Abstract
Background
Angiostrongylus cantonensis (Ac), or the rat lungworm, is a major cause of eosinophilic meningitis. Humans are infected by ingesting the 3rd stage larvae from primary hosts, snails, and slugs, or paratenic hosts. The currently used molecular test is a qPCR assay targeting the ITS1 rDNA region (ITS1) of Ac.
Methods
In silico design of a more sensitive qPCR assay was performed based on tandem repeats predicted to be the most abundant by the RepeatExplorer algorithm. Genomic DNA (gDNA) of Ac were used to determine the analytical sensitivity and specificity of the best primer/probe combination. This assay was then applied to clinical and environmental samples.
Results
The limit of detection of the best performing assay, AcanR3990, was 1 fg (the DNA equivalent of 1/100 000 dilution of a single 3rd stage larvae). Out of 127 CDC archived CSF samples from varied geographic locations, the AcanR3990 qPCR detected the presence of Ac in 49/49 ITS1 confirmed angiostrongyliasis patients, along with 15/73 samples previously negative by ITS1 qPCR despite strong clinical suspicion for angiostrongyliasis. Intermediate hosts (gastropods) and an accidental host, a symptomatic horse, were also tested with similar improvement in detection observed. AcanR3990 qPCR did not cross-react in 5 CSF from patients with proven neurocysticercosis, toxocariasis, gnathostomiasis, and baylisascariasis. AcanR3990 qPCR failed to amplify genomic DNA from the other related Angiostrongylus species tested except for Angiostrongylus mackerrasae (Am), a neurotropic species limited to Australia that would be expected to present with a clinical syndrome indistinguishable from Ac.
Conclusion
These results suggest AcanR3990 qPCR assay is highly sensitive and specific with potential wide applicability as a One Health detection method for Ac and Am.
Keywords: PCR, Angiostrongylus, eosinophilia, meningitis
The AcanR3990 qPCR is designed to target repeated elements in the A. cantonensis genome, resulting in an assay with 100% specificity for human Angiostrongylus infection, improved clinical sensitivity, and the highest analytical sensitivity for A. cantonensis reported to date (1 fg).
The nematode Angiostrongylus cantonensis (Ac) is a major etiologic agent of eosinophilic meningitis in both humans and animals occurring autochthonously on all inhabited continents [1–11]. Disease occurs following transmission of Ac larvae from an intermediate host, a slug or snail, or from one of the many possible paratenic hosts, to an accidental host [10, 12–16]. The developing larvae attempt to mimic the migration pathway in the primary host, the rat, wherein they invade the central nervous system (CNS) leading, most commonly, to meningitis with an eosinophilic predominant pleocytosis [13, 17]. Acute symptoms range in severity from mild headache to coma, but the infection has been generally thought to be a benign, self-resolving process [18, 19]. Recently, there has been renewed interest in understanding the natural history and environmental epidemiology of this parasitic infection because of the increasing evidence of related severe sequelae (eg, cognitive deficits and death) [20–22].
Molecular techniques have been employed to improve both clinical diagnosis and environmental sampling. Currently, the most widely used and sensitive molecular assay is a quantitative polymerase chain reaction (qPCR) using primers and a probe designed to detect a portion of the internal transcribed spacer region 1 (ITS1) of Ac [23]. Even with this improved diagnostic method, samples from patients with clinical histories highly suspicious for Ac meningitis have been found to be negative by the ITS1 assay. Also, many positive tests by ITS1 qPCR had cycle numbers near the limit of detection. Thus, there appears to be a need for assays with improved analytical and clinical sensitivity.
RepeatExplorer is a bioinformatic tool previously shown to identify highly repetitive genomic DNA targets that, when used as the basis for qPCR assays, has provided improved clinical sensitivity and specificity as well as analytical sensitivity in a number of helminth infections [24–26]. Herein, we describe the development and initial performance evaluation of AcanR3990, a bioinformatically-informed highly sensitive and specific target for the identification of Ac DNA in relevant clinical and environmental samples.
METHODS
In Silico Assay Design
Sequence Read Archive files from BioProjects PRJNA533181 and PRJNA350391 were downloaded and fed into a bioinformatic pipeline to trim, clean, and pair the reads. Then RepeatExplorer1 algorithm was utilized to detect the most often repeated contigs. [24] The top 13 contigs were analyzed and primer/probe combinations were identified using PrimerQuest (Integrated DNA Technologies Iowa). (Supplementary Table 1) Further evaluation focused on 1 primer/probe combination, AcanR3990 (Forward primer: 5’-AAACTGTTGCTTTCGAAGCTATG-3’; reverse primer: 5’-GCGCAAATCTGACGTTCTTG-3’; Probe: 5’ 6-FAM/ACA TGA AAC /ZEN/ACC TCA AAT GTG CTT CGA /3’ IABkFQ/).
qPCR Performed at the National Institutes of Health (NIH)
AcanR3990 qPCR was performed in a 10 µL reaction using primers and a probe at the final concentrations of 500 nM and 250 nM, respectively, 1 × TaqMan Fast Advanced Master Mix (ThermoFisher Scientific, Waltham, MA) and 1 μL of extracted template DNA. A subset of the samples was tested by the NIH using 5 µL of unaltered CSF as template. Thermocycling (40 cycles) was performed on an ABI Viia 7 with the following cycling conditions: 95 °C for 20 sec followed by 40 °C for 1 sec and 60 °C for 20 sec. The ITS1 qPCR assay used the previously described primer/probe combination and reaction conditions [23].
AcanR3990 qPCR Validation at NIH
Archived Angiostrongylus cantonensis helminths were provided by the Centers for Disease Control and Prevention (CDC) to the NIH for use in initial validation studies as were samples previously tested at the CDC reference laboratory. The set of CSF samples used for initial validation at NIH included 16 samples (initially 17 samples, but 1 had degraded in storage). Eight of these had tested positive by the ITS1 qPCR at CDC, while 5 had tested negative by the same assay. Three were specificity controls: 1 each from patients with cysticercosis (confirmed by serology), toxocariasis (confirmed by serology), and gnathostomiasis (parasitologically confirmed).
Transfer of AcanR3990 qPCR to Laboratories Around the World
In addition to NIH, 4 institutions contributed to this study by testing the AcanR3990 qPCR on samples at their disposal: CDC on human CSF samples (n = 111); University of Hawaii on Ac 3rd stage larvae, tissue from 3 gastropods, and various samples from an equine veterinary case; and University of Sydney and University of Veterinary and Pharmacaeutical Sciences Brno on adult worms of 5 other Angiostrongylus species. Because of the use of different equipment, the 4 laboratories adapted the AcanR3990 primers and probes to their previously established methods. DNA extraction techniques and qPCR reaction conditions can be found in Supplementary Table 2.
Analytical Sensitivity Testing
Genomic DNA was extracted from an Ac adult worm isolated from the pulmonary arteries of a wild rat captured on Hawaii Island in 2009. The concentration of DNA in the initial extraction was measured by Qubit fluorometry (ThermoFisher Scientific) and serially 10-fold diluted down to 1 ag/μL. The AcanR3990 qPCR was performed on descending dilutions to find the lowest concentration of DNA that was reliably detected. Isolation and quantification of 3rd stage larvae from P. martensii was performed via previously published protocol. [27] Genomic DNA was extracted using the Qiagen Dneasy animal tissue protocol, per manufacturer’s instructions. Final elution volume was 100 μL.
Statistical Analyses
For the purpose of conveniently comparing the performance of ITS1 qPCR vs AcanR3990 qPCR, an agreement table was created in which ITS1 qPCR was assumed to be the gold standard. Under this assumption, the relative sensitivity was calculated as the number of samples amplified by both ITS1 qPCR and AcanR3990 qPCR divided by the number amplified by ITS1 qPCR alone. The relative specificity was calculated as the number of samples not amplified by both ITS1 qPCR and AcanR3990 qPCR divided by the number not amplified by ITS1 qPCR alone.
RESULTS
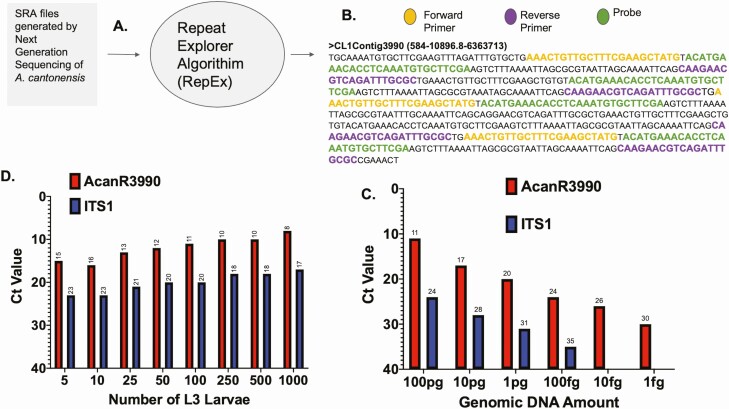
The RepeatExplorer algorithm performed on Sequence Read Archive (SRA) data from genomic sequencing of Ac provided information on repetitive sequences within the Ac genome of which the 13 most repeated sequences were chosen to evaluate for assay design (Figure 1A, Supplementary Table 1) with the readout for contig 3990 depicted in Figure 1B. AcanR3990 comes from a region of satellite DNA predicted to be highly repetitive, although the exact nature and location of the repetitive element cannot be determined from the current Ac genome assembly. As can be seen, within this 584 bp contig, there are 2 exact repeats of 107 bases separated by an additional target region. Thus, based on the primer/probe system in the context of the above repeats, detection is further augmented by multimer amplification. Moreover, the AcanR3990 repeat sequence was only found in Ac and not in other related Angiostrongylus sp. available in either GenBank or in Wormbase Parasite.
Figure 1.
AcanR3990 design and initial evaluation. RepeatExplorer algorithm used on National Center for Biotechnology Information Sequence Read Archive files from the Angiostrongylus cantonensis (Ac) genomes (Panel A) identified highly repeated target sequences. Panel B shows the repetitive sequence contig 3990 with the primer/probe sequences highlighted. Limits of detection of qPCR assay based on AcanR3990 (red) and ITS1 (blue) (Panel C). qPCR data from increasing numbers of Ac 3rd stage larvae in head to head comparison of AcanR3990 (red) and ITS1 (blue) (Panel D). Nonstandard Abbreviations: A. cantonensis, Angiostrongylus cantonensis; Ac, Angiostrongylus cantonensis; CDC, Centers for Disease Control and Prevnetion; Ct, cycle threshold; gDNA, genomic DNA; ITS1, internal transcribed spacer region 1; NGS, next generation sequencing; qPCR, quantitative polymerase chain reaction.
The initial primer/probe screen suggested the assay targeting contig 3990 (thus AcanR3990 qPCR) would perform the best with an analytical sensitivity of ~1 fg. In comparison, ITS1 qPCR had an analytical sensitivity of ~100 fg, making the AcanR3990 qPCR ~100 times more sensitive when assessed by Ac genomic DNA dilution series (Figure 1C). At any given Ac gDNA dilution, the AcanR3990 qPCR detected a specific gDNA concentration at consistently lower Ct values than did ITS1 qPCR. A similar increase in sensitivity afforded by AcanR3990 could be seen when the assay was used to detect increasing numbers of Ac 3rd stage larvae. For any given number of larvae there was a Ct difference of ~10 between the Ct for the target of AcanR3990 and that of ITS1 (~1000-fold increase in sensitivity by cycle time as calculated by 2(CtITS1-Ct AcanR3990); Figure 1D).
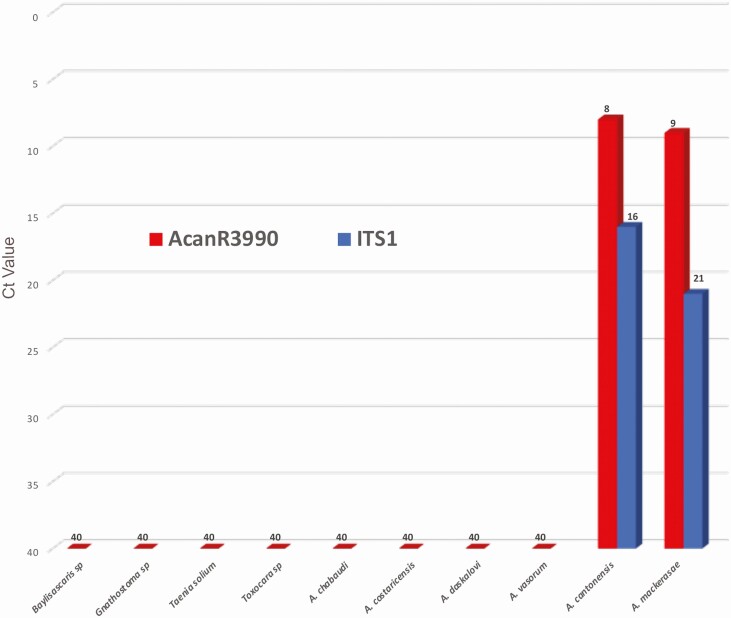
The analytical specificity of AcanR3990 was next evaluated. Using genomic DNA from Angiostrongylus chabaudi, Angiostrongylus costaricensis, Angiostrongylus daskalovi, and Angiostrongylus vasorum (see Supplementary Table 2), there was no amplification of any of these closely related species using AcanR3990. When gDNA of Angiostrongylus mackerrasae (Am) was tested by qPCR, both AcanR3990- and ITS1-based assays were positive for this species. Archived human CSF samples from patients known to be infected by Gnathostoma sp., Toxocara sp., Taenia solium, and Baylisascaris sp. were all negative by the AcanR3990 qPCR (Figure 2) as well as by the ITS1 qPCR. Thus, the relative specificity for AcanR3990 was 100%.
Figure 2.
Assessment for cross-reactivity of AcanR3990. Ct values are shown as obtained by AcanR3990 against genomic DNA from other Angiostrongylus species and cerebrospinal fluid samples from cerebral infections caused by other parasites. Angiostrongylus mackerasae and Angiostrongylus cantonensis genomic DNA were tested by both AcanR3990 and ITS1 for comparison. Abbreviations: Ct, cycle threshold; sp, species.
To test the utility of the AcanR3990 qPCR in detection of Ac DNA in human CSF samples, 127 archived CSF samples previously sent to the CDC for diagnostic evaluation of eosinophilic meningitis were assessed using AcanR3990. Previously, 49/127 were found to be positive using the ITS1 qPCR. The AcanR3990 qPCR yielded positive results in each of the 49 cases previously identified, resulting in a relative sensitivity of 100%. In line with the improved analytical sensitivity, the AcanR3990 qPCR found 15 additional samples positive (Table 1). The 15 additional positive samples were collected from 14 patients, of which at least 12 had clinical and epidemiologic histories consistent with angiostrongyliasis. Interestingly, CSF from 3 patients (2000–1, 2010–5, and 2010–6) were found to be positive for AcanR3990 at first lumbar puncture (4–10 days after onset of symptoms), whereas those samples were negative for ITS1. The AcanR3990 qPCR yielded positive results on all CSF collected throughout the illness from these 3 patients as well as from patient 2000–2, whereas ITS1 qPCR results were only intermittently positive (Table 1). Additionally, on a smaller subset of the above samples, testing unprocessed CSF and DNA extracted from CSF by the AcanR3990 qPCR returned similar results. (Supplementary Figure 1).
Table 1.
Clinical and Epidemiologic Characteristics of Selected Human Cerebrospinal Fluid Samples Tested by ITS1 and AcanR3990 qPCRs
| Subject | Location of Presentation | Exposure History | Clinical Symptoms | Days after Onset of Symptoms | ITS1 qPCR Results (Ct) | AcanR3990 qPCR Results (Ct) | CSF Eosino-philia |
|---|---|---|---|---|---|---|---|
| 2000–1 | Illinois | Travel to Jamaica | Blurry vision, paresthesias, headache | 10 | negative | positive (30) | 0% |
| 16 | positive (38) | positive (29) | 19% | ||||
| 20 | negative | positive (28) | 10% | ||||
| 33 | positive (33) | positive (20) | 32% | ||||
| 2000–2 | Illinois | Travel to Jamaica | 16 | positive (36) | positive (28) | 11% | |
| 23 | positive (38) | positive (29) | 18% | ||||
| 34 | negative | positive (29) | 47% | ||||
| 2009-1 | Utah | Travel to Hawai‘i | Headache, paresthesias, dysathesias | 7 | negative | positive (27) | 40% |
| 2009-2 | Hawai‘i | Paresthesias, headache, paralysis | 14 | negative | positive (27) | 28% | |
| 2009-3 | Hawai‘i | Generalized pain | 15 | negative | positive (25) | 57% | |
| 2010–5 | Hawai‘i | Ate a snail | Myalgia, headache | 6 | negative | positive (30) | 15% |
| 15 | positive (29) | positive (19) | 27% | ||||
| 2010–6 | Hawai‘i | Fever, bulging fontanelle | 4 | negative | positive (28) | 18% | |
| Hawai‘i | 27 | positive (27) | positive (17) | 10% | |||
| 2011–2 | Hawai‘i | Headache, neck pain | 5 | negative | positive (27) | 21% | |
| 2011–4 | Hawai‘i | Headache, paresthesias, nausea, shoulder pain, sensitive to light and sound | 16 | negative | positive (31) | 65% | |
| A | Florida | Ate a snail | Severe headache, neck stiffness, nausea, vomiting | N/A | negative | positive (27) | 61% |
| B | Virginia | Travel to India | Headache | 23 | negative | positive (28) | 14% |
| C | Guadeloupe | N/A | Prostration, mutism, meningoencephalitis, fever, coma | 1 | negative | positive (30) | 21% |
| D | Canada | N/A | N/A | N/A | negative | positive (33) | N/A |
| E | California | N/A | N/A | N/A | negative | positive (36) | N/A |
Abbreviations: CSF, cerebral spinal fluid; Ct, cycle threshold; N/A, not applicable; qPCR, quantitative PCR.
Bold values indicate the specimens negative by ITS1 but positive by AcanR3990.
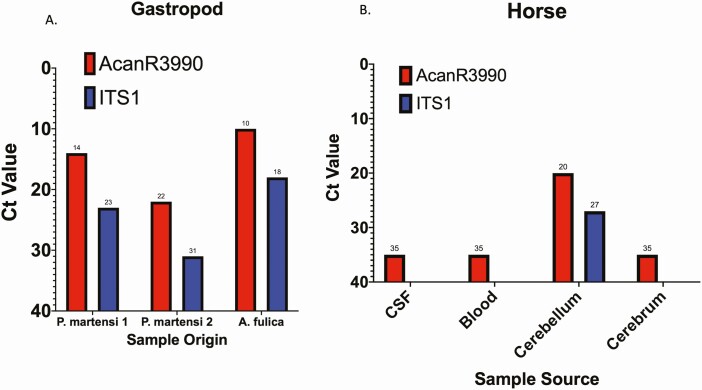
Testing of Ac-infected gastropods by both ITS1 and AcanR3990 qPCR showed a ~10-fold improvement in Ct values, respectively, as seen for 2 samples from Parmarion martensi and 1 from Achatina fulica (Figure 3A).
Figure 3.
Ct values of gastropod and veterinary samples tested by AcanR3990 and ITS1. Panel A shows Ct values from field-collected gastropod tissue as part of environmental sampling for Ac tested by AcanR3990 (red) and ITS1(blue) qPCR. Panel B shows results of testing various samples from a horse suspected of having neuroangiostrongyliasis by AcanR3990 (red) and ITS1(blue) qPCR.
Tissue and blood samples from a Hawaiian horse with clinical disease consistent with Ac infection were tested for the presence of Ac DNA (Figure 3B). While the ITS1-based qPCR failed to amplify Ac DNA in each of the listed samples except cerebellum, AcanR3990 qPCR successfully identified Ac DNA in CSF, cerebellum, cerebrum, and blood (Figure 3B).
DISCUSSION
The expansion of high-quality genomic next generation sequencing (NGS) data available for analysis has greatly benefited the field of molecular diagnostics by allowing a priori selection of potentially highly sensitive assay targets based on an understanding of the intrinsic properties of qPCR. The study presented herein adds to the accumulating evidence of the robustness of using highly repetitive sequences derived from NGS data to develop bioinformatically informed qPCR assays.
The AcanR3990 repeated sequence appears largely to be tandemly arrayed within the genome. While it is impossible to determine with accuracy the number of times this contig is repeated due to the fundamentals of the RepeatExplorer algorithm, the abundance can be estimated at 1.3% of the Ac genome by considering the contig length, depth of coverage, and total size of Ac genome. The AcanR3990 qPCR described here provides improved diagnostic sensitivity when compared to the ITS1 qPCR without any loss of species-specificity. The predicted improvement in sensitivity was supported by a screen of CSF samples from patients with suspected angiostrongyliasis wherein 21% of CSF samples reported as negative by ITS1 qPCR were found to be positive by AcanR3990 qPCR. The CSF results also suggest that AcanR3990 qPCR will become positive sooner and remain more consistently positive during the course of infection [28]. A notable result is the CSF sample positive by AcanR3990 qPCR from patient A that is the first documented autochthonous human case of Ac in Florida, confirming the risk of human infection suggested by previous environmental studies of Ac transmission in primary hosts[2, 8] and in a clinical case in a zoo-kept primate [29]. The collected clinical data also suggest that AcanR3990 can be positive in the absence of CSF eosinophilia (Table 1, patient 2000–1), which highlights absence of CSF eosinophilia is not sufficiently sensitive to exclude angiostrongyliasis given the appropriate clinical presentation and exposure history [28].
These data suggest that AcanR3990 qPCR is a method potentially well suited to One Health approaches given the improved sensitivity for detection of Ac infection. Epidemiologic and environmental surveys could benefit from improved molecular assays (Figure 3) wherein the scale of intermediate and accidental host involvement might have been underestimated previously. Examples of improved understanding of the epidemiology of a helminth infection brought about by a RepeatExplorer-based qPCR design exist in the literature already for soil-transmitted helminthes [25, 30]. Furthermore, drawing on the veterinary data, the positive blood sample from the infected horse (Figure 3B) suggests that it may be possible to detect DNA in the blood of Ac-infected individuals. Thus, the potential for a laboratory diagnosis of angiostrongyliasis without the need for a lumbar puncture warrants further investigation. Indeed, laboratory infection of wild caught rats has shown that Ac DNA can be detected in the blood by PCR at specific times during the infection cycle, including at stages prior to the migration of adult worms to the pulmonary vasculature [31]. Also, the AcanR3990 qPCR performed equally well using unprocessed CSF (ie, no DNA extraction; Supplemental Figure 1) in comparison to standard DNA extraction techniques. These data suggest that faster processing and therefore shorter turnaround time may be achievable.
This study is limited by the relatively small number of human and veterinary clinical cases. Also, the full clinical history of each case tested for ITS1 and AcanR3990 is unknown, and so we are unable to independently assess the clinical likelihood of Ac infection. Thus, the diagnostic sensitivity of the AcanR3990 could not be determined in this study. The diagnostic specificity is likely to be high since no cross-reactivity was detected, with the exception of Am. The fact that Am was detected by both AcanR3990 qPCR and ITS1 qPCR suggests that these 2 species are phylogenetically similar; the genomic characterization of Am is ongoing and will be required to understand the interrelationship between Ac and Am [32]. Most recently, the analysis of the mitochondrial genome suggests Am is a species distinct from Ac [32]. Regardless of the relationship between the 2 species, differentiation of Ac and Am would not be likely to lead to differences in clinical management of human or animal cases (although Am has never been proven as a cause of human infection). Epidemiological surveys would be affected only in the narrow geographic range of Am (Australia). Additional species such as Angiostrongylus malaysiensis (another neurotropic Angiostrongylus sp.) should be further evaluated.
Despite these limitations, the data suggest that AcanR3990 is a highly specific and sensitive assay for diagnosis of Ac infection (and Am where the geographic range overlaps). The successful application of the assay using various reaction conditions in the different laboratories participating in this study indicates that the assay is robust and transferable to the laboratories around the world interested in Ac infection.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Author Contributions. The authors have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
Acknowledgments. The authors thank Dr Lisa Wood (Veterinary Associates, Kamuela, Hawai“i) for providing the clinical veterinary samples used in this study. The samples of A. chabaudi and A. daskalovi were kindly provided by Calin M. Gherman, those of A. costaricensis by Domenico Otranto.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The use of trade marked products were for experimental purposes only and are not an endorsement of such products.
Financial support. This work was supported, in part, by the Division of Intramural Research (DIR), NIAID. Funding for part of this study was provided by the Hawai“i State Legislature, and the Daniel K. Inouye College of Pharmacy, University of Hawai“i, Hilo, HI. The Czech team was supported by the grant SEA-EUROPE JFS 2019–053.
Potential conflicts of interest. The authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest.
References
- 1.Rael RC, Peterson AC, Ghersi-Chavez B, Riegel C, Lesen AE, Blum MJ. Rat lungworm infection in rodents across post-Katrina New Orleans, Louisiana, USA. Emerg Infect Dis 2018; 24:2176–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stockdale Walden HD, Slapcinsky JD, Roff S, et al. . Geographic distribution of Angiostrongylus cantonensis in wild rats (Rattus rattus) and terrestrial snails in Florida, USA. PLoS One 2017; 12:e0177910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng J, He ZP, Zhang S, et al. . Phylogeography of Angiostrongylus cantonensis (Nematoda: Angiostrongylidae) in southern China and some surrounding areas. PLoS Negl Trop Dis 2017; 11:e0005776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarvi SI, Quarta S, Jacquier S, et al. . High prevalence of Angiostrongylus cantonensis (rat lungworm) on eastern Hawai’i Island: a closer look at life cycle traits and patterns of infection in wild rats (Rattus spp.). PLoS One 2017; 12:e0189458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guerino LR, Pecora IL, Miranda MS, et al. . Prevalence and distribution of Angiostrongylus cantonensis (Nematoda, Angiostrongylidae) in Achatina fulica (Mollusca, Gastropoda) in Baixada Santista, São Paulo, Brazil. Rev Soc Bras Med Trop 2017; 50:92–8. [DOI] [PubMed] [Google Scholar]
- 6.Waugh CA, Lindo JF, Lorenzo-Morales J, Robinson RD. An epidemiological study of A. cantonensis in Jamaica subsequent to an outbreak of human cases of eosinophilic meningitis in 2000. Parasitology 2016; 143:1211–7. [DOI] [PubMed] [Google Scholar]
- 7.Foster CE, Nicholson EG, Chun AC, et al. . Angiostrongylus cantonensis infection: a cause of fever of unknown origin in pediatric patients. Clin Infect Dis 2016; 63:1475–8. [DOI] [PubMed] [Google Scholar]
- 8.Stockdale-Walden HD, Slapcinsky J, Qvarnstrom Y, McIntosh A, Bishop HS, Rosseland B. Angiostrongylus cantonensis in introduced gastropods in Southern Florida. J Parasitol 2015; 101:156–9. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Alonso A, Abreu-Yanes E, Feliu C, et al. . Intermediate hosts of Angiostrongylus cantonensis in Tenerife, Spain. PLoS One 2015; 10:e0120686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qvarnstrom Y, Bishop HS, da Silva AJ. Detection of rat lungworm in intermediate, definitive, and paratenic hosts obtained from environmental sources. Hawaii J Med Public Health 2013; 72:63–9. [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen Y, Rossi B, Argy N, et al. . Autochthonous case of Eosinophilic meningitis caused by Angiostrongylus cantonensis, France, 2016. Emerg Infect Dis 2017; 23:1045–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Courdurier J, Guillon J, Malarde L, Laigret J, Desmoulins G, Schollhammer. Demonstration of the cycle of Angiostrongylus cantonensis in the laboratory. Observations on this cycle and anatomo-pathology caused by this parasite in various laboratory animals. Bull Soc Pathol Exot Filiales 1964; 57:1255–62. [PubMed] [Google Scholar]
- 13.Alicata JE, Loison G, Cavallo A. Parasitic meningoencephalitis experimentally produced in a monkey with larvae of Angiostrongylus cantonensis. J Parasitol 1963; 49:156–7. [PubMed] [Google Scholar]
- 14.Radomyos P, Tungtrongchitr A, Praewanich R, et al. . Occurrence of the infective stage of Angiostrongylus cantonensis in the yellow tree monitor (Varanus bengalensis) in five Provinces of Thailand. Southeast Asian J Trop Med Public Health 1994; 25:498–500. [PubMed] [Google Scholar]
- 15.Asato R, Taira K, Nakamura M, Kudaka J, Itokazu K, Kawanaka M. Changing epidemiology of Angiostrongyliasis cantonensis in Okinawa prefecture, Japan. Jpn J Infect Dis 2004; 57:184–6. [PubMed] [Google Scholar]
- 16.Ash LR. The occurrence of Angiostrongylus cantonensis in frogs of New Caledonia with observations on paratenic hosts of metastrongyles. J Parasitol 1968; 54:432–6. [PubMed] [Google Scholar]
- 17.Weinstein PP, Rosen L, Laqueur GL, Sawyer TK. Angiostrongylus cantonensis infection in rats and rhesus monkeys, and observations on the survival of the parasite in vitro. Am J Trop Med Hyg 1963; 12:358–77. [DOI] [PubMed] [Google Scholar]
- 18.Chotmongkol V, Sawadpanitch K, Sawanyawisuth K, Louhawilai S, Limpawattana P. Treatment of eosinophilic meningitis with a combination of prednisolone and mebendazole. Am J Trop Med Hyg 2006; 74:1122–4. [PubMed] [Google Scholar]
- 19.Thanaviratananich S, Thanaviratananich S, Ngamjarus C. Corticosteroids for parasitic eosinophilic meningitis. Cochrane Database Syst Rev 2012; 10:CD009088. [DOI] [PubMed] [Google Scholar]
- 20.McAuliffe L, Fortin Ensign S, Larson D, et al. . Severe CNS angiostrongyliasis in a young marine: a case report and literature review. Lancet Infect Dis 2019; 19:e132–42. [DOI] [PubMed] [Google Scholar]
- 21.Evans-Gilbert T, Lindo JF, Henry S, Brown P, Christie CD. Severe eosinophilic meningitis owing to Angiostrongylus cantonensis in young Jamaican children: case report and literature review. Paediatr Int Child Health 2014; 34:148–52. [DOI] [PubMed] [Google Scholar]
- 22.Morton NJ, Britton P, Palasanthiran P, et al. . Severe hemorrhagic meningoencephalitis due to Angiostrongylus cantonensis among young children in Sydney, Australia. Clin Infect Dis 2013; 57:1158–61. [DOI] [PubMed] [Google Scholar]
- 23.Qvarnstrom Y, Xayavong M, da Silva AC, et al. . Real-time polymerase chain reaction detection of Angiostrongylus cantonensis DNA in cerebrospinal fluid from patients with eosinophilic meningitis. Am J Trop Med Hyg 2016; 94:176–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novák P, Neumann P, Pech J, Steinhaisl J, Macas J. RepeatExplorer: a Galaxy-based web server for genome-wide characterization of eukaryotic repetitive elements from next-generation sequence reads. Bioinformatics 2013; 29:792–3. [DOI] [PubMed] [Google Scholar]
- 25.Pilotte N, Papaiakovou M, Grant JR, et al. . Improved PCR-based detection of soil transmitted helminth infections using a next-generation sequencing approach to assay design. PLoS Negl Trop Dis 2016; 10:e0004578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Connell EM, Harrison S, Dahlstrom E, Nash T, Nutman TB. A novel, highly sensitive quantitative polymerase chain reaction assay for the diagnosis of subarachnoid and ventricular neurocysticercosis and for assessing responses to treatment. Clin Infect Dis 2020; 70:1875–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howe K, Kaluna L, Lozano A, et al. . Water transmission potential of Angiostrongylus cantonensis: larval viability and effectiveness of rainwater catchment sediment filters. PLoS One 2019; 14:e0209813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slom TJ, Cortese MM, Gerber SI, et al. . An outbreak of eosinophilic meningitis caused by Angiostrongylus cantonensis in travelers returning from the Caribbean. N Engl J Med 2002; 346:668–75. [DOI] [PubMed] [Google Scholar]
- 29.Duffy MS, Miller CL, Kinsella JM, de Lahunta A. Parastrongylus cantonensis in a nonhuman primate, Florida. Emerg Infect Dis 2004; 10:2207–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilotte N, Maasch JRMA, Easton AV, Dahlstrom E, Nutman TB, Williams SA. Targeting a highly repeated germline DNA sequence for improved real-time PCR-based detection of Ascaris infection in human stool. PLoS Negl Trop Dis 2019; 13:e0007593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jarvi SI, Pitt WC, Farias ME, et al. . Detection of Angiostrongylus cantonensis in the blood and peripheral tissues of wild Hawaiian Rats (Rattus rattus) by a quantitative PCR (qPCR) assay. PLoS One 2015; 10:e0123064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valentyne H, Spratt DM, Aghazadeh M, Jones MK, Slapeta J. The mitochondrial genome of Angiostrongylus mackerrasae is distinct from A. cantonensis and A. malaysiensis. Parasitology 2020;147:681–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.