Abstract
One of the major clinical features of COVID-19 is a hyperinflammatory state, which is characterized by high expression of cytokines (such as IL-6 and TNF-α), chemokines (such as IL-8) and growth factors and is associated with severe forms of COVID-19. For this reason, the control of the “cytokine storm” represents a key issue in the management of COVID-19 patients. In this study we report evidence that the release of key proteins of the COVID-19 “cytokine storm” can be inhibited by mimicking the biological activity of microRNAs. The major focus of this report is on IL-8, whose expression can be modified by the employment of a molecule mimicking miR-93-5p, which is able to target the IL-8 RNA transcript and modulate its activity. The results obtained demonstrate that the production of IL-8 protein is enhanced in bronchial epithelial IB3-1 cells by treatment with the SARS-CoV-2 Spike protein and that IL-8 synthesis and extracellular release can be strongly reduced using an agomiR molecule mimicking miR-93-5p.
Keywords: COVID-19, Spike protein, Interleukin-8, Mir-93-5p, Gene therapy, MicroRNA
Abbreviations: RT-qPCR, reverse transcription quantitative polymerase chain reaction; miR/miRNA, microRNA; S-protein, SARS-CoV-2 Spike protein; IL, interleukin
1. Introduction
A major new challenge for global health is the dramatic pandemic caused by the severe acute respiratory syndrome coronavirus (SARS-CoV-2), responsible of COVID-19 (Corona Virus Disease-2019) [1], [2]. The exponential increase of the number of severe COVID-19 cases and associated deaths has required awareness and proactive actions with respect to diagnosis and possible therapeutic treatments [3], [4]. Despite the fact that vaccination against SARS-CoV-2 is considered a key approach to prevent virus spread within the population and to limit the clinical manifestation of COVID-19 [5], [6], [7], novel therapeutic protocols, drugs and management actions are urgently needed, also considering the increase of the number of SARS-CoV-2 mutants [8], [9], [10].
COVID-19 is characterized by two major clinical phases [11]. The first is the SARS-CoV-2 viral infection of target cells and tissues, leading to important clinical manifestations and complications, such as pulmonary failure [12], [13], [14]. The second phase is a deep inflammatory state, known as “cytokine storm”, caused by activation of pro-inflammatory genes, such as NF-kB, STAT-3, IL-6, IL-8, IL-1β, G-CSF [15], [16], [17], [18], [19], [20], [21], [22]. The COVID-19 cytokine storm is initiated by the attachment of the SARS-CoV-2 Spike protein to the membrane-bound form of ACE2 (angiotensin-converting enzyme 2), followed by hyperactivation of NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) [23], [24]. This hyperactivation of NF-κB in the lungs strongly contributes to the induction of the cytokine storm with subsequent ARDS (acute respiratory distress syndrome) [24], frequently observed in severe COVID-19 patients. The pharmacological approaches for treating ARDS need of novel anti-inflammatory reagents and therapeutic strategies, also considering that COVID-19 patients might respond differently to anti-inflammatory treatments [25], [26], [27], [28], [29], [30], [31], [32]. The impact of anti-SARS-CoV-2 pharmacological strategies and of anti-inflammatory protocols is clear, as recently reviewed in translating IL-6 biology into effective treatments [33].
In addition to a transcriptional regulation of the expression of pro-inflammatory genes [34], [35], post transcriptional regulation has been also proposed, involving epigenetic changes, some of which associated with microRNAs control [36], [37], [38], [39], [40].
MicroRNAs are short (about 20–23 nucleotides in length) non-coding RNAs that play a very important role in the post-transcriptional control of gene expression, through recognition of miRNA binding sites present in the target mRNA sequences (mainly in the 3′-UTR region) [41], [42]. In this respect, we have recently shown that microRNA miR-93-5p is involved in regulation of IL-8 (interleukin-8) gene expression [43]. The IL-8 chemokine is a key mediator associated with inflammation and is deeply involved in neutrophil recruitment and degranulation [20], [35], [44]. With respect to COVID-19, IL-8 is an important player of the “cytokine storm” and, therefore, it can be considered a molecular target of anti-inflammatory therapy in the management of COVID-19 patients [30]. The direct regulation of IL-8 mRNA by miR-93-5p in IB3-1 cells was demonstrated by Fabbri et al. using a luciferase vector containing the 3′-UTR region of the IL-8 mRNA, a luciferase vector deleted of the IL-8 3′-UTR region or a vector containing an IL-8 3′-UTR region in which the miR-93-5p site was mutagenized [43]. Following this study, the involvement of miR-93-5p in regulation of IL-8 was confirmed in other reports [45], [46], [47]. This was the reason for selecting IL-8 as the major focus of our study.
The main objective of this investigation was to study if the cytokine upregulation following SARS-CoV-2 infection is associated with dysregulation of miR-93-5p. The second objective of the study was to verify whether pharmacological regulation of pro-inflammatory genes by delivery of a miR-93-5p agomiR might be considered for the development of future anti-COVID-19 therapeutic protocols. As a model system we used bronchial epithelial IB3-1 cells [48] exposed to SARS-CoV-2 Spike protein. MicroRNA levels were analyzed by RT-qPCR, while cytokine/chemokine expression was analyzed by RT-qPCR and Bio-Plex analysis.
2. Materials and methods
2.1. Materials
AgomiR-93-5p and control sequences were obtained by Ambion (Thermo Fischer Scientific, Waltham, Massachusetts, USA). SARS-Cov-2 Spike recombinant glycoprotein (ab49046) was purchased by Abcam (Cambridge, UK).
2.2. Cell culture conditions
The IB3-1 cell line [48] was cultured in humidified atmosphere of 5% CO2/air in LHC-8 medium (Thermo Fischer Scientific) supplemented with 5% fetal bovine serum (FBS, Biowest, Nuaillé, France) in the absence of gentamycin. To verify the effect on proliferation, cell growth was monitored by determining the cell number/ml using a Z2 Coulter Counter (Coulter Electronics, Hialeah, FL, USA).
2.3. Stimulation of cells with SARS-CoV-2 Spike protein
SARS-CoV-2 Spike protein (139 KDa; stock concentration = 7.2 μM in 9% urea, 0.32% Tris-HCl pH 7.2, 50% glycerol) was diluted in 200 µl of LHC-8 medium to achieve the final concentrations used to treat IB3-1 cells. Briefly, cells seeded at 50% of confluence were treated with Spike protein (5–50 nM) and incubated for 30 min at 4 °C, then for 30 min at 37 °C, according with the protocol published by Wang et al. [49]. After this incubation, LHC-8 medium supplemented with 5% (final concentration) FBS was added to a final 500 μl volume and the cultures were further incubated at 37 °C and for 24 h. The S-protein buffer, used at a volume corresponding to the S-protein concentrations used, was found unable to stimulate the expression of IL-8 gene in treated IB3-1 cells.
2.4. PremiRNA transfection procedure
Transfection procedure of agomiR-93 in IB3-1 cells was performed following manufacturer’s instruction. Transfection was performed in 24-well plates, two hours before the stimulation with SARS-Cov-2 Spike protein. Cells transfection was performed using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen, Thermo Fischer Scientific) accordingly to manufacturer’s instruction, with 100 nM of hsa-miR-93-5p miRNA precursor (PM10951, Ambion, Thermo Fisher Scientific). MicroRNA Precursor Negative Control #1 (AM17110 Ambion, Thermo Fisher Scientific) was used as negative control [50].
2.5. RNA extraction
Cultured cells were trypsinized (0.05% trypsin and 0.02% EDTA; Sigma-Aldrich, St. Louis, Missouri, USA) and collected by centrifugation at 1,200 rpm for 8 min at 4 °C, washed twice with DPBS 1X (Gibco, Thermo Fischer Scientific) and lysed with Tri-Reagent (Sigma Aldrich), according to manufacturer’s instructions. The isolated RNA was washed once with cold 75% ethanol, dried and dissolved in nuclease free pure water before use. Obtained RNA was stored at −80 °C until use [51].
2.6. Quantitative analyses of miRNAs
MicroRNA relative quantification was performed using real-time RT-qPCR and miRNA-specific primers and probes (reported in Table 1 ) obtained from Applied Biosystems (Thermo Fischer Scientific). Reverse transcriptase (RT) reactions were performed using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Thermo Fischer Scientific) according to the manufacturer’s protocol. All RT reactions, including no-template controls and RT-minus controls, were run in duplicate using TaqMan Universal PCR Master Mix, no AmpErase UNG 2X (Applied Biosystems, Thermo Fischer Scientific) and CFX96 Touch Real-Time PCR Detection System (BioRad, Hercules, CA, USA), as described [52]. Data were collected and analyzed using Bio-Rad CFX Manager Software Version 1.7 (Bio-Rad). The relative expression was calculated using the comparative cycle threshold method and, as reference, sequences snRNA U6 and hsa-let-7c were used to normalize samples [53].
Table 1.
List of assays employed for miRNA detection.
| miRNA name | Assay ID |
|---|---|
| hsa-miR-93-5p | 001090 |
| hsa U6 snRNA | 001973 |
| hsa-let-7c-5p | 000379 |
2.7. Quantitative analyses of Interleukin mRNA
For IL-8 mRNA analysis 500 ng of total RNA were reverse transcribed to cDNA using the Taq-Man Reverse Transcription PCR Kit and random hexamers (Applied Biosystems, Thermo Fischer Scientific) in a final reaction volume of 50 µl. Real-time-qPCR experiments were carried out using assay composed by a primer pair and a fluorescently labeled 5′ nuclease probe purchased from IDT (Integrated DNA Technologies, Coralville, Iowa, USA; Assays ID: Hs.PT.58.38869678.g). Two µl of complementary DNA (cDNA) were amplified, in presence of 2X PrimeTime Gene Expression Master Mix for 40 PCR cycles using CFX96 Touch Real-Time PCR Detection System (Bio-Rad). Relative expression was calculated using the comparative cycle threshold method (ΔΔCT method) and the endogenous control: human β-actin, GAPDH and RPL13A were used as normalizer. Negative controls (no template cDNA and RT-minus control) were also run in every experimental plate to assess specificity and to rule out contamination. RT-qPCR reactions were performed in duplicate for both target and normalizer gene [50].
2.8. Analysis of proteins released from cells into culture supernatants
Proteins released into culture supernatants were measured using Bio-Plex Human Cytokine 27-plex Assay (Bio-Rad) as suggested by the manufacturer. The assay allows the multiplexed quantitative measurement of 27 cytokines/chemokines (including: FGF basic, Eotaxin, G-CSF, GM-CSF, IFN-γ, IL-1β, IL-1ra, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p70), IL-13, IL-15, IL-17A, IP-10, MCP-1 (MCAF), MIP-1α, MIP-1β, PDGF-BB, RANTES, TNF-α, VEGF) in a single well. 50 μl of cytokine standards or samples (diluted supernatants recovered from IB3-1 cells) were incubated with 50 μl of anticytokine conjugated beads in 96-well filter plate for 30 min at room temperature with shaking. Plate was washed by vacuum filtration three times with 100 μl of Bio-Plex Wash Buffer, 25 μl of diluted detection antibody were added, to each wells and plate was incubated for 30 min at room temperature with shaking. After three filter washes, 50 μl of streptavidin–phycoerythrin were added, and plate was incubated for 10 min at room temperature with shaking. Finally, plate was washed by vacuum filtration three times, beads were suspended in Bio-Plex Assay Buffer, plate was read by Bio-Rad 96-well plate reader. Data were collected and analyzed by the Bio-Plex Manager Software (Bio-Rad) [54].
2.9. Western blotting
Twenty micrograms of total proteic extract were denatured for 5 min at 98 °C and loaded on SDS polyacrylamide gel in Tris-glycine buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). The electrotransfer to 0.2 μm nitrocellulose membrane was performed overnight at 360 mA and 4 °C in electrotransfer buffer (25 mM Tris, 192 mM glycine, 10% methanol). Obtained membranes were pre-stained in Ponceau S solution (Sigma-Aldrich, St. Louis, MI. USA) to verify the transfer and incubated in 25 mL of blocking buffer for 1 h at room temperature. After three washes in TBST 1X (Tris Buffered Saline-Tween) membranes were incubated in primary antibody against NFκB p50/p105 (Cat: GTX133711, GeneTex, Irvine, CA, USA) overnight at 4 °C. The day after, the membranes were washed in TBST 1X and incubated for 1 h at room temperature, with an appropriate horseradish peroxidase-conjugated secondary antibody (anti-rabbit Igg HRP conjugated, Cat: 7074P3, 1:2000, Cell Signaling, Danvers, MA, USA). The primary antibody against β-actin (Cat: 4970S, Cell Signaling) was used as normalization control.
2.10. Statistics
In order to detect significance of the observed effects, results have been expressed as mean ± standard errors (SEM) and comparison among groups was made by using analysis of variances (ANOVA) with Dunnett's test for comparison with a single control. Statistical significance was defined as significant (p < 0.05) and highly significant (p < 0.01).
3. Results
We first briefly characterized the effects of SARS-CoV-2 Spike protein on the secretome profile and IL-8 gene expression in the human bronchial epithelial IB3-1 cell line. Then, we verified the Spike-mediated effects on the microRNA miR-93-5p, known to be a key regulator of IL-8 gene expression. Finally, we verified the effects of an agomiR-93 on Spike-mediated IL-8 upregulation.
3.1. SARS-CoV-2 Spike protein (S-protein) mediates a sharp increase of IL-8 mRNA and protein in IB3-1 cells
When the analysis of the secretome profile of the human bronchial epithelial IB3-1 cell line is performed, it is clearly evident that production of cytokines, chemokines and growth-factors is differential and the most abundant protein in IB3-1 supernatant is IL-8 (Fig. 1 A). In this work we considered to study the 15 proteins found to display the highest concentrations: PDGF, IL-6, IL-8, IL-9, IL-10, IL-15, FGF, G-CSF, GM-CSF, IFN-γ, IP-10, MCP-1, MIP-1β, RANTES, VEGF. In order to obtain information about the possible use of the “miRNA mimicking” approach to interfere with SARS-CoV-2 mediated induction of pro-inflammatory genes, we first analyzed the effects of exposure of IB3-1 cells to SARS-CoV-2 Spike protein (S-protein).
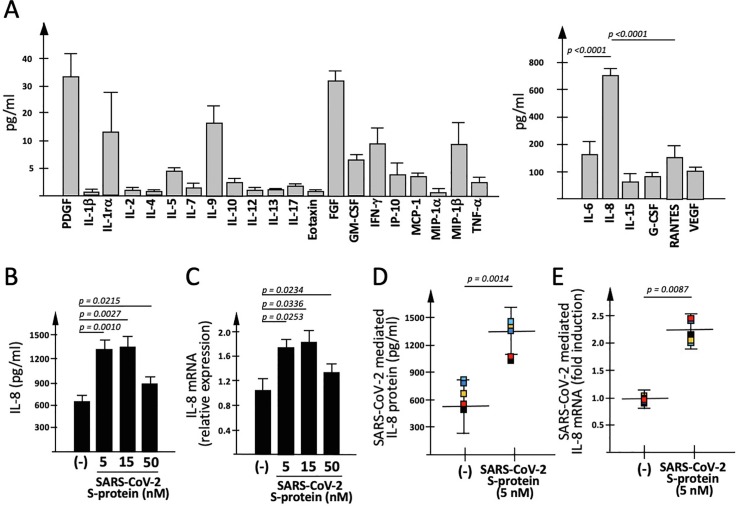
Fig. 1.
Effects of SARS-CoV-2 Spike protein on IB3-1 secretome. (A) Differential expression of cytokines, chemokines and growth factors in IB3-1 cells (Bio-Plex analysis). Medium was harvested after 48 h culture, starting from 30% confluence seeding cells (results are the average of 9 independent experiments). The significance of the difference between IL-6 and IL-8 content is shown. (B, C) Effects on IL-8 gene expression of 24 h exposure to SARS-CoV-2 Spike protein (S-protein) of IB3-1 cells (results are the average of 3 independent experiments). Release of IL-8 was quantified by Bio-plex analysis (B), IL-8 mRNA accumulation by RT-qPCR (C). (D,E) Summary of the effects of S-protein on IL-8 release (D) and IL-8 mRNA accumulation (E) by treated IB3-1 cells (results are the average of 5 independent experiments).
The data obtained studying IL-8 gene expression are shown in Fig. 1B–D. Fig. 1B shows that exposure to 5 nM SARS-CoV-2 S-protein is sufficient to induce a significant increase of released IL-8. The analysis conducted on the other proteins are shown in Supplementary Fig.S1 and demonstrate a differential effect of SARS-CoV-2 on the different proteins. S-protein induced increase was found for IL-6 and G-CSF, while no significant changes in IL-15, RANTES and VEGF release were observed after exposure of IB3-1 cells to 5 nM and 15 nM S-protein. Fully in agreement with secretome data (Fig. 1B), the exposure of IB3-1 cells to S-protein was associated with an increase on IL-8 mRNA content within cells (Fig. 1C). Observation of IL-8 up-regulation in S-protein exposed IB3-1 cells was highly reproducible, as shown by analyzing the data obtained by 5 different independent experiments and analyzing the extent of the S-protein mediated increase (pg/ml) of released IL-8 protein (Fig. 1D; p = 0.0014) and the fold induction of IL-8 mRNA (Fig. 1E; p = 0.0087). Moreover, the effects of SARS-CoV-2 S-protein on IL-8 expression were confirmed in other cell lines, such as the A549 [55] and Calu-3 [56] cell lines (Supplementary Fig.S2). Furthermore, when RT-qPCR analysis using β-actin as internal control was compared with those obtained using as internal control GAPDH and RPL13A sequences, similar results were obtained (Supplementary Fig.S3A).
3.2. SARS-CoV-2 Spike mediated induction of IL-8 is associated with inhibition of microRNA miR-93-5p
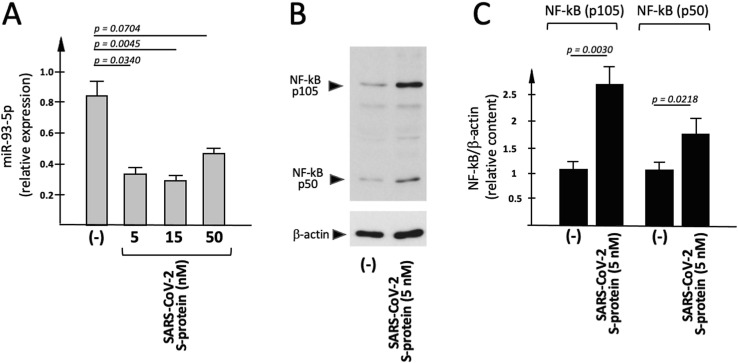
Since no comprehensive information is available in the literature on possible effects of SARS-CoV-2 on microRNAs, we tested whether the exposure to the Spike protein has effects on miR-93-5p. The microRNA miR-93-5p was selected in consideration of the fact that this microRNA has been firmly demonstrated to control IL-8 gene expression [43], [44], [45], [46], [47]. Among the molecular basis of this effects, at least two mechanisms of action have been proposed and strongly supported by experimental evidences: (a) miR-93-5p can directly bind the 3'-UTR sequence of the IL-8 mRNA [43] and (b) miR-93-5p suppresses Toll–like receptor 4 (TLR4), an upstream regulator of the nuclear factor NF–κB signaling pathway [57]. This is clearly of interest, since NF-κB is a transcriptional regulator of the IL-8 gene [35]. Fig. 2 A shows that miR-93-5p content is reduced in IB3-1 cells exposed to 5 nM SARS-CoV-2 Spike protein. Moreover, in Fig. 2 (B,D) the effect of exposure of IB3-1 to SARS-CoV-2 S-protein is shown by performing Western Blotting analysis of cellular extracts. Both NF-κB p105 and NF-κB p50 proteins significantly increased (respectively: 2.0 and 1.7 fold; p = 0.0218 for NF-κB p105 and p = 0.0030 for NF-κB p50) following treatment with 5 nM S-protein. The endogenous content of β-actin was analyzed as internal control.
Fig. 2.
Effects of SARS-CoV-2 Spike protein on miR-93-5p content and NF-κB p105/p50 production. (A) IB3-1 cells were exposed for 24 h to S-protein. RNA was isolated and miR-93-5p content quantified by RT-qPCR. (B,C) Proteins were purified from IB3-1 cells treated as described in (A), and Western blotting performed (see the Methods section) using antibodies recognizing NF-κB p105/p50 and β-actin. (B) Representative results indicating S-protein mediated upregulation of NF-κB; (C) quantitative analysis. The relative expression (average values ± SD) of NF-κB p105/p50 is indicated (n = 3).
In conclusion, increased expression of IL-8 (analyzed by Bio-Plex assay and RT-qPCR) is accompanied to a decreased production of miR-93-5p and an increased expression of the IL-8 gene regulator NF-κB in IB3-1 cells exposed to SARS-CoV-2 S-protein.
3.3. The miR-93-5p dependency of the genes producing the IB3-1 secretome
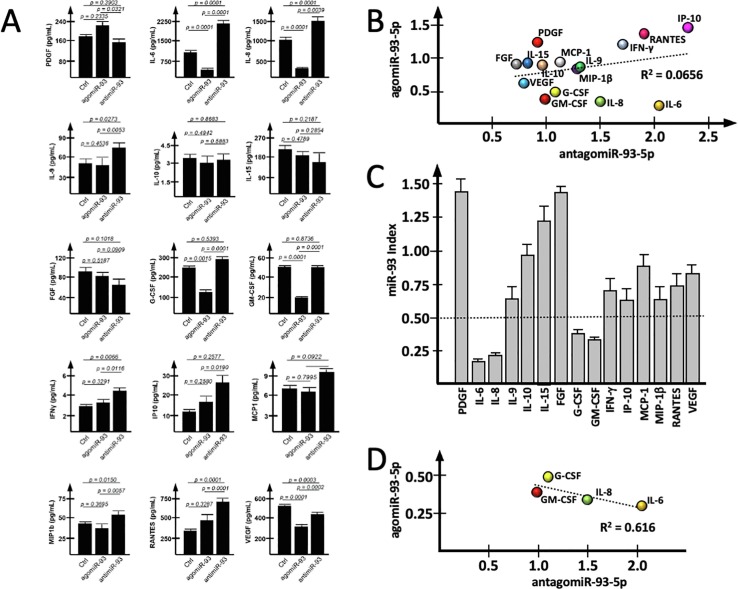
In order to have information about the possible regulation of IB3-1 secretome proteins by miR-93-5p, IB3-1 cells were treated in parallel with agomiR-93-5p and an antagomiR-93-5p molecule. After 48-hours treatment, medium was collected and Bio-Plex analysis was performed. Data concerning the 15 proteins highly expressed in IB3-1 cells (see Fig. 1A) are reported in Fig. 3 A. In the presence of an effect of miR-93-5p on gene expression, we were expecting an increased expression following transfection with the antagomiR-93-5p, associated with a down-regulation following transfection with the agomiR-93-5p. The data obtained demonstrated that this does not occur for all the analyzed proteins. In fact, when we tried to correlate the effects of the agomiR-93 with the effects of antimiR-93, a R2 = 0.0656 was obtained (Fig. 3B). This is also confirmed by a complementary analysis comparing the miR-93-5pINDEX, which is proposed to reflects the direct dependency from miR-93-5p activity, based on the determination of the treated/untreated fold values as follows: miR-93-5pINDEX = fold (agomiR treatment)/fold (antagomiR treatment). Following this algoritm, we expected low values of the miR-93-5pINDEX for the genes whose expression was regulated by miR-93-5p. Data regarding the miR-93-5pINDEX are shown in Fig. 3C and strongly suggest that the expression of IL-6, IL-8, G-CSF and GM-CSF are all regulated by miR-93-5p. Supporting this conclusion, when data of the transfections with the agomiR-93 and the antagomiR-93 were correlated, a higher level of correlation was found (R2 = 0.616) (Fig. 3D).
Fig. 3.
Effects of miR-93-5p mimic or inhibitor on the IB3-1 secretome. (A) Effects on selected secretome proteins of IB3-1 cells transfection with the agomiR-93-5p or the antagomiR-93-5p, respectively mimicking (the agomiR) or interfering (the antagomiR) with the biological functions of miR-93-5p. (B) Correlation between the effects of the agomiR-93-5p and the antimiR-93-5p on unselected secretome proteins. (C) miR-93-5pINDEX, calculated with the algoritm: fold (pre-miR-93 treatment)/fold (anti-miR-93 treatment). Low values of miR-93-5pINDEX are expected for genes responsive to miR-93-5p effects. (D) Correlation between effects of the agomiR-93-5p and the antimiR-93-5p on secretome proteins displaying a miR-93-5pINDEX lower than 0.5.
With respect to IL-8 gene expression, the differential effects on IL-8 mRNA following treatment with the agomiR-93 and the antagomiR-93 were reproducibly obtained (p < 0.01) using β-actin, GAPDH and RPL13A as housekeeping sequences (Supplementary Fig. S3B).
3.4. Treatment of IB3-1 cells with the miR-93-5p agomiR reverses IL-8 upregulation induced by SARS-CoV-2 Spike protein
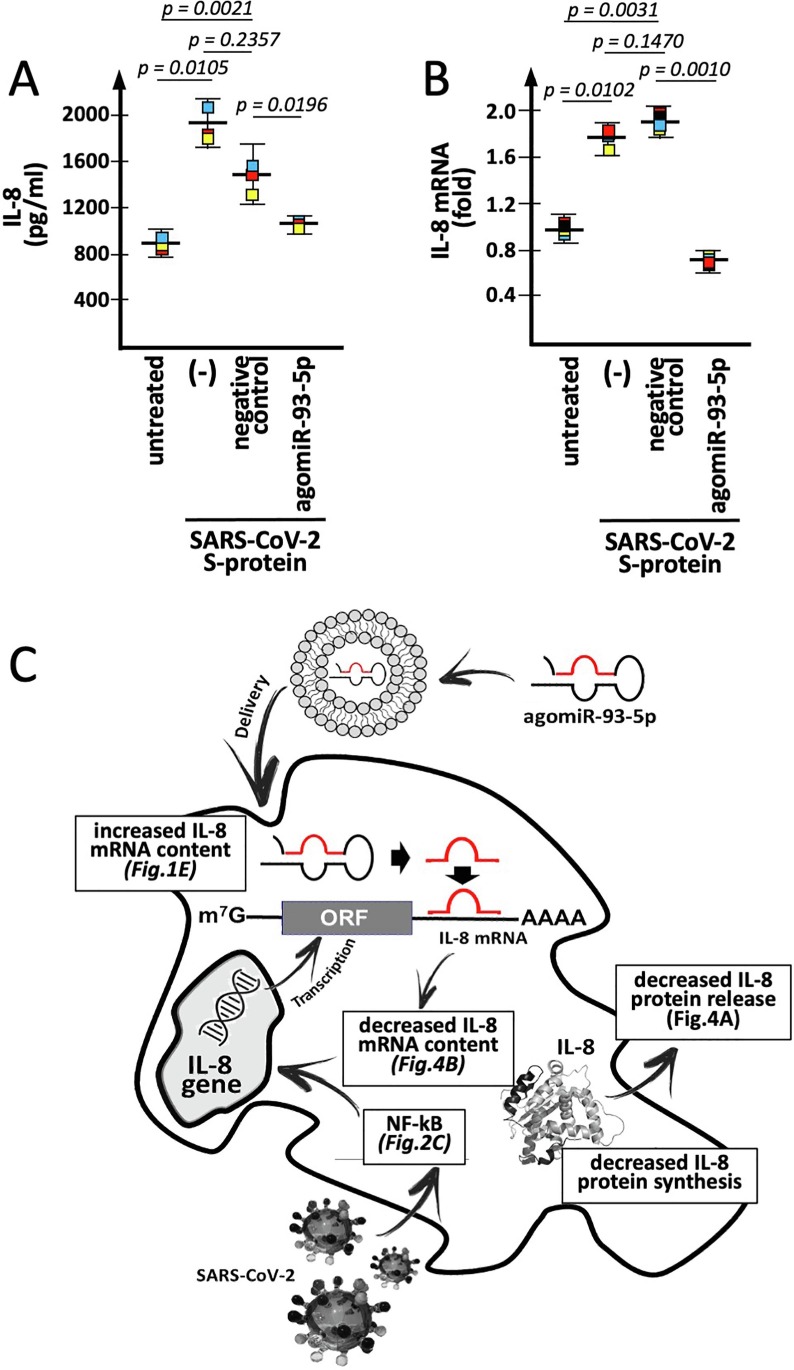
In order to verify whether transfection with a miR-93-5p mimic leads to a decrease of SARS-CoV-2 Spike-mediated increase of IL-8 gene expression, S-protein treated IB3-1 cells were transfected with 100 nM of lipofectamine-delivered premiR-93-5p. Data presented in Fig. 4 (A and B) show that the transfection is associated with sharp inhibition of IL-8 extracellular release (Fig. 4A) and that this is well in agreement with the miR-93-5p mediated inhibition of the S-protein induced increase of IL-8 mRNA, analyzed by RT-qPCR (Fig. 4B). When data obtained after transfection of IB3-1 cells with the agomiR-93 and the negative control are compared, it can be observed a significant differential effect, i.e. the inhibition of the S-protein induced increase of IL-8 release (Fig. 4A) and IL-8 mRNA content (Fig. 4B). These data were found to be highly significant (p < 0.01), while no significant differences were found when data concerning cells treated with the negative control are compared to paired control S-protein induced samples. Interestingly, values of released IL-8 protein and IL-8 mRNA content found in S-protein exposed, agomiR-93-5p treated IB3-1 cells (unlike cells treated with the negative control molecule) approached values found in untreated control IB3-1 unexposed to SARS-CoV-2 S-protein.
Fig. 4.
Effects of agomiR-93 in S-protein-induced IB3-1 cells: IL-8 gene expression. (A, B) Effects of agomiR-93 or negative control on 5 nM S-protein-treated IB3-1 cells. IL-8 protein release (A) and IL-8 mRNA content (B) were analyzed respectively by Bio-Plex analysis and RT-qPCR. In panel B the fold mRNA content was determined with respect to untreated IB3-1 cells. (C) Pictorial description outlining the proposed model illustrating the effects of the agomiR-93-5p on the post-transcriptional expression of IL-8 gene (in parenthesis the experimental evidence sustaining the model).
As expected, when the results obtained when the effects on IL-8 mRNA of the agomiR-93 are compared with those of a negative control sequence, no significant difference was found using β-actin, GAPDH and RPL13A as housekeeping sequences (Supplementary Fig. 3C).
3.5. Treatment of IB3-1 cells with a miR-93-5p agomiR: Effects on other secretomic proteins.
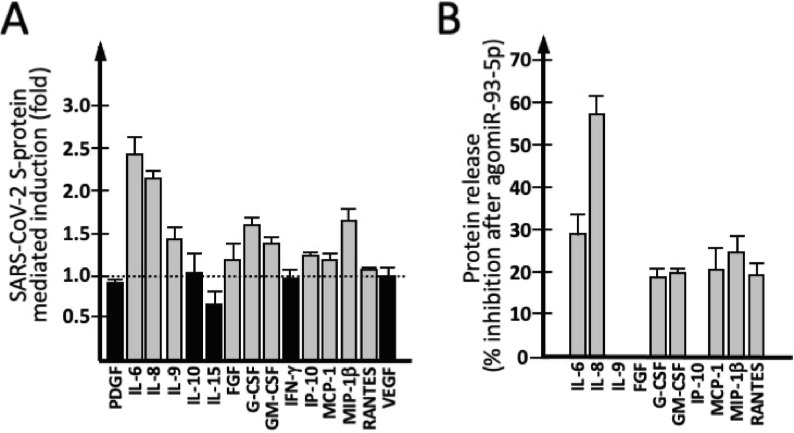
In order to verify whether transfection with a miR-93-5p mimic (premiR-93-5p) leads to a decrease of other secretomic proteins, we analyzed the Bio-Plex data focusing on the 15 secretomic proteins displaying high expression level in IB3-1 cells (see Fig. 1A). As depicted in Fig. 5 A, exposure to SARS-CoV-2 Spike protein was associated with a sharp increase of release of IL-6, IL-8, C-CSF and GM-CSF (fold > 1.5), a moderate increase of IL-9, MIP-1β (fold between 1.25 and 1.5) and no significant increase for the remaining secreted proteins. Interestingly, the proteins found to be induced by the SARS-CoV-2 S-proteins are all known to participate to the COVID-19 “cytokine storm”. Moreover, in S-protein exposed IB3-1 cells transfected with agomiR-93-5p, reduction of IL-6, IL-8, G-CSF and GM-CSF was found (Fig. 5B). Interestingly, this is in agreement with the miR-93-5pINDEX of IL-6, IL-8, G-CSF and GM-CSF, discussed in panel C of Fig. 2.
Fig. 5.
Effects of agomiR-93-5p on S-protein-induced IB3-1 cells: cytokines, chemokines and growth factors. (A) Effects of IB3-1 cells exposure to 5 nM S-protein on released cytokines, chemokines and growth factors. IL-6, IL-8, IL-9, FGF, G-CSF, GM-CSF, IP-10, MCP-1, MIP-1β and RANTES are increased in S-protein exposed cells (grey boxes). (B) AgomiR-93 mediated inhibition of release of protein induced by SARS-CoV-2 S-protein (see panel A). No agomiR-93-5p mediated inhibition was observed for IL-9, FGF and IP-10.
4. Discussion
The potential targeting or mimicking of microRNAs has been proposed as a possible therapeutic strategy for SARS-CoV-2 infection and COVID-19 management [58], [59], [60], [61], [62]. Direct targeting of SARS-CoV-2 miRNA binding sites might be proposed [59], [63]; alternatively, the retrieval of miRNA levels which are deeply altered by SARS-CoV-2 infection might be considered [59]; a further strategy is to employ miRNA therapeutics to modify the expression of cellular mRNA relevant in the COVID-19 clinical features [64], [65], [66], [67], [68], [69], [70]. In this respect, COVID-19 is characterized, in addition to the SARS-CoV-2 viral infection of target cells and tissues, by a deep inflammatory state, known as “cytokine storm”, caused by activation of several pro-inflammatory genes, such as NF-κB, STAT-3, IL-6, IL-8, IL-1β, G-CSF [15], [16], [17], [18], [19], [20], [21], [22].
The proposed study is a proof-of-principle that the release of key proteins of the COVID-19 “cytokine storm” [16], [18] can be strongly inhibited by mimicking the biological activity of microRNAs regulating the respective mRNAs. While the major focus of this study is on IL-8, the data obtained allow to identify miR-93-5p responsive pro-inflammatory genes, whose expression can be modified by treatment of the cells with an agomiR-93-5p.
The control of the “cytokine storm” is a major issue in the management of COVID-19 patients [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], [33], [71], [72], [73]. This hyperinflammatory activity is associated with severe forms of COVID-19 and poor prognosis of COVID-19 patients [74], [75], [76], [77], [78], [79], [80], [81], [82]. For instance, Del Valle et al. found that high serum IL-6, IL-8 and TNF-α levels at the time of hospitalization are strong and independent predictors of patient survival. In this study, a rapid multiplex cytokine assay was implemented in order to measure serum IL-6, IL-8, TNF-α and IL-1β in a large cohort of hospitalized COVID-19 patients (n = 1484) [26]. It was found that high serum IL-6, IL-8 and TNF-α levels at the time of hospitalization were strong and independent predictors of patient survival. In respect to IL-8, survival curves based on each cytokine measured, after multiple variable adjustments for sex, age, race/ethnicity, smoking, CKD (Chronic Kidney Disease), hypertension, asthma and CHF (Congestive Heart Failure) sustained the concept that high IL-8 serum levels are associated with low survival probability. These and similar observations sustain the hypothesis that anti-inflammatory compounds and protocols are highly needed [83].
Concerning this issue, targeting IL-8 has been proposed in several studies as well as in clinical trials [83], [84], [85], [86], [87]. For instance, NCT04347226 (“Anti-Interleukin-8 (Anti-IL-8) for Patients With COVID-19”) [88], is aimed at verifying whether neutralizing IL-8 with the anti-IL8 monoclonal antibody BMS-986253 [89] can help to improve the health condition of recruited COVID-19 patients. NCT04347226 is a single center, randomized, open-label, phase 2 trial to evaluate the time-to-improvement following treatment with anti-IL-8 therapy (BMS-986253) compared to standard of care in hospitalized patients with COVID-19 respiratory disease. It should be underlined that at present there are no FDA approved medications that improve the chance of survival in patients diagnosed with COVID-19.
The results presented in this study sustain the concept that the release of IL-8 in SARS-CoV-2 induced IB3-1 cells can be strongly inhibited by transfection of the cells with agomiR-93-5p. This was demonstrated by the data obtained following quantification of released IL-8, as well as accumulation IL-8 mRNA.
Since the control of the “cytokine storm” is a major issue in the management of COVID-19 patients [16], [17], [18], [19], [20], [21], [22], [23], [24], [25], [26], [27], our study could stimulate research activity that can contribute to the development of protocols useful to control hyperinflammatory state associated with SARS-CoV-2 infection. Interestingly, despite the fact that our study was focused on IL-8, the agomiR-93-5p was found to inhibit also the release of IL-6, IL-8, G-CSF and GM-CSF, all of them participating to the cytokine storm. This finding should stimulate further studies on these proteins, all of them potential targets of agomiR-93-5p activity (in agreement the miR-93-5pINDEX values depicted in Fig. 3C). In addition, the possible effects of the agomiR-93-5p on the TRL4/NF-κB network should be in the future studied, considering that the TLR4/NF-κB signaling pathway is a potential target for treatment of critical stage of COVID-19 patients [90], [91], [92].
A possible limit of the present study is the fact that (a) only one cell line was studied; (b) no effects of the agomiR-93-5p were found on other pro-inflammatory proteins known to be activated in the COVID-19 “cytokine storm” and (c) the combination with other agents modulating IL-8 expression has not been considered. As far as the first point, we should emphasize that we consider our study just a proof-of-principle that should be confirmed and extended by further experimental efforts. This is feasible project, since we have found that Spike exposure induces change in the expression of pro-inflammatory genes also in other cells lines, such as Calu-3 and A549 cells (Supplementary Fig.S2).
Concerning the second point, we could propose to identify additional miRNAs targeting and regulating other pro-inflammatory genes activated during the COVID-19 “cytokine storm” but not responding to agomiR-93-5p inhibition. In any case, the differential response of pro-inflammatory genes to the agomiR-93-5p support the specificity of the observed effects. Finally, as found as in other experimental model systems, the combined treatments using small chemical molecules and oligonucleotides might be a reasonable approach to limit side effects and increase the biological activity. Our study might contribute to these forthcoming projects.
Funding
This work is funded by the MUR-FISR COVID-miRNAPNA Project (FISR2020IP_04128).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.intimp.2021.108201.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019, novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen G., Wu D., Guo W., Cao Y., Huang D., Wang H., Wang T., Zhang X., Chen H., Yu H., Zhang X., Zhang M., Wu S., Song J., Chen T., Han M., Li S., Luo X., Zhao J., Ning Q. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Invest. 2020;130:2620–2629. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jia S., Li Y., Fang T. System dynamics analysis of COVID-19 prevention and control strategies. Environ. Sci. Pollut. Res. Int. 2021:1–14. doi: 10.1007/s11356-021-15902-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaur S.P., Gupta V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karpiński T.M., Ożarowski M., Seremak-Mrozikiewicz A., Wolski H., Wlodkowic D. The 2020 race towards SARS-CoV-2 specific vaccines. Theranostics. 2020;11(2021):1690–1702. doi: 10.7150/thno.53691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty C., Sharma A.R., Bhattacharya M., Sharma G., Saha R.P., Lee S.S. Ongoing clinical trials of vaccines to fight against COVID-19 pandemic. Immune Netw.. 2021;21 doi: 10.4110/in.2021.21.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L.L., Lu L., Choi C.Y., Cai J.P., Tsoi H.W., Chu A.W., Ip J.D., Chan W.M., Zhang R.R., Zhang X., Tam A.R., Lau D.P., To W.K., Que T.L., Yip C.C., Chan K.H., Cheng V.C., Yuen K.Y., Hung I.F., To K.K. Impact of SARS-CoV-2 variant-associated RBD mutations on the susceptibility to serum antibodies elicited by COVID-19 infection or vaccination. Clin. Infect. Dis. 2021 doi: 10.1093/cid/ciab656. [DOI] [PubMed] [Google Scholar]
- 9.Forchette L., Sebastian W., Liu T. A comprehensive review of COVID-19 virology, vaccines, variants, and therapeutics. Curr. Med. Sci. 2021:1–15. doi: 10.1007/s11596-021-2395-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Rong, Liu Jun, Zhang Hui. The challenge of emerging SARS-CoV-2 mutants to vaccine development. J. Genet. Genomics. 2021;48(2):102–106. doi: 10.1016/j.jgg.2021.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pascarella G., Strumia A., Piliego C., Bruno F., Del Buono R., Costa F., Scarlata S., Agrò F.E. COVID-19 diagnosis and management: a comprehensive review. J. Intern. Med. 2020;288:192–206. doi: 10.1111/joim.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bharat A., Querrey M., Markov N.S., Kim S., Kurihara C., Garza-Castillon R., Manerikar A., Shilatifard A., Tomic R., Politanska Y., Abdala-Valencia H., Yeldandi A.V., Lomasney J.W., Misharin A.V., Budinger G.R.S. Lung transplantation for patients with severe COVID-19. Sci. Transl. Med. 2020;12:4282. doi: 10.1126/scitranslmed.abe4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsai P.H., Lai W.Y., Lin Y.Y., Luo Y.H., Lin Y.T., Chen H.K., Chen Y.M., Lai Y.C., Kuo L.C., Chen S.D., Chang K.J., Liu C.H., Chang S.C., Wang F.D., Yang Y.P. Clinical Manifestation and Disease Progression in COVID-19 Infection. J. Chin. Med. Assoc. 2020;84:3–8. doi: 10.1097/JCMA.0000000000000463. [DOI] [PubMed] [Google Scholar]
- 14.King C.S., Sahjwani D., Brown A.W., Feroz S., Cameron P., Osborn E., Desai M., Djurkovic S., Kasarabada A., Hinerman R., Lantry J., Shlobin O.A., Ahmad K., Khangoora V., Aryal S., Collins A.C., Speir A., Nathan S. Outcomes of mechanically ventilated patients with COVID-19 associated respiratory failure. PLoS One. 2020;15 doi: 10.1371/journal.pone.0242651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J. COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhi S., Nabipour I. COVID-19 cytokine storm: The anger of inflammation. Cytokine. 2020;133 doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirano T., Murakami M. COVID-19: A new virus, but a familiar receptor and cytokine release syndrome. Immunity. 2020;52:731–733. doi: 10.1016/j.immuni.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi Y., Ge Y., Wu B., Zhang W., Wu T., Wen T., Liu J., Guo X., Huang C., Jiao Y., Zhu F., Zhu B., Cui L. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 (COVID-19) in China. J. Infect. Dis. 2020;222:746–754. doi: 10.1093/infdis/jiaa363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mehta P., Porter J.C., Manson J.J., Isaacs J.D., Openshaw P.J.M., McInnes I.B., Summers C., Chambers R.C. Therapeutic blockade of granulocyte macrophage colony-stimulating factor in COVID-19-associated hyperinflammation: challenges and opportunities. Lancet Respir. Med. 2020;S2213-S2600:30267–30268. doi: 10.1016/S2213-2600(20)30267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aggarwal A., Baker C.S., Evans T.W, Haslam P.L. G-CSF and IL-8 but not GM-CSF correlate with severity of pulmonary neutrophilia in acute respiratory distress syndrome. Eur. Respir. J. 2000;15(5):895–901. doi: 10.1034/j.1399-3003.2000.15e14.x. [DOI] [PubMed] [Google Scholar]
- 21.Crisafulli S., Isgrò V., La Corte L., Atzeni F., Trifirò G. Potential Role of Anti-interleukin (IL)-6 Drugs in the Treatment of COVID-19: Rationale, Clinical Evidence and Risks. BioDrugs. 2020;34:415–422. doi: 10.1007/s40259-020-00430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soy M., Keser G., Atagündüz P., Tabak F., Atagündüz I., Kayhan S. Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment. Clin. Rheumatol. 2020;39:2085–2094. doi: 10.1007/s10067-020-05190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulchandani R., Lyngdoh T., Kakkar A.K. Deciphering the COVID-19 cytokine storm: Systematic review and meta-analysis. Eur. J. Clin. Invest. 2020;14 doi: 10.1111/eci.13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hojyo S., Uchida M., Tanaka K., Hasebe R., Tanaka Y., Murakami M., Hirano T. How COVID-19 induces cytokine storm with high mortality. Inflamm. Regen. 2020;40:37. doi: 10.1186/s41232-020-00146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bahrami M., Kamalinejad M., Latifi S.A., Seif F., Dadmehr M. Cytokine storm in COVID-19 and parthenolide: preclinical evidence. Phytother. Res. 2020;34:2429–2430. doi: 10.1002/ptr.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D.M. Del Valle, S. Kim-Schulze, H. Hsin-Hui, N.D. Beckmann, S. Nirenberg, B. Wang, Y. Lavin, T. Swartz, D. Madduri, A. Stock, T. Marron, H. Xie, M.K. Patel, O. van Oekelen, A. Rahman, P. Kovatch, J. Aberg, E. Schadt, S. Jagannath, M. Mazumdar, A. Charney, A. Firpo-Betancourt, D.R. Mendu, J. Jhang, D. Reich, K. Sigel, C. Cordon-Cardo, M. Feldmann, S. Parekh, M. Merad, S. Gnjatic. An inflammatory cytokine signature helps predict COVID-19 severity and death. medRxiv (2020) 10.1101/2020.05.28.20115758.
- 27.Rodrigues-Diez R.R., Tejera-Muñoz A., Marquez-Exposito L., Rayego-Mateos S., Sanchez L.S., Marchant V., Santamaria L.T., Ramos A.M., Ortiz A., Egido J. M, Ruiz-Ortega, Statins: Could an old friend help the fight against COVID-19? Br. J. Pharmacol. 2020;177:4873–4886. doi: 10.1111/bph.15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y.F., Bai C., He F., Xie Y., Zhou H. Review on the potential action mechanisms of Chinese medicines in treating Coronavirus Disease 2019 (COVID-19) Pharmacol. Res. 2020;158 doi: 10.1016/j.phrs.2020.104939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheong D.H.J., Tan D.W.S., Wong F.W.S., Tran T. Anti-malarial drug, artemisinin and its derivatives for the treatment of respiratory diseases. Pharmacol. Res. 2020;158 doi: 10.1016/j.phrs.2020.104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Q., Pan W., Li R., Liu B., Li C., Xie Y., Wang Z., Zhao J., Jiang H., Huang J., Shi Y., Dai J., Zheng K., Li X., Yang Z. Liu Shen capsule shows antiviral and anti-inflammatory abilities against novel coronavirus SARS-CoV-2 via suppression of NF-κB signaling pathway. Pharmacol. Res. 2020;158 doi: 10.1016/j.phrs.2020.104850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosch-Barrera J., Martin-Castillo B., Buxó M., Brunet J., Encinar J.A., Menendez J.A. Silibinin and SARS-CoV-2: dual targeting of host cytokine storm and virus replication machinery for clinical management of COVID-19 Patients. J. Clin. Med. 2020;9:E1770. doi: 10.3390/jcm9061770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.C. Pelaia, C. Tinello, A. Vatrella, G. De Sarro, G. Pelaia, Lung under attack by COVID-19-induced cytokine storm: pathogenic mechanisms and therapeutic implications, Ther. Adv. Respir. Dis. 14 (2020) 1753466620933508. 10.1177/1753466620933508. [DOI] [PMC free article] [PubMed]
- 33.Choy Ernest H., De Benedetti Fabrizio, Takeuchi Tsutomu, Hashizume Misato, John Markus R., Kishimoto Tadamitsu. Translating IL-6 biology into effective treatments. Nat. Rev. Rheumatol. 2020;16(6):335–345. doi: 10.1038/s41584-020-0419-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borgatti Monica, Bezzerri Valentino, Mancini Irene, Nicolis Elena, Dechecchi Maria Cristina, Lampronti Ilaria, Rizzotti Paolo, Cabrini Giulio, Gambari Roberto. Induction of IL-6 gene expression in a CF bronchial epithelial cell line by Pseudomonas aeruginosa is dependent on transcription factors belonging to the Sp1 superfamily. Biochem. Biophys. Res. Commun. 2007;357(4):977–983. doi: 10.1016/j.bbrc.2007.04.081. [DOI] [PubMed] [Google Scholar]
- 35.Bezzerri V., Borgatti M., Finotti A., Tamanini A., Gambari R., Cabrini G. Mapping the transcriptional machinery of the IL-8 gene in human bronchial epithelial cells. J. Immunol. 2011;187(2011):6069–6081. doi: 10.4049/jimmunol.1100821. [DOI] [PubMed] [Google Scholar]
- 36.Bardin Pauline, Marchal-Duval Emmeline, Sonneville Florence, Blouquit-Laye Sabine, Rousselet Nathalie, Le Rouzic Philippe, Corvol Harriet, Tabary Olivier. O, Tabary, Small RNA and transcriptome sequencing reveal the role of miR-199a-3p in inflammatory processes in cystic fibrosis airways. J. Pathol. 2018;245(4):410–420. doi: 10.1002/path.2018.245.issue-410.1002/path.5095. [DOI] [PubMed] [Google Scholar]
- 37.Oglesby Irene K., Vencken Sebastian F., Agrawal Raman, Gaughan Kevin, Molloy Kevin, Higgins Gerard, McNally Paul, McElvaney Noel G., Mall Marcus A., Greene Catherine M. miR-17 overexpression in cystic fibrosis airway epithelial cells decreases interleukin-8 production. Eur. Respir. J. 2015;46(5):1350–1360. doi: 10.1183/09031936.00163414. [DOI] [PubMed] [Google Scholar]
- 38.Bhattacharyya S., Balakathiresan N.S., Dalgard C., Gutti U., Armistead D., Jozwik C., Srivastava M., Pollard H.B., Biswas R. Elevated miR-155 promotes inflammation in cystic fibrosis by driving hyperexpression of interleukin-8. J. Biol. 2011;286:11604–11615. doi: 10.1074/jbc.M110.198390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malemud C.J. MicroRNAs and Osteoarthritis. Cells. 2018;7:92. doi: 10.3390/cells7080092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zong D., Liu X., Li J., Ouyang R., Chen P. The role of cigarette smoke-induced epigenetic alterations in inflammation. Epigenetics Chromatin. 2019;12:65. doi: 10.1186/s13072-019-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He L., Hannon G.L. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 42.Alvarez-Garcia I., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 43.Fabbri E., Borgatti M., Montagner G., Bianchi N., Finotti A., Lampronti I., Bezzerri V., Dechecchi M.C., Cabrini G. R, Gambari, Expression of microRNA-93 and Interleukin-8 during Pseudomonas aeruginosa-mediated induction of proinflammatory responses. Am. J. Respir. Cell. Mol. Biol. 2014;50:1144–1155. doi: 10.1165/rcmb.2013-0160OC. [DOI] [PubMed] [Google Scholar]
- 44.Harada A., Sekido N., Akahoshi T., Wada T., Mukaida N., Matsushima K. Essential involvement of interleukin-8 (IL-8) in acute inflammation. J. Leukocyte Biol. 1994;56:559–564. doi: 10.1002/jlb.56.5.559. [DOI] [PubMed] [Google Scholar]
- 45.Hübner M., Moellhoff N., Effinger D., Hinske C.L., Hirschberger S., Wu T., Müller M.B., Strauß G., Kreth F.W., Kreth S. MicroRNA-93 acts as an “anti-inflammatory tumor suppressor” in glioblastoma. Neurooncol. Adv. 2020;2:vdaa047. doi: 10.1093/noajnl/vdaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabbri E., Brognara E., Montagner G., Ghimenton C., Eccher A., Cantù C., Khalil S., Bezzerri V., Provezza L., Bianchi N., Finotti A., Borgatti M., Moretto G., Chilosi M., Cabrini G., Gambari R. Regulation of IL-8 gene expression in gliomas by microRNA miR-93. BMC Cancer. 2015;15:661. doi: 10.1186/s12885-015-1659-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hua Q., Chen Y., Liu Y., Li M., Diao Q., Xue H., Zeng H., Huang L., Jiang Y. Circular RNA 0039411 Is Involved in Neodymium Oxide-induced Inflammation and Antiproliferation in a Human Bronchial Epithelial Cell Line via Sponging miR-93-5p. Toxicol. Sci. 2019;17:69–81. doi: 10.1093/toxsci/kfz074. [DOI] [PubMed] [Google Scholar]
- 48.P.L. Zeitlin, L. Lu, J.S. Rhim, G. Cutting, G. Stetten, K.A. Kieffer, R. Craig, W.B. Guggino, A cystic fibrosis bronchial epithelial cell line: immortalization by adeno-12-SV40 infection. Am. J. Respir. Cell Mol. Biol. 4 (1991) 313-319. 10.1165/ajrcmb/4.4.313. [DOI] [PubMed]
- 49.Wang W., Ye L., Ye L., Li B., Gao B., Zeng Y., Kong L., Fang X., Zheng H., Wu Z. Y, She, Up-regulation of IL-6 and TNF-alpha induced by SARS-coronavirus spike protein in murine macrophages via NF-kappaB pathway. Virus Res. 2007;128:1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gasparello J., Fabbri E., Bianchi N., Breveglieri G., Zuccato C., Borgatti M., Gambari R., Finotti A. BCL11A mRNA Targeting by miR-210: A Possible Network Regulating γ-Globin Gene Expression. Int. J. Mol. Sci. 2017;18:2530. doi: 10.3390/ijms18122530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gasparello Jessica, Gambari Laura, Papi Chiara, Rozzi Andrea, Manicardi Alex, Corradini Roberto, Gambari Roberto, Finotti Alessia. High Levels of Apoptosis Are Induced in the Human Colon Cancer HT-29 Cell Line by Co-Administration of Sulforaphane and a Peptide Nucleic Acid Targeting miR-15b-5p. Nucleic Acid Ther. 2020;30(3):164–174. doi: 10.1089/nat.2019.0825. [DOI] [PubMed] [Google Scholar]
- 52.Brognara E., Fabbri E., Montagner G., Gasparello J., Manicardi A., Corradini R., Bianchi N., Finotti A., Breveglieri G., Borgatti M., Lampronti I., Milani R., Dechecchi M.C., Cabrini G., Gambari R. High levels of apoptosis are induced in human glioma cell lines by co-administration of peptide nucleic acids targeting miR-221 and miR-222. Int. J. Oncol. 2016;48:1029–1038. doi: 10.3892/ijo.2015.3308. [DOI] [PubMed] [Google Scholar]
- 53.Gasparello J., Lomazzi M., Papi C., D'Aversa E., Sansone F., Casnati A., Donofrio G., Gambari R., Finotti A. Efficient Delivery of MicroRNA and AntimiRNA Molecules Using an Argininocalix[4]arene Macrocycle. Mol. Ther. Nucleic Acids. 2019;18:748–763. doi: 10.1016/j.omtn.2019.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cervellati F., Muresan X.M., Sticozzi C., Gambari R., Montagner G., Forman H.J., Torricelli C., Maioli E., Valacchi G. Comparative effects between electronic and cigarette smoke in human keratinocytes and epithelial lung cells. Toxicol. In Vitro. 2014;28(5):999–1005. doi: 10.1016/j.tiv.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu J., Xu X., Jiang L., Dua K., Hansbro P.M., Liu G. SARS-CoV-2 induces transcriptional signatures in human lung epithelial cells that promote lung fibrosis. Respir. Res. 2020;21:182. doi: 10.1186/s12931-020-01445-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Park Byoung Kwon, Kim Dongbum, Park Sangkyu, Maharjan Sony, Kim Jinsoo, Choi Jun-Kyu, Akauliya Madhav, Lee Younghee, Kwon Hyung-Joo. Differential Signaling and Virus Production in Calu-3 Cells and Vero Cells upon SARS-CoV-2 Infection. Biomol. Ther. (Seoul) 2021;29(3):273–281. doi: 10.4062/biomolther.2020.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao H., Xiao D., Gao L., Li X. MicroRNA-93 contributes to the suppression of lung inflammatory responses in LPS-induced acute lung injury in mice via the TLR4/MyD88/NF-kappaB signaling pathway. Int. J. Mol. Med. 2020;46:561–570. doi: 10.3892/ijmm.2020.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ying H., Ebrahimi M., Keivan M., Khoshnam S.E., Salahi S., Farzaneh M. miRNAs; a novel strategy for the treatment of COVID-19. Cell Biol. Int. 2021 doi: 10.1002/cbin.11653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hu J., Stojanović J., Yasamineh S., Yasamineh P., Karuppannan S.K., Hussain Dowlath M.J., Serati-Nouri H. The potential use of microRNAs as a therapeutic strategy for SARS-CoV-2 infection. Arch. Virol. 2021:1–24. doi: 10.1007/s00705-021-05152-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chauhan Neeraj, Jaggi Meena, Chauhan Subhash C., Yallapu Murali M. COVID-19: fighting the invisible enemy with microRNAs. Expert Rev. Anti. Infect. Ther. 2021;19(2):137–145. doi: 10.1080/14787210.2020.1812385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.El-Nabi S.H., Elhiti M., El-Sheekh M. A new approach for COVID-19 treatment by micro-RNA. Med. Hypotheses. 2020;143 doi: 10.1016/j.mehy.2020.110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alam T., Lipovich L. miRCOVID-19: Potential Targets of Human miRNAs in SARS-CoV-2 for RNA-Based Drug Discovery. Noncoding RNA. 2021;7:18. doi: 10.3390/ncrna7010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barreda-Manso M.A., Nieto-Díaz M., Soto A., Muñoz-Galdeano T., Reigada D., Maza R.M. In Silico and In Vitro Analyses Validate Human MicroRNAs Targeting the SARS-CoV-2 3'-UTR. Int. J. Mol. Sci. 2021;22:6094. doi: 10.3390/ijms22116094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lukiw W.J. microRNA Heterogeneity, Innate-Immune Defense and the Efficacy of SARS-CoV-2 Infection-A Commentary. Noncoding RNA. 2021;7:37. doi: 10.3390/ncrna7020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matarese A., Gambardella J., Sardu C., Santulli G. miR-98 Regulates TMPRSS2 Expression in Human Endothelial Cells: Key Implications for COVID-19. Biomedicines. 2020;8:462. doi: 10.3390/biomedicines8110462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mishra R., Banerjea A.C. SARS-CoV-2 Spike Targets USP33-IRF9 Axis via Exosomal miR-148a to Activate Human Microglia. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.656700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mone P., Gambardella J., Wang X., Jankauskas S.S., Matarese A., Santulli G. miR-24 Targets the Transmembrane Glycoprotein Neuropilin-1 in Human Brain Microvascular Endothelial Cells. Noncoding RNA. 2021;7:9. doi: 10.3390/ncrna7010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Meng F., Siu G.K., Mok B.W., Sun J., Fung K.S.C., Lam J.Y., Wong N.K., Gedefaw L., Luo S., Lee T.M.H., Yip S.P., Huang C.L. Viral MicroRNAs Encoded by Nucleocapsid Gene of SARS-CoV-2 Are Detected during Infection, and Targeting Metabolic Pathways in Host Cells. Cells. 2021;10:1762. doi: 10.3390/cells10071762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gambardella J., Coppola A., Izzo R., Fiorentino G., Trimarco B., Santulli G. Role of endothelial miR-24 in COVID-19 cerebrovascular events. Crit. Care. 2021;25:306. doi: 10.1186/s13054-021-03731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Milenkovic D., Ruskovska T., Rodriguez-Mateos A., Heiss C. Polyphenols could prevent SARS-CoV-2 infection by modulating the expression of miRNAs in the host cells. Aging Dis. 2021;12:1169–1182. doi: 10.14336/AD.2021.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Choudhary S., Sharma K., Singh H., Silakari O. The interplay between inflammatory pathways and COVID-19: A critical review on pathogenesis and therapeutic options. Microb. Pathog. 2020;150 doi: 10.1016/j.micpath.2020.104673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Molinaro R., Pasto A., Taraballi F., Giordano F., Azzi J.A., Tasciotti E., Corbo C. Biomimetic Nanoparticles Potentiate the Anti-Inflammatory Properties of Dexamethasone and Reduce the Cytokine Storm Syndrome: An Additional Weapon against COVID-19? Nanomaterials (Basel) 2020;10:2301. doi: 10.3390/nano10112301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kaye A.G., Siegel R. The efficacy of IL-6 inhibitor Tocilizumab in reducing severe COVID-19 mortality: a systematic review. PeerJ. 2020;8 doi: 10.7717/peerj.10322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen Y., Wang J., Liu C., Su L., Zhang D., Fan J., Yang Y., Xiao M., Xie J., Xu Y., Li Y., Zhang S. IP-10 and MCP-1 as biomarkers associated with disease severity of COVID-19. Mol. Med. 2020;26:97. doi: 10.1186/s10020-020-00230-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Song Yujing, Ye Yuxuan, Su Shiuan-Haur, Stephens Andrew, Cai Tao, Chung Meng-Ting, Han Meilan K., Newstead Michael W., Yessayan Lenar, Frame David, Humes H. David, Singer Benjamin H., Kurabayashi Katsuo. A digital protein microarray for COVID-19 cytokine storm monitoring. Lab Chip. 2021;21(2):331–343. doi: 10.1039/D0LC00678E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Amiri-Dashatan N., Koushki M., Ghorbani F., Naderi N. Increased inflammatory markers correlate with liver damage and predict severe COVID-19: a systematic review and meta-analysis. Gastroenterol. Hepatol. Bed Bench. 2020;13:282–291. PMID: 33244370. [PMC free article] [PubMed] [Google Scholar]
- 77.Pandolfi L., Fossali T., Frangipane V., Bozzini S., Morosini M., D'Amato M., Lettieri S., Urtis M., Di Toro A., Saracino L., Percivalle E., Tomaselli S., Cavagna L., Cova E., Mojoli F., Bergomi P., Ottolina D., Lilleri D., Corsico A.G., Arbustini E., Colombo R., Meloni F. Broncho-alveolar inflammation in COVID-19 patients: a correlation with clinical outcome. BMC Pulm. Med. 2020;20:301. doi: 10.1186/s12890-020-01343-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Angioni R., Sánchez-Rodríguez R., Munari F., Bertoldi N., Arcidiacono D., Cavinato S., Marturano D., Zaramella A., Realdon S., Cattelan A., Viola A., Molon B. Age-severity matched cytokine profiling reveals specific signatures in Covid-19 patients. Cell Death Dis. 2020;11:957. doi: 10.1038/s41419-020-03151-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Xiaolei, Liu Yang, Li Junming, Sun Longhua, Yang Jibin, Xu Fei, Zhou Jing, Wan Lagen, Xu Xinping, Le Aiping, Zhang Wei. Immune characteristics distinguish patients with severe disease associated with SARS-CoV-2. Immunol. Res. 2020;68(6):398–404. doi: 10.1007/s12026-020-09156-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Burke H., Freeman A., Cellura D.C., Stuart B.L., Brendish N.J., Poole S., Borca F., Phan H.T.T., Sheard N., Williams S., Spalluto C.M., Staples K.J., Clark T.W., Wilkinson T.M.A. REACT COVID investigators, Inflammatory phenotyping predicts clinical outcome in COVID-19. Respir. Res. 2020;21:245. doi: 10.1186/s12931-020-01511-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ghazavi A., Ganji A., Keshavarzian N., Rabiemajd S., Mosayebi G. Cytokine profile and disease severity in patients with COVID-19. Cytokine. 2020;137 doi: 10.1016/j.cyto.2020.155323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mandel M., Harari G., Gurevich M., Achiron A.M. Cytokine prediction of mortality in COVID19 patients. Cytokine. 2020;134 doi: 10.1016/j.cyto.2020.155190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Amigues I., Pearlman A.H., Patel A., Reid P., Robinson P.C., Sinha R., Kim A.H., Youngstein T., Jayatilleke A., Konig M. Coronavirus disease 2019, investigational therapies in the prevention and treatment of hyperinflammation. Expert Rev. Clin. Immunol. 2020;16:1185–1204. doi: 10.1080/1744666X.2021.1847084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Varchetta S., Mele D., Oliviero B., Mantovani S., Ludovisi S., Cerino A., Bruno R., Castelli A., Mosconi M., Vecchia M., Roda S., Sachs M., Klersy C., Mondelli M.U. Unique immunological profile in patients with COVID-19, Cell. Mol. Immunol. 2020:1–9. doi: 10.1038/s41423-020-00557-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z., Wang Y., Vilekar P., Yang S.P., Gupta M., Oh M.I., Meek A., Doyle L., Villar L., Brennecke A., Liyanage I., Reed M., Barden C., Weaver D.F. Small molecule therapeutics for COVID-19: repurposing of inhaled furosemide. PeerJ. 2020;8 doi: 10.7717/peerj.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McElvaney Oliver J., McEvoy Natalie L., McElvaney Oisín F., Carroll Tomás P., Murphy Mark P., Dunlea Danielle M., Ní Choileáin Orna, Clarke Jennifer, O’Connor Eoin, Hogan Grace, Ryan Daniel, Sulaiman Imran, Gunaratnam Cedric, Branagan Peter, O’Brien Michael E., Morgan Ross K., Costello Richard W., Hurley Killian, Walsh Seán, de Barra Eoghan, McNally Cora, McConkey Samuel, Boland Fiona, Galvin Sinead, Kiernan Fiona, O’Rourke James, Dwyer Rory, Power Michael, Geoghegan Pierce, Larkin Caroline, O’Leary Ruth Aoibheann, Freeman James, Gaffney Alan, Marsh Brian, Curley Gerard F., McElvaney Noel G. Characterization of the Inflammatory Response to Severe COVID-19 Illness. Am. J. Respir. Crit. Care Med. 2020;202(6):812–821. doi: 10.1164/rccm.202005-1583OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Crisci C.D., Ardusso L.R.F., Mossuz A., Müller L. A precision medicine approach to SARS-CoV-2 pandemic management. Curr. Treat. Options Allergy. 2020:1–19. doi: 10.1007/s40521-020-00258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Anti-Interleukin-8 (Anti-IL-8) for Patients With COVID-19. https://clinicaltrials.gov/ct2/show/NCT04347226 (2020).
- 89.Bilusic M., Heery C.R., Collins J.M., Donahue R.N., Palena C., Madan R.A., Karzai F., Marté J.L., Strauss J., Gatti-Mays M.E., Schlom J., Gulley J.L. Phase I trial of HuMax-IL8 (BMS-986253), an anti-IL-8 monoclonal antibody, in patients with metastatic or unresectable solid tumors. J. Immunother. Cancer. 2019;7:240. doi: 10.1186/s40425-019-0706-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Aboudounya M.M., Heads R.J. COVID-19 and Toll-Like Receptor 4 (TLR4): SARS-CoV-2 May Bind and Activate TLR4 to Increase ACE2 Expression, Facilitating Entry and Causing Hyperinflammation. Mediators Inflamm. 2021:8874339. doi: 10.1155/2021/8874339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kaushik D., Bhandari R., Kuhad A. TLR4 as a therapeutic target for respiratory and neurological complications of SARS-CoV-2. Expert Opin. Ther. Targets. 2021;25:491–508. doi: 10.1080/14728222.2021.1918103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kircheis R., Haasbach E., Lueftenegger D., Heyken W.T., Ocker M., Planz O. NF-κB Pathway as a Potential Target for Treatment of Critical Stage COVID-19 Patients. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.598444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.