Abstract
Background
Laboratory confirmation of early Lyme borreliosis (LB) is challenging. Serology is insensitive during the first days to weeks of infection, and blood polymerase chain reaction (PCR) offers similarly poor performance. Here, we demonstrate that detection of Borrelia burgdorferi (B.b.) cell-free DNA (cfDNA) in plasma can improve diagnosis of early LB.
Methods
B.b. detection in plasma samples using unbiased metagenomic cfDNA sequencing performed by a commercial laboratory (Karius Inc) was compared with serology and blood PCR in 40 patients with physician-diagnosed erythema migrans (EM), 28 of whom were confirmed to have LB by skin biopsy culture (n = 18), seroconversion (n = 2), or both (n = 8). B.b. sequence analysis was performed using investigational detection thresholds, different from Karius’ clinical test.
Results
B.b. cfDNA was detected in 18 of 28 patients (64%) with laboratory-confirmed EM. In comparison, sensitivity of acute-phase serology using modified 2-tiered testing (MTTT) was 50% (P = .45); sensitivity of blood PCR was 7% (P = .0002). Combining B.b. cfDNA detection and MTTT increased diagnostic sensitivity to 86%, significantly higher than either approach alone (P ≤ .04). B.b. cfDNA sequences matched precisely with strain-specific sequence generated from the same individual’s cultured B.b. isolate. B.b. cfDNA was not observed at any level in plasma from 684 asymptomatic ambulatory individuals. Among 3000 hospitalized patients tested as part of clinical care, B.b. cfDNA was detected in only 2 individuals, both of whom had clinical presentations consistent with LB.
Conclusions
This is the first report of B.b. cfDNA detection in early LB and a demonstration of potential diagnostic utility. The combination of B.b. cfDNA detection and acute-phase MTTT improves clinical sensitivity for diagnosis of early LB.
Keywords: Lyme, Borrelia burgdorferi, metagenomics, cell-free DNA, next-generation sequencing
Cell-free DNA derived from B. burgdorferi was detected in 64% of plasma samples from patients with laboratory-confirmed erythema migrans using next-generation sequencing. Combining this technique with acute-phase modified 2-tiered serologic testing resulted in 86% diagnostic sensitivity and 100% specificity.
Two-tiered serologic testing remains the most widely used laboratory tool to support the diagnosis of Lyme borreliosis (LB). This approach yields high specificity, and sensitivity is high among patients with advanced LB who have been infected for months to years [1]. Sensitivity is poor in early infection, however, because a detectable antibody response takes time to develop. For example, conventional 2-tiered serologic testing (using an enzyme-linked immunosorbent assay [ELISA] followed by immunoblots) is falsely negative at the time of initial clinical presentation in 70%–80% of patients with localized erythema migrans (EM), a skin rash that is often present during early LB [1]. Serologic testing is also limited by the frequent persistence of anti–Borrelia burgdorferi (B.b.) immunoglobulin M (IgM) and immunoglobulin G (IgG) antibodies for months to years after infection, despite effective treatment [2].
Potentially, these limitations could be addressed by a test that directly detects the infectious agent [3]. Thus far, this has remained an elusive goal. Although B.b. can often be detected using culture or polymerase chain reaction (PCR) of skin biopsies taken from EM lesions [4], this approach is impractical for routine use. Blood PCR has not been very useful; the sensitivity of whole-blood or plasma PCR in patients with EM is usually in the 30%–50% range [5–9], and blood PCR is seldom positive in patients with later manifestations of LB [10]. Similarly, cerebrospinal fluid PCR is poorly sensitive in patients with Lyme neuroborreliosis [11, 12], leaving PCR analysis of joint fluid/synovial tissue from patients with suspected Lyme arthritis as the only clinically useful PCR application.
Here, we report the findings of a study performed in collaboration with colleagues at Karius Inc (Redwood City, California) to analyze plasma samples from patients with physician-diagnosed EM using Karius’ quantitative microbial cell-free DNA (cfDNA) detection method. This direct detection method applies unbiased (“shotgun”) metagenomic sequencing to human plasma samples for identification and quantitation of microbial sequence reads [13]. Rigorous analytical and clinical validation has established this method’s ability to detect cfDNA from a panel of >1000 bacteria, DNA viruses, fungi, and eukaryotic parasites [13]. This is the first report of its performance in the diagnosis of early LB via detection of B.b. sensu stricto cfDNA.
MATERIALS AND METHODS
Patients and Control Subjects
Forty-two patients presenting in the summer of 2015 with ≥1 skin lesion meeting the Centers for Disease Control and Prevention (CDC) surveillance case definition of EM [14] were enrolled by investigators on Nantucket Island (T. J. L.) and in Rhode Island (N. S. D.) under a previous research protocol [15]. Among them, 40 patients provided consent for future use of their samples in other research studies and were included in the present study. The parent and current studies were approved by the Partners Healthcare Human Research Committee. At initial presentation, a clinical history was taken and a physical examination was performed. A 2-mm punch biopsy was taken from the leading edge of the skin lesion for Borrelia culture [16]. Ethylenediaminetetraacetic acid (EDTA)–whole blood, EDTA-plasma, and serum samples were collected and stored at −80°C. After completion of standard antimicrobial therapy for EM, most enrolled patients returned for a follow-up assessment 3–6 weeks after the initial visit, during which convalescent-phase serum samples were collected.
In addition to patients with physician-diagnosed EM, 3 groups of control subjects were included in the study: (1) T. J. L. enrolled 3 ambulatory subjects on Nantucket during the summer of 2015, who complained of constitutional symptoms but had no skin rash on physical examination. None of these 3 subjects received antimicrobial therapy, and all had improvement of symptoms at their follow-up visit. Baseline and follow-up blood samples were collected as described above. (2) EDTA-plasma samples were collected by Karius from 684 ambulatory, asymptomatic subjects between 2015 and 2019 as controls for clinical validation of the Karius test. (3) EDTA-plasma samples were submitted to Karius for clinical testing, as ordered by treating physicians, from 3000 hospitalized patients suspected of having acute or chronic infection.
Laboratory Analysis of Collected Samples
Borrelia Culture
Cultures were performed as described previously [16]. If spirochetes were detected, B.b. PCR targeting the flaB gene [17] was performed on pelleted microbes for confirmation. Isolated B.b. strains were analyzed by whole-genome sequencing (Supplementary Methods) as a part of an ongoing B.b. sequencing effort (J. E. L., K. S., J. A. B., Pardis C. Sabeti, and others) and made available prepublication.
Serologic Testing
Serum samples were analyzed using the C6 B.b. ELISA (Immunetics/Oxford), the ZEUS B.b. IgG/IgM ELISA (Zeus Scientific), the ZEUS VlsE1/pepC10 IgG/IgM ELISA (Zeus Scientific), and IgM and IgG ViraStripe ladder immunoblots (Viramed AG) interpreted according to CDC criteria [18]. Conventional and modified 2-tiered testing algorithms [19] were applied to classify each sample as seropositive or seronegative, as described in Tables 1 and 2.
Table 1.
Clinical Sensitivity of Borrelia burgdorferi Cell-free DNA Detection Compared With Standard Diagnostic Methods in Patients With a Clinical Diagnosis of Erythema Migrans
| No. (%) Positive | |||||
|---|---|---|---|---|---|
| Patients | Borrelia burgdorferi Cell-free DNA | Borrelia PCR (Whole Blood) | Acute-phase Serology | ||
| CTTTa | MTTT Algorithm 1b | MTTT Algorithm 2c | |||
| All patients with suspected EM (N = 40) | 19 (48) | 3 (8)d | 12 (30) | 18 (45) | 18 (45) |
| Laboratory confirmed (n = 28) | 18 (64) | 2 (7)e | 10 (36) | 14 (50) | 14 (50) |
| Unconfirmed (n = 12f) | 1 (8) | 1 (8)g | 2 (17) | 4 (33) | 4 (33) |
Abbreviations: CTTT, conventional 2-tiered testing; EM, erythema migrans; MTTT, modified 2-tiered testing; PCR, polymerase chain reaction.
aSeropositivity by CTTT required a positive or equivocal whole-cell sonicate enzyme-linked immunosorbent assay (ELISA) (Zeus B. burgdorferi immunoglobulin G [IgG]/immunoglobulin M [IgM] ELISA) and a positive IgM or IgG immunoblot (ViraStripe, Viramed AG). If only the IgM immunoblot was positive, the individual was considered CTTT seropositive only if symptoms were present for <1 month.
bSeropositivity by MTTT algorithm 1 required a positive or equivocal result in both the Zeus B. burgdorferi IgG/IgM ELISA and the Immunetics/Oxford C6 Lyme ELISA.
cSeropositivity by MTTT algorithm 2 required a positive or equivocal result in both the Zeus VlsE/pepC10 IgG/IgM ELISA and the Zeus B. burgdorferi IgG/IgM ELISA.
dOne sample was PCR positive; 2 additional samples were PCR equivocal.
eOne sample was PCR positive; the other sample was PCR equivocal.
fTwo patients’ skin cultures were contaminated with skin flora and were discontinued before the complete incubation period.
gThis sample was PCR equivocal.
Table 2.
Clinical Sensitivity of Borrelia burgdorferi Cell-free DNA Detection Combined with Acute-Phase 2-Tiered Serologic Testing in Patients With a Clinical Diagnosis of Erythema Migrans
| No. (%) Positive | |||
|---|---|---|---|
| Patients | B.b. cfDNA or CTTTa | B.b. cfDNA or MTTT Algorithm 1b | B.b. cfDNA or MTTT Algorithm 2c |
| All patients with suspected EM (N = 40) | 24 (60) | 28 (70) | 28 (70) |
| Laboratory confirmed (n = 28) | 22 (79) | 24 (86) | 24 (86) |
| Unconfirmed (n = 12d) | 2 (17) | 4 (33) | 4 (33) |
Abbreviations: B.b., Borrelia burgdorferi; cfDNA, cell-free DNA; CTTT, conventional 2-tiered testing; EM, erythema migrans; MTTT, modified 2-tiered testing.
aSeropositivity by CTTT required a positive or equivocal whole-cell sonicate enzyme-linked immunosorbent assay (ELISA) (Zeus B. burgdorferi immunoglobulin G [IgG]/ immunoglobulin M [IgM] ELISA) and a positive IgM or IgG immunoblot (ViraStripe, Viramed AG). If only the IgM immunoblot was positive, the individual was considered CTTT seropositive only if symptoms were present for <1 month.
bSeropositivity by MTTT algorithm 1 required a positive or equivocal result in both the Zeus B. burgdorferi IgG/IgM ELISA and the Immunetics/Oxford C6 Lyme ELISA.
cSeropositivity by MTTT algorithm 2 required a positive or equivocal result in both the Zeus VlsE/pepC10 IgG/IgM ELISA and the Zeus B. burgdorferi IgG/IgM ELISA.
dTwo skin cultures were contaminated with skin flora and were discontinued before the complete incubation period.
Blood PCR for B.b. DNA and Auxiliary Testing for Other Pathogens
These tests were performed at reference laboratories (Supplementary Methods).
Microbial cfDNA Detection and Analysis
Frozen aliquots of EDTA-plasma were sent to Karius for processing and analysis. Microbial cfDNA was extracted from 250 μL of EDTA-plasma, next-generation sequencing libraries were prepared, and sequencing was performed on an Illumina NextSeq 500. Sequence reads identified as human were removed, and remaining reads were aligned to a curated pathogen database. Per routine for the Clinical Laboratory Improvement Amendments–certified laboratory diagnostic test, any of >1000 microorganisms in the Karius Test Pathogen List found to have statistically significant levels of cell-free DNA in plasma, relative to real-time negative controls, were reported in molecules of cfDNA/μL (MPM) of plasma as previously described [13]. If the number of cfDNA sequence reads derived from a particular microorganism was too low to reach the predefined statistical threshold, the microorganism was not reported among the test results. However, for all samples included in the present study (both case and control samples), a relaxed statistical threshold was applied for B.b., such that B.b. would be reported among the test results if cfDNA derived from this microorganism was observed at any level (investigational use only).
Statistical Methods
Differences between proportions were considered statistically significant if the 2-tailed P value was <.05 as determined using McNemar test.
RESULTS
B.b. cfDNA Detection in Patients With Physician-diagnosed EM
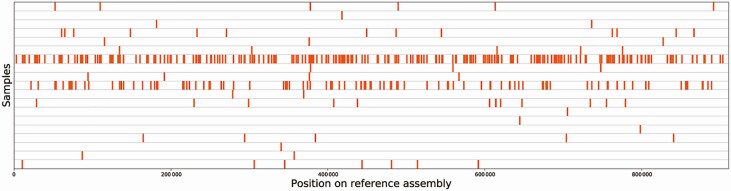
Among 40 patients in this category, B.b. cfDNA was detected in 19 patients (48%; Table 1). Alignment of B.b. cfDNA sequence reads to the reference genome (B.b. strain B31) resulted in a uniform distribution of alignments spanning the B.b. linear chromosome (Figure 1). The number of B.b.-specific reads ranged from 1 to 253 per unique patient sample (Supplementary Table 1). When the absolute number of B.b. reads per sample was converted to MPM, the range was <1 to 185 MPM, with a median of 2 MPM and an interquartile range of 4.
Figure 1.
Alignment of cell-free DNA reads to Borrelia burgdorferi (B.b.) reference genome. Sequence reads from microbial cell-free DNA aligning to assembly accession GCF_000008685.2 (B.b. strain B31) were plotted along the genome, with each row representing a unique patient sample (n = 19). Alignments were uniform across the genome and were of high percentage identity to the reference. None of the reads that aligned to B.b. aligned to any assembly outside of the Borrelia genus.
In comparison, a commercial pan-Borrelia whole-blood PCR assay (Supplementary Methods) was positive or equivocal in 3 of 40 samples from the same cohort (8%; P = .0004; Table 1). When the pan-Borrelia PCR-positive (n = 1) or PCR-equivocal (n = 2) DNA extracts were further analyzed using B.b. species–specific and Borrelia miyamotoi–specific PCR assays, none tested positive. Serologic testing for LB using the conventional 2-tiered testing (CTTT) algorithm (a whole-cell sonicate ELISA followed by immunoblots) was positive in 12 of 40 acute-phase serum samples from the same patients (30%; P = .15 for the comparison with B.b. cfDNA detection). Using a modified serologic testing algorithm involving a whole-cell sonicate (WCS) ELISA followed by a C6 ELISA (MTTT 1), without the use of immunoblots, 18 of 40 patients were seropositive in the acute phase of illness (45%; P = 1.0 when compared with B.b. cfDNA detection). A second MTTT algorithm, using a VlsE/pepC10 ELISA followed by a WCS ELISA (MTTT 2), also demonstrated seroreactivity in 18 of 40 patients (45%) using acute-phase serum samples (Table 1).
B.b. cfDNA Detection in Patients With Laboratory-confirmed EM
Among 40 patients with physician-diagnosed EM, the diagnosis was confirmed in 28 (70%) either by recovering B.b. in skin biopsy culture (n = 18), by demonstrating seroconversion between acute- and convalescent-phase serum samples (n = 2), or both (n = 8). B.b. cfDNA was detected in 18 of 28 such patients (64%; Table 1). In comparison, using a commercial pan-Borrelia whole-blood PCR assay, 2 of 28 samples were positive or equivocal (7%; P = .0002). Whereas both samples were negative when further analyzed using a B.b.-specific PCR assay, both were positive for B.b. cfDNA. Acute-phase serologic testing using the CTTT algorithm was positive in 10 of 28 patients (36%; P = .08 for the comparison with B.b. cfDNA detection). Using either MTTT algorithm 1 or 2, acute-phase serology was positive in 14 of 28 patients (50%; P = .45 compared with B.b. cfDNA detection; Table 1).
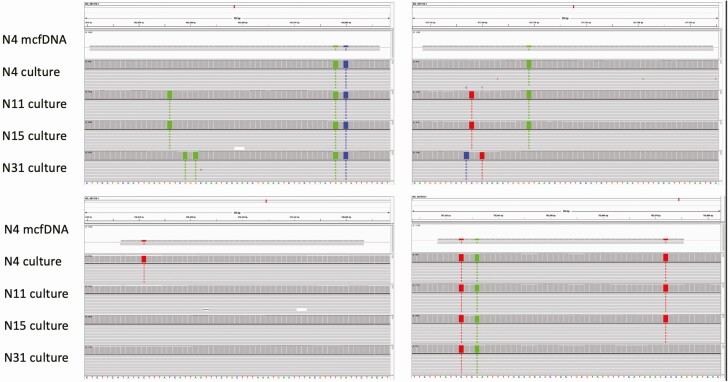
We aligned B.b. cfDNA sequence reads from individual patients against whole-genome sequence reads generated from the same individual’s cultured B.b. isolate, in cases where cfDNA sequencing yielded sufficient B.b.-specific reads for such an analysis. In multiple individuals, B.b. cfDNA sequences from plasma precisely matched strain-specific genomic sequence of the B.b. isolate cultured from that individual’s skin biopsy (Figure 2), meaning the paired reads shared single-nucleotide polymorphisms (SNPs) not present in other strains, or lacked SNPs found in other strains.
Figure 2.
Alignment of Borrelia burgdorferi (B.b.) cell-free DNA (cfDNA) reads to strain-specific genome sequence from matched culture isolates. The B.b. cfDNA sequence reads match with reads from the source patient’s infecting B.b. strain and differ from reads obtained from other study patients’ cultured strains. Four informative B.b. cfDNA reads from patient N4’s plasma sample, along with sequence reads from representative B.b strains isolated from skin biopsy specimens, are shown aligned to the B31 reference genome. Abbreviation: mcfDNA, microbial cell-free DNA.
Combination of B.b. cfDNA Detection With 2-Tiered Serologic Testing
Among 40 patients with physician-diagnosed EM, 24 (60%) were positive for B.b. cfDNA detection and/or were seropositive in the acute phase of illness using the CTTT algorithm (Table 2). The combined sensitivity of 60% was significantly higher compared with acute-phase CTTT alone (30%; P = .002) and approached significance when compared with B.b. cfDNA detection alone (48%; P = .07). When acute-phase serologic testing using either MTTT algorithm 1 or 2 was combined with B.b. cfDNA detection, 28 patients (70%) were positive. The combined sensitivity of 70% was significantly higher than either MTTT algorithm 1 or 2 alone (45% using either algorithm; P = .004) or B.b. cfDNA detection alone (48%; P = .008).
Among patients with laboratory-confirmed EM, 22 of 28 (79%) were positive using B.b. cfDNA detection and/or were seropositive in the acute phase of illness using CTTT (Table 2). This test combination was significantly more sensitive compared with acute-phase CTTT alone (36% sensitivity; P = .002), but not compared with B.b. cfDNA detection alone (64%; P = .13). When combining B.b. cfDNA detection with acute-phase MTTT, 24 of 28 (86%) of patients were positive by 1 or both diagnostic approaches, exceeding the sensitivity of acute-phase MTTT alone (50% using either algorithm 1 or 2; P = .004) or B.b. cfDNA detection alone (64%; P = .04).
B.b. cfDNA Detection in Control Subjects
In a retrospective review of 3000 hospitalized patients who recently underwent microbial cfDNA testing at Karius for clinical care, B.b. cfDNA was detected in 2 individuals, with concentrations of 9 and 195 MPM. Both patients had clinical features consistent with acute early LB (A. A. A., personal communication). Also, in a cohort of 684 asymptomatic ambulatory control subjects, sequence reads aligning with B.b. reference genomes were not detected at any concentration. We also analyzed 3 control samples collected from ill ambulatory subjects without rash on Nantucket (see Methods); none of the 3 seroconverted and all 3 were negative for B.b. cfDNA.
Non-B.b. Microbial cfDNA Detection in Cases and Controls
Among 40 patients with physician-diagnosed EM, cfDNA from microorganisms other than B.b. was detected in the plasma of 12 (30%). In most cases, the reads aligned with genomic DNA from commensal flora of the human skin, gut, or oropharynx (Supplementary Table 1), although some likely pathogens were also detected (eg, Helicobacter pylori). In 2 patients with laboratory-confirmed EM, cfDNA derived from Rickettsia felis, a flea-transmitted pathogen, was detected at concentrations of 405 and 55 MPM. To investigate this finding, PCR assays capable of detecting R. felis were performed at CDC using genomic DNA extracts prepared from whole-blood samples of both patients. The PCR results were negative, however, and serologic tests for R. felis infection were not available to adjudicate the discrepancy.
Among 3 control samples collected from ill, ambulatory subjects without rash on Nantucket in whom LB was not suspected clinically, 1 produced sequence reads from Francisella tularensis (52 MPM). The sample was collected from a 62-year-old woman presenting in July 2015 with fever/chills, headache, malaise, fatigue, myalgias, and arthralgias. When the cfDNA result was obtained, we performed F. tularensis serology. Results were negative (titer <1:128) using serum collected at the time of initial presentation, but strongly positive (titer = 1:2048) using a sample collected 32 days later, confirming the diagnosis of acute tularemia.
DISCUSSION
This study demonstrates that B.b. is detectable using unbiased metagenomic sequencing of cfDNA in acute-phase plasma samples from patients with EM, the most common manifestation of early LB. The cfDNA sequence reads were uniformly distributed across the B.b. reference genome, indicating that the findings are not artifactual. Moreover, B.b. cfDNA sequence reads precisely matched the strain-specific genome sequence generated from the B.b. isolate cultured from the same individual, proving that the B.b. cfDNA sequence detected in plasma was derived from infecting B.b. spirochetes and confirming the validity of this diagnostic approach.
Detection rates of B.b. cfDNA were higher in the subcategory of patients with laboratory-confirmed EM, compared with those in whom physician-diagnosed EM could not be confirmed by skin biopsy culture or seroconversion. Speculatively, this may be due to differences in B.b. organism burden between the 2 groups, or may reflect incorrect clinical diagnosis in some of the unconfirmed cases. At the time of clinical presentation, EM lesions often do not have central clearing or a classical “bullseye” appearance [20, 21] and other entities can look similar.
B.b. cfDNA detection also appears to complement acute-phase serologic testing as a diagnostic tool in early LB. The combination of modified 2-tiered serologic testing and B.b. cfDNA detection produced significantly higher sensitivity compared with either method alone—reaching 86% in patients with laboratory-confirmed EM. Potentially, this phenomenon could be explained by variability in duration of infection prior to testing; it is often the case that infectious agents are detected directly in blood at a higher rate prior to development of a humoral immune response. There was no significant difference in the mean duration of EM prior to presentation between the groups of patients with discordant results (positive B.b. cfDNA detection but negative MTTT, and vice versa; data not shown), although this measurement does not capture the incubation period and relies on patient self-reporting.
Detection of B.b. cfDNA was significantly more sensitive compared with blood PCR performed at a commercial reference laboratory, both in the cohort with physician-diagnosed EM and in the subset with laboratory-confirmed EM. The relatively low yield of blood PCR in early- or late-stage LB is well described, whereas our findings suggest that cfDNA metagenomic sequencing is a more promising direct pathogen detection method. Among patients with laboratory-confirmed EM, B.b. cfDNA detection was 64% sensitive, compared with 7% using blood PCR. Notably, this was achieved without prior capture/enrichment of B.b. DNA targets from the plasma samples, and without optimization of the Karius microbial cfDNA sequencing method to increase detection of B.b. DNA. Adding these steps is expected to improve sensitivity further [3], and future studies will address this.
It is unlikely that the difference in sensitivity between B.b. cfDNA detection and blood PCR is explained by differences in the volume of primary sample analyzed. For B.b. cfDNA detection, each DNA extraction was performed on 250 µL of EDTA-plasma, whereas for PCR the starting sample volume was 500 µL of EDTA-whole blood, which should translate to at least 250 µL of plasma on average. (As an extracellular bacterium, B.b. is present in the plasma fraction of blood.) More likely, the results are attributable to another methodological difference: B.b. PCR methods rely on detection of specific, intact DNA targets that are present in limited quantity within each spirochete (typically 1 copy per organism), whereas cfDNA detection is unbiased and can reveal an organism’s presence if informative DNA sequence is produced from anywhere in the genome, even if the DNA is fragmented.
Besides B.b., 2 other vector-borne pathogens were detected by metagenomic sequencing in our study. Francisella tularensis cfDNA reads were detected in plasma from 1 ill ambulatory control subject, and this finding’s significance was confirmed by demonstration of seroconversion using an F. tularensis microagglutination assay. Rickettsia felis cfDNA reads were detected in 2 patients with laboratory-confirmed EM, but subsequent efforts to confirm R. felis infection using whole-blood PCR assays were unsuccessful. Unfortunately, a validated serologic assay for R. felis is not available to assess for seroconversion in these patients, and it is possible that blood PCR assays for R. felis are simply less sensitive compared with microbial cfDNA detection (as with Borrelia blood PCR). Overall, the clinical significance of this finding remains uncertain, although it warrants future investigation. While human infection with R. felis is most common in sub-Saharan Africa, cases have been documented in Europe and the United States [22]. Our findings raise questions about the potential for unappreciated exposure to this emerging pathogen in temperate climates, and the significance of its detection in patients with concurrent infections.
A potential limitation of our study is the use of relaxed criteria for reporting B.b. among microorganisms detected using cfDNA metagenomic sequencing. To control for environmental contamination, the routine Karius test (offered for clinical diagnostic purposes) contrasts the quantity of sequence reads observed for each microbe against the quantity observed in a set of negative control samples; microorganisms are only reported if a statistically significant difference between the 2 quantities is observed. While this approach is critical for achieving high analytical specificity across a broad spectrum of microbes, not all microbes pose the same level of risk. For B.b., we hypothesized that the usual statistical threshold was not needed, as B.b. is not a known environmental contaminant, nor does it exhibit high homology with other free-living contaminants. As such, incidental B.b. cfDNA detection in human plasma is assumed to be highly unlikely. Thus, in a modification of the clinical Karius test method made for investigational use only, we considered B.b.-specific reads to be reportable in any quantity for cases and control subjects, and dispensed with the statistical threshold (only for B.b.). This resulted in higher sensitivity for B.b. detection than would have been achieved with the clinical version of the test. The validity of this approach is supported by our retrospective review of results from 684 asymptomatic control subjects and 3000 hospitalized patients, which demonstrated that B.b. cfDNA reads were not detected in any quantity except in 2 hospitalized individuals, both of whom had a clinical syndrome compatible with early LB.
Our study is also limited by its focus on patients with EM, without inclusion of patients with other LB manifestations (or other vector-borne infections). Although blood PCR techniques have proven insensitive for the diagnosis of acute neuroborreliosis or Lyme arthritis, we have demonstrated that B.b. cfDNA detection has higher sensitivity than blood PCR in EM, and its potential use in the diagnosis of noncutaneous LB should be investigated. Indeed, previous studies have shown that microbial cfDNA sequencing is capable of detecting pathogen DNA in plasma from patients with deep-seated infection at distant sites [23–26]. Also, our detection of F. tularensis in a symptomatic control subject suggests that microbial cfDNA detection for diagnosis of tick-borne illnesses more broadly may have utility. Future studies will address whether this single platform could be applied as a robust syndromic test for direct detection of a wide array of pathogens in patients with suspected tick-borne illness.
In summary, unbiased metagenomic sequencing applied to acute-phase plasma samples can detect B.b. cfDNA in patients with EM. This approach was significantly more sensitive compared with whole-blood PCR, both in the full cohort of patients with physician-diagnosed EM and in the subset of patients with laboratory-confirmed EM. When B.b. cfDNA detection was combined with acute-phase serologic testing using MTTT algorithms, 86% of patients with laboratory-confirmed EM were positive by 1 or both diagnostic methods, demonstrating that prompt, timely, and accurate test results can potentially be obtained in early LB by pairing a direct detection method with an indirect detection method at the time of initial clinical presentation. Technical modifications to the B.b. cfDNA detection assay will be pursued in an attempt to further improve sensitivity.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors are grateful to Dr Sandor E. Karpathy, PhD (Division of Vector-Borne Diseases, National Center for Emerging and Zoonotic Infectious Diseases, Centers for Disease Control and Prevention) for performing the blood polymerase chain reaction assays for Rickettsia felis. We also thank Dr Sharjeel Ahmad, MD of OSF Healthcare for providing clinical information about 2 control subjects in whom B. burgdorferi cfDNA was detected.
Financial support. This work was undertaken as an unfunded collaboration between the academically affiliated authors and Karius, Inc, a for-profit company. Sample collection was undertaken as part of a previous study with funding from the Bay Area Lyme Foundation and the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (award number 1R21AI119457-01).
Potential conflicts of interest. J. A. B. has received research support from Zeus Scientific, bioMérieux, Immunetics, Alere, DiaSorin, the Bay Area Lyme Foundation (BALF), and the NIAID (award number 1R21AI119457-01) for LB-related research projects, and also reports personal fees from T2 Biosystems, DiaSorin, and Roche Diagnostics, outside the submitted work. N. R. P. has received funding from BALF and NIAID (award number 1R21AI119457-01) for LB-related research. J. E. L. has received payments from Sherlock Biosciences for consulting services. K. S. has served as a paid consultant for T2 Biosystems and Roche Diagnostics and has received research support from the Massachusetts General Hospital Executive Committee on Research and the National Institutes of Health (award number K01AR062098). L. B., A. A. A., D. K. H., S. B., T. B., D. H., and C. H. are employed by Karius, Inc. A. A. A. is an attending physician at Boston Children’s Hospital but did not participate in the care of any of the patients cited in this study (adult patients obtained through Massachusetts General Hospital). A. A. A.’s work at Boston Children’s Hospital has no bearing on his role or contribution as a Medical Director at Karius toward this work. T. A. B. reports U.S. patents US20150133391A1, US20170016048A1 (pending), US20170275691A1, and US20190256891A1 (pending), unrelated to the submitted work. All other authors report no potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Steere AC, McHugh G, Damle N, Sikand VK. Prospective study of serologic tests for Lyme disease. Clin Infect Dis 2008; 47:188–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feder HM Jr, Gerber MA, Luger SW, Ryan RW. Persistence of serum antibodies to Borrelia burgdorferi in patients treated for Lyme disease. Clin Infect Dis 1992; 15:788–93. [DOI] [PubMed] [Google Scholar]
- 3.Schutzer SE, Body BA, Boyle J, et al. Direct diagnostic tests for Lyme disease. Clin Infect Dis 2019; 68:1052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowakowski J, Schwartz I, Liveris D, et al. , Lyme Disease Study Group . Laboratory diagnostic techniques for patients with early Lyme disease associated with erythema migrans: a comparison of different techniques. Clin Infect Dis 2001; 33:2023–7. [DOI] [PubMed] [Google Scholar]
- 5.Liveris D, Schwartz I, McKenna D, et al. Comparison of five diagnostic modalities for direct detection of Borrelia burgdorferi in patients with early Lyme disease. Diagn Microbiol Infect Dis 2012; 73:243–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liveris D, Schwartz I, McKenna D, et al. Quantitation of cell-associated borrelial DNA in the blood of Lyme disease patients with erythema migrans. Eur J Clin Microbiol Infect Dis 2012; 31:791–5. [DOI] [PubMed] [Google Scholar]
- 7.Coulter P, Lema C, Flayhart D, et al. Two-year evaluation of Borrelia burgdorferi culture and supplemental tests for definitive diagnosis of Lyme disease. J Clin Microbiol 2005; 43:5080–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones KL, Glickstein LJ, Damle N, Sikand VK, McHugh G, Steere AC. Borrelia burgdorferi genetic markers and disseminated disease in patients with early Lyme disease. J Clin Microbiol 2006; 44:4407–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Snyder JL, Giese H, Bandoski-Gralinski C, et al. T2 magnetic resonance assay-based direct detection of three Lyme disease-related Borrelia species in whole-blood samples. J Clin Microbiol 2017; 55:2453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pícha D, Moravcová L, Zdárský E, Maresová V, Hulínský V. PCR in Lyme neuroborreliosis: a prospective study. Acta Neurol Scand 2005; 112:287–92. [DOI] [PubMed] [Google Scholar]
- 11.Dumler JS. Molecular diagnosis of Lyme disease: review and meta-analysis. Mol Diagn 2001; 6:1–11. [DOI] [PubMed] [Google Scholar]
- 12.Avery RA, Frank G, Eppes SC. Diagnostic utility of Borrelia burgdorferi cerebrospinal fluid polymerase chain reaction in children with Lyme meningitis. Pediatr Infect Dis J 2005; 24:705–8. [DOI] [PubMed] [Google Scholar]
- 13.Blauwkamp TA, Thair S, Rosen MJ, et al. Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 2019; 4:663–74. [DOI] [PubMed] [Google Scholar]
- 14.Council of State and Territorial Epidemiologists. Revised national surveillance case definition for Lyme disease.2007. Available at: https://cdn.ymaws.com/www.cste.org/resource/resmgr/PS/07-ID-11.pdf. Accessed 24 February 2020.
- 15.Branda JA, Strle K, Nigrovic LE, et al. Evaluation of modified 2-tiered serodiagnostic testing algorithms for early Lyme disease. Clin Infect Dis 2017; 64:1074–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaz A, Glickstein L, Field JA, et al. Cellular and humoral immune responses to Borrelia burgdorferi antigens in patients with culture-positive early Lyme disease. Infect Immun 2001; 69:7437–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liveris D, Schwartz I, Bittker S, et al. Improving the yield of blood cultures from patients with early Lyme disease. J Clin Microbiol 2011; 49:2166–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Centers for Disease Control and Prevention. Recommendations for test performance and interpretation from the Second National Conference on Serologic Diagnosis of Lyme Disease. MMWR Morb Mortal Wkly Rep 1995; 44: 590–1. [PubMed] [Google Scholar]
- 19.Mead P, Petersen J, Hinckley A. Updated CDC recommendation for serologic diagnosis of Lyme disease. MMWR Morb Mortal Wkly Rep 2019; 68:703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith RP, Schoen RT, Rahn DW, et al. Clinical characteristics and treatment outcome of early Lyme disease in patients with microbiologically confirmed erythema migrans. Ann Intern Med 2002; 136:421–8. [DOI] [PubMed] [Google Scholar]
- 21.Wormser GP, Masters E, Nowakowski J, et al. Prospective clinical evaluation of patients from Missouri and New York with erythema migrans-like skin lesions. Clin Infect Dis 2005; 41:958–65. [DOI] [PubMed] [Google Scholar]
- 22.Angelakis E, Mediannikov O, Parola P, Raoult D. Rickettsia felis: the complex journey of an emergent human pathogen. Trends Parasitol 2016; 32:554–64. [DOI] [PubMed] [Google Scholar]
- 23.Farnaes L, Wilke J, Ryan Loker K, et al. Community-acquired pneumonia in children: cell-free plasma sequencing for diagnosis and management. Diagn Microbiol Infect Dis 2019; 94:188–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong DK, Blauwkamp TA, Kertesz M, Bercovici S, Truong C, Banaei N. Liquid biopsy for infectious diseases: sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn Microbiol Infect Dis 2018; 92:210–3. [DOI] [PubMed] [Google Scholar]
- 25.Vudatha V, Ranson M, Blair L, Ahmed AA. Rapid detection of bacille Calmette-Guérin-associated mycotic aortic aneurysm using novel cell-free DNA assay. J Vasc Surg Cases Innov Tech 2019; 5:143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rossoff J, Chaudhury S, Soneji M, et al. Noninvasive diagnosis of infection using plasma next-generation sequencing: a single-center experience. Open Forum Infect Dis 2019; 6:ofz327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.