Abstract
The COVID-19 pandemic caused by the coronavirus SARS-CoV-2 has become a serious challenge for medicine and science. Analysis of the molecular mechanisms associated with the clinical manifestations and severity of COVID-19 has identified several key points of immune dysregulation observed in SARS-CoV-2 infection. For diabetic patients, factors including higher binding affinity and virus penetration, decreased virus clearance and decreased T cell function, increased susceptibility to hyperinflammation, and cytokine storm may make these patients susceptible to a more severe course of COVID-19 disease. Metabolic changes induced by diabetes, especially hyperglycemia, can directly affect the immunometabolism of lymphocytes in part by affecting the activity of the mTOR protein kinase signaling pathway. High mTOR activity can enhance the progression of diabetes due to the activation of effector proinflammatory subpopulations of lymphocytes and, conversely, low activity promotes the differentiation of T-regulatory cells. Interestingly, metformin, an extensively used antidiabetic drug, inhibits mTOR by affecting the activity of AMPK. Therefore, activation of AMPK and/or inhibition of the mTOR-mediated signaling pathway may be an important new target for drug therapy in COVID-19 cases mostly by reducing the level of pro-inflammatory signaling and cytokine storm. These suggestions have been partially confirmed by several retrospective analyzes of patients with diabetes mellitus hospitalized for severe COVID-19.
Keywords: COVID-19, Immunometabolism, Lymphocytes, Metformin
1. Introduction
Dysregulation of the immune system caused by coronavirus SARS-CoV-2 is one of the main causes of severe COVID-19 disease. Several comorbidity pathologies including diabetes mellitus can result in a more severe course of COVID-19 that leads to higher mortality [1], [2]. For example, changes in the immunometabolism of lymphocytes, as is observed in diabetes, directly affect the course of the disease and thus point out the importance of glycemic control in people with COVID-19 infection. In this aspect, metformin might be the most attractive drug that can be use to simultaneously prevent hyperglycemia and affect the immunometabolism of lymphocytes via its effects on the mTOR signaling pathway. However, it is still unclear how beneficial metformin might be for patients with COVID-19. On the one hand, metformin might cause some additional complications [3]. On another hand, metformin is the drug with the lowest mortality rate among all available antidiabetic drugs [4]. Metformin modulates the activity of the PI3K/AKT/mTOR signaling network by targeting AMP-activated protein kinase (AMPK), and thereby regulate the metabolism and balance between key subpopulations of immune cells involved in the pathogenesis of COVID-19. In this review, we provide evidence of the significant benefits of metformin for diabetic patients with coronavirus infection making it the most promising drug for pathogenetic therapy. Currently, various other studies are also ongoing to investigate ways to decrease disease severity and mortality among COVID-19 patients. These include the possible beneficial effects of anticoagulant therapy and potential interventions that improve endothelial function, such as RAS inhibitors, statins, and antioxidants. The present review begins with this question. How does metformin influence endothelial function/dysfunction, inflammation, and thrombotic events in diabetic/hyperglycemic COVID-19 patients?
2. Immune dysregulation in COVID-19
2.1. The recognition of SARS-COV-2 by components of the innate immune system
Infection by the new coronavirus SARS-CoV-2 leads to the development of COVID-19 disease that is a serious challenge to medicine and science. However, the amount of information about the mechanisms associated with immune protection against SARS-CoV-2 or provoked immunopathology is increasing exponentially. Accumulation of these data is important for appropriate diagnostic and therapeutic strategies [5]. Analysis of the molecular and immune mechanisms associated with the clinical manifestations and severity of COVID-19 have revealed several key features of immune dysregulation observed during SARS-CoV-2 infection.
When penetrating into cells, viral RNA is sensed by the innate immune system using pattern recognition receptors (PRR) of three classes: endosomal Toll-like receptors (TLR), RIG-I-like receptors (RLR), and NOD-similar receptors (NLR) [6]. Among TLRs, TLR-7 and -8 recognize the single-stranded RNA of SARS-COV-2. Cytosolic sensors, such as MDA5 and RIG-I, can detect intermediates of intracellular replication of the virus, subsequently activating the nuclear factor kappa B (NF-κB) and the interferon regulatory factor 3/7 (IRF3/7). NF-κB promotes the transcription of systemic proinflammatory cytokines such as tumor necrosis factor alpha (TNF-α), IL-1β, and IL-6. IRF3/7 promotes the production of interferon, which, in turn, activates the JAK1/TYK2-STAT1/2 cascade [7]. Transcription factors STAT1 and STAT2 form a complex with the interferon regulatory factor 9 (IRF9), which is translocated to the nucleus to induce transcription of interferon-stimulated genes (ISG). The degree of NLR involvement in SARS-COV-2 recognition is supported by the high levels of circulating IL-1β, IL-1RA, and IL-18 [8]. NLRs induce the assembly of the NLRP3-inflammasome, which activates caspase-1, IL-1β, IL-18, and also induces a hyperinflammatory form of cell death - pyroptosis, by splitting of pore-forming gasdermin D [9]. The cytosolic sensor RIG-I can also induce the formation of an inflammasome that causes caspase-1 activation and subsequent secretion of IL-1β, IL-18, and pyroptosis [10].
2.2. Evasion strategies of the innate immune receptors in SARS-CoV-2 detection
Coronaviruses have developed several strategies to avoid recognition by the innate immune system. The low genomic level of CpGs islands prevents the destruction of viral RNA by zinc-finger antiviral protein (ZAP) [11]. Processing and 5′-end capping protection of viral RNA limits its degradation and blocks recognition by cytosolic PRR. The formation of a replicase-transcriptase complex (RTC) or replication organelle that binds to the endoplasmic reticulum and the Golgi apparatus may protect the virus during maturation [12]. Finally, SARS-CoV-2 evolved glycans to mask immunogenic epitopes of viral proteins. For example, subunits S1 and S2 of the adhesion protein contain 22 glycan groups. Other structural, non-structural, and auxiliary proteins are also modified by phosphorylation, palmitoylation, glycosylation, SUMOylation, and ADP-ribosylation [13]. In addition to PPR recognition evasion strategies, the virus also has several strategies that prevent IFN type I/III production by cells. One of the non-structural proteins, nsp1, is the real "cellular sabotage"; it causes production slowdown of interferons by an infected cell and at the same time forces the cell to work for the virus and produce more viral proteins. This is achieved by physically closing the ribosome channel into which the mRNA usually enters [14]. Nsp1, 4, and 6 and ORF6 prevent STAT-1/2 signaling, while nsp 10, 13, and 16 protect viral RNA by preventing recognition by IFITs, RIG-I, or MDA-5.
2.3. The adaptive immunity in COVID-19
The number of white blood cells may vary in patients with COVID-19, but the more common observation is leukocytosis and lymphopenia associated with increased disease severity [15]. Lymphopenia predicts the disease severity of COVID-19. The etiology and mechanisms of lymphopenia in patients with COVID-19 are unknown, but there are several hypotheses of its occurrence. The virus can directly infect T cells to cause their death by apoptosis, necrosis, or pyroptosis [16]. The number of cytokines secreted by infected lung macrophages or epithelial cells can cause apoptosis of T cells, or prevent their proliferation (IL-10) [17]. Finally, lymphopenia may be due to the redistribution of cells of the immune system with the accumulation of lymphocytes in the lungs or lymphoid organs [18].
Phenotypic and functional changes of T cells in COVID-19 are manifested by increased expression of activated T cell markers such as HLA-DR, CD25, CD44, CD38, or CD69 [19]. Regardless of the severity of COVID-19 activation of CD8, T cells prevail among the cell types [20]. Experimental data show increased expression of programmed cell death 1 (PD-1) protein, which is associated with depletion of T cells, higher expression of various co-stimulating and inhibiting molecules such as OX-40 and CD137, CTLA-4 and NKG2a [21]. A decrease in the number of CD28 and CD8 T cells has been reported, as well as an increase in the frequency of PD-1/TIM3/CD8 positive T cells in ICU patients [22]. The expression of most of these markers is higher in CD8 than in CD4 T cells, and the levels tend to increase in severe cases compared to mild cases, which may be related to differences in viral load. Cell functionality is impaired in CD4 and CD8 T cells of severe patients. CD8 T cells of these patients are characterized by decreased cytotoxicity, lever expression of Cd107a (degranulation rate), and reduced production of granzyme B (Gznb) [23]. Data produced from RNAseq by Liao and colleagues showed that CD8 T cells of severe COVID-19 patients express higher levels of the cytotoxic genes Granzyme A, B, and K [24]. Thus, T cells in severe COVID-19 appear to be more activated and may tend to decrease in amount due to continuous expression of inhibition markers such as PD-1 and TIM-3. Moreover, an increased amount of T follicular helper cell (TFH) levels of effector molecules such as Gzma, Gznb, and perforin with decreased levels of inhibition markers were found in convalescent patients [25]. SARS-CoV-2-specific CD4+ T cells generally have a Th1 or TFH phenotype. TFH differentiation is impaired in some severe COVID-19 patients to support the absence of germ centers in the spleen and lymph nodes [26].
Despite the fast increase in the number of publications, several questions related to T-cell immunity to SARS-CoV-2 remain unresolved. What factors regulate the strength and effectiveness of the antiviral T-cell response? What is the lifespan of memory T cells and is the severe course of COVID 19 associated with impaired development of memory T cells specific to SARS-CoV-2? Do existing cross-reactive memory T cells help in cases of seasonal coronavirus? What predominant type of T-cell response elicited by vaccines would be the best predictor of protection against disease after exposure to the virus?
Abnormalities of humoral immunity, also affecting the severity of COVID-19, can manifest as antibody-dependent enhancement (ADE). During ADE, antibodies with low affinity or low concentration, instead of neutralization, opsonize viral particles and promote FcγR-mediated internalization by lung epithelial cells and infiltration of monocytes and DCs, which can worsen the course of COVID-19 [27].
T cell-mediated immunity is crucial for the effective clearance of SARS-CoV-2 infection. In severe and critical COVID-19 patients, peripheral blood T cells are significantly decreased and show a phenotype of hyperactivation/exhaustion relative to controls. A complex pattern of T cell response to SARS-CoV-2 infection has been demonstrated, but inferences regarding population level immunity are hampered by significant methodological limitations and heterogeneity between studies. In contrast to antibody responses, population-level surveillance of the T cell response is unlikely to be feasible in the near term. Focused evaluation in specific sub-groups, including vaccine recipients, should be prioritized. However, large-scale studies of T cell immunity are needed using technologies such as Elispot and T-detect.
3. Comorbidity of diabetes mellitus and COVID-19
3.1. Mechanisms of increased susceptibility of diabetic patients to severe COVID-19
Recent relevant studies have reported several mechanisms that have been proposed to explain the increased susceptibility of diabetic patients to severe COVID-19 disease [28], [29], [30]. Higher binding affinity and penetration of the virus to cells decreased viral clearance efficiency, reduced T-cell function, increased susceptibility to hyperinflammation, and cytokine storm are among most possible contributors [31]. Partially defective phagocytosis of macrophages, neutrophils, and monocytes was shown in patients with diabetes, disorders of neutrophil chemotaxis, bactericidal activity, and innate cellular-mediated immunity [32]. Even short-term hyperglycemia suppressed the innate and adaptive immune response.
ACE2 protein plays a key linking role between COVID-19 and diabetes mellitus [33]. Treatment of patients with type 1 or type 2 diabetes mellitus with ACE2 inhibitors and angiotensin II receptor blockers increases the expression of ACE2 in the renal and cardiovascular systems [34]. However, there is insufficient evidence to support the hypothesis that switching people from ACE inhibitors to other drugs can reduce the risk of infection and the severity of COVID-19. Increased expression of ACE2 was observed in the lungs, kidneys, heart, and pancreas [35], which possibly contributes to more intensive virus penetration in diabetic animals. Treatments with hypoglycemic agents such as thiazolidinediones and glucagon-like peptide-1 receptor agonists also increased ACE2 expression [36]. Randomized studies showed that higher expression of ACE2 in the lung of diabetic patients increased susceptibility to SARS-CoV-2 infection [37]. The genome-wide association study of patients with T2D showed that this type was causally associated with increased expression of ACE2. Another study showed that diabetic patients had increased levels of furin, a cellular protease that cleaves the S1 and S2 domains of the SARS-CoV-2 spike protein possibly contributing to virus entry [38].
Another potential reason for the increased risk of severe COVID-19 in patients with diabetes may be related to cytokine storm because patients with diabetes suffer from persistent low-level chronic inflammation, contributing to a hyperinflammatory response [39]. Besides, increased production of advanced glycation end products (AGEs) in diabetic patients may increase the basal production of pro-inflammatory cytokines TNF-α, IL-1β, IL-6, and IL-8. Patients with diabetes associated with COVID-19, despite a significantly lower absolute number of lymphocytes in peripheral blood, have a higher absolute number of neutrophils compared to patients without diabetes [40].
Poor glycemic control is also a risk factor for disease progression in patients with COVID-19. Several retrospective multicenter studies have shown that patients with well-controlled blood glucose levels had lower mortality rates [41]. Fasting blood glucose at admission, regardless of previous diabetes diagnosis, was an independent predictor of critical illness, death, or poor outcome in patients hospitalized with COVID-19 [42].
3.2. Interaction between COVID-19 and the AMPK/mTOR signaling pathway in diabetes mellitus
AMPK is a key physiological energy sensor. Its activity is mostly regulated by glucose availability. Under conditions of diabetic hyperglycemia, the activity of the AMPK signaling pathways is changed. Transmission of signals through AMPK modulates pathways of cellular metabolism, growth, and proliferation to maintain cellular energy homeostasis [43]. AMPK becomes activated to stimulate glucose uptake in skeletal muscle and fatty acid oxidation in the adipose tissue when the cellular energy level is low. Several studies have emphasized the dysregulation of AMPK signaling in diabetes and metabolic syndrome [44], [45]. The mTOR (mammalian target of rapamycin) kinase pathway is one of the main interacting signaling pathways affected by AMPK. AMPK suppresses the activity of the TOR pathway mostly by phosphorylation of TSC2 or Raptor [46]. The mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2) have different properties. The two subtypes are composed of different sets of protein binding partners [47]. mTORC1 regulates protein synthesis and cell size and consists of mTOR, mLST8, and the rapamycin-sensitive adapter protein of mTOR (Raptor) [48]. mTORC2 is formed by mTOR, mSIN1, mLST8, and the rapamycin-insensitive Rictor subunit and regulates cell survival and cytoskeleton organization [49]. Inhibition of AMPK causes chronic hyperactivation of mTORC1 leading to insulin resistance and progression of complications caused by diabetes in patients [50]. Moreover, the activity of the mTOR pathway might be modulated by specific drugs that make them possible co-treatments in diabetic patients [51], [52], [53].
4. Lymphocyte immunometabolism and its changes in diabetes
4.1. The key metabolic pathways of immune cells
Over the last decade, there has been a significant breakthrough in the study of lymphocyte metabolism, generally called immunometabolism. Lymphocytes use several different metabolic pathways to generate the required ATP energy to support survival and proliferation. Glycolysis, TCA cycle, pentose-phosphate pathway, oxidation of fatty acids, fatty acids synthesis, and amino acid metabolism are key metabolic pathways that play crucial roles in the differentiation and survival of immune cells. However, these pathways can possess different activities in certain types of immunocytes.
Activated T lymphocytes and macrophages have a high affinity for glucose and use significant amounts of glucose [54]. The use of glycolysis inhibitors such as 2-deoxy-D-glucose prevents the activation of macrophages in vitro and blocks inflammation in vivo [55]. Activated immune cells use glycolysis extensively despite its relatively low efficiency in ATP. Many growth signaling pathways, involving phosphoinositide 3-kinases (PI3K) and mitogen-activated protein kinases (MAPKs), promote glycolysis. Thus, it is the dominant pathway in rapidly proliferating cells. Several studies have shown activated glycolysis in LPS-activated macrophages and dendritic cells, activated NK cells, and T- and B-lymphocytes [56]. Effector T lymphocytes show increased glycolytic activity after activation, especially Th17, Th1, and Th2 cells, as well as activated effector CD8+ T lymphocytes [57]. Increased activity of mTOR correlates with increased glycolysis and the generation of T-regulatory cells (Treg) [58]. However, factors that initiate glycolysis during lymphocyte activation are only partially described. For example, bacterial LPS causes activation of hypoxia-inducible factor 1α (HIF1α), an important transcription factor for the induction of several glycolytic enzymes [59]. NFκB (nuclear factor kappa-light-chain-enhancer of activated B cells) affects the activation of phosphofructokinase 2 (PFK2), a master regulator of glycolysis. LPS can also rapidly activate glycolysis in dendritic cells due to activation of TBK1 kinase and/or inhibition of NFκB ε (IKKε) kinase and hexokinase 2 [60]. An important mechanism for enhancing glycolytic activity in LPS-activated macrophages is the induction of pyruvate kinase type M2 (PKM2). 2-Deoxy-D-glucose promotes the reprogramming of Th17 into Treg cells [61]. These studies emphasize the relationship between metabolism and lymphocyte phenotype. Another interesting aspect of glycolysis induction in activated immunocytes is the role of the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH). It has been shown that in Th1-cells GAPDH binds to mRNA, which encodes one of the main Th1-dependent effector cytokines, interferon-γ (IFNγ), inhibiting its translation [62]. Once glycolysis is activated, GAPDH dissociates from IFNγ mRNA, allowing it to be translated. In macrophages, another glycolytic enzyme, hexokinase 1, can interact with the NLRP3 inflammasome, causing its activation and induce inflammation.
The tricarboxylic acid cycle (TCA cycle) is used most actively in dormant cells or non-proliferating cells for energy generation. The cycle is functional in most T-lymphocytes, but it is used more actively by CD8+-memory T-lymphocytes [63]. Effector T-cells are more prone to glycolysis. The influence of TCA pathway on the differentiation of macrophages has been extensively studied. In M2 macrophages the tricarboxylic acid cycle is active, while in M1 it is partially blocked in two places - after citrate and after succinate. Excessive accumulation of citrate promotes the production of nitric oxide, prostaglandins, and itaconic acid, which provides direct bactericidal effects on microorganisms such as Salmonella enterica and Mycobacterium tuberculosis [64].
NADPH is mostly produced in the pentose phosphate pathway. It is used by macrophages and neutrophils to generate ROS. NADPH is necessary for activation of dendritic cell secretion of cytokines [65]. Pathway induction is observed in LPS-activated macrophages, and the key enzyme for macrophage polarization is CARKL (carbohydrate kinase-like protein) [66]. This enzyme limits the flow through the pentose phosphate pathway and is highly active in M2 macrophages. If CARKL is suppressed, macrophages acquire an M1-like phenotype.
Fatty acid oxidation (FAO) provides acetyl-CoA, NADH, and FADH2 that are used for energy production. FAO is one of the key pathways in the regulation of adaptive and innate immune responses. Unlike glycolysis, which is often used by effectors and rapidly proliferating cells, oxidation of fatty acids is active in cells that are not pro-inflammatory and that demonstrate increased lifespan. M2 macrophages, Treg cells, and memory T cells are among those cell types. Thus, the increase in FAO is observed in Treg lymphocytes and reduced in effector Th1, Th2 і Th17-cells [67]. Treg cells possess an enhanced expression of genes involved in the oxidation of fatty acids, including Cpt1a (carnitine palmitoyltransferase 1a), compared with Th17 lymphocytes [68]. Effector T lymphocytes show a decrease in FAO activity during activation. FAO plays an important role in the generation of long-lived CD8 T-memory cells. Stimulation of memory CD8+ T cells with IL-15 enhances the expression of the Cpt1a and oxidation of fatty acids, which leads to increased survival of this population.
The metabolic pathway of fatty acids synthesis (FAS) allows cells to produce lipids that are necessary for cell growth and rapid proliferation. The activity of this pathway is partially regulated by mTOR signaling, which promotes FAS by regulating key enzymes responsible for de novo lipid synthesis, including SREBP (Sterol regulatory element-binding proteins), FASN (Fatty acid synthase), and ACC (Acetyl-CoA carboxylase) [48]. In contrast to the oxidation of fatty acids, FAS promotes the generation of pro-inflammatory subpopulations of immune cells, and activates innate and adaptive immune responses. Several studies have shown that LPS and cytokines initiate an increase in FAS in macrophages [69]. FAS provides a connection between innate and adaptive immunity through the regulation of DC functions. Thus, TLR indirect activation of DC is accompanied by an increase in FAS. This metabolic pathway is important for the proliferation of T and B lymphocytes after their activation through antigenic receptors. T-specific deletion of acetyl-CoA carboxylase 1 (ACC1), a regulatory enzyme of FAS, leads to a decrease in the number of antigen-specific CD8 T-lymphocytes [70]. The balance between effector and T-regulatory cells is also partially dependent on the synthesis of fatty acids. Pharmacological or genetic inhibition of ACC1 in CD4 T-lymphocytes has shown that FAS is required for Th17 differentiation but not for Treg functions [71].
Amino acid metabolism is mostly regulated by mTOR. Some amino acids play an important role in the immunometabolism of lymphocytes. For example, glutamine and arginine are required for the activation of T and B lymphocytes [72]. Glutamine metabolism also regulates the balance between the effector and Treg cells. Genetic knockout of the ASCT2 transporter protein (which is responsible for the uptake of glutamine in T lymphocytes) leads to impaired generation and function of Th1 and Th17 reduced expression of mTORC1 but does not affect Treg [73]. Arginine metabolism plays a leading role in the regulation of the phenotype of M1 and M2 macrophages [74], whereas tryptophan is required for the proliferation of T lymphocytes.
4.2. Lymphocyte as a sensor of metabolism
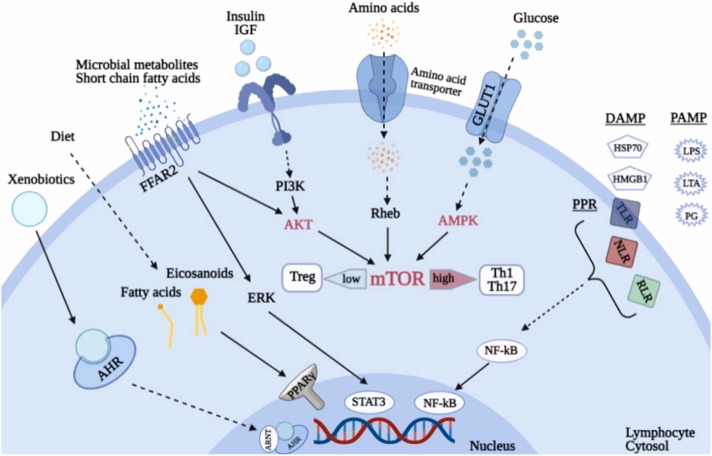
Glycolysis, pentose phosphate pathway, and FAS are extensively used by Th1-, Th2-, Th17- effector cells and M1-macrophages. T-regulatory, CD8 memory T-cells and M2-macrophages mostly use tricarboxylic acids and oxidation of fatty acids for energy production. Changes in the metabolism of various amino acids can affect the generation of both effector and Treg lymphocytes [75]. The TOR pathway in lymphocytes integrates signals of amino acid availability and growth factors and is one of the central regulators of cell proliferation and survival of this cell type [76]. AMP-activated protein kinase (AMPK) receives signals from a key glucose sensor and is a regulator of cellular energy balance. Receptors activated by PPARγ peroxisome proliferators include pattern-recognizing innate immune receptors (TLR, NLR, RLR, etc.), sensors of extracellular ATP purinergic receptors P2XR, sensors of xenobiotic aryl-hydrocarbon receptors (AHR), receptors of short-chain fatty acid FFAR2, ligands for which are microbial metabolites such as butyrate, acetate and propionate [77] ( Fig. 1).
Fig. 1.
Lymphocytes as sensors of metabolic changes. Notation keys: mTOR – target of rapamycin, AMPK – AMP-activated protein kinase, PPARγ – receptors activated by peroxisome proliferators, Glut 1 – glucose transporters type 1, P2XR – purinergic receptors, FFAR2 – short-chain fatty acid receptors, AHR – aryl-hydrocarbon receptors, PRR – pattern-recognizing receptors of innate immunity, PAMP – pathogen-associated molecular patterns, DAMP – damage- associated molecular patterns.
4.3. mTOR as a regulator of lymphocyte immunometabolism in diabetes
Changes in metabolism induced by hyperglycemia in diabetes can directly affect the immunometabolism of lymphocytes [78], [79]. T cells express several glucose transporters including Glut 1 [80]. Prodiabetogenic Th1 and Th17 cells that cause stroke are characterized by a high level of Glut1 expression and extensively use glycolysis for energy production [81]. By contrast, Treg suppressors have low Glut1 expression and a high rate of oxidative metabolism [82]. Immune disorders lead to the development of type 1 diabetes and hyperglycemia, and thereby increase autoimmune attack, leading to the formation of a “vicious circle” [83]. mTOR protein kinase is an important regulator of lymphocyte immunometabolism and exists as a subunit of the intracellular multimolecular signaling complexes, mTORC1 and mTORC2 [84]. As part of these complexes, mTOR integrates both intracellular and extracellular signals and serves as one of the master regulator of metabolism, growth, proliferation, and survival of lymphocytes and other cells. Moreover, mTOR may be inhibited by metformin via AMPK [85]. High mTOR activity may cause the progression of diabetes due to the activation of effector proinflammatory subpopulations of lymphocytes. However, its low activity promotes the differentiation of Treg by blocking insulitis [86].
5. Metformin and COVID-19
5.1. Pleiotropic effects of metformin and the signaling pathway of metformin – AMPK – mTOR – SARS-CoV-2
Metformin was initially used as an anti-influenza agent and the decrease in blood glucose was only one of its side effects [87]. The many pleiotropic effects of metformin and its widespread use in medicine today prompted scientists to call it the 21st-century aspirin [88]. However, the initial discovery of the antiviral effects of metformin prompted us to consider the possibility of its use as one of the drugs to combat the SARS-CoV-2 virus. Along with antidiabetic effects metformin possesses other beneficial actions for an organism including lifespan extension [89].
It was previously reported that diabetic COVID-19 patients showed a more severe disease course as compared to nondiabetic counterparts [90]. Using metformin as a glucose-lowering agent results in anti-inflammatory and immunomodulatory effects. Moreover, using metformin for COVID-19 therapy improved insulin sensitivity and led to a reduction in blood glucose levels of the patients [28] that, in turn, reduced the risk of SARS-CoV-2 infections [91].
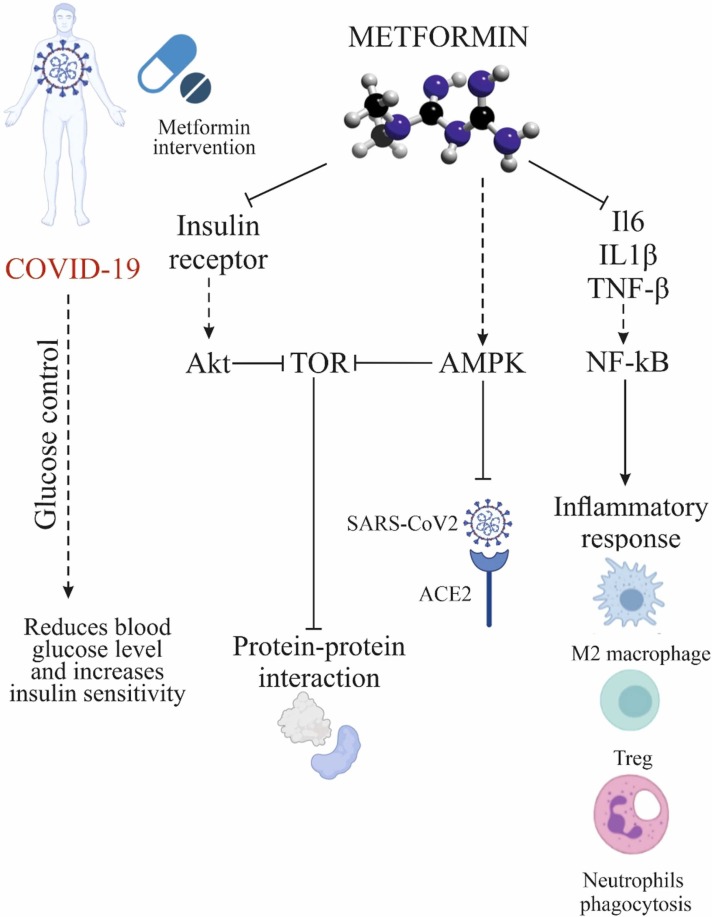
Metformin activates AMPK in hepatocytes, causing its phosphorylation. This is the main mechanism by which the beneficial effects metformin on glucose and lipid metabolism are mediated [92]. Metformin activates AMPK via hepatic kinase B1 (LKB1), inhibiting the mTOR pathway. It also indirectly attenuates AKT activation through phosphorylation of insulin receptor substrate 1 (IRS-1). This leads to inhibition of the mTOR signaling cascade. In addition, the PI3K/AKT/mTOR network plays an important role in MERS-CoV infection progression [93]. Since metformin inhibits the same pathway, it would be interesting to decipher its role concerning SARS-CoV-2.
There are several possible mechanisms of the positive effects of metformin upon COVID-19 infection. In addition to controlling glucose levels, reducing weight, and decreasing insulin resistance, metformin improves the immune response and reduces inflammation by promoting the formation of M2 macrophages and T regulatory and CD8+ memory T cells [94]. It also reduces the expression of genes encoding cytokines and chemokines associated with the inflammatory response [95]. Metformin also affects the composition of the microbiota and thus reduces inflammation [96]. The induction of autophagy by metformin also contributes to the elimination of pathogens and the control of inflammation [97]. Metformin stimulates AMPK activity and modifies the activity of catalase and superoxide dismutase. Serum of patients with COVID-19 have elevated levels of NETs (neutrophil extracellular traps), and though NETs are useful in protecting the host from pathogens. Overproduction of NETs can trigger an inflammatory cascade associated with cytokine storm and human acute respiratory distress syndrome (ARDS) [98]. Metformin reduces the number of neutrophils and neutrophil/lymphocyte ratio in diabetic patients, reduces NETosis in diabetic patients and prediabetes regardless of glucose control [99]. Animal studies have shown that metformin reduces cardiomyocyte damage after myocardial infarction by reducing cardiac remodeling and myocardial neutrophil activity [100]. Metformin also reduces neutrophil and macrophage infiltration in hyperoxia-induced lung damage [101]. Zhang and colleagues found a potential role of AMPK in the regulation of expression and stability of ACE2. The authors demonstrated that metformin increases the stability of ACE2 by phosphorylation of Ser680 of ACE2 in human umbilical vein endothelial cells and human embryonic kidney cells, which leads to conformational and functional changes in the ACE2 receptor. These changes can lead to decreased binding between the ACE2 receptor and the binding domain of the SARS-CoV-2 receptor (RBD) due to steric hindrance by the addition of phosphate groups, which reduces the infectivity of SARS-CoV-2. Moreover, metformin-induced AMPK-mediated phosphorylation of ACE2 improves the stability of ACE2 by inhibiting its ubiquitination and proteasome degradation [102]. Activation of AMPK-mediated signaling by metformin treatment that is associated with an increase in ACE2 levels and stability suppresses the inflammatory response by reducing the release of proinflammatory cytokines via inhibiting macrophage activation and NF-κB signaling [103].
The main immunometabolic effects of metformin are undoubtedly mediated by its effect on mTOR, which reduces the level of pro-inflammatory signaling and cytokine storm. Thus, metformin may be useful in controlling COVID-19 but most proposed mechanisms are currently still theoretical. In contrast, some authors suggested the possible risk of lactic acidosis in cases of multiple organ failure [104].
Another potential mechanism of the antiviral action of metformin is blocking the release of SARS-CoV-2 from the endosome. Vacuolar ATPase (V-ATPase) and endosomal Na+/H+ exchangers (eNHE) are important membrane compartments for pH regulation in endosomes. Several studies have shown that metformin can directly affect eNHE and/or V-ATPase, causing the suppression of viral infection by increased pH in endosomes [105], [106]. Additionally, metformin may prevent the development of pulmonary fibrosis associated with COVID-19 [106]. All these data support the notion that metformin may be useful adjuvant therapy for patients in the acute, chronic, and even recovery phases of COVID-19 ( Fig. 2).
Fig. 2.
Mechanisms of the positive effects of metformin to decrease COVID-19 severity.
The use of metformin rather than insulin in patients with diabetes and COVID-19 might be also relevant. Yu and co-authors have determined that insulin treatment can be a possible trigger for mortality in patients with COVID-19 and diabetes [107]. Paradoxically, insulin injections to diabetic COVID-19 patients were associated with a higher mortality rate rather than hyperglycemia. One of the explanations for these effects is that insulin can stimulate the maturation of pro-IL-1β via the NLRP3-inflammasome in activated macrophages [108]. SARS-CoV-2 activates the NLRP3 inflammasome and thus may enhance the action of insulin. Therefore, treatment alternatives to insulin should be considered. Yu et al. performed additional analyzes and compared insulin with other antidiabetic treatments in patients with COVID-19 [107]. Interestingly, in patients receiving insulin, the mortality rate was significantly higher than in patients receiving any other antidiabetic treatment. Even patients who received insulin alone had higher mortality, compared to those treated with insulin in combination with other antidiabetic drugs, despite the higher initial levels of glucose and HbA1c in the latter group. Although metformin may theoretically increase the risk of lactic acidosis, it is still an attractive alternative.
Inhibition of complex I of the mitochondrial electron transport chain (ETC) by metformin treatment led to reduced generation of reactive oxygen species (ROS) [91]. Moreover, similar to metformin-related mTORC1 signaling suppression, ETC inhibition also results in the suppression of host–viral protein interactions that benefit from reducing viral replication and maturation [106].
During illness, renal function should be carefully monitored because of the high risk of chronic kidney disease or acute kidney injury. Nevertheless, among antidiabetic drugs, metformin is the most tolerated and has only mild side effects.
5.2. Retrospective analysis of observations of metformin use in diabetic patients hospitalized with severe COVID-19
Several retrospective analyzes of patients with T2DM hospitalized for severe COVID-19 showed that metformin use may be associated with reduced mortality [4], [109], [110], [111], [112], [113], [114], [115], [116], [117], [142]. A beneficial effect (significant or not) was observed in almost all studies with an overall reduction in mortality of 25% ( Table 1).
Table 1.
Characteristics of the observational studies comparing death rates in metformin users vs non-users among patients with type 2 diabetes hospitalized for COVID-19.
| No. | Author | Subjects | Mortality (Metformin vs non Metformin) (n) | Odds Ratio M-H (Mantel–Haenszel test), Fixed, 95% CI |
|---|---|---|---|---|
| 1 | Abu-Jamous et al. | 411 | 4 vs 94 | 0.19 (0.05–0.70) |
| 2 | Cariou et al. | 1317 | N/A | 0.59 (0.42–0.84) |
| 3 | Bramante et al. | 6256 | 394 (17.8%) vs 791 (21.3%) | OR 0.802 (0.701, 0.917) |
| 4 | Chen et al. | 120 | 4 (9.30%) vs 15 (19.48%) | N/A |
| 5 | Crouse et al. | 239 | 8 (19%) vs 34 (81%) | OR 0.38 (0.17, 0.87) p = 0.0221 |
| 6 | Kim et al. | 235 | N/A | 0.36 (0.10–1.23) p = 0.10 |
| 7 | Luo et al. | 283; 104 vs 179 | 3 (2.9%) vs 22 (12.3%) | 4.36 ((1.22–15.59) p = 0.02 |
| 8 | Philipose et al. | 159 | 45 (22.6) | 1.39 (0.84–2.16) |
| 9 | Cheng et al. | 1213; 678 vs 535 | N/A | 1.65 (0.71,3.86) p = 0.27 |
| 10 | Khunti et al. | 1,800,005 | Mortality rate per 1000 person (9.9 vs 12.9) | 0.77 (0.73–0.81) |
The study of Khunti and colleagues involves a national cohort of 2.85 million people with type 2 diabetes. Current data demonstrated a statistically lower risk of COVID-19-related mortality in patients prescribed metformin and a higher risk of COVID-19-related mortality in patients prescribed insulin, supporting findings from previous smaller studies [114]. This large study provides the strong evidence of the true effect of metformin treatment because they provide excellent control of confounding. However, on our mind, there are some important factors not taken into account. First, the presence of confounding factors such as age, other comorbid conditions of patients, and other medications which are taken by patients that can affect the relationship between metformin and the mortality of COVID-19 still need to be considered. Second, study does not state the information regarding dosage and duration of metformin treatment in their patients. Smaller retrospective studies from the USA, China, and France have all reported a lower or neutral risk of COVID-19-related mortality in people previously or currently prescribed metformin. CORONADO, a nationwide observational study, demonstrated a decreased risk of death in the patients with T2D admitted for COVID-19 under metformin treatment [118]. Luo and colleagues compared the outcome of metformin users and nonusers in hospitalized COVID-19 patients with diabetes in Wuhan, China. This retrospective analysis demonstrated that antidiabetic treatment with metformin was associated with decreased mortality compared with diabetics not receiving metformin [111]. However, Luo and colleagues did not take into account the duration and complications of diabetes, the prevalence and treatment of obesity, study groups were not well balanced regarding the prevalence of comorbidities and antiviral treatments [119]. Hence, to better analyze the value of treatment with metformin for COVID-19 more studies are needed.
Several clinical studies are currently being conducted using metformin for the treatment of COVID-19 such as the “COVIDOUT – Outpatient treatment of COVID-19 with Metformin” (NCT04510194; Phase II/III; 750 participants; age 30–85 years). The current clinical trial aims to investigate whether metformin as a widely used anti-inflammatory drug could prevent inflammation caused by COVID-19. It was suggested that lower cases of hospitalization or mortality from the infection occurred in individuals taking metformin. Moreover, daily treatment of non-hospitalized adults with SARS-CoV-2 by metformin at a dose 1500 mg can prevent hypoxia and emergency department utilization. The study demonstrated prevention of disease progression in COVID-19 and improvement of viral load and C-reactive protein (CRP).
A Phase II trial “Pilot Study into the Use of Metformin and Low Dose Naltrexone (LDN) for Patients with Coronavirus Disease 2019 (COVID-19) – Assessment of Short and Long Term Effects” (NCT04604678; 80 participants, age 30–70 years) describes additional cases of patients with active COVID-19 who were treated with LDN and metformin and obtained clinical benefits. It was of interest to note that using a combination of metformin (1500 mg/day) and LDN (4.5 mg/day) led to a significant reduction of symptoms and disease severity, recovery time, hospitalization rates, and mortality in COVID-19 patients. Based on these case reports, the patients experienced significant clinical improvement after 1, 2, and 4 weeks after initiation of treatment.
Another Phase II trial, the “Adaptive Study for Efficacy and Safety of Metformin Glycinate for the Treatment of Patients with Metabolic Syndrome (MS) and Type-2 Diabetes Mellitus, Hospitalized with Severe Acute Respiratory Syndrome Secondary to SARS-CoV-2 (randomized, double-blind)” (NCT04626089) investigates the metformin glycinate as a potential treatment in COVID-19 patients who suffer from metabolic syndrome or type 2 diabetes. The investigators evaluate the efficacy and safety of metformin glycinate at a dose of 620 mg twice daily in 20 participants at the age of 18 years and older. The study demonstrated normalization of fever and oxygen saturation under metformin glycinate treatment. A similar trial is being conducted (NCT04625985; Phase II; 20 participants, 18 years and older) to discover the efficacy and safety of metformin glycinate at a dose 620 mg twice daily in hospitalized patients with Severe Acute Respiratory Syndrome secondary to SARS-CoV2.
Nevertheless, the results of these studies should be interpreted with caution. Individuals not taking metformin may relate to patients with contraindications to metformin prescription, such as old age, renal failure, and/or cardiovascular disease. On the other hand, metformin is more commonly used in obese patients with T2DM, and obesity is associated with a higher risk of severe COVID-19 infection and a higher mortality rate [120]. Naturally, conclusions about the effect of metformin therapy can be made only based on double-blind randomized controlled trials, which are unlikely in the context of COVID-19. Given the low cost of metformin, pharmaceutical companies are unlikely to be interested in planning a study to demonstrate the benefits of metformin for clinical outcomes associated with COVID-19. Also, Bramante et al. reported gender differences — the reduction in mortality among patients taking metformin was very significant in women, but not observed in men [110]. According to the authors, this gender-dependent difference is due to the larger decrease in TNF-α caused by metformin in women than in men and suggests that metformin provides protection against COVID-19 due to TNF-α-mediated effects [110]. A recent study identified 332 protein interactions between SARS-CoV-2 proteins and human proteins using mass spectrometric analysis [121] and showed that metformin can affect interactions between viral Nsp7 protein and human NDUFA2, as well as the viral protein Orf9c and human NDUFAF1 or NDUFB9, and thus has antiviral activity.
The results of these studies are consistent with previous observations that showed significantly lower mortality in patients treated with metformin, compared to those who did not take metformin during bacterial infections such as sepsis and tuberculosis [122]. It has also been found that metformin is associated with significantly lower mortality in patients with respiratory conditions such as chronic obstructive pulmonary disease [123]. The potential antiviral effects of metformin have been shown in several studies. Thus, metformin suppresses dengue virus infection by restoring AMPK activity, reduces the replication of Coxsackie B3 virus (CVB3), and protects mice from CVB3-induced myocarditis [124]. Metformin treatment also suppresses hepatitis B virus (HBV) replication by repressing genes associated with viral transcription, including LRH1, PPARα, and HNF4α [125]. In HCV and T2DM patients, the effects of metformin on sustained viral response are controversial, however, metformin reduces the incidence of hepatocellular carcinoma, possibly by activating type I INF signaling against HCV by activating AMPK [126]. Metformin is the first-line drug for the treatment of type 2 diabetes and insulin resistance in people living with HIV, but metformin may also play a role in the pathogenesis of HIV [127]. An independent study in China showed that metformin inhibits the phosphorylation of NF-κB/p65 to suppress CD54 expression in CD4+ T cells, which is associated with the progression of the disease in people living with HIV [128]. Metformin may also reduce inflammation through the intestinal microbiota, by increasing the number of goblet cells and mucin production, mediated by an increase in the number of Akkermansia muciniphila and bacteria producing of short-chain fatty acids, such as Bacteroides and Butyricimonas [129]. Several clinical studies investigating the relationship between intestinal microbiota and metformin treatment were conducted in people with T2DM [130] and confirmed the relationship between glucose tolerance and metformin modulated intestinal microbiota. In addition to A. muciniphila, people treated with metformin had larger populations of intestinal microbes, including Butyrivibrio, Bifidobacterium bifidum, and Megasphaera, which are known to produce short-chain fatty acids. Regulation of intestinal microbiota by increasing auspicious types such as Akkermansia spp., may enhance the antidiabetic effect of metformin [131]. On the other hand, modifications of the microbiota caused by metformin can affect immune function.
Metformin showed direct antimycobacterial effects and controlled the growth of drug-resistant bacterial strains, increased the production of mitochondrial reactive oxygen species (mROS), and promoted the fusion of phagosomes and lysosomes [132]. Retrospective analysis of patients with tuberculosis who took metformin showed a lower mortality rate than patients who did not take metformin [133]. Laboratory research using cells or model organisms shows that metformin may be effective against many other pathogens, including Trypanosoma cruzi, Trichinella spiralis, Staphylococcus aureus, and Pseudomonas aeruginosa [134]. Interestingly, metformin affects the level of antibody production during vaccination against influenza, which also depends on the age of the vaccinated [135]. While many COVID-19 vaccines are still in the development stage, it is important to note that the response to the vaccine also decreases with age. The effects of metformin in vaccinated to determine the potential mechanisms of strengthening of defense reactions in the elderly remains to be seen.
5.3. An effect of metformin on immune cells and cytokine storm
Metformin suppresses mTOR signaling by AMPK-dependent or -independent mechanisms [136]. Moreover, metformin may regulate other pathways related to inflammation and autoimmunity, including NF-κB and mitogen-activated protein kinase (MAPK)/c-Jun NH2-terminal kinase (JNK) [137]. The consequences of AMPK activation may explain many of the effects of metformin on immune homeostasis. Indeed, after the activation of immune cells, even those that have strong anti-inflammatory properties, are subject to metabolic reprogramming [138]. A subset of inflammatory cells such as neutrophils, M1-macrophages, Th17, and effector Th1-cells predominantly produce ATP through glycolysis, whereas cells of anti-inflammatory origin, i.e. memory T cells, regulatory T cells (Tregs), and M2 macrophages, promote the production of mitochondrial ATP [138]. In this context, AMPK activation promotes the oxidation of substrates in mitochondria, thereby limiting the glycolytic capacity of cells [139]. mTOR-deficient T cells cannot differentiate into Th1, Th2, or Th17 effector cells while retaining their ability to differentiate into Treg [140].
Administration of metformin to mice with experimental autoimmune encephalomyelitis (EAE, a T-cell model of multiple sclerosis) leads to a slower progression of the disease, a decrease in the infiltration of the CNS by inflammatory cells, and the expression of inflammatory cytokines IFN-γ, TNF-α, IL-17, IL-1β, and IL-6. T cells isolated from mice treated with metformin exhibit reduced expression of IFN-γ and IL-17 and the T-box and RORγt transcription factors that affect Th1 and Th17 differentiation, respectively [141]. Metformin also increased the number of Tregs in animals with EAE by inhibiting the mTOR pathway. Similar effects of metformin has been shown in animal models of rheumatoid arthritis (RA) [142], reducing the severity of bone destruction, production of inflammatory cytokines, and T-cells expressing RORγt. The ability of metformin to reduce the level of pro-inflammatory signaling through the AMPK/mTOR pathway has also been confirmed in colitis induced by dextran sodium sulfate (DSS), as well as various models of systemic lupus erythematosus [143]. In patients with type 2 diabetes, there is a decrease in the number of export of CD127+ and CD132+ naive T cells from the thymus and metformin restores this indicator [144]. In addition, 12 weeks of metformin treatment reduced IL-17 levels in patients with diabetes.
In primary peritoneal macrophages of mice, metformin treatment suppressed lipopolysaccharide (LPS)-induced expression of TNF-α and IL-6 in a dose-dependent manner [145]. In mice kept on a high-fat diet (HFD-model 2 diabetes) metformin treatment resulted in decreased serum levels of IL-6 and TNF-α and led to AMPK-mediated modulation of macrophage polarization with a shift towards the anti-inflammatory phenotype M2 [146]. In patients with impaired glucose tolerance, the addition of metformin for 12 weeks reduced the LPS-induced production of TNF-α and IL-6 by peripheral blood monocytes [147]. Similarly, in patients with impaired fasting glucose treated with simvastatin, metformin supplementation reduced LPS-induced production of TNF-α, IL-1β, IL-6, IL-8, and MCP-1 [147]. Also, the formation of NETs plays an important role in the pathogenesis of COVID-19 and type 2 diabetes. NETs can even be experimentally induced in vitro by exposing human peripheral blood leukocytes to high glucose concentrations [148]. Increased formation of NETs is observed in both of these pathologies, and this process can be reduced by treatment with metformin [149]. In patients with prediabetes, metformin treatment reduces the concentration of NETs regardless of glycemic control [150].
Some studies show a detrimental effect of metformin on the host immune response. Interferons such as IFN-α are essential for priming the T cell response against viral pathogens such as SARS-CoV-2 [41]. A study by Saenwongsa and colleagues reported that metformin treatment in patients with T2DM inhibits the expression of IFN-α in human peripheral blood mononuclear cells (PBMCs) via the mTORC1 pathway [148]. In the same study, patients with T2DM prescribed with metformin or glibenclamide had a delayed and reduced humoral immune longevity to influenza viruses after vaccination [151]. Similarly, metformin reduced Type I interferon-stimulated genes in CD4+ T cells from human PBMCs after IFN-α stimulation [152].
6. Conclusion and future perspectives
This literature analysis shows that the cytokine storm induced by SARS-CoV-2 significantly worsens the prognosis in patients with diabetes due to dysregulation of the AMPK/mTOR signaling pathway. Therefore, activation of AMPK and/or inhibition of the mTOR-mediated signaling pathway may be an important new target for drug development in COVID-19 therapy. In this aspect, metformin is of considerable interest. However, despite several potential positive effects, some issues need further clarification, For example, how does metformin affect other comorbidity conditions, in particular the risk of adverse cardiopulmonary outcomes in COVID-19? Will metformin treatment be clinically beneficial in patients with COVID-19 who do not suffer from diabetes or obesity? How safe is metformin therapy for people infected with SARS-CoV-2 who have not previously taken it?
While effective vaccines are at the forefront in the fight against COVID-19, drugs routinely used for other pathological conditions must not be overlooked in terms of their potential efficacy to treat this disease. The antidiabetic drug metformin, in addition to its glucose-lowering effect, has potential antiviral, cardioprotective, vasculoprotective, immunomodulatory, and anti-inflammatory effects and hence could be repurposed for the treatment of COVID-19. Furthermore, metformin is considered to be safe, is well-tolerated, has minimal side effects, a low cost, and can be made readily available to COVID-19 patients who may benefit from metformin intervention.
CRediT authorship contribution statement
Olexandr Kamyshnyi: Conceptualization, Writing – original draft. Victoriya Matskevych: Writing – original draft. Tetyana Lenchuk: Writing – original draft. Olha Strilbytska: Writing – original draft, Writing – review & editing. Kenneth Storey: Writing – review & editing. Oleh Lushchak: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Conflict of interest statement
The authors have no conflict of interest to declare.
Acknowledgments
This work was partially supported by a grant from the National Science Foundation of Ukraine (#2020.01/0157).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.biopha.2021.112230.
Appendix A. Supplementary material
Supplementary material.
.
References
- 1.Fleming N., Sacks L.J., Pham C.T., Neoh S.L., Ekinci E.I. An overview of COVID-19 in people with diabetes: pathophysiology and considerations in the inpatient setting. Diabet. Med. 2021;38 doi: 10.1111/dme.14509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamyshnyi A., Krynytska I., Matskevych V., Marushchak M., Lushchak O. Arterial hypertension as a risk comorbidity associated with COVID-19 pathology. Int. J. Hypertens. 2020;2020 doi: 10.1155/2020/8019360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puig-Domingo M., Marazuela M., Giustina A. COVID-19 and endocrine diseases. A statement from the European Society of Endocrinology. Endocrine. 2020;68:2–5. doi: 10.1007/s12020-020-02294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cariou B., Hadjadj S., Wargny M., Pichelin M., Al-Salameh A., Allix I., Amadou C., Arnault G., Baudoux F., Bauduceau B., Borot S., Bourgeon-Ghittori M., Bourron O., Boutoille D., Cazenave-Roblot F., Chaumeil C., Cosson E., Coudol S., Darmon P., Disse E., Ducet-Boiffard A., Gaborit B., Joubert M., Kerlan V., Laviolle B., Marchand L., Meyer L., Potier L., Prevost G., Riveline J., Robert R., Saulnier P., Sultan A., Thébaut J., Thivolet C., Tramunt B., Vatier C., Roussel R., Gautier J., Gourdy P., CORONADO investigators Phenotypic characteristics and prognosis of inpatients with COVID-19 and diabetes: the CORONADO study. Diabetologia. 2020;63:1500–1515. doi: 10.1007/s00125-020-05180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paces J., Strizova Z., Smrz D., Cerny J. COVID-19 and the immune system. Physiol. Res. 2020;69:379–388. doi: 10.33549/physiolres.934492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Onomoto K., Onoguchi K., Yoneyama M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell Mol. Immunol. 2021;18:1–17. doi: 10.1038/s41423-020-00602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carty M., Guy C., Bowie A.G. Detection of viral infections by innate immunity. Biochem. Pharmacol. 2021;183 doi: 10.1016/j.bcp.2020.114316. [DOI] [PubMed] [Google Scholar]
- 8.Root-Bernstein R. Innate receptor activation patterns involving TLR and NLR synergisms in COVID-19, ALI/ARDS and sepsis cytokine storms: a review and model making novel predictions and therapeutic suggestions. Int. J. Mol. Sci. 2021;22:2108. doi: 10.3390/ijms22042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.López-Reyes A., Martinez-Armenta C., Espinosa-Velázquez R., Vázquez-Cárdenas P., Cruz-Ramos M., Palacios-Gonzalez B., Gomez-Quiroz L.E., Martínez-Nava G.A. NLRP3 inflammasome: the stormy link between obesity and COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.570251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen K., Xiao F., Hu D., Ge W., Tian M., Wang W., Pan P., Wu K., Wu J. SARS-CoV-2 nucleocapsid protein interacts with RIG-I and represses RIG-mediated IFN-beta production. Viruses. 2020;13 doi: 10.3390/v13010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia X. Extreme genomic CpG deficiency in SARS-CoV-2 and evasion of host antiviral defense. Mol. Biol. Evol. 2020;37:2699–2705. doi: 10.1093/molbev/msaa094. 10.1093/molbev/msaa094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double‐membrane vesicles. mBio. 2013;4:e00524–e00613. doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Stürzel C.M., Fröhlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;15:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falck-Jones S., Vangeti S., Yu M., Falck-Jones R., Cagigi A., Badolati I., Österberg B., Lautenbach M.J., Åhlberg E., Lin A., Lepzien R., Szurgot I., Lenart K., Hellgren F., Maecker H., Sälde J., Albert J., Johansson N., Bell M., Loré K., Färnert A., Smed-Sörensen A. Functional monocytic myeloid-derived suppressor cells increase in blood but not airways and predict COVID-19 severity. J. Clin. Invest. 2021;131 doi: 10.1172/JCI144734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan S., Wu G. Is lymphopenia different between SARS and COVID-19 patients? FASEB J. 2021;35 doi: 10.1096/fj.202002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Braun J., Loyal L., Frentsch M., Wendisch D., Georg P., Kurth F., Hippenstiel S., Dingeldey M., Kruse B., Fauchere F., Baysal E., Mangold M., Henze L., Lauster R., Mall M.A., Beyer K., Röhmel J., Voigt S., Schmitz J., Miltenyi S., Demuth I., Müller M.A., Hocke A., Witzenrath M., Suttorp N., Kern F., Reimer U., Wenschuh H., Drosten C., Corman V.M., Giesecke-Thiel C., Sander L.E., Thiel A. SARS-CoV-2-reactive T cells in healthy donors and patients with COVID-19. Nature. 2020;587:270–274. doi: 10.1038/s41586-020-2598-9. [DOI] [PubMed] [Google Scholar]
- 20.Shrotri M., van Schalkwyk M.C.I., Post N., Eddy D., Huntley C., Leeman D., Rigby S., Williams S.V., Bermingham W.H., Kellam P., Maher J., Shields A.M., Amirthalingam G., Peacock S.J., Ismail S.A. T cell response to SARS-CoV-2 infection in humans: a systematic review. PLoS One. 2021;16 doi: 10.1371/journal.pone.0245532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., Xie C., Ma K., Shang K., Wang W., Tian D.S. Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Cheng L., Li J., Wang X., Wang F., Liu L., Amit I., Zhang S., Zhang Z. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 25.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B., Tong S.Y.C., Lewin S.R., Kedzierska K. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duan Y.Q., Xia M.H., Ren L., Zhang Y.F., Ao Q.L., Xu S.P., Kuang D., Liu Q., Yan B., Zhou Y.W., Chu Q., Liu L., Yang X.P., Wang G.P. Deficiency of Tfh cells and germinal center in deceased COVID-19 patients. Curr. Med. Sci. 2020;40:618–624. doi: 10.1007/s11596-020-2225-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wan Y., Shang J., Sun S., Tai W., Chen J., Geng Q., He L., Chen Y., Wu J., Shi Z., Zhou Y., Du L., Li F. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J. Virol. 2020;94:e02015–e02019. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samuel S.M., Varghese E., Büsselberg D. Therapeutic potential of metformin in COVID-19: reasoning for its protective role. Trends Microbiol. 2021 doi: 10.1016/j.tim.2021.03.004. (S0966–842X(21)00063–00069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zangiabadian M., Nejadghaderi S.A., Zahmatkesh M.M., Hajikhani B., Mirsaeidi M., Nasiri M.J. The efficacy and potential mechanisms of metformin in the treatment of COVID-19 in the diabetics: a systematic review. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.645194. 10.3389/fendo.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varghese E., Samuel S.M., Liskova A., Kubatka P., Büsselberg D. Diabetes and coronavirus (SARS-CoV-2): molecular mechanism of metformin intervention and the scientific basis of drug repurposing. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muniyappa R., Gubbi S. COVID-19 pandemic, corona viruses, and diabetes mellitus. Am. J. Physiol. Endocrinol. Metab. 2020;318:E736–E741. doi: 10.1152/ajpendo.00124.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrelli A., Atkinson M.A., Pietropaolo M., Giannoukakis N. Modulation of leukocytes of the innate arm of the immune system as a potential approach to prevent the onset and progression of type 1 diabetes. Diabetes. 2021;70:313–322. doi: 10.2337/dbi20-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma R.C.W., Holt R.I.G. COVID-19 and diabetes. Diabet. Med. 2020;37:723–725. doi: 10.1111/dme.14300. 10.1111/dme.14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batchu S.N., Kaur H., Yerra V.G., Advani S.L., Kabir M.G., Liu Y., Klein T., Advani A. Lung and kidney ACE2 and TMPRSS2 in renin angiotensin system blocker treated comorbid diabetic mice mimicking host factors that have been linked to severe COVID-19. Diabetes. 2020;70 doi: 10.2337/db20-0765. (db200765–771) [DOI] [PubMed] [Google Scholar]
- 35.Fignani D., Licata G., Brusco N., Nigi L., Grieco G.E., Marselli L., Overbergh L., Gysemans C., Colli M.L., Marchetti P., Mathieu C., Eizirik D.L., Sebastiani G., Dotta F. SARS-CoV-2 receptor angiotensin I-converting enzyme type 2 (ACE2) is expressed in human pancreatic beta-cells and in the human pancreas microvasculature. Front. Endocrinol. 2020;11 doi: 10.3389/fendo.2020.596898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang L., Karakiulakis G., Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection. Lancet Respir. Med. 2020;8 doi: 10.1016/S2213-2600(20)30116-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao S., Lau A., So H.C. Exploring diseases/traits and blood proteins causally related to expression of ACE2, the putative receptor of SARS-CoV-2: a Mendelian randomization analysis highlights tentative relevance of diabetes-related traits. Diabetes Care. 2020;43:1416–1426. doi: 10.2337/dc20-0643. [DOI] [PubMed] [Google Scholar]
- 38.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Across H.L.H. Speciality Collaboration, UK COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo W., Li M., Dong Y., Zhou H., Zhang Z., Tian C., Qin R., Wang H., Shen Y., Du K., Zhao L., Fan H., Luo S., Hu D. Diabetes is a risk factor for the progression and prognosis of COVID-19. Diabetes Metab. Res. Rev. 2020;36 doi: 10.1002/dmrr.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Izzi-Engbeaya C., Distaso W., Amin A., Yang W., Idowu O., Kenkre J.S., Shah R.J., Woin E., Shi C., Alavi N., Bedri H., Brady N., Blackburn S., Leczycka M., Patel S., Sokol E., Toke-Bjolgerud E., Qayum A., Abdel-Malek M., Hope D.C.D., Oliver N.S., Bravis V., Misra S., Tan T.M., Hill N.E., Salem V. Adverse outcomes in COVID-19 and diabetes: a retrospective cohort study from three London teaching hospitals. BMJ Open Diabetes Res. Care. 2021;9 doi: 10.1136/bmjdrc-2020-001858. (e001858) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soliman A.T., Prabhakaran Nair A., Al Masalamani M.S., De Sanctis V., Abu Khattab M.A., Alsaud A.E., Sasi S., Ali E.A., Ola A.H., Iqbal F.M., Nashwan A.J., Fahad J., El Madhoun I., Yassin M.A. Prevalence, clinical manifestations, and biochemical data of type 2 diabetes mellitus versus nondiabetic symptomatic patients with COVID-19: a comparative study. Acta Biomed. 2020;91 doi: 10.23750/abm.v91i3.10214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szewczuk M., Boguszewska K., Kaźmierczak-Barańska J., Karwowski B.T. The role of AMPK in metabolism and its influence on DNA damage repair. Mol. Biol. Rep. 2020;47:9075–9086. doi: 10.1007/s11033-020-05900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Juszczak F., Caron N., Mathew A.V., Declèves A.E. Critical role for AMPK in metabolic disease-induced chronic kidney disease. Int. J. Mol. Sci. 2020;21:7994. doi: 10.3390/ijms21217994. PMID: 33121167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haye A., Ansari M.A., Rahman S.O., Shamsi Y., Ahmed D., Sharma M. Role of AMP-activated protein kinase on cardio-metabolic abnormalities in the development of diabetic cardiomyopathy: a molecular landscape. Eur. J. Pharmacol. 2020;888 doi: 10.1016/j.ejphar.2020.173376. [DOI] [PubMed] [Google Scholar]
- 46.Dasgupta B., Chhipa R.R. Evolving lessons on the complex role of AMPK in normal physiology and cancer. Trends Pharmacol. Sci. 2016;37:192–206. doi: 10.1016/j.tips.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sukumaran A., Choi K., Dasgupta B. Insight on transcriptional regulation of the energy sensing AMPK and biosynthetic mTOR pathway genes. Front. Cell Dev. Biol. 2020;8:671. doi: 10.3389/fcell.2020.00671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lushchak O., Strilbytska O., Piskovatska V., Storey K.B., Koliada A., Vaiserman A. The role of the TOR pathway in mediating the link between nutrition and longevity. Mech. Ageing Dev. 2017;164:127–138. doi: 10.1016/j.mad.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 49.Jacinto E. Amplifying mTORC2 signals through AMPK during energetic stress. Sci. Signal. 12. 2019;12 doi: 10.1126/scisignal.aax5855. (eaax5855) [DOI] [PubMed] [Google Scholar]
- 50.Yao F., Zhang M., Chen L. 5’-Monophosphate-activated protein kinase (AMPK) improves autophagic activity in diabetes and diabetic complications. Acta Pharm. Sin. B. 2016;6:20–25. doi: 10.1016/j.apsb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaiserman A.M., Lushchak O.V., Koliada A.K. Anti-aging pharmacology: promises and pitfalls. Ageing Res. Rev. 2016;31:9–35. doi: 10.1016/j.arr.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Piskovatska V., Strilbytska O., Koliada A., Vaiserman A., Lushchak O. Health benefits of anti-aging drugs. Subcell. Biochem. 2019;91:339–392. doi: 10.1007/978-981-13-3681-2_13. [DOI] [PubMed] [Google Scholar]
- 53.Piskovatska V., Strilbyska O., Storey K.B., Vaiserman A.M., Lushchak O. mTOR pharmacology. Encycl. Biomed. Gerontol. 2019:447–454. doi: 10.1016/B978-0-12-801238-3.62134-7. [DOI] [Google Scholar]
- 54.Palmer C.S., Cherry C.L., Sada-Ovalle I., Singh A., Crowe S.M. Glucose metabolism in T cells and monocytes: new perspectives in HIV pathogenesis. EBioMedicine. 2016;6:31–41. doi: 10.1016/j.ebiom.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O'Neill L.A., Pearce E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michalek R.D., Gerriets V.A., Jacobs S.R., Macintyre A.N., MacIver N.J., Mason E.F., Sullivan S.A., Nichols A.G., Rathmell J.C. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gubser P.M., Bantug G.R., Razik L., Fischer M., Dimeloe S., Hoenger G., Durovic B., Jauch A., Hess C. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 2013;14:1064–1072. doi: 10.1038/ni.2687. [DOI] [PubMed] [Google Scholar]
- 58.Wei J., Long L., Yang K., Guy C., Shrestha S., Chen Z., Wu C., Vogel P., Neale G., Green D.R., Chi H. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 2016;17:277–285. doi: 10.1038/ni.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H., Zheng L., Gardet A., Tong Z., Jany S.S., Corr S.C., Haneklaus M., Caffrey B.E., Pierce K., Walmsley S., Beasley F.C., Cummins E., Nizet V., Whyte M., Taylor C.T., Lin H., Masters S.L., Gottlieb E., Kelly V.P., Clish C., Auron P.E., Xavier R.J., O'Neill L.A.J. Succinate is an inflammatory signal that induces IL 1β through HIF 1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huynh A., DuPage M., Priyadharshini B., Sage P.T., Quiros J., Borges C.M., Townamchai N., Gerriets V.A., Rathmell J.C., Sharpe A.H., Bluestone J.A., Turka L.A. Control of PI(3) kinase in Treg cells maintains homeostasis and lineage stability. Nat. Immunol. 2015;16:188–196. doi: 10.1038/ni.3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Newton R., Priyadharshini B., Turka L.A. Immunometabolism of regulatory T cells. Nat. Immunol. 2016;17:618–625. doi: 10.1038/ni.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chang C.H., Curtis J.D., Maggi L.B., Jr, Faubert B., Villarino A.V., O'Sullivan D., Ching-Cheng Huang S., van der Windt G.J.W., Blagih J., Qiu J., Weber J.D., Pearce E.J., Jones R.G., Pearce E.L. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Sullivan D., van der Windt G.J.W., Ching-Cheng Huang S., Curtis J.D., Chang C., Buck M.D., Qiu J., Smith A.M., Lam W.Y., DiPlato L.M., Hsu F., Birnbaum M.J., Pearce E.J., Pearce E.L. Memory CD8+ T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. 2014;41:75–88. doi: 10.1016/j.immuni.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O., Binz T., Wegner A., Tallam A., Rausell A., Buttini M., Linster C.L., Medina E., Balling R., Hiller K. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Everts B., Amiel E., Ching-Cheng Huang S., Smith A.M., Chang C., Lam W.Y., Redmann V., Freitas T.C., Blagih J., van der Windt G.J.W., Artyomov M.N., Jones R.G., Pearce E.L., Pearce E.J. TLR-driven early glycolytic reprogramming via the kinases TBK1 IKKε supports the anabolic demands of dendritic cell activation. Nat. Immunol. 2014;15:323–332. doi: 10.1038/ni.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Haschemi A., Kosma P., Gille L., Evans C.R., Burant C.F., Starkl P., Knapp B., Haas R., Schmid J.A., Jandl C., Amir S., Lubec G., Park J., Esterbauer H., Bilban M., Brizuela L., Pospisilik J.A., Otterbein L.E., Wagner O. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012;15:813–826. doi: 10.1016/j.cmet.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Newton R., Priyadharshini B., Turka L.A. Immunometabolism of regulatory T cells. Nat. Immunol. 2016;17:618–625. doi: 10.1038/ni.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gerriets V.A., Kishton R.J., Nichols A.G., Macintyre A.N., Inoue M., Ilkayeva O., Winter P.S., Liu X., Priyadharshini B., Slawinska M.E., Haeberli L., Huck C., Turka L.A., Wood K.C., Hale L.P., Smith P.A., Schneider M.A., MacIver N.J., Locasale J.W., Newgard C.B., Shinohara M.L., Rathmell J.C. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Invest. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feingold K.R., Shigenaga J.K., Kazemi M.R., McDonald C.M., Patzek S.M., Cross A., Moser A., Grunfeld C. Mechanisms of triglyceride accumulation in activated macrophages. J. Leukoc. Biol. 2012;92:829–839. doi: 10.1189/jlb.1111537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J., Walsh M.C., Hoehn K.L., James D.E., Wherry E.J., Choi Y. Regulator of fatty acid metabolism, acetyl coenzyme a carboxylase 1, controls T cell immunity. J. Immunol. 2014;192:3190–3199. doi: 10.4049/jimmunol.1302985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berod L., Friedrich C., Nandan A., Freitag J., Hagemann S., Harmrolfs K., Sandouk A., Hesse C., Castro C.N., Bähre H., Tschirner S.K., Gorinski N., Gohmert M., Mayer C.T., Huehn J., Ponimaskin E., Abraham W., Müller R., Lochner M., Sparwasser T. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 2014;20:1327–1333. doi: 10.1038/nm.3704. [DOI] [PubMed] [Google Scholar]
- 72.Carr E.L., Kelman A., Wu G.S., Gopaul R., Senkevitch E., Aghvanyan A., Turay A.M., Frauwirth K.A. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakaya M., Xiao Y., Zhou X., Chang J., Chang M., Cheng X., Blonska M., Lin X., Sun S. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity. 2014;40:692–705. doi: 10.1016/j.immuni.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rath M., Muller I., Kropf P., Closs E.I., Munder M. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front. Immunol. 2014;5:532. doi: 10.3389/fimmu.2014.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pacella I., Piconese S. Immunometabolic checkpoints of Treg dynamics: adaptation to microenvironmental opportunities and challenges. Front. Immunol. 2019;27:1889. doi: 10.3389/fimmu.2019.01889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shyer J.A., Flavell R.A., Bailis W. Metabolic signaling in T cells. Cell Res. 2020;30:649–659. doi: 10.1038/s41422-020-0379-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zherebiatiev A.S., Kamyshny A.M. Expression of aryl hydrocarbon receptor and ATG16L1 protein in experimental oxazolone-induced colitis in rats. Fiziolohichnyĭ Zh. 2015;61:57–64. doi: 10.15407/fz61.05.057. [DOI] [PubMed] [Google Scholar]
- 78.Degen A.S., Krynytska I.Y., Kamyshnyi A.M. Changes in the transcriptional activity of the entero-insular axis genes in streptozotocin-induced diabetes and after the administration of TNF-α non-selective blockers. Endocr. Regul. 2020;54:160–171. doi: 10.2478/enr-2020-0019. [DOI] [PubMed] [Google Scholar]
- 79.Putilin D.A., Evchenko S.Y., Fedoniuk L.Y., Tokarskyy O.S., Kamyshny O.M., Migenko L.M., Andreychyn S.M., Hanberher I.I., Bezruk T.O. The influence of metformin to the transcriptional activity of the mTOR and FOX3 genes in parapancreatic adipose tissue of streptozotocin-induced diabetic rats. J. Med. Life. 2020;13:50–55. doi: 10.25122/jml-2020-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Palmer C.S., Hussain T., Duette G., Weller T.J., Ostrowski M., Sada-Ovalle I., Crowe S.M. Regulators of glucose metabolism in CD4+ and CD8+ T cells. Int. Rev. Immunol. 2015;25:1–12. doi: 10.3109/08830185.2015.1082178. [DOI] [PubMed] [Google Scholar]
- 81.Michalek R.D., Gerriets V.A., Jacobs S.R., Macintyre A.N., MacIver N.J., Mason E.F., Sullivan S.A., Nichols A.G., Rathmell J.C. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 2011;186:3299–3303. doi: 10.4049/jimmunol.1003613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Basu S., Hubbard B., Shevach E.M. Foxp3-mediated inhibition of Akt inhibits Glut1 (glucose transporter 1) expression in human T regulatory cells. J. Leukoc. Biol. 2015;97:279–283. doi: 10.1189/jlb.2AB0514-273RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Putilin D.A., Kamyshnyi A.M. Changes of glut1, mTOR and AMPK1α gene expression in pancreatic lymph node lymphocytes of rats with experimental diabetes mellitus. Med. Immunol.) 2016;18:339–346. doi: 10.15789/1563-0625-2016-4-339-346. [DOI] [Google Scholar]
- 84.Pollizzi K.N., Patel C.H., Sun I.H., Oh M.H., Waickman A.T., Wen J., Delgoffe G.M., Powell J.D. mTORC1 and mTORC2 selectively regulate CD8⁺ T cell differentiation. J. Clin. Invest. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hardie D.G., Ashford M.L. AMPK: regulating energy balance at the cellular and whole body levels. Physiology. 2014;29:99–107. doi: 10.1152/physiol.00050.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chapman N.M., Chi H. mTOR signaling, Tregs and immune modulation. Immunotherapy. 2014;6:1295–1311. doi: 10.2217/imt.14.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Salvatore T., Pafundi P.C., Morgillo F., Di Liello R., Galiero R., Nevola R., Marfella R., Monaco L., Rinaldi L., Adinolfi L.E., Sasso F.C. Metformin: an old drug against old age and associated morbidities. Diabetes Res. Clin. Pract. 2020;160 doi: 10.1016/j.diabres.2020.108025. [DOI] [PubMed] [Google Scholar]
- 88.Romero R., Erez O., Hüttemann M. Metformin, the aspirin of the 21st century: its role in gestational diabetes mellitus, prevention of preeclampsia and cancer, and the promotion of longevity. Am. J. Obstet. Gynecol. 2017;217:282–302. doi: 10.1016/j.ajog.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Piskovatska V., Storey K.B., Vaiserman A.M., Lushchak O. The use of metformin to increase the human healthspan. Adv. Exp. Med. Biol. 2020;1260:319–332. doi: 10.1007/978-3-030-42667-5_13. [DOI] [PubMed] [Google Scholar]
- 90.Singh A.K., Gillies C.L., Singh R., Singh A., Chudasama Y., Coles B., Seidu S., Zaccardi F., Davies M.J., Khunti K. Prevalence of co-morbidities and their association with mortality in patients with COVID-19: a systematic review and meta-analysis. Diabetes Obes. Metab. 2020;22:1915–1924. doi: 10.1111/dom.14124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Scheen A.J. Obésité et risque de COVID-19 sévère [Obesity and risk of severe COVID-19] Rev. Med. Suisse. 2020;16:1115–1119. [PubMed] [Google Scholar]
- 92.Zhou G., Myers R., Li Y. Role of AMP-activated protein kinase in mechanism of metformin action. J. Clin. Invest. 2001;108:1167–1174. doi: 10.1172/JCI200113505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kindrachuk J., Ork B., Hart B.J. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob. Agents Chemother. 2015;59:1088–1099. doi: 10.1128/AAC.03659-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schuiveling M., Vazirpanah N., Radstake T., Zimmermann M., Broen J.C.A. Metformin, a new era for an old drug in the treatment of immune mediated disease? Curr. Drug Targets. 2018;19:945–959. doi: 10.2174/1389450118666170613081730. [DOI] [PubMed] [Google Scholar]
- 95.Ba W., Xu Y., Yin G., Yang J., Wang R., Chi S., Wang Y., Li C. Metformin inhibits pro-inflammatory responses via targeting nuclear factor-kappaB in HaCaT cells. Cell Biochem. Funct. 2019;37:4–10. doi: 10.1002/cbf.3367. [DOI] [PubMed] [Google Scholar]
- 96.Ouyang J., Isnard S., Lin J., Fombuena B., Marette A., Routy B., Chen Y., Routy J.P. Metformin effect on gut microbiota: insights for HIV-related inflammation. AIDS Res. Ther. 2020;17:10. doi: 10.1186/s12981-020-00267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yew W.W., Chang K.C., Chan D.P., Zhang Y. Metformin as a host-directed therapeutic in tuberculosis: is there a promise? Tuberculosis. 2019;115:76–80. doi: 10.1016/j.tube.2019.02.004. [DOI] [PubMed] [Google Scholar]
- 98.Bendib I., de Chaisemartin L., Granger V. Neutrophil extracellular traps are elevated in patients with pneumonia-related acute respiratory distress syndrome. Anesthesiology. 2019;130:581–591. doi: 10.1097/ALN.0000000000002619. [DOI] [PubMed] [Google Scholar]
- 99.Menegazzo L., Scattolini V., Cappellari R. The antidiabetic drug metformin blunts NETosis in vitro and reduces circulating NETosis biomarkers in vivo. Acta Diabetol. 2018;55:593–601. doi: 10.1007/s00592-018-1129-8. [DOI] [PubMed] [Google Scholar]
- 100.Soraya H., Rameshrad M., Mokarizadeh A., Garjani A. Metformin attenuates myocardial remodelling and neutrophil recruitment after myocardial infarction in rat. Bioimpacts. 2015;5:3–8. doi: 10.15171/bi.2015.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang J., Dong J., Martin M., He M., Gongol B., Marin T.L. AMP-activated protein kinase phosphorylation of angiotensin-converting enzyme 2 in endothelium mitigates pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2018;198:509–520. doi: 10.1164/rccm.201712-2570OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bangi S., Barve R., Qamar A. Protective effects of CVD and DM medications in SARS-CoV-2 infection, SN Compr. Clin. Med. 2020:1–3. doi: 10.1007/s42399-020-00452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sharma L., Chang D., Cruz C.S.Dela. Does inflammation help during COVID-19? ERJ Open Res. 2020;6 doi: 10.1183/23120541.00557-2020. (00557-2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kow C.S., Hasan S.S. Metformin use amid coronavirus disease 2019 pandemic. J. Med. Virol. 2020;92:2401–2402. doi: 10.1002/jmv.26090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Eaton A.F., Merkulova M., Brown D. The H(+)ATPase (V-ATPase): from proton pump to signaling complex in health and disease. Am. J. Physiol. Cell Physiol. 2020;320:392. doi: 10.1152/ajpcell.00442.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Esam Z. A proposed mechanism for the possible therapeutic potential of metformin in COVID-19. Diabetes Res. Clin. Pract. 2020;167 doi: 10.1016/j.diabres.2020.108282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yu B., Li C., Sun Y., Wang D.W. Insulin treatment is associated with increased mortality in patients with COVID-19 and type 2 diabetes. Cell Metab. 2021;33:65–77. doi: 10.1016/j.cmet.2020.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dror E., Dalmas E., Meier D.T., Wueest S., Thévenet J., Thienel C., Timper K., Nordmann T.M., Traub S., Schulze F., Item F., Vallois D., Pattou F., Kerr-Conte J., Lavallard V., Berney T., Thorens B., Konrad D., Böni-Schnetzler M., Donath M.Y. Postprandial macrophage-derived IL-1β stimulates insulin, and both synergistically promote glucose disposal and inflammation. Nat. Immunol. 2017;18:283–292. doi: 10.1038/ni.3659. [DOI] [PubMed] [Google Scholar]
- 109.Bramante C., Ingraham N., Murray T., Marmor S., Hoversten S., Gronski J. Observational study of metformin and risk of mortality in patients hospitalized with Covid-19. medRxiv. 2020 doi: 10.1101/2020.06.19.20135095. (2020.06.19.20135095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen Y., Yang D., Cheng B., Chen J., Peng A., Yang C. Clinical characteristics and outcomes of patients with diabetes and COVID-19 in association with glucose-lowering medication. Diabetes Care. 2020;43:1399–1407. doi: 10.2337/dc20-0660. [DOI] [PubMed] [Google Scholar]
- 111.Luo P., Qiu L., Liu Y., Liu X.L., Zheng J.L., Xue H.Y., Liu W.H., Liu D., Li J. Metformin treatment was associated with decreased mortality in COVID-19 patients with diabetes in a retrospective analysis. Am. J. Trop. Med. Hyg. 2020;103:69–72. doi: 10.4269/ajtmh.20-0375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Crouse A.B., Grimes T., Li P., Might M., Ovalle F., Shalev A. Metformin use is associated with reduced mortality in a diverse population with COVID-19 and diabetes. Front. Endocrinol. 2020 doi: 10.1101/2020.07.29.20164020. (medRxiv, 2020.07.29.20164020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang B., Van Oekelen O., Mouhieddine T.H., Del Valle D.M., Richter J., Cho H.J. A tertiary center experience of multiple myeloma patients with COVID-19: lessons learned and the path forward. J. Hematol. Oncol. 2020;13:94. doi: 10.1186/s13045-020-00934-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Khunti K., Knighton P., Zaccardi F., Bakhai C., Barron E., Holman N., Kar P., Meace C., Sattar N., Sharp S., Wareham N.J., Weaver A., Woch E., Young B., Valabhji J. Prescription of glucose-lowering therapies and risk of COVID-19 mortality in people with type 2 diabetes: a nationwide observational study in England. Lancet Diabetes Endocrinol. 2021;9:293–303. doi: 10.1016/S2213-8587(21)00050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.B. Abu-Jamous, A. Anisimovich, J. Baxter, L. Mackillop, M.P. Vizcaychipi, A. McCarthy, R.T. Khan, Associations of comorbidities and medications with COVID-19 outcome: a retrospective analysis of real-world evidence data, medRxiv, 2020.08.20.20174169, doi: 10.1101/2020.08.20.20174169.
- 116.Z. Philipose, N. Smati, C.S.J. Wong, K. Aspey, M.A. Mendall, Obesity, old age and frailty are the true risk factors for COVID-19 mortality and not chronic disease or ethnicity in Croydon, MedRxiv, 2020, 2020, doi: 10.1101/2020.08.12.20156257.
- 117.Cheng X., Xin S., Chen Y., Li L., Chen W., Li W., Zhou B., Li C., Gong Y., Li F., Duan P., Zhou X. Effects of metformin, insulin on COVID-19 patients with pre-existed type 2 diabetes: a multicentral retrospective study. Life Sci. 2021;275 doi: 10.1016/j.lfs.2021.119371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lalau J.D., Al-Salameh A., Hadjadj S., Goronflot T., Wiernsperger N., Pichelin M., Allix I., Amadou C., Bourron O., Duriez T., Gautier J.F., Dutour A., Gonfroy C., Gouet D., Joubert M., Julier I., Larger E., Marchand L., Marre M., Meyer L., Olivier F., Prevost G., Quiniou P., Raffaitin-Cardin C., Roussel R., Saulnier P.J., Seret-Begue D., Thivolet C., Vatier C., Desailloud R., Wargny M., Gourdy P., Cariou B., CORONADO investigators Metformin use is associated with a reduced risk of mortality in patients with diabetes hospitalised for COVID-19. Diabetes Metab. 2020;47 doi: 10.1016/j.diabet.2020.101216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fysekidis M., Cohen R., Al-Salameh A. More studies are needed on the link between metformin and decreased mortality in diabetic COVID-19 patients. Am. J. Trop. Med. Hyg. 2020;103:1337–1338. doi: 10.4269/ajtmh.20-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scheen A.J., Marre M., Thivolet C. Prognostic factors in patients with diabetes hospitalized for COVID-19: findings from the CORONADO study and recent reports. Diabetes Metab. 2020;46:265–271. doi: 10.1016/j.diabet.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Singh K.K., Chaubey G., Chen J.Y., Suravajhala P. Decoding SARS-CoV-2 hijacking of host mitochondria in COVID-19 pathogenesis. Am. J. Physiol. Cell Physiol. 2020;319:C258–C267. doi: 10.1152/ajpcell.00224.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhang M., He J.Q. Impacts of metformin on tuberculosis incidence and clinical outcomes in patients with diabetes: a systematic review and meta-analysis. Eur. J. Clin. Pharmacol. 2020;76:149–159. doi: 10.1007/s00228-019-02786-y. [DOI] [PubMed] [Google Scholar]
- 123.Ho T.W., Huang C.T., Tsai Y.J., Lien A.S., Lai F., Yu C.J. Metformin use mitigates the adverse prognostic effect of diabetes mellitus in chronic obstructive pulmonary disease. Respir. Res. 2019;20:69. doi: 10.1186/s12931-019-1035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Xie W., Wang L., Dai Q., Yu H., He X., Xiong J., Sheng H., Zhang D., Xin R., Qi Y., Hu F., Guo S., Zhang K. Activation of AMPK restricts coxsackievirus B3 replication by inhibiting lipid accumulation. J. Mol. Cell Cardiol. 2015;85:155–167. doi: 10.1016/j.yjmcc.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 125.Honda M., Shirasaki T., Terashima T., Kawaguchi K., Nakamura M., Oishi N., Wang X., Shimakami T., Okada H., Arai K., Yamashita T., Sakai Y., Yamashita T., Mizukoshi E., Kaneko S. Hepatitis B virus (HBV) core-related antigen during nucleos(t)ide analog therapy is related to intra-hepatic HBV replication and development of hepatocellular carcinoma. J. Infect. Dis. 2016;213:1096–1106. doi: 10.1093/infdis/jiv572. [DOI] [PubMed] [Google Scholar]
- 126.Malik F., Mehdi S.F., Ali H., Patel P., Basharat A., Kumar A., Ashok F., Stein J., Brima W., Malhotra P., Roth J. Is metformin poised for a second career as an antimicrobial? Diabetes Metab. Res. Rev. 2018;34 doi: 10.1002/dmrr.2975. [DOI] [PubMed] [Google Scholar]
- 127.Lamarca K., García Sarasola A., Vidal F., Domingo P. Drug therapies for HIV-related metabolic disorders. Expert. Opin. Pharmacother. 2016;17:1327–1338. doi: 10.1080/14656566.2016.1187133. [DOI] [PubMed] [Google Scholar]
- 128.Chen X., Chen H., Zhang Z., Fu Y., Han X., Zhang Y., Xu J., Ding H., Cui H., Dong T., Shang H., Jiang Y. Elevated CD54 expression renders CD4+ T cells susceptible to natural killer cell-mediated killing. J. Infect. Dis. 2019;220:1892–1903. doi: 10.1093/infdis/jiz413. [DOI] [PubMed] [Google Scholar]
- 129.Shin N.R., Lee J.C., Lee H.Y., Kim M.S., Whon T.W., Lee M.S., Bae J.W. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 130.Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S., Prifti E., Vieira-Silva S., Gudmundsdottir V., Pedersen H.K., Arumugam M., Kristiansen K., Voigt A.Y., Vestergaard H., Hercog R., Costea P.I., Kultima J.R., Li J., Jørgensen T., Levenez F., Dore J., MetaHIT consortium, Nielsen H.B., Brunak S., Raes J., Hansen T., Wang J., Ehrlich S.D., Bork P., Pedersen O. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.de la Cuesta-Zuluaga J., Mueller N.T., Corrales-Agudelo V., Velasquez-Mejia E.P., Carmona J.A., Abad J.M., Escobar J.S. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid-producing microbiota in the gut. Diabetes Care. 2017;40:54–62. doi: 10.2337/dc16-1324. [DOI] [PubMed] [Google Scholar]
- 132.Naicker N., Sigal A., Naidoo K. Metformin as host-directed therapy for TB treatment: scoping review. Front. Microbiol. 2020;11:435. doi: 10.3389/fmicb.2020.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Degner N.R., Wang J.Y., Golub J.E., Karakousis P.C. Metformin use reverses the increased mortality associated with diabetes mellitus during tuberculosis treatment. Clin. Infect. Dis. 2018;66(2):198–205. doi: 10.1093/cid/cix819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brima W., Eden D.J., Mehdi S.F., Bravo M., Wiese M.M., Stein J., Almonte V., Zhao D., Kurland I., Pessin J.E., Zima T., Tanowitz H.B., Weiss L.M., Roth J., Nagajyothi F. The brighter (and evolutionarily older) face of the metabolic syndrome: evidence from Trypanosoma cruzi infection in CD-1 mice. Diabetes Metab. Res. Rev. 2015;31:346–359. doi: 10.1002/dmrr.2636. 10.1002/dmrr.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Agarwal D., Schmader K.E., Kossenkov A.V., Doyle S., Kurupati R., Ertl H.C.J. Immune response to influenza vaccination in the elderly is altered by chronic medication use. Immun. Ageing. 2018;15:19. doi: 10.1186/s12979-018-0124-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kalender A., Selvaraj A., Kim S.Y., Gulati P., Brule S., Viollet B., Kemp B.E., Bardeesy N., Dennis P., Schlager J.J., Marette A., Kozma S.C., Thomas G. Metformin, independent of AMPK, inhibits mTORC1 in a Rag GTPase-dependent manner. Cell Metab. 2010;11:390–401. doi: 10.1016/j.cmet.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Saleiro D., Platanias L.C. Intersection of mTOR and STAT signaling in immunity. Trends Immunol. 2015;36:21–29. doi: 10.1016/j.it.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pearce E.L., Pearce E.J. Metabolic pathways in immune cell activation and quiescence. Immunity. 2013;38:633–643. doi: 10.1016/j.immuni.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.O’Neill L.A., Hardie D.G. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 140.Delgoffe G.M., Kole T.P., Zheng Y., Zarek P.E., Matthews K.L., Xiao B., Worley P.F., Kozma S.C., Powell J.D. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Paintlia A.S., Mohan S., Singh I. Combinatorial effect of metformin and lovastatin impedes T-cell autoimmunity and neurodegeneration in experimental autoimmune encephalomyelitis. J. Clin. Cell Immunol. 2013;4(10) doi: 10.4172/2155-9899.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kang K.Y., Kim Y.K., Yi H., Kim J., Jung H.R., Kim I.J., Cho J., Park S., Kim H., Ju J.H. Metformin downregulates Th17 cells differentiation and attenuates murine autoimmune arthritis. Int. Immunopharmacol. 2013;16:85–92. doi: 10.1016/j.intimp.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 143.Yin Y., Choi S.C., Xu Z., Zeumer L., Kanda N., Croker B.P., Morel L. Glucose oxidation is critical for CD4+ T cell activation in a mouse model of systemic lupus erythematosus. J. Immunol. 2016;196:80–90. doi: 10.4049/jimmunol.1501537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dworacki G., Urazayev O., Bekmukhambetov Y., Iskakova S., Frycz B.A., Jagodzinski P.P., Dworacka M. Thymic emigration patterns in patients with type 2 diabetes treated with metformin. Immunology. 2015;146:456–469. doi: 10.1111/imm.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kim J., Kwak H.J., Cha J.Y., Jeong Y.S., Rhee S.D., Kim K.R., Cheon H.G. Metformin suppresses lipopolysaccharide (LPS)-induced inflammatory response in murine macrophages via activating transcription factor-3 (ATF-3) induction. J. Biol. Chem. 2014;289:23246–23255. doi: 10.1074/jbc.M114.577908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Jing Y., Wu F., Li D., Yang L., Li Q., Li R. Metformin improves obesity-associated inflammation by altering macrophages polarization. Mol. Cell Endocrinol. 2018;461:256–264. doi: 10.1016/j.mce.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 147.Krysiak R., Gdula-Dymek A., Okopien B. Monocyte-suppressing effect of high-dose metformin in fenofibrate-treated patients with impaired glucose tolerance. Pharmacol. Rep. 2013;65:1311–1316. doi: 10.1016/S1734-1140(13)71489-0. [DOI] [PubMed] [Google Scholar]
- 148.Menegazzo L., Ciciliot S., Poncina N., Mazzucato M., Persano M., Bonora B., Albiero M., de Kreutzenberg S.V., Avogaro A., Fadini G.P. NETosis is induced by high glucose and associated with type 2 diabetes. Acta Diabetol. 2015;52:497–503. doi: 10.1007/s00592-014-0676-x. [DOI] [PubMed] [Google Scholar]
- 149.Wong S.L., Demers M., Martinod K., Gallant M., Wang Y., Goldfine A.B., Kahn C.R., Wagner D.D. Diabetes primes neutrophils to undergo NETosis, which impairs wound healing. Nat. Med. 2015;21:815–819. doi: 10.1038/nm.3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Menegazzo L., Scattolini V., Cappellari R., Bonora B.M., Albiero M., Bortolozzi M., Romanato F., Ceolotto G., de Kreutzeberg S.V., Avogaro A., Fadini G.P. The antidiabetic drug metformin blunts NETosis in vitro and reduces circulating NETosis biomarkers in vivo. Acta Diabetol. 2018;55:593–601. doi: 10.1007/s00592-018-1129-8. [DOI] [PubMed] [Google Scholar]
- 151.Saenwongsa W., Nithichanon A., Chittaganpitch M., Buayai K., Kewcharoenwong C., Thumrongwilainet B., Butta P., Palaga T., Takahashi Y., Ato M., Lertmemongkolchai G. Metformin-induced suppression of IFN-α via mTORC1 signalling following seasonal vaccination is associated with impaired antibody responses in type 2 diabetes. Sci. Rep. 2020;10:3229. doi: 10.1038/s41598-020-60213-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Titov A.A., Baker H.V., Brusko T.M., Sobel E.S., Morel L. Metformin inhibits the type 1 IFN response in human CD4+ T cells. J. Immunol. 2019;203:338–348. doi: 10.4049/jimmunol.1801651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.