Abstract
The alarming pandemic situation of Coronavirus infectious disease COVID-19, caused by the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), has become a critical threat to public health. The unexpected outbreak and unrealistic progression of COVID-19 have generated an utmost need to realize promising therapeutic strategies to fight the pandemic. Drug repurposing-an efficient drug discovery technique from approved drugs is an emerging tactic to face the immediate global challenge. It offers a time-efficient and cost-effective way to find potential therapeutic agents for the disease. Artificial Intelligence-empowered deep learning models enable the rapid identification of potentially repurposable drug candidates against diseases. This study presents a deep learning ensemble model to prioritize clinically validated anti-viral drugs for their potential efficacy against SARS-CoV-2. The method integrates the similarities of drug chemical structures and virus genome sequences to generate feature vectors. The best combination of features is retrieved by the convolutional neural network in a deep learning manner. The extracted deep features are classified by the extreme gradient boosting classifier to infer potential virus–drug associations. The method could achieve an AUC of 0.8897 with 0.8571 prediction accuracy and 0.8394 sensitivity under the fivefold cross-validation. The experimental results and case studies demonstrate the suggested deep learning ensemble system yields competitive results compared with the state-of-the-art approaches. The top-ranked drugs are released for further wet-lab researches.
Keywords: COVID-19 drug repurposing, Deep learning, Convolutional neural network, XGBoost, SARS-coV-2, Antiviral drugs
1. Introduction
Coronavirus disease 2019 (COVID-19) is a highly contagious and pathogenic respiratory illness that created a dreadful situation worldwide, affecting people’s lives and causing many deaths. The causative agent for the disease, severe acute respiratory syndrome coronavirus-2, SARS-CoV-2 (previously 2019-novel coronavirus, 2019-nCoV), is an enveloped positive-strand RNA virus with mammalian hosts [1], [2]. They are classified under the Coronaviridae family and Betacoronavirus genus. SARS-CoV-2 is the seventh coronavirus known to infect humans, and they are genetically very similar to SARS (severe acute respiratory syndrome) and MERS (Middle East respiratory syndrome) coronaviruses [3], [4]. With a genome of 80–160 nm in length and 27–32 kb in size, coronaviruses are the biggest among known RNA viruses [5], [6]. The ongoing COVID-19 pandemic has highlighted the urgency to develop, test, and deploy new drugs and therapeutics. However, designing a novel drug from scratch is very tough and tedious, and thus impractical to combat the global challenge of the SARS-CoV-2 pandemic. One efficient way to face this challenge is to effectively screen clinically approved drugs for their anti-viral activity against SARS-CoV-2 for repurposing.
Drug repurposing (DR) is the deployment of already approved drugs for different indications other than the drug’s original therapeutic application. Since the de novo drug discovery is high-cost, lengthy, and laborious, DR has become a promising strategy to combat newly emerging diseases [7], [8]. This effective drug discovery technique can quickly identify potential therapeutic agents for difficult to treat diseases like COVID-19 [9]. DR based on biological experiments is generally a high-risk and high-investment process [10]. The availability of huge biological and structural databases and high-performance computing empowered computational DR as an alternative to experimental approaches to identify the most efficacious drugs against specific diseases in a short time [11], [12], [13].
In the present study, we propose a deep learning ensemble approach, DLEVDA, capable of identifying novel virus–drug associations to combat the rapidly evolving pandemic of COVID-19. Ensemble methods integrate the potential of multiple classifiers, thus yield models with enhanced predictive power and credibility. The proposed approach combines the pairwise similarities of drug chemical structures and virus genome sequences to build classification features. The generated samples were fed to the convolutional neural network for learning intricate input patterns. The learned abstract features were used to train the extreme gradient boosting classifier, which infers promising candidate drugs against SARS-CoV-2 infections. We conducted fivefold cross-validation (CV) to assess the efficacy of DLEVDA; The model achieved an AUC of 0.8897 with 0.8571 prediction accuracy and 0.8394 sensitivity. The comparison results with the state-of-the-art methods and competing classifiers reveal the predictive power of the approach. Experiments were conducted with distinct datasets. We could confirm the majority of the predicted results with existing literature which indicates the robustness of the model in prediction.
The rest of the paper is organized as follows: Section 2 discusses a review of recent works in drug repositioning to identify potential therapeutic drugs targeting covid-19. Section 3 describes the data preparation and architectural details of DLEVDA. Section 4 presents and analyzes the results of the proposed method. Section 5 discusses the results and Section 6 concludes the paper with future directions.
2. Related works
Based on the method, computational DR applied to infer anti-viral drugs against COVID-19 falls mainly into two categories: network-based and machine learning-based. Network-based DR methods represent drugs, diseases, and other biological entities like proteins, genes, etc., as network nodes and their associations as edges between nodes. Zhou et al. [14] suggested a method that identifies new anti-viral drugs against covid-19 by analyzing the association networks related to the Human coronaviruses (HCoVs) and drugs with their target proteins in the human protein–protein interaction (PPI) network. They collected the target proteins related to the HCoVs and generated the HCoV-host protein subnetwork. By computing the network proximity among the drug’s target proteins and HCoV-related proteins, potential anti-SARS-CoV-2 drugs were identified. Fiscon et al. [15] proposed a method that identifies new anti-viral drugs against covid-19 from already approved drugs. They quantified the relationships between drug targets and disease-associated genes using a network similarity-based approach to identify repurposable anti-SARS-CoV-2 drugs. The method utilized a drug–target network based on the drugs and their target proteins and a disease–gene network based on the diseases and their associated genes for prediction. The idea behind the algorithm is that a drug will be effective for a specific disease if the drug targets and the disease genes are nearby in the constructed network. Their algorithm calculated the network proximity value between each drug and disease to prioritize appropriate drug candidates against SARS-CoV-2. Meng et al. [16] proposed a method that predicts new anti-viral drugs against SARS-CoV-2 using similarity constrained probabilistic matrix factorization. They utilized drug chemical structure and virus genomic sequence-based similarities and known drug–virus relationships for prediction. They applied probabilistic matrix factorization on the drug–virus relationship matrix by introducing similarity constraints for drugs and viruses in the factorization process.
Adhami et al. [17] suggested a method that infers novel therapeutic drugs against COVID-19 by identifying the causal genes behind it. They retrieved the PPI network corresponding to the human proteins interacting with SARS-CoV-2 from the String database [18]. They identified 7 clusters of proteins that are deeply linked to SARS-CoV-2 and retrieved the genes and associated miRNAs related to the identified protein clusters. Finally, they acquired the drug targeting gene modules from the DGIdb [19] and then rebuilt the drug–gene network for the obtained protein modules. Next, they implemented a network-oriented drug repositioning method using computational bioinformatics tools to identify novel anti-viral drugs to fight COIVD-19. Peng et al. [20] presented an approach that predicts novel anti-viral drugs against COVID from FDA-approved drugs by utilizing drug chemical structure similarities, virus genome sequence-based similarities, and known drug–virus relationships. By integrating this data, they built a heterogeneous network and applied the random walk with restart algorithm [21] on the built network to identify new anti-viral drugs against SARS-CoV-2. The algorithm predicts the association score between SARS-CoV-2 and each drug in the dataset.
Though artificial intelligence-based researches are very active in tackling the Covid-19 epidemic, few articles are concerned with DR. Beck et al. [22] suggested a method that identifies currently available drugs that can interact with the proteins of SARS-CoV-2. They trained a pre-trained drug–target interaction prediction deep learning model [23], with the samples made of drug SMILES (Simplified Molecular Input Line Entry System) strings and amino acid sequences to infer new drug–virus associations. Ke et al. [24] implemented a deep learning method that prioritizes known drugs for their efficacy in fighting against SARS-CoV-2. They trained the model with two datasets; one with the approved drugs against viruses like SARS-CoV, influenza virus, etc., and the other with the confirmed protease inhibitors. They tested the efficacy of the identified drugs using in vitro cell-based assays. With the obtained results, they retrained their model and finally built a model that could identify efficacious drugs against COVID-19. Both of these studies did not evaluate their models quantitatively, and so they do not have exactly comparable results. Systematic deep learning or ensemble machine learning techniques are not applied in the field of virus–drug association prediction. Based on the status of the studies mentioned above, we are proposing a deep learning ensemble approach to infer novel virus–drug associations for identifying promising drug candidates against SARS-CoV-2.
3. Materials and methods
3.1. Datasets
This section describes the preparation of data used in the study:
Drug–virus associations: Experimentally verified virus–drug relationships are obtained from various literature through text mining technology. The dataset contains 455 human drug–virus associations between 219 drugs and 34 viruses. A binary matrix is to represent the drug–virus associations. is set as 1 if the drug has an association with virus ; otherwise .
Intra-drug similarities: The intra-drug similarities were quantified based on the chemical structures of drugs. The drug chemical structures were acquired from DrugBank [25] by adopting the SMILES format [26]. The Molecular Access System (MACCS) fingerprints of drugs were computed using Open Babel v2.3.1 [27]. The similarity between the two drugs was measured using the Tanimoto index [28] based on their MACCS fingerprints. The Tanimoto index between two drugs can be defined as:
where and represent the number of bits set in the corresponding drug fingerprints, and represents the number of bits that are set in both the fingerprints.
Intra-virus similarities: The intra-virus similarities were measured based on the virus genome sequences. The virus genome nucleotide sequences belonging to the human hosts were acquired from National Center for Biotechnology Information, NCBI [29]. Their pairwise sequence similarities were computed with Multiple Alignment using Fast Fourier Transform, MAFFT version 7 [30], a multiple sequence alignment tool. The known drug–virus relationships and the pairwise similarities among drugs and viruses were acquired from [16]; We consider it the benchmark dataset for this study.
3.2. Methods
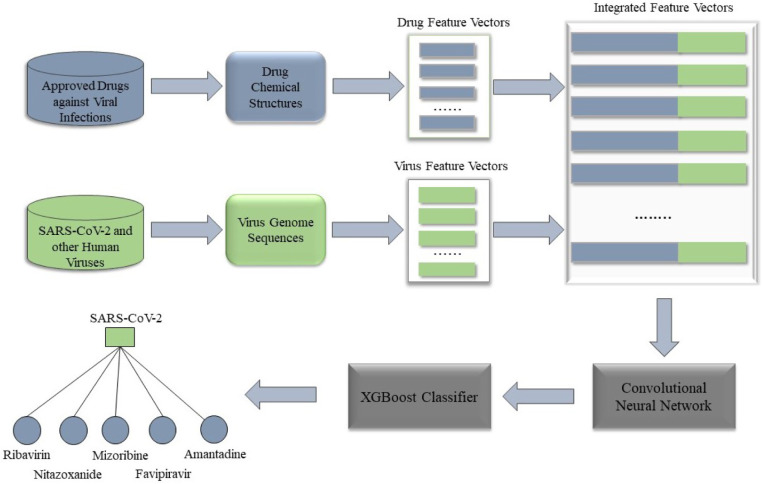
In this research, we presented an ensemble machine learning method that identifies new virus–drug relationships utilizing the drug, virus pairwise similarities, and known drug–virus interactions. The overall idea of DLEVDA is narrated in Fig. 1. The proposed approach includes two main segments: convolutional neural network (CNN) and Extreme Gradient Boosting (XGBoost). DLEVDA first generated feature vectors for each drug–virus pair in the dataset by considering the drug and virus pairwise similarities. The feature vector (, ) for the drug–virus pair (, ) can be represented by
where denotes the chemical structure-based similarities of the drug to all other drugs, denotes the genomic sequence-based similarities of the virus to all other viruses, and denotes the concatenation operation. In more detail, we defined [ , , ,… …… ] and [,……… …….… ], where denotes the pairwise similarity between the and drugs, is the pairwise similarity between the and viruses, and , the total number of drugs and viruses respectively in the dataset. Altogether, there were samples of length ; each corresponds to a drug–virus relationship. The associated labels were picked from the relationship matrix . The label was set to one if there was a confirmed interaction between the corresponding drug and virus; otherwise, to zero. The samples with label one formed the positive set. Next, random samples from unconfirmed interactions were selected and created the negative set such that the ratio of positive and negative samples was 1:1. There is a chance for unknown positive interactions among the chosen negative samples, but the probability, , is very less when compared to the total unknown interactions in the dataset. Finally, the positive and negative sets were integrated to generate the training set, which comprised 910 samples. With CNN, the intricate patterns of the samples were extracted and fed to the XGBoost classifier to identify novel drug candidates against SARS-CoV-2 and other viruses in the dataset.
Fig. 1.
The workflow of DLEVDA to prioritize potential virus–drug associations. With the integrated features of viruses and drugs as input, DLEVDA comprises two essential components: CNN-based feature learning and XGBoost-based classification.
3.2.1. Convolutional neural network for feature extraction
CNN is a deep learning algorithm proposed by Lecun et al. [31] that consists of three essential layers — convolutional layer, subsampling layer, and fully connected layer. Convolutional layers are the primary building blocks of CNN which are capable of capturing hidden patterns from the raw input data. CNN comprises two fundamental sections: feature extraction and classification. Feature extraction is achieved by multiple convolutions and subsampling layers and classification by fully connected layers. CNNs were effectively utilized for feature learning and classification in prediction problems for identifying the relationships between diseases, drugs, microRNAs, circular RNAs, etc. [32], [33], [34]. In DLEVDA, we employed CNN for extracting the sophisticated input patterns from the concatenated drug–virus feature vectors in a deep learning manner. We performed multiple convolution operations on the input samples using different kernels to generate the activation map. The activation map at layer can be described as:
where denotes the activation function, the convolution kernel at layer k, the offset vector, and the convolution operation. To compress data and minimize overfitting, the subsampling layer is used. The sampling formula at the subsampling layer can be expressed as:
We employed max-pooling at the subsampling layer, which retains the most prominent feature at each filter area. The CNN was trained to decrease the loss function of the network. The training samples were sent to the CNN to capture the significant features. To get our best model, we tuned the CNN hyper-parameters through several experiments. We implemented the convolution operation by using 16 filters of 116 size. At the subsampling layer, we set the filter size to 12. We used rectified linear unit, Relu [35], as the activation function at the convolution and fully connected layers and the sigmoid function at the output layer. The model was implemented using binary cross-entropy as the error function and Adam as the optimizer. To prevent overfitting, dropout layers [36] are added with convolution and hidden layers. Finally, the learned latent representations after numerous convolution and pooling operations are retrieved for identifying the potential virus–drug relationships.
3.2.2. Extreme gradient boosting based classification
XGBoost is a classification algorithm founded by Chen and Guestrin [37] which works under the framework of gradient boosting. In XGBoost, classification, and regression trees, CART, is created in sequential form. The basic idea is to continuously reduce the residual of the prior model in the gradient direction to get a new model. The algorithm employs multiple regularization parameters, including LASSO (L1) and Ridge (L2), which help prevent overfitting and improve performance.
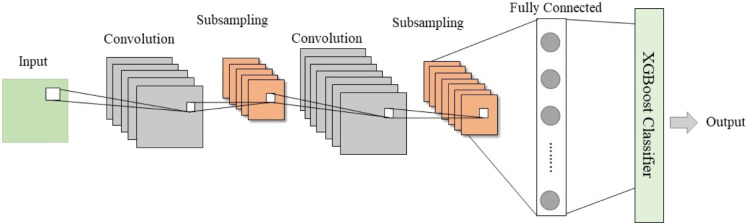
XGBoost has been successfully applied for binary classification problems such as microRNA/lncRNA-disease association predictions [38], [39], prediction of hot spots in protein–DNA binding interfaces [40], protein submitochondrial localization prediction [41], etc. This study established a deep learning model in which XGBoost was employed to perform the task of classification. The feature vectors obtained after convolution and pooling operations from the CNN were a dense, high-level representation of the original samples. We trained the XGBoost classifier with the training set features learned by CNN; the trained model could predict the correlation score for each unverified drug–virus pair in the dataset. The samples with scores above the threshold were considered as potential virus–drug associations, and they were released for future biological tests. We optimized XGBoost hyperparameters through grid search and set the values for parameters such as n_estimators, max_depth, and learning_rate to 150,8 and 0.1, respectively. Fig. 2. depicts the basic structure of the CNN-XGBoost model.
Fig. 2.
The basic architecture of the CNN-XGBoost model.
4. Results
4.1. Performance evaluation
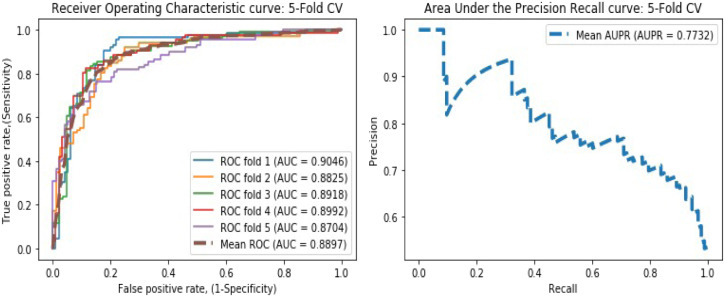
We conducted the fivefold CV to evaluate the predictive performance of DLEVDA in identifying new virus–drug relationships. In k-fold CV, the training set is separated into random subsets of uniform size. At each fold, the model was trained with k-1 subsets and validated with the remaining subset. The process was repeated k times until each subset was validated once, and the mean was taken as the end result. In the predicted results, known virus–drug relationships with correlation scores beyond the threshold were treated as true positives (TP), and lower than the threshold were treated as false negatives (FN). Likewise, unknown relationships with correlation scores lower than the threshold were treated as true negatives (TN), and beyond the threshold were treated as false positives (FP). We plotted the Receiver Operating Characteristic curve (ROC) [42], [43] by measuring the true positive rate and the false positive rate at different cut-offs, and the model obtained an Area Under the ROC curve (AUC) [44] of 0.8897. We further assessed the predictive capability of DLEVDA by quantifying other statistical parameters such as accuracy, sensitivity, specificity, F1-score, PPV (positive predictive value), NPV (negative predictive value), and Matthews’s correlation coefficient (MCC), and the result is shown in Table 1. In addition, we computed the Area Under the Precision–Recall curve (AUPR) [45] as another kind of evaluation metric. The ROC and Precision–Recall (PR) curves based on the fivefold CV are shown in Fig. 3. To diminish the deviations from randomly partitioned samples, we implemented fivefold CV twenty times, and the performances were averaged.
Table 1.
Cross-validation results of DLEVDA based on fivefold CV experiment.
| Method | Accuracy | Sensitivity | Specificity | F1-score | PPV | NPV | MCC | AUC | AUPR |
|---|---|---|---|---|---|---|---|---|---|
| Fivefold | 0.8571 | 0.8394 | 0.8624 | 0.8432 | 0.8563 | 0.8667 | 0.7337 | 0.8897 | 0.7732 |
Fig. 3.
ROC and PR curves obtained by DLEVDA under the fivefold CV experiment.
Additionally, we evaluated the model performance by implementing it on an independent test set. We generated the independent test set by randomly choosing 20% samples from the training set such that it contains equal positive and negative samples. The remaining training set samples were partitioned into five random subsets of roughly equal size, which were used as the training and validation sets for the fivefold CV. Next, the validated model is trained with the whole samples in the training set, excluding the independent test set. The trained model is used to predict the correlation score for the samples in the independent test set. The experimental results based on the independent test set are summarized in Table 2.
Table 2.
Cross-validation results of DLEVDA based on the independent test set.
| Method | Accuracy | Sensitivity | Specificity | F1-score | PPV | NPV | MCC | AUC | AUPR |
|---|---|---|---|---|---|---|---|---|---|
| Independent test set | 0.8635 | 0.8418 | 0.8692 | 0.8316 | 0.8602 | 0.8701 | 0.7432 | 0.8926 | 0.7624 |
4.2. Comparison with previous studies
We evaluated the predictive performance of DLEVDA by comparing it to related approaches. Existing researches for identifying repurposable anti-viral drugs against COVID-19 was rare as the COVID-19 researches were mainly focused on sequence data of viruses. We compared DLEVDA with other methods predicting virus–drug relationships such as the similarity constrained probabilistic matrix factorization, SCPMF [16], and virus–drug association prediction based on random walk with restart, VDA-RWR [20], using the same dataset we used. We further evaluated our model by comparing it to other association prediction approaches such as IMCMDA [46], NCPMDA [47], and SAEROF [48]. These three models achieved robust performances in their respective applications. IMCMDA was applied to identify new miRNA-disease associations based on the inductive matrix completion algorithm. NCPMDA identified novel diseases associated with miRNAs based on Network Consistency Projection. SAEROF was applied to predict novel drug–disease relationships utilizing sparse autoencoder and rotation forest. We compared DLEVDA with these models using the same dataset used in our study. The performance was evaluated based on the fivefold CV, and the results are depicted in Table 3. From the table, it is evident that DLEVDA outperformed other methods with high robustness.
Table 3.
Comparison of DLEVDA with related approaches, through the AUC scores under fivefold CV experiment.
| Fivefold | DLEVDA | SCPMF | VDA-RWR | IMCMDA | NCPMDA | SAEROF |
|---|---|---|---|---|---|---|
| AUC score | 0.8897 | 0.8631 | 0.8501 | 0.6423 | 0.6711 | 0.7935 |
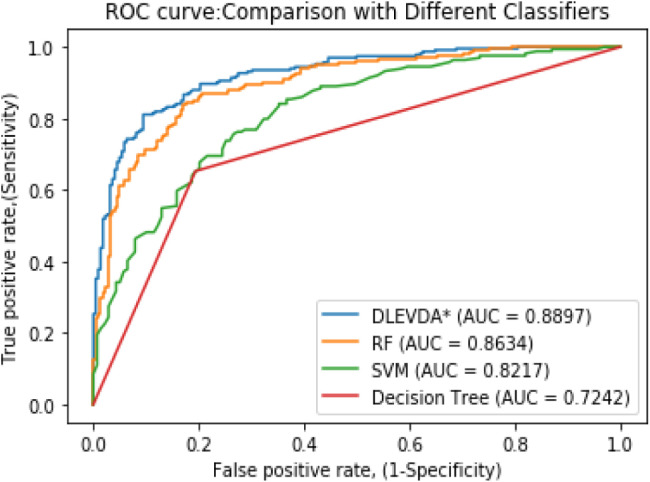
4.3. Comparison with different classifiers
To further assess the efficacy of DLEVDA, we compared the model performance with other state-of-the-art classifiers such as random forest (RF), support vector machine (SVM), and decision tree under fivefold CV. In order to assure the fairness of the experiment, we adopted the same feature construction and feature extraction methods during comparison. We could achieve AUCs of 0.8897, 0.8634, 0.8217, and 0.7242 for DLEVDA, RF, SVM, and decision tree classifiers, respectively. Fig. 4 plots a comparison of ROC curves generated by these classifiers. Next, we compared the model performance by implementing these classifiers without employing CNN for feature extraction. The experiments yielded 0.8169, 0.8201, 0.7628, and 0.6581 for XGBoost, RF, SVM, and decision tree classifiers, respectively. From the results, it can be seen that deep learning-based feature retrieval improved the classification results significantly. These experimental results with both the raw and learned high-level features demonstrate the predictive power of DLEVDA in identifying novel virus–drug relationships.
Fig. 4.
Comparison of DLEVDA with the competing classifiers under the fivefold CV experiment.
4.4. Comparison with other datasets
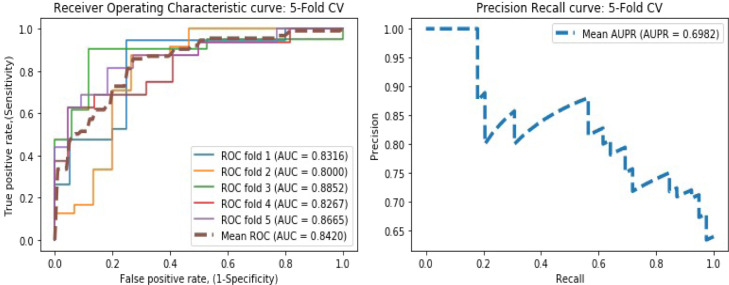
To test the influence of various datasets in DLEVDA, we implemented it with another dataset that consists of 96 drug–virus associations between 78 drugs and 11 viruses. In this dataset, 12 viruses akin to SARS-CoV-2 were considered, and their genome sequence-based information was obtained from the NCBI database. Their pairwise similarities were computed using MAFFT version 7. The drugs associated with these viruses were acquired from Drugbank, NCBI, and PubMed databases, and their structural similarities were computed. We conducted fivefold CV, and DLEVDA yielded mean values of 0.8039, 0.7554, 0.7783, 0.7017, 0.7647, 0.7805, 0.6505, 0.8420, and 0.6982 for accuracy, sensitivity, specificity, F1-score, PPV, NPV, MCC, AUC, and AUPR, respectively. The corresponding ROC and PR curves are plotted in Fig. 5. The model performance is slightly low compared to the benchmark dataset used in this study as the number of training data is significantly less. We downloaded this dataset from the supplementary material associated with the paper [20].
Fig. 5.
ROC and PR curves obtained by DLEVDA on a different drug–virus association dataset under the fivefold CV experiment.
4.5. Case studies
To further validate the predictive capability of DLEVDA, we conducted case studies on the top-predicted results. For this, we trained the model with all known drug–virus relationships and predicted correlation scores for all unknown drug–virus pairs in the dataset. The predicted correlation scores were sorted in descending order with the corresponding virus–drug relationships. Specifically, we ranked the top predicted drugs associated with the SARS-CoV-2, Tab. 4. Out of twelve top-predicted drugs, 9 of them could be validated by recent literature. For example, ribavirin, the top-ranked drug against COVID-19, is an anti-viral drug used to treat Hepatitis C and some viral hemorrhagic fevers. It inhibits the replication of RNA viruses and has been applied for treating COVID-19 patients [49], [50], [51]. Nitazoxanide, the second top-predicted candidate drug, boosts the host’s anti-viral response by upregulating the host interferon and impedes virus replication [52]. It has been proved that Nitazoxanide can prevent SARS-CoV-2 infections at a reduced micromolar concentration and has been recommended for clinical trials to treat COVID-19 [53], [54], [55]. The fourth-ranked drug favipiravir is one of the anti-viral agents considered in numerous clinical trials to combat COVID-19. Favipiravir is a purine nucleic acid analog with a broad spectrum of anti-RNA virus activities [56], [57], [58], [59]. In addition, among the top twelve predicted drugs against SARS-CoV-2, many are undergoing clinical trials [60], [61]. These results reveal the efficacy and credibility of DLEVDA in identifying repurposable ant-viral drugs against COVID-19 and other emerging infectious diseases (see Table 4).
Table 4.
The top twelve predicted anti-viral drugs against SARS-CoV-2 with their evidence in the literature.
| Rank | Drug | Evidence (PMID) |
|---|---|---|
| 1 | Ribavirin | 32227493, 32149772, 33689451 |
| 2 | Nitazoxanide | 32020029, 32568620, 33031085 |
| 3 | Mizoribine | Unconfirmed |
| 4 | Favipiravir | 32346491, 32246834, 33176367 |
| 5 | Amantadine | 32361028, 32571606 |
| 6 | N4-Hydroxycytidine | Unconfirmed |
| 7 | Quinacrine | 33477376 |
| 8 | Zanamivir | 32511320 |
| 9 | Maribavir | 32147628 |
| 10 | Chloroquine | 32145363, 32074550, 32203437 |
| 11 | Clevudine | Unconfirmed |
| 12 | EIDD-2801 | 33561864 |
5. Discussion
Prioritization of clinically validated drugs for their anti-viral efficacy is urgent for the rapid clinical trials against COVID-19. This research proposed an ensemble deep learning architecture to infer promising preclinical drug candidates to treat SARS-CoV-2 infections. The proposed architecture comprised two essential segments of CNN-based feature learning and XGBoost-based classification. The CNN was trained with the feature vectors constructed based on drug chemical structures and virus genomic sequences. Then, the learned high-level features were classified with the XGBoost classifier to identify novel candidate drugs to combat COVID-19 and other viral infectious diseases.
There are many factors attributed to the efficient performance of DLEVDA. Ensemble approaches yield excellent results by integrating the potency of multiple classifiers. CNN is a powerful feature extractor that automatically learns high-quality features from the raw input data. However, a large amount of training data is required by CNN to prevent overfitting [62]. The available training data for this research is limited. So, we employed XGBoost for the task of classification. XGBoost utilizes multicore CPU parallel computing to enhance performance. It combines software and hardware optimization strategies to produce more accurate results with lesser computing resources. The incorporation of a regularized model makes the classifier unique [37], [63]. However, we examine that XGBoost is still unclear for feature extraction. In addition, the potential of a single classifier may not be sufficient to meet the perfection required for many biological problems. In DLEVDA, the integration of CNN and XGBoost classifiers produced more accurate and robust results.
The network-oriented DR techniques have the drawback that the network needs to be reconstructed whenever a new drug or disease is added to the dataset [14], [15], [16], [17], [20]. Many of the network-oriented DR strategies cannot be applied to drugs with no confirmed disease interactions or diseases with no confirmed drug interactions in the dataset. DLEVDA can be applied to drugs (diseases) for which no confirmed disease(drug) associations; Hence, we can apply the model to predict potential drugs for emerging diseases like COVID-19. Similar to other machine learning methods, DLEVDA can quickly adapt to changes. When newly discovered drugs, diseases, or drug–virus associations are identified, they can be easily included in the dataset after similarity computation. In addition, DLEVDA incorporated multiple biological data, including the complete genome sequences of viruses and chemical structures of drugs for feature construction. Above all, artificial intelligence-empowered DR is low-cost, fast, and effective and can minimize failures in clinical trials. The limitation is that DLEVDA requires positive and negative samples for training. But it is tough to acquire the actual negative samples. We built the negative set by picking samples from unconfirmed drug–virus relationships at random. There is a possibility for unconfirmed positive interactions in the constructed negative set, even if the probability is low. Besides, the known drug–virus associations available for the study are limited. We believe the performance of DLEVDA can be improved further as more drug–virus associations are discovered. Since similarity scores play a crucial part in predictive performance, it is required to further investigate the kinds of features bundled up for similarity computation.
The overall time complexity of DLEVDA can be expressed as , where the first and second components represent the time complexities of CNN-based feature learning and XGBoost-based classification, respectively, with training samples [37], [64]. In the first component, denotes the number of convolution layers, the number of epochs, the number of input channels, the spatial size of the kernel, the number of kernels, and the spatial size of the output feature map of the layer. In the second component, represents the total number of trees, the maximum depth of the tree, and the number of non-missing entries in the training data. From the equation, it can be inferred that the complexity of DLEVDA depends on the complexity of CNN-based feature learning. The complexity of pooling and fully connected layers are not involved in this formulation. These layers may take 5%–10% computational time.
6. Conclusion
In summary, this study proposed an efficacious deep learning ensemble model for rapid identification of candidate repurposable drugs to fight against SARS-CoV-2 infections. We carried out extensive experiments and case studies to measure the efficacy of the developed system. The comparison results with the state-of-the-art methods demonstrated an improvement over the existing techniques evaluated under the same condition. Experiments performed with different machine learning classifiers using both the raw and deep features reveal the robustness of the model. The case studies could identify many drugs under clinical trials, which indicate the promising performance of DLEVDA to identify highly credible candidates for experimental analysis. However, the use of randomly chosen negative samples and the limited number of experimentally confirmed virus–drug associations are some of the limitations of the model. All the top predicted drugs against COVID-19 are released for further researches. We believe these drug candidates provide a meaningful reference to support clinicians. In the future, the proposed model can be extended to the next level for predicting the collective effect of a set of drugs against SARS-CoV-2 and other viruses.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Implementation details
We implemented DLEVDA with the python toolkits Scikit-learn and Keras library [65], [66].
Informed consent
Informed consent has been derived from all the participants.
Funding
No funding received.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Data availability
The source code and dataset of DLEVDA are available at https://github.com/Deepthi-K523/DLEVDA.
References
- 1.Hui D.S., Azhar E.I., Madani T.A., Ntoumi F., Kock R., Dar O., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—The latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberfeld B., Achanta A., Carpenter K., Chen P., Gilette N.M., Langat P., et al. SnapShot: Covid-19. Cell. 2020;181(4):954. doi: 10.1016/j.cell.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pal M., Berhanu G., Desalegn C., Kandi V. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020;12(3) doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin A.R., Erdogan A., Agaoglu P.M., Dineri Y., Cakirci A.Y., Senel M.E., et al. 2019 novel coronavirus (COVID-19) outbreak: a review of the current literature. EJMO. 2020;4(1):1–7. [Google Scholar]
- 6.Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Ag. 2020;55(3) doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pushpakom S., Iorio F., Eyers P.A., Escott K.J., Hopper S., Wells A., et al. Drug repurposing: progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019;18(1):41–58. doi: 10.1038/nrd.2018.168. [DOI] [PubMed] [Google Scholar]
- 8.Li J., Zheng S., Chen B., Butte A.J., Swamidass S.J., Lu Z. A survey of current trends in computational drug repositioning. Brief. Bioinform. 2016;17(1):2–12. doi: 10.1093/bib/bbv020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dotolo S., Marabotti A., Facchiano A., Tagliaferri R. A review on drug repurposing applicable to COVID-19. Brief. Bioinform. 2021;22(2):726–741. doi: 10.1093/bib/bbaa288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avorn J. The $2.6 billion pill–methodologic and policy considerations. N. Engl. J. Med. 2015;372(20):1877–1879. doi: 10.1056/NEJMp1500848. [DOI] [PubMed] [Google Scholar]
- 11.Gns H.S., Saraswathy G.R., Murahari M., Krishnamurthy M. An update on Drug Repurposing: Re-written saga of the drug’s fate. Biomed. Pharmacotherapy. 2019;110:700–716. doi: 10.1016/j.biopha.2018.11.127. [DOI] [PubMed] [Google Scholar]
- 12.Lippmann C., Kringel D., Ultsch A., Loetsch J. Computational functional genomics-based approaches in analgesic drug discovery and repurposing. Pharmacogenomics. 2018;19(9):783–797. doi: 10.2217/pgs-2018-0036. [DOI] [PubMed] [Google Scholar]
- 13.Ahsan M.A., Liu Y., Feng C., Zhou Y., Ma G., Bai Y., Chen M. Bioinformatics resources facilitate understanding and harnessing clinical research of SARS-CoV-2. Brief. Bioinform. 2021 doi: 10.1093/bib/bbaa416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Y., Hou Y., Shen J., et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiscon G., Conte F., Farina L., Paci P. SaveRUNNER: A network-based algorithm for drug repurposing and its application to COVID-19. PLoS Comput. Biol. 2021;17(2) doi: 10.1371/journal.pcbi.1008686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng Y., Jin M., Tang X., Xu J. Drug repositioning based on similarity constrained probabilistic matrix factorization: COVID-19 as a case study. Appl. Soft Comput. 2021;103 doi: 10.1016/j.asoc.2021.107135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adhami M., Sadeghi B., Rezapour A., Haghdoost A.A., MotieGhader H. Repurposing novel therapeutic candidate drugs for coronavirus disease-19 based on protein-protein interaction network analysis. BMC Biotechnol. 2021;21(1):1–11. doi: 10.1186/s12896-021-00680-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D., et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2018;47(D1):D607–13. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wagner A.H., et al. DGidb 2.0: mining clinically relevant drug–gene interactions. Nucleic Acids Res. 2016;44(D1):D1036–44. doi: 10.1093/nar/gkv1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng L., Shen L., Xu J., Tian X., Liu F., Wang J., et al. Prioritizing anti-viral drugs against SARS-CoV-2 by integrating viral complete genome sequences and drug chemical structures. Sci. Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-83737-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Valdeolivas A., Tichit L., Navarro C., Perrin S., Odelin G., Levy N., et al. Random walk with restart on multiplex and heterogeneous biological networks. Bioinformatics. 2019;35(3):497–505. doi: 10.1093/bioinformatics/bty637. [DOI] [PubMed] [Google Scholar]
- 22.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available anti-viral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin B., Park S., Kang K., Ho J.C. Machine Learning for Healthcare Conference. PMLR; 2019. Self-attention based molecule representation for predicting drug-target interaction; pp. 230–248. [Google Scholar]
- 24.Ke Y.Y., Peng T.T., Yeh T.K., Huang W.Z., Chang S.E., Wu S.H., et al. Artificial intelligence approach fighting COVID-19 with repurposing drugs. Biomed. J. 2020;43(4):355–362. doi: 10.1016/j.bj.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Law V., Knox C., Djoumbou Y., Jewison T., Guo A.C., Liu Y., et al. DrugBank 4.0: shedding new light on drug mefigolism. Nucleic Acids Res. 2014;42(D1):D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Öztürk H., Ozkirimli E., Özgür A. A comparative study of SMILES-based compound similarity functions for drug-target interaction prediction. BMC Bioinformatics. 2016;17(1):1–11. doi: 10.1186/s12859-016-0977-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Boyle N.M., Banck M., James C.A., Morley C., Vandermeersch T., Hutchison G.R. Open Babel: An open chemical toolbox. J. Cheminf. 2011;3(1):1–14. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bajusz D., Rácz A., Héberger K. Why is tanimoto index an appropriate choice for fingerprint-based similarity calculations? J. Cheminf. 2015;7(1):1–13. doi: 10.1186/s13321-015-0069-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wheeler D.L., Chappey C., Lash A.E., Leipe D.D., Madden T.L., Schuler G.D., et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2000;28(1):10–14. doi: 10.1093/nar/28.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeCun Y., Boser B., Denker J.S., Henderson D., Howard R.E., Hubbard W., Jackel L.D. Backpropagation applied to handwritten zip code recognition. Neural Comput. 1989;1(4):541–551. [Google Scholar]
- 32.Peng J., Hui W., Li Q., Chen B., Hao J., Jiang Q., et al. A learning-based framework for miRNA-disease association identification using neural networks. Bioinformatics. 2019;35(21):4364–4371. doi: 10.1093/bioinformatics/btz254. [DOI] [PubMed] [Google Scholar]
- 33.Wang L., You Z.H., Huang Y.A., Huang D.S., Chan K.C. An efficient approach based on multi-sources information to predict circRNA–disease associations using deep convolutional neural network. Bioinformatics. 2020;36(13):4038–4046. doi: 10.1093/bioinformatics/btz825. [DOI] [PubMed] [Google Scholar]
- 34.Deepthi K., Jereesh A.S. An ensemble approach based on multi-source information to predict drug-MiRNA associations via convolutional neural networks. IEEE Access. 2021;9:38331–38341. [Google Scholar]
- 35.V. Nair, G.E. Hinton, Rectified linear units improve restricted boltzmann machines, in: Icml, 2010.
- 36.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014;15(1):1929–1958. [Google Scholar]
- 37.T. Chen, C. Guestrin, Xgboost: A scalable tree boosting system, in: Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining, 2016, pp. 785–794.
- 38.Chen X., Huang L., Xie D., Zhao Q. EGBMMDA: extreme gradient boosting machine for MiRNA-disease association prediction. Cell Death Dis. 2018;9(1):1–16. doi: 10.1038/s41419-017-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Ye F., Xiong D., Gao X. LDNFSGB: prediction of long non-coding rna and disease association using network feature similarity and gradient boosting. BMC Bioinformatics. 2020;21(1):1–27. doi: 10.1186/s12859-020-03721-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li K., Zhang S., Yan D., Bin Y., Xia J. Prediction of hot spots in protein–DNA binding interfaces based on supervised isometric feature mapping and extreme gradient boosting. BMC Bioinformatics. 2020;21(13):1–10. doi: 10.1186/s12859-020-03683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu B., Qiu W., Chen C., Ma A., Jiang J., Zhou H., Ma Q. SubMito-Xgboost: predicting protein submitochondrial localization by fusing multiple feature information and eXtreme gradient boosting. Bioinformatics. 2020;36(4):1074–1081. doi: 10.1093/bioinformatics/btz734. [DOI] [PubMed] [Google Scholar]
- 42.Mandrekar J.N. Receiver operating characteristic curve in diagnostic test assessment. J. Thoracic Oncol. 2010;5(9):1315–1316. doi: 10.1097/JTO.0b013e3181ec173d. [DOI] [PubMed] [Google Scholar]
- 43.Kumar R., Indrayan A. Receiver operating characteristic (ROC) curve for medical researchers. Indian Pediatr. 2011;48(4):277–287. doi: 10.1007/s13312-011-0055-4. [DOI] [PubMed] [Google Scholar]
- 44.Bradley A.P. The use of the Area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997;30(7):1145–1159. [Google Scholar]
- 45.Boyd K., Eng K.H., Page C.D. Joint European Conference on Machine Learning and Knowledge Discovery in Databases. Springer; Berlin, Heidelberg: 2013. Area under the precision–recall curve: point estimates and confidence intervals; pp. 451–466. [Google Scholar]
- 46.Chen X., Wang L., Qu J., Guan N.N., Li J.Q. Predicting miRNA-disease association based on inductive matrix completion. Bioinformatics. 2018;34(24):4256–4265. doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
- 47.Gu C., Liao B., Li X., Li K. Network consistency projection for human miRNA-disease associations inference. Sci. Rep. 2016;6:36054. doi: 10.1038/srep36054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang H.J., Huang Y.A., You Z.H. SAEROF: an ensemble approach for large-scale drug-disease association prediction by incorporating rotation forest and sparse autoencoder deep neural network. Sci. Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-61616-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khalili J.S., Zhu H., Mak N.S.A., Yan Y., Zhu Y. Novel coronavirus treatment with ribavirin: Groundwork for an evaluation concerning COVID-19. J. Med. Virol. 2020;92(7):740–746. doi: 10.1002/jmv.25798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeng Y.M., Xu X.L., He X.Q., Tang S.Q., Li Y., Huang Y.Q., et al. Comparative effectiveness and safety of ribavirin plus interferon-alpha, lopinavir/ritonavir plus interferon-alpha, and ribavirin plus lopinavir/ritonavir plus interferon-alpha in patients with mild to moderate novel coronavirus disease 2019: study protocol. Chinese Med. J. 2020;133(9):1132–1134. doi: 10.1097/CM9.0000000000000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unal M.A., Bitirim C.V., Summak G.Y., Bereketoglu S., Cevher Zeytin I., Besbinar O., et al. Ribavirin shows anti-viral activity against SARS-CoV-2 and downregulates the activity of TMPRSS2 and the expression of ACE2 in vitro. Can. J. Physiol. Pharmacol. 2021;99(5):449–460. doi: 10.1139/cjpp-2020-0734. [DOI] [PubMed] [Google Scholar]
- 52.Jasenosky L.D., Cadena C., Mire C.E., Borisevich V., Haridas V., Ranjbar S., et al. The FDA-approved oral drug nitazoxanide amplifies host anti-viral responses and inhibits Ebola virus. Iscience. 2019;19:1279–1290. doi: 10.1016/j.isci.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., et al. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30(3):269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Naik V.R., Munikumar M., Ramakrishna U., Srujana M., Goudar G., Naresh P., et al. Remdesivir (GS-5734) as a therapeutic option of 2019-nCOV main protease–in silico approach. J. Biomol. Struct. Dyn. 2020:1–14. doi: 10.1080/07391102.2020.1781694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calderón J.M., Flores M.D.R.F, Coria L.P., Garduño J.C.B., Figueroa J.M., Contretas M.J.V., et al. Nitazoxanide against COVID-19 in three explorative scenarios. J. Infect. Dev. Countries. 2020;14(09):982–986. doi: 10.3855/jidc.13274. [DOI] [PubMed] [Google Scholar]
- 56.Du Y.X., Chen X.P. Favipiravir: pharmacokinetics and concerns about clinical trials for 2019-nCoV infection. Clin. Pharmacol. Ther. 2020;108(2):242–247. doi: 10.1002/cpt.1844. [DOI] [PubMed] [Google Scholar]
- 57.Cai Q., Yang M., Liu D., Chen J., Shu D., Xia J., et al. Experimental treatment with favipiravir for COVID-19: an open-label control study. Engineering. 2020;6(10):1192–1198. doi: 10.1016/j.eng.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen C., Huang J., Cheng Z., Wu J., Chen S., Zhang Y., et al. 2020. Favipiravir versus arbidol for COVID-19: a randomized clinical trial. MedRxiv. [Google Scholar]
- 59.Ghasemnejad-Berenji M., Pashapour S. Favipiravir and COVID-19: a simplified summary. Drug Res. 2020 doi: 10.1055/a-1296-7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong L., Hu S., Gao J. Discovering drugs to treat coronavirus disease 2019 (COVID-19) Drug Discov. Therapeutics. 2020;14(1):58–60. doi: 10.5582/ddt.2020.01012. [DOI] [PubMed] [Google Scholar]
- 61.Tarighi P., Eftekhari S., Chizari M., Sabernavaei M., Jafari D., Mirzabeigi P. A review of potential suggested drugs for coronavirus disease (COVID-19) treatment. Eur. J. Pharmacol. 2021 doi: 10.1016/j.ejphar.2021.173890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pasupa K., Sunhem W. A comparison between shallow and deep architecture classifiers on small dataset. 2016 8th International Conference on Information Technology and Electrical Engineering; ICITEE; IEEE; 2016. pp. 1–6. [Google Scholar]
- 63.Ma J., Yu Z., Qu Y., Xu J., Cao Y. Application of the XGBoost machine learning method in PM2. 5 prediction: A case study of shanghai. Aerosol Air Qual. Res. 2020;20(1):128–138. [Google Scholar]
- 64.K. He, J. Sun, Convolutional neural networks at constrained time cost, in: Proceedings of the IEEE conference on computer vision and pattern recognition, 2015, pp. 5353–5360.
- 65.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 66.Chollet F., et al. GitHub; 2015. Keras. https://github.com/fchollet/keras. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The source code and dataset of DLEVDA are available at https://github.com/Deepthi-K523/DLEVDA.