Abstract
Background
Norovirus is a leading cause of epidemic acute gastroenteritis (AGE), with most outbreaks occurring during winter. The majority of outbreaks are caused by GII.4 noroviruses; however, data to support whether this is true for sporadic medically attended AGE are limited. Therefore, we sought to compare the clinical characteristics and seasonality of GII.4 vs non-GII.4 viruses.
Methods
Children aged 15 days -17 years with AGE symptoms were recruited from the outpatient, emergency department, and inpatient settings at Vanderbilt Children’s Hospital, Davidson County, Nashville, Tennessee, from December 2012 -November 2015. Stool specimens were tested using qRT-PCR for GI and GII noroviruses and subsequently genotyped by sequencing a partial region of the capsid gene.
Results
A total of 3705 patients were enrolled, and stool specimens were collected and tested from 2885 (78%) enrollees. Overall, 636 (22%) samples were norovirus-positive, of which 567 (89%) were GII. Of the 460 (81%) genotyped GII-positive samples, 233 (51%) were typed as GII.4 and 227 (49%) as non-GII.4. Compared with children with non-GII.4 infections, children with GII.4 infections were younger, more likely to have diarrhea, and more likely to receive oral rehydration fluids. Norovirus was detected year-round and peaked during winter.
Conclusions
Approximately 40% of sporadic pediatric norovirus AGE cases were caused by GII.4 norovirus. Children infected with GII.4 had more severe symptoms that required more medical care. Seasonal variations were noticed among different genotypes. These data highlight the importance of continuous norovirus surveillance and provide important information on which strains pediatric norovirus vaccines should protect against.
Keywords: norovirus, surveillance, sporadic, genotypes, acute gastroenteritis
GII.4 noroviruses are the most common genotype to cause medically attended pediatric acute gastroenteritis infections in Davidson County, Tennessee. This genotype was associated with more severe disease compared with other genotypes.
Norovirus is a major cause of acute gastroenteritis (AGE) outbreaks [1] and is associated with recurrent infections throughout life among all age groups [1, 2]. After the introduction of rotavirus vaccine led to a substantial reduction in pediatric rotavirus AGE, norovirus has emerged as the leading cause of pediatric AGE, with more than 1 million pediatric healthcare visits attributed to norovirus infections every year in the United States [1, 3–5]. Recent studies from the New Vaccine Surveillance Network documented norovirus as the leading cause of AGE hospitalization and emergency department (ED) visits in children aged <5 years, and they also reported that GII.4 was the most common genotype among these medically attended pediatric infections [5]. Noroviruses are genetically and antigenically diverse single-stranded RNA viruses that can now be classified into at least 10 genogroups (GI–GX), of which GI and GII viruses cause the majority of infections in humans. Genogroups are further subdivided into at least 49 genotypes based on complete capsid protein sequences [6], with GII.4 viruses being responsible for the majority of outbreaks over the last 15 years worldwide [7–10]. New GII.4 strains typically emerge every 2–4 years and replace previous predominant strains, often causing more severe AGE [11].
Since several norovirus vaccines are currently under development [12–14], understanding the burden and severity of norovirus disease, including which genotypes predominate, is important [5]. Identifying which genotypes could be associated with more severe presentations that warrant hospitalization in young children compared with the genotypes that are associated with milder presentation that can be managed in the outpatient (OP) settings will provide essential information. Therefore, we compared the clinical characteristics and distribution among norovirus genotypes in children aged <18 years who presented with AGE symptoms and sought medical care in 3 medical settings over 3 consecutive seasons. We also compared demographics, clinical characteristics, and genotype distributions between norovirus-positive AGE cases and healthy controls (HC).
METHODS
Study Design
We retrospectively analyzed a cohort of norovirus-positive children who were part of a 3-year, prospective, active, population-based AGE surveillance study [15]. Children who resided in Davidson County, Tennessee, and presented to Monroe Carrell Jr Children’s Hospital at Vanderbilt University Medical Center (VUMC) in Nashville, Tennessee, were enrolled from 3 settings: OP, ED, and inpatient (IP) between 1 December 2012 and 30 November 2015 [16]. AGE was defined as diarrhea (≥3 episodes of loose stools within 24 hours) and/or vomiting (≥1 episode in 24 hours) within 10 days of enrollment [5].
Study Population
AGE Cases
Children aged 15 days–17 years with AGE who presented to ED or OP clinic settings and admitted children aged 15 days–10 years were approached and enrolled if eligible and if the parent or guardian consented. Patients were excluded if they had a noninfectious cause of diarrhea, were immunocompromised, previously enrolled for the same AGE episode (defined as ≤3 symptom-free days), or transferred from another hospital after 48 hours of admission. Children enrolled in the ED and OP settings were reclassified as IP if they were hospitalized for their illness and had no alternative diagnosis noted upon medical chart review.
Healthy Controls
Children aged 15 days–17 years who resided in Davidson County were approached during scheduled well-child visits at the VUMC pediatric OP clinic. HC were frequency-matched by age, race/ethnicity, and the time of enrollment based on AGE case patients who provided a stool sample, with a case-control ratio of 2:1 in the first 2 study years and 3:1 in the third study year. Children were deemed ineligible if they reported acute respiratory infection symptoms within 3 days of enrollment, AGE symptoms within 14 days of enrollment, or clinical immunodeficiency.
After obtaining informed written consent from a parent or guardian, demographic and clinical data were collected through parent/guardian interviews, and chart reviews were performed to determine outcome data. Institutional review board approval was obtained from the Centers for Disease Control and Prevention [2], Tennessee Department of Health (TDOH), and VUMC.
Specimen Collection and Testing
Whole stool specimens were collected within 5 and 10 days of enrollment for HC and AGE cases, respectively. Those who provided a stool sample outside this window were excluded. Norovirus testing was performed by TDOH using quantitative reverse-transcription polymerase chain reaction as previously described [10, 15, 17]. Norovirus-positive specimens were genotyped by sequencing a partial region of the capsid gene [15, 18].
Data Analyses
Baseline Demographics and Clinical Characteristics
Descriptive statistics were summarized as frequency (percentage), median (interquartile range), or mean (standard deviation) where appropriate. Only typable norovirus GII strains were included in strain analyses; GI, mixed GI/GII, and nontypable GII viruses were excluded from the analyses. GII.4 and non-GII.4 genotypes were compared using the Pearson χ2 test for categorical variables. The 2-sample t test, allowing unequal variances or 1-way analysis of variance, was used for continuous variables. A significance level of 0.05 (2-tailed) was used for all analyses. All statistical analyses were performed using Stata version 15.0 (StataCorp, College Station, TX). Cycle threshold (Ct) values were used as a proxy for viral load, with lower Ct values representing higher viral loads. Ct values of GII.4 viruses were compared to values of non-GII.4 viruses.
Regression Analysis Model
Among children infected with GII norovirus, a multivariable logistic model with robust standard errors was used to compare the odds of oral rehydration fluid use across predictors of interest, including age, gender, race, ethnicity, prematurity, current or history of breastfeeding, daycare/preschool/school attendance, viral codetection, norovirus genogroup (GII.4 vs non-GII.4), and norovirus Ct values.
RESULTS
Study Population
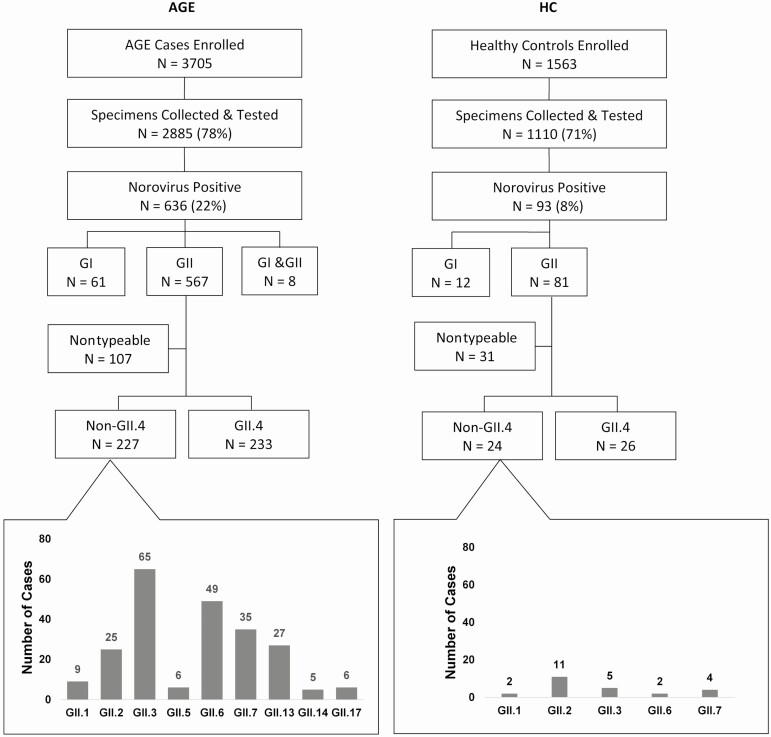
Over the 3-year study period, 5434 AGE cases were eligible for enrollment, of which 3705 (68%) were enrolled (Figure 1). Of those enrolled, 2885 (78%) had stool collected and viral test results available; 636 (22%) tested positive for norovirus (Figure 1). Among the norovirus-positives, 567 (89%) tested positive for GII, 61 (10%) for GI, and 8 (1%) were mixed GI/GII infections (Figure 1). The median age of symptomatic children infected with norovirus was 19 months (interquartile range [IQR], 10.7–44.2); 50% were male, 64% were white, and 45% were Hispanic. Of the 636 norovirus-positive patients, 350 (55%) were enrolled from the ED, 236 (37%) from the OP setting, and 50 (8%) from the IP setting.
Figure 1.
Study enrollment algorithm for AGE cases and HC and results of norovirus protein capsid sequencing. Abbreviations: AGE, acute gastroenteritis; HC, healthy control.
For HC, 1563 were enrolled and 1110 (71%) had stool collected and viral test results available; 93 (8.4%) tested positive for norovirus; 81 (87%) were GII and 12 (13%) were GI (Figure 1). The median age of norovirus-positive HC was 14 months (IQR, 7.5–22.6), 57% were male, 54% were white, and 44% were Hispanic.
Clinical and Demographic Characteristics of GII.4 vs Non-GII.4 Infections
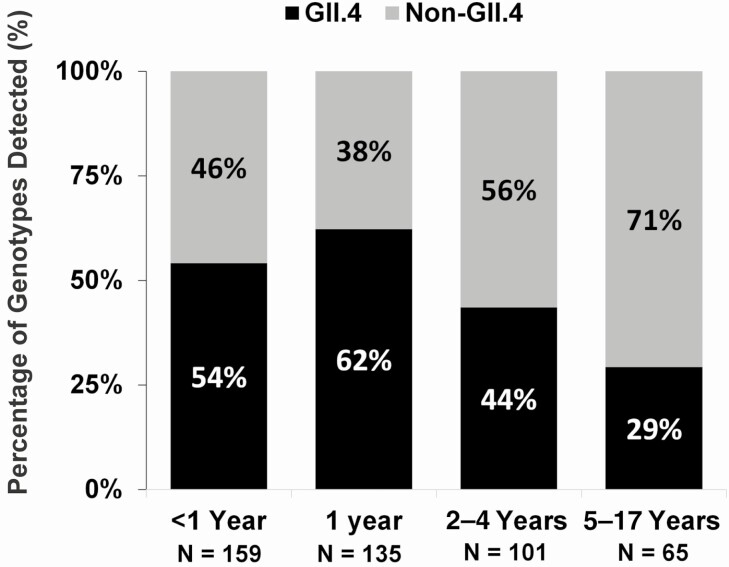
Among 567 AGE cases with a GII norovirus-positive specimen, 107 (19%) could not be typed, 233 (41%) were typed as GII.4, and 227 (40%) were typed as non-GII.4 (Figure 1). Children infected with GII.4 norovirus were younger, more likely to receive oral rehydration fluids, and less likely to attend daycare/preschool/school than those infected with non-GII.4 norovirus (P < .05 for all variables; Table 1). When comparing Ct values, GII.4 infections had lower mean Ct values compared with non-GII.4 infections (20.35 ± 4.22 vs 21.16 ± 4.455, P < .001). There was no difference in the frequency of codetection of other viruses between the 2 groups (GII.4, 6% vs non-GII.4,10%; P = .140). Figure 2 displays the age groups by genotype, with similar distribution by genotype in children aged <5 years. However, nearly three-quarters of the older children had non-GII.4 genotypes. Regardless of genotype, most children infected with GII norovirus were managed in the ED. Moreover, there was no significant difference between the detection of GII.4 (53% ED, 38% OP, 9% IP) and non-GII.4 (53% ED, 40% OP, 7% IP) among the 3 clinical settings (P = .58).
Table 1.
Characteristics of Patients With Acute Gastroenteritis and Typeable GII Genotype Detection
| Non-GII.4 (n = 227) | GII.4 (n = 233) | P Value | |
|---|---|---|---|
| Demographics | |||
| Age, mean ± standard deviation, mo | 36.07 ± 37.30 | 26.58 ± 35.18 | .005 a |
| Age, median (interquartile range), mo | 21 (10–52) | 15 (10–26) | |
| Sex, male | 107 (47) | 124 (53) | .192b |
| White | 143 (63) | 156 (67) | .084b |
| Black/African American | 72 (32) | 56 (24) | |
| Other/None/Mixed | 12 (5) | 21 (9) | |
| Hispanic or Latino | 103 (45) | 111 (48) | .626b |
| Preterm | 16 (7) | 19 (8) | .905b |
| Daycare/Preschool/School | 88 (39) | 52 (23) | <.001 b |
| Current or history of breastfeeding | 176 (78) | 171/232 (74) | .340b |
| Antibiotics during admission | 10/225 (4) | 5 (2) | .167b |
| Symptoms at presentation | |||
| Duration of illness, days | 2.47 ± 1.82 | 2.72 ± 1.83 | .153a |
| Fever | 112 (49) | 100 (43) | .167b |
| Duration, days | 1.96 ± 1.49 | 1.90 ± 1.29 | .728a |
| Vomitingc | 212 (93) | 227 (97) | .038 b |
| Duration, days | 1.82 ± 1.17 | 2.02 ± 1.33 | .088a |
| Episodes | 6.03 ± 4.24 | 6.61 ± 5.25 | .208a |
| Diarrheac | 128 (56) | 174/232 (75) | <.001 b |
| Duration, days | 2.43 ± 1.65 | 2.55 ± 1.85 | .552a |
| Episodes | 5.08 ± 4.22 | 5.95 ± 5.44 | .138 |
| Severity | |||
| Intensive care unit admission | 0 | 1/23 (4) | .398b |
| Dehydration (skin test)d | 15/216 (7) | 12/220 (5) | .519b |
| Oral rehydration fluids | 101 (44) | 139 (60) | .001b |
| IV rehydration before admission | 7 (3) | 2/231 (1) | .087b |
| IV rehydration during admission | 29/226 (13) | 35 (15) | .498b |
Categorical data are n (%), continuous data are mean ± standard deviation, and median (interquartile range).
Abbreviation: IV, intravenous.
at test.
bPearson χ2 test.
cYes/no question for any episode.
dFor the skin test, the child’s abdominal skin was pulled for 1 second. The interviewer measured the time for the skin fold to retract. A child was considered dehydrated if their skin retracted slowly (ie, skin fold remains visible for 1–2 seconds).
Figure 2.
Frequency of GII.4 vs non-GII.4 in children with acute gastroenteritis by age group.
GII.4 patients had a higher frequency of vomiting and diarrhea than non-GII.4 patients (P = .038 and P < .001, respectively; Table 1). When we compared the distribution of AGE symptom presentations, non-GII.4 patients had a higher frequency of presenting with vomiting only, fever and vomiting, and fever and diarrhea compared with GII.4 children (P < .05 for all 3 variables; Supplementary Figure 1). However, no significant differences in the duration of diarrhea, vomiting, and fever between the 2 groups were noted (Table 1).
Disease Severity
On multivariable logistic regression, GII.4 norovirus genotype infection was the only predictor directly associated with increased odds of oral rehydration fluid use (odds ratio, 1.727; 95% confidence interval, 1.177–2.533; P = .005; Table 2).
Table 2.
Coefficient Estimates From Logistic Regression Models to Evaluate Association Between Variables Associated With Oral Rehydration Fluid Use
| Oral Rehydration Fluid | |||
|---|---|---|---|
| Odds Ratio | 95% Confidence Interval | P Value | |
| GII.4 norovirus genotype | 1.727 | 1.177–2.533 | .005 |
| Age, mo | 0.994 | .988–1.00 | .070 |
| Male | 0.845 | .574–1.242 | .392 |
| White | 1.348 | .803–2.261 | .258 |
| Hispanic or Latino | 0.606 | .32–1.617 | .969 |
| Prematurity | 0.865 | .434–1.724 | .681 |
| Current or history of breastfeeding | 0.885 | .565–1.384 | .591 |
| Daycare/Preschool/School | 1.003 | .620–1.622 | .990 |
| Viral codetection | 1.083 | .532–2.204 | .826 |
| Cycle threshold values | 0.998 | .955–1.043 | .921 |
Comparison of GII.4 and Non-GII.4 Seasonality
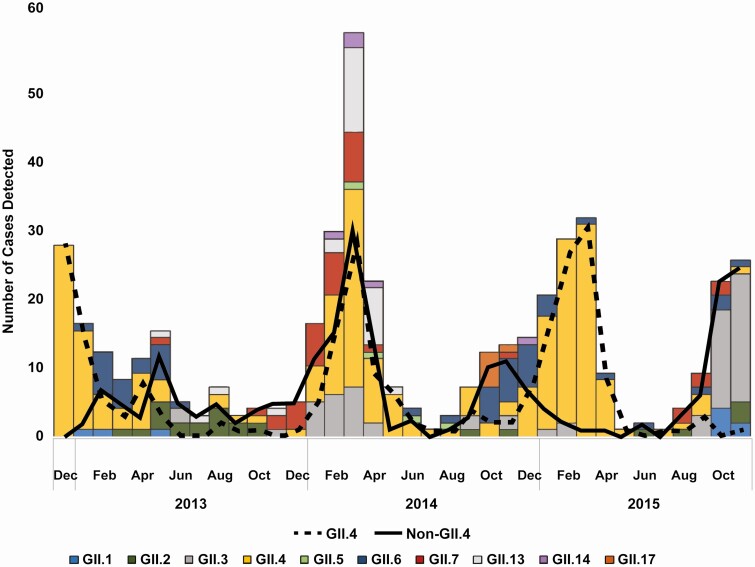
Norovirus was detected year-round during the study period (Figure 3). GII.4 noroviruses were not detected in several summer months but had significant peaks during winter months in each study year. In contrast, non-GII.4 viruses were detected throughout the year, with multiple peaks observed over time (Figure 3). In order, GII.3, GII.6, GII.7, GII.13, and GII.2 were the most common non-GII.4 genotypes detected over the 3-year study period (Figures 1 and 3).
Figure 3.
Seasonal patterns of norovirus GII.4 vs non-GII.4 and all GII genotypes during 2012–2015.
Norovirus-positive AGE Cases vs Controls
In HC, 50/81 (62%) GII norovirus infections were typable, of those, 26/50 (52%) were GII.4. The most common non-GII.4 in HC was GII.2 (46%; Figure 1 and Supplementary Figure 2). Among all children infected with GII norovirus, HC had higher Ct values compared with AGE cases, indicating a lower viral load (24.2 ± 4.4 vs 20.6 ± 4.2, P < .001). AGE cases were more likely to attend daycare/preschool/school (34% vs 15%, P = .001) and were older (34 ± 39.5 vs 21 ± 22.3, P < .001) compared with HC, but there was no difference in sex, race, ethnicity, prematurity, current or history of breastfeeding, or viral codetection between the 2 groups.
DISCUSSION
Of the pediatric sporadic norovirus medically attended AGE cases, norovirus GII.4 accounted for nearly 40%. Our findings are consistent with those from other published studies from the United States that also noted the predominance of GII.4 viruses both in outbreak and nonoutbreak settings [5, 18–20]. Children infected with GII.4 also had more diarrhea and vomiting and were more likely to receive oral rehydration fluids compared with children infected with other non-GII.4 genotypes. In particular, GII.4 norovirus infection was independently associated with increased odds of oral rehydration fluid use, even with adjusting for other risk factors, including age, breastfeeding, and Ct values. Our data suggest that GII.4 viruses are associated with more severe disease compared with other genotypes.
The greater severity of GII.4 norovirus infections is also consistent with previous studies [21–23]. For instance, a Finnish study of children aged <2 years found that children with GII.4 infections had a higher overall AGE severity score compared with those infected with other genotypes [22]. Moreover, in norovirus outbreaks in the United States, GII.4 viruses were more likely to be associated with a more severe outcome [24], including in subsequent seasons [25–28]. Factors that may explain the higher severity of illness associated with GII.4 in our cohort include younger age and higher viral loads (eg, lower Ct values). In fact, several studies noted that a higher viral load was associated with more severe symptoms [29, 30]. In addition, GII.4 strains may be intrinsically more virulent due to increased breadth of ligand binding on cell surfaces and their rapid evolution that favors evasion of the host immune response [23, 24, 31, 32].
Interestingly, we also noted age differences among children with a known GII genotype. For example, the frequency of GII.4 decreased inversely with age. Non-GII.4 genotypes also represented nearly three-quarters of children in the 5– 17-year age group, which suggests a degree of immune protection elicited by previous GII.4 infections. These age differences were also found in a study from Hong Kong that reported that GII.4 viruses accounted for the majority of AGE cases in children aged <5 years, while non-GII.4 viruses were more prevalent in older age groups, including younger adults [33]. Further surveillance studies in other cohorts are needed to determine whether these findings are consistent across diverse geographic and clinical settings. Although GII.4 viruses are the most commonly detected genotype, non-GII.4 viruses are cocirculating, including year-round, and the particular genotypes vary in different years. We noted seasonal differences for several non-GII.4 viruses that showed a higher prevalence during summer and autumn, while GII.4 viruses had a clear winter seasonality [10] as has been reported previously [34]. In addition, we also found that GII.3, GII.6, and GII.7 followed by GII.2 and GII.13 were the most common non-GII.4 viruses. In the same study mentioned above from Hong Kong, GII.17 was the most common genotype in 2014/2015 and GII.2 in 2016/2017 [33]. Another study from Thailand, in which the investigators tested fecal/rectal swab samples from both adult and pediatric hospitalized patients with AGE, reported that 2/3 of patients with a positive norovirus test were aged <5 years and that GII.17, GII.3, and GII.6 were the most common non-GII.4 genotypes [35]. Among medically attended norovirus-infected children in the United States during 2009–2011, GII.12 was the most common genotype second only to GII.4 [5]. These differences highlight the dynamic nature of norovirus molecular epidemiology and indicate a need for continued molecular strain surveillance, ideally using dual typing of strains [10] in order to identify newly emerging strains and their impact [18, 33, 36–38].
GII.4 was the most common genotype in HC followed by GII.2 viruses. In contrast, GII.3 was most prevalent second to GII.4 in AGE cases. In addition, compared with HC, AGE cases had higher viral loads for all genotypes, consistent with what we found in our previous study [15], demonstrating that higher viral loads correlate with symptomatic infections and likely contribute to a higher likelihood of viral transmission [30]. Compared with HC, AGE patients were older and more likely to attend daycare/preschool/school. The latter finding suggests an increased risk of exposure to norovirus in daycare/preschool/school in nonoutbreak settings [39–41]. The impact of norovirus genotype and age on transmission is unclear and warrants further investigation [24].
Our study has several limitations and strengths. Several children in the older age groups did not provide stool samples and were excluded from the study. Of those who provided a stool sample and tested positive for norovirus, we were not able to genotype all samples. Using a multivariate logistic regression model, Ct values were the only independent predictor of typing success, which explains the typing failure of samples with a high Ct value (ie, lower viral load provides insufficient RNA for downstream sequencing). We included only patients from Monroe Carrell Jr Children’s Hospital at VUMC–Davidson County, and thus our findings may not be generalizable to other geographic regions. However, our study is unique in that we included children infected with norovirus from 3 clinical settings (OP, ED, and IP), which generates a more representative sample of children with medically attended AGE, accounting for the potential differences in severity, causative agents, and variable management in different clinical settings. In addition, due to our large sample size, we were able to note significant clinical and demographic differences among norovirus genotypes. Also, nearly 80% of the children enrolled provided a stool sample, and nearly 3000 stool samples were tested.
In summary, GII.4 norovirus was consistently the most common genotype in children who presented with AGE and caused a more severe illness. Seasonal variations were noticed among different genotypes. These data might help in strain selection for candidate norovirus vaccines and also highlight the importance of continuous surveillance of norovirus genotypes in different geographic regions for vaccine formulation.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the New Vaccine Surveillance Network, our clinical trial associates, and the families that participated in this study.
Financial support. This work was supported by the Centers for Disease Control and Prevention (CDC) Cooperative (grant U01IP001063) and [UL1 TR000445] from the National Center for Advancing Translational Sciences at the National Institute of Health.
Potential conflicts of interest. Z. H., L. S., J. C., and N. H. report grants from the CDC. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Hall AJ, Eisenbart VG, Etingüe AL, Gould LH, Lopman BA, Parashar UD. Epidemiology of foodborne norovirus outbreaks, United States, 2001–2008. Emerg Infect Dis 2012; 18:1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Available at: https://www.cdc.gov/norovirus/index.html. Accessed 2 February 2020.
- 3.Ahmed SM, Hall AJ, Robinson AE, et al. Global prevalence of norovirus in cases of gastroenteritis: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14:725–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wikswo ME, Kambhampati A, Shioda K, Walsh KA, Bowen A, Hall AJ. Outbreaks of acute gastroenteritis transmitted by person-to-person contact, environmental contamination, and unknown modes of transmission—United States, 2009–2013. MMWR Surveill Summ 2015; 64:1–16. [DOI] [PubMed] [Google Scholar]
- 5.Payne DC, Vinjé J, Szilagyi PG, et al. Norovirus and medically attended gastroenteritis in US children. N Engl J Med 2013; 368:1121–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chhabra P, de Graaf M, Parra GI, et al. Updated classification of norovirus genogroups and genotypes. J Gen Virol 2019; 100:1393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson C, Bargatze RF, Goodwin R, Mendelman PM. Norovirus virus-like particle vaccines for the prevention of acute gastroenteritis. Expert Rev Vaccines 2013; 12:155–67. [DOI] [PubMed] [Google Scholar]
- 8.Atmar RL, Estes MK. Norovirus vaccine development: next steps. Expert Rev Vaccines 2012; 11:1023–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vinjé J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol 2015; 53:373–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cannon JL, Barclay L, Collins NR, et al. Genetic and epidemiologic trends of norovirus outbreaks in the United States from 2013 to 2016 demonstrated emergence of novel GII.4 recombinant viruses. J Clin Microbiol 2017; 55:2208–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee BE, Pang XL. New strains of norovirus and the mystery of viral gastroenteritis epidemics. CMAJ 2013; 185:1381–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robilotti E, Deresinski S, Pinsky BA. Norovirus. Clin Microbiol Rev 2015; 28:134–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattison CP, Cardemil CV, Hall AJ. Progress on norovirus vaccine research: public health considerations and future directions. Expert Rev Vaccines 2018; 17:773–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aliabadi N, Lopman BA, Parashar UD, Hall AJ. Progress toward norovirus vaccines: considerations for further development and implementation in potential target populations. Expert Rev Vaccines 2015; 14:1241–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Halasa N, Piya B, Stewart LS, et al. The changing landscape of pediatric viral enteropathogens in the post-rotavirus vaccine era. Clin Infect Dis 2020; ciaa100. doi: 10.1093/cid/ciaa100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Payne DC, Staat MA, Edwards KM, et al. Active, population-based surveillance for severe rotavirus gastroenteritis in children in the United States. Pediatrics 2008; 122:1235–43. [DOI] [PubMed] [Google Scholar]
- 17.Chhabra P, Payne DC, Szilagy PG, et al. Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. J Infect Dis 2013; 208:790–800. [DOI] [PubMed] [Google Scholar]
- 18.Vega E, Barclay L, Gregoricus N, Shirley SH, Lee D, Vinjé J. Genotypic and epidemiologic trends of norovirus outbreaks in the United States, 2009 to 2013. J Clin Microbiol 2014; 52:147–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez MD, Langley LC, Buchan BW, et al. Multicenter evaluation of the Xpert norovirus assay for detection of norovirus genogroups I and II in fecal specimens. J Clin Microbiol 2016; 54:142–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siebenga JJ, Vennema H, Zheng DP, et al. Norovirus illness is a global problem: emergence and spread of norovirus GII. 4 variants, 2001–2007. J Infect Dis 2009; 200:802–12. [DOI] [PubMed] [Google Scholar]
- 21.Desai R, Hembree CD, Handel A, et al. Severe outcomes are associated with genogroup 2 genotype 4 norovirus outbreaks: a systematic literature review. Clin Infect Dis 2012; 55:189–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huhti L, Szakal ED, Puustinen L, et al. Norovirus GII-4 causes a more severe gastroenteritis than other noroviruses in young children. J Infect Dis 2011; 203:1442–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friesema IH, Vennema H, Heijne JC, et al. Differences in clinical presentation between norovirus genotypes in nursing homes. J Clin Virol 2009; 46:341–4. [DOI] [PubMed] [Google Scholar]
- 24.Burke RM, Shah MP, Wikswo ME, et al. The norovirus epidemiologic triad: predictors of severe outcomes in US norovirus outbreaks, 2009–2016. J Infect Dis 2018; 219:1364–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leshem E, Wikswo M, Barclay L, et al. Effects and clinical significance of GII.4 Sydney norovirus, United States, 2012-2013. Emerg Infect Dis 2013; 19:1231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yen C, Wikswo ME, Lopman BA, Vinje J, Parashar UD, Hall AJ. Impact of an emergent norovirus variant in 2009 on norovirus outbreak activity in the United States. Clin Infect Dis 2011; 53:568–71. [DOI] [PubMed] [Google Scholar]
- 27.Hall AJ, Vinjé J, Lopman B, et al. Updated norovirus outbreak management and disease prevention guidelines. MMWR Recomm Rep 2011; 60:1–15. [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. Norovirus activity— United States, 2006–2007. MMWR Morb Mortal Wkly Rep 2007; 56:842. [PubMed] [Google Scholar]
- 29.Kabue JP, Meader E, Hunter PR, Potgieter N. Norovirus prevalence and estimated viral load in symptomatic and asymptomatic children from rural communities of Vhembe district, South Africa. J Clin Virol 2016; 84:12–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shioda K, Barclay L, Becker-Dreps S, et al. Can use of viral load improve norovirus clinical diagnosis and disease attribution? Open Forum Infect Dis 2017; 4: ofx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lindesmith LC, Donaldson EF, Lobue AD, et al. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Med 2008; 5:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Donaldson EF, Lindesmith LC, Lobue AD, Baric RS. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol Rev 2008; 225:190–211. [DOI] [PubMed] [Google Scholar]
- 33.Chan MCW, Kwok K, Zhang LY, et al. Bimodal seasonality and alternating predominance of norovirus GII. 4 and non-GII. 4, Hong Kong, China, 2014–2017. Emerg Infect Dis 2018; 24:767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng DP, Widdowson MA, Glass RI, Vinjé J. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. J Clin Microbiol 2010; 48:168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chuchaona W, Chansaenroj J, Wanlapakorn N, Vongpunsawad S, Poovorawan YRecombinant GII. Pe-GII. 4 norovirus, Thailand, 2017–2018. Emerg Infect Dis 2019; 25:1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vega E, Barclay L, Gregoricus N, Williams K, Lee D, Vinjé J. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerg Infect Dis 2011; 17:1389–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan MC, Leung TF, Chung TW, et al. Virus genotype distribution and virus burden in children and adults hospitalized for norovirus gastroenteritis, 2012–2014, Hong Kong. Sci Rep 2015; 5:11507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathew S, Alansari K, Smatti MK, Zaraket H, Al Thani AA, Yassine HM. Epidemiological, molecular, and clinical features of norovirus infections among pediatric patients in Qatar. Viruses 2019; 11:400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harris JP, Lopman BA, O’Brien SJ. Infection control measures for norovirus: a systematic review of outbreaks in semi-enclosed settings. J Hosp Infect 2010; 74:1–9. [DOI] [PubMed] [Google Scholar]
- 40.Bitler EJ, Matthews JE, Dickey BW, Eisenberg JN, Leon JS. Norovirus outbreaks: a systematic review of commonly implicated transmission routes and vehicles. Epidemiol Infect 2013; 141:1563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Isakbaeva ET, Bulens SN, Beard RS, et al. Norovirus and child care: challenges in outbreak control. Pediatr Infect Dis J 2005; 24:561–3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.