Abstract
To study repair of DNA double-strand breaks (DSBs) in mammalian chromosomes, we designed DNA substrates containing a thymidine kinase (TK) gene disrupted by the 18-bp recognition site for yeast endonuclease I-SceI. Some substrates also contained a second defective TK gene sequence to serve as a genetic donor in recombinational repair. A genomic DSB was induced by introducing endonuclease I-SceI into cells containing a stably integrated DNA substrate. DSB repair was monitored by selection for TK-positive segregants. We observed that intrachromosomal DSB repair is accomplished with nearly equal efficiencies in either the presence or absence of a homologous donor sequence. DSB repair is achieved by nonhomologous end-joining or homologous recombination, but rarely by nonconservative single-strand annealing. Repair of a chromosomal DSB by homologous recombination occurs mainly by gene conversion and appears to require a donor sequence greater than a few hundred base pairs in length. Nonhomologous end-joining events typically involve loss of very few nucleotides, and some events are associated with gene amplification at the repaired locus. Additional studies revealed that precise religation of DNA ends with no other concomitant sequence alteration is a viable mode for repair of DSBs in a mammalian genome.
Chromosomes suffer a variety of types of damage that may result in loss or alteration of genetic information, or death, if left unrepaired. The double-strand break (DSB) is one type of damage that can arise spontaneously or that may be induced by a variety of agents (1, 3, 37, 49). Several pathways for DSB repair in eukaryotes have been described. DSB repair in the yeast Saccharomyces cerevisiae is generally viewed as occurring almost exclusively through homologous recombination (HR) when a homolog is available (8, 9, 11, 20, 27). In one model for recombinational repair, termed DSBR (30, 41), broken DNA ends are degraded to form a double-stranded gap. This is followed by invasion of an unbroken homologous template by a broken end, which primes DNA synthesis that ultimately leads to repair of the gap with information contributed by the homologous template. In this model, recombination is conservative and proceeds through the formation of two Holliday junctions, which may be resolved to generate either a crossover or a noncrossover product. A distinct type of pathway termed single-strand annealing (SSA) (15) has also been proposed for recombination between repeated sequences. In SSA, DNA ends are degraded bidirectionally from a DSB by a strand-specific exonuclease until complementary (homologous) sequences are exposed. Following annealing of the complementary sequences, nonhomologous DNA tails are trimmed, single-strand gaps are repaired, and nicks are ligated to produce recombination products that look like crossovers. SSA is nonconservative in the sense that sequence information between the annealed complementary sequences is lost.
In contrast to yeast, mammalian cells have been reported to be efficient at joining any two noncognate DNA ends with essentially no requirement for sequence homology (5, 10, 32, 33, 36) in a process often referred to as nonhomologous end-joining (NHEJ). Some studies have suggested that NHEJ in higher eukaryotes may be facilitated by the annealing of short complementary sequences one or a few bases in length at or near the DNA termini (5, 19, 23–25, 28, 32, 33, 43). Since this process is reminiscent of SSA, NHEJ events involving the annealing of short complementary sequences have been referred to as micro-SSA (42). (Throughout this paper, we use the term micro-SSA specifically to refer to a type of nonconservative NHEJ event that is facilitated by the annealing of short complementary sequences that are exposed by strand degradation, or unwinding, originating from a DSB. This definition does not include religation, in the absence of strand degradation, of complementary single-strand overhang sequences generated by a staggered DSB.)
In recent years, we (19) and several other groups (4, 7, 14, 22, 31, 35, 36, 42) have developed experimental systems using the rare-cutting endonuclease I-SceI from S. cerevisiae to induce a specific DSB within a mammalian chromosome in order to study the repair of the induced DSB. Although there have been differences in some of the details of the reported results from such studies, it has been demonstrated that DSBs in mammalian chromosomes can be repaired by either HR or NHEJ and that DSBs stimulate HR as well as illegitimate genetic rearrangements in mammalian chromosomes. There has been little or no information presented on what factors may influence the balance between DSB-induced HR and NHEJ, nor has there been any reported investigation of how often a genomic DSB may be precisely religated with no concomitant alteration of the DNA sequence.
To assess the last two issues and investigate several other aspects of intrachromosomal repair of DSBs in mammalian cells, we introduced DNA constructs containing a herpes simplex virus (HSV) thymidine kinase (TK) gene disrupted by a recognition site for yeast endonuclease I-SceI into TK-deficient mouse cells. Some constructs additionally contained a second TK gene sequence to potentially act as a genetic donor in recombinational repair of an I-SceI-induced DSB. A DSB was introduced into each DNA substrate by treatment with I-SceI after the DNA substrate had stably integrated into the genome. DSB repair was monitored by selecting for cells in which a functional TK gene had been reconstructed.
In this report we draw some comparisons between spontaneous and DSB-induced HR in mammalian chromosomes and we present evidence that precise religation is a viable pathway for healing a genomic DSB. Our studies contribute to an expanding paradigm in which multiple pathways function to achieve DSB repair in a mammalian genome while often, but not always, producing minimal change to the genome.
MATERIALS AND METHODS
Cell culture.
Mouse L cells and derivatives deficient in TK were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum, 0.1 mM minimal essential medium nonessential amino acids (GIBCO), and 50 μg of gentamicin sulfate/ml. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2. The parental cell line used in this study has been reported to express functional p53 (40).
Plasmid substrates.
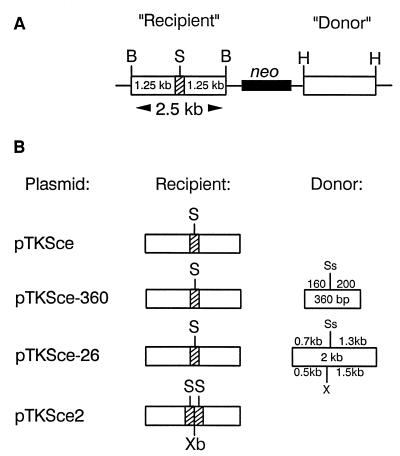
Plasmids (Fig. 1) are based on the vector pJS-1 (13, 17), which is a derivative of pSV2neo (38). Plasmid pTK1 (19) contains the wild-type HSV type 1 (HSV-1) (strain F) TK gene on a 2.5-kb BamHI fragment inserted into the unique BamHI site of the vector. Into the SstI site at position 963 of the TK gene coding region (numbering as described by Wagner et al. [44]) was inserted a double-stranded oligonucleotide containing the 18-bp recognition sequence for yeast endonuclease I-SceI. Insertion of the oligonucleotide introduced a frameshift (net gain of 22 nucleotides), inactivating the TK gene. Insertion of the oligonucleotide also resulted in a duplication of the 4-bp SstI overhang sequence (5′-AGCT-3′) flanking the I-SceI recognition site (see Fig. 2). The plasmid containing the TK gene disrupted by the I-SceI recognition site was named pTKSce (Fig. 1B) and has been described previously as pTK1-Sce (19).
FIG. 1.
DNA substrates for DSB repair. (A) Schematic of a generic substrate. For simplicity, the DNA construct is shown in linear form as if linearized at the unique ClaI site in the vector. Inserted between two BamHI (B) sites is a 2.5-kb DNA fragment containing a TK gene disrupted by a 22-bp oligonucleotide (stippled segment) containing the 18-bp recognition site for yeast endonuclease I-SceI (S). The TK gene is referred to as “recipient” since it is intended to receive information in the recombinational repair of an I-SceI-induced DSB. Some substrates also contain a TK gene sequence inserted between two HindIII (H) sites to act as a genetic donor in recombinational DSB repair. In all substrates, the orientation of the TK gene coding sequences is from left to right. All constructs also contain the neo gene, transcribed from right to left. (B) Schematics of specific DNA substrates. Only recipient and donor (if any) TK gene sequences are shown for each substrate, but all DNA constructs have the general configuration shown in panel A. The 360-bp donor TK gene sequence on pTKSce-360 has 5′ and 3′ truncations of coding sequences. The 2-kb donor sequence on pTKSce-26 contains a complete TK gene disrupted by an 8-bp XhoI linker insertion (X). Indicated on the two donor sequences is the position of the SstI site (Ss) corresponding to the site into which the I-SceI oligonucleotide was inserted in the recipient TK gene sequence. The 2.5-kb recipient sequence on pTKSce2 is disrupted by a 47-bp oligonucleotide containing two I-SceI sites flanking an XbaI (Xb) site. See Materials and Methods for further details.
FIG. 2.
Intrachromosomal DSB repair products generated in the absence of a homologous donor sequence. Presented at the top of the figure is the parental DNA sequence of the I-SceI recognition site insertion (uppercase letters) within the TK gene (lowercase letters) on pTKSce (Fig. 1B). Sites of staggered cleavage by I-SceI are indicated by arrowheads, and the 4-bp repeats flanking the I-SceI site are underlined. Presented below the parental sequence are nucleotide sequences determined for products of intrachromosomal DSB repair recovered from 51 independent HAT-resistant clones derived from cell line Sce-3 following the introduction of I-SceI into cells. All repair products displayed small deletions ranging from 1 to 22 bp that restored the TK gene reading frame. No., number of HAT-resistant clones.
Plasmid pTKSce-360 (Fig. 1B) is identical to pTKSce except that it contains a 360-bp donor fragment (nucleotides 805 to 1165) of the HSV-1 TK gene inserted into the unique HindIII site of pJS-1.
The mutant 26 HSV-1 TK gene contains an 8-bp XhoI linker inserted after nucleotide 737 and has been described previously (17). Plasmid pTKSce-26 (Fig. 1B) is identical to pTKSce except that it contains a 2.0-kb fragment harboring the mutant 26 HSV-1 TK gene inserted at the HindIII site.
Plasmid pTKSce2 (Fig. 1B) is identical to pTKSce except that the TK gene contains a 47-bp oligonucleotide containing two I-SceI recognition sites inserted into the SstI site.
Assay for intrachromosomal DSB repair.
To establish stably transfected experimental cells lines, the appropriate DNA construct was linearized with ClaI and introduced into cells by electroporation (19). Stable transformants were isolated following selection in G418, 200 μg of active drug/ml, as previously described (46). Plasmid pCMV-I-SceI (obtained from M. Jasin), which encodes endonuclease I-SceI under control of the cytomegalovirus promoter and which is expressible in mouse cells (34), was introduced into cell lines as follows. Cells (5 × 106 or 107) and 10 μg of pCMV-I-SceI were resuspended at room temperature in 800 μl of phosphate-buffered saline (PBS) and were electroporated in a Bio-Rad Gene Pulser set to 1,000 V and 25 μF. Following electroporation, cells were plated at a density of 106 cells per 75-cm2 flask. After 2 days of incubation at 37°C under no selection, medium was changed to hypoxanthine-aminopterin-thymidine (HAT) medium to select for TK-positive clones. Colonies were counted and harvested 14 days later.
In experiments involving cell lines containing pTKSce-26, it was necessary to reduce the background of TK-positive segregants produced by spontaneous intrachromosomal HR between the TK gene sequences on the integrated copy of pTKSce-26 in order to measure the effect of the I-SceI-induced DSB. This was accomplished by growing cells in DMEM supplemented with 5 μg of trifluorothymidine (TFT) per ml for 5 days followed by 2 days of growth in DMEM supplemented with 150 μM thymidine to dilute the intracellular pools of TFT. Cells were then electroporated with pCMV-I-SceI and selected in HAT medium as described above except that a supplement of 150 μM thymidine was maintained in the medium throughout selection in HAT.
In all experiments, mock electroporations of cells in PBS alone were performed as controls.
PCR and sequence analysis of products of intrachromosomal DSB repair.
A DNA segment encompassing the original location of the I-SceI site was amplified by PCR from the genome of HAT-resistant clones with primer AL-1 (5′-CCAGCGTCTTGTCATTGGCG-3′), nucleotides 308 through 327 of the coding strand of the HSV-1 TK gene, and primer s1134 (5′-CGGTGGGGTATCGACAGAGT-3′), nucleotides 1786 through 1767 of the noncoding TK gene strand. PCR products were sequenced directly without cloning with a Sequence, version 2.0, kit (Amersham) following treatment of PCR products with a PCR product presequencing kit (Amersham).
RESULTS
Experimental design.
To study DSB repair within mammalian chromosomes, cell lines were derived from mouse L TK-negative fibroblasts containing a variety of DNA substrates (Fig. 1) stably integrated in the genome. Each substrate contained a TK gene disrupted by an inserted oligonucleotide containing one or more copies of the 18-bp recognition sequence for endonuclease I-SceI. Some DNA substrates contained an additional defective TK gene sequence to serve as a potential donor in recombinational DSB repair. Endonuclease I-SceI was introduced into cells to induce a DSB within the integrated substrate, and HAT selection was subsequently applied in order to recover clones in which a functional TK gene had been reconstructed via DSB repair. The gain of TK gene function required correction of the frameshift caused by the inserted oligonucleotide but not necessarily the precise reconstruction of the wild-type TK gene sequence. The 360-bp TK gene donor on pTKSce-360 was deleted at both the 5′ and 3′ ends, and so gene conversions, but not crossovers, could produce a functional TK gene. For pTKSce-26, both gene conversions and crossovers could produce a functional TK gene.
Intrachromosomal DSB repair in the absence of an homologous genetic donor.
Stably transfected cell lines containing one or more copies of pTKSce (Fig. 1B) were isolated. One such cell line, designated Sce-3, previously described (19) contains a single integrated copy of pTKSce. Endonuclease I-SceI or plasmid pCMV-I-SceI was electroporated into Sce-3 cells, and HAT-resistant colonies were selected (Table 1). Electroporation with the expression plasmid pCMV-I-SceI, rather than endonuclease I-SceI itself, was a more efficient means for inducing DSB repair events (Table 1), and so pCMV-I-SceI was used in subsequent experiments. DNA sequences surrounding the I-SceI-induced DSB were amplified by PCR from 51 HAT-resistant clones, and nucleotide sequences were determined (Fig. 2). Fifty of the 51 clones displayed small deletions of 1 to 19 nucleotides (Fig. 2). Each deletion restored the reading frame of the TK gene. Smaller deletions were more common than larger ones, with one-base deletions comprising 23 of the 51 (45%) recovered clones. Deletions extended in either one or both directions from the DSB site. Three clones (classes 13 and 14; Fig. 2) displayed a net deletion of one nucleotide that was one position removed from the actual I-SceI cut site, and two of these clones (class 14; Fig. 2) displayed an A-to-G mutation of the nucleotide immediately upstream from the deleted nucleotide. The three exceptional clones (classes 13 and 14) may have been produced by resection of several nucleotides from the DSB in conjunction with the addition of a nucleotide to a DNA terminus. Of the 51 clones analyzed, only one (class 12; Fig. 2) displayed a wild-type TK gene sequence with a restored SstI site (5′-GAGCTC-3′). The rarity of recovery of a wild-type TK gene sequence from cell line Sce-3 indicated that micro-SSA involving annealing of the 4-bp (5′-AGCT-3′) repeat flanking the I-SceI site (see top of Fig. 2) was not a common mode for intrachromosomal DSB repair.
TABLE 1.
Repair of a genomic DSB in the absence or presence of a homologous donor
| Donor and cell linea | Electroporationb
|
Total no. of HAT-resistant colonies | Frequency of HAT-resistant colonies (10−5)c | |
|---|---|---|---|---|
| Construct used | No. | |||
| No donor | ||||
| Sce-3 | I-SceI | 42 | 157 | 0.03 |
| pCMV-I-SceI | 10 | 650 | 0.65 | |
| Mock | 13 | 0 | 0 | |
| 360-bp donor | ||||
| 360-1 | pCMV-I-SceI | 20 | 2,600 | 2.6 |
| Mock | 5 | 0 | 0 | |
| 360-2 | pCMV-I-SceI | 20 | 2,000 | 2.0 |
| Mock | 5 | 0 | 0 | |
| 2-kb donor | ||||
| 26-1 | pCMV-I-SceI | 5 | 673 | 2.7 |
| Mock | 5 | 101 | 0.4 | |
| 26-2 | pCMV-I-SceI | 8 | 1,156 | 2.9 |
| Mock | 8 | 58 | 0.14 | |
Cell line Sce-3 contains an integrated copy of pTKSce (Fig. 1B), cell lines 360-1 and 360-2 each contain an integrated copy of pTKSce-360 (Fig. 1B), and cell lines 26-1 and 26-2 each contain an integrated copy of pTKSce-26 (Fig. 1B).
Cell line Sce-3 was electroporated with 150 U of I-SceI endonuclease or 5 μg of pCMV-I-SceI per electroporation. Other cell lines were electroporated with 10 μg of pCMV-I-SceI per electroporation. Mock electroporations were with PBS alone.
Total number of HAT-resistant colonies recovered divided by total number of cells electroporated.
Intrachromosomal DSB repair in the presence of a closely linked 360-bp homologous sequence.
Two cell lines designated 360-1 and 360-2, each stably transfected with a single copy of TKSce-360 (Fig. 1B), were electroporated with pCMV-I-SceI, and HAT-resistant colonies were selected (Table 1). Genomic DNA was isolated from a total of 41 DSB-induced HAT-resistant clones. DNA samples from 13 HAT-resistant clones each from cell lines 360-1 and 360-2 were examined on a Southern blot following digestion with BamHI and SstI (data not shown), and it was learned that none of the clones displayed a restored SstI site expected for a wild-type TK gene sequence. From the genomic DNA isolated from the remaining 15 HAT-resistant clones, a TK gene fragment which encompassed the original location of the I-SceI site was amplified by PCR. Incubation of the 15 PCR products with SstI revealed that all products were resistant to cleavage with SstI (data not shown), indicating that a wild-type TK gene had not been restored in any of the HAT-resistant clones. Thus, in total, none of the 41 HAT-resistant clones analyzed had arisen from either HR or micro-SSA since either repair pathway would have produced a wild-type TK gene. The nucleotide sequence surrounding the position of the I-SceI-induced DSB was determined for six HAT-resistant clones (Fig. 3), and each revealed a 1-bp deletion at the DSB site equivalent to the class 1 repair product in Fig. 2. From our analyses, we inferred that intrachromosomal DSB repair in the presence of a closely linked 360-bp homologous sequence is accomplished almost exclusively by NHEJ.
FIG. 3.
Intrachromosomal NHEJ products generated in the presence of a 360-bp or 2.0-kb homologous donor. Presented at the top of the figure is the parental DNA sequence of the I-SceI recognition site insertion (uppercase letters) within the recipient TK gene (lowercase letters) on pTKSce-360 and pTKSce-26 (Fig. 1B). Sites of staggered cleavage by I-SceI are indicated by arrowheads, as are the 4-bp repeats flanking the I-SceI site (underlining). Presented below the parental sequence are nucleotide sequences determined for the products of intrachromosomal NHEJ products recovered from 6 independent HAT-resistant clones derived from cell lines 360-1 and 360-2 and from 11 independent HAT-resistant clones derived from cell lines 26-1 and 26-2 following the introduction of I-SceI into cells. All NHEJ repair products displayed small deletions ranging from 1 to 16 bp that restored the TK gene reading frame. It should be noted (see Table 2 and text) that some HAT-resistant clones derived from lines 26-1 and 26-2 displayed wild-type TK gene sequences as products of HR but are not shown here. No., number of HAT-resistant clones.
Intrachromosomal DSB repair in the presence of a closely linked 2-kb homologous sequence.
Two cell lines designated 26-1 and 26-2, each containing a single integrated copy of pTKSce-26, were electroporated with pCMV-I-SceI, and HAT-resistant colonies were selected (Table 1). For cell line 26-1, pCMV-I-SceI brought about more than a 6-fold induction of colonies relative to mock electroporations, while for cell line 26-2 induction was about 20-fold (Table 1). Genomic DNA was isolated from 12 HAT-resistant clones derived from line 26-1 and from 30 HAT-resistant clones derived from line 26-2 after transfection with pCMV-I-SceI. The DNA samples were digested with various combinations of BamHI, HindIII, XhoI, and SstI and subjected to Southern blotting analysis using a TK gene-specific probe (Fig. 4A). Results of the Southern blotting analysis are summarized below and in Table 2. (Additional digestions not shown in Fig. 4A were used to confirm the identity of some bands.)
FIG. 4.
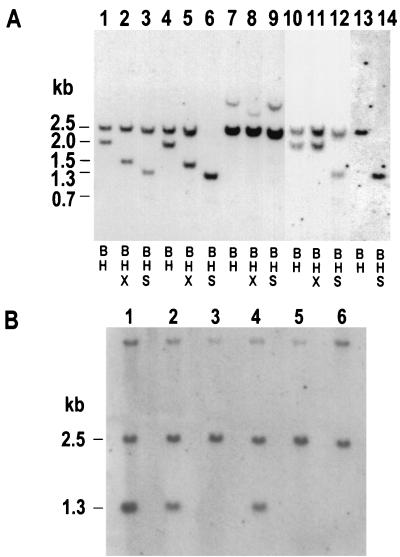
Representative Southern blotting analysis of HAT-resistant clones derived from cell line 26-2. (A) DNA samples (8 μg each) isolated from HAT-resistant clones were digested with various combinations of BamHI (B), XhoI (X), and SstI (S), as indicated below the lanes, and were displayed on a Southern blot by using a TK gene-specific probe. Digestions from each individual clone are presented in three adjacent lanes (for example, lanes 1 to 3), with the exception of the clone presented in lanes 13 and 14. Clones displayed in lanes 1 to 9, 13, and 14 were recovered from cell line 26-2 following transfection of pCMV-I-SceI into cells. The HAT-resistant clone displayed in lanes 10 to 12 arose spontaneously (following a mock transfection). In total, 42 DSB-induced and 20 spontaneous HAT-resistant clones from cell lines 26-1 and 26-2 were analyzed by Southern blotting. See text for details. (B) DNA samples (8 μg each) from six clones exhibiting an aberrant restriction pattern similar to that exhibited by the clone in lanes 7 to 9 in panel A were digested simultaneously with BamHI and I-SceI and displayed on a Southern blot by using a TK gene-specific probe.
TABLE 2.
Summary of Southern blotting analysis of HAT-resistant segregants from cell lines 26-1 and 26-2
| Event type and cell line | No. of HAT-resistant segregants analyzed | No. of HAT-resistant segregants displaying:
|
||
|---|---|---|---|---|
| HR | NHEJ | Amplificationa | ||
| DSB-induced | ||||
| 26-1 | 12 | 7b | 5 | 0 |
| 26-2 | 30 | 5c | 12 | 13 |
| Spontaneous | ||||
| 26-2 | 20 | 20d | 0 | 0 |
Thirteen HAT-resistant segregants from cell line 26-2 displayed an amplification of sequences in the vicinity of the I-SceI-induced DSB, in conjunction with NHEJ. These 13 clones are distinct from the 12 segregants from cell line 26-2 listed under NHEJ. See text for details.
Among the recombinants, six were gene conversions of the I-SceI recognition sequence insertion and one was a crossover.
Among the recombinants, all five were gene conversions of the I-SceI recognition sequence insertion.
Among the recombinants, 18 were gene conversions of the I-SceI recognition sequence insertion, 1 was a gene conversion of the XhoI linker insertion, and 1 was a crossover.
Six of 12 clones isolated from cell line 26-1 and 5 of 30 clones from line 26-2 displayed the patterns shown in Fig. 4A, lanes 4 to 6. These 11 clones retained both the 2.5-kb BamHI fragment and the 2.0-kb HindIII fragment (Fig. 4A, lane 4) present in the parental DNA construct, pTKSce-26 (see Fig. 1 for the origin of fragments). The 2.0-kb HindIII sequence retained the XhoI linker insertion and was cleavable by XhoI into predicted 1.5- and 0.5-kb fragments (Fig. 4A, lane 5). (The 0.5-kb fragment is not visible.) The 2.5-kb BamHI fragment which originally contained the I-SceI-disrupted TK gene was cleaved in half by SstI (Fig. 4A, lane 6). The 2.0-kb HindIII fragment was also cleavable by SstI into expected 1.3- and 0.7-kb fragments (Fig. 4A, lane 6), although the 0.7-kb fragment is difficult to see on the blot. Most notably, restoration of the SstI site on the 2.5-kb BamHI fragment was diagnostic for reconstruction of a wild-type TK gene. Since only 1 of 51 HAT-resistant clones recovered in the absence of a homologous donor sequence and none of the 41 HAT-resistant clones recovered in the presence of a 360-bp homologous donor contained a wild-type TK gene sequence (see above), it was clear that the wild-type TK gene sequences recovered from cell lines 26-1 and 26-2 had arisen as products of HR or, more specifically, gene conversion eliminating the I-SceI recognition site on the broken TK gene sequence. One clone from cell line 26-1 displayed the pattern seen in Fig. 4A, lanes 13 and 14. This clone displayed a single fragment upon digestion with BamHI and HindIII (lane 13), which was cleavable with SstI (lane 14) and most likely arose from a crossover between the two TK genes on pTKSce-26.
Five HAT-resistant clones isolated from cell line 26-1 and 12 HAT-resistant clones from cell line 26-2 displayed the pattern shown in Fig. 4A, lanes 1 to 3. These clones contained a 2.5-kb BamHI fragment that resisted cleavage with SstI (lane 3) but retained a 2.0-kb HindIII fragment (lane 1) that was cleavable with XhoI (lane 2), suggesting no change to the parental HindIII fragment (Fig. 1B). Such a pattern was expected for NHEJ of the DNA termini at the I-SceI-induced DSB. The sequence in the vicinity of the I-SceI site was determined for 11 of these putative NHEJ events (Fig. 3), confirming that each of the HAT-resistant clones displayed a small deletion at the original location of the I-SceI site, consistent with NHEJ. Seven of the 11 HAT-resistant clones analyzed, comprising classes 1, 2, 13, and 15 in Fig. 3, displayed a deletion of only a single nucleotide, which was similar to the observation of a predominance of single-nucleotide deletions for NHEJ in the absence of any donor (Fig. 2). Repair classes 1, 2, and 13 (Fig. 3) also had been recovered in the absence of a donor (Fig. 2). The unique class 15 clone (Fig. 3) was interesting in that the single deleted G was two nucleotides away from the closest DNA terminus produced by cleavage with I-SceI. Repair classes 16 through 18 (Fig. 3) were not previously recovered in the absence of a donor (Fig. 2). Class 18 (Fig. 3) displayed an A-to-T mutation at the position immediately downstream from the 16-bp deletion and may have arisen from the addition of the aberrant T residue to a DNA terminus.
Thirteen HAT-resistant segregants from cell line 26-2 displayed a pattern similar to that shown in Fig. 4A, lanes 7 to 9. These clones displayed a loss of the 2.0-kb HindIII fragment with the concomitant gain of a novel fragment of about 3.9 kb as well as a marked increase in the hybridization signal of the 2.5-kb BamHI fragment (Fig. 4A, lane 7). These clones displayed various degrees of amplification of the 2.5-kb BamHI fragment, with phosphorimager analysis indicating that some clones contained as many as six copies of the BamHI fragment (data not shown). The 2.5-kb BamHI fragments in these clones were insensitive to digestion with XhoI (lane 8) or SstI (lane 9). Additional Southern blotting and sequencing analysis (not shown) corroborated that these clones had undergone NHEJ as well as an amplification of a portion of the integrated pTKSce-26 construct. In several clones, some copies of the 2.5-kb BamHI fragment were cleavable with I-SceI while the remaining copies resisted such cleavage (Fig. 4B, lanes 1, 2, and 4).
Southern blotting analysis was also performed on 20 HAT-resistant clones recovered following mock electroporations of cell line 26-2. These clones had presumably arisen from spontaneous HR events, and this supposition was confirmed by Southern blotting analysis (Table 2). One clone, displayed in Fig. 4A, lanes 10 to 12, displayed a 2.0-kb HindIII fragment that resisted cleavage with XhoI (lane 11) and a 2.5-kb BamHI fragment resistant to cleavage with SstI (lane 12). This clone likely arose from a gene conversion eliminating the XhoI linker insertion mutation in the TK gene on the 2.0-kb HindIII fragment on pTKSce-26 (Fig. 1B). One spontaneously arising HAT-resistant clone displayed a pattern similar to that for the clone displayed in Fig. 4A, lanes 13 and 14, and likely arose from a crossover between TK genes. The remaining 18 HAT-resistant clones all displayed a pattern similar to that displayed by the clone in Fig. 4A, lanes 4 to 6, and likely arose from a gene conversion eliminating the I-SceI recognition site insertion mutation from the TK gene on the 2.5-kb BamHI fragment, restoring the SstI site. Of note was the absence among the 20 spontaneously arising HAT-resistant clones of any clone displaying the aberrant pattern shown in Fig. 4A, lanes 7 to 9. This strongly suggested that the aberrant rearrangements and amplifications seen in some HAT-resistant clones recovered after transfection with pCMV-I-SceI were indeed DSB induced.
Precise religation as a mode of DSB repair.
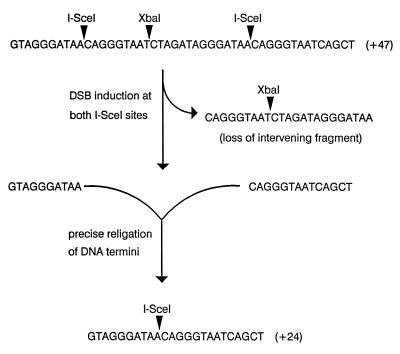
The experiments described above indicated that intrachromosomal DSB repair can be accomplished by either NHEJ or by HR if a potential genetic donor has a suitable amount of homology with the broken sequence. It remained unclear how often DSB repair is accomplished by simple, precise religation of DNA termini with no alteration of sequence information. To address this issue we established cell lines containing a single integrated copy of pTKSce2 (Fig. 1B). At the SstI site of the TK gene in pTKSce2 was inserted a 47-bp insert containing two I-SceI sites flanking an XbaI site (Fig. 5). As illustrated in Fig. 5, cleavage at the two I-SceI sites followed by ligation of the outermost pair of sticky ends produced a functional TK gene containing a 24-bp insert harboring a single I-SceI site. Four cell lines containing a single copy of pTKSce2 and designated Sce2-1, Sce2-2, Sce2-3, and Sce2-4 were electroporated with pCMV-I-SceI, and HAT-resistant colonies were selected (Table 3). DNA was isolated from several HAT-resistant clones from each cell line (Table 3) and initially analyzed by amplifying a TK gene segment encompassing the position of the original I-SceI sites by PCR and testing the PCR products for sensitivity to cleavage by XbaI since, as indicated in Fig. 5, precise religation removes the XbaI site. Clones that lost the XbaI site were further analyzed by DNA sequencing. As indicated in Table 3, despite the wide range in the overall frequencies of recovery of HAT-resistant colonies from the four cell lines, a significant portion (ranging from 33 to 65%) of HAT-resistant clones isolated from all four cell lines had arisen via precise religation and the majority of clones that had lost the XbaI site had undergone precise religation. All clones analyzed retained one I-SceI site (data not shown), indicating that degradation from DSBs was limited. The fact that every clone retained an I-SceI site also suggested that precise religation was a frequent event since, in any given clone, had two I-SceI-induced DSBs existed simultaneously and had degradation from both DSBs occurred prior to religation (NHEJ), then both I-SceI sites would have been lost. Southern blotting analysis (not shown) revealed no additional rearrangements or amplifications in clones that underwent precise religation.
FIG. 5.
Substrate for monitoring precise religation. Shown at the top of the figure is the 47-bp oligonucleotide inserted into the TK gene on pTKSce2. The oligonucleotide causes a frameshift mutation in the TK gene and contains two I-SceI recognition sites (5′-TAGGGATAACAGGGTAAT-3′) flanking an XbaI site (5′-TCTAGA-3′). The actual sites of cleavage by I-SceI and XbaI on the top strand of the construct are indicated. As illustrated, simultaneous cleavage of the substrate at both I-SceI sites followed by loss of the intervening 23-bp fragment and precise religation of the two outermost I-SceI sticky ends produces a functional, and thus recoverable, TK gene containing a 24-bp insert harboring a single I-SceI site.
TABLE 3.
Precise religation as a mode of intrachromosomal DSB repair
| Cell line | No. of electroporations | Frequency of HAT-resistant colonies (10−4)a | No. of HAT-resistant colonies analyzed | XbaI site loss (%)b | Precise religation (%)c |
|---|---|---|---|---|---|
| Sce2-1 | 3 | 2.2 | 39 | 44 | 36 |
| Sce2-2 | 1 | 2.4 | 17 | 65 | 65 |
| Sce2-3 | 1 | 0.068 | 15 | 33 | 27 |
| Sce2-4 | 1 | 0.032 | 6 | 50 | 33 |
Number of HAT-resistant colonies divided by total number of cells electroporated.
Percentage of analyzed HAT-resistant clones that lost the XbaI site between the I-SceI sites (Fig. 5), based on PCR analysis.
Percentage of analyzed HAT-resistant clones that had undergone precise religation, based on nucleotide sequence determination.
DISCUSSION
In this report, we examined several aspects of DSB repair in mammalian chromosomes. We have demonstrated that mammalian cells enjoy several pathways for healing DSBs, while producing little or no concomitant genetic alteration.
In studies of intrachromosomal DSB repair using cell line Sce-3 containing pTKSce (Fig. 1B), only 1 of 51 HAT-resistant clones analyzed in detail displayed a wild-type TK gene sequence (Fig. 2, class 12), suggesting that micro-SSA is not an important pathway for intrachromosomal DSB repair in our cell lines. Production of a wild-type TK gene by micro-SSA required that DNA termini be degraded back by as much as 13 bp to the left and 17 bp to the right of the DSB in order to expose the complementary 4-bp SstI overhang sequences. Fifty of 51 HAT-resistant segregants from line Sce-3 displayed small deletions that were produced by loss of fewer than 13 bp from either side of the DSB, with loss of only 1 bp being the most commonly recovered deletion (Fig. 2). In recent experiments in which we monitored intrachromosomal DSB repair by selecting for mutations involving loss of TK gene function (16), we also recovered a preponderance of small deletions. We thus infer that intrachromosomal DSB repair in our cell lines often proceeds with little strand degradation from the DNA termini and that NHEJ is not dependent on the uncovering of sequence microhomologies. Our precise religation experiments, discussed below, imply that terminal microhomologies may play a role in DSB repair in our cell lines as long as strand degradation is not required to expose the complementary sequences.
Experiments with cell lines 360-1 and 360-2 containing pTKSce-360, with a 360-bp donor, corroborated the relative infrequency of micro-SSA (involving nucleotide loss) as a means for intrachromosomal DSB repair. None of the 41 HAT-resistant clones contained a wild-type TK gene sequence, indicating that none of the clones were produced by micro-SSA. The six HAT-resistant clones from cell lines 360-1 and 360-2 that were sequenced each displayed a 1-bp deletion (Fig. 3), again indicating that often only a small degree of degradation of DNA termini occurs prior to intrachromosomal NHEJ. The lack of any wild-type TK gene sequence among the 41 HAT-resistant clones analyzed also revealed that none of the clones had arisen from HR. The lack of intrachromosomal DSB repair by HR in the presence of a 360-bp donor suggests that the length of homology shared by recipient and donor sequences on one or both sides of the DSB may have been insufficient for intrachromosomal HR and that perhaps the general homology length requirements observed for spontaneous HR (18, 47) are preserved for DSB-induced recombination. In related preliminary work, we have been unable to detect DSB induction of homeologous recombination between two TK genes having 80% homology (16), suggesting that requirements for a high degree of homology observed for spontaneous recombination (18, 46, 47) may not be compromised during DSB repair.
In light of reports (5, 19, 23–25, 28, 32, 33, 43) that NHEJ in eukaryotes may be facilitated by small (1- to 5-bp) homologies, it seems striking that in our present study only one of a total of 92 recovered NHEJ events (combining results from cell lines Sce-3, 360-1, and 360-2) was associated with a deletion spanning the 4-bp homologous repeats flanking the I-SceI site to produce a wild-type TK gene sequence (Fig. 2, class 12). This result suggests that in our system the joining of noncognate DNA ends occurs more rapidly than strand degradation and/or homology searching. It was fortuitous that, due to the nature of the TK protein, accurate reconstruction of wild-type TK gene sequence at the site of the I-SceI DSB was not required for restoration of TK gene function, and thus we could recover almost any NHEJ event associated with a small deletion as long as the correct TK gene reading frame had been restored. In some other reports (4, 7, 22, 31, 35, 42) recovery of certain NHEJ events may have been restricted by stringent selection for the accurate reconstruction of a wild-type marker gene sequence.
Experiments with cell lines 26-1 and 26-2 containing pTKSce-26 demonstrated that, in the presence of a suitably long donor sequence, intrachromosomal DSB repair in mammalian cells can indeed be accomplished by HR as reported by others (4, 7, 14, 22, 31, 35, 36, 42). Seven of 12 and 5 of 30 HAT-resistant segregants from 26-1 and 26-2, respectively, were produced by HR (Table 2). Since electroporation with pCMV-I-SceI induced the overall number of HAT-resistant colonies recovered from cell lines 26-1 and 26-2 about 7-fold and 20-fold, respectively, it was clear that a significant portion of the induced colonies were produced by HR. However, we note that while DSBs can stimulate intrachromosomal HR, the presence of a closely linked homologous partner sequence does not stimulate DSB repair, as evidenced by the similar frequencies of recovery of DSB-induced HAT-resistant segregants from the various cell lines listed in Table 1. This observation would appear to represent a fundamental difference between DSB repair in higher and lower eukaryotes since DSB repair in yeast has been found to be highly dependent on the presence of an HR partner (8, 11, 20).
In previous studies (19, 45) we were unable to recover any DSB-induced gene targeting in mouse L cells by using DNA constructs similar to the ones used in this study despite the fact that in some targeting experiments the transfected DNA molecule had over 8 kb of homology with the broken locus. Our collective results may reflect a fundamental difference between the rate-limiting step and/or mechanism for gene targeting and intrachromosomal HR in our cell lines. We note that others (4, 6, 7, 35) have reported substantial stimulation of gene targeting by a genomic DSB introduced by I-SceI, and these differing results may reflect differences in the cell lines or DNA sequences used.
Only one of the analyzed DSB-induced HR events recovered from cell lines 26-1 and 26-2 following transfection with pCMV-I-SceI was a crossover event (Fig. 4, lanes 13 and 14), while the remaining 11 HR events analyzed appeared to be gene conversions correcting the I-SceI recognition site insertion mutation. A preferential induction of gene conversions (versus crossovers) by DSBs has been previously observed (42) and may be mechanistically related to the recently reported (31) suppression of DSB-induced translocations. Suppression of DSB-induced crossovers and translocations may be viewed as additional safeguards against deleterious genetic rearrangements. We have also observed a marked preferential recovery of gene conversions during spontaneous HR in our system (2, 17, 48, 50, 51) (Table 2), so it is not clear that there is need to invoke a distinct mechanism for the reduction of crossover specifically during DSB-induced recombination. It is reasonably clear, however, that DSBs do not induce gene conversions exclusively, since others (6) have observed a substantial induction of intrachromosomal crossovers between direct repeats when substrates designed specifically to recover intrachromosomal crossovers were used.
Richardson et al. (31) proposed a mechanism for preferential recovery of gene conversions during DSB repair by suggesting that DSB-induced HR is coupled to replication. The amplification events that we recovered (Fig. 4) also suggest a possible coupling of replication to DSB repair since it seems likely that amplification results from reiterated replication through all or part of the integrated construct. Others have also observed sequence duplications or amplifications that appeared to be associated with repair of I-SceI-induced DSBs (22, 29, 31), and it has been reported that DNA breakage can play a critical role in initiating gene amplifications in general (12, 21, 26). One intriguing and speculative possibility is that a DSB may function as a cryptic replication origin. In unpublished work (16) we have observed that illegitimate integration of transfected plasmid sequences into a genomic DSB occurs preferentially at replication origins on the plasmid. One might imagine that such integration is mediated by proteins that have affinity for replication origins as well as DSBs, raising the prospect that a replication complex may be able to assemble on a DSB. The cell line used in this study has been reported to express functional p53 (40), and it is not yet clear what role p53 plays in the observed amplifications or in any of the other DSB repair events we have recovered.
We have found that precise religation with no alteration of sequence is a viable pathway for repair of DSBs in a mammalian genome (Table 3). It would seem that such a mode for repair provides an effective and simple means for preventing unwanted rearrangements during repair. The benefit of an efficient pathway for precise religation is all the more apparent if reported estimates of nine genomic DSBs per mammalian cell per day (1) are accurate. Our findings are also similar to the recent report (39) of the addition of new telomeric sequences directly onto the end of a broken chromosome without the loss of a single nucleotide. Because our scheme for recovery of precise religation events required that two DSBs exist simultaneously, it is likely that our results represent an underestimate of the frequency of simple religation. We recognize that our substrate for precise religation includes 4-bp complementary overhang sequences which possibly facilitate religation. Complementary overhangs would be generated in naturally occurring DSBs only if staggered breaks were made. In any case, our experiments raise the real possibility that multiple, and perhaps many, rounds of breakage and religation occur prior to the occurrence of HR or NHEJ in these and other studies. Therefore, the true frequency with which a DSB is repaired in a mammalian cell by HR or NHEJ remains elusive.
Our present work and reports from others (4, 5, 14, 19, 22, 31, 35, 36, 42) make it reasonably clear that mammals have evolved several pathways for repairing DSBs while producing minimal change to the genome. At the same time, observations made by us and by others (22, 29, 31) of DSB-induced sequence amplification hint at the potential for deleterious genomic rearrangement in response to damage. In consideration of the association of cancer with altered cellular responses to DNA damage and the genesis of genomic instability, it is of considerable interest to explore how various modes of DSB repair may be influenced by genetic mutations that have been implicated in carcinogenesis.
ACKNOWLEDGMENTS
We thank Barbara Criscuolo Waldman for helpful comments on the manuscript.
This work was supported by grants from the American Cancer Society (NP-949) and the National Institute of General Medical Sciences (GM47110) to A.S.W.
REFERENCES
- 1.Bernstein C, Bernstein H. Aging, sex, and DNA repair. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 2.Bollag R J, Waldman A S, Liskay R M. Homologous recombination in mammalian cells. Annu Rev Genet. 1989;23:199–225. doi: 10.1146/annurev.ge.23.120189.001215. [DOI] [PubMed] [Google Scholar]
- 3.Camerini-Otero R D, Hsieh P. Homologous recombination proteins in prokaryotes and eukaryotes. Annu Rev Genet. 1995;29:509–552. doi: 10.1146/annurev.ge.29.120195.002453. [DOI] [PubMed] [Google Scholar]
- 4.Choulika A, Perrin A, Dujon B, Nicolas J-F. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1963–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derbyshire M K, Epstein L H, Young C S H, Munz P L, Fishel R. Nonhomologous recombination in human cells. Mol Cell Biol. 1994;14:156–169. doi: 10.1128/mcb.14.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliot B, Richardson C, Winderbaum J, Nickoloff J A, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairhead C, Dujon B. Consequences of unique double-strand breaks in yeast chromosomes: death or homozygosis. Mol Gen Genet. 1993;240:170–178. doi: 10.1007/BF00277054. [DOI] [PubMed] [Google Scholar]
- 9.Haber J E. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 10.Jackson S P, Jeggo P A. DNA double-strand break repair and V(D)J recombination: involvement of DNA-PK. Trends Biochem Sci. 1995;20:412–415. doi: 10.1016/s0968-0004(00)89090-8. [DOI] [PubMed] [Google Scholar]
- 11.Kramer K M, Brock J A, Bloom K, Moore J K, Haber J E. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuo M T, Sen S, Hittelman W N, Hsu T C. Chromosomal fragile sites and DNA amplification. Biochem Pharmacol. 1998;56:7–13. doi: 10.1016/s0006-2952(98)00040-9. [DOI] [PubMed] [Google Scholar]
- 13.Letsou A, Liskay R M. Effect of the molecular nature of mutation on the efficiency of intrachromosomal gene conversion in mouse cells. Genetics. 1987;117:759–769. doi: 10.1093/genetics/117.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang F, Han M, Romanienko P J, Jasin M. Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA. 1998;95:5172–5177. doi: 10.1073/pnas.95.9.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin F-L, Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol Cell Biol. 1990;10:103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin, Y., and A. S. Waldman. Unpublished data.
- 17.Liskay R M, Stachelek J L, Letsou A. Homologous recombination between repeated chromosomal sequences in mouse cells. Cold Spring Harbor Symp Quant Biol. 1984;49:183–189. doi: 10.1101/sqb.1984.049.01.021. [DOI] [PubMed] [Google Scholar]
- 18.Lukacsovich T, Waldman A S. Suppression of intrachromosomal gene conversion in mammalian cells by small degrees of sequence divergence. Genetics. 1999;151:1559–1568. doi: 10.1093/genetics/151.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lukacsovich T, Yang D, Waldman A S. Repair of a specific double-strand break generated within a mammalian chromosome by yeast endonuclease I-SceI. Nucleic Acids Res. 1994;22:5649–5657. doi: 10.1093/nar/22.25.5649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore J K, Haber J E. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 21.Morgan W F, Day J P, Kaplan M I, McGhee E M, Limoli C L. Genomic instability induced by ionizing radiation. Radiation Res. 1996;146:247–258. [PubMed] [Google Scholar]
- 22.Moynahan M E, Jasin M. Loss of heterozygosity induced by a chromosomal double-strand break. Proc Natl Acad Sci USA. 1997;94:8988–8993. doi: 10.1073/pnas.94.17.8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicolas A L, Young C S H. Characterization of DNA end-joining in a mammalian cell nuclear extract: junction formation is accompanied by nucleotide loss, which is limited and uniform but not site specific. Mol Cell Biol. 1994;14:170–180. doi: 10.1128/mcb.14.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicolas A L, Munz P L, Young C S H. A modified single-strand annealing model best explains the joining of DNA double-strand breaks in mammalian cells and cell extracts. Nucleic Acids Res. 1995;23:1036–1043. doi: 10.1093/nar/23.6.1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.North P, Ganesh A, Thaker J. The rejoining of double-strand breaks in DNA by human cell extracts. Nucleic Acids Res. 1990;18:6205–6210. doi: 10.1093/nar/18.21.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paulson T G, Almasan A, Brody L L, Wahl G M. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J R, Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1991. pp. 407–521. [Google Scholar]
- 28.Pfeiffer P, Thode S, Hanke J, Vielmetter W. Mechanisms of overlap formation in nonhomologous DNA end-joining. Mol Cell Biol. 1994;14:888–895. doi: 10.1128/mcb.14.2.888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pipiras E, Coquelle A, Bieth A, Debatisse M. Interstitial deletions and intrachromosomal amplification initiated from a double-strand break targeted to a mammalian chromosome. EMBO J. 1998;17:325–333. doi: 10.1093/emboj/17.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Resnick M A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976;59:97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- 31.Richardson C, Moynahan M E, Jasin M. Double-strand break repair by interchromosomal recombination: suppression of chromosomal translocation. Genes Dev. 1998;12:3831–3842. doi: 10.1101/gad.12.24.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roth D, Wilson J. Illegitimate recombination in mammalian cells. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C.: American Society for Microbiology; 1988. pp. 621–653. [Google Scholar]
- 33.Roth D B, Wilson J. Nonhomologous recombination in mammalian cells: role of short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rouet P, Smith F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rouet P, Smith F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14:8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sargent R G, Brenneman M A, Wilson J H. Repair of site-specific double-strand breaks in a mammalian chromosome by homologous and illegitimate recombination. Mol Cell Biol. 1997;17:267–277. doi: 10.1128/mcb.17.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shinohara A, Ogawa T. Homologous recombination and the roles of double-strand breaks. Trends Biol Sci. 1995;20:387–391. doi: 10.1016/s0968-0004(00)89085-4. [DOI] [PubMed] [Google Scholar]
- 38.Southern P S, Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early promoter. J Mol Appl Genet. 1982;1:327–341. [PubMed] [Google Scholar]
- 39.Sprung C N, Reynolds G E, Jasin M, Murnane J P. Chromosome healing in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1999;96:6781–6786. doi: 10.1073/pnas.96.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sun X, Shimizu H, Yamamoto K-I. Identification of a novel p53 promoter element involved in genotoxic stress-inducible p53 gene expression. Mol Cell Biol. 1995;15:4489–4496. doi: 10.1128/mcb.15.8.4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 42.Taghian D G, Nickoloff J A. Chromosomal double-strand breaks induce gene conversion at high frequency in mammalian cells. Mol Cell Biol. 1997;17:6386–6393. doi: 10.1128/mcb.17.11.6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thode S, Schafer A, Pfeiffer P, Vielmetter W. A novel pathway of DNA end-to-end joining. Cell. 1990;60:921–928. doi: 10.1016/0092-8674(90)90340-k. [DOI] [PubMed] [Google Scholar]
- 44.Wagner M J, Sharp J A, Summers W C. Nucleotide sequence of the thymidine kinase gene of herpes simplex virus type 1. Proc Natl Acad Sci USA. 1981;78:1441–1445. doi: 10.1073/pnas.78.3.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Waldman, A. S., and T. Lukacsovich. Unpublished data.
- 46.Waldman A S, Liskay R M. Differential effects of base-pair mismatch on intrachromosomal versus extrachromosomal recombination in mouse cells. Proc Natl Acad Sci USA. 1987;84:5340–5344. doi: 10.1073/pnas.84.15.5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waldman A S, Liskay R M. Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol Cell Biol. 1988;8:5350–5357. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waldman A S, Waldman B C. Stimulation of intrachromosomal homologous recombination in mammalian cells by an inhibitor of poly(ADP-ribosylation) Nucleic Acids Res. 1991;19:5943–5947. doi: 10.1093/nar/19.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wood R D. DNA repair in eukaryotes. Annu Rev Biochem. 1996;65:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 50.Yang D, Waldman A S. Fine-resolution analysis of products of intrachromosomal homeologous recombination in mammalian cells. Mol Cell Biol. 1997;17:3614–3628. doi: 10.1128/mcb.17.7.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, D., and A. S. Waldman. Unpublished data.