Abstract
Background
Hemophagocytic lymphohistiocytosis (HLH) is a life-threatening condition of immune dysregulation. Children often suffer from primary genetic forms of HLH, which can be triggered by infection. Others suffer from secondary HLH as a complication of infection, malignancy, or rheumatologic disease. Identifying the exact cause of HLH is crucial, as definitive treatment for primary disease is hematopoietic stem cell transplant. Adenoviruses have been associated with HLH but molecular epidemiology data are lacking.
Methods
We describe the clinical and virologic characteristics of 5 children admitted with adenovirus infection during 2018–2019 who developed HLH or HLH-like illness. Detailed virologic studies, including virus isolation and comprehensive molecular typing were performed.
Results
All patients recovered; clinical management varied but included immunomodulating and antiviral therapies. A genetic predisposition for HLH was not identified in any patient. Adenovirus isolates were recovered from 4/5 cases; all were identified as genomic variant 7d. Adenovirus type 7 DNA was detected in the fifth case. Phylogenetic analysis of genome sequences identified 2 clusters—1 related to strains implicated in 2016–2017 outbreaks in Pennsylvania and New Jersey, the other related to a 2009 Chinese strain.
Conclusions
It can be challenging to determine whether HLH is the result of an infectious pathogen alone or genetic predisposition triggered by an infection. We describe 5 children from the same center presenting with an HLH-like illness after onset of adenovirus type 7 infection. None of the patients were found to have a genetic predisposition to HLH. These findings suggest that adenovirus 7 infection alone can result in HLH.
Keywords: adenovirus, hemophagocytic lymphohistiocytosis, cidofovir, children, hematopoietic stem cell transplant
We describe clinical and virologic characteristics of five children with hemophagocytic lymphohistiocytosis (HLH) and hyperinflammation after infection with adenovirus 7d. These findings raise interesting questions regarding the contribution of the pathogen versus the host in HLH in young children.
Hemophagocytic lymphohistiocytosis (HLH) is a life-threatening process of immune dysregulation and ineffective immune response that is diagnosed based on clinical signs and laboratory abnormalities [1]. In most cases of HLH, there is an inciting event such as an infection, malignant process, or disease flare of a rheumatologic condition. Infants and young children frequently suffer from primary or familial HLH due to mutations in genes responsible for the cytotoxic function of T lymphocytes or natural killer cells [1, 2]. In other patients, a specific genetic predisposition is not identified, and these events are classified as secondary HLH. As we enter the age of genomic medicine, this dichotomy has become blurred. For example, there are infants who clearly have familial HLH based on a strong family history and recurrent disease triggered by mild viral illness but have no identifiable genetic lesion.
Accurate classification of primary or secondary HLH is paramount when making management decisions, as current treatment guidelines recommend hematopoietic stem cell transplantation (HSCT) as definitive therapy for primary HLH [1]. Treatment of patients with secondary HLH is focused on treating the inciting event and dampening immune dysregulation. When considering the management of HLH, host factors are the primary drivers for estimating the likelihood of primary HLH. Primary HLH is more common in infants and young children, and experts often recommend HSCT for neonates, even in the absence of an identified genetic predisposition and regardless of the antecedent pathogen.
Human adenoviruses (HAdV) typically cause mild and self-limiting infections of the respiratory tract, gastrointestinal tract, and conjunctiva [3]. Interestingly, HAdV infections have been associated with HLH in neonates, children, and adults [4–6]. Although some reports describe the associated HAdV type, there has been in general limited molecular detail for HAdV-associated HLH in children. The family Adenoviridae features significant genetic diversity, with over 90 genotypes infecting humans classified into 7 species displaying distinct pathobiological characteristics [7]. Understanding the molecular characteristics of HAdV strains associated with HLH could provide value in deciphering potential pathogen contributions to HLH.
We describe 5 cases of HAdV-associated HLH or HLH-like illness in children, with detailed molecular characterization of the detected viruses. This additional information is considered in the context of the complex clinical decision-making process for managing HLH in the absence of genetic predisposition data.
CASE PATIENTS
Five patients were admitted to our institution with HAdV infection followed by a hyper-inflammatory syndrome consistent with HLH during the 2018–2019 respiratory virus season.
Case 1
A previously healthy 15-month-old girl was admitted with 5 days of fever and severe respiratory distress. A respiratory virus quantitative polymerase chain reaction (RV-qPCR) panel performed on admission on a nasopharyngeal swab was positive for HAdV, and a serum specimen processed for HAdV DNA qPCR was also positive (Table 1). She developed a clinical syndrome consistent with HLH (Table 2). She received high-dose dexamethasone (10 mg/m2 daily) per HLH-2004 [1], but etoposide was not administered given her known HAdV infection. She received a single 1g/kg dose of intravenous immunoglobulin (IVIG). Antiviral therapies such as cidofovir were not administered given concerns for toxicity and limited effectiveness data. Repeat ferritin levels improved after receipt of dexamethasone, and she never required any additional HLH-directed therapy, including etoposide. Even in the absence of antiviral therapy, her plasma HAdV DNA levels decreased, as indicated by increasing PCR cycle threshold (Ct) values. She was discharged and completed her steroid course as an outpatient. She has not had relapse or other disease-related complications for over 1 year after discharge.
Table 1.
Virology Findings on Clinical Specimens Available Reported Cases
| Case | Specimen Type | Collection Time | HAdV qPCR Ct | Virus Isolation/RFLP Variant | Molecular Type | Sequence Data (GenBank No.) |
|---|---|---|---|---|---|---|
| 1 | Serum | Admission | 21.0 | No | H 7 | HVR: MN822238 |
| Plasma | Day 5 | 36.34 | No | … | … | |
| Plasma | Day 10 | 43.93 | No | … | … | |
| 2 | NP | Admission | 19.08 | Yes/HAdV-7d | P7H7F7 | WGS: MN422292 |
| Plasma | Day 3 | 27.14 | No | … | … | |
| Plasma | Day 5 | 25.05 | No | … | … | |
| Plasma | Day 7 | 22.61 | No | H 7 | HVR: MN822239 | |
| Serum | Day 9 | 23.08 | No | H 7 | HVR: MN822240 | |
| Plasma | Day 11 | 25.51 | No | … | … | |
| Serum | Day 14 | 31.28 | No | … | … | |
| Plasma | Day 17 | 27.62 | No | … | … | |
| Plasma | Day 24 | 39.31 | No | … | … | |
| 3 | ET | Admission | 18.01 | Yes/HAdV-7d | P7H7F7 | WGS: MN422293 |
| Plasma | Day 5 | 25.1 | No | … | … | |
| Plasma | Day 7 | 25.15 | No | H 7 | HVR: MN822241 | |
| Plasma | Day 9 | 26.23 | No | … | … | |
| Plasma | Day 11 | 31.09 | No | … | … | |
| Plasma | Day 14 | 41.84 | No | … | … | |
| Plasma | Day 17 | 42.92 | No | … | … | |
| 4 | NP | Admission | 25.45 | Yes/HAdV-7d | P7H7F7 | WGS: MN422294 |
| NP | Day 6 | 16.02 | Yes/HAdV-7d | … | … | |
| Plasma | Day 6 | 21.21 | No | H 7 | HVR: MN822242 | |
| 5 | ET | Admission | 22.17 | Yes/HAdV-7d | P7H7F7 | WGS: MN422295 |
| Serum | Day 7 | 20.10 | No | … | … | |
| Plasma | Day 11 | 29.59 | No | H 7 | HVR: MN822243 | |
| NP | Day 13 | 30.37 | No | … | … | |
| NP | Day 21 | 38.13 | No | … | … | |
| ET | Day 28 | 39.94 | No | … | … |
Abbreviations: Ct, cycle threshold; ET, endotracheal aspirate; F, fiber; H, hexon; HAdV, human adenovirus; HVR, hexon hypervariable region; NP, nasopharyngeal aspirate or swab; P, penton base; qPCR, quantitative polymerase chain reaction; RFLP, restriction fragment length polymorphism; WGS, whole genome sequence.
Table 2.
Clinical Characteristics Documented in 5 Cases of HLH-Like Disease Associated With HAdV-7d Infection
| Case 1 | Case 2‡ | Case 3‡ | Case 4 | Case 5 | |
|---|---|---|---|---|---|
| Age | 15mo | 2mo | 2mo | 17mo | 16y |
| Sex | Female | Male | Male | Male | Male |
| Comorbidities | None | Prematurity | Prematurity | Stuve-Weidemann syndrome | Chronic respiratory failure |
| Clinical Features | |||||
| Fever | Present | Present | Present | Present | Present |
| Hepatomegaly | Present | Present | Present | Present | Not Present |
| Splenomegaly | Present | Present | Present | Present | Not Present |
| Respiratory failure | Present | Present | Present | Present | Present |
| Laboratory Values During Initial Evaluation for HLH | |||||
| Absolute neutrophil count | 190 | 11 150 | 4790 | 1680 | 1840 |
| Hemoglobin (mg/dL) | 8.9 | 8.8 | 7.8 | 7.0 | 8.6 |
| Platelet count (cells/L) | 64 000 | 107 000 | 85 000 | 52 000 | 125 000 |
| Ferritin level (ng/mL) a | 16 600 | 5968 | 12 399 | 11 688 | TNP |
| Soluble IL-2R level (pg/mL) a | 7160 | 11 900 | 15 390 | 5850 | TNP |
| Fibrinogen (mg/dL) a | 208 | 106 | 144 | 141 | TNP |
| Triglyceride level (mg/dL) a | 445 | 94 | 430 | 445 | TNP |
| CD107a mobilization (% positive) a | 17.6% | 3% | 5% | 8% | TNP |
| CXCL9 (pg/mL) a | TNP | 1928 | 1465 | 4299 | TNP |
| IFN-γ (pg/mL) a | 34 | 12 | 61 | 31 | TNP |
| IL-6 (pg/mL) a | 59 | 28 | TNP | 200 | TNP |
| IL-10 (pg/mL) a | 88 | 84 | 52 | 70 | TNP |
| NK cell perforin expression (% positive) a | 77% | 64% | 74% | 88% | TNP |
| NK cell granzyme B expression (% positive) a | 77% | 97% | 92% | 97% | TNP |
| Hemophagocytosis in bone marrow | Present | TNP | TNP | TNP | TNP |
| Transaminitis | Present | Present | Present | Present | Present |
| Coagulopathy | Absent | Present | Present | Present | Present |
| HLH criteria metb | 8/8 | 7/8 | 7/8 | 6/8 | 2/8 |
| Treatment | |||||
| Antiviral therapies? | IVIG ×1 | IVIG ×2 | IVIG ×1 | Nonec | None |
| Cidofovir | |||||
| Brincidofovir | |||||
| HLH treatment? | Dexamethasone | Dexamethasone | Dexamethasone | Nonec | None |
| Etoposide |
Abbreviations: ARDS, acute respiratory distress syndrome; HAdV, human adenovirus; HLH, hemophagocytic lymphohistiocytosis; IL, interleukin; INF, interferon; IVIG, intravenous immunoglobulin; NK, natural killer; TNP, test not performed.
‡, Identical twins.
aReference ranges: ferritin, 10.0–99.9 ng/mL; soluble IL-2R, ≤1033 pg/mL; fibrinogen, 172–471 mg/dL; triglyceride level, 27–125 mg/dL;CD107a mobilization, 11–35%; CXCL9 < 121 pg/mL; IFN-γ, ≤5 pg/mL; IL-6, ≤5 pg/mL; IL-10, ≤18 pg/mL; NK cell perforin expression, 87–95%; NK cell granzyme B expression, 80–98%.
bSee Henter et al [1].
cCase 4 was not treated for HLH or HAdV infection, but did receive a dose of IVIG due to hypogammaglobulinemia, as well as pulse methylprednisolone for ARDS.
Case 2
A 2-month-old boy, the product of a twin gestation born prematurely at 35 weeks gestational age, was admitted with 3 days of fever and acute respiratory failure. A nasopharyngeal aspirate (NPA) was sent for RV-qPCR panel testing and was positive for HAdV (Table 1). Plasma HAdV DNA qPCR was also positive. He developed circulatory shock, acute respiratory distress syndrome (ARDS), and was diagnosed with HLH (Table 2). High-dose dexamethasone therapy was given at a dose of 10 mg/m2 daily per HLH-2004 [1]. IVIG was administered at a dose of 1 g/kg. Etoposide was initially held due to his known HAdV infection but was administered after his clinical status worsened. HAdV DNA was detected in multiple plasma and serum samples collected up to 24 days from admission. Cidofovir, dosed at 5 mg/kg once weekly with hyperhydration, was administered. Due to issues with fluid overload, he was ultimately given enteral brincidofovir (Chimerix, Durham, NC, USA) through an expanded access protocol. His laboratory parameters of HLH and his plasma HAdV DNA viral loads improved over the course of 2 weeks with this regimen. His clinical course was complicated by severe lung disease and chronic respiratory failure, which ultimately required a tracheostomy. He has not had relapse of HLH, now 1 year from initial presentation, but remains admitted with chronic respiratory failure.
Case 3
A 2-month-old boy, the identical twin of case 2, was admitted on the same day as his sibling with 2 days of fever and acute respiratory failure. An endotracheal aspirate was sent for RV-qPCR panel testing, which was positive for HAdV (Table 1). Plasma HAdV DNA qPCR testing was also positive. After several days, he developed clinical signs and laboratory values consistent with HLH (Table 2). High-dose dexamethasone was given at a dose of 10 mg/m2 daily per HLH-2004 [1], but etoposide was held due to confirmed HAdV infection. He was given IVIG at a dose of 1 g/kg but not antiviral therapy. He showed significant clinical improvement, with down-trending ferritin levels and clearance of his plasma HAdV load after 1 week of treatment. He was discharged and completed his steroid course. Given the concern for an unidentified genetic predisposition for HLH, he underwent an unrelated allogeneic cord blood transplantation.
Case 4
A 17-month-old boy with Stuve-Weidemann syndrome complicated by autonomic dysfunction was transferred to the PICU with 5 days of fever and worsening respiratory distress. A RV-qPCR panel of a NPA was positive for HAdV on admission and again 6 days later. A plasma sample also tested positive for HAdV at this time (Table 1). Pulse therapy with methylprednisolone dosed at 1 g/kg was initiated, but he worsened and ultimately developed ARDS requiring extracorporeal membrane oxygenation (ECMO). At this time, his clinical and laboratory assessments were consistent with HLH (Table 2), but his severe presentation was attributed to his underlying genetic syndrome. He did not receive any HLH-directed or antiviral therapy. He was noted to have hypogammaglobulinemia and received IVIG at a dose of 500 mg/kg. He improved and was able to be de-cannulated from the ECMO circuit after 1 week but suffered from chronic respiratory failure necessitating tracheostomy. He has not had relapse of HLH, now 11 months from initial presentation.
Case 5
A 16-year-old boy with complex past medical history, including history of anoxic brain injury and chronic respiratory failure with tracheostomy dependence, was admitted with 1 day of fever and respiratory distress. A RV-qPCR panel of a NPA was positive for HAdV (Table 1). His respiratory status worsened, and he developed ARDS. Plasma HAdV DNA qPCR testing was positive. Additional laboratory testing revealed pancytopenia, hepatitis, and coagulopathy, and elevated C-reactive protein (CRP) at 38.9 mg/dL. He did not have splenomegaly or hepatomegaly. As his clinical state was thought to be the result of disseminated adenovirus disease, he did not undergo laboratory testing for HLH (Table 2) and did not receive HLH-directed therapy. His HAdV infection was managed symptomatically and without antiviral therapy. He continued to have daily temperatures >39°C for >4 weeks but slowly improved, with resolution of his HAdV DNAemia. He was ultimately discharged back to his long-term care facility.
Cases 1–4 underwent testing for Epstein-Barr virus (EBV) by qPCR. Case 1 tested positive but below the limit of quantification of the assay; cases 2–4 tested negative. Cases 2–4 also underwent testing for cytomegalovirus (CMV) by qPCR with negative results. Genetic assessment, including whole human exome sequencing of cases 2–4, did not reveal any known genetic variants associated with primary HLH. Other than cases 2 and 3, there was no history of family history of HLH, death in infancy, or other inflammatory disorder.
METHODS
Laboratory Diagnosis of Adenovirus Infection
For qPCR-based detection of HAdV at CHOP’s Infectious Disease Diagnostics Laboratory, DNA was extracted from 200 µL of each clinical specimen (serum, plasma, nasopharyngeal secretions, or endotracheal aspirate) using an automated MagNA Pure LC instrument and total nucleic acid isolation kit (Roche Diagnostics, Indianapolis, IN, USA). Diagnostic testing for a panel of respiratory viruses including RSV, parainfluenza virus 1–3, adenoviruses, metapneumovirus, rhinoviruses, and coronaviruses was performed on respiratory specimens using in-house, laboratory-derived TaqMan® qPCR assays. HAdV testing of serum or plasma specimens was performed using an in-house qPCR assay targeting a conserved region of the hexon gene, as previously described [8]. For respiratory viruses, a Ct < 40 cycles was considered positive. For the dedicated serum or plasma HAdV qPCR, a Ct < 45 cycles was considered positive. In-house qPCR assays to detect EBV and CMV were also performed on plasma or serum specimens.
Virus Isolation and Molecular Typing
At Lovelace Respiratory Research Institute all HAdV-positive specimens were inoculated onto monolayers of A549 cells for virus isolation and further characterization. For molecular typing and next generation whole genome sequencing (WGS), intracellular viral DNA was purified from A549 cells infected with virus isolates. Initial typing at the species level was accomplished by analysis of BamHI digestion profiles, as previously described [9].
Viral DNA was processed for next generation WGS on an Illumina MiSeq at the Wadsworth Center NYSDOH, as previously described [10]. Contiguous sequences were de novo-assembled using SPAdes 3.5 [11] in the BaseSpace sequence hub (Illumina, Inc., San Diego, CA, USA). Annotated genomic sequences were deposited in GenBank under accession numbers MN422292-MN422295. For molecular typing of those specimens not yielding an isolate in culture, total DNA was extracted from 200 µL of specimen using a NUCLISENS easyMAG instrument (bioMerieux, Durham, NC, USA) and used as template for PCR amplification of a portion of the hexon gene comprising hypervariable regions (HVRs) 1–6, using the primers and cycling conditions developed by Okada and colleagues [12], and DreamTaq PCR master mix (ThermoFisher Scientific, Grand Island, NY, USA). Sanger sequencing of amplicons was performed on a 3130 analyzer (ThermoFisher Scientific) using the amplification primers. Forward and reverse sequences were trimmed and aligned in Sequencher 5.3. The resulting sequences were trimmed to the same length to include HVRs 1–5, identified by BLAST analysis and deposited in GenBank under accession numbers MN822238–MN822243.
Genome typing of sequenced strains was conducted by in silico restriction enzyme analysis (REA) using the panel of endonucleases implemented by Li and Wadell [13] to discriminate HAdV-7 genomic variants.
Genomic Sequence Data Analysis
For phylogenetic analysis, genomic sequences of clinical isolates were aligned with a selection of sequences available from GenBank for HAdV-7 strains circulating in the United States since 2014 and also for HAdV-7 strains detected in the United States and other countries in association with outbreaks or isolated cases of severe ARDS using MAFFT version 7 [14]. A phylogenetic tree was constructed in MEGA 6.0 using the Neighbor-Joining method with 500 bootstrap replicates [15]. In silico REA was carried out using Geneious v6.1.6 (Biomatters, Ltd, New Zealand).
RESULTS
A summary of the virologic findings on available clinical specimens in presented in Table 1. Virus isolates were readily recovered from the respiratory specimens available from patients 2 to 5. Restriction enzyme analysis of viral DNA extracted from infected A549 cells conclusively identified the 4 isolated strains as corresponding to genome type 7d [13]. Whole genome sequence for the 4 respiratory isolates and in silico digestion with a panel of endonucleases confirmed the results of in-gel analysis.
Virus isolates could not be recovered from the available plasma or serum specimens despite the relatively high viral load present in many of them. Most of these specimens were toxic to the cell monolayers and thus incompatible with the process. These specimens were processed for molecular typing. HAdV-7 DNA was detected in several plasma specimens, confirming that the DNAemia was associated with the same virus. Although no viral isolates could be recovered from the serum or plasma samples from patient 1, HAdV-7 DNA of identical partial hexon gene sequence was detected in the serum collected on admission.
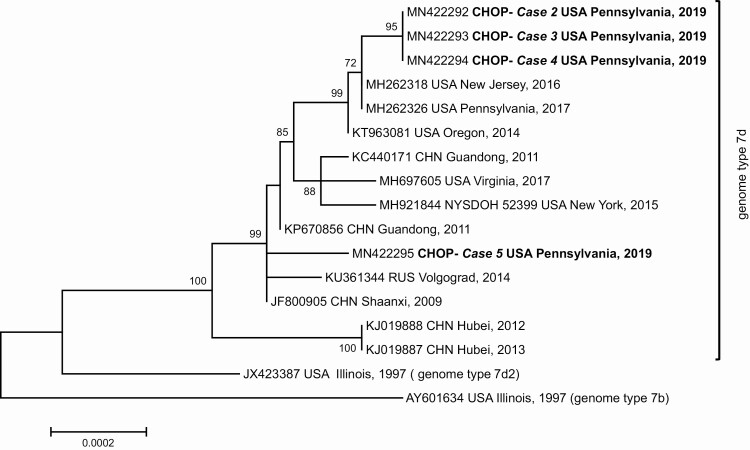
Phylogenetic analysis of genomic sequences obtained for the respiratory isolates of patients 2–5 and previously characterized strains of genome type 7d shows the strains isolated from the 4 cases falling into 2 distinct clades (Figure 1). The first includes the viral isolates identified from cases 2–4, which are closely related to strains circulating in New Jersey and Pennsylvania in 2016 and 2017 [16, 17]. The second includes the strain isolated from case 5, which is more closely related to strains isolated in China in 2009 and in Russia in 2014 [18, 19].
Figure 1.
Phylogenetic analysis of whole genome sequences obtained from the respiratory isolates of the case patients and those available for previously characterized strains of genome type 7d isolated since 2009. Abbreviations: CHN, China; CHOP, The Children’s Hospital of Philadelphia; RUS, Russia.
DISCUSSION
We describe the clinical and virologic characteristics of 4 cases of pediatric HLH and 1 case of hyperinflammatory illness associated with HAdV-7 infection during the 2018–2019 respiratory virus season. In 4 cases the virus was isolated from respiratory specimens and typed as genomic variant HAdV-7d. Virus typing in the 5th case went as far as hexon molecular type with the identification of HAdV-7.
Analysis of passive surveillance data found that HAdV-7 was the 5th-most frequently identified HAdV type in the United States between 2003 and 2016 [20]. HAdV-7 has been associated with outbreaks and isolated cases of severe respiratory illness worldwide [21–25], and various genomic variants have been described [15]. The circulation of variants 7b, 7d2, and 7h has been documented in the United States over the last 2 decades [26–28]. Genomic variant 7d was first detected in 2014 in Oregon and has since been detected in cases of acute respiratory disease in various settings, including colleges and rehabilitation facilities [18, 19, 29]. Previous phylogenetic analysis of WG sequences of HAdV-7d strains identified multiple events of introduction of this variant into the United States, most likely from East Asia where it has caused outbreaks of severe respiratory disease since 2009 [29]. The isolates from cases 2–4 are closely related to those described in association with recent outbreaks in New Jersey [16, 17]. The isolate in case 5 was more closely related to a strain isolated in China during a 2009 outbreak of severe adenovirus pneumonia in infants [19].
The severe clinical phenotypes reported in this case series are similar to those previously described for infections with various HAdV-7 genomic variants and, more recently, for HAdV-7d-associated disease [16, 17, 24, 26, 30]. Symptoms consistent across these reports include fever, pneumonia with respiratory failure, shock, and hepatitis. All patients in our series also had laboratory evidence of hyperinflammation, including elevated CXCL9, interferon (IFN)-γ, and interleukin (IL)-6 in patients 1–4 (Table 2), and elevated CRP in patient 5. The management of HLH in these patients varied but included immune-modulating corticosteroid therapy with only 1 patient receiving directed antiviral therapy. All patients cleared HAdV despite receiving immunosuppressive therapy.
HAdV infections, in particular those by species B, have previously been associated with hyperinflammatory syndromes [31, 32]. In a series of 38 pediatric patients with HAdV infection, elevated levels of IL-6, IL-8, and tumor necrosis factor (TNF)-α were associated with hypoperfusion, septic shock, and increased mortality [31]. Direct comparison to our patients is not possible as HAdV typing and HLH-directed laboratory assessment were not reported, but the description of a severe illness state is consistent with our observations. A case of HAdV-7-associated HLH was recently reported in a 3-year-old previously healthy boy in Pittsburgh, Pennsylvania. This child had severe respiratory failure requiring ECMO [33] and was treated with IVIG, methylprednisolone, and IL-1 blockade. After lack of improvement in his viral load, he received cidofovir. The patient ultimately survived, and an immunological evaluation did not reveal a genetic predisposition to HLH. Detailed HAdV molecular characterization data were not available.
Current treatment guidelines for HLH recommend HSCT for patients with a confirmed genetic predisposition for HLH. In patients with primary HLH, HSCT is a life-saving intervention as recurrence of HLH after future infection events is expected. This is particularly relevant for younger children and infants that sustain an infection-associated HLH event early in life; many infants with familial HLH who have relapsed or progressive disease succumb to HLH prior to HSCT. The odds of death from disease exceed transplant-related mortality. However, not all genetic mutations underlying primary HLH are known, and many experts still recommend HSCT for HLH in infants without an identified genetic mutation, especially as mutations in noncoding regions of the genome have been reported to cause familial HLH [34, 35]. There are clearly infants with no genetic predisposition who develop HLH and undergo unnecessary HSCT, which can have significant morbidity and mortality. Further investigation into the pathogen may provide additional information to guide the classification of primary or secondary HLH and the decision to consider HSCT.
EBV and CMV are frequently associated with the onset of HLH in children [36], but neither virus was detected in the presented cases. It is plausible that some pathogens are more likely than others to induce secondary HLH in the absence of a genetic predisposition. The molecular data obtained for our case series suggest that HAdV-7d can be particularly virulent to the infected host. Furthermore, the young age of some of our patients suggests that the severe clinical presentation may be the result of timing of exposure to HAdV rather than a host predisposition. Importantly, no underlying genetic predisposition for HLH was identified in our patients or in the case reported by Alcamo et al [32].
These data support the importance of pursuing adenovirus typing to guide clinical decisions. A neonate or young child with HLH known to be associated with HAdV-7, more specifically HAdV-7d, and with negative genetic testing may not need referral for HSCT. Nonetheless, the association of HAdV-7d with HLH in our case series only infers causality. At present, it is not known whether different genomic variants of a given HAdV type or different HAdV types differ in their ability to trigger a hyperinflammatory response. This issue warrants further investigation as such evidence may serve to inform the clinician that the inflammatory state is likely a result of the pathogen and thus reduce the concern for an unidentified predisposition in the host.
In conclusion, the described 5 cases of pediatric HLH or HLH-like illness associated with HAdV-7 raise interesting questions about the contribution of the pathogen to the clinical presentation of HLH in young children, particularly when no genetic predisposition is identified. The development of clinically available ex vivo cellular assays to assess the proclivity of a particular pathogen to induce exacerbated cytokine production may further help distinguish the pathogen’s role in the pathogenesis of this hyperinflammatory disorder.
Notes
Acknowledgments. The authors thank Ms Anjinette Lewis at LRRI for technical assistance with virus isolation and genome typing.
Financial support. This work was supported in part by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (T32 HD060550-10) to W. R. O. The Immune Dysregulation Program is supported by the Children’s Hospital of Philadelphia Frontier Programs.
Potential conflicts of interest. E. M. B. has received grants from AB2 Bio Ltd, and NIH/NIAID. B. T. F. has received grants from Merck and Pfizer and has served on a data safety monitoring board for Astellas Pharma Inc. D. T. T. serves on advisory boards for Janssen, Amgen, La Roche, and Sobi. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Henter JI, Horne A, Arico M, et al. HLH-2004: diagnostic and therapeutic guidelines for hemophagocytic lymphohistiocytosis. Pediatr Blood Cancer 2007; 48:124–31. [DOI] [PubMed] [Google Scholar]
- 2.Janka GE. Familial and acquired hemophagocytic lymphohistiocytosis. Annu Rev Med 2012; 63:233–46. [DOI] [PubMed] [Google Scholar]
- 3.Lynch JP 3rd, Kajon AE. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin Respir Crit Care Med 2016; 37:586–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Censoplano N, Gorga S, Waldeck K, Stillwell T, Rabah-Hammad R, Flori H. Neonatal adenovirus infection complicated by hemophagocytic lymphohistiocytosis syndrome. Pediatrics 2018; 141:475–80. [DOI] [PubMed] [Google Scholar]
- 5.Seidel MG, Kastner U, Minkov M, Gadner H. IVIG treatment of adenovirus infection-associated macrophage activation syndrome in a two-year-old boy: case report and review of the literature. Pediatr Hematol Oncol 2009; 20:445–51. [PubMed] [Google Scholar]
- 6.Mellon G, Henry B, Aoun O, et al. Adenovirus related lymphohistiocytic hemophagocytosis: case report and literature review. J Clin Virol 2016; 78:53–6. [DOI] [PubMed] [Google Scholar]
- 7.Houldcroft CJ, Beale MA, Sayeed MA, Qadri F, Dougan G, Mutreja A. Identification of novel adenovirus genotype 90 in children from Bangladesh. Microb Genom 2018; 4:e000221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heim A, Ebnet C, Harste G, Pring-Akerblom P. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol 2003; 70:228–39. [DOI] [PubMed] [Google Scholar]
- 9.Kajon AE, Erdman DD. Assessment of genetic variability among subspecies B1 human adenoviruses for molecular epidemiology studies. In: Wold WSM, Tollefson AE. Adenovirus methods and protocols: volume 2: ad proteins, RNA lifecycle, host interactions, and phylogenetics. Totowa, NJ: Humana Press, 2007:335–55. [DOI] [PubMed] [Google Scholar]
- 10.Kajon AE, Lamson DM, Bair CR, et al. Adenovirus type 4 respiratory infections among civilian adults, Northeastern United States, 2011–2015(1). Emerg Infect Dis 2018; 24:201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012; 19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okada M, Ogawa T, Kubonoya H, Yoshizumi H, Shinozaki K. Detection and sequence-based typing of human adenoviruses using sensitive universal primer sets for the hexon gene. Arch Virol 2007; 152:1–9. [DOI] [PubMed] [Google Scholar]
- 13.Li QG, Wadell G. Analysis of 15 different genome types of adenovirus type 7 isolated on five continents. J Virol 1986; 60:331–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30:772–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 2004; 101:11030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Killerby ME, Rozwadowski F, Lu X, et al. Respiratory illness associated with emergent human adenovirus genome type 7d, New Jersey, 2016–2017. Open Forum Infect Dis 2019; 6:ofz017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rozwadowski F, Caulcrick-Grimes M, McHugh L, et al. Fatalities associated with human adenovirus type 7 at a substance abuse rehabilitation facility—New Jersey, 2017. MMWR Morbid Mortal Wkly Rep 2018; 67:371–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yatsyshina SB, Ageeva MR, Deviatkin AA, et al. Complete genome sequence of human adenovirus 7 associated with fatal adult pneumonia. Genome Announc 2016; 4:e01204–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang L, Wang L, Tan X, Xu W. Adenovirus serotype 7 associated with a severe lower respiratory tract disease outbreak in infants in Shaanxi Province, China. Virol J 2011; 8:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binder AM, Biggs HM, Haynes AK, et al. Human adenovirus surveillance—United States, 2003–2016. MMWR Morb Mortal Wkly Rep 2017; 66:1039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao S, Wan C, Ke C, et al. Re-emergent human adenovirus genome type 7d caused an acute respiratory disease outbreak in Southern China after a twenty-one year absence. Sci Rep 2014; 4:7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erdman DD, Xu W, Gerber SI, et al. Molecular epidemiology of adenovirus type 7 in the United States, 1966–2000. Emerg Infect Dis 2002; 8:269–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerber SI, Erdman DD, Pur SL, et al. Outbreak of adenovirus genome type 7d2 infection in a pediatric chronic-care facility and tertiary-care hospital. Clin Infect Dis 2001; 32:694–700. [DOI] [PubMed] [Google Scholar]
- 24.Scott MK, Chommanard C, Lu X, et al. Human adenovirus associated with severe respiratory infection, Oregon, USA, 2013–2014. Emerg Infect Dis 2016; 22:1044–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cui X, Wen L, Wu Z, et al. Human adenovirus type 7 infection associated with severe and fatal acute lower respiratory illness and nosocomial transmission. J Clin Microbiol 2015; 53:746–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kajon AE, Ison MG. Severe infections with human adenovirus 7d in 2 adults in family, Illinois, USA, 2014. Emerg Infect Dis 2016; 22:730–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan MA, Gray GC, Smith B, McKeehan JA, Hawksworth AW, Malasig MD. Large epidemic of respiratory illness due to adenovirus types 7 and 3 in healthy young adults. Clin Infect Dis 2002; 34:577–82. [DOI] [PubMed] [Google Scholar]
- 28.Gray GC, Setterquist SF, Jirsa SJ, DesJardin LE, Erdman DD. Emergent strain of human adenovirus endemic in Iowa. Emerg Infect Dis 2005; 11:127–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kajon AE, Lamson DM, St George K. Emergence and re-emergence of respiratory adenoviruses in the United States. Curr Opin Virol 2019; 34:63–9. [DOI] [PubMed] [Google Scholar]
- 30.Munoz FM, Piedra PA, Demmler GJ. Disseminated adenovirus disease in immunocompromised and immunocompetent children. Clin Infect Dis 1998; 27:1194–200. [DOI] [PubMed] [Google Scholar]
- 31.Mistchenko AS, Diez RA, Mariani AL, et al. Cytokines in adenoviral disease in children: association of interleukin-6, interleukin-8, and tumor necrosis factor alpha levels with clinical outcome. J Pediatr 1994; 124:714–20. [DOI] [PubMed] [Google Scholar]
- 32.Takahashi I, Takahashi T, Tsuchida S, et al. Pulse methylprednisolone therapy in type 3 adenovirus pneumonia with hypercytokinemia. Tohoku J Exp Med 2006; 209:69–73. [DOI] [PubMed] [Google Scholar]
- 33.Alcamo AM, Wolf MS, Alessi LJ, et al. Successful use of cidofovir in an immunocompetent child with severe adenoviral sepsis. Pediatrics 2020; 145:e20191632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meeths M, Chiang SC, Wood SM, et al. Familial hemophagocytic lymphohistiocytosis type 3 (FHL3) caused by deep intronic mutation and inversion in UNC13D. Blood 2011; 118:5783–93. [DOI] [PubMed] [Google Scholar]
- 35.Tesi B, Lagerstedt-Robinson K, Chiang SC, et al. Targeted high-throughput sequencing for genetic diagnostics of hemophagocytic lymphohistiocytosis. Genome Med 2015; 7:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dropulic LK, Cohen JI. Severe viral infections and primary immunodeficiencies. Clin Infect Dis 2011; 53:897–909. [DOI] [PMC free article] [PubMed] [Google Scholar]