22–3.92) | .099 | 0.62 (0.62-0.62) | 3.16 (2.15–3.95) | .100 |
| Ureaplasma urealyticum | ||||||
| Detected, n (%) | 28 (25) | 28 (23) | .878 | 26 (25) | 15 (26) | 1.000 |
| Log10 concentration, median (IQR),d copies/mL | 3.23 (2.37–4.00) | 3.19 (2.00–4.10) | .731 | 3.10 (2.37–3.96) | 3.84 (2.05–4.20) | .607 |
n = 8 missing herpes simplex virus and adenovirus test results.
Abbreviations: IQR, interquartile range; MSM, men who have sex with men only; MSW, men who have sex with women only; NGU, nongonococcal urethritis; qPCR, quantitative polymerase chain reaction.
aRestricted to participants who tested negative or had missing data for Chlamydia trachomatis and Mycoplasma genitalium by nucleic acid amplification test and for adenovirus, herpes simplex virus, and Trichomonas vaginalis by qPCR.
bFisher’s exact test (binary variables), Wilcoxon rank-sum test (continuous variables). *Significance at the ɑ = 0.05 level.
cn = 3 missing Mycoplasma penetrans, Haemophilus influenzae, Ureaplasma urealyticum, and Trichomonas vaginalis test results. n = 10 missing herpes simplex virus and adenovirus test results.
dAmong participants with the bacterium detected.
eIncludes 2 transgender women who have sex with men only.
Characteristics associated with HI, MP, and UU were evaluated separately by sex of sex partners (Supplementary Tables 4–6). No sociodemographic (age, race, ethnicity, education), sexual behavior (number sex partners, sexual exposures, condom use), or STD history (NGU, GC, CT) characteristics were associated with either HI or MP. With 2 exceptions, none of these characteristics were associated with UU. Among MSM, UU was significantly associated with insertive anal sex (77% vs 54%, P = .001) and inversely associated with consistent condom use (8% vs 25%, P = .026). Among MSW, UU was significantly less common in those with HI (P = .031).
Given the absence of associations with other pathogens and potential risk factors, no multivariable models were developed.
DISCUSSION
We identified significant associations between the presence and quantities of HI and MP with NGU and idiopathic NGU. The microbiota in men with and without NGU were heterogeneous and MSM had different community types than MSW. Men with NGU had bacterial communities that were less diverse than men without NGU, suggesting that potential pathogens tend to dominate the bacterial community.
There have been suggestions that HI may be an infrequent cause of NGU [1]. This bacterium has been isolated in culture from men with urethral symptoms [27–29] but many studies did not include men without NGU. A case-control study of NGU among Australian MSW noted that 2.5% of cases were HI-positive by culture but did not find a significant association with NGU [30]. Molecular approaches have also been used to evaluate whether HI is associated with NGU. A case-control study of 73 asymptomatic controls and 211 NGU cases attending STD clinics in Sweden noted that HI was detected in 5% of men with acute NGU compared with 0% of controls, but the association was not significant [12]. In our study, HI was detected by targeted qPCR in 14% of men with NGU and 27% of men with idiopathic NGU, and in 2% of controls. Both studies recruited men from STD clinics, but differences in study population or PCR assay conditions may have contributed to the differential observations. As we continue to investigate the role of HI in NGU, an important consideration is determining how HI is acquired. Haemophilus influenzae is a common commensal of the upper respiratory tract [31]. Unprotected oral sex may be 1 mode of transmission [5, 32], but insertive oral sex was not associated with HI detection among both MSM and MSW. However, most men reported engaging in oral sex and additional factors likely influence the acquisition of HI. From a treatment standpoint, there is limited information on the effect of antibiotics on HI in the genital tract. Antimicrobial susceptibility testing of HI strains from the urogenital tract suggest some strains may be resistant to azithromycin and tetracyclines, antibiotics typically used to treat urethritis [33, 34]. Longitudinal studies examining the association between HI and NGU with careful collection of sexual behavior data are needed to further understand acquisition, transmission, and treatment outcomes. Given the suggested link between HI and pelvic inflammatory disease in women [35], MSW should be a priority population for these investigations.
Some studies have detected MP in MSM, but none have associated MP with NGU. Mycoplasma penetrans was originally isolated from urine samples of MSM living with human immunodeficiency virus (HIV) [36]. A small study that collected urethral, oral, and rectal samples from 10 MSM with NGU and 18 MSM without NGU noted that this bacterium was present in all 3 body sites by targeted PCR [37]. More recently, MP has been detected in urogenital specimens from MSM, but not MSW, in a study of Chinese men with HIV [38]. Frølund et al [13] noted a predominance of MP in 1 man with idiopathic NGU. Here, we noted that MP was detected in 8 MSM (dominant in 6 of the 8) and 1 MSW by broad-range PCR and sequencing, and in 17 MSM and 4 MSW by targeted qPCR. As expected, targeted qPCR was more sensitive in detecting this bacterium.
Detection of HI and MP accounted for an additional 47 cases of NGU in our study, reducing the number of idiopathic NGU cases from 113 to 66 men. Overall, most men in whom we identified an etiological agent had a single infection (90.8%), while 8.7% were coinfected with 2 known or suspected pathogens. Jordan et al [6] recently assessed the prevalence of CT, MG, TV, and UU in men enrolled in the Idiopathic Urethritis Men’s Project and found that 88% of men had a monoinfection. Important differences between the 2 studies are that the study by Jordan et al included UU as a known pathogen and they did not evaluate relationships with UU quantity; however, this bacterium was not associated with NGU in their population [6] or in ours.
Our study had several strengths. First, we included 2 study populations (MSM and MSW) who may have different etiologies for NGU. Few studies have evaluated the urethral microbiota and potential sexually transmitted infection (STI) agents in MSM, a group at highest risk of STI; this study fills this critical gap. Second, strict objective definitions to classify cases and controls helped facilitate clear identification of 2 potential etiologic agents of NGU. For cases, we required that men have either urethral symptoms or visible discharge on examination and urethral inflammation with 5 or more PMNs/HPF, which is the conventional, widely used cutoff for diagnosis of NGU. Current treatment guidelines from the Centers for Disease Control and Prevention recommend 2 or more PMNs/HPF as the threshold for inflammation for diagnosing NGU [39]. Our findings may not be generalizable to men with a lower level of inflammation. One study found that the prevalence of CT and MG was significantly higher in men with more than 9 PMNs/HPF [40], indicating that lowering the cutoff may not be required. In addition, we stipulated that controls had no urethral symptoms or discharge on clinical examination and less than 5 PMNs/HPF, which ensured separation between cases and controls. Third, we classified idiopathic NGU cases in a systematic manner by testing for previously known pathogens CT and MG, and uncommon but confirmed ones including TV, HSV, and adenovirus. Fourth, we used a sequential PCR approach to identify and validate associations of MP and HI with NGU. Moreover, our bioinformatics pipeline and reference set enabled taxonomic placement of 96% of sequence reads to the species level.
Our study also had limitations. First, men in our study were from a single STD clinic in Seattle and these findings may not be generalizable to other populations. Second, the strict definitions of cases and controls excluded men with asymptomatic NGU and low-grade inflammation. However, until more is known about the consequences of asymptomatic NGU, our population represents the most clinically relevant group. Third, this is a case-control study with measures from a single point in time. Longitudinal studies with repeated observations will enable better understanding of the temporal changes in bacterial concentrations, how colonization may be impacted by sexual behavior, and how bacteria are associated with signs and symptoms of urethritis. Fourth, there was insufficient DNA to generate sequences for 104 samples and we were unable to investigate bacteria that may be associated with NGU in those men.
In conclusion, we identified 2 new potential causes of male urethritis, MP and HI, which together accounted for an additional 18.8% of NGU cases. Future work should aim to understand mode of transmission, natural history, and treatment outcomes for these bacteria.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author contributions. Manuscript concept: S. S., L. C. C., D. N. F., L. E. M. Clinical data collection: L. C. C., J. L. M., M. S. L., M. R. G., L. E. M. Laboratory data generation: S. S., M. M. M., D. D., M.-L. H., O. O. S., K. R. J., D. N. F. Data analysis and interpretation: S. S., L. C. C., K. A. T., N. G. H., S. P., J. P. H., D. N. F., L. E. M. Drafting of the manuscript: S. S., D. N. F., L. E. M. Critical revision of the manuscript: S. S., L. C. C., K. A. T., N. G. H., M. M. M., J. L. M., D. D., M. S. L., S. P., M.-L. H., O. O. S., K. R. J., J. P. H., D. N. F., L. E. M.
Acknowledgments. The authors thank the study participants and the Public Health–Seattle & King County STD Clinic staff; Gina Leipertz, Tashina Robinson, and Sarah Romano for expert study coordination; and Anna Unutzer and Nandita Somayaji for data entry assistance. We also thank Patricia Totten’s laboratory for performing some M. genitalium testing on the Hologic DTS system. We appreciate Susan Strenk’s efforts in developing the Trichomonas vaginalis assay.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (grant numbers U19 AI113173 and R01 AI110666; to L. E. M. and D. N. F.). L. C. C. was supported by the National Institutes of Health (grant number TL1 TR002318 for trainee support). Study data were collected and managed using Research Electronic Data Capture (REDCap) tools hosted at the University of Washington Institute of Translational Health Sciences and supported by the National Institutes of Health (grant number UL1TR002319). K. A. T. was supported in part by a National Institutes of Health grant awarded to the University of Washington, Seattle, from the National Institutes of Health (grant number P30 AI027757).
Potential conflicts of interest. L. E. M. received donations of test kits and reagents from Hologic during the conduct of the study, and research funding, speaker’s fees, and conference support from Hologic, outside the submitted work. L. E. M. also has a patent pending to the University of Washington. D. N. F. reports royalties from Becton, Dickinson and Company, outside the submitted work, and a patent pending for Diagnosis of NGU, related to this work. M. R. G. reports grants from Hologic, outside the submitted work. J. P. H. reports grants from the National Institutes of Health, outside the submitted work. S. S. reports a patent pending for diagnosis of NGU related to this work. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Martin DH. Urethritis in males. In: Holmes KK, Sparling PF, Stamm WE, et al. , eds. Sexually transmitted diseases. 4th ed. New York: McGraw Hill, 2008:987–1016. [Google Scholar]
- 2.Ito S, Tsuchiya T, Yasuda M, Yokoi S, Nakano M, Deguchi T. Prevalence of genital mycoplasmas and ureaplasmas in men younger than 40 years-of-age with acute epididymitis. Int J Urol 2012; 19:234–8. [DOI] [PubMed] [Google Scholar]
- 3.Pennisi M, Perdue J, Roulston T, Nicholas J, Schmidt E, Rolfs J. An overview of reactive arthritis. JAAPA 2019; 32:25–8. [DOI] [PubMed] [Google Scholar]
- 4.Holmes KK, Stamm WE, Sobel JD. Lower genital tract syndromes in women. In: Holmes KK, Sparling PF, Stamm WE, et al. , eds. Sexually transmitted diseases. 4th ed. New York: McGraw-Hill, 2008:987–1016. [Google Scholar]
- 5.Bradshaw CS, Tabrizi SN, Read TR, et al. . Etiologies of nongonococcal urethritis: bacteria, viruses, and the association with orogenital exposure. J Infect Dis 2006; 193:336–45. [DOI] [PubMed] [Google Scholar]
- 6.Jordan SJ, Toh E, Williams JA, et al. . Aetiology and prevalence of mixed-infections and mono-infections in non-gonococcal urethritis in men: a case-control study. Sex Transm Infect 2020; 96:306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manhart LE, Gillespie CW, Lowens MS, et al. . Standard treatment regimens for nongonococcal urethritis have similar but declining cure rates: a randomized controlled trial. Clin Infect Dis 2013; 56:934–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rane VS, Fairley CK, Weerakoon A, et al. . Characteristics of acute nongonococcal urethritis in men differ by sexual preference. J Clin Microbiol 2014; 52:2971–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anagrius C, Loré B, Jensen JS. Mycoplasma genitalium: prevalence, clinical significance, and transmission. Sex Transm Infect 2005; 81:458–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwebke JR, Rompalo A, Taylor S, et al. . Re-evaluating the treatment of nongonococcal urethritis: emphasizing emerging pathogens—a randomized clinical trial. Clin Infect Dis 2011; 52:163–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wetmore CM, Manhart LE, Lowens MS, et al. . Demographic, behavioral, and clinical characteristics of men with nongonococcal urethritis differ by etiology: a case-comparison study. Sex Transm Dis 2011; 38:180–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frølund M, Lidbrink P, Wikström A, Cowan S, Ahrens P, Jensen JS. Urethritis-associated pathogens in urine from men with Non-gonococcal urethritis: a case-control study. Acta Derm Venereol 2016; 96:689–94. [DOI] [PubMed] [Google Scholar]
- 13.Frølund M, Wikström A, Lidbrink P, et al. . The bacterial microbiota in first-void urine from men with and without idiopathic urethritis. PLoS One 2018; 13:e0201380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wetmore CM, Manhart LE, Lowens MS, et al. . Ureaplasma urealyticum is associated with nongonococcal urethritis among men with fewer lifetime sexual partners: a case-control study. J Infect Dis 2011; 204:1274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chambers LC, Morgan JL, Lowens MS, et al. . Cross-sectional study of urethral exposures at last sexual episode associated with non-gonococcal urethritis among STD clinic patients. Sex Transm Infect 2019; 95:212–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corey L, Huang ML, Selke S, Wald A. Differentiation of herpes simplex virus types 1 and 2 in clinical samples by a real-time Taqman PCR assay. J Med Virol 2005; 76:350–5. [DOI] [PubMed] [Google Scholar]
- 17.Kuypers J, Wright N, Ferrenberg J, et al. . Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J Clin Microbiol 2006; 44:2382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munch MM, Chambers LC, Manhart LE, et al. . Optimizing bacterial DNA extraction in urine. PLoS One 2019; 14:e0222962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khot PD, Ko DL, Hackman RC, Fredricks DN. Development and optimization of quantitative PCR for the diagnosis of invasive aspergillosis with bronchoalveolar lavage fluid. BMC Infect Dis 2008; 8:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srinivasan S, Hoffman NG, Morgan MT, et al. . Bacterial communities in women with bacterial vaginosis: high resolution phylogenetic analyses reveal relationships of microbiota to clinical criteria. PLoS One 2012; 7:e37818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vancutsem E, Soetens O, Breugelmans M, Foulon W, Naessens A. Modified real-time PCR for detecting, differentiating, and quantifying Ureaplasma urealyticum and Ureaplasma parvum. J Mol Diagn 2011; 13:206–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin W, Shi P, Feng R, Li H. Variable selection in regression with compositional covariates. Biometrika 2014; 101:785–97. [Google Scholar]
- 24.Bowie WR, Wang SP, Alexander ER, et al. . Etiology of nongonococcal urethritis. Evidence for Chlamydia trachomatis and Ureaplasma urealyticum. J Clin Invest 1977; 59:735–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deguchi T, Yoshida T, Miyazawa T, et al. . Association of Ureaplasma urealyticum (biovar 2) with nongonococcal urethritis. Sex Transm Dis 2004; 31:192–5. [DOI] [PubMed] [Google Scholar]
- 26.Yoshida T, Ishiko H, Yasuda M, et al. . Polymerase chain reaction-based subtyping of Ureaplasma parvum and Ureaplasma urealyticum in first-pass urine samples from men with or without urethritis. Sex Transm Dis 2005; 32:454–7. [DOI] [PubMed] [Google Scholar]
- 27.Deza G, Martin-Ezquerra G, Gómez J, Villar-García J, Supervia A, Pujol RM. Isolation of Haemophilus influenzae and Haemophilus parainfluenzae in urethral exudates from men with acute urethritis: a descriptive study of 52 cases. Sex Transm Infect 2016; 92:29–31. [DOI] [PubMed] [Google Scholar]
- 28.Ito S, Hatazaki K, Shimuta K, et al. . Haemophilus influenzae isolated from men with acute urethritis: its pathogenic roles, responses to antimicrobial chemotherapies, and antimicrobial susceptibilities. Sex Transm Dis 2017; 44:205–10. [DOI] [PubMed] [Google Scholar]
- 29.Sturm AW. Haemophilus influenzae and Haemophilus parainfluenzae in nongonococcal urethritis. J Infect Dis 1986; 153:165–7. [DOI] [PubMed] [Google Scholar]
- 30.Iser P, Read TH, Tabrizi S, et al. . Symptoms of non-gonococcal urethritis in heterosexual men: a case control study. Sex Transm Infect 2005; 81:163–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farley MM, Stephens DS, Brachman PS Jr, Harvey RC, Smith JD, Wenger JD; CDC Meningitis Surveillance Group. Invasive Haemophilus influenzae disease in adults: a prospective, population-based surveillance. Ann Intern Med 1992; 116:806–12. [DOI] [PubMed] [Google Scholar]
- 32.Barbee LA, Khosropour CM, Dombrowski JC, Manhart LE, Golden MR. An estimate of the proportion of symptomatic gonococcal, chlamydial and non-gonococcal non-chlamydial urethritis attributable to oral sex among men who have sex with men: a case-control study. Sex Transm Infect 2016; 92:155–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deguchi T, Ito S, Hatazaki K, et al. . Antimicrobial susceptibility of Haemophilus influenzae strains isolated from the urethra of men with acute urethritis and/or epididymitis. J Infect Chemother 2017; 23:804–7. [DOI] [PubMed] [Google Scholar]
- 34.Magdaleno-Tapial J, Valenzuela-Oñate C, Giacaman-von der Weth MM, et al. . Haemophilus species isolated in urethral exudates as a possible causative agent in acute Urethritis: a study of 38 cases. Actas Dermosifiliogr 2019; 110:38–42. [DOI] [PubMed] [Google Scholar]
- 35.Paavonen J, Westrom L, Eschenbach D. Pelvic inflammatory disease. In: Holmes KK, Sparling PF, Stamm WE, et al. , eds. Sexually transmitted diseases. 4th ed. New York: McGraw Hill Medical, 2008. [Google Scholar]
- 36.Taylor-Robinson D, Furr PM. Update on sexually transmitted mycoplasmas. Lancet 1998; 351(Suppl 3):12–5. [DOI] [PubMed] [Google Scholar]
- 37.Taylor-Robinson D, Gilroy CB, Keane FE. Detection of several Mycoplasma species at various anatomical sites of homosexual men. Eur J Clin Microbiol Infect Dis 2003; 22:291–3. [DOI] [PubMed] [Google Scholar]
- 38.Jian-Ru W, Bei W, Hao C, Jin-Shui X, Xi-Ping H. Mycoplasmas in the urine of HIV-1 infected men. Epidemiol Infect 2012; 140:1141–6. [DOI] [PubMed] [Google Scholar]
- 39.Workowski KA, Bolan GA; Centers for Disease Control and Prevention . Sexually transmitted diseases treatment guidelines, 2015. MMWR Recomm Rep 2015; 64:1–137. [PMC free article] [PubMed] [Google Scholar]
- 40.Moi H, Hartgill U, Skullerud KH, Reponen EJ, Syvertsen L, Moghaddam A. Microscopy of stained urethral smear in male Urethritis; which cutoff should be used? Sex Transm Dis 2017; 44:189–94. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.