Abstract
Background
Repeated exposure to malaria infections could protect against symptomatic progression as people develop adaptive immunity to infections acquired over time.
Methods
We investigated how new, recurrent, and persistent Plasmodium falciparum infections were associated with the odds of developing symptomatic compared with asymptomatic malaria. Using a 14-month longitudinal cohort in Western Kenya, we used amplicon deep sequencing of 2 polymorphic genes (pfama1 and pfcsp) to assess overlap of parasite genotypes (represented by haplotypes) acquired within an individual’s successive infections. We hypothesized infections with novel haplotypes would increase the odds of symptomatic malaria.
Results
After excluding initial infections, we observed 534 asymptomatic and 88 symptomatic infections across 186 people. We detected 109 pfcsp haplotypes, and each infection was classified as harboring novel, recurrent, or persistent haplotypes. Incident infections with only new haplotypes had higher odds of symptomatic malaria when compared with infections with only recurrent haplotypes [odds ratio (OR): 3.24; 95% confidence interval (CI), 1.20–8.78], but infections with both new and recurrent haplotypes (OR: 0.64; 95% CI: 0.15–2.65) did not. Assessing persistent infections, those with mixed (persistent with new or recurrent) haplotypes (OR: 0.77; 95% CI: 0.21–2.75) had no association with symptomatic malaria compared with infections with only persistent haplotypes. Results were similar for pfama1.
Conclusions
These results confirm that incident infections with only novel haplotypes were associated with increased odds of symptomatic malaria compared with infections with only recurrent haplotypes but this relationship was not seen when haplotypes persisted over time in consecutive infections.
Keywords: falciparum malaria, asymptomatic, adaptive immunity
Using high-resolution genotyping of over 800 P falciparum infections collected in a 14-month Kenyan cohort, we observed that exposure to new genotypes increased the risk of symptomatic malaria, and this risk was attenuated by recurrent and persistent genotypes.
Plasmodium falciparum causes more than 200 million clinical malaria cases annually [1]. Many of these infections occur in young children, who are more likely to develop symptomatic malaria compared with adults [2–4]. This age-dependent risk of symptomatic disease is thought to be due to repeated exposure to P falciparum that produces adaptive, disease-controlling immune responses [5–9]. The targets and mechanisms of this naturally acquired, antidisease immunity remain largely obscure.
In the absence of measurable immune correlates, the contours of functional clinical immunity to disease have been inferred from patterns of disease risk and parasite genetics. Specifically, the dependence of antidisease immunity on the gradual accumulation of functional responses to genetically diverse parasites has been supported by studies reporting that symptomatic malaria is often associated with the presence of parasite genotypes that were unobserved in prior infections [10–15]; this suggests that symptomatic malaria results from new infections that exploit gaps in immunologic memory. These studies, though, have been limited in scope and follow-up [12–15], resolution of genotyping approach [10–15], and an inability to partition effects of parasite genotypes between newly acquired and persistent infections, which collectively limit generalizability of findings. Furthermore, most [10, 12–15] have interrogated neutral parasite genes that do not encode targets of functional immunity, which limits causal inference of immunologic mechanisms. A clearer understanding of the influence of parasite genetic diversity on disease risk would inform the development of polyvalent vaccines.
To explore how specific P falciparum infections acquired over time influence the risk of symptomatic malaria, we investigated the association between P falciparum genotypes and an individual’s risk of symptomatic infection using a 14-month longitudinal cohort in a high-transmission setting in Western Kenya. We classified each person’s infections as harboring novel, recurrent, or persistent parasites on the basis of amplicon deep sequencing of 2 diverse parasite genes that encode targets of known functional immunity at the liver (circumsporozoite protein, pfcsp) and blood (apical membrane antigen-1, pfama1) stages, and analyzed associations between haplotype categories and odds of symptomatic malaria. We hypothesized that, compared with infections harboring parasite genotypes observed within a person’s prior infections, those harboring hitherto-unobserved haplotypes would be associated with increased likelihood of symptomatic malaria.
Materials and Methods
Study Population and Sample Collection
From June 2017 to July 2018, we followed a cohort in Webuye, Western Kenya, consisting of people aged 1 to 85 years residing in 38 households radially sampled in 3 villages [16] in an area of high malaria transmission primarily by Anopheles gambiae. Asymptomatic P falciparum infections were detected by active case detection using monthly dried blood spot (DBS) collection, in which parasites were detected by real-time polymerase chain reaction (qPCR). Symptomatic malaria infections were passively detected whereby participants experiencing malaria-like symptoms were tested for malaria using a rapid diagnostic test (RDT; Carestart Malaria HRP2 Pf from Accessbio) [17] and had a DBS collected. RDT-positive participants were treated with Artemether-Lumefantrine.
Sample Processing
Parasite genotyping has been previously described [18]. Briefly, genomic DNA from DBS was tested in duplicate for P falciparum parasites using a qPCR assay targeting the P falciparum pfr364 motif [19, 20], P falciparum-positive samples were genotyped at pfama1 and pfcsp using PCR amplification and sequencing on an Illumina MiSeq [21, 22]. Reads were quality-filtered and mapped to the 3D7 reference sequences for pfama1 and pfcsp [22–24] before haplotype inference using DADA2 (version 1.8) in R (version 4.0.2) [25, 26], and haplotypes were filtered to mitigate false discovery risk using previously validated criteria [27]. The output was a catalog of all pfcsp and pfama1 unique haplotypes in each qPCR-positive infection for each person. Sequences are available through GenBank (PRJNA646940).
Exposure and Outcome Assessment
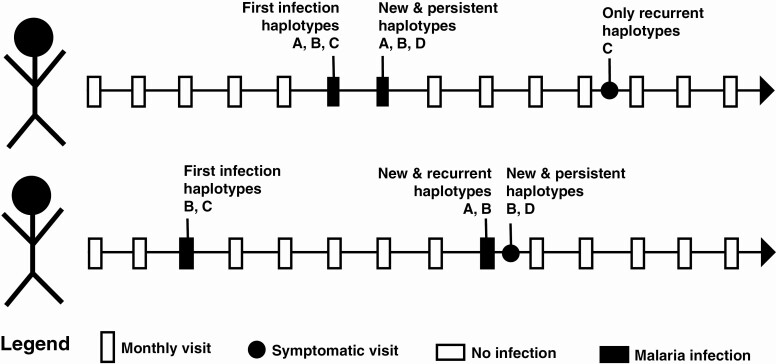
For every person, we classified each parasite haplotype in each of their infections as: (1) new, a haplotype not previously observed in that person during the study period; (2) recurrent, one previously observed in that person but not in the most recent DBS; or (3) persistent, a haplotype previously observed in the most recent DBS (Figure 1). For the main exposure, we used these haplotype classifications to categorize each infection; categories were assigned independently for pfama1 and pfcsp.
Figure 1.
Haplotype categorization throughout participant follow-up. Two hypothetical scenarios illustrate how the malaria haplotypes that participants acquired over time were categorized as new, recurrent, or persistent. Visits in which participants did not have an actively detected asymptomatic malaria infection at a monthly visit are indicated by a white box.
For the main outcome, each P falciparum infection was classified as asymptomatic or symptomatic. An asymptomatic infection was P falciparum-positive by qPCR in a person lacking symptoms during a monthly follow-up visit. A symptomatic infection was P falciparum-positive by RDT and qPCR in a person with at least 1 malaria symptom (ie, fever, aches, vomiting, diarrhea, chills, cough, or congestion) during a sick visit. Infections were excluded from outcome ascertainment if they occurred within 14 days of taking Artemether-Lumefantrine or were the person’s first infection during the study.
We assessed odds of symptomatic malaria as a function of haplotypes in 2 distinct types of malaria infections: (1) incident infections, where none of the haplotypes in the infection were previously observed in the participant’s most recent DBS or (2) persistent infections, where at least 1 haplotype persisted between consecutive DBS collections, excluding infections where participants had a symptomatic infection, were prescribed antimalarials, and had another infection with persistent haplotypes within 30 days following the initial infection.
Comparing Odds of Symptomatic Malaria Among Incident Infections
Among incident infections, we conducted a multilevel logistic regression comparing the odds of having a symptomatic compared to an asymptomatic infection among people infected with (1) only new haplotypes; (2) new and recurrent haplotypes; or (3) only recurrent haplotypes (Equation).
| (1) |
The model included a participant-level random intercept and controlled for confounding covariates (Figure S1): participant age (≤15 or >15 years), number of prior malaria infections during the study (≤3 or >3 infections), transmission season (≤50 or >50 mosquitoes collected in the prior 14 days across study site), and multiplicity of infection (≤2 or >2 haplotypes). Categorization thresholds were determined by functional form assessment. Differences in model covariates stratified by symptomatic status were compared using the Pearson’s χ 2 test.
We evaluated effect measure modification by age on the multiplicative scale by computing multilevel logistic regression models stratified by age category (≤15 or >15 years). Direction of effect and 95% confidence intervals were compared across age-stratified models. The log-likelihood ratio test compared output from an adjusted multilevel logistic regression with an interaction term between age and the haplotype categories to results from the model in the Equation.
Comparing Odds of Symptomatic Malaria Among Persistent Infections
We next focused on infections harboring persistent haplotypes, which were defined as haplotypes also observed in testing immediately before the episode. To do this, we restricted the data set to only infections with persistent haplotypes occurring within 30 days of a prior asymptomatic infection and classified the second infection within each of these pairs based on presence or absence of additional haplotypes: (1) only persistent; (2) new and persistent; (3) recurrent and persistent; or (4) new, recurrent, and persistent. We compared between categories the number of days since previous asymptomatic infection using the Kruskal-Wallis χ 2 test.
We then collapsed these persistent categories into infections with mixed types of haplotypes compared with only persistent haplotypes and computed a multilevel logistic regression similar to Equation 1. Model covariates and effect measure modification by age were evaluated as discussed previously.
To assess potential for presymptomatic infections, we analyzed pairs of symptomatic infections that had a preceding asymptomatic infection less than 30 days prior; among these asymptomatic-symptomatic pairings, we compared the time interval from asymptomatic to symptomatic infection between the symptomatic infections that did or did not harbor persistent haplotypes using a Kruskal-Wallis χ 2 test. Statistical analyses were performed using R (version 4.0.2) [26].
Ethical Review
The study was approved by the ethical review boards of Moi University (2017/36), Duke University (Pro00082000), and the University of North Carolina at Chapel Hill (19-1273). All study participants provided written informed consent or assent (for children).
RESULTS
Haplotype Classification and Infection Categorization
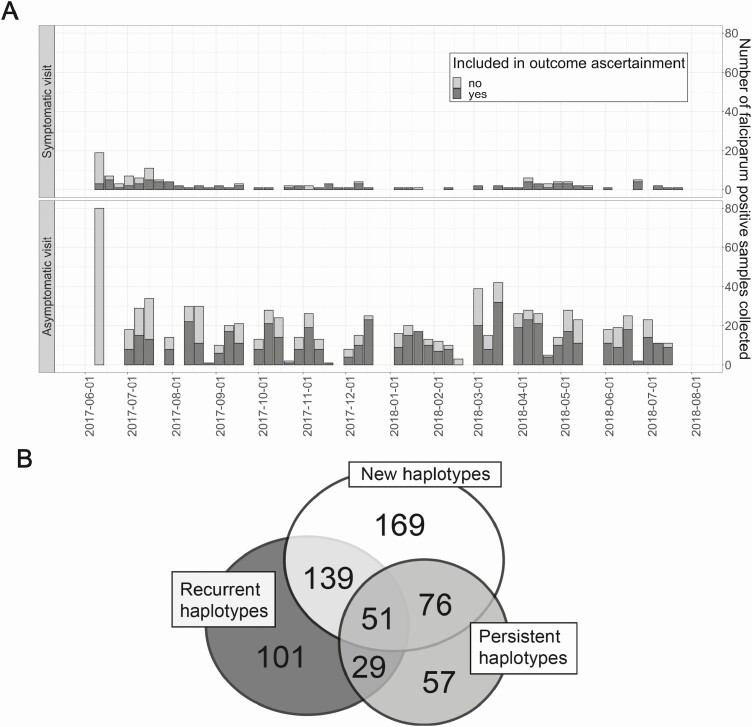
Over 14 months, we recorded 902 asymptomatic and 137 symptomatic P falciparum infections (Figure 2A). After parasite sequencing, we obtained genotypes for 861 P falciparum infections among 239 people, with a range from 1 to 14 infections during the study period (mean: 3.8). From these data, events meeting criteria for analysis as outcomes consisted of 622 infections (534 asymptomatic and 88 symptomatic) harboring 109 pfcsp haplotypes across 186 people; 435 infections (69.9%) harbored new haplotypes, 320 (51.4%) harbored recurrent haplotypes, and 213 (34.2%) had persistent haplotypes (Figure 2B). A plurality of infections (27.2%) harbored only new haplotypes (Table 1). Results for pfama1 are recorded in the supplement (Table S1, Figure S2).
Figure 2.
Total number of Plasmodium falciparum infection types and categorization of pfcsp haplotypes within these infections. A, Asymptomatic and symptomatic P falciparum-positive samples were captured during 14 months of sampling. Symptomatic infections were captured during as-needed sick visits and asymptomatic infections during monthly visits. A person’s initial infection is light gray. Subsequent infections for that person were used for outcome ascertainment (dark gray). B, Overlap of pfcsp haplotype categories across all symptomatic and asymptomatic P falciparum infections (N = 622). Numbers indicate the number of infections that had haplotypes within each category: new, recurrent, or persistent.
Table 1.
Distribution of Covariates Across Infections With Haplotype Results for pfcsp
| Infection Typea | |||||||
|---|---|---|---|---|---|---|---|
| Incident Infections (N = 409) | Persistent Infections Occurring With 30 Days (N = 139) | ||||||
| All (N = 622) | Asymptomatic Infections (N = 358) | Symptomatic Infections (N = 51) | P Value | Asymptomatic Infections (N = 109) | Symptomatic Infections (N = 30) | P Value | |
| Haplotype category, N (%) | <.001b | .002b | |||||
| Only new | 169 (27.2) | 133 (37.2) | 36 (70.6) | … | … | ||
| New and recurrent | 139 (22.3) | 134 (37.4) | 5 (9.8) | … | … | ||
| Only recurrent | 101 (16.2) | 91 (25.4) | 10 (19.6) | … | … | ||
| Persistent + ≥1 new or recurrent | 156 (25.1) | … | … | 86 (78.9) | 13 (43.3) | ||
| Only persistent | 57 (9.2) | … | … | 23 (21.1) | 17 (56.7) | ||
| Age, N (%) | .016b | 1.000b | |||||
| ≤15 years | 408 (65.6) | 213 (59.5) | 42 (82.4) | 76 (69.7) | 23 (76.7) | ||
| >15 years | 214 (34.4) | 145 (40.5) | 9 (17.6) | 33 (30.3) | 7 (23.3) | ||
| Number of prior malaria infectionsc, N (%) | 1.000b | .801b | |||||
| ≤3 | 425 (68.3) | 252 (70.4) | 40 (78.4) | 65 (59.6) | 23 (76.7) | ||
| >3 | 197 (31.7) | 106 (29.6) | 11 (21.6) | 44 (40.4) | 7 (23.3) | ||
| Transmission seasond, N (%) | .004b | .755b | |||||
| Low | 387 (62.2) | 245 (68.4) | 22 (43.1) | 70 (64.2) | 14 (46.7) | ||
| High | 235 (37.8) | 113 (31.6) | 29 (56.9) | 39 (35.8) | 16 (53.3) | ||
| Multiplicity of infection, N (%) | .022b | <.001b | |||||
| 1–2 pfcsp haplotypes | 317 (51.0) | 200 (55.9) | 40 (78.4) | 29 (26.6) | 21 (70.0) | ||
| >2 pfcsp haplotypes | 305 (49.0) | 158 (44.1) | 11 (21.6) | 80 (73.4) | 9 (30.0) | ||
Abbreviations: IQR, interquartile range; NE, not evaluated
aIncident infections were defined as Plasmodium falciparum infections in which none of the haplotypes in the infection were previously observed in the participant’s most recent dried blood spot. Persistent infections were defined as those in which at least 1 haplotype persisted between consecutive dried blood spot collections, excluding infections where participants had a symptomatic infection, were prescribed antimalarials, and had another infection with persistent haplotypes within 30 days following the initial infection. Some persistent infections occurred greater than 30 days apart; these were excluded from the persistent infection analysis.
bPearson’s χ 2 test with Bonferroni correction for repeated measures for 6 infections.
cDuring their participation in the cohort before the event.
dLow: ≤50 mosquitoes collected in the two weeks prior; high: > 50 mosquitoes.
Analysis of Symptomaticity in Incident Infections
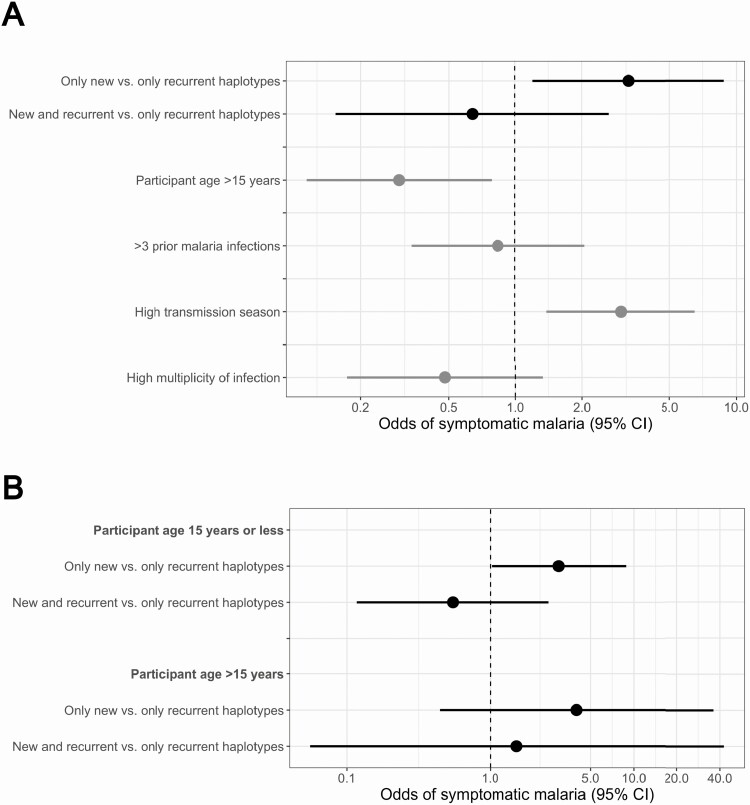
We first assessed if the presence of new haplotypes influenced odds of symptomatic malaria among 409 incident (358 asymptomatic and 51 symptomatic) infections consisting of: (1) only new (N = 169); (2) new and recurrent (N = 139); or (3) only recurrent (N = 101) pfcsp haplotypes. Among incident infections, symptomatic infections were more likely to consist of only new haplotypes, occur in children, arise during the high malaria transmission season, and have a lower multiplicity of infection (Tables 1, S1). Compared with infections composed of only recurrent pfcsp haplotypes, odds of symptomatic malaria were similar in those with both new and recurrent haplotypes (Odds Ratio [OR]: 0.64; 95% confidence interval [CI]: 0.15–2.65) but significantly higher for those harboring only new haplotypes (OR: 3.24; 95% CI: 1.20–8.78) (Figure 3A). Results were similar but not statistically significant for pfama1 (Figure S3). In age-stratified models, the association was similar in children ≤15 years (OR: 3.01; 95% CI: 1.02–8.84) and adults >15 years (OR: 4.00; 95% CI: 0.44–36.08), indicating, along with similarity in model fit between models with and without an interaction term for age (P = .996 by log-likelihood ratio test), that age did not modify the association between haplotype classification and symptoms (Figure 3B).
Figure 3.
Incident infections: comparison of odds of symptomatic malaria between infections harboring new versus recurrent pfcsp haplotypes. A, Multilevel logistic regression results for the odds of symptomatic malaria comparing (1) only new versus only recurrent (black) and (2) new and recurrent versus only recurrent (black) pfcsp haplotypes. Dots indicate the odds ratios and lines the 95% confidence intervals. B, Assessment of effect measure modification on symptomatic disease by age. Adjusted multilevel logistic regression models comparing the odds of developing symptomatic malaria between (1) only new versus only recurrent and (2) new and recurrent versus only recurrent haplotypes were computed conditioned on age category. Dots indicate the odds ratios and lines the corresponding 95% confidence intervals.
Analysis of Symptomaticity in Persistent Infections
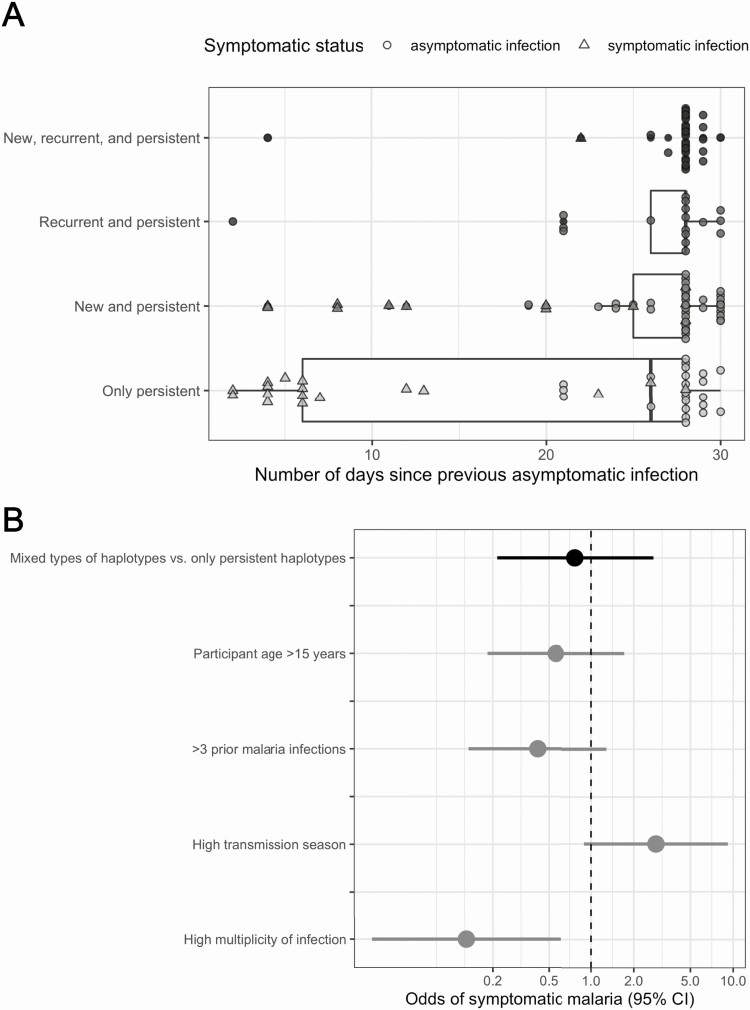
Restricting the data set to consecutive infections occurring within 30 days, persistent pfcsp haplotypes were identified in 139 infections (109 asymptomatic and 30 symptomatic) categorized into those containing: (1) only persistent (N = 40); (2) new and persistent (N = 49); (3) recurrent and persistent (N = 17); or (4) new, recurrent, and persistent (N = 33) pfcsp haplotypes. The number of days since previous asymptomatic infection differed between haplotype categories (P = .023 by Kruskal-Wallis χ 2 test), owing to a cluster of symptomatic infections consisting solely of persistent haplotypes with very small intervals (Figure 4A). A total of 66.7% of symptomatic infections with persistent haplotypes occurred within 14 days (N = 20/30). Results were similar for pfama1 (Figure S4).
Figure 4.
Persistent infections: comparison of odds of symptomatic malaria between infections harboring mixed (persistent mixed with new or recurrent) versus only persistent pfcsp haplotypes that occurred within 30 days. A, Distribution of the number of days since previous asymptomatic infection for malaria infections with persistent pfcsp haplotypes occurring within 30 days. Current infections were categorized into: (1) only persistent; (2) new and persistent; (3) recurrent and persistent; and (4) new, recurrent, and persistent. Current asymptomatic infections were represented by circles and symptomatic ones by triangles. B, Adjusted multilevel logistic regression results for the odds of symptomatic malaria comparing consecutive infections occurring within 30 days with mixed (persistent mixed with new or recurrent) types of haplotypes versus only persistent haplotypes (black). Dots represent odds ratios and lines the corresponding 95% confidence intervals.
To test whether the acquisition of new or recurrent haplotypes affected the odds of symptomatic compared with asymptomatic malaria among people with a background of persistent parasite haplotypes, we collapsed these categories into having only persistent pfcsp haplotypes or having mixed types of haplotypes (persistent haplotypes + at least 1 new or recurrent haplotype). Compared with infections with only persistent pfcsp haplotypes, the acquisition of additional haplotypes (either new or recurrent) was not associated with symptoms (OR: 0.77; 95% CI: 0.21–2.75) (Figure 4B). Results were similar using pfama1 (Figure S5). Owing to small sample sizes, we could not assess effect measure modification of these associations by age.
The shorter time interval between infections with persistent pfcsp haplotypes (median: 8; range, 2–28) compared with that for those without persistent haplotypes (median: 21.5; range, 4–30) (p value 0.022 by Kruskal-Wallis χ 2 test) suggested that some of these persistent infections could have been presymptomatic (Figures S6, S7). Sensitivity analyses removing potential presymptomatic infections could not be conducted due to data sparsity.
Discussion
In a high-transmission setting in Western Kenya, incident P falciparum infections composed of parasite haplotypes that were hitherto unobserved within an individual increased that person’s odds of symptomatic malaria. In contrast, the appearance of new haplotypes in a person who was already infected with persistent haplotypes did not increase the odds of symptoms. Collectively, our results are consistent with a model of antidisease immunity in which genetically distinct parasites can overcome immunity and cause disease in incident infections, but this ability is attenuated by the presence of persistent, tolerated parasites.
Compared with infections with haplotypes a person experienced previously during the 14-month study, we found incident infections with only new haplotypes increased odds of symptomatic malaria more than 3-fold. These results are consistent with the phenomena that partial variant-specific immunity is acquired over time to provide antidisease protection, and extend the findings of prior studies that report an increased risk of symptomatic malaria when infected with novel haplotypes [10–15]. Notably, our findings resulted from approaches that overcame limitations in these studies, including small sample sizes with brief follow-up [12–14], infrequent sampling [14, 15], genotyping approaches with high failure rates [10], and an inability to capture multiclonal genotypes [10–15]. Specifically, genotyping approaches that use PCR-restricted fragment length polymorphism to detect size variants [10–15] capture only 30% of the unique clones present compared with amplicon deep sequencing [28]. Under these approaches, complex infections common in high-transmission areas [29] are incompletely captured and can be incorrectly classified as new or recurrent. Using fine-scale genotypes created by the more sensitive amplicon deep sequencing method [28], we more definitively partitioned the distinct effects of new or recurrent haplotypes within incident infections. Our results suggest that symptomatic malaria among frequently infected residents of a high-transmission setting is associated with the acquisition of blood-stage parasites to which a person has been hitherto unexposed.
Surprisingly, this increased risk of symptomatic disease with new parasite haplotypes was attenuated when new haplotypes were mixed with recurrent ones. Because this analysis was restricted to incident infections, this could not be attributed to the “persistence” of recurrent haplotypes, suggesting the acquisition of recurrent parasite strains may mediate the disease-causing effects of new haplotypes. Attenuation could result from cross-reacting immune recognition of recurrent parasites that enhances parasite clearance or diminishes immune activation [9, 30], with competition between haplotypes that reduces pathogenesis [31] or alternate mechanisms. Also surprising, and in contrast to a prior report [10], we observed an increased risk of symptomatic malaria when new haplotypes were present both in adults and in children. The ability to register this effect is likely from use of a sensitive genotyping method that could capture diverse clones in polygenomic infections, which are more common in adults. Additionally, the enhanced though partial control of malaria transmission in Kenya over decades may have mitigated the durability of disease-controlling responses. The presence of this risk in adults supports an age-independent mechanism for this phenomenon, despite the common assumption that by reaching adulthood one has acquired durable immunity to diverse parasites.
In contrast to incident infections, persistent infections were not likely to be symptomatic when supplemented by new or recurrent parasite haplotypes. Asymptomatic infections with persistent haplotypes could be presymptomatic and, thus, not greatly impacted by the acquisition of additional haplotypes; this has been observed in previous work in which many asymptomatic infections later became symptomatic [32, 33] and is supported by the short time intervals we observed between asymptomatic-symptomatic infection pairs with persistent haplotypes. Alternatively, the presence of persistent haplotypes may limit immune responses [30] or the efficiency of establishing superinfections [34]; this would be consistent with the original meaning of “premunition,” wherein contemporaneous infection confers resistance to superinfection [35]. Regardless of mechanism, these results illustrate the importance of distinguishing between recurrent and persistent haplotypes in incident and persistent infections, which has previously not been done [10–15, 33]. Moreover, future work could assess variability in haplotype within-host competition [31], virulence of specific haplotypes [36], and host immune responses to more directly measure how new, recurrent, and persistent haplotypes affect symptomatic malaria risk.
Broadly, our results highlight not only the role that recurrent and persistent haplotypes have in reducing odds of symptomatic disease, but also the critical influence of parasite genetic diversity on this relationship. In population-based studies, reduced transmission can increase and shift the severity of disease [37], possibly by reduced acquisition of antidisease immunity in childhood. In our study, such antidisease immunity manifested in incident infections such that symptomaticity was prevented by the presence of recurrent haplotypes. Because these recurrent haplotypes require exposure to prior diverse infections, reduced exposure would increase the likelihood that incident infections are composed of new haplotypes and likely to manifest symptoms. However, if reduced transmission is accompanied by reductions in parasite genetic diversity, as has been reported in several settings [38, 39], even with fewer prior infections, the per-infection likelihood that a parasite will harbor recurrent haplotypes would remain high and thereby attenuate symptoms. Future studies could explore whether specific haplotypes at disease-mediating loci differentially modify the risk of malaria and furnish targets for surveillance.
The study had limitations. Although amplicon deep sequencing was a sensitive method for identifying different malaria infections [28], it might not have captured all genetically distinct infections that occurred during the study. To account for this, we compared results across 2 unlinked parasite gene targets, pfama1 and pfcsp. We did not observe malaria infections that participants acquired before the study; misclassifying a haplotype as new when it might have been present in an individual before the study would bias results toward the null. Additionally, persistent infections were possibly presymptomatic. Future studies could have more frequent longitudinal sampling to distinguish between asymptomatic and presymptomatic infections.
In conclusion, infections harboring novel haplotypes increased the likelihood of symptomatic malaria in incident infections, but not when acquired in the presence of persistent infections. Future research could explore the immunological mechanisms by which new haplotypes change the risk of symptomatic malaria when compared to recurrent or persistent haplotypes.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors thank the study households for their participation in this study and also are appreciative of the study implementation skills of project manager and field technicians in Webuye and Eldoret: J. Kipkoech Kirui, I. Khaoya, L. Marango, E. Mukeli, E. Nalianya, J. Namae, L. Nukewa, E. Wamalwa, and A. Wekesa. We thank A. Nantume and J. Saelens (each of Duke University) for their help with laboratory sample and data processing.
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (R21AI126024 to W. P. O. and R01AI146849 to W. P. O. and S. M. T.). This work was also partially supported by a Triangle Center for Evolutionary Medicine (TriCEM) Graduate Student Award.
Potential conflicts of interest. The authors have no conflicts of interest to declare. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.World malaria report 2019. Geneva: World Health Organization; 2019. Licence: CC BY-NC-SA 3.0 IGO. Available at: https://www.who.int/publications/i/item/9789241565721. [Google Scholar]
- 2.Newell K, Kiggundu V, Ouma J, et al. . Longitudinal household surveillance for malaria in Rakai, Uganda. Malar J 2016; 15:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sondén K, Doumbo S, Hammar U, et al. . Asymptomatic multiclonal plasmodium falciparum infections carried through the dry season predict protection against subsequent clinical malaria. J Infect Dis 2015; 212:608–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adomako-Ankomah Y, Chenoweth MS, Durfee K, et al. . High Plasmodium falciparum longitudinal prevalence is associated with high multiclonality and reduced clinical malaria risk in a seasonal transmission area of Mali. PLoS One 2017; 12:e0170948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsh K, Howard RJ. Antigens induced on erythrocytes by P. falciparum: expression of diverse and conserved determinants. Science 1986; 231:150–3. [DOI] [PubMed] [Google Scholar]
- 6.Giha HA, Theander TG, Staalsø T, et al. . Seasonal variation in agglutination of Plasmodium falciparum-infected erythrocytes. Am J Trop Med Hyg 1998; 58:399–405. [DOI] [PubMed] [Google Scholar]
- 7.Gupta S, Day KP. A strain theory of malaria transmission. Parasitol Today 1994; 10:476–81. [DOI] [PubMed] [Google Scholar]
- 8.Ntoumi F, Bakoua D, Fesser A, Kombo M, Vouvoungui JC, Koukouikila Koussounda F. Characterization of asymptomatic Plasmodium falciparum infection and its risk factors in pregnant women from the Republic of Congo. Acta Trop 1995; 153:111–5. [DOI] [PubMed] [Google Scholar]
- 9.Smith T, Felger I, Tanner M, Beck HP. Premunition in Plasmodium falciparum infection: insights from the epidemiology of multiple infections. Trans R Soc Trop Med Hyg 1999; 93(Suppl 1):59–64. [DOI] [PubMed] [Google Scholar]
- 10.Buchwald AG, Sixpence A, Chimenya M, et al. . Clinical implications of asymptomatic plasmodium falciparum infections in Malawi. Clin Infect Dis 2019; 68:106–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takala SL, Coulibaly D, Thera MA, et al. . Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci Transl Med 2009; 1:2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Contamin H, Fandeur T, Rogier C, et al. . Different genetic characteristics of Plasmodium falciparum isolates collected during successive clinical malaria episodes in Senegalese children. Am J Trop Med Hyg 1996; 54:632–43. [DOI] [PubMed] [Google Scholar]
- 13.Kun JF, Missinou MA, Lell B, et al. . New emerging Plasmodium falciparum genotypes in children during the transition phase from asymptomatic parasitemia to malaria. Am J Trop Med Hyg 2002; 66:653–8. [DOI] [PubMed] [Google Scholar]
- 14.Babiker HA, Abdel-Muhsin AM, Ranford-Cartwright LC, Satti G, Walliker D. Characteristics of Plasmodium falciparum parasites that survive the lengthy dry season in eastern Sudan where malaria transmission is markedly seasonal. Am J Trop Med Hyg 1998; 59:582–90. [DOI] [PubMed] [Google Scholar]
- 15.Roper C, Richardson W, Elhassan IM, et al. . Seasonal changes in the Plasmodium falciparum population in individuals and their relationship to clinical malaria: a longitudinal study in a Sudanese village. Parasitology 1998; 116 (Pt 6):501–10. [DOI] [PubMed] [Google Scholar]
- 16.O’Meara WP, Simmons R, Bullins P, et al. . Mosquito exposure and malaria morbidity; a micro-level analysis of household mosquito populations and malaria in a population-based longitudinal cohort in western Kenya. J Infect Dis 2019; 27708:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.AccessBio. CareStart Malaria.2019; Available at: http://www.accessbio.net/eng/products/products01_02.asp. Accessed 14 January 2021.
- 18.Sumner KM, Freedman E, Abel L, et al. . Genotyping cognate Plasmodium falciparum in humans and mosquitoes to estimate onward transmission of asymptomatic infections. Nat Commun 2021; 12:909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 1995; 52:565–8. [DOI] [PubMed] [Google Scholar]
- 20.Taylor SM, Sumner KM, Freedman B, Mangeni JN, Obala AA, Prudhomme O’Meara W. Direct estimation of sensitivity of Plasmodium falciparum rapid diagnostic test for active case detection in a high-transmission community setting. Am J Trop Med Hyg 2019; 101:1416–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Illumina. Illumnia miseq.2018. Available at: https://www.illumina.com/systems/sequencing-platforms/miseq.html. Accessed 5 December 2018.
- 22.Nelson CS, Sumner KM, Freedman E, et al. . High-resolution micro-epidemiology of parasite spatial and temporal dynamics in a high malaria transmission setting in Kenya. Nat Commun 2019; 10:5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gardner MJ, Hall N, Fung E, et al. . Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 2002; 419:498–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall N, Pain A, Berriman M, et al. . Sequence of Plasmodium falciparum chromosomes 1, 3-9 and 13. Nature 2002; 419:527–31. [DOI] [PubMed] [Google Scholar]
- 25.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016; 13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Core Team. R: a language and environment for statistical computing.2020. Available at: https://www.r-project.org/.
- 27.Early AM, Daniels RF, Farrell TM, et al. . Detection of low-density Plasmodium falciparum infections using amplicon deep sequencing. Malar J 2019; 18:219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juliano JJ, Porter K, Mwapasa V, et al. . Exposing malaria in-host diversity and estimating population diversity by capture-recapture using massively parallel pyrosequencing. Proc Natl Acad Sci U S A 2010; 107:20138–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levitt B, Obala A, Langdon S, Corcoran D, O’Meara WP, Taylor SM. Overlap extension barcoding for the next generation sequencing and genotyping of plasmodium falciparum in individual patients in Western Kenya. Sci Rep 2017; 7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boutlis CS, Yeo TW, Anstey NM. Malaria tolerance–for whom the cell tolls? Trends Parasitol 2006; 22:371–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Read AF, Taylor LH. The ecology of genetically diverse infections. Science 2001; 292:1099–102. [DOI] [PubMed] [Google Scholar]
- 32.Njama-Meya D, Kamya MR, Dorsey G. Asymptomatic parasitaemia as a risk factor for symptomatic malaria in a cohort of Ugandan children. Trop Med Int Health 2004; 9:862–8. [DOI] [PubMed] [Google Scholar]
- 33.Nsobya SL, Parikh S, Kironde F, et al. . Molecular evaluation of the natural history of asymptomatic parasitemia in Ugandan children. J Infect Dis 2004; 189:2220–6. [DOI] [PubMed] [Google Scholar]
- 34.Portugal S, Carret C, Recker M, et al. . Host-mediated regulation of superinfection in malaria. Nat Med 2011; 17:732–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sergent ED, Parrot L. L’immunité, la prémunition et la résistance innée. Arch l’Institut Pasteur d’Algérie 1935; 13:279. [Google Scholar]
- 36.Chaorattanakawee S, Nuchnoi P, Hananantachai H, et al. . Sequence variation in Plasmodium falciparum merozoite surface protein-2 is associated with virulence causing severe and cerebral malaria. PLoS One 2018; 13:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snow RW, Marsh K. The consequences of reducing transmission of Plasmodium falciparum in Africa. Adv Parasitol 2002; 52:235–64. [DOI] [PubMed] [Google Scholar]
- 38.Daniels RF, Schaffner SF, Wenger EA, et al. . Modeling malaria genomics reveals transmission decline and rebound in Senegal. Proc Natl Acad Sci U S A 2015; 112:7067–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nkhoma SC, Nair S, Al-Saai S, et al. . Population genetic correlates of declining transmission in a human pathogen. Mol Ecol 2013; 22:273–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.