Abstract
The rapid, global dispersion of SARS-CoV-2 has led to the emergence of a diverse range of variants. Here, we describe how the mutational landscape of SARS-CoV-2 has shaped HLA-restricted T cell immunity at the population level during the first year of the pandemic. We analyzed a total of 330,246 high-quality SARS-CoV-2 genome assemblies, sampled across 143 countries and all major continents from December 2019 to December 2020 before mass vaccination or the rise of the Delta variant. We observed that proline residues are preferentially removed from the proteome of prevalent mutants, leading to a predicted global loss of SARS-CoV-2 T cell epitopes in individuals expressing HLA-B alleles of the B7 supertype family; this is largely driven by a dominant C-to-U mutation type at the RNA level. These results indicate that B7-supertype-associated epitopes, including the most immunodominant ones, were more likely to escape CD8+ T cell immunosurveillance during the first year of the pandemic.
Keywords: SARS-CoV-2, mutation, epitope, T cell, HLA, supertype, peptide, variant, COVID-19
Graphical abstract
Hamelin et al. investigated the global mutation landscape of SARS-CoV-2 by interrogating 330,246 SARS-CoV-2 sequences from GISAID. The dominant C→U mutation type was found to diversify the repertoire of experimentally validated SARS-CoV-2 CD8+ T cell epitopes in an HLA-supertype-dependent manner. Notably, the prevalent removal of proline was predicted to preferentially abrogate epitopes presented by the B7 HLA supertype. This model lays a foundation for testing the impact of SARS-COV-2 mutants on T cell escape in an HLA-dependent manner.
Introduction
As of September 2021, the COVID-19 pandemic, caused by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to upward 4.6 million deaths and 222 million confirmed cases worldwide (https://coronavirus.jhu.edu/map.html), making vaccine development and deployment an urgent necessity (Callaway, 2020). As a result of unprecedent efforts, vaccines have been developed and licensed within a 1-year time frame and are currently being widely distributed for mass vaccination (Krammer, 2020).
A clear understanding of the natural protective immune response against SARS-CoV-2 is essential for the development of vaccines that can trigger lifelong immunologic memory to prevent COVID-19 (Sette and Crotty, 2021; Stephens and McElrath, 2020). Since the start of the pandemic, numerous studies have investigated the association between COVID-19 clinical outcomes and SARS-CoV-2-specific antibodies and T cell immunity (Altmann and Boyton, 2020; Le Bert et al., 2020; Braun et al., 2020; Grifoni et al., 2020a; Long et al., 2020a, 2020b; Meckiff et al., 2020; Moderbacher et al., 2020; Sekine et al., 2020; Weiskopf et al., 2020). Memory may be a concern for SARS-CoV-2-specific antibodies, as they were recently shown to be present in convalescent COVID-19 patients in a highly heterogeneous manner (Dan et al., 2021) and, in some cases, observed to be undetectable just a few months post-infection (Seow et al., 2020). In contrast, an increasing number of studies point CD4+ and CD8+ T cells as key regulators of disease severity (Liao et al., 2020; Moderbacher et al., 2020; Schub et al., 2020; Weiskopf et al., 2020; Zhou et al., 2020). Studies of convalescent COVID-19 patients have also shown broad and strong CD4+ and CD8+ memory T cells induced by SARS-COV-2, suggesting that T cells may provide robust and long-term protection (Dan et al., 2021; Peng et al., 2020). Similar observations have been made for the most closely related human coronavirus, SARS-CoV, for which T cells have been detected 11 years (Ng et al., 2016) and 17 years (Le Bert et al., 2020) after the initial infection, whereas antibodies were noted to be undetectable after 2–3 years (Liu et al., 2006; Tang et al., 2011; Wu et al., 2007). Thus, vaccines designed to produce robust T cell responses are likely to be important for eliciting lifelong immunity against COVID-19 in the general population.
To investigate how T cells could contribute to long-term vaccine effectiveness, precise knowledge about SARS-CoV-2 T-cell-specific epitopes is of paramount importance (Liu et al., 2020). To this end, bioinformatics tools were developed to predict T-cell-specific epitopes during the early phase of the pandemic (Grifoni et al., 2020b). A comprehensive map of epitopes recognized by CD4+ and CD8+ T cell responses across the entire SARS-CoV-2 viral proteome was also recently reported (Tarke et al., 2021a). The structural proteins spike (S), nucleocapsid (N), and membrane (M) were shown to be rich sources of immunodominant HLA-associated epitopes, accounting for a large proportion of the total CD4+ and CD8+ T cell response in the context of a broad set of HLA alleles (Tarke et al., 2021a). As of May 2021, ∼700 HLA-class-I-restricted SARS-CoV-2-derived epitopes have been experimentally validated (https://www.mckayspcb.com/SARS2TcellEpitopes/) (Quadeer et al., 2021).
T cell epitopes that have been mapped across the entire SARS-CoV-2 viral proteome are reference peptides that are unmutated because they have been predicted from the sequence of the original SARS-CoV-2 that emerged from Wuhan, China (Grifoni et al., 2020b). However, analyses of unprecedented numbers of SARS-CoV-2 genome assemblies available from large-scale efforts have shown that SARS-CoV-2 is accumulating an array of mutations across the world, leading to the circulation and transmission of thousands of variants around the globe at various frequencies, and hence, contributing to the global genomic diversification of SARS-CoV-2 (van Dorp et al., 2020a; Korber et al., 2020; Laamarti et al., 2020; Mercatelli and Giorgi, 2020; Mercatelli et al., 2021; Popa et al., 2020). This extensive diversification has resulted in widespread variants such as B.1.1.7 (alpha), B.1.351 (beta), and B.1.617.2 (delta) (Cherian et al., 2021; Frampton et al., 2021; Tegally et al., 2021). Although the delta lineage was not yet present in the human population during the first year of the pandemic, it is of the utmost importance to continually interrogate the relationship between emerging SARS-CoV-2 variants and the adaptive immune system (Tarke et al., 2021b). In addition, it is important to highlight here that the pool of mutations observed in SARS-CoV-2 sequences were shown to be associated with a remarkably high proportion of cytidine-to-uridine (C-to-U) changes that were hypothesized to be induced by members of the APOBEC RNA-editing enzyme family (van Dorp et al., 2020a; Di Giorgio et al., 2020; Klimczak et al., 2020; Kosuge et al., 2020; Li et al., 2020; Matyášek and Kovařík, 2020; Rice et al., 2020; Simmonds, 2020; Wang et al., 2020). Since shown for other viruses (Grant and Larijani, 2017; Monajemi et al., 2014), we reasoned that the putative action of such host enzymes during the first year of the pandemic could lead to the large-scale escape from immunodominant and protective SARS-CoV-2-specific T cell responses, thereby potentially compromising their effectiveness to control the virus at the population scale.
In this study, we report a comprehensive study of the global genetic diversity of SARS-CoV-2 to expose the impact of mutation bias on epitope presentation and HLA-restricted T cell response within the first year of the pandemic, from December 2019 to December 2020. More specifically, we asked the following questions: (1) what are the impact of SARS-CoV-2 prevalent mutations detected across the global human population on the repertoire of validated SARS-CoV-2 T cell targets, with specific emphasis on CD8+ T cell epitopes? and (2) are mutational patterns in the genomic and proteomic composition of SARS-CoV-2 indicative of disrupted (or enhanced) epitope presentation and T cell immunity in human populations? By answering these questions, we provide a theoretical framework to understand how SARS-CoV-2 mutants have shaped T cell immunity to evade effective T cell immune responses at the population level during the first year of the pandemic, i.e., without mass-vaccination-induced immune pressure on viral evolution and adaptation.
Results
The global diversity of SARS-CoV-2 genomes influences the repertoire of T cell targets
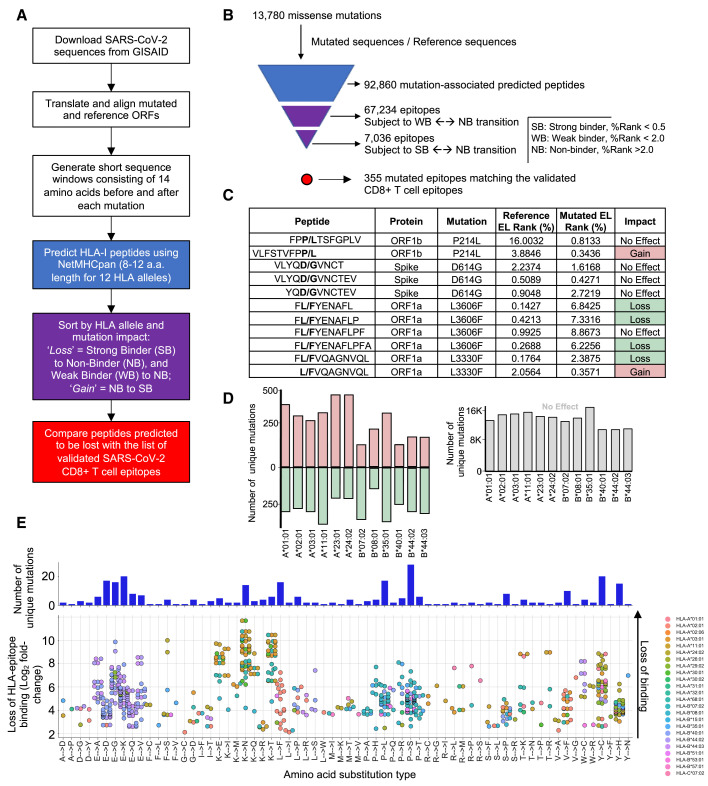
As of May 2021, nearly 1.7 million complete SARS-CoV-2 genome assemblies are publicly available via the Global Initiative on Sharing All Influenza Data (GISAID) repository. In the context of this large-scale effort, we performed a global analysis of SARS-CoV-2 genomes to assess whether mutations that emerged during the first year of the pandemic could disrupt HLA binding of clinically relevant SARS-CoV-2 CD8+ T cell epitopes. First, we identified missense mutations by aligning 330,246 high-quality consensus SARS-CoV-2 genomic sequences (GISAID; December 31st, 2020, prior to mass vaccination) to the reference sequence, Wuhan-1 SARS-CoV-2 genome (Figure 1 ). We found a total of 13,780 mutations identified in at least 4 SARS-CoV-2 genomes/individuals from GISAID, including 1,721 unique amino acid mutations in the S protein, with D614G as the most frequent one (94%) (Korber et al., 2020) (Tables S1 and S2). Next, we implemented a bioinformatics pipeline to assess the impact of these mutations on HLA binding for 620 unique SARS-CoV-2 HLA class-I epitopes that were recently reported to trigger a CD8+ T cell response in acute or convalescent COVID-19 patients (Quadeer et al., 2021; Tarke et al., 2021a) (see STAR Methods). On average, we found that the predicted binding affinity of 181 of these SARS-CoV-2 epitopes (30%) for common HLA-I alleles was reduced by ∼100-fold (Table S3; Figure 1). It is also apparent that mutations negatively impacted the HLA binding affinity of 56 (31%) and 19 (10%) CD8+ T cell epitopes located in the immunodominant S and N proteins, respectively (Figures 2A and 2B). Notably, a gap in the N protein, composed of a serine-rich region, is associated with higher mutation rate and a marked lack of predicted T cell epitopes and response (Figure 2B). Epitopes located in the RBD vaccine locus were also impacted by mutations (Figure 2C).
Figure 1.
Impact of SARS-CoV-2 mutations on CD8+ T cell epitopes
(A) Bioinformatic pipeline for the prediction of SARS-CoV-2 mutated class I peptides associated to 12 common HLA alleles.
(B) Pyramidal graph showing the number of (1) missense mutations in SARS-CoV-2 genomes, (2) predicted class I mutated peptides, (3) predicted class I peptides subject to Weak Binder (WB) to non-binder (NB) and strong binder (SB) to NB transition (epitope loss category), and (4) predicted class-I mutated peptides matching reference CD8+ T cell epitopes that have been experimentally validated.
(C) Representative examples of predicted class-I mutated peptides and the impact of the identified amino acid mutation (bold) on peptide binding to a given HLA-I allele. Reference and mutated EL (eluted ligand) rank (%) generated by NetMHCpan 4.1 EL is indicated for individual predictions. Gain = NB to SB (pale red); loss = SB to NB (pale green).
(D) Left panel: number of unique mutations leading to “gain” or “loss” of class-I peptides for the indicated HLA-I alleles. Right panel: number of unique mutations showing no effect on peptide binding for the indicated HLA-I alleles.
(E) Frequency of amino acid substitution types leading to loss of HLA binding for experimentally validated SARS-CoV-2 CD8+ T cell epitopes (from Quadeer et al., 2021). Mutations considered were those detected in more than 4 individuals (GISAID) and predicted to lead to a strong loss of HLA-epitope binding for common HLA-I alleles. Top: number of unique missense mutations for various amino acid substitution types. Bottom: Log2 fold change (mutated/reference) of predicted loss of HLA-epitope binding (NetMHCpan4.1 %Rank) for the various amino acid substitution types. Each dot represents an epitope pair (mutated/reference). Color indicates HLA-I alleles affected by the mutations.
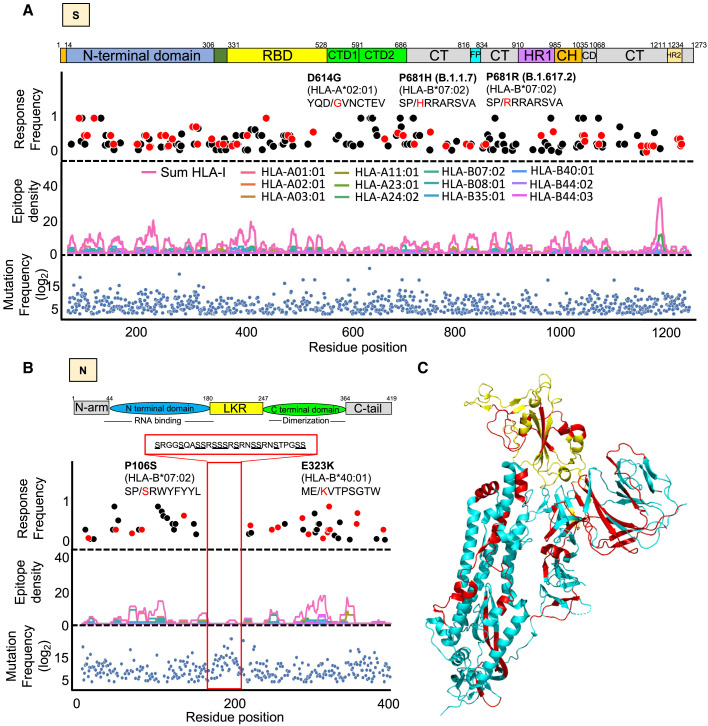
Figure 2.
Distribution of CD8+ T cell epitopes and their mutated variants across the immunodominant spike (S) and nucleocapsid (N) antigens
(A and B) Lower panel: blue dots showing all mutations that occurred in at least 4 SARS-CoV-2 genomes (GISAID). Middle panel: epitope density showing the overlap of HLA class-I epitopes predicted within the 1st percentile for 12 queried HLA-I molecules. Upper panel: dots showing the frequency of CD8+ T cell response as determined from multiple studies aggregated in Quadeer et al. (2021). Red dots are mutated epitopes wherein the mutation event led to a predicted loss of binding. Sequences of specific epitopes are shown with the mutant amino acid in red. The red box in the N protein highlights a serine-rich region associated with no T cell response, low epitope density, and high mutation frequency.
(C) 3D structure of the S glycoprotein (Moderna vaccine) and highlighted in yellow is the receptor binding domain (Pfizer vaccine). Shown in red are mutated epitopes wherein mutation events led to a predicted loss of HLA binding.
Loss of epitope binding for commonly expressed HLA class-I molecules was validated in vitro for a subset of representative SARS-CoV-2 epitopes (Figure S1). Of relevance, we found that the common D614G mutation in the S protein is linked to a 15-fold decrease in the binding affinity for the mutated HLA-A∗02:01 epitope YQGVNCTEV when compared with the reference/unmutated epitope YQDVNCTEV (Figures S1A and S1B). Our analysis also identified a mutation in the HLA-B∗07:02-restricted N105 epitope SPRWYFYYL, which is one of the most immunodominant SARS-CoV-2 epitope (Ferretti et al., 2020; Kared et al., 2021; Saini et al., 2021; Schulien et al., 2021; Sekine et al., 2020; Tarke et al., 2021a). Although relatively rare (found in only two genomes), the mutation in the N105 epitope consists of P→S at anchor residue position P2 (P106S: SPRWYFYYL → SSRWYFYYL) (Figure 2B) and is predicted to decrease HLA epitope binding by 47-fold (Figure 4D), thereby likely reducing the breadth of the immune response in B∗07:02 individuals carrying this mutation. Moreover, our global analysis validated the presence of two previously reported CD8+ T cell mutated epitopes (i.e., GLMWLSYFI → GFMWLSYFI, found in 38 genomes, and MEVTPSGTWL → MKVTPSGTWL, found in 23 genomes), which were shown to lose binding to HLA-A∗02:01 and -B∗40:01, respectively, in addition to disrupt epitope-specific CD8+ T cell response in COVID-19 patients (Figure S2) (Agerer et al., 2021). Together, these results demonstrate that mutations driving the global genomic diversity of SARS-CoV-2 can drastically disrupt HLA binding of clinically relevant CD8+ T cell epitopes, including epitopes encoded by the immunodominant S and N antigens, therefore affecting epitope-specific T cell responses in COVID-19 patients.
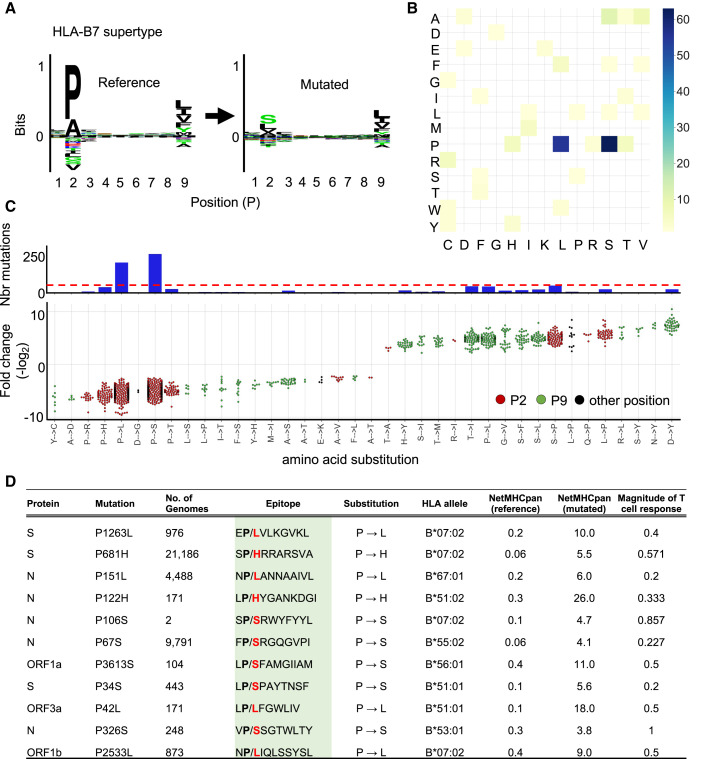
Figure 4.
Mutation of P at the anchor residue position for B7 supertype-associated epitopes
(A) (Left panel) Motif view of SARS-CoV-2 reference peptides predicted to bind B7 supertype molecules (HLA-B∗07:02, -B∗35:03, -B42:02, -B∗5101, -B∗53:01, -B∗54:01, -B∗55:01, -B∗56:01, and -B∗67:01). (Right panel) Motif view of the corresponding mutated peptides.
(B) Heatmap showing the frequency of specific amino acid substitutions between reference and mutated peptides.
(C) Graph showing the number of mutations (upper panel; y axis) leading to specific amino acid substitutions (x axis) at anchor residue positions P2 (red dots) and P9 (green dots) or elsewhere (black dots). Dotted red line indicate the cutoff used to define dominant substitutions. The lower panel shows fold changes for individual amino acid substitutions.
(D) Experimentally validated CD8+ T cell epitopes (from Quadeer et al., 2021) that are affected by the lost of a P residue. Mutated epitopes encoded by S, N, open reading frame (ORF) 1a, 1b, and 3a are shown as representative examples. Effect of the P→X substitutions on predicted epitope-binding affinities (NetMHCpan 4.1 %Rank) is shown. Data of magnitude of T cell response for reference epitopes were obtained from Quadeer et al. (2021).
In addition to mutations leading to a loss of HLA epitope binding, we identified a significant number of mutations predicted to enhance the presentation of peptides by their respective HLA molecules, leading to a “gain” of binding (Figures 1C, 1D, and S3). Because the unmutated epitopes are predicted to be non-HLA binders, these mutations were not searched against the list of known validated epitopes, which consist of strong-HLA-binding reference epitopes. Whether SARS-CoV-2 mutations predicted to increase HLA epitope binding can enhance T cell responses to control the virus in COVID-19 patients remains to be determined experimentally.
Amino acid mutational biases shape the global diversity of SARS-CoV-2 proteomes
While analyzing the impact of the mutational landscape of SARS-CoV-2 on experimentally validated CD8+ T cell epitopes, we observed that specific mutation types were over-represented while others were under-represented (Figures 1E, S1C, and S1D). For instance, we found that 31% of the prevalent mutations (i.e., found in >100 genomes) predicted to abrogate the presentation of experimentally validated CD8+ T cell epitopes (Quadeer et al., 2021) led to the removal of proline residues (Pro→X) (Figure S1C). These observations led to the hypothesis that the disproportionate presence of certain mutation types among mutations predicted to disrupt peptide presentation could originate from biases in the proteome of SARS-CoV-2 mutants. To further investigate whether specific amino acid mutational biases could be observed globally in the proteome of SARS-CoV-2 mutants, we asked whether certain amino acid residues were preferentially removed from or introduced into the global proteomic diversity of SARS-CoV-2, thereby potentially diversifying CD8+ T cell epitopes in a systematic manner.
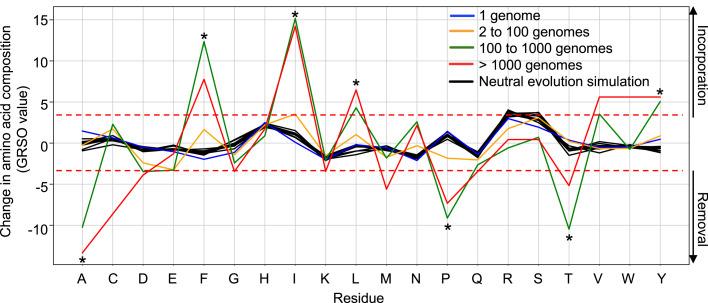
To test this, we computed all residue substitutions (amino acid removed and introduced) found in SARS-CoV-2 proteomes and calculated global residue substitution output (GRSO) values, i.e., the percentage difference in overall amino acid composition for individual amino acids (see STAR Methods for details). GRSO values were computed for mutations found at various frequencies in GISAID (i.e., found in only 1 genome, 2 to 100 genomes, 100 to 1,000 genomes, and >1,000 genomes) (Figure 3 ). Distinct mutational patterns at the amino acid level were observed among mutations detected in more than 100 genomes/individuals (Figure 3), referred to in this study as “prevalent mutations” (see STAR Methods and Table S2). Among those mutations, the amino acids alanine (A), proline (P), and threonine (T) were preferentially removed by 10.2% (p = 1.2 × 10−13), 9.1% (p = 1.6 × 10−15), and 10.5% (p = 1.3 × 10−14), respectively. In contrast, phenylalanine (F), isoleucine (I), leucine (L), and tyrosine (Y) were preferentially introduced by 13.4% (p = 2.0 × 10−17), 15.2% (p = 2.4 × 10−17), 4.3% (p = 6.3 × 10−11), and 5.0% (p = 7.0 × 10−14), respectively (Figure 3). Statistical significance of these GRSO values was assessed by generating simulated samples of 1,000 SARS-CoV-2 genomes evolving under neutrality (n = 10 replicates) using the SANTA-SIM algorithm (Jariani et al., 2019) (see STAR Methods for details). Of note, mutations that were detected in 2 to 100 individuals appeared significantly more neutral, with none of the mutational patterns enriched above the selected cutoff values (fold-change >4; p value < 1 × 10−11). Thus, our results show that specific amino acid residues were preferentially removed or introduced in the proteome of SARS-CoV-2 mainly by prevalent mutations. Therefore, we introduce the notion that the global diversity of SARS-CoV-2 proteomes is shaped by specific amino acid mutational biases. Such biased amino acid compositions generated by prevalent mutations may have a systematic impact on epitope processing and presentation to shape SARS-CoV-2 T cell immunity in human populations. To address this systematic impact, all downstream analyses described in this study were performed from the set of 1,933 prevalent mutations (identified in >100 genomes) listed in Table S2.
Figure 3.
Global amino acid mutational biases in SARS-CoV-2 proteomes
A total of 330,246 SARS-CoV-2 genomes were translated into protein sequences and analyzed for the identification of any amino acid mutational bias. Amino acid residues (x axis) that were removed and introduced in SARS-CoV-2 variants are presented by negative and positive percentage difference in overall amino acid composition (GRSO values; y axis), respectively. Analysis of mutational biases was performed for mutations occurring at various frequencies: 1 genome (blue line), 2 to 100 genomes (yellow line), 100 to 1,000 genomes (green line), and more than 1,000 genomes (red line). Simulations of neutral evolution simulation (random mutations; black lines) were performed using the SANTA-SIM algorithm and serve as control for assessing the statistical significance of the observed pattern for individual amino acid residues. The dotted red lines show the cutoff values (fold-change >4; p value < 1×10−11) that were used to define the residues that were preferentially removed or introduced (asterisk).
Prominent removal of proline residues leads to a predicted global loss of epitopes presented by HLA-B7 supertype molecules
The association of peptides with the binding groove of HLA molecules largely relies on the presence of anchor residues, also known as peptide-binding motifs (Falk et al., 1991). Hundreds of different peptide-binding motifs have been reported over the last decades (Gfeller and Bassani-Sternberg, 2018). Overlapping binding motifs are qualified as “HLA supertypes” on the basis of their main anchor specificity (Greenbaum et al., 2011; Sidney et al., 2008). Of relevance here, proline acts as a critical anchor residue at position P2 for epitopes presented by HLA-B7 (B7) supertype molecules, which include a wide range of commonly expressed HLA-B alleles in humans, i.e., HLA-B∗07, -B∗15, -B∗35, -B∗42, -B∗51, -B∗53, -B∗54, -B∗55, -B∗56, -B∗67, and B∗78 (Sidney et al., 2008). In fact, the B7 supertype covers ∼35% of the human population (Franciscodos et al., 2015). Hence, we reasoned that the global removal of proline residues observed in the proteome of prevalent SARS-CoV-2 mutants (Figure 3) could drastically compromise T cell epitope binding to B7 supertype molecules, thereby potentially interfering with SARS-CoV-2 T cell immunity in a relatively large proportion of the human population.
Due to the preferential removal of proline by prevalent mutations, we investigated the extent at which proline residues were substituted at anchor binding position P2 and, consequently, resulted in loss of epitopes presented by B7 supertype molecules. To answer this, we performed the following four steps: (1) we applied NetMHCpan 4.1 (Reynisson et al., 2020) using the reference and mutated SARS-CoV-2 genomes to generate a list of all possible reference/mutated peptide pairs (8–11 mers) predicted to bind 16 common HLA-B types that belong to the B7 supertype family (Figure S4B). (2) We analyzed all reference/mutated peptide pairs, along with their differential predicted binding affinities to quantitatively identify HLA strong binder (SB) to non-binder (NB) transitions [(SB) NetMHCpan %rank < 0.5 to (NB) NetMHCpan %rank >2]. (3) We categorized all peptide pairs based on the mutation type (amino acid X → amino acid Y) and the position of the mutation within the peptide sequence. (4) Lastly, we quantified the number of reference/mutated peptide pairs and the associated fold-change in predicted binding affinity for each category. Our results show that prevalent mutations predicted to impact the presentation of peptides by the B7 supertype are dominated by P→L (p = 8.6 × 10−35) and P→S (p = 3.4 × 10−24) substitutions at anchor residue position P2 (Figures 4 A and 4B). Reference/mutated peptide pairs from these categories were the most abundant, with >250 mutated peptides per category (Figure 4C). P→L and P→S mutations resulted, on average, in a 61-fold reduction in predicted HLA binding affinity for a representative set of clinically validated CD8+ T cell epitopes (Figure 4D).
In addition to the dominant P→S/L substitution type, other P→X substitutions were observed, including in variants of concern. For instance, our most recent analysis (August 2021) of mutations found in the pangolin B.1.1.7 variant (alpha) showed that the P681H mutation found in the spike protein led to disrupted association of the reference epitope SPRRARSVA for several HLA-B7 types. In fact, the P-to-H substitution resulted in a strong loss of epitope binding predicted for 7/16 HLA-B7 types tested. Notably, the more recent B.1.617.2 (delta) variant was also found to disrupt the same epitope SPRRARSVA via a proline-to-arginine mutation in the spike protein (Spike:P681R) (Figure 2A). Thus, our results strongly suggest that biased substitutions of proline residues in the proteome of SARS-CoV-2 shapes the repertoire of epitopes presented by B7 supertype, including epitopes encoded by the genome of the B.1.1.7 and B.1.617.2 variants. This finding lets us to propose that mutation biases found in SARS-CoV-2 may contribute to CD8+ T cell epitope escape in a B7 supertype-dependent manner.
The mutational landscape of SARS-CoV-2 enables disruption or enhancement of epitope presentation in an HLA-supertype-dependent manner
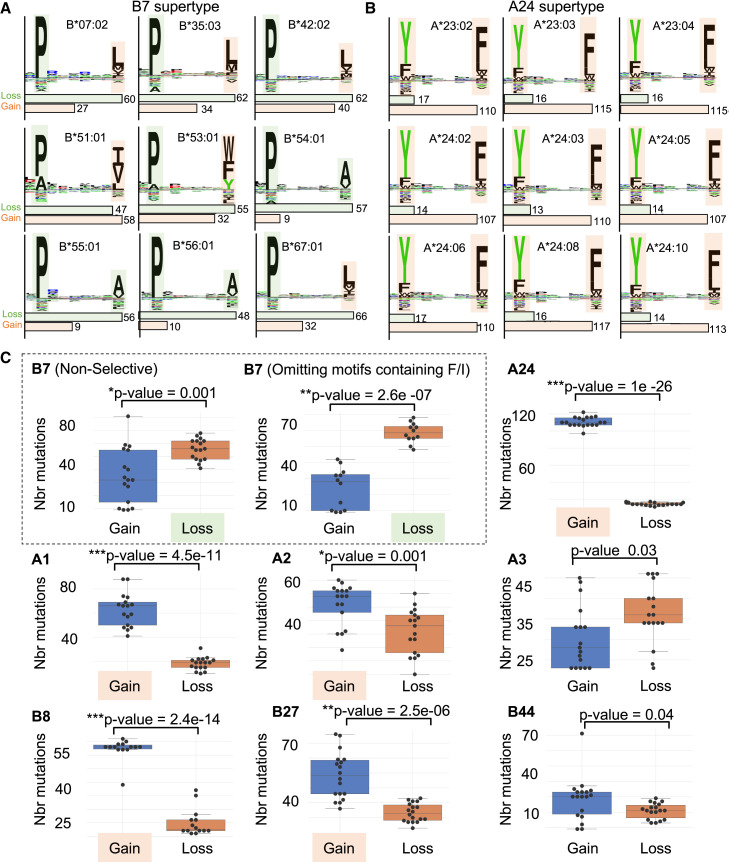
We found that specific amino acid residues were preferentially removed (proline, alanine, and threonine) or introduced (isoleucine, phenylalanine, leucine, and tyrosine) in SARS-CoV-2 proteomes (Figure 3). Most of these amino acids act as key epitope anchor residues for multiple HLA class-I supertypes (Figure S4). For instance, phenylalanine and tyrosine are key anchor residues for all known A∗24 alleles of the A24 supertype family, whereas proline is known to play a critical role in the anchoring of epitopes to alleles of the B7 supertype family (Figure 5 ). Therefore, one would expect the introduction of phenylalanine and tyrosine in SARS-CoV-2 proteomes to facilitate peptide presentation by A24, whereas the removal of proline would disrupt peptide presentation by B7. With this concept in mind, we hypothesized that the distinct amino acid mutational biases found throughout prevalent SARS-CoV-2 mutations could systematically mold epitope presentation in an HLA-supertype-dependent manner.
Figure 5.
Loss or gain of SARS-CoV-2 mutated epitopes for different HLA class-I supertypes
(A and B) Motif views showing established epitope-binding motifs for different HLA-I alleles that belong to the HLA-B7 (A) and HLA-A24 (B) supertype family. Shaded squares highlight anchor residues that are preferentially removed (pale green) or introduced (pale orange) in SARS-CoV-2 proteomes (related to Figure 3), respectively. Histograms below the motif views indicate the number of frequent mutations (identified in at least 100 individuals) leading to the loss or gain of epitopes.
(C) “Gain/loss plots” showing number of mutations (y axis) leading to a significant loss (pale green) or gain (pale orange) of epitopes for different HLA class-I supertypes. Each black dot represents the number of mutations associated with gain and loss of epitopes for a given HLA-I allele. Between 14 to 19 alleles per supertype (Figure S4) were used to generate the graphs and p values (∗p ≤ 0.001, ∗∗p < 1e−5, ∗∗∗p < 1e−10).
In order to compare supertypes with each other, we generated a “gain/loss plot” for each supertype assessed (Figure 5C). Gain/loss plot were generated by computing the number of mutations that resulted in “gain” or “loss” of epitopes for representative class-I alleles selected for each supertype (see STAR Methods for details). “Gain” was assigned for mutated epitopes that were predicted to transit from non-HLA binders (NetMHCpan %rank >2) to strong HLA binders (NetMHCpan %rank < 0.5), whereas “loss” was assigned for mutated epitopes that were predicted to transit from strong HLA binders to non-HLA binders. Our analysis shows that most supertypes preferentially gain new epitopes as a result of SARS-CoV-2 mutations: A1 (p = 4.5 × 10−11), A2 (p = 0.001), A24 (p = 1.0 × 10−26), B8 (p = 2.4 × 10−14), B27 (p = 2.5 × 10−6). Preferential loss of epitopes was only shown to be statistically significant for B7 supertype (p = 0.0012). Note that we explain the relatively low statistical value obtained for B7 supertype by the presence of isoleucine and phenylalanine (preferentially introduced in SARS-CoV-2 proteomes; see Figure 3) at anchor residue P9 for certain HLA types (namely HLA-B∗51:01 and HLA-B∗53:01) (Figure 5A). In fact, omitting motifs containing isoleucine or phenylalanine increased the significance of epitope lost versus gained (p = 2.6 × 10−7) (Figure 5C). Together, our results show that the amino acid mutational biases that feature the global diversity of SARS-CoV-2 proteomes can positively or negatively affect binding affinities of mutated epitopes for a wide range of HLA class-I molecules in a supertype-dependent manner.
The C-to-U point mutation bias largely drives diversification of SARS-CoV-2 T cell epitopes
Next, we sought to better understand the genetic determinants that drive the association between epitope presentation and the amino acid mutational biases found in the SARS-CoV-2 population. To this end, we analyzed the abundance of all the possible nucleotide mutation types (i.e., A-to-C, A-to-G, A-to-U, C-to-A, C-to-G, C-to-U, etc.). This analysis indicates that C-to-U is the most common mutation type (43%), followed by G-to-U (28%), as well as A-to-G, G-to-A, and U-to-C
(from 9.7% to 11.6%) (Figure S5A), in line with observations made by others (Di Giorgio et al., 2020; Klimczak et al., 2020; Kosuge et al., 2020; Li et al., 2020; Matyášek and Kovařík, 2020; Rice et al., 2020; Simmonds, 2020; Wang et al., 2020).
Next, we aimed to determine the contribution of these different nucleic acid mutation types to the global mutational pattern observed at the amino acid level in Figure 3. To do so, we generated simulated population samples of 1,000 SARS-CoV-2 genomes using SANTA-SIM (Jariani et al., 2019), applying various extents of mutational biases corresponding to the two most common mutation types observed (i.e., C-to-U and G-to-U). The resulting simulated viral populations were then analyzed to elucidate the global amino acid mutational pattern engendered by these simulated nucleic acid point mutation biases and whether they recapitulate the observed patterns. Indeed, our data show that the mutational pattern resulting from the simulated C-to-U bias very closely mimicked the mutational pattern observed in the real-life dataset (Figure 6 A). Namely, the in silico introduction of a C-to-U mutation bias resulted in the preferential removal of alanine, proline, and threonine, by 6.7% (p = 5.1 × 10−11), 6.9% (p = 1.2 × 10−11), and 8% (p = 4.8 × 10−12), respectively, as well as the introduction of isoleucine and phenylalanine by 8.2% (p = 1.3 × 10−8) and 5.2% (p = 4.3 × 10−11), respectively (Figure 6A). The G-to-U mutation bias also contributed to the introduction of isoleucine and phenylalanine (Figure S5B). Together, these results show that the predominant C-to-U point mutations largely contribute to shaping the global proteomic diversity of SARS-CoV-2.
Figure 6.
The C-to-U point mutation bias largely drives the diversity of SARS-CoV-2 proteomes and CD8+ T cell epitopes
(A) Comparison of global amino acid mutational patterns generated from real-life versus simulated SARS-CoV-2 genomes. Amino acid residues (x axis) that were removed (y axis; negative values) and introduced (y axis; positive values) in real-life (red line) versus simulated (black, blue, and green lines) SARS-CoV-2 are presented by percentage difference in overall amino acid composition (y axis; GRSO values), respectively. Evolution of SARS-CoV-2 was simulated by introducing various extents of C-to-U biases, i.e., ×1, ×15, and ×20 (n = 10). The red line shows the pattern obtained from mutations identified in more than 100 SARS-CoV-2 genomes, related to Figure 3.
(B) (Top) Pie chart showing the proportion of nucleotide substitution types from the list of validated CD8+ T cell epitopes in Quadeer et al. (2021). (Bottom) Pie chart showing the proportion of nucleotide substitution types from the list of validated CD8+ T cell epitopes that belong to the B7 supertype family in Quadeer et al. (2021).
(C) Schematic illustrating the C-to-U-mediated epitope escape model. The observed P-to-S substitution in the immunodominant SPRWYLFYYL epitope from the N antigen is shown as an example.
Given the significant impact of the C-to-U point mutation bias on the amino acid content of SARS-CoV-2 proteomes, we reasoned that C-to-U could be the main driver shaping the repertoire and diversification of SARS-CoV-2 T cell targets in human populations, including targets presented by the particularly interesting B7 supertype molecules. To investigate this, we used all the SARS-CoV-2 CD8+ T cell epitopes that were experimentally validated using peripheral blood mononuclear cells (PBMC) of acute and convalescent COVID-19 patients (Quadeer et al., 2021; Tarke et al., 2021a) and matched them with their corresponding nucleic acid sequence found in reference/mutated genome pairs. We then calculated the frequency of the various mutation types (i.e., A-to-C, A-to-G, A-to-U, C-to-A, C-to-G, C-to-U, etc.) coding for the mutated form of those experimentally validated CD8+ T cell epitopes. We found that C-to-U and G-to-U were the two main mutation types leading to mutated epitopes, both accounting for 37% of all mutation types among prevalent mutations (>100 individuals) (Figure 6B). In addition, our data show that 62% of the prevalent mutations predicted to disrupt the presentation of epitopes by HLA alleles for the B7 supertype were found to derive from the C-to-U mutation type (Figure 6B). These results strongly suggest that the dominant C-to-U point mutation bias found among prevalent SARS-CoV-2 mutants has the potential to contribute to shaping the repertoire of SARS-CoV-2 T cell epitopes in B7 supertype individuals across human populations. Collectively, our study lets us to propose the model that C-to-U editing enzymes play a fundamental role in shaping the mutational landscape dynamics of SARS-CoV-2 CD8+ T cell targets in humans (Figure 6C), and hence, may contribute to molding T cell immunity against COVID-19 at the population level.
Discussion
Mutations contribute to the genetic diversity of SARS-CoV-2 and shape the progression of the COVID-19 pandemic (van Dorp et al., 2020b, 2020a; Popa et al., 2020). T cells are key players controlling COVID-19 disease severity. Therefore, determining whether and how the mutational landscape of SARS-CoV-2 shapes HLA-restricted T cell responses is fundamentally important. Traditionally, most studies have investigated how viral mutations are shaped by T cell response in the context of HLA-typed cohort patients. This type of approach sought to determine the evolutionary relationship between HLA genotypes and variants of long-standing viruses such as HIV-1 (Brumme et al., 2007; Kawashima et al., 2009) and influenza (Woolthuis et al., 2016). In the case of a novel virus such as SARS-CoV-2, such a relationship remains to be established and does not constitute the scope of our work. Here, we rationalized that an alternative approach to interrogating SARS-CoV-2 epitope-associated variants is by investigating the global genomic and proteomic diversity of SARS-CoV-2 for any outstanding mutational biases, and then, assessing the relationship between such biases and epitope presentation for a broad set of HLA alleles. In other words, in this study, we did not seek to understand how viral mutations are shaped by T cell immunity but rather to understand how mutational biases in SARS-CoV-2 may have shaped T cell immunity at the population level during the first year of the pandemic. This approach was possible thanks to an unprecedented number of SARS-CoV-2 genome sequences available for downstream analysis. Our approach is universal and could be applied to other epidemic or pandemic viruses in the future, given the development of distinct, prevalent mutational biases. Our global approach has led to several conclusions to help understand how the increasing genomic diversity of SARS-CoV-2 may shape T cell immunity in human populations. Our findings have important implications that are discussed below in the context of disease severity, viral evolution, and vaccine resistance.
In this study, we found that prevalent SARS-CoV-2 mutations are governed by defined mutational patterns, with C-to-U being a predominant mutation type, as previously shown by others (Di Giorgio et al., 2020; Klimczak et al., 2020; Kosuge et al., 2020; Li et al., 2020; Matyášek and Kovařík, 2020; Rice et al., 2020; Simmonds, 2020; Wang et al., 2020). In fact, we show that the C-to-U mutation bias in SARS-CoV-2 genomes has a remarkably intimate relationship with the observed amino acid mutational biases, indicating that C-to-U mutations largely contribute to the global proteomic diversity of SARS-CoV-2. Moreover, we show that this mutational bias leads to the preferential substitution of proline residues with leucine or serine residues in the P2 anchor position of SARS-CoV-2 CD8+ T cell epitopes, and hence, drastically compromise epitope binding to B7 supertype molecules. These molecules, which represent ∼35% of the human population, preferentially bind epitopes with proline at P2 (Franciscodos et al., 2015). Therefore, the C-to-U mutational bias observed among prevalent mutants may partially disrupt SARS-CoV-2 T cell immunity in a very significant proportion of the human population. Noteworthy, this impact of C-to-U mutations on B7-dependent epitope escape was somehow predictable. In fact, proline residues originate from codons that are highly rich in C, whereas serine and leucine residues originate from codons that are rich in U. One could therefore predict, at least to some extent, that a strong C-to-U bias would lead to proline-to-leucine or proline-to-serine substitutions. Thus, this study highlights the impact of viral mutational biases and codon usage in shaping the diversity of CD8+ T cell targets. The impact of the loss of several B7 epitopes on the immune response of an individual, however, remains unclear.
In this study, we observed that proline→X mutations were more enriched among prevalent mutations (>100 genomes) predicted to abrogate the presentation of experimentally validated CD8+ T cell epitopes than across the global mutation landscape of SARS-CoV-2 proteomes (31% and 9.1%, respectively). These two percentages are in fact indicative of different phenomena. The former reflects the susceptibility of certain HLA alleles to specific mutational patterns (the removal of proline in this case), whereas the latter reflects the overall mutational biases observed across SARS-CoV-2 proteomes. This noticeable difference may suggest that certain mutation types play a particularly important role in HLA-type-dependent cytotoxic T lymphocyte (CTL) escape. This concept becomes evident when considering the 13 common alleles investigated in this study. The detrimental impact of proline→X mutations on the presentation of peptides by B7 alleles is reflected in the higher proportion of proline→X mutations (31%) leading to the loss of epitopes. This being said, it is important to realize that we do not make the claim that the presence of proline-to-leucine or proline-to-serine mutations in the SARS-CoV-2 proteomes depend on patients being B7 supertype positive or that the B7 supertype drives the evolution of proline-to-leucine/serine mutations. We do, however, demonstrate that the prevalent mutations currently in circulation are enriched for proline-to-leucine/serine, and our in silico predictions suggest that the high occurrence of this mutation type leads to widespread hinderance of epitope presentation in B7-supertype-positive individuals.
A key question to address is to what extent does the C-to-U bias drive SARS-CoV-2 evolution and adaptation over the course of the ongoing pandemic. As proposed by others, the most likely explanation for the observed C-to-U bias is the action of the host-mediated RNA-editing APOBEC enzymes, a family of cytidine deaminases that catalyze deamination of cytidine to uridine in RNA (van Dorp et al., 2020a; Di Giorgio et al., 2020; Kosuge et al., 2020; Olson et al., 2018; Salter et al., 2016). In this regard, APOBEC activity has been shown to broadly drive viral evolution and diversity, including in human immunodeficiency virus (HIV) (Albin et al., 2010; Cuevas et al., 2015; Haché et al., 2008; Jern et al., 2009; Peretti et al., 2018; Sadler et al., 2010; Wood et al., 2009). In fact, APOBEC-induced mutations driving the evolution and diversification of HIV-1 were shown to have an intimate relationship with T cell immunity (Kim et al., 2014; Wood et al., 2009). Those studies have shown that the impact of APOBEC-induced mutations may result in either a decrease or increase of CD8+ T cell recognition and that the direction of this response is dictated by the HLA context (Casartelli et al., 2010; Grant and Larijani, 2017; Kim et al., 2014; Monajemi et al., 2014; Squires et al., 2015; Wood et al., 2009). This is very much in line with our findings. Indeed, we showed that amino acid mutation biases in SARS-CoV-2 proteomes generally positively affect epitope binding for various HLA class-I supertypes, and most strikingly for A24, whereas B7 is the only supertype that is consistently negatively affected by the mutation biases given the markable loss of proline residues in SARS-CoV-2 proteomes. Together, our results raise the important hypothesis that host-mediated RNA-editing systems shape the repertoire of SARS-CoV-2 T cell epitopes in a positive and negative HLA-dependent manner.
Another question is whether populations of B7 supertype individuals represent an advantageous reservoir for the virus to evolve toward more transmissible variants. As the genetic diversity of the SARS-CoV-2 population continue to increase, and as new variants emerge, our global analysis suggests that the probability for SARS-CoV-2 epitopes to escape CD8+ T cell immunosurveillance is higher in B7 individuals compared with A24 individuals. In fact, mutated epitopes are predicted to be unfavorably and favorably presented by B7 and A24 supertypes, respectively (Figure 5). The supertype dependency is important here because it suggests that T cell responses are shaped differently across different human populations in response to infection by mutated forms of SARS-CoV-2. For instance, the predicted model lets us hypothesize that, within the first year of the pandemic (from December 2019 to December 2020), human populations expressing the A24 supertype at higher frequency (e.g., >90% of people in specific geographical regions in Taiwan) may likely mount a T cell response upon infection by mutated forms of SARS-CoV-2 that will not be as readily disrupted by mutation events, in comparison with individuals expressing the B7 supertype (i.e., ∼35% of the human population) (Franciscodos et al., 2015). Interestingly, a recent computational study corroborated the propensity of HLA-B∗07:02 to lose epitopes due to SARS-CoV-2 variants (Nersisyan et al., 2021). Our proposed model may therefore act as a contributing factor addressing the global diversity of immunological responses against SARS-CoV-2 variants as the pandemic progresses. Several studies have indeed interrogated associations between HLA alleles and COVID-19 disease severity (Naemi et al., 2021; Pisanti et al., 2020; Tomita et al., 2020) as well as mutations and T cell evasion (Agerer et al., 2021; Geers et al., 2021; Motozono et al., 2021). However, to the best of our knowledge, this is the first study that proposes a connection between mutation biases, differential presentation of epitope variants (HLA supertype dependent), and variability in host responses to SARS-CoV-2 infection, all in the context of the continuously expanding genomic diversity of SARS-CoV-2 mutants. Additionally, the current study establishes a basis for investigating CTL-escape in the context of HLA (super)types strategically selected based on the diversification patterns of SARS-CoV-2.
With regard to the variants of concern, we noted that the B.1.1.7 (alpha) variant was predicted to lose the B7-supertype-associated, experimentally validated epitope SP/HRRARSVA as a result of a proline-to-histidine substitution. The B.1.617.2 (delta) variant was in fact also predicted to lead to the loss of the same epitope via a proline-to-arginine substitution (SP/RRRARSVA). As the B.1.617.2 variant has become the most widespread SARS-COV-2 lineage globally since July 2021, it would be of interest to experimentally interrogate the impact of this variant in the activation of CTLs in B7+ individuals. Although our study does not demonstrate that the disproportionate loss of proline across the SARS-CoV-2 mutation landscape is the cause for the increased infectivity of the discussed variants of concern, we propose that it may be a contributing factor in the context of certain populations. In this regard, while genomic surveillance is ongoing in different regions of the world, measuring the level of transmission of the B.1.1.7 and B.1.617.2 variants within geographical regions of the world with low B7 population densities and high A24 population densities (in Asia) or the opposite trend (in Sub-Saharan Africa) (http://www.allelefrequencies.net/top10freqs.asp) may provide insights into this concern. As new variants of concern continue to emerge and as new epitope data are continuously being generated (Grifoni et al., 2021), another interesting avenue would be to study the mutational patterns of those emerging variants and assess whether and how the potential loss of B7-associated epitopes in those specific variants impact T cell response in infected patients. Understanding the impact of losing several subdominant B7-associated epitopes versus one single immunodominant epitope could also be investigated in the context of those variants. In this regard, a particular attention was allocated in our study to the B∗07:02-restricted N105 epitope SPRWYFYYL. This epitope is of high interest as its immunodominance was experimentally demonstrated in many independent studies (Ferretti et al., 2020; Kared et al., 2021; Saini et al., 2021; Schulien et al., 2021; Sekine et al., 2020; Tarke et al., 2021a). Precisely, we found a rare mutation consisting of P→S at P2 of this epitope (SPRWYFYYL → SSRWYFYYL). Its occurrence was predicted to result in the complete abrogation of binding of the epitope to B∗07:02, thereby likely reducing the breadth of the immune response in individuals carrying this mutation. As such, we advise the community to carefully monitor this mutation in subsequent months. Moreover, it is also possible that B7 individuals respond less efficiently to the currently available vaccines, as genetic variants promoting B7 escape might favorably emerge in the future. The B7 supertype could therefore potentially represent a biomarker of vaccine resistance.
In summary, our study shows that mutation biases in the SARS-CoV-2 population diversify the repertoire of SARS-CoV-2 T cell targets in humans in an HLA-supertype-dependent manner. Hence, we provide a foundation model to help understand how SARS-CoV-2 may continue to mutate over time to shape T cell immunity at a global population scale. The proposed process will likely continue to influence the evolution and diversification of SARS-CoV-2 lineages as the virus is under tremendous pressure to adapt in response to mass vaccination.
Limitations and future directions
Our analyses focused on class-I molecules for which predictors are established to be more accurate in comparison with class II. HLA-C and non-classical HLA were not included in this study. Predictions were performed on the most common HLA class-I alleles and rare HLA alleles were not included. Study has been performed using the GISAID dataset available in December 31, 2020, i.e., first year of the pandemic, before mass vaccination. Our epitope binding results rely on in silico predictions using a method that has been widely benchmarked but is designed to predict peptide presentation rather than immunogenicity. Follow up experiments would need to be performed to further validate the proposed model. Priority follow up studies are (1) to investigate T cell response to SARS-CoV-2 mutants in large cohorts of B7 supertype-positive versus negative patients, and (2) to determine the direct role of APOBEC family proteins in modulation of SARS-CoV-2-specific T cell immunity. Moreover, this study lays the foundation to understand the evolutionary dynamics of pandemic viruses with a time 0/no vaccine-induced immune pressure start point. Employing SARS-CoV-2 as model provides an opportunity in future studies to look at the dynamic of the relationship between mutational patterns and HLA-restricted T cell immunity in real time. Kinetic analyses using the latest GISAID dataset, which includes 1.7M SARS-CoV-2 genomes as of May 2021, may lead to additional insights in this regard.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| Synthetic peptides | TC Peptide Lab | tcpeptidelab.com |
| Deposited data | ||
| Wuhan-Hu-1 RNA isolate | NCBI nuccore database | NCBI: NC_045512.2 |
| Structure of SARS-CoV-2 Spike Protein Trimer | Xiong et al., 2020 | PDB: 6ZP2 |
| GISAID | Freunde von GISAID e.V. | https://www.gisaid.org/ |
| Experimentally Validated SARS-CoV-2 T cell epitopes | Quadeer et al., 2021 | https://www.mckayspcb.com/SARS2TcellEpitopes/ |
| Supplemental information | Hamelin et al. | DOI: 10.5281/zenodo.5520066 |
| Software and algorithms | ||
| netMHCpan 4.1 | Reynisson et al., 2020 | https://services.healthtech.dtu.dk/service.php?NetMHCpan-4.1 |
| Python (v3.7) | Python Software Foundation | https://www.python.org/ |
| Santa-Sim | Jariani et al., 2019 | https://github.com/santa-dev/santa-sim |
| CoVescape | In-house algorithm | DOI: 10.5281/zenodo.5493359 |
Resource availability
Lead contact
Further information and requests should be directed to the lead contact, Dr. Etienne Caron (etienne.caron@umontreal.ca).
Materials availability
This study did not generate new materials.
Method details
Identification of SARS-CoV-2 mutations
All SARS-CoV-2 nucleotide sequences were acquired from the GISAID on 31/12/2021. A total of 330,246 SARS-CoV-2 sequences spanning 143 countries were acquired and analyzed. All sequences isolated from animals (including viral RNA isolated from bat, pangolin, mink, cat and tiger) were removed from the list and only high-quality sequences were further analysed. Consensus sequences were aligned to the reference sequence, Wuhan-1 (NC_045512.2) using minimap2 2.17-r974. All mapped sequences were then merged back with all others in a single alignment bam file. The variant calling was done using bcftools mpileup v1.91 in a haploid calling mode. Sequences were processed by batches of 1000 to overcome technical issues with very low-frequency variants. With the variant calling obtained for each batch, vcf-merge (from the vcftools suite) was used to merge all the variant calls across the entire dataset. A total of 24,220 variants in at least two consensus sequences were identified. Mutations appearing in only one genome were excluded as they are likely enriched for sequencing errors. A list of all missense mutations considered in our analyses is provided in Table S1. The 1,933 prevalent mutations observed in more than 100 genomes are also clearly shown in Table S2.
Prediction of mutated and reference CD8+ T-cell epitopes
Prediction of CD8+ T cell epitopes was carried out using netMHCpan 4.1 EL (Reynisson et al., 2020). For each unique missense mutation, short sequence windows consisting of 14 amino acids on either side of the mutation site were generated, containing either the reference or mutated amino acid. Working from the resulting 29-residue sequence windows (mutation +/- 14 residues), 811mers were predicted against the 12 most frequent HLA alleles within the global population (HLA-A∗01:01, HLA-A∗02:01, HLA-A∗03:01, HLA-A∗11:01, HLA-A∗23:01, HLA-A∗24:02, HLA-B∗07:02, HLA-B∗08:01, HLA-B∗35:01, HLA-B∗40:01, HLA-B∗44:02, and HLAB∗44:03). Briefly, the NetMHCpan 4.1 EL method relies on a neural network trained on both binding affinity as well as eluted ligand data to produce a likelihood score for a peptide to be an eluted ligand for the indicated HLA types. The likelihood score consists of a percentile rank (%rank) wherein predicted (weak) binders obtain a %rank below 2.0, whereas strong binder (SB) obtain a %rank below 0.5. Using this ranking system, only mutation-containing peptides where the mutated and/or the reference peptide were ranked as SB were considered for further analyses. Mutations causing percentile ranks to transition from strong HLA-binder (SB, netMHCpan %Rank < 0.5) to HLA non-binders (NB, netMHCpan %Rank > 2.0) were considered as leading to ‘Loss of binding’. Mutations causing predicted binding affinities to transition from NB to SB were considered as leading to ‘Gain of binding’. Selection of clinically validated CD8+ T-Cell epitopes
A list of validated CD8+ T Cell epitopes presented by both HLA-A and -B molecules were downloaded from https://www.mckayspcb.com/SARS2TcellEpitopes/ (as of January 2021). This database, developed by Dr. Matthew R. McKay and his team, contains compiled and catalogued validated T-cell epitope-HLA pairs from 13 studies aimed at identifying immunogenic SARSCOV-2 T-cell epitopes.
In vitro HLA-peptide binding assays
Peptide binding to class I HLA molecules was quantitatively measured using classical competition assays based on the inhibition of binding of a high affinity radiolabeled peptide to purified HLA molecules, as detailed elsewhere (Sidney et al., 2013). Briefly, HLA molecules were purified from lysates of EBV transformed homozygous cell lines by affinity chromatography by repeated passage over Protein A Sepharose beads conjugated with the W6/32 (anti-HLA-A, -B, -C) antibody, following separation from HLA-B and -C molecules by pre-passage over a B1.23.2 (antiHLA B, C) column. Protein purity, concentration, and the effectiveness of depletion steps was monitored by SDS-PAGE and BCA assay. Peptide affinity for respective class I molecules was determined by incubating 0.1-1 nM of radiolabeled peptide at room temperature with 1 μM to 1 nM of purified HLA in the presence of a cocktail of protease inhibitors and 1 μM B2microglobulin. Following a two-day incubation, HLA bound radioactivity was determined by capturing MHC/peptide complexes on W6/32 antibody coated Lumitrac 600 plates (Greiner Bioone, Frickenhausen, Germany). Bound cpm was measured using the TopCount (Packard Instrument Co., Meriden, CT) microscintillation counter. The concentration of peptide yielding 50% inhibition of the binding of the radiolabeled peptide was calculated. Under the conditions utilized, where [label]<[MHC] and IC50 ≥ [MHC], the measured IC50 values are reasonable approximations of the true Kd values. Each competitor peptide was tested at six different concentrations covering a 100,000-fold dose range, and in three or more independent experiments. As a positive control for inhibition, the unlabeled version of the radiolabeled probe was also tested in each experiment.
SANTA-SIM simulations
We simulated SARS-CoV-2 genomes with SANTA-SIM, using the consensus sequence WuhanHu-1 as input sequence available at https://www.ncbi.nlm.nih.gov/nuccore/MN908947.3. Each simulation was run with a population size of 10,000 individual viral sequences evolving for 1000 generations, and analyses were conducted on random samples of 1,000 viral sequences. Following Huddelston et.al. (Huddleston et al., 2020) who used SANTA-SIM to simulate influenza A/H3N2 that has a yearly substitution rate approximately twice as high as SARS-CoV-2 [∼48,824 substitutions/year (https://nextstrain.org/flu/seasonal/h3n2/ha/2y?l=clock) vs. ∼24.5 substitution/year (https://nextstrain.org/ncov/global?l=clock)], we chose 400 generations/year, with the mutation rate per position per generation set to 2.04E-6 (yearly substitution rate/(generations in one year ∗ genome size)). The transition bias was set to 3.0 for baseline simulations. To evaluate the impact of specific substitution biases, additional simulations were conducted using a substitution matrix with scores set to 1.0 of transversions, 3.0 for transitions, and biases ranging from 4.0 to 20.0 for the targeted substitution. We generated 10 replicates for all simulated scenarios, except for C-to-U where we made 100 replicates to better assess statistical significance.
Determination of amino acid mutational patterns
Mutational biases were identified by calculating the overall change in amino acid composition caused by the mutational landscape of SARS-CoV-2 for each individual amino acid, referred in the main text as ‘global residue substitution output’ (GRSO). For this analysis, all mutations found globally in at least 4 GISAID entries were analysed together. Preferential introduction or removal of amino acids was determined by comparing the overall amino acid composition in reference residues vs mutated residues throughout the mutation pool, resulting in a percentile difference in amino acid composition. As such, for amino acid X, the % difference was calculated according to the following formula:
This analysis took into consideration the number of unique mutations. Therefore, to consider mutational biases in the context of mutation frequencies, the analysis described above was conducted separately for mutations occurring in a single GISAID entry (expected to be enriched for errors); 2-10 GISAID entries; 11-99 GISAID entries; and 100 or more GISAID entries. As a negative control, the SANTA SIM algorithm was used to simulate the neutral evolution of 1000 SARS-CoV-2 genomes (baseline simulations, N = 10 replicates). This control was used to calculate the statistical significance of the observed biases, by way of a One-Sample T-Test.
Prediction of mutation impacts on peptide presentation in the context of HLA supertypes
Reference/mutated peptide pairs for which the differential predicted binding affinities led to transitions from strong HLA binder (SB) to non-HLA binder (NB) [(SB) NetMHCpan %rank < 0.5 to (NB) NetMHCpan %rank >2] or from NB to SB, were identified, catalogued and analyzed as described above. Binding affinities were predicted for representative HLA types from several major HLA supertypes (A1, A2, A3, A24, B7, B8, B27, B44), as defined by Sydney et al. We then categorized all reference/mutated peptide pairs on the basis of their 1) mutation type (amino acid X → amino acid Y) and 2) the position of the mutation in the peptide sequence. Finally, we quantified the number of reference/mutated peptide pairs and the associated average fold change in predicted binding affinity for each category. P-values were generated for each category by performing a two-tailed independent T-Test between the fold changes in binding affinity associated with mutation type A at position X, and all fold changes in binding affinity associated with position X.
Assessing the contribution of nucleic acid mutation types to the global amino acid mutational patterns
To assess the contribution of various nucleic acid mutation types to the observed amino acid mutational patterns, we first determined the respective contributions of each nucleic acid mutation type to the global mutation landscape. We then selected the five most abundant mutation types [C→U (41%), G→U (18%), A→G, G→A, U→C (9.7-11.6%)] and assessed their individual impacts on amino acid mutational patterns using the simulation algorithm SANTA SIM as follows:
For each mutation type, we simulated the evolution of 1000 SARS-CoV-2 genomes over 1000 generations (N = 10 replicates) with varying degrees of biases (the coefficient used to determine the extent of the biases was exploratively set to ‘x4’, ‘x8’, ‘x15’, and ‘x20’) (Figure S5A). Because the input coefficient does not have a linear relationship with the abundance of the mutation type observed in the simulation output, we used the simulations with all four parameter values (x4, x8, x15, x20) in order to identify the simulation parameter that most closely reflected observations in real-life SARS-CoV-2 data. The coefficient for the ratio of X → Y nucleic acid mutation type to all other mutation types was generated using the following formula:
Finally, all amino acid mutations were identified for the output of each simulation, as described above. To determine statistical significances, simulated mutational biases (at the amino acid level) were compared to a neutral evolution as a negative control (N = 10 replicates) by way of twotailed independent T-Test.
Statistical analysis
A Two-tailed One-Sample T-Test was used to assess the statistical significance of the observed mutational biases against the neutral simulations (N = 10 replicates). A Two-tailed Independent T-Test assuming different variances was used to assess the statistical significances of 1) the simulated biased SARS-CoV-2 evolution, 2) the gain/loss plots in the context of supertypes, and 3) the statistical significance associated with the average fold change in %rank associated with each position-specific amino acid mutation type in the supertype analysis.
Acknowledgments
We acknowledge and thank GISAID as well as all contributing laboratories for giving access to their SARS-CoV-2 genome sequences. We also thank Drs. Alessandro Sette, John Sidney, and Alba Grifoni (La Jolla Institute for Immunology, USA) for helpful discussions. This study was supported by funding from the Fonds de Recherche du Québec – Santé (FRQS), the Cole Foundation, CHU Sainte-Justine, the Charles-Bruneau Foundations, Canada Foundation for Innovation, IVADO COVID19 Rapid Response grant (CVD19-030), the Montreal Heart Institute Foundation, the National Sciences and Engineering Research Council (NSERC) (#RGPIN-2020-05232), and the Canadian Institutes of Health Research (CIHR) (#174924). K.A.K. is a recipient of IVADO’s postdoctoral scholarship (#4879287150). D.F. is a BioTalent awardee. E.C. and J.H. are FRQS Junior 1 research scholars.
Author contributions
Conceptualization, D.J.H., J.H., and E.C.; data curation and bioinformatic analysis, D.J.H., D.F., J.-C.G., F.M., K.A.K., and P.K.; formal analysis, D.J.H. and D.F.; investigation, D.J.H., D.F., J.S., J.-C.G., K.A.K., J.D.D., F.S., P.K., I.S., H.D., S.P., J.H., and E.C.; writing – original draft, D.J.H. and E.C.; writing – review & editing, D.J.H., D.F., J.S., J.-C.G., F.M., K.A.K., P.K., J.D.D., F.S., I.S., M.A.S., H.S., H.D., S.P., J.H., and E.C.; supervision, J.H. and E.C.; funding acquisition, J.H. and E.C.
Declaration of interests
Jana Schockaert and Sofie Pattijn are employees of ImmunXperts, a Nexelis Group Company.
Published: October 5, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cels.2021.09.013.
Supplemental information
SARS-CoV-2 mutations at the nucleic and amino acid level are indicated. Number of genomes carrying mutation show the frequency of individual mutations among all SARS-CoV-2 variants.
Epitopes were downloaded from https://www.mckayspcb.com/SARS2TcellEpitopes/ (as of January 2021). This database has effectively catalogued all SARS-CoV-2 CD8+ epitopes validated by 18 separate studies. See Quadeer et al. (2021) for details.
Table of authors from the Originating laboratories responsible for obtaining the specimens and the Submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which this research is based.
Data and code availability
-
•
Source data statement. This paper analyzes existing, publicly available data. All sequence data used are available from The Initiative for Sharing All Influenza Data (GISAID), at https://gisaid.org/. The user agreement for GISAID does not permit redistribution of sequences, but researchers can register to get access to the dataset. A GISAID acknowledgment table containing a full list of the laboratories and authors who contributed to the extensive GISAID SARS-CoV-2 genome database queried in this study is available in supplementary materials as Table S5.
-
•
Code statement. All original code has been deposited at https://github.com/CaronLab/CoVescape and is publicly available as of the date of publication. DOIs are listed in the Key Resources Table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Agerer B., Koblischke M., Gudipati V., Montaño-Gutierrez L.F., Smyth M., Popa A., Genger J.-W., Endler L., Florian D.M., Mühlgrabner V., et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8+ T cell responses. Sci. Immunol. 2021;6:eabg6461. doi: 10.1126/sciimmunol.abg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albin J.S., Haché G., Hultquist J.F., Brown W.L., Harris R.S. Long-term restriction by APOBEC3F selects human immunodeficiency virus type 1 variants with restored Vif function. J. Virol. 2010;84:10209–10219. doi: 10.1128/JVI.00632-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann D.M., Boyton R.J. SARS-CoV-2 T cell immunity: specificity, function, durability, and role in protection. Sci. Immunol. 2020;5:eabd6160. doi: 10.1126/sciimmunol.abd6160. [DOI] [PubMed] [Google Scholar]
- Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., et al. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- Brumme Z.L., Brumme C.J., Heckerman D., Korber B.T., Daniels M., Carlson J., Kadie C., Bhattacharya T., Chui C., Szinger J., et al. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007;3:e94. doi: 10.1371/journal.ppat.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway E. The race for coronavirus vaccines: a graphical guide. Nature. 2020;580:576–577. doi: 10.1038/d41586-020-01221-y. [DOI] [PubMed] [Google Scholar]
- Casartelli N., Guivel-Benhassine F., Bouziat R., Brandler S., Schwartz O., Moris A. The antiviral factor APOBEC3G improves CTL recognition of cultured HIV-infected T cells. J. Exp. Med. 2010;207:39–49. doi: 10.1084/jem.20091933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian S., Potdar V., Jadhav S., Yadav P., Gupta N., Das M., Rakshit P., Singh S., Abraham P., Panda S., NIC Team SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9:1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas J.M., Geller R., Garijo R., López-Aldeguer J., Sanjuán R. Extremely high mutation rate of HIV-1 in vivo. PLoS Biol. 2015;13:e1002251. doi: 10.1371/journal.pbio.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063. doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio S.D., Martignano F., Torcia M.G., Mattiuz G., Conticello S.G. Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2. Sci. Adv. 2020;6:eabb5813. doi: 10.1126/sciadv.abb5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Richard D., Tan C.C.S., Shaw L.P., Acman M., Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020;11:5986. doi: 10.1038/s41467-020-19818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., Owen C.J., Pang J., Tan C.C.S., Boshier F.A.T., et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect. Genet. Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franciscodos R.S., Buhler S., Nunes J.M., Bitarello B.D., França G.S., Meyer D., Sanchez-Mazas A. HLA supertype variation across populations: new insights into the role of natural selection in the evolution of HLA-A and HLA-B polymorphisms. Immunogenetics. 2015;67:651–663. doi: 10.1007/s00251-015-0875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk K., Rötzschke O., Stevanović S., Jung G., Rammensee H.-G. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351:290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W., et al. Unbiased screens show CD8+ T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton D., Rampling T., Cross A., Bailey H., Heaney J., Byott M., Scott R., Sconza R., Price J., Margaritis M., et al. Genomic characteristics and clinical effect of the emergent SARS-CoV-2 B.1.1.7 lineage in London, UK: a whole-genome sequencing and hospital-based cohort study. Lancet Infect. Dis. 2021;21:1246–1256. doi: 10.1016/S1473-3099(21)00170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geers D., Shamier M.C., Bogers S., Hartog G. den, Gommers L., Nieuwkoop N.N., Schmitz K.S., Rijsbergen L.C., van Osch J.A.T., Dijkhuizen E., et al. SARS-CoV-2 variants of concern partially escape humoral but not T-cell responses in COVID-19 convalescent donors and vaccinees. Sci. Immunol. 2021;6:eabj1750. doi: 10.1126/sciimmunol.abj1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller D., Bassani-Sternberg M. Predicting antigen presentation-what could we learn From a million peptides? Front. Immunol. 2018;9:1716. doi: 10.3389/fimmu.2018.01716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant M., Larijani M. Evasion of adaptive immunity by HIV through the action of host APOBEC3G/F enzymes. AIDS Res. Ther. 2017;14:44. doi: 10.1186/s12981-017-0173-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum J., Sidney J., Chung J., Brander C., Peters B., Sette A. Functional classification of class II human leukocyte antigen (HLA) molecules reveals seven different supertypes and a surprising degree of repertoire sharing across supertypes. Immunogenetics. 2011;63:325–335. doi: 10.1007/s00251-011-0513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Vita R., Peters B., Crotty S., Weiskopf D., Sette A. SARS-CoV-2 human T cell Epitopes: adaptive immune response against COVID-19. Cell Host Microbe. 2021;29:1076–1092. doi: 10.1016/j.chom.2021.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Sidney J., Zhang Y., Scheuermann R.H., Peters B., Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27:671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haché G., Shindo K., Albin J.S., Harris R.S. Evolution of HIV-1 isolates that use a novel Vif-independent mechanism to resist restriction by human APOBEC3G. Curr. Biol. 2008;18:819–824. doi: 10.1016/j.cub.2008.04.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J., Barnes J.R., Rowe T., Xu X., Kondor R., Wentworth D.E., Whittaker L., Ermetal B., Daniels R.S., McCauley J.W., et al. Integrating genotypes and phenotypes improves long-term forecasts of seasonal influenza A/H3N2 evolution. Elife. 2020;9:e60067. doi: 10.7554/eLife.60067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jariani A., Warth C., Deforche K., Libin P., Drummond A.J., Rambaut IV A., Matsen F.A., Theys K. SANTA-SIM: simulating viral sequence evolution dynamics under selection and recombination. Virus Evol. 2019;5:vez003. doi: 10.1093/ve/vez003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P., Russell R.A., Pathak V.K., Coffin J.M. Likely role of APOBEC3G-mediated G-to-A mutations in HIV-1 evolution and drug resistance. PLoS Pathog. 2009;5:e1000367. doi: 10.1371/journal.ppat.1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H.R., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P., et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Invest. 2021;131:e145476. doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y., Pfafferott K., Frater J., Matthews P., Payne R., Addo M., Gatanaga H., Fujiwara M., Hachiya A., Koizumi H., et al. Adaptation of HIV-1 to human leukocyte antigen class I. Nature. 2009;458:641–645. doi: 10.1038/nature07746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.-Y., Lorenzo-Redondo R., Little S.J., Chung Y.-S., Phalora P.K., Maljkovic Berry I.M., Archer J., Penugonda S., Fischer W., Richman D.D., et al. Human APOBEC3 induced mutation of human immunodeficiency virus Type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 2014;10:e1004281. doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimczak L.J., Randall T.A., Saini N., Li J.-L., Gordenin D.A. Similarity between mutation spectra in hypermutated genomes of rubella virus and in SARS-CoV-2 genomes accumulated during the COVID-19 pandemic. PLoS One. 2020;15:e0237689. doi: 10.1371/journal.pone.0237689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., et al. Tracking changes in SARS-CoV-2 Spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182:812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosuge M., Furusawa-Nishii E., Ito K., Saito Y., Ogasawara K. Point mutation bias in SARS-CoV-2 variants results in increased ability to stimulate inflammatory responses. Sci. Rep. 2020;10:17766. doi: 10.1038/s41598-020-74843-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Laamarti M., Alouane T., Kartti S., Chemao-Elfihri M.W., Hakmi M., Essabbar A., Laamarti M., Hlali H., Bendani H., Boumajdi N., et al. Large scale genomic analysis of 3067 SARS-CoV-2 genomes reveals a clonal geo-distribution and a rich genetic variations of hotspots mutations. PLoS One. 2020;15:e0240345. doi: 10.1371/journal.pone.0240345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bert N.L., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- Li Y., Yang X., Wang N., Wang H., Yin B., Yang X., Jiang W. Mutation profile of over 4500 SARS-CoV-2 isolations reveals prevalent cytosine-to-uridine deamination on viral RNAs. Future Microbiol. 2020;15:1343–1352. doi: 10.2217/fmb-2020-0149. [DOI] [PubMed] [Google Scholar]
- Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- Liu G., Carter B., Bricken T., Jain S., Viard M., Carrington M., Gifford D.K. Computationally optimized SARS-CoV-2 MHC class I and II vaccine formulations predicted to target human haplotype distributions. Cell Syst. 2020;11:131–144.e6. doi: 10.1016/j.cels.2020.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Fontanet A., Zhang P.H., Zhan L., Xin Z.T., Baril L., Tang F., Lv H., Cao W.-C. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis. 2006;193:792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K., Liao P., Qiu J.-F., Lin Y., Cai X.-F., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- Matyášek R., Kovařík A. Mutation patterns of human SARS-CoV-2 and bat RaTG13 coronavirus genomes are strongly biased Towards C>U transitions, indicating rapid evolution in their hosts. Genes (Basel) 2020;11:761. doi: 10.3390/genes11070761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meckiff B.J., Ramírez-Suástegui C., Fajardo V., Chee S.J., Kusnadi A., Simon H., Eschweiler S., Grifoni A., Pelosi E., Weiskopf D., et al. Imbalance of regulatory and cytotoxic SARS-CoV-2-reactive CD4+ T cells in COVID-19. Cell. 2020;183:1340–1353.e16. doi: 10.1016/j.cell.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Giorgi F.M. Geographic and genomic distribution of SARS-CoV-2 mutations. Front. Microbiol. 2020;11:1800. doi: 10.3389/fmicb.2020.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli D., Triboli L., Fornasari E., Ray F., Giorgi F.M. Coronapp: a web application to annotate and monitor SARS-CoV-2 mutations. J. Med. Virol. 2021;93:3238–3245. doi: 10.1002/jmv.26678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moderbacher C.R., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monajemi M., Woodworth C.F., Zipperlen K., Gallant M., Grant M.D., Larijani M. Positioning of APOBEC3G/F mutational hotspots in the human immunodeficiency virus genome favors reduced recognition by CD8+ T cells. PLoS One. 2014;9:e93428. doi: 10.1371/journal.pone.0093428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motozono C., Toyoda M., Zahradnik J., Saito A., Nasser H., Tan T.S., Ngare I., Kimura I., Uriu K., Kosugi Y., et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe. 2021;29:1124–1136.e11. doi: 10.1016/j.chom.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naemi F.M.A., Al-Adwani S., Al-Khatabi H., Al-Nazawi A. Association between the HLA genotype and the severity of COVID-19 infection among South Asians. J. Med. Virol. 2021;93:4430–4437. doi: 10.1002/jmv.27003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nersisyan S., Zhiyanov A., Shkurnikov M., Tonevitsky A. T-CoV: a comprehensive portal of HLA-peptide interactions affected by SARS-CoV-2 mutations. Nucleic Acids Res. 2021 doi: 10.1093/nar/gkab701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng O.-W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.-J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson M.E., Harris R.S., Harki D.A. APOBEC enzymes as targets for virus and cancer therapy. Cell Chem. Biol. 2018;25:36–49. doi: 10.1016/j.chembiol.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peretti A., Geoghegan E.M., Pastrana D.V., Smola S., Feld P., Sauter M., Lohse S., Ramesh M., Lim E.S., Wang D., et al. Characterization of BK polyomaviruses from kidney transplant recipients suggests a role for APOBEC3 in driving in-host virus evolution. Cell Host Microbe. 2018;23:628–635.e7. doi: 10.1016/j.chom.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisanti S., Deelen J., Gallina A.M., Caputo M., Citro M., Abate M., Sacchi N., Vecchione C., Martinelli R. Correlation of the two most frequent HLA haplotypes in the Italian population to the differential regional incidence of Covid-19. J. Transl. Med. 2020;18:352. doi: 10.1186/s12967-020-02515-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa A., Genger J.W., Nicholson M.D., Penz T., Schmid D., Aberle S.W., Agerer B., Lercher A., Endler L., Colaço H., et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci. Transl. Med. 2020;12:eabe2555. doi: 10.1126/scitranslmed.abe2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadeer A.A., Ahmed S.F., McKay M.R. Landscape of epitopes targeted by T cells in 852 individuals recovered from COVID-19: Meta-analysis, immunoprevalence, and web platform. Cell Rep. Med. 2021;2:100312. doi: 10.1016/j.xcrm.2021.100312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynisson B., Alvarez B., Paul S., Peters B., Nielsen M. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice A.M., Morales A.C., Ho A.T., Mordstein C., Mühlhausen S., Watson S., Cano L., Young B., Kudla G., Hurst L.D. Evidence for strong mutation bias towards, and selection against, U content in SARS-CoV-2: implications for vaccine design. Mol. Biol. Evol. 2020;38:67–83. doi: 10.1093/molbev/msaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler H.A., Stenglein M.D., Harris R.S., Mansky L.M. APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis. J. Virol. 2010;84:7396–7404. doi: 10.1128/JVI.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Amaya Hernandez S.P.A., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8+ T cell activation in COVID-19 patients. Sci. Immunol. 2021;6:eabf7550. doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter J.D., Bennett R.P., Smith H.C. The APOBEC protein family: united by structure, divergent in function. Trends Biochem. Sci. 2016;41:578–594. doi: 10.1016/j.tibs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schub D., Klemis V., Schneitler S., Mihm J., Lepper P.M., Wilkens H., Bals R., Eichler H., Gärtner B.C., Becker S.L., et al. High levels of SARS-CoV-2 specific T-cells with restricted functionality in severe courses of COVID-19. JCI Insight. 2020;5:e142167. doi: 10.1172/jci.insight.142167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8+ T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- Sekine T., Perez-Potti A., Rivera-Ballesteros O., Strålin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K.J.A., Hemmings O., O’Byrne A., Kouphou N., Galao R.P., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Peters B., Frahm N., Brander C., Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidney J., Southwood S., Moore C., Oseroff C., Pinilla C., Grey H.M., Sette A. Measurement of MHC/peptide interactions by gel filtration or monoclonal antibody capture. Curr. Protoc. Immunol. 2013;100:18.3.1–18.3.36. doi: 10.1002/0471142735.im1803s100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. Rampant C→U hypermutation in the genomes of SARS-CoV-2 and other coronaviruses: causes and consequences for their short- and long-term evolutionary trajectories. mSphere. 2020;5:e00408. doi: 10.1128/mSphere.00408-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires K.D., Monajemi M., Woodworth C.F., Grant M.D., Larijani M. Impact of APOBEC mutations on CD8+ T cell recognition of HIV epitopes varies depending on the restricting HLA. J. Acquir. Immune Defic. Syndr. 2015;70:172–178. doi: 10.1097/QAI.0000000000000689. [DOI] [PubMed] [Google Scholar]
- Stephens D.S., McElrath M.J. COVID-19 and the path to immunity. JAMA. 2020;324:1279–1281. doi: 10.1001/jama.2020.16656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Quan Y., Xin Z.-T., Wrammert J., Ma M.-J., Lv H., Wang T.-B., Yang H., Richardus J.H., Liu W., Cao W.-C. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: A six-year follow-up study. J. Immunol. 2011;186:7264–7268. doi: 10.4049/jimmunol.0903490. [DOI] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R.da S., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2:100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarke A., Sidney J., Methot N., Yu E.D., Zhang Y., Dan J.M., Goodwin B., Rubiro P., Sutherland A., Wang E., et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2:100355. doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Tomita Y., Ikeda T., Sato R., Sakagami T. Association between HLA gene polymorphisms and mortality of COVID-19: an in silico analysis. Immun. Inflamm. Dis. 2020;8:684–694. doi: 10.1002/iid3.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Hozumi Y., Zheng Y.-H., Yin C., Wei G.-W. Host immune response driving SARS-CoV-2 evolution. Viruses. 2020;12:1095. doi: 10.3390/v12101095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., et al. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5:eabd2071. doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood N., Bhattacharya T., Keele B.F., Giorgi E., Liu M., Gaschen B., Daniels M., Ferrari G., Haynes B.F., McMichael A., et al. HIV evolution in early infection: selection pressures, patterns of insertion and deletion, and the impact of APOBEC. PLoS Pathog. 2009;5:e1000414. doi: 10.1371/journal.ppat.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolthuis R.G., Dorp C.H. van, Keşmir C., Boer R.J. de, Boven M. van. Long-term adaptation of the influenza A virus by escaping cytotoxic T-cell recognition. Sci. Rep. 2016;6:33334. doi: 10.1038/srep33334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L.-P., Wang N.-C., Chang Y.-H., Tian X.-Y., Na D.-Y., Zhang L.-Y., Zheng L., Lan T., Wang L.-F., Liang G.-D. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007;13:1562–1564. doi: 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., To K.K.-W., Wong Y.C., Liu L., Zhou B., Li X., Huang H., Mo Y., Luk T.Y., Lau T.T.-K., et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SARS-CoV-2 mutations at the nucleic and amino acid level are indicated. Number of genomes carrying mutation show the frequency of individual mutations among all SARS-CoV-2 variants.
Epitopes were downloaded from https://www.mckayspcb.com/SARS2TcellEpitopes/ (as of January 2021). This database has effectively catalogued all SARS-CoV-2 CD8+ epitopes validated by 18 separate studies. See Quadeer et al. (2021) for details.
Table of authors from the Originating laboratories responsible for obtaining the specimens and the Submitting laboratories where genetic sequence data were generated and shared via the GISAID Initiative, on which this research is based.
Data Availability Statement
-
•
Source data statement. This paper analyzes existing, publicly available data. All sequence data used are available from The Initiative for Sharing All Influenza Data (GISAID), at https://gisaid.org/. The user agreement for GISAID does not permit redistribution of sequences, but researchers can register to get access to the dataset. A GISAID acknowledgment table containing a full list of the laboratories and authors who contributed to the extensive GISAID SARS-CoV-2 genome database queried in this study is available in supplementary materials as Table S5.
-
•
Code statement. All original code has been deposited at https://github.com/CaronLab/CoVescape and is publicly available as of the date of publication. DOIs are listed in the Key Resources Table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.