Abstract
Background
Evolutionary analyses of well-annotated human immunodeficiency virus (HIV) sequence data can provide insights into viral transmission patterns and associated factors. Here, we explored the transmission dynamics of the HIV-1 subtype B epidemic across the San Diego (US) and Tijuana (Mexico) border region to identify factors that could help guide public health policy.
Methods
HIV pol sequences were collected from people with HIV in San Diego County and Tijuana between 1996–2018. A multistep phylogenetic approach was used to characterize the dynamics of spread. The contributions of geospatial factors and HIV risk group to the local dynamics were evaluated.
Results
Phylogeographic analyses of the 2034 sequences revealed an important contribution of local transmission in sustaining the epidemic, as well as a complex viral migration network across the region. Geospatial viral dispersal between San Diego communities occurred predominantly among men who have sex with men, with central San Diego being the main source (34.9%) and recipient (39.5%) of migration events. HIV migration was more frequent from San Diego county towards Tijuana than vice versa. Migrations were best explained by the driving time between locations.
Conclusions
The US-Mexico border may not be a major barrier to the spread of HIV, which may stimulate coordinated transnational intervention approaches. Whereas a focus on central San Diego has the potential to avert most spread, the substantial viral migration independent of central San Diego shows that county-wide efforts will be more effective. Combined, this work shows that epidemiological information gleaned from pathogen genomes can uncover mechanisms that underlie sustained spread and, in turn, can be a building block of public health decision-making.
Keywords: HIV, phylogeography, Bayesian discrete phylogeography, generalized linear model
Discrete migration models using human immunodeficiency virus (HIV) sequences show San Diego as a major hub for HIV US-Mexico border region spread, particularly in men who have sex with men. In Mexico, viral migration was more intense towards Tijuana.
San Diego County has the third highest number of human immunodeficiency virus (HIV) cases in California, with an estimated 13 200 persons living with HIV (PWH) [1, 2]. The state of Baja California in Mexico, which includes the city of Tijuana, has the eighth highest HIV prevalence in Mexico, with 9300 PWH, nearly all living in and around Tijuana [3]. Together, these cities constitute the San Diego–Tijuana border region, among the largest metropolitan areas in North America, with approximately 6 million residents. Further, the border between San Diego and Tijuana is the busiest land border-crossing area in the world with 43 million registered crossings in 2018 [4].
The HIV epidemic in the San Diego–Tijuana border region is complex. In contrast to other parts of Mexico and the United States that have epidemics in which the major risk group are men who have sex with men (MSM), the HIV epidemic along the border is more distributed among risk populations, including MSM, persons who inject drugs (PWID), persons who have transactional sex, and Mexican and Central American migrants [5–10]. Hence, the San Diego–Tijuana border region is at the crossroad of multiple risk groups, which leads to transmissions between risk groups [11], possibly enhancing the spread of HIV [12].
Genetic sequence data of pathogens are increasingly used to investigate the transmission dynamics of infectious diseases. This is possible because pathogens evolve as they spread, meaning that their genome contains a genetic imprint of past transmission events. By statistically analyzing this trail of mutations using phylogenetic models, we can detect linkages among infections in time and space that may not be evident otherwise, and gain insights into the processes that govern the spatiotemporal spread among individuals. This holds the potential to support the planning, implementation, and evaluation of public health practices and response. More specifically, understanding the transmission dynamics by identifying the geographic locations and risk groups associated with viral dispersal can help direct effective prevention and surveillance efforts [13–18]. Here, we use a discrete phylogeographic approach, leveraging the evolutionary signal present in HIV sequences, to explore the patterns and trends in the HIV epidemics of the San Diego–Tijuana border region.
METHODS
Compilation of Data Sets for Phylogenetic Inference
Our data set was compiled using all available HIV subtype B partial pol sequences, the predominant circulating subtype in the United States and Mexico [19, 20] and samples obtained in convenience-based efforts from participants enrolled (1) in the San Diego Primary Infection Resource Consortium (SD PIRC) from 1996 to 2018; and (2) in Tijuana as part of the Mexican HIV Drug Resistance Surveillance Network studies (76%) [10, 21] and the Enlaces and El Cuete research studies (24%) [22] from 2008 to 2018. For all participants, sociodemographic information was collected by a counselor or technician at the time of blood sample donation, and included the date of collection, gender, age, and HIV transmission risk (and communities for San Diego). See Supplementary Figure S1 for a map of Tijuana and the communities where SD PIRC participants resided, and the Supplementary Material for cohorts’ descriptions.
All studies were revised and approved by the University of California San Diego Human Research Protections Program or the National Institute of Respiratory Diseases (Mexico City) Ethics Review Board.
Phylogenetic Inference
Transmission networks that best approximate the epidemic dynamics were identified following Cuypers et al [23]. For the identified networks, the spread across risk groups was reconstructed jointly with the geographical migration history in the BEAST 1.10.5 software package [24]. Transmission risks were defined as follows: heterosexual (HTS), MSM, heterosexual people who inject drugs (PWID-HTS), MSM who inject drugs (PWID-MSM), bisexual people, and other (Supplementary Table S1). The missing data for the risk group variable in the Tijuana cohort were accommodated for in the ancestral reconstructions as sampling uncertainty [25]. To accommodate for the different sampling periods of the cohorts, sensitivity analyses were performed in which all sequences from the SD PIRC cohort sampled prior to the inclusion of sequences from Tijuana were excluded (ie, prior to 2008; n = 406). We refer to these analyses as “time-filtered.” A Generalized Linear Model (GLM) extension of the discrete trait model implemented in BEAST 1.10.5 [26] was used to investigate the potential contribution of location-associated variables to the dispersal rates among San Diego County communities and Tijuana.
Details of the methods are provided in the Supplementary Methods.
RESULTS
Population Characteristics
The San Diego–Tijuana data set included a total of 2034 partial pol sequences and associated sociodemographic data collected from 1996–2018 in San Diego (n = 806, including 49.7% sampled after 2008) and 2008–2018 in Tijuana (n = 1228). Enrolled PWH were predominantly male (83.5%; 1445/1730 PWH with available data), and reported an MSM risk (62%; 1004/1621 PWH with available data). Compared to PWH from Tijuana, PWH from San Diego were significantly more likely to be male (97% vs 71.8%, respectively; P < .01) and to report an MSM risk (including MSM also reporting injecting drug use; 93% vs 31.2%, respectively; P < .01; Supplementary Table S1). PWH from San Diego lived across 30 communities, while participants from Mexico were assigned to Tijuana as a single municipality (Supplementary Table S2; Supplementary Figure S1).
Preliminary Phylogenetic Analysis and Down-Sampling
A set of 33 637 HIV-1 subtype B pol sequences from 60 countries across the world, collected between 1992 and 2018, was combined with the San Diego–Tijuana data set. Using a branch support threshold of 0.9 based on a Shimodaira Hasegawa (SH) procedure, 104 highly supported clusters corresponding to independent introductions of HIV-1 B lineages into the San Diego/Tijuana area were identified (see Supplementary Tables S2 and S3). Of these clades, 58 (55.8%) included only sequences from PWH living in Tijuana, 28 (26.9%) comprised only sequences from persons living in San Diego County, and the remaining 18 clades (17.3%) included PWH living in both San Diego and Tijuana. Of these, clades that could not inform on the between-location movement (ie, size <3 or with sequences from a single location) were excluded from further analysis. This down-sampling left 31 clades, of which 15 (48.4%) included sequences from both San Diego County and Tijuana. Using a SH support threshold ≥0.7, the final data set consisted of 41 clades, 19 (46.3%) of which included sequences from both sides of the border (Supplementary Figure S2). Sequences within the SH ≥0.9 clades made up 75% of the sequences within clades identified through the relaxed branch support threshold of 0.7.
Discrete Phylogeographic Inferences
Transmission Dynamics Across San Diego County and Tijuana
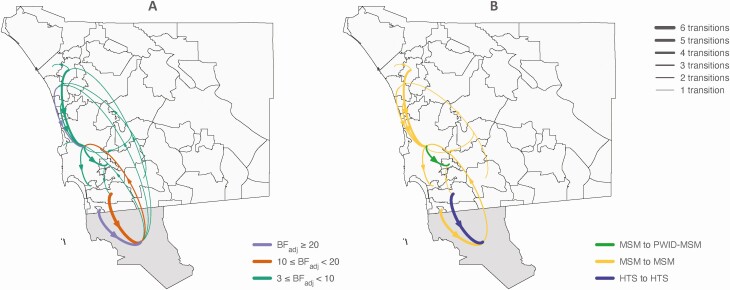
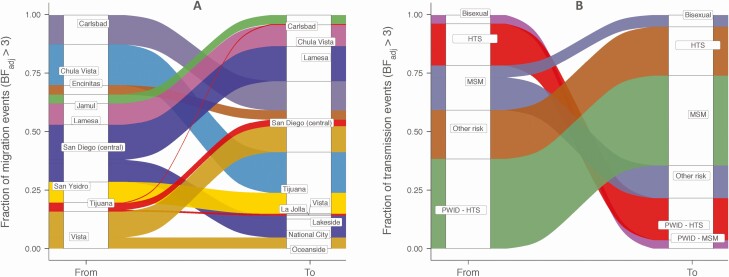
Phylogeographic analyses of the 2034 sequences revealed a complex viral migration history across the region, with support for links between San Diego communities and reciprocal migration between San Diego and Tijuana (Table 1; Figures 1A and 2A). As the level of statistical support for a particular migration link does not inform on the relative importance of that pathway, the estimated number of expected migration events for the subset of well-supported movements was quantified (Figures 1A and 2A). This analysis showed that, on average, 69.7% of migration events occurred within San Diego County, while 30.3% were of cross-border events (Figure 2A). Central San Diego was a major hub of viral migration within San Diego County, as migration from central San Diego to other San Diego communities accounted for 34.9% of all migration events (BF ≥3). Central San Diego was also the dominant destination of virus migration within San Diego County (39.5%), suggesting it was acting as the gravitational center of the San Diego epidemic. Moreover, our model also revealed that migration from San Diego County, mainly from the border communities of Chula Vista and San Ysidro, which are adjacent to and less than 3 miles from the international border, towards Tijuana was more frequent than migration from Tijuana to San Diego (mainly towards La Jolla and central San Diego). Using the more conservative SH clade support threshold of ≥0.7 yielded similar findings (Supplementary Figure S3A; Supplementary Table S4). To evaluate the potential impact of the discrepancies in the sampling period for the SD PIRC cohort (starting in 1996) and Tijuana cohort (starting in 2008), we repeated the analyses after excluding sequences from the SD PIRC cohort collected prior to 2008 (n = 406). The signal for more intense migration from San Diego towards Tijuana was robust to this time filtering (Supplementary Figure S4).
Table 1.
Overview of the Well-Supported Migration Links Across Locations and Risk Groups
| From | To | BF | BFadj | BF | BFadj |
|---|---|---|---|---|---|
| Complete San Diego cohort | Time-filtered San Diego cohort | ||||
| Location | |||||
| Carlsbad | San Diego (central) | 64.04 | 27 | 110.74 | 20.04 |
| Chula Vista | Tijuana | 43.99 | 11.34 | 27.82 | 4.42 |
| Encinitas | San Diego (central) | 15.03 | 3.88 | … | … |
| San Ysidro | Tijuana | 44.46 | 32.43 | 98.68 | 6.64 |
| Tijuana | Vista | 7.5 | 4.08 | 4.78 | 3.59 |
| Jamul | Carlsbad | 39.65 | 3.09 | 29.9 | 3.63 |
| Tijuana | Carlsbad | 4.29 | 3.11 | 5.47 | 3.89 |
| Lamesa | Chula Vista | 14.25 | 3.22 | 16.21 | 4.02 |
| Tijuana | La Jolla | 5.25 | 3.13 | … | … |
| San Diego (central) | Lakeside | 6.39 | 3.08 | 4.45 | 3.50 |
| San Diego (central) | Lamesa | 26.41 | 6.81 | 35.71 | 5.83 |
| San Diego (central) | National City | 16.94 | 4.67 | … | … |
| Vista | Oceanside | 21.21 | 3.99 | 19.66 | 3.41 |
| Tijuana | San Diego (central) | 12.5 | 10.76 | 18.57 | 5.24 |
| Vista | San Diego (central) | 26.41 | 4.97 | 34.54 | 4.47 |
| Escondido | San Ysidro | … | … | 5.12 | 3.50 |
| National City | San Diego (central) | … | … | 17.31 | 3.25 |
| Tijuana | Bonita | … | … | 4.57 | 3.91 |
| Lakeside | Escondido | … | … | 9.53 | 5.56 |
| San Diego (central) | Escondido | … | … | 5.98 | 4.64 |
| Spring Valley | La Jolla | … | … | 9.11 | 6.52 |
| Lamesa | Lakeside | … | … | 3.17 | 3.62 |
| Risk group | |||||
| HTS | PWID-HTS | 33.36 | 16.16 | 47.78 | 26.23 |
| PWID-HTS | MSM | 22.7 | 5.07 | 16.1 | 4.26 |
| Other risk | HTS | 13.19 | 5.7 | 40.04 | 5.15 |
| Bisexual | PWID-MSM | 8.68 | 3.18 | 7.67 | 3.06 |
| MSM | Other risk | 5.89 | 3.04 | … | … |
| MSM | Bisexual | 3.78 | 4.06 | … | … |
| HTS | PWID-MSM | … | … | 4.78 | 3.47 |
| MSM | PWID-MSM | … | … | 8.44 | 85.85 |
| MSM | HTS | … | … | 6.67 | 3.86 |
| PWID-MSM | HTS | … | … | 4.36 | 3.51 |
| Other risk | PWID-MSM | … | … | 3.86 | 4.28 |
The BFs were obtained using a model averaging procedure (Bayesian stochastic search variable selection) [43]. We refer to the Supplementary Methods for details on how the BFadj, which is a more realistic measure of support compared to the “default” BF [44], is obtained. BFadj support of 3 was considered as the lower bound for consideration [45]. Results based on clades identified with SH ≥0.7 are presented in Supplementary Table S4. Abbreviations: BF, Bayes factor; BFadj, adjusted Bayes factor; HTS, heterosexual; MSM, men who have sex with men; PWID, people who inject drugs; SH, Shimodaira Hasegawa.
Figure 1.
Lineage dispersal events between locations (ie, San Diego communities and the city of Tijuana) and between risk groups. A) The thickness of the arrows reflects the average number of inferred migration events between locations, and the color of the arrows indicates the corresponding BFadj support. B) For all migration events between locations with BFadj ≥3, the thickness of the arrows reflects the average number of inferred migration events within or between risk groups, and the color of the arrows indicates the group mixing patterns. Results were obtained from discrete models including clades with SH branch support ≥0.9. Tijuana is colored in darker gray. See also Supplementary Figure S5 for results from discrete models including clades with SH branch support ≥0.7. Abbreviations: BFadj, adjusted Bayes factor; HTS, heterosexual; MSM, men who have sex with men; PWID, people who inject drugs; SH, Shimodaira Hasegawa.
Figure 2.
(A) Relative contribution of the various migration links to the spread of HIV-1 subtype B in the San Diego–Tijuana area and (B) the relative contribution of risk groups to the spread of HIV-1 B in San Diego and the city of Tijuana. A) We present the results from the discrete phylogeographic analysis including clades with SH support ≥0.9. The Sankey plot represents the average proportion of migration events from each source location (“from”) toward the recipient location (“to”). The left side of the plot shows the origin location, and the right side of the plot shows the destination location. We here only report migration events associated with a BFadj support ≥3. All corresponding BFs are presented in Table 1. B) Results are based on the clade identification using SH branch support ≥0.9 and accounting for migration links associated with a BFadj ≥3. The Sankey plot represents the proportion of migration events from each source risk group (“from”) toward the recipient risk group (“to”). Results from the discrete phylogeographic reconstruction based on clades with SH support ≥0.7 are presented in Supplementary Figure S3. All corresponding BFs are presented in Table 1. Colors were chosen to visually clearly distinguish the different types of migration events and have no specific meaning. Abbreviations: BF, Bayes factor; BFadj, adjusted Bayes factor; HIV, human immunodeficiency virus; HTS, heterosexual; MSM, men who have sex with men; PWID, people who inject drugs; SH, Shimodaira Hasegawa.
Transmission Dynamics Between Risk Groups
As each sequence represents a unique PWH, the internal branches in the phylogeny can be assumed to encompass at least 1 migration event [27]. Hence, the inferred risks at the start and end nodes of internal branches can be used to assess the spread within and between risk groups. Among the 31 clades included in the model, MSM accounted for 30.3% of individuals from Tijuana and 92% of PWH living in San Diego county. This was consistent with the local population characteristics (MSM representing 28.7% and 88.3% of PWH from Tijuana and San Diego, respectively). Of the links between risk groups that were significant (adjusted Bayes factor [BFadj] ≥3; Table 1; Supplementary Table S4), those from HTS toward PWID-HTS and from PWID-HTS toward MSM were robust to the cluster identification threshold and the time filtering. Combined, they represent, on average, between 52.0% and 60.8% of all transmissions between risk groups across these analyses (Figure 2; Supplementary Figures S5 and S6; Supplementary Table S5). Transmission from bisexual people toward PWID-MSM was also consistently recovered, but this contributes only little to the mixing between risk groups (range, 1.4–5.1%). MSM and people that reported “other risk” are consistently identified as sources of spread, but spread to each risk group varies with the cluster identification threshold and time filtering. Their contribution to the overall spread also varies widely (the ranges for MSM and other risk are 11.9–24.1% and 3.2–20.9%, respectively).
As the geographical and risk group migration processes were simultaneously inferred, the association between the patterns of spread among locations and risk groups can be probed. Considering only internal branches that accommodate a migration event for SH ≥0.9 and BFadj ≥3, we observed that MSM were the major source risk group (on average, in 70% of the migration events between locations; Figure 1B). Viral migration from San Ysidro and Chula Vista toward Tijuana was among heterosexuals and MSM, respectively. In contrast, migration events from Tijuana towards central San Diego were associated with transmission among MSM. These findings are similar for the model with an SH threshold ≥0.7 (Supplementary Figure S5B).
Estimating Correlates of Viral Migration
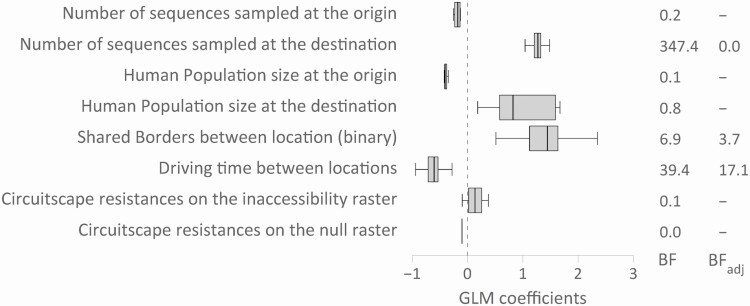
A phylogeography-based GLM analysis was used to investigate the association of potential explanatory variables to the dispersal of HIV across San Diego communities and Tijuana. Given the high degree of collinearity between the population size and number of HIV cases per administrative area, only population size was kept as a predictor in the final model. Here, shorter driving time (BF = 39.4) and shared borders (BF = 6.9) were both associated with the frequency of viral migration. These associations were robust to sampling imbalances (BFadj = 18.4 and 3.7, respectively), suggesting that the movement of HIV between communities in the region is driven mainly by access and proximity (Figure 3). A preliminary analysis clearly indicated that no interaction is expected between both predictors.
Figure 3.
Predictors of migration rates between locations. The box plots report the contribution of each predictor when included in the model. We also report BF support associated with each predictor considered in the GLM, and the BFadj when ≥3. Abbreviations: BF, Bayes factor; BFadj, adjusted Bayes factor.
Discussion
This study focused on the San Diego–Tijuana border region, which encompasses the busiest land border crossing in the Western hemisphere. We found strong support for at least 104 independent transmission networks of HIV-1 subtype B within San Diego County and Tijuana. The majority (71/104; 68.3%) of these clades were comprised of sequences from a single community (central San Diego, n = 13; Tijuana, n = 57) and did not contribute to cross-border viral migration. This suggests that HIV transmission in San Diego County and Tijuana is mainly sustained by local transmission.
Many HIV prevention programs in large cities are focused on the downtown areas, as this approach is thought to most effectively contain viral spread [28], and this is also the first-phase focus of the Ending the HIV Epidemic program [29]. The high proportion of clades with sequences only from downtown San Diego indicates that this strategy makes sense. Furthermore, as transmissions towards other San Diego communities often originated from downtown San Diego (Figure 2A), this approach could also help reduce geographic spread at larger scales. On the flip side, the intensity of these intervention efforts is often significantly reduced in the surrounding suburban communities. San Diego County includes several larger population centers that each have varying numbers of prevalent and incident cases. Virus migration from these suburban communities towards downtown San Diego (Figure 2A) underscores the need for prevention programs (eg, Ending the HIV Epidemic) to be vigilant for infections in all communities across a region, as infections in these peripheral communities can seed new chains of transmission in higher-risk communities and populations [30].
We also uncovered bidirectional viral migration across the San Diego–Tijuana border, which is predominantly from San Diego toward Tijuana. This indicates that controlling infections in San Diego has the potential to also positively affect the HIV burden in Tijuana and, to a lesser extent, vice versa. Specifically, we found evidence for cross-border viral transmission linking the communities of San Ysidro and Chula Vista (San Diego County communities close to the US-Mexico border) and Tijuana. The San Ysidro port of entry is among the busiest land border crossings in the world, with tens of thousands of daily commuters traveling from Tijuana to jobs in San Diego, and US residents working in maquiladoras, purchasing services, or seeking entertainment in Tijuana [31]. San Ysidro is tied closely to Mexico, as 93% of San Ysidro residents were Hispanic and, of those over 5 years old, 87% spoke Spanish [32]. Chula Vista, the second largest city in the San Diego County and also very close to the international border, was also identified as a source of HIV spread to Tijuana. These results show that in spite of anti-HIV programs targeting the border/border towns and local cross-border collaborative initiatives [22, 33], the US-Mexico border does not act as a major barrier to the spread of HIV. This should be a stimulant for more comprehensive efforts, including more coordinated cross-border collaboration. The latter can take the form of a binational border registry to help providers on both sides of the border manage patients that often move between countries. Maintaining these individuals in care with viral suppression is likely to reduce transmission in the region [34]. Data sharing could also be aimed at improved epidemic monitoring; for example, to allow for the timelier identification of growing transmission clusters [35, 36].
Viral migration between communities in the San Diego–Tijuana border region is negatively associated with driving time between communities. This indicates that progress in reducing the infectivity of PWH in 1 area will be most felt in nearby locations. Further analyses evaluating how viral migration into and out of a community is impacted by interventions (eg, screening for acute and early infection) delivered to those communities will be of great interest. With respect to this, promising developments are being made to accommodate such analyses in an online framework [37].
Understanding mixing patterns in transmission risk may also help in understanding which groups are at a greater risk of disassortative transmission (ie, transmission between different risk groups), potentially seeding new outbreaks [11, 38]. We found many links between risk groups (Figure 2; Supplementary Figure S5; Supplementary Table S5). This shows that the border region is a “melting pot” where different types of transmission networks are bridged, which is in line with previous findings [8, 39]. Furthermore, the joint spatial and risk group ancestral reconstructions revealed that viral dispersal within San Diego County occurred almost exclusively among MSM and MSM that also report injecting drug use. Migration events across the San Diego–Tijuana border towards Tijuana were confined within MSM or heterosexual networks, and viral migration towards San Diego was between MSM (Figure 1B; Supplementary Figure S5). Combined, this indicates that the risk group intermixing occurs almost uniquely within communities, and suggests that non-MSM, non-HTS risk groups do not drive longer-distance spread in this border region.
Some aspects of the spread process in the border region could not be captured by our analyses. In particular, the absence of high-resolution sampling location information for the Tijuana municipality implies that the inter-neighborhood spread in Tijuana could not be investigated. Like the general SD PIRC cohort [40, 41], the geo-annotated subset of the SD PIRC cohort reflects the demographics of the HIV population in San Diego (Supplementary Table S1), with a large majority of MSM and a sampling by community that is proportional to the local HIV prevalence [42]. In contrast, determining how well the Tijuana cohort represents the general HIV population in Tijuana is more challenging (see also Supplementary Material). Nonetheless, the Tijuana cohort reflects the gender and risk group characteristics of the local HIV epidemic, including a larger proportion of women (Supplementary Table S1). For the Tijuana general HIV population (and hence also our sample thereof), fear of discrimination and repression likely underlie reporting biases in disclosing risk behaviors; stigmatized risk groups (including MSM and PWID) are likely underreported and, consequently, our results likely underestimate their role in disassortative transmission. Bisexual people are grouped under “other risk” in the San Diego cohort, and because of this their role too is likely underestimated.
CONCLUSION
In conclusion, our analysis of HIV migration across space and risk groups points to an uneven intensity of bidirectional viral migration across the US-Mexico border. We also identified central San Diego as a central hub in the regional geographic spread, and corroborate the importance of MSM in the movement of HIV across the region. The approaches used to gain these insights have the potential to become standard instruments in the public health response toolbox.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Table S1. Population characteristics. Note that PWID who did not report MSM as risk category are assumed to be heterosexual (ie, HTS and PWID).
Supplementary Table S2. Distribution of sequences per location in the original data set (with a total of 2034 sequences) and the final set based on clades with a Shimodaira Hasegawa (SH) support ≥0.9 and 0.7.
Supplementary Table S3. Distribution of sequences per risk group in the original data set (with a total of 2034 sequences) and the final set based on clades with a Shimodaira Hasegawa (SH) support ≥0.9 and 0.7. The same risk group abbreviations as in Supplementary Table S1 are used. MSM: Men who have Sex with Men; PWID: Person Who Inject Drugs.
Supplementary Table S4. Bayes Factor support for all migration events across risk groups. BFadj of 3 was considered as the lower bound for consideration. Results based on clades identified with SH ≥0.7. Results based on clades identified with SH ≥0.9 are presented in Table 1.
Supplementary Table S5. Overview of the average contribution of the various risk groups to disassortative transmission. The contribution is expressed as the percentage of the total number of between-risk group migration events of each particular link. BFadj support of 3 was considered as the lower bound for consideration [45]; when no value is given, there was no BFadj ≥3 support for that particular link between risk groups. Links between risk groups that are always well supported are highlighted in orange. Risk groups that are always well supported as a source of infections in other risk groups, but for which the risk group to which they spread HIV varies across the analyses, are highlighted in blue.
Supplementary Figure S1. Map of communities in San Diego county and Tijuana, Mexico. Areas from where samples were available are highlighted in light gray.
Supplementary Figure S2. Schematic description of the down-sampling procedure. The identification of well-supported clades that capture the San Diego–Tijuana epidemic dynamics was done following Cuypers et al [23]. After their identification, these clades were further down-sampled while preserving all information on the between-location dynamics. For this, we delineated clusters (*) of sequences sampled from the same administrative area (ie, Tijuana or San Diego community), from which 1 randomly selected sequence was kept [14]. See the Methods section for details. SH: Shimodaira Hasegawa.
Supplementary Figure S3. (A) Relative importance of migration events between communities in San Diego and the city of Tijuana and (B) relative importance of migration events between risk groups in San Diego and the city of Tijuana. A) Results from the discrete phylogeographic analysis including clades with Shimodaira Hasegawa (SH) support ≥0.7. The Sankey plot represents the proportion of migration events from each source location (“from”) toward the recipient location (“to”). Left side of the plot shows the source location and right side of the plot shows the destination location. We here only report migration events associated with an adjusted Bayes factor (BFadj) ≥3. B) Results from the discrete phylogeographic analysis including clades with Shimodaira Hasegawa (SH) support ≥0.7. The Sankey plot shows the proportion of migration events from each source risk group toward the recipient risk group. Left side of the plot shows the source risk group (“from”) and right side of the plot shows the destination risk group (“to”). We here only report migration events associated with an adjusted Bayes factor (BFadj) ≥3. Corresponding results from the discrete phylogeographic reconstruction based on clades with SH support ≥0.9 are presented in Figure 2.
Supplementary Figure S4. Number of introductions into San Diego from Tijuana and into Tijuana from San Diego per year based on the time-filtered analysis.
Supplementary Figure S5. Migration events between locations (ie, San Diego communities and the city of Tijuana) and between risk groups. A) The thickness of the arrows reflects the average number of inferred migration events between locations and the color of the arrows indicate the corresponding adjusted Bayes factor (BFadj) support. B) For all migration events between locations with BFadj ≥3, the thickness of the arrows reflects the average number of inferred migration events within or between risk groups and the color of the arrows indicates the group mixing patterns. Results were obtained from discrete models including clades with Shimodaira Hasegawa (SH) support ≥0.7. The Tijuana area is colored in darker gray. See also Figure 1 for results from discrete models including clades with Shimodaira Hasegawa (SH) branch support ≥0.9.
Supplementary Figure S6. Relative importance of migration events between risk groups in San Diego and the city of Tijuana after excluding sequences from the San Diego PIRC cohort collected prior to 2008. We present the results from the discrete phylogeographic analysis including clades with Shimodaira Hasegawa (SH) support ≥0.9 (A) and 0.7 (B). The Sankey plot shows the proportion of migration events from each source risk group toward the recipient risk group. Left side of the plot shows the source risk group (“from”) and right side of the plot shows the destination risk group (“to”). We here only report migration events associated with an adjusted Bayes factor (BFadj) ≥3.
Supplementary Figure S7. Variables tested as predictors of migration rates across locations. (1) The numbers of sequences sampled at the origin/destination were included in the GLM to account for the potential impact of sampling biases within the analysis [26]. (2) The human population size obtained from the United States Bureau of Statistics [32] and the National Institute of Statistics and Geography of Mexico (INEGI) [46] and (3) the number of HIV cases [3, 47]. We also included a binary shared border predictor, a measure of road network connectivity (travel time solely through the road network), as well as pairwise resistance measures computed with the program Circuitscape on a null raster (uniform raster with all accessible cell values set to “1,” tested to account for spatial distance [48]) and on an inaccessibility raster (reflecting the travel time toward nearest cities of at least 50 000 people).
Notes
Disclaimer. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by grants from the National Institutes of Health (San Diego Center for AIDS Research [CFAR]) and the Mexican Government (Comisión de Equidad y Género de las Legislaturas LX-LXI y Comisión de Igualdad de Género de la Legislatura LXII de la Cámara de Diputados de la República Mexicana to G. R.-T.); Consejo Nacional de Ciencia y Tecnología (grant numbers CONACyT SALUD-2017-01-289725 and CONACyT 303079 to S. A.-R.); and NVIDIA Corporation, which donated the Titan V graphics processing unit (GPU) used for this research.
Potential conflicts of interest. B. V. is supported by a postdoctoral grant of the Fonds Wetenschappelijk Onderzoek–Vlaanderen (grant number 12U7118N). S. J. L. acknowledges support from the National Institutes of Health (NIH; grant numbers AI106039 [R24] and MH100974 [R01]); non-financial support in the form of antiretroviral medication from Gilead Sciences during the conduct of the study; and personal fees and non-financial support from Gilead Sciences outside the submitted work. S. D. was supported by the Fonds Wetenschappelijk Onderzoek (Belgium) and is currently funded by the Fonds National de la Recherche Scientifique (Belgium). A. C. was supported by the NIH (grant number AI131971 [R21]). S. R. M. is supported by the NIH (grant numbers AI106039 [R24] and R01 AI35992; PI: Wertheim). H. A. P. and T. P. are supported by the NIH (grant number R01DA037811). H. A. P. reports grants from the National Institute on Drug Abuse during the conduct of the study and grants from Gilead Sciences outside the submitted work. M. H. reports grants from Gilead and Pfizer outside the submitted work. All other authors report no potential conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Health and Human Services Agency County of San Diego. HIV/AIDS epidemiology report 2015. San Diego, California: County of San Diego, 2016. [Google Scholar]
- 2.AIDSVu. AIDSVu: an interactive online mapping tool that visualizes the impact of the HIV epidemic on communities across the United States. Available at: https://aidsvu.org/. Accessed 30 March 2019.
- 3.Centro Nacional para la Prevención y el Control del VIH y el SIDA (CENSIDA). Vigilancia epidemiológica de casos de HIV/AIDS en México, Registro Nacional de Casos de SIDA. Washington, DC: Bureau Of Transportation Statistics, U.S. Department of Transportation. Accessed 11 November 2019. [Google Scholar]
- 4.United States Department of Transportation. Bureau of transportation statistics (BTS). Available at: https://www.bts.gov/topics/national-transportation-statistics. Accessed 15 April 2019.
- 5.Goldenberg S, Silverman J, Engstrom D, Bojorquez-Chapela I, Strathdee S. “Right here is the gateway”: mobility, sex work entry and HIV risk along the Mexico-U.S. border. Int Migr 2014; 52:26–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pitpitan EV, Rocha-Jimenez T, Salazar M, Chavarin C, Magis-Rodriguez C. A mixed methods analysis of the venue-related social and structural context of drug use during sex among male clients of female sex workers in Tijuana, Mexico. AIDS Behav 2020; 24:724–37. doi: 10.1093/ve/vey043 [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Martinez-Donate AP, Simon NE, et al. Risk behaviours for HIV infection among travelling Mexican migrants: the Mexico-US border as a contextual risk factor. Glob Public Health 2017; 12:65–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta SR, Wertheim JO, Brouwer KC, et al. HIV transmission networks in the San Diego–Tijuana border region. EBioMedicine 2015; 2:1456–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marks C, Zúñiga ML. CAM practices and treatment adherence among key subpopulations of HIV+ Latinos receiving care in the San Diego–Tijuana border region: a latent class analysis. Front Public Health 2019; 7:179. doi: 10.1093/ve/vey043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García-Morales C, Tapia-Trejo D, Quiroz-Morales VS, et al. ; HIV Drug Resistance MexNet Group. HIV pretreatment drug resistance trends in three geographic areas of Mexico. J Antimicrob Chemother 2017; 72:3149–58. [DOI] [PubMed] [Google Scholar]
- 11.Le Vu S, Ratmann O, Delpech V, et al. HIV-1 transmission patterns in men who have sex with men: insights from genetic source attribution analysis. AIDS Res Hum Retroviruses 2019; 35:805–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esbjörnsson J, Mild M, Audelin A, et al. ; SPREAD (Strategy to Control SPREAD of HIV Drug Resistance) /European Society for translational Antiviral Research (ESAR) Programme. HIV-1 transmission between MSM and heterosexuals, and increasing proportions of circulating recombinant forms in the Nordic Countries. Virus Evol 2016; 2:vew010. doi: 10.1093/ve/vew010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gräf T, Vrancken B, Maletich Junqueira D, et al. Contribution of epidemiological predictors in unraveling the phylogeographic history of HIV-1 subtype C in Brazil. J Virol 2015; 89:12341–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez AB, Vrancken B, Chueca N, et al. Increasing importance of European lineages in seeding the hepatitis C virus subtype 1a epidemic in Spain. Euro Surveill 2019; 24:1800227. doi: 10.2807/1560-7917.ES.2019.24.9.1800227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia Q, Wertheim JO, Braunstein SL, Misra K, Udeagu CC, Torian LV. Use of molecular HIV surveillance data and predictive modeling to prioritize persons for transmission-reduction interventions. AIDS 2019; 34:459-67. doi: 10.1097/QAD.0000000000002452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little SJ, Kosakovsky Pond SL, Anderson CM, et al. Using HIV networks to inform real time prevention interventions. PLoS One 2014; 9:e98443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wertheim JO, Kosakovsky Pond SL, Little SJ, De Gruttola V. Using HIV transmission networks to investigate community effects in HIV prevention trials. PLoS One 2011; 6:e27775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vrancken B, Cuypers L, Pérez AB, et al. Cross-country migration linked to people who inject drugs challenges the long-term impact of national HCV elimination programmes. J Hepatol 2019; 71:1270–2. [DOI] [PubMed] [Google Scholar]
- 19.Hemelaar J, Gouws E, Ghys PD, Osmanov S. Isolation W-UNfH, characterisation. Global trends in molecular epidemiology of HIV-1 during 2000–2007. AIDS (London, England) 2011; 25:679–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avila-Ríos S, García-Morales C, Garrido-Rodríguez D, et al. National prevalence and trends of HIV transmitted drug resistance in Mexico. PLoS One 2011; 6:e27812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ávila-Ríos S, García-Morales C, Valenzuela-Lara M, et al. ; HIVDR MexNet Group . HIV-1 drug resistance before initiation or re-initiation of first-line ART in eight regions of Mexico: a sub-nationally representative survey. J Antimicrob Chemother 2019; 74:1044–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mehta SR, Chaillon A, Gaines TL, et al. Impact of public safety policies on human immunodeficiency virus transmission dynamics in Tijuana, Mexico. Clin Infect Dis 2018; 66:758–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuypers L, Vrancken B, Fabeni L, et al. Implications of hepatitis C virus subtype 1a migration patterns for virus genetic sequencing policies in Italy. BMC Evol Biol 2017; 17:70. doi: 10.1186/s12862-017-0913-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 2018; 4:vey016. doi: 10.1093/ve/vey016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scotch M, Tahsin T, Weissenbacher D, et al. Incorporating sampling uncertainty in the geospatial assignment of taxa for virus phylogeography. Virus Evol 2019; 5:vey043. doi: 10.3389/fpubh.2019.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemey P, Rambaut A, Bedford T, et al. Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog 2014; 10:e1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero-Severson E, Skar H, Bulla I, Albert J, Leitner T. Timing and order of transmission events is not directly reflected in a pathogen phylogeny. Mol Biol Evol 2014; 31:2472–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ávila-Ríos S, García-Morales C, Valenzuela-Lara M, et al. HIV-1 drug resistance before initiation or re-initiation of first-line ART in eight regions of Mexico: a sub-nationally representative survey. J Antimicrob Chemother 2019; 74:1044–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.HIV.gov. What is ending the HIV epidemic: a plan for America? Available at: https://www.hiv.gov/federal-response/ending-the-hiv-epidemic/overview. Accessed 9 August 2020.
- 30.Mehta SR, Murrell B, Anderson CM, et al. Using HIV sequence and epidemiologic data to assess the effect of self-referral testing for acute HIV infection on incident diagnoses in San Diego, California. Clin Infect Dis 2016; 63: 101–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The General Services Administration. San Ysidro land port of entry. Available at: https://www.gsa.gov/about-us/regions/welcome-to-the-pacific-rim-region-9/land-ports-of-entry/san-ysidro-land-port-of-entry. Accessed 30 August 2019.
- 32.US Census Bureau. American factfinder–results from San Diego County–California. Available at: https://www.census.gov/quickfacts/table/PST045215/06073. Accessed 30 August 2019.
- 33.Strathdee SA, Magis-Rodriguez C, Mays VM, Jimenez R, Patterson TL. The emerging HIV epidemic on the Mexico-U.S. border: an international case study characterizing the role of epidemiology in surveillance and response. Ann Epidemiol 2012; 22:426–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith LR, Patterson TL, Magis-Rodriguez C, et al. Engagement in the HIV care continuum among key populations in Tijuana, Mexico. AIDS Behav 2016; 20:1017–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poon AF, Gustafson R, Daly P, et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study. Lancet HIV 2016; 3:e231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ragonnet-Cronin M, Hodcroft EB, Wertheim JO. Understanding disclosed and cryptic HIV transmission risk via genetic analysis: what are we missing and when does it matter? Curr Opin HIV AIDS 2019; 14:205–12. [DOI] [PubMed] [Google Scholar]
- 37.Gill MS, Lemey P, Suchard MA, Rambaut A, Baele G. Online Bayesian phylodynamic inference in BEAST with application to epidemic reconstruction. Mol Biol Evol 2020; 37:1832–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoenigl M, Chaillon A, Kessler HH, et al. Characterization of HIV transmission in South-East Austria. PLoS One 2016; 11:e0151478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehta SR, Chaillon A, Gaines TL, et al. Impact of public safety policies on human immunodeficiency virus transmission dynamics in Tijuana, Mexico. Clin Infect Dis 2018; 66:758–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hoenigl M, Chaillon A, Morris SR, Little SJ. HIV infection rates and risk behavior among young men undergoing community-based testing in San Diego. Sci Rep 2016; 6:25927. Available at: https://doi.org/10.1038/srep25927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.County of San Diego. HIV/AIDS epidemiology report, 2015. 04/2016. [Google Scholar]
- 42.Chaillon A, Hoenigl M, Freitas L, et al. Optimizing screening for HIV. Open Forum Infect Dis 2020; 7:ofaa024. Available at: https://doi.org/10.1093/ofid/ofaa024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Comput Biol 2009; 5:e1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chaillon A, Gianella S, Dellicour S, et al. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest 2020; 130:1699–712. doi: 10.1172/JCI134815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc 1995; 90:773–95. [Google Scholar]
- 46.National Institute of Statistics and Geography. Mexico population census. Available at: http://www3.inegi.org.mx/sistemas/scitel/Default?ev=5. Accessed December 2018.
- 47.San Diego County Epidemiology Unit. HIV/AIDS epidemiology report Available at: http://www.sandiegocounty.gov/hhsa/programs/phs/hiv_aids_epidemiology_unit/reports_and_statistics.html. Accessed 25 August 2019.
- 48.Dellicour S, Vrancken B, Trovão NS, Fargette D, Lemey P. On the importance of negative controls in viral landscape phylogeography. Virus Evol 2018; 4:vey023. doi: 10.1093/ve/vey023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.