Abstract
Background
Conventional blood cultures were compared to plasma cell-free DNA–based 16S ribosomal RNA (rRNA) gene polymerase chain reaction (PCR)/next-generation sequencing (NGS) for detection and identification of potential pathogens in patients with sepsis.
Methods
Plasma was prospectively collected from 60 adult patients with sepsis presenting to the Mayo Clinic (Minnesota) Emergency Department from March through August 2019. Results of routine clinical blood cultures were compared to those of 16S rRNA gene NGS.
Results
Nineteen (32%) subjects had positive blood cultures, of which 13 yielded gram-negative bacilli, 5 gram-positive cocci, and 1 both gram-negative bacilli and gram-positive cocci. 16S rRNA gene NGS findings were concordant in 11. For the remaining 8, 16S rRNA gene NGS results yielded discordant detections (n = 5) or were negative (n = 3). Interestingly, Clostridium species were additionally detected by 16S rRNA gene NGS in 3 of the 6 subjects with gastrointestinal sources of gram-negative bacteremia and none of the 3 subjects with urinary sources of gram-negative bacteremia. In the 41 remaining subjects, 16S rRNA gene NGS detected at least 1 potentially pathogenic organism in 17. In 15, the detected microorganism clinically correlated with the patient’s syndrome. In 17 subjects with a clinically defined infectious syndrome, neither test was positive; in the remaining 7 subjects, a noninfectious cause of clinical presentation was identified.
Conclusions
16S rRNA gene NGS may be useful for detecting bacteria in plasma of septic patients. In some cases of gram-negative sepsis, it may be possible to pinpoint a gastrointestinal or urinary source of sepsis based on the profile of bacteria detected in plasma.
Keywords: molecular diagnostics, sepsis, metagenomic sequencing, 16S rRNA gene sequencing, cell-free DNA
In patients with sepsis and negative blood cultures, plasma cell-free DNA–based, 16S rRNA gene polymerase chain reaction/next-generation sequencing may be useful to identify pathogenic bacteria.
(See the Editorial Commentary by Rello and Alonso-Tarrés on pages 1173–5.)
Sepsis is the second leading cause of noncardiac death in the United States and its incidence has increased over the past several decades [1]. Mortality in patients with sepsis and septic shock remains high despite extensive research in this field [2]. Blood cultures are the most frequent microbiologic test performed in patients with suspected sepsis. However, they have limitations, including slow turnaround time and low yield. Historically, only about 30% of septic patients have a causative organism identified, even in cases with high clinical suspicion for infection, limiting targeted and maximally effective treatment [3]. Other non-culture-based diagnostic tests, including serologic and most molecular tests, are limited by their ability to detect a single to a handful of pathogens. Rapid detection of pathogens has the potential to alter therapeutic management and ultimately reduce inappropriate antimicrobial use and associated side effects, potentially improving patient outcomes. Over the last decade, several small studies utilizing molecular diagnostics have been conducted in patients with sepsis; however, large studies need to be done to inform the utilization of these novel techniques [4–6].
Cell-free DNA (cfDNA), which refers to fragments of freely circulating DNA, is found in almost all body fluids. cfDNA was first described by Mandel and Metais in 1948 and has now found its use in detection of fetal aneuploidies and transplant rejection as well as in noninvasive diagnosis of malignancies [7–10]. More recently, studies evaluating the role of cfDNA-based next-generation sequencing (NGS) in the diagnosis of infections, including infective endocarditis, and atypical bacterial and mycobacterial infections, among other infection types, have been published; however, more data are required to inform clinical utilization of these techniques [11–15].
We compared traditional blood cultures to plasma cfDNA-based 16S ribosomal RNA gene polymerase chain reaction (PCR)/next-generation sequencing (16S rRNA gene NGS)—a targeted metagenomic sequencing approach—for detection and identification of bacteria in septic patients.
MATERIALS AND METHODS
Study Overview and Subjects
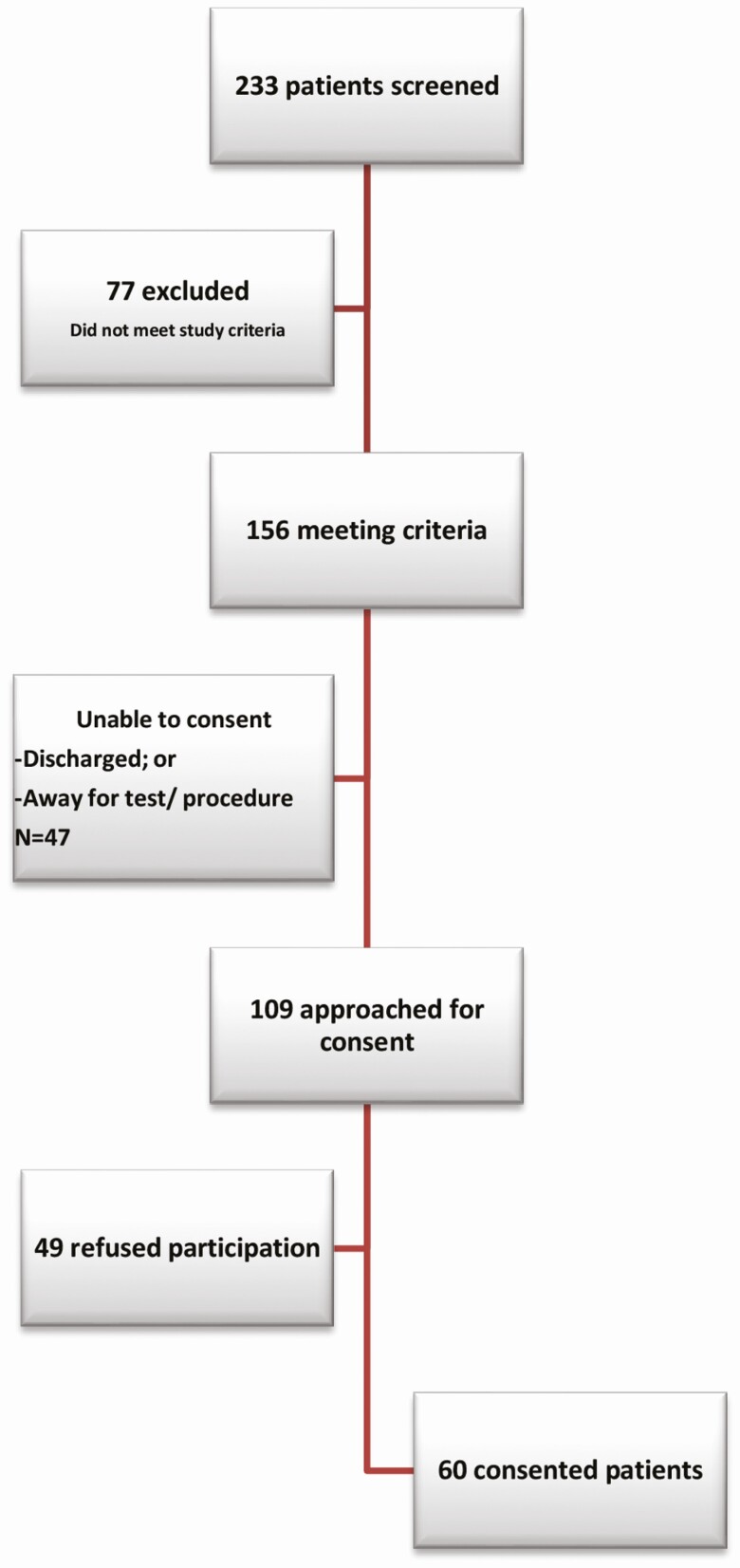
This is a single-center noninterventional prospective study in which plasma samples were collected from 60 patients who presented to the Mayo Clinic emergency department from March through August 2019. Adult patients (≥18 years old) were screened if upon presentation they had suspected infection as defined by the need to obtain blood cultures and receipt of antibiotics and were admitted to the hospital. Samples for 16S rRNA NGS were obtained at the same time as blood cultures in patients with clinical suspicion of sepsis in the emergency department. Following sample collection, the investigator reviewed the patient’s medical record to determine if the patient met study criteria. Patients with sepsis or septic shock according to new sepsis definitions (Sepsis-3) were included [16]. Specimens from patients who did not meet study criteria were discarded. Patients who met study criteria were approached for consent. If individuals were unable to provide consent (eg, due to sedation for mechanical ventilation or encephalopathy), a legally authorized representative was approached for informed consent. If consent was not obtained, the specimen was discarded. This study was approved by the Mayo Clinic Institutional Review Board.
Sample Collection and Processing
Blood Cultures
The standard blood culture included 2 sets, with each set consisting of 2 BD BACTEC Plus Aerobic/F bottles and 1 BD BACTEC Lytic Anaerobic/F bottle (Becton Dickinson, Heidelberg, Germany) with each bottle inoculated with a targeted 10 mL of whole blood. Blood culture bottles were incubated for 5 days in the Division of Clinical Microbiology’s Clinical Bacteriology Laboratory, Mayo Clinic, on the BD BACTEC FX platform.
DNA Extraction/Purification
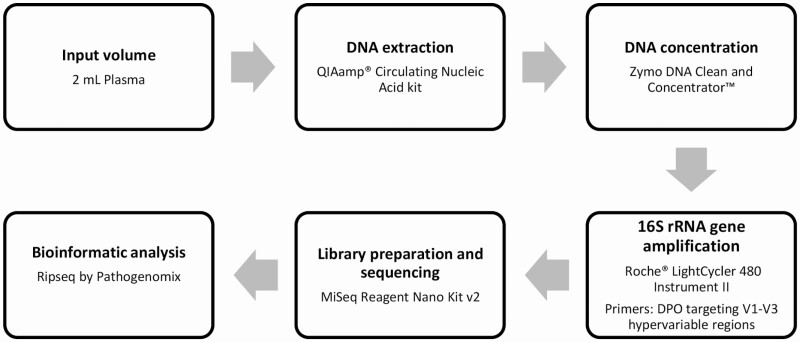
Sample preparation and NGS analysis are depicted in Figure 1. Ten milliliters of whole blood was processed to plasma in the Mayo Clinic Central Clinical Laboratory/Central Processing within 4 hours of obtaining the sample, by centrifugation for 10 minutes at 1900g at 4°C. DNA extraction and purification was performed using 2 mL plasma with the Qiagen (Hilden, Germany) QIAamp Circulating Nucleic Acid kit. The following modifications to the manufacturer’s protocol were implemented: An additional 150 µL elution buffer was applied to the QIAamp Mini membrane after initial elution with 150 µL of elution buffer, yielding 300 µL of eluted DNA. The Zymo DNA Clean and Concentrator-5 kit was then used to concentrate the eluted DNA to 60 µL following the manufacturer’s protocol.
Figure 1.
Workflow showing sample preparation and next-generation sequencing analysis. Abbreviation: DPO, dual priming oligonucleotide; rRNA, ribosomal RNA.
16S rRNA Gene NGS
Quantitative real-time PCR and amplification of the 16S rRNA gene was conducted on a Roche LightCycler 480 Instrument II with SYBR Green DNA detection using dual priming oligonucleotides targeting the V1–V3 hypervariable (forward primer 5′-AGAGTTTGATCMTGGCTCAIIIIIAACGC-3′ and reverse primers reverse (1) 5′-CGGCTGCTGGCAIIIAITTDGC-3′ and reverse primers 5′-CGGCTGCTGGCAIIIAITTDGC-3′ and 5′-CGGCTGCTGGCAIIIAITTDGT-3′) regions of the 16S rRNA gene. PCR thermal cycling and sequencing conditions were as previously described [17]. Library preparation was done using the MiSeq Reagent Nano Kit version 2. The process included an adapter PCR using a forward primer (5′TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGAGAGTTTGATCMTGGCTCAG) and reverse primer (5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG CGGCTGCTGGCA-3′) and the following cycling conditions: initial incubation at 95°C for 3 minutes followed by 25 cycles of 30 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C with final incubation at 72°C for 5 minutes. PCR product was then purified and quantified. Index PCR was done utilizing Illumina Nextera Unique Dual Indexes using KAPA HiFi HotStart ReadyMix with the following cycling conditions: initial incubation at 95°C for 3 minutes followed by 8 cycles of 30 seconds at 95°C, 30 seconds at 55°C, and 30 seconds at 72°C, with final incubation at 72°C for 5 minutes. The library was then quantified, normalized, and pooled to a final library concentration of 1.3–1.7 ng/µL. The MiSeq Reagent Kit version 2 Nano (Illumina, San Diego, California) was used (approximately 28 hours of run time). On average, each run consisted of 50 samples. Bioinformatic analysis was performed using the Ripseq NGS web application by Pathogenomix, Inc (Santa Cruz, California). The Ripseq database contains clinically relevant bacteria and is based on information from GenBank. Only bacteria with ≥98% sequence identify to sequences in the database were included in the analysis. Bacteria previously described as pathogenic or clinically relevant were considered significant, whereas those that are well-known contaminants were excluded from the analysis. Processing time for the entire process of 16S rRNA NGS was approximately 72 hours.
RESULTS
Between March and August 2019, 233 patients who presented to the emergency department were screened; 109 were approached for consent and 60 were included in the study (Figure 2). The mean patient age was 63 years and 68% were male. The mean leukocyte count was 13.7 × 103 cells/µL. The median duration of hospitalization was 5 days. Overall, 35% (n = 21) required admission to the intensive care unit (ICU) with a median duration of ICU stay of 2 days; 57% (n = 12) of these patients required vasopressors. Comorbidities included hypertension (n = 15 [25%]), ischemic heart disease (n = 9 [15%]), chronic kidney disease (n = 6 [10%]), active malignancy (n = 23 [38%]), organ transplantation (n = 5 [8%]), and cirrhosis (n = 2 [3%]). Underlying infectious syndromes included respiratory (n = 15 [25%]), abdominal (n = 13 [22%]), urinary tract (n = 5 [8%]), catheter/device related (n = 3 [5%]), skin/soft tissue (n = 10 [17%]), and other/unknown (n = 2 [3%]) infections, and neutropenic fever (n = 4 [7%]). Thirteen percent (n = 8) of cases in which infection was suspected on admission were eventually diagnosed as having a noninfectious condition.
Figure 2.
Patient disposition.
Blood cultures were positive in 32% (n = 19) of the patients, whereas 16S rRNA gene NGS detected a potentially pathogenic organism in 47% (n = 28). Among those with positive blood cultures, 16S rRNA gene NGS had concordant results in 58% (n = 11), whereas in 8 cases, 16S rRNA gene NGS was either negative (n = 3) or yielded discordant identifications (n = 5) compared with blood cultures.
Samples With Positive Blood Cultures
Nineteen (32%) subjects had positive blood cultures, of which 68% (n = 13) yielded gram-negative bacilli and 26% (n = 5) gram-positive cocci; 1 individual had combined Escherichia coli and Staphylococcus aureus bacteremia. 16S rRNA gene NGS results were concordant with blood cultures in 11 (8 gram-negative bacilli, 2 gram-positive cocci, and the polymicrobial case). For the remaining 8, 16S rRNA gene NGS showed discordant identifications or was negative.
Concordant 16S rRNA Gene NGS Results
In 11 cases, 16S rRNA gene NGS results were concordant with blood cultures (Table 1). All except 1 represented monomicrobial bacteremia. Most patients in this subset had gram-negative bacteremia (72%). In 4 cases, bacteremia was from a urinary source, 5 from a gastrointestinal source, and 2 from a central line– or cardiac device–associated infection. In those with a gastrointestinal source of bacteremia, additional fecal bacteria were detected by 16S rRNA gene NGS in 4 of 5 cases, compared to none of those with urinary tract infection as a source of bacteremia. Most patients (4 of 5) with an abdominal source of bacteremia received anaerobic coverage as part of their antimicrobial regimen. In 2 patients with bacteremia from a urinary source, organisms traditionally considered urinary tract colonizers were detected as well, including Lactobacillus crispatus in the case with Klebsiella pnueumoniae bacteremia and Lactococcus lactis in the case with E. coli bacteremia. In the patient with E. coli and S. aureus bacteremia, 16S rRNA gene NGS identified both organisms. This was a case of febrile neutropenia with a rectal abscess; 16S rRNA gene NGS additionally identified Clostridium species. Time to blood culture positivity was variable (3–92 hours) with a median time of positivity of 12 hours. More than half of the patients (n = 7 [64%]) required ICU care with a median duration of ICU stay of 1.5 days and a median duration of hospitalization of 5 days. Three of these patients also required vasopressors.
Table 1.
Concordant Culture and 16S Ribosomal RNA Gene Polymerase Chain Reaction Followed by Next-Generation Sequencing Results
| Blood Culture Results | Bottles Positive | Time to Positivity | 16S rRNA Gene PCR Followed by NGS Results | Clinical Syndrome Causing Sepsis |
|---|---|---|---|---|
| Klebsiella oxytoca/Raoultella ornithinolytica | 3/3, 3/3 | 3 h | Enterobacterales, Clostridium species | Chronic fistulizing Crohn disease |
| K. pneumoniae | 2/3, 1/3 | 13 h | Enterobacterales, Lactobacillus crispatus | UTI with bacteremia |
| Escherichia coli | 1/3, 0/3 | 92 h | Enterobacterales, Allobaculum stercoricanis | Necrotizing pancreatitis, abdominal fluid collection |
| E. coli | 3/3, 3/3 | 11 h | Enterobacterales, Lactococcus lactis | UTI with bacteremia |
| E. coli | 1/3, 0/3 | 28 h | Enterobacterales | Refractory cholangitis |
| E. coli, Staphylococcus aureus complex | E. coli 3/3, 1/3 S. aureus complex 2/3, 1/3 | 9 h (E. coli) 10 h (S. aureus) | Enterobacterales, S. aureus complex, Clostridium species | Neutropenic fever with rectal abscess |
| Citrobacter freundii complex | 3/3, 3/3 | 12 h | C. freundii, Clostridium species | Abdominal abscess |
| Pseudomonas aeruginosa | 1/3, 0/3 | 17 h | Pseudomonas aeruginosa, Haemophilus parainfluenzae | Allograft pyelonephritis |
| K. oxytoca/R. ornithinolytica | 3/3, 3/3 | 11 h | Enterobacterales | UTI |
| Staphylococcus warneri | 3/3, 3/3 | 13 h | Staphylococcus warneri | CLABSI |
| S. aureus complex | 2/2, 2/2 | 10 h | S. aureus complex | Cardiac device–associated infection |
Abbreviations: CLABSI, central line–associated bloodstream infection; NGS, next-generation sequencing; PCR, polymerase chain reaction; rRNA, ribosomal RNA; UTI, urinary tract infection.
Discordant 16S rRNA Gene NGS Results
Eight patients had positive blood cultures with negative (n = 3) or discordant (n = 5)16S rRNA gene NGS results. Among these cases, 5 were gram-negative and 3 gram-positive bacteremias (Table 2). Pseudomonas aeruginosa was not detected by 16S rRNA gene NGS in 2 patients. In 1 of these patients, 1 of 6 bottles was positive, and infection was from a urinary source, whereas in the other, 4 of 6 bottles were positive and the source was pneumonia. In a patient with E. coli bacteremia secondary to ascending cholangitis, 16S rRNA gene NGS was negative for E. coli despite 6 of 6 bottles being positive; however, Clostridium species was detected. In a patient with Proteus mirabilis bacteremia from a urinary source, 2 of 2 bottles were positive with a time to positivity of 21 hours; however, 16S rRNA gene NGS was negative. In a patient with neutropenic fever, 1 of 6 bottles were positive for Fusobacterium nucleatum with a time to positivity of 60 hours, whereas 16S rRNA gene NGS detected Bacteroides vulgatus. Gram-positive organisms not detected by 16S rRNA gene NGS included Staphylococcus epidermidis, Streptococcus pyogenes, and Enterococcus faecalis. In the patient with E. faecalis bacteremia secondary to liver abscess and another with S. pyogenes bacteremia related to abdominal wall cellulitis, all 6 blood culture bottles were positive. In both cases, 16S rRNA gene NGS detected Enterobacterales. 16S rRNA gene NGS failed to detect S. epidermidis in a case of central line–associated S. epidermidis bacteremia.
Table 2.
Discordant Culture and 16S Ribosomal RNA Gene Polymerase Chain Reaction Followed by Next-Generation Sequencing Results
| Blood Culture Results | Bottles Positive (Set 1, Set 2) | Time to Positivity, h | 16S rRNA Gene PCR Followed by NGS Result | Clinical Syndrome Causing Sepsis |
|---|---|---|---|---|
| Staphylococcus epidermidis | 2/3, 1/3 | 11 | Negative | CLABSI |
| Streptococcus pyogenes | 3/3, 3/3 | 10 | Enterobacterales | Cellulitis |
| Enterococcus faecalis | 3/3, 3/3 | 10 | Enterobacterales | Polymicrobial liver abscess |
| Pseudomonas aeruginosa | 2/3, 2/3 | 16 | Negative | Pneumonia |
| P. aeruginosa | 2/3, 2/3 | 20 | Enterobacterales | Unknown source of bacteremia |
| Fusobacterium nucleatum | 1/3, 0/3 | 60 | Bacteroides vulgatus | Neutropenic fever |
| Escherichia coli | 3/3, 3/3 | 10 | Clostridium species | Proctitis |
| Proteus mirabilis | 2/2a | 21 | Negative | UTI, septic thrombophlebitis |
Abbreviations: CLABSI, central line–associated bloodstream infection; NGS, next-generation sequencing; PCR, polymerase chain reaction; rRNA, ribosomal RNA; UTI, urinary tract infection.
aOnly 2 bottles were obtained in this case.
Blood Culture Negative and 16S rRNA Gene NGS-Positive Results
In 17 cases, blood cultures were negative whereas 16S rRNA gene NGS was positive (Table 3). In 1 of these cases, the eventual diagnosis was noninfectious (cardiogenic shock) with 16S rRNA gene NGS detecting Enterobacterales. In another case with diabetic foot infection, multiple pathogenic organisms (Streptococcus agalactiae, Enterobacter cloacae, Streptococcus salivarius, and Staphylococcus lugdunensis) were detected by 16S rRNA gene NGS. In 2 patients with Clostridioides difficile infection, anaerobic gastrointestinal bacteria (Prevotella bivia and B. vulgatus, 1 in each of the subjects) were detected. In 3 patients with abdominal sources of infection, pathogenic enteric organisms were detected; Clostridium species in pancolitis, Fusobacterium periodonticum in cholecystitis, and Enterobacterales in abdominal abscess.
Table 3.
Cases With Negative Blood Culture and Positive 16S Ribosomal RNA Gene Polymerase Chain Reaction Followed by Next-Generation Sequencing Results
| 16S rRNA Gene NGS Results | Clinical Syndrome |
|---|---|
| Fusobacterium periodonticum | Cholecystitis |
| Enterobacterales | Abdominal abscess |
| Streptococcus species | Neutropenic fever |
| Clostridium saudiense | Pancolitis |
| Bacteroides vulgatus | Clostridioides difficile colitis |
| Prevotella bivia | C. difficile colitis, pneumonia |
| Legionella pneumophila | Legionella pneumophila serotype 1 pneumonia |
| Streptococcus agalactiae, Enterobacter cloacae, Streptococcus salivarius, Staphylococcus lugdunensis | Diabetic foot infection |
| Fusobacterium nucleatum | Pneumonia |
| F. nucleatum, Capnocytophaga species | Necrotizing pneumonia |
| Enterobacterales | Pneumonia with cystic fibrosis exacerbation |
| Enterobacterales | Pneumonia |
| Stenotrophomonas maltophilia | Pneumonia |
| S. maltophilia | Pneumonia |
| Acidovorax species | Newly diagnosed acute myelogenous leukemia, fever with functional neutropenia |
| Clostridium species | |
| Enterobacterales | Cardiogenic shock |
| Enterobacterales | Probable septic arthritis |
Abbreviations: NGS, next-generation sequencing; rRNA, ribosomal RNA.
In a patient with Legionella pneumophila serotype 1 pneumonia diagnosed by Legionella urine antigen, 16S rRNA gene NGS detected L. pneumophila. In 6 patients with pneumonia, Enterobacterales (n = 2), Stenotrophomonas maltophilia (n = 2), and F. nucleatum (n = 2) were detected. In a case of septic arthritis in which operative cultures were negative, Enterobacterales were detected by 16S rRNA gene NGS. Six (35%) of the patients with negative blood cultures and positive 16S rRNA gene NGS required an ICU stay, with 2 requiring vasopressors.
Negative Blood Cultures and 16S rRNA Gene NGS Results
In 24 cases, neither 16S rRNA gene NGS nor blood cultures were positive. The final diagnoses in these cases included skin/soft tissue or musculoskeletal infection (n = 8 [33%]), pneumonia (n = 7 [29%]), intra-abdominal infection (n = 2 [8%]), and noninfectious conditions (n = 7 [33%]).
DISCUSSION
In this study, 16S rRNA gene NGS identified a potentially pathogenic organism in 47% (n = 28) of cases compared to 32% (n = 19) with blood cultures. A novel finding was the ability of NGS to possibly point to the source of sepsis (abdominal or genitourinary) in several cases based on the profile of identified organisms. Anaerobic gastrointestinal tract organisms were detected in 4 patients with sepsis from gastrointestinal sources, with genitourinary organisms detected in 2 patients with urinary tract infections [18]. Additionally, in 1 patient with E. coli bacteremia from an abdominal source, Allobaculum stercoricanis was identified; this organism has previously been isolated from dog feces [19]. Interestingly, the involved patient had a dog. The gastrointestinal tract is the most common source of anaerobic bacteremia, and patients with gram-negative bacillary bacteremia from gastrointestinal sources are frequently given anaerobic antibiotic coverage [20, 21]. Interestingly, in 2 patients with bacteremia secondary to urinary sources, potential genitourinary organisms, which are usually considered colonizers of the urinary tract, were detected (Lactococcus lactis and Lactobacillus crispatus) [22]. It is unclear if these organisms contributed to sepsis or if there is a need for targeted treatment; however, the findings suggest that if detected, these organisms may point to a genitourinary source of infection.
Four patients had febrile neutropenia; 16S rRNA gene NGS detected S. aureus, Enterobacterales, and Clostridium species in a patient with S. aureus and E. coli bacteremia, B. vulgatus in a patient with F. nucleatum bacteremia, Streptococcus species in a case with negative blood cultures, and Acidovorax plus Clostridium species in another case with negative blood cultures. Hematologic diseases are known to increase the risk of anaerobic bacteremia [23]. Streptococci are among the most common causes of bacteremia in patients with febrile neutropenia [24–27]. Acidovorax species has previously been identified in a patient with hematologic malignancy as well as in another case without malignancy [28, 29], although it may alternatively have represented an assay contaminant.
In 2 cases with C. difficile colitis, anaerobic organisms (B. vulgatus and P. bivia) were detected. Secondary bloodstream infections after C. difficile have been reported, most commonly due to Candida species followed by Enterobacterales [30–32]. No such cases of secondary bloodstream infection were found in this cohort. In 1 patient with non–C. difficile pancolitis, Clostridium species was identified by 16S rRNA gene NGS; this may have been the result of microbial translocation into the blood in the context of mucosal damage.
In 1 patient with sepsis secondary to a diabetic foot infection, multiple pathogenic organisms were detected by 16S rRNA gene NGS (S. agalactiae, E. cloacae, S. salivarius, and S. lugdunensis). Diabetic foot infections are frequently polymicrobial and the organisms detected by 16S rRNA gene NGS potentially reflect microbial pathogens in infected tissue [33]. This patient underwent debridement after receiving several days of broad-spectrum antimicrobials, at which time, tissue cultures were negative.
A limitation of blood cultures is the low yield of fastidious organisms, such as Legionella species. Legionella species are typically not isolated from routine blood cultures unless blind subcultures are performed; sequencing may be more useful in these cases [34]. In 1 case of sepsis secondary to severe pneumonia from L. pneumophila serotype 1, L. pneumophila was detected from the patient’s plasma by 16S rRNA gene NGS. In a mouse model, Legionella PCR was positive in blood samples up to 8 days after infection compared to 4 days in bronchoalveolar lavage fluid [35]. However, in humans, previous studies have shown low sensitivity of Legionella PCR from blood [36, 37]. cfDNA-based molecular testing directly from plasma could be a noninvasive way for diagnosing legionellosis in cases with a high suspicion of disease when urine testing is negative; however, further studies need to be done to better assess the sensitivity and specificity of such an approach.
In 2 patients with negative 16S rRNA gene NGS findings and positive blood cultures (P. aeruginosa and F. nucleatum), only 1 of 6 bottles were positive; it is unclear to what degree bacterial burden affects results of 16S rRNA gene NGS as there were 3 incidences in which 16S rRNA gene NGS was able to identify organisms found in single-positive blood culture bottles.
Enterobacterales were detected in 2 patients with sepsis secondary to pneumonia. Community-acquired pneumonia secondary to gram-negative organisms is uncommon, so it is unclear if Enterobacterales were the etiological agents responsible for pneumonia, or a result of microbial translocation in the setting of sepsis [38, 39].
Several cases were missed by 16S rRNA gene NGS as well, including gram-positive and gram-negative bacteria. Among those that were 16S rRNA gene NGS negative and culture positive, 2 had low-grade bacteremia; however, 16S rRNA gene NGS was negative in some cases with all or most blood culture bottles positive. In 3 cases with discordant 16S rRNA gene NGS results in which a member of the Enterobacterales was detected by 16S rRNA gene NGS, an underlying infectious syndrome was polymicrobial liver abscess (E. faecalis bacteremia), pneumonia (P. aeruginosa bacteremia), and abdominal cellulitis (S. pyogenes bacteremia). In a patient with neutropenic fever and Fusobacterium bacteremia, although 16S rRNA gene NGS did not detect F. nucleatum, B. vulgatus was detected. Both organisms are part of gastrointestinal microbiota, pointing to possible microbial mucosal translocation in the setting of neutropenia.
PCR can be falsely negative for a variety of reasons, including the need for effective lysis of microbes, interference of human DNA, or other inhibitory substances in blood. Additionally, cfDNA has a short half-life and delays in processing can lead to falsely negative results. The overall sensitivity of 16S rRNA gene NGS from plasma is limited by the amount of microbial DNA in the volume of plasma tested (which is substantially less than the volume of blood cultured in blood cultures), whereas specificity is limited by microbial contaminants from reagents and the environment [40–42]. Due to limited sensitivity of most molecular assays performed directly from blood, none has the ability to replace blood cultures; there is a need to optimize sensitivity.
16S rRNA gene NGS–based approaches may offer advantages, including the ability to detect multiple organisms; however, molecular approaches do not differentiate between viable and nonviable organisms, which may be beneficial in the context of antecedent antimicrobial therapy. Also, current molecular approaches such as that described here are limited by their inability to provide antimicrobial susceptibility results and can therefore only serve as a supplement to culture-based methods. Larger studies need to be done in diverse patient populations to better understand these modalities.
Our ability to obtain delayed informed consent was a significant strength of the study since it allowed us to compare samples collected at the same time for blood culture and 16S rRNA gene NGS, thereby limiting bias. This approach was used because pre-procedural consent was not practical in this population for a number of reasons. In patients with sepsis, antibiotics need to be administered within an hour of presentation, providing a short window to obtain informed consent directly from the patient or a legally authorized representative of the patient. The blood draw required for the intended study population is time sensitive as results may change after the initiation of antibiotics. Also, if blood samples are obtained after initiation of antibiotics, there is not a fair comparison of molecular methods with the blood cultures. Residual blood is not always readily available and delays in processing to plasma may affect the cfDNA concentrations. Patients with sepsis are usually critically ill and may not be able to provide informed consent at the time of presentation. Also, some patients require mechanical ventilation, which makes informed consent difficult to obtain in the intended study population in a timely manner. A large proportion of patients with sepsis may have encephalopathy or altered mentation at the time of presentation. This emphasizes that the study results are only generalizable to scenarios where samples for 16S rRNA gene NGS are collected at the same time as blood culture and not as an afterthought when blood cultures return negative.
There are important limitations to this study. As shotgun metagenomic sequencing was not performed, viral, parasitic, and fungal organisms were not identifiable. The small sample size does not allow definitive conclusions; future studies will be necessary to confirm the described findings. Enterobacterales were not identified to a genus- or species-level in most cases and it is to be determined if these represent pathogens or contamination. Despite these limitations, results of this study suggest that findings of 16S rRNA gene NGS can potentially point to the source of bacteremia in some cases. Finally, a large proportion of samples did not yield a positive blood culture result but were positive by 16S rRNA gene NGS. Even though several organisms detected by NGS were conceivably pathogenic, some identified species have been described as contaminants. Defining the ground truth in such scenarios can be challenging.
A targeted metagenomic 16S rRNA gene–based sequencing approach performed on plasma may be useful for detecting bacteria in sepsis and may hypothetically point to the source of bacteremia in some cases, based on detection of gastrointestinal or genitourinary bacteria.
Supplementary Material
Notes
Acknowledgments. The authors are grateful for assistance from the study coordinators Katelyn Reed, Lina Hines, and Kaitlin Schwartz from the Mayo Clinic Research and Innovation Office, as well as the Mayo Clinic Emergency Department staff, for supporting this study.
Financial support. R. B. is supported in part by the National Institute of Allergy and Infectious Diseases (grant number UM1 AI104681).
Potential conflicts of interest. R. P. has received grants from ContraFect, TenNor Therapeutics Limited, Hylomorph, and Shionogi, and is a consultant to Curetis, Specific Technologies, Next Gen Diagnostics, PathoQuest, Selux Diagnostics, 1928 Diagnostics, PhAST, and Qvella; monies are paid to Mayo Clinic. R. P. is also a consultant to Netflix; in addition, R. P. has a patent on Bordetella pertussis/parapertussis polymerase chain reaction issued, a patent on a device/method for sonication with royalties paid by Samsung to Mayo Clinic, and a patent on an anti-biofilm substance issued. R. P. receives an editor’s stipend from the Infectious Diseases Society of America, and honoraria from the National Board of Medical Examiners (NBME), UpToDate, and the Infectious Diseases Board Review Course. All other authors report no potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Zhang YZ, Singh S. Antibiotic stewardship programmes in intensive care units: why, how, and where are they leading us. World J Crit Care Med 2015; 4:13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luhr R, Cao Y, Söderquist B, Cajander S. Trends in sepsis mortality over time in randomised sepsis trials: a systematic literature review and meta-analysis of mortality in the control arm, 2002–2016. Critical Care Lond Engl 2019; 23:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burillo A, Bouza E. Use of rapid diagnostic techniques in ICU patients with infections. BMC Infect Dis 2014; 14:593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grumaz S, Grumaz C, Vainshtein Y, et al. Enhanced performance of next-generation sequencing diagnostics compared with standard of care microbiological diagnostics in patients suffering from septic shock. Crit Care Med 2019; 47:e394–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenner T, Decker SO, Grumaz S, et al. Next-generation sequencing diagnostics of bacteremia in sepsis (Next GeneSiS-Trial). Medicine 2018; 97:e9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rossoff J, Chaudhury S, Soneji M, et al. Non-invasive diagnosis of infection using plasma next-generation sequencing: a single center experience. Open Forum Infect Dis 2019; 6:ofz327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allyse MA, Wick MJ. Noninvasive prenatal genetic screening using cell-free DNA. JAMA 2018; 320:591–2. [DOI] [PubMed] [Google Scholar]
- 8.Schütz E, Fischer A, Beck J, et al. Graft-derived cell-free DNA, a noninvasive early rejection and graft damage marker in liver transplantation: a prospective, observational, multicenter cohort study. PLoS Med 2017; 14:e1002286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bloom RD, Bromberg JS, Poggio ED, et al. ; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators . Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol 2017; 28:2221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vlaminck ID, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med 2014; 6:241ra77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah P, Ruffin F, Seng H, et al. Direct detection and quantification of bacterial cell-free DNA in patients with infective endocarditis (IE) using the Karius plasma next generation sequencing (NGS) test. Open Forum Infect Dis 2018; 5:S12–12. [Google Scholar]
- 12.Kondo M, Dalai S, Westblade L, Venkatasubrahmanyam S, Eisenberg N, Marks KM. Diagnosis and genotyping of Coxiella burnetii causing endocarditis in a patient with prosthetic pulmonary valve replacement (PVR) using next-generation sequencing (NGS) of plasma. Open Forum Infect Dis 2018; 5:S11–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nomura J, Rieg G, Bluestone G, et al. Rapid detection of invasive Mycobacterium chimaera infection by using a novel plasma-based next-generation sequencing assay. Open Forum Infect Dis 2017; 4:S174–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hogan CA, Yang S, Garner OB, et al. Clinical impact of metagenomic next-generation sequencing of plasma cell-free DNA for the diagnosis of infectious diseases: a multicenter retrospective cohort study. Clin Infect Dis 2021; 72:239–45. [DOI] [PubMed] [Google Scholar]
- 15.Branda JA, Lemieux JE, Blair L, et al. Detection of Borrelia burgdorferi cell-free DNA in human plasma samples for improved diagnosis of early Lyme borreliosis. Clin Infect Dis 2021; 73:e2355–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016; 315:801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kommedal Ø, Simmon K, Karaca D, Langeland N, Wiker HG. Dual priming oligonucleotides for broad-range amplification of the bacterial 16S rRNA gene directly from human clinical specimens. J Clin Microbiol 2012; 50:1289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mylotte JM, Tayara A, Goodnough S. Epidemiology of bloodstream infection in nursing home residents: evaluation in a large cohort from multiple homes. Clin Infect Dis 2002; 35:1484–90. [DOI] [PubMed] [Google Scholar]
- 19.Greetham HL, Gibson GR, Giffard C, et al. Allobaculum stercoricanis gen. nov., sp. nov., isolated from canine feces. Anaerobe 2004; 10:301–7. [DOI] [PubMed] [Google Scholar]
- 20.Lombardi DP, Engleberg NC. Anaerobic bacteremia: incidence, patient characteristics, and clinical significance. Am J Med 1992; 92:53–60. [DOI] [PubMed] [Google Scholar]
- 21.Lassmann B, Gustafson DR, Wood CM, Rosenblatt JE. Reemergence of anaerobic bacteremia. Clin Infect Dis 2007; 44:895–900. [DOI] [PubMed] [Google Scholar]
- 22.Martinez RM, Hulten KG, Bui U, Clarridge JE 3rd. Molecular analysis and clinical significance of Lactobacillus spp. recovered from clinical specimens presumptively associated with disease. J Clin Microbiol 2014; 52:30–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funada H, Matsuda T. Bacteremia caused by the Bacteroides fragilis group in patients with hematologic diseases. Jpn J Clin Oncol 1995; 25:86–90. [PubMed] [Google Scholar]
- 24.Feld R, DePauw B, Berman S, Keating A, Ho W. Meropenem versus ceftazidime in the treatment of cancer patients with febrile neutropenia: a randomized, double-blind trial. J Clin Oncol 2000; 18:3690–8. [DOI] [PubMed] [Google Scholar]
- 25.Winston DJ, Lazarus HM, Beveridge RA, et al. Randomized, double-blind, multicenter trial comparing clinafloxacin with imipenem as empirical monotherapy for febrile granulocytopenic patients. Clin Infect Dis 2001; 32:381–90. [DOI] [PubMed] [Google Scholar]
- 26.Del Favero A, Menichetti F, Martino P, et al. ; Gruppo Italiano Malattie Ematologiche dell’Adulto (GIMEMA) Infection Program . A multicenter, double-blind, placebo-controlled trial comparing piperacillin-tazobactam with and without amikacin as empiric therapy for febrile neutropenia. Clin Infect Dis 2001; 33:1295–301. [DOI] [PubMed] [Google Scholar]
- 27.Cordonnier C, Buzyn A, Leverger G, et al. ; Club de Réflexion sur les Infections en Onco-Hématologie . Epidemiology and risk factors for gram-positive coccal infections in neutropenia: toward a more targeted antibiotic strategy. Clin Infect Dis 2003; 36:149–58. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Moore JE, Millar BC, et al. Improved laboratory diagnosis of bacterial and fungal infections in patients with hematological malignancies using PCR and ribosomal RNA sequence analysis. Leuk Lymphoma 2004; 45:1637–41. [DOI] [PubMed] [Google Scholar]
- 29.Shetty A, Barnes RA, Healy B, Groves P. A case of sepsis caused by Acidovorax. J Infect 2005; 51:e171–2. [DOI] [PubMed] [Google Scholar]
- 30.Falcone M, Russo A, Iraci F, et al. Risk factors and outcomes for bloodstream infections secondary to Clostridium difficile infection. Antimicrob Agents Chemother 2016; 60:252–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amit S, Mishali H, Kotlovsky T, Schwaber MJ, Carmeli Y. Bloodstream infections among carriers of carbapenem-resistant Klebsiella pneumoniae: etiology, incidence and predictors. Clin Microbiol Infect 2015; 21:30–4. [DOI] [PubMed] [Google Scholar]
- 32.Guastalegname M, Russo A, Falcone M, Giuliano S, Venditti M. Candidemia subsequent to severe infection due to Clostridium difficile: is there a link? Clin Infect Dis 2013; 57:772–4. [DOI] [PubMed] [Google Scholar]
- 33.Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes 2013; 62:923–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rihs JD, Yu VL, Zuravleff JJ, Goetz A, Muder RR. Isolation of Legionella pneumophila from blood with the BACTEC system: a prospective study yielding positive results. J Clin Microbiol 1985; 22:422–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aoki S, Hirakata Y, Miyazaki Y, et al. Detection of Legionella DNA by PCR of whole-blood samples in a mouse model. J Med Microbiol 2003; 52:325–9. [DOI] [PubMed] [Google Scholar]
- 36.Murdoch DR, Walford EJ, Jennings LC, et al. Use of the polymerase chain reaction to detect Legionella DNA in urine and serum samples from patients with pneumonia. Clin Infect Dis 1996; 23:475–80. [DOI] [PubMed] [Google Scholar]
- 37.Avni T, Bieber A, Green H, Steinmetz T, Leibovici L, Paul M. Diagnostic accuracy of PCR alone and compared to urinary antigen testing for detection of Legionella spp.: a systematic review. J Clin Microbiol 2016; 54:401–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arancibia F, Bauer TT, Ewig S, et al. Community-acquired pneumonia due to gram-negative bacteria and Pseudomonas aeruginosa: incidence, risk, and prognosis. Arch Intern Med 2002; 162:1849–58. [DOI] [PubMed] [Google Scholar]
- 39.Thau M, Falkowski NR, Branton W, et al. Evidence of gut translocation in sepsis: burden and diversity of bacterial DNA in blood predicts organ failure in septic patients. Am J Resp Crit Care Med 2018; 197:A4179. [Google Scholar]
- 40.Long Y, Zhang Y, Gong Y, et al. Diagnosis of sepsis with cell-free DNA by next-generation sequencing technology in ICU patients. Arch Med Res 2016; 47:365–71. [DOI] [PubMed] [Google Scholar]
- 41.Lusk RW. Diverse and widespread contamination evident in the unmapped depths of high throughput sequencing data. PLoS One 2014; 9:e110808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014; 12:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.