Abstract
Between September 1995 and August 1998, the incidence and diversity of the main human rotavirus genotypes (G1, G2, G3, and G4 and P[8], P[4], P[6], and P[9]) among Irish children were determined by using established and adapted reverse transcriptase PCR-based genotyping methods. From a total of 193 rotavirus-positive specimens collected from nine hospitals we successfully identified the P type in 182 (94%) of the samples and the G type in 165 (85.5%) of the samples. Only four samples could not be assigned a G or P type. Two P types existed in Ireland, P[8] (78%) and P[4] (16%), and their relative incidence varied over the 3 years of this study. No P[6] or P[9] types were detected. G1 was the most predominant G type (55%), and the incidences of G2, G3, and G4 isolates were 15.5, 1, and 11%, respectively. Three percent of the samples tested had a mixed G type. A P and G type was assigned to 158 (81.8%) of samples. Of the typeable samples, G1 P[8] was the most prevalent (65%), whereas G2 P[4] (17%), G3 P[8] (1%), G4 P[8] (12%), and mixed types (all G1/ G4 P[8]) (4%) were detected less frequently. In the third year a significant genotypic shift from G1 P[8] to G2 P[4] and G4 P[8] was observed. During the study, we noticed that the inclusion of random primers during cDNA synthesis greatly increased the specificity of the PCR typing assays. No correlation was seen between the contributing hospitals and a specific genotype. In conclusion, the coverage of infection given by the recently licensed tetravalent vaccine would be significantly high in Ireland, although future monitoring of genotypic changes among Irish isolates should be encouraged.
Rotavirus is the single most important cause of infantile gastroenteritis worldwide, affecting an estimated 130 million infants and causing 873,000 deaths every year (2). The rotavirus genome consists of 11 segments of double-stranded RNA which can be separated by polyacrylamide gel electrophoresis into a characteristic electrophoretic pattern that is useful for diagnostic and epidemiological purposes (4). Each genomic segment codes for at least one protein (3). Two immunogenic surface viral proteins (VP), VP4 and VP7, are significant for serotype classification and are coded for by gene segments 4 and either segments 7, 8, or 9, respectively. To date, 10 VP7 antigenic types (G types) and VP4 types (P types) have been identified in viruses recovered from humans (10). The nomenclature for G genotypes and serotypes are identical (followed by an open number), whereas a P genotype is denoted by closed brackets and serotype is indicated by an open number. Only the genotype pairs G1 P[8], G2 P[4], G3 P[8], and G4 P[8] are important causes of diarrhea among infants worldwide, accounting for 95.9% of all typeable strains (6). Many techniques have been developed to determine the serotypes of rotaviruses, including enzyme immunoassays (20), hybridization techniques (13), and reverse transcriptase PCR (RT-PCR) (5, 7). These various techniques have recently been critically evaluated (9).
The genotype of human rotavirus correlates well with serotype specificity, and the application of RT-PCR technology has been instrumental in generating information on the genetic diversity of rotavirus strains. Amplification of VP4 (5) and VP7 (7) gene fragments by using serotype-specific primers has given researchers the ability to closely examine genetic and inferred serotypic differences among isolated viruses (15, 17, 18, 19, 21).
The extremely high morbidity and mortality associated with rotaviruses emphasizes the need to develop an effective vaccine (1, 10). The suitability of a vaccine for a particular population can only be evaluated by examining the molecular characteristics of a representative number of circulating strains. To our knowledge, no such detailed analysis of the prevalence of rotavirus genotypes in Ireland has been undertaken. The objective of this research was to carry out a retrospective genotypic study on a representative number of rotavirus samples (193) which may reflect the incidence and diversity of the four main genotypes in Ireland. The P and G types of rotavirus-positive specimens obtained from nine contributing hospitals were determined by using both established and adapted RT-PCR-based techniques.
MATERIALS AND METHODS
Specimen collection and RNA analysis.
A total of 193 rotavirus stool specimens (deemed positive by a latex agglutination assay; Biomerieux, Marcy l’Etoile, France) were collected from nine Irish hospitals from September 1995 to August 1998 (Table 1) and stored at −20°C until analyzed.
TABLE 1.
Hospital source and assigned P and G genotypes of rotaviruses recovered over a 3-year period in Ireland
| Source | No. of G and P types (% of total)
|
|||||
|---|---|---|---|---|---|---|
| G1 [P8] | G2 [P4] | G3 [P8] | G4 [P8] | G1/G4 [P8] | Total | |
| Ballinasloe | 34 (68) | 7 (14) | 1 (2) | 7 (14) | 1 (2) | 50 |
| Tralee | 28 (59) | 15 (32) | 0 | 2 (4) | 2 (4) | 47 |
| Crumlin | 20 (71) | 0 | 0 | 5 (18) | 3 (11) | 28 |
| Mercy Hospital | 12 (63) | 5 (26) | 0 | 2 (7) | 0 | 19 |
| Galway | 3 (50) | 2 (33) | 0 | 1 (17) | 0 | 6 |
| Mullingar | 1 (33) | 0 | 0 | 2 (66) | 0 | 3 |
| Limerick | 2 (100) | 0 | 0 | 0 | 0 | 2 |
| Bon Secour | 2 (100) | 0 | 0 | 0 | 0 | 2 |
| Dublin | 1 (100) | 0 | 0 | 0 | 0 | 1 |
| Total | 103 (65) | 29 (18) | 1 (0.5) | 19 (12) | 6 (4) | 158 |
A 10 to 40% (wt/vol) suspension of fecal matter was made in diethyl pyrocarbonate-treated sterile distilled water (DEPC water) (14). A 500-μl aliquot was mixed with an equal volume of Freon (BDH Lab Supplies, Poole, England) and centrifuged at 12,000 rpm for 10 min in an Eppendorf centrifuge. The clarified supernatant was mixed with an equal volume of Ultraspec RNA isolation reagent (Biotecx Laboratories, Houston, Tex.), and 5% chloroform was added; the mixture was then vortexed for 30 s and left on ice for 15 min with intermittent mixing. The samples were centrifuged for 15 min at 12,000 rpm, and the supernatant was decanted and mixed with an equal volume of ice-cold isopropanol. The RNA was left to precipitate at −20°C for at least 15 min, centrifuged, washed, dried, and resuspended in 50 μl of DEPC water.
RNA samples were routinely run on 1.5% agarose Tris-borate gels at 100 V for 1 h to check for RNA quality and on 10% Tris-borate-EDTA (TBE) polyacrylamide gels at 200 V for 6 h to examine electropherotypes. Both gel types were visualized under UV illumination after ethidium bromide (EtBr) staining.
VP4 and VP7 cDNA synthesis.
A separate reverse transcription (RT) was employed for VP4 and VP7 analysis based on the previously determined conditions used by Gentsch et al. and Gouvea et al. (5, 7). Briefly, 5 μl of extracted double-stranded RNA was denatured with 3.5 μl of dimethylsulfoxide (DMSO) (BDH Lab Supplies) and boiled for 5 min. The samples were then immediately cooled on ice and used as template for an RT reaction. Cooled template (8.5 μl) was added to an RT mix consisting of 4 μl of 5× RT buffer (250 mM Tris-HCl, 200 mM KCl, 25 mM MgCl2, 2.5% Tween 20 [vol/vol], pH 8.3) (Boehringer Mannheim, East Sussex, England), 2 μl of 100 mM dithiothreitol, 0.5 μl of a deoxynucleoside triphosphate (dNTP) mix (dATP, dCTP, dGTP, and dTTP; each 10 mM), 1 μl of RNasin, and either 100 ng of con3 (VP4 5′) primer (5) or 100 ng of Beg9 (VP7 5′) primer (7). Finally, 1 μl of Expand RT (Boehringer Mannheim) was added, and the 20-μl reaction mixture was incubated at 42°C for 1 h.
Randomly primed cDNA synthesis.
More recently, the RT reaction was modified and improved by adding 200 ng of random hexamer primer p(dN)6 (Boehringer Mannheim) to the RT mix, which facilitates the random priming of all denatured rotavirus RNA template instead of the con3 and Beg9 primers.
Primers.
The primers used in this study were as follows: Beg9, GGCTTTAAAAGAGAGAATTTCCGTCTGG; End9, GGTCACATCATACAATTCTAATCTAAG; G1, aBT1, CAAGTACTCAAATCAATGATGG; G2, aCT2, CAATGATATTAACACATTTTCTGTG; G3, aET3, CGTTTGAAGAAGTTGCAACAG; and G4, aDT4, CGTTTCTGGTGAGGAGTTG (7); con3, TGGCTTCGCCATTTTATAGACA; con2, ATTTCGGACCATTTATAACC; P[8], 1-T1, TCTACTTGGATAACGTGC; P[4], 2-T1, CTATTGTTAGAGGTTAGAGTC; P[6], 3-T1, TGTTGATTAGTTGGATTCAA; and P[9], 4-T1, TGAGACATGCAATTGGAC (5).
VP7 genotyping.
Determination of the VP7 genotype was based on the system employed by Gouvea et al. (7). For each sample, approximately 2 ng of cDNA template was added to a PCR Eppendorf tube containing 5 μl of 10× NH4 reaction buffer [160 mM (NH4)2SO4; 670 mM Tris-HCl, pH 8.8; 0.1% Tween], 1.5 mM Mg, 0.2 mM concentrations of dATP, dCTP, dGTP, and dTTP, 4% DMSO, 150 ng of Beg9 and 150 ng of End9 primers for full-length products or else End9 and 100 ng each of G1 to G4 type-specific primers (aBT1, aCT2, aET3, and aDT4) for direct typing analysis. The reaction mix was brought up to 49.75 μl with H2O, and subsequently 1 U of Taq polymerase was added to each reaction tube. The tubes were overlaid with mineral oil and placed in a thermocycler for 28 cycles of PCR (94°C for 1 min, 42°C for 2 min, and 72°C for 2 min), followed by a 5-min incubation at 72°C. A nested PCR with 5 μl of full-length template in the same reaction buffer with 150 ng of End9 and 100 ng of G1 to G4 primers for 20 cycles was carried out for samples which were difficult to type. PCR products were analyzed on a 2% TBE agarose gel and viewed under UV illumination after EtBr staining. To confirm the specificity of the primers, four random VP7 PCR products were gel purified (Qiagen GmbH, Hilden, Germany), cloned into pGEMT (Promega, Madison, Wis.), and partially sequenced (data not shown).
VP4 genotyping.
The VP4 genotyping was a modification of the technique used by Gentsch et al. (5). Briefly, 2 ng of cDNA template from the RT reaction mixture was added to a similar PCR mix as described above, except that a different set of primers and amplification conditions was used. A total of 150 ng of con3 and con2 was used for full-length VP4 synthesis or else 150 ng of the con3 primer and 100 ng of each type-specific primer (1-T1, 2-T1, 3-T1, and 4-T1) (for direct genotyping analysis) was added to a PCR tube with the same buffer, with Mg, dNTP, DMSO, and Taq concentrations as described above for the VP7 genotyping. Thirty cycles of PCR were carried out (94°C for 1 min, 50°C for 2 min, and 72°C for 2 min), followed by a 5-min incubation at 72°C. For samples that were difficult to type, a nested PCR was carried out by using 1 to 5 μl of full-length template with 150 ng of con3 and 100 ng of 1-T1/4-T1 for 20 cycles. To confirm the specificity of the primers, five full-length random VP4 PCR products were gel purified (Qiagen), cloned into pGEMT (Promega), and partially sequenced.
Cell culturing.
Four standard rotavirus strains, Wa (G1 P[8]), DS1 (G2 P[4]), P (G3 P[8]), and ST3 (G4 P[8]), used as controls during these experiments were propagated in MA104 cells cultured as described previously (9). RNA extraction and RT-PCR were carried out for these strains as described above.
RESULTS
A total of 193 fecal samples (rotavirus positive by latex agglutination) were collected from nine Irish hospitals over a 3-year period. RNA was extracted from these samples and analyzed for their P and G types by using established (5, 7) and adapted RT-PCR-based genotyping methods.
Determination of P type.
The P type (VP4 associated) was successfully determined in 182 (94%) of the clinical samples (Table 2). A P type was assigned after PCR with P[8], P[4], P[6], and P[9]-specific primers carried out on rotavirus cDNA template or PCR product. Only two P types were shown to be prevalent in Ireland: P[8] (78%) and P[4] (16%). Eleven samples (6%) were untypeable. The incidence of each type was seen to vary from year to year. P[8] occurred most frequently during the first 2 years of this study (93 and 98%, respectively), whereas in year 3 the incidence of the P[4] types increased (34%) (Table 2). No P[6]- or P[9]-type viruses were detected during the survey. RNA template from tissue culture-adapted strains Wa, DS1, P, and ST3 were used as positive controls.
TABLE 2.
Distribution of rotavirus G and P types over 3 years in Ireland
| Yr | No. of G types (%)
|
No. of P types (%)
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G1 | G2 | G3 | G4 | Mixa | NT | Total | P8 | P4 | P6 | P9 | NT | Total | |
| Sept. 95–Aug. 96 | 12 (44) | 2 (8) | 0 | 1 (4) | 0 | 12 (44) | 27 | 25 (93) | 2 (7) | 0 | 0 | 0 | 27 |
| Sept. 96–Aug. 97 | 63 (79) | 0 | 0 | 6 (7) | 3 (4) | 8 (10) | 80 | 78 (98) | 0 | 0 | 0 | 2 (2) | 80 |
| Sept. 97–Aug. 98 | 31 (36) | 28 (32) | 2 (2) | 14 (16) | 3 (4) | 8 (9) | 86 | 48 (56) | 29 (34) | 0 | 0 | 9 (10) | 86 |
| Total | 106 | 30 | 2 | 21 | 6 | 28 | 193 | 151 | 31 | 0 | 0 | 11 | 193 |
Mixed G types G1 and G4; NT, not typeable.
Determination of G type.
The G type (VP7 associated) was successfully determined in 165 (85.5%) of all samples. The slightly lower efficiency relative to P typing can be partially attributed to the RNA degradation from some of the earlier isolates, since the P-typing assays were completed first. The overall incidence for G typing was G1 (55%), G2 (15.5%), G3 (1%), G4 (11%), and mixed (3%) (Table 2). G1- to G4-type-specific PCR products from a random selection of clinical isolates over the 3 years are shown with controls in Fig. 1. The incidences of each type in year 1 were as follows: G1 (44%), G2 (8%), and G4 (4%). Twelve samples (44%) from this early period remain untypeable. Only G1 (79%), G4 (7.5%), and G1/G4 mixed types (4%) were detected in the second year, but a dramatic shift in the incidence and diversity of prevailing types occurred in year 3. The incidence of G1 types dropped to 31 (40%), whereas those of G2 and G4 isolates increased to 28 (32%) and 14 (18%), respectively. G3 isolates (2%) were also detected for the first time, and mixed types (all G1/G4) occurred in 4% of the samples (Table 2). RNA template from the tissue culture-adapted strains described above was used as a positive control during RT-PCR.
FIG. 1.
Composite photograph which shows a representative agarose gel depicting VP7-positive PCR products from clinical isolates and positive controls from tissue culture-adapted strains. Lanes: 1 and 13, 100-bp ladder (Boehringer Mannheim); 2, G1 isolate (Ballinasloe); 3, G1 isolate (Tralee); 4, G2 isolate (Tralee); 5, G2 isolate (Mercy Hospital); 6, G2 isolate (Galway); 7, G2 isolate (Ballinasloe); 8, G4 isolate (Crumlin); 9, Wa; 10, DS1; 11, P; 12, ST3.
Correlation of G and P types.
A G and P type was assigned to 158 (81.8%) of 193 samples by using the procedures outlined. More recently, in our laboratory a method which utilizes random hexamer primers in the RT mix was used in a single-tube cDNA synthesis reaction. A subsequent PCR performed by using only con3 and VP4-specific primers amplified a P-type-specific product from the cDNA template. A second PCR from the same RT mix performed with End9 and VP7-type-specific primers amplified a G-type-specific product. By this method no full-length PCR product is formed or required for a nested reaction. A comparison between random and conventional RT primers is exhibited in Fig. 2.
FIG. 2.
Composite photograph showing the PCR products of random primed (bottom panel) versus conventionally primed (top panel) template during cDNA synthesis. Note that the gels depicted are representative examples of the two techniques and are not directly comparable.
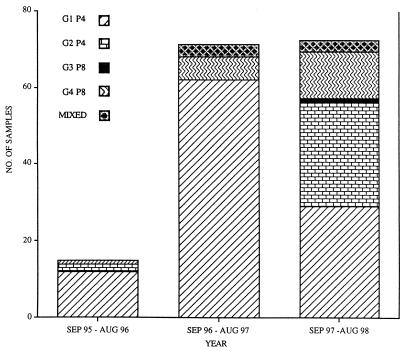
During the typing assays it was observed that a specific G type could always be correlated to a P type; namely, G1, G3, and G4 always coexisted with P[8], and G2 was always associated with P[4]. The incidence of each genotype varied considerably for the three years (Fig. 3). Overall, G1 P[8] was recorded as the most common (65% of all doubly typed viruses). The other types, namely, G2 P[4], G3 P[8], G4 P[8], and mixed types (all G1/G4 P[8]) were less frequent (17, 1, 12, and 4%, respectively). No bias was observed between hospital and genotype, and it was noted that with the exception of Mullingar, G1 P[8] was the most predominant type collected from each of the Irish hospitals (Table 1).
FIG. 3.
Graphic illustration showing the incidence and diversity of rotavirus genotypes for 3 years in Ireland.
For the winter period of the third year we observed a marked increase in the number of rotavirus samples sent to our laboratory. This correlated with a noticeable shift in genotype from G1 P[8] to G2 P[4] and G4 P[8]. This is confirmed by data shown in Fig. 3, which shows that 93% of G2 P[4] types and 63% of G4 P[8] types were collected during the third year of this study.
Electropherotypes.
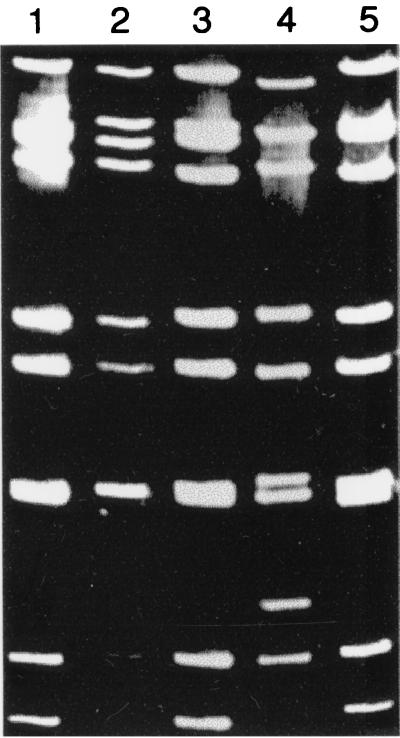
Throughout the study, G1 and G4 rotaviruses were always associated with “long” electropherotypes whereas G2 electropherotypes were “shorter” (Fig. 4). During the study it was noted that the pattern of the electropherotype suggested but did not confirm a particular genotype, as described by Sethi et al. (16).
FIG. 4.
Photograph which shows different RNA electropherotypes isolated from clinical specimens during this study. Lanes: 1, G4 isolate; 2, G4 isolate; 3, G4 isolate; 4, G2 isolate; 5, G1 isolate.
DISCUSSION
The objective of this study was to ascertain for the first time the P and G genotypes of circulating rotaviruses isolated from infected hospitalized children in Ireland. Genotyping is a well established and recognized epidemiological tool for examining strain diversity, and the correlation between genotype and serotype is well understood. The significance of determining serotypes of circulating rotaviruses has become increasingly recognized (21), and RT-PCR has been shown to be the most sensitive assay for determining genotypes (9). A rotavirus tetravalent vaccine that has recently been licensed specifically targets G1 to G4 serotypes, and it is essential to determine the predicted efficiency of this vaccine for each country before it is used.
The Republic of Ireland has a population of over 3.5 million and is served by over 50 medical microbiology laboratories. Sixteen of these routinely test pediatric patients (1 to 48 months) for rotavirus, and positive specimens were donated by over one half of these hospital laboratories. A total of 193 samples were collected from nine hospitals nationwide. Only four samples could not be assigned a G or a P type, which suggests that the incidence of unusual genotypes among Irish isolates is rare. The efficiency of the primers was determined by cloning and partially sequencing randomly chosen PCR products. As expected, in all cases the primers amplified the correct sequence (data not shown). After the P-typing assays it was discovered that our Irish results match those observed in studies from seven other countries involving 500 samples (6), in that two main P types prevail, namely, P[4] and P[8]. It was also observed that their relative incidence over the 3 years varied. P[8] types were detected almost exclusively for the first 2 years, whereas a notable shift occurred in year 3, when P[4] became more prevalent (38%) (Table 2). It is interesting that no P[9] or P[6] types were observed, even though the incidence of P[6] in recent American (12), Brazilian (18), and Indian (11) surveys seems to highlight this strain as a significant emerging genotype.
Of the samples tested, 165 were assigned a G type. As expected from similar surveys carried out worldwide, G1 was the most predominant type detected for each of the 3 years in Ireland, even though G2 emerged as a significant type in the third year (Table 2). This seems to indicate a notable relative shift in the prevalence of circulating viruses, which should be monitored over the coming years. It is interesting that this bias away from G1 in December 1997 correlated with a simultaneous marked increase in the number of samples received by our laboratory (data not shown). This may suggest a greater susceptibility to emerging genotypes by Irish infants. Six mixed-type strains (all G1/G4) were recovered from patients in the last 2 years, and just two G3 isolates were detected during this survey. It is interesting that non-G1 to G4 genotypes are prevalent in other countries: G9 was the most predominant type reported in India (11), and G5 was the third most reported G type in Brazil (6).
Both G and P genotypes were assigned to 158 (81.8%) of collected viruses. Consistent with the findings of numerous reports, G1 P[8] was by far the most common type, while G2 P[4] and G4 P[8] were less prevalent. However, in the third year of this study the number of G1 P[8] isolates recovered was reduced by over 50% and an almost equal number of G1 P[8] and G2 P[4] viruses coexisted in Ireland (Fig. 3). This would seem to indicate a significant genotypic shift, which will be of major importance for future studies carried out in Ireland. Although noteworthy, this is not an unusual phenomenon, as periodic changes in prevailing genotypes have previously been reported (8). Even though a disparity exists between sample numbers and source, it appears that a correlation did not exist between the hospital source and a particular genotype. Each genotype appeared to be evenly distributed among the collection centers around the country (Table 1).
In conclusion, genotyping by RT-PCR is an extremely sensitive technique (∼50,000 times more so than enzyme immunoassays) (19) and is adaptable to accommodating new and emerging types. However, the method is 10 times more expensive than other methods (9), and the RNA used must be of a reputable quality. We found that the inclusion of random primers in the RT mix greatly reduced the appearance of nonspecific amplification during PCR and also halved the number of RT reactions required. This method was introduced in the last year of the study and, although more efficient, it was not able to detect previously untypeable samples. The detection of rotaviral genotypes for the first time in Irish children and the indication that the prevalance of certain genotypes may change over a rotavirus season is significant and mirrors observations from studies in other temperate climates (8). If the tetravalent vaccine becomes available in Ireland, it appears that the coverage against infection would be significantly high, although monitoring of the genotypic changes among circulating viruses should be encouraged over the coming years.
ACKNOWLEDGMENTS
This research was funded in part by grant aid under the food subprogramme of the operational programme for industrial development administered by the Department of Agriculture and Food and was supported by national and EU funds.
We greatly appreciate the cooperation and help of the Irish hospitals listed in this study, particularly Gerry Christie FAMLS and staff at Tralee General hospital. Strains used for cell culturing were generously donated by Paul Masendycz (9).
REFERENCES
- 1.Barnes G. Rotavirus vaccines. Acta Paed Sin. 1998;39:17–20. [PubMed] [Google Scholar]
- 2.Cimons M. Rotavirus vaccine seems headed for approval. ASM News. 1998;64:6. [Google Scholar]
- 3.Donelli G, Superti F. The rotavirus genus. Comp Immunol Microbiol Infect Dis. 1994;17:305–320. doi: 10.1016/0147-9571(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 4.Estes M, Cohen J. Rotavirus: gene structure and function. Microbiol Rev. 1989;53:410–449. doi: 10.1128/mr.53.4.410-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gentsch J, Glass R, Woods P, Gouvea V, Gorziglia M, Flores J, Das B, Bhan M. Identification of group A rotavirus gene 4 types by PCR. J Clin Microbiol. 1992;30:1365–1373. doi: 10.1128/jcm.30.6.1365-1373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gentsch J, Woods P, Ramachandran M, Das B, Levite J, Alfieri A, Kumar R, Bhan M, Glass R. Review of G and P typing results from a global collection of rotavirus strains: implications for vaccine development. J Infect Dis. 1996;174:S30–S36. doi: 10.1093/infdis/174.supplement_1.s30. [DOI] [PubMed] [Google Scholar]
- 7.Gouvea V, Glass R, Woods P, Taniguchi K, Clark H F, Forrester B, Fang Z-Y. PCR amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276–282. doi: 10.1128/jcm.28.2.276-282.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haffejee I. Epidemiology of rotavirus infections: a global perspective. J Pediatr Gastroenterol Nutr. 1995;20:275–286. doi: 10.1097/00005176-199504000-00003. [DOI] [PubMed] [Google Scholar]
- 9.Masendycz P, Palombo E, Gorrell R, Bishop R. Comparison of enzyme immunoassay, polymerase chain reaction and type-specific cDNA probe techniques for identification of group A rotavirus gene 4 types. J Clin Microbiol. 1997;35:3104–3108. doi: 10.1128/jcm.35.12.3104-3108.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matson, D. 1997, posting date. Virologic properties of rotavirus. [Online.] http://rotavirus.com/docs_html/virologic_prop_of_rotavir.html. [28 March 1999, last date accessed.]
- 11.Ramachandran M, Das B, Vij A, Kumar R, Bhambal S, Kesari N, Rawat K, Bahl L, Thakur S, Woods P, Glass R, Bhan M, Gentsch J. Unusual diversity of human rotavirus G and P genotypes in India. J Clin Microbiol. 1996;34:436–439. doi: 10.1128/jcm.34.2.436-439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran M, Gentsch J, Parashar U, Jin S, Woods P, Holmes J, Kirkwood C, Bishop R, Greenberg H, Urasawa S, Gerna G, Coulson B, Taniguchi K, Bresee J, Glass R The National Rotavirus Strain Surveillance System Collaborating Laboratories. Detection and characterization of novel rotavirus strains in the United States. J Clin Microbiol. 1998;36:3223–3229. doi: 10.1128/jcm.36.11.3223-3229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasool N, Larralde G, Gorziglia M. Determination of human rotavirus VP4 using serotype specific cDNA probes. Arch Virol. 1993;133:275–282. doi: 10.1007/BF01313768. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 15.Santos N, Riepenhoff-Tally M, Clark H, Offit P, Gouvea V. VP4 genotyping of human rotaviruses in the United States. J Clin Microbiol. 1994;32:205–208. doi: 10.1128/jcm.32.1.205-208.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sethi S, Olive D, Strannegard O, Al-Nakib W. Molecular epidemiology of human rotavirus infections based on genome segment variations in viral strains. J Med Virol. 1988;26:249–259. doi: 10.1002/jmv.1890260305. [DOI] [PubMed] [Google Scholar]
- 17.Silberstein I, Shulman L M, Mendelson E, Shif I. Distribution of both rotavirus VP4 genotypes and VP7 serotypes among hospitalized and nonhospitalized Israeli children. J Clin Microbiol. 1995;33:1421–1422. doi: 10.1128/jcm.33.5.1421-1422.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timetesky M, Santos N, Gouvea V. Survey of rotavirus G and P types associated with human gastroenteritis in Sao Paolo, Brazil, from 1986 to 1992. J Clin Microbiol. 1994;32:2262–2264. doi: 10.1128/jcm.32.10.2622-2624.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ushijima H, Mukoyama A, Hasegawa A, Nishimura S, Konishi K, Bosu K. Serotyping of human rotaviruses in the Tokyo area (1990–1993) by enzyme immunoassay with monoclonal antibodies and by reverse transcription and polymerase chain reaction amplification. J Med Virol. 1994;44:162–165. doi: 10.1002/jmv.1890440208. [DOI] [PubMed] [Google Scholar]
- 20.Woods P, Gentsch J, Gouvea V, Mata L, Simhon A, Santosham M, Bai Z, Urasawa S, Glass R. Distribution of serotypes of human rotavirus in different populations. J Clin Microbiol. 1992;30:781–785. doi: 10.1128/jcm.30.4.781-785.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu H, Taniguchi K, Wakasugi F, Ukae S, Chiba S, Ohseto M, Hasegawa A, Urasawa T, Urasawa S. Survey on the distribution of the gene 4 alleles of human rotaviruses by polymerase chain reaction. Epidemiol Infect. 1994;112:615–622. doi: 10.1017/s0950268800051311. [DOI] [PMC free article] [PubMed] [Google Scholar]