Abstract
Coronavirus disease 2019 (COVID-19) continues to exact a devastating global toll. Ascertaining the factors underlying differential susceptibility and prognosis following viral exposure is critical to improving public health responses. We propose that gut microbes may contribute to variation in COVID-19 outcomes. We synthesise evidence for gut microbial contributions to immunity and inflammation, and associations with demographic factors affecting disease severity. We suggest mechanisms potentially underlying microbially mediated differential susceptibility to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). These include gut microbiome-mediated priming of host inflammatory responses and regulation of endocrine signalling, with consequences for the cellular features exploited by SARS-CoV-2 virions. We argue that considering gut microbiome-mediated mechanisms may offer a lens for appreciating differential susceptibility to SARS-CoV-2, potentially contributing to clinical and epidemiological approaches to understanding and managing COVID-19.
Keywords: COVID-19, microbiota, immunity, immunological dark matter, inflammation, modelling
The host microbiome as a contributor to immunological dark matter
The emergence of SARS-CoV-2, which causes COVID-19, has triggered a pandemic with devastating global consequences. Here, we evaluate how the gut microbiome (see Glossary) may act as a driver of COVID-19 risk and contribute to COVID-19 outcomes. Several risk factors contribute to the risk of COVID-19 disease severity [1,2], and COVID-19 symptoms range from mild and transient to prolonged illness that may involve pronounced inflammation and death. The striking variation in outcomes suggests that sociodemographic or biological factors characteristic of individual hosts or populations may modify the course of SARS-CoV-2 infection. Recently, dynamic causal modelling [3] has been applied to epidemiological patterns. This approach emphasises differential susceptibility, which suggests that there are populations that are resistant or less susceptible to infection [4., 5., 6., 7.] and, by implication, populations that are more susceptible and at greater risk of virulent infection. The specific causes of differential susceptibility to SARS-CoV-2 remain under investigation and, because many of these causes are presently unobserved or ‘hidden’, they have been referred to as immunological dark matter [6,8]. The goals of understanding host–virus interactions and effectively treating infections both require knowledge of the factors contributing to outcome heterogeneity. Accounting for as much of this heterogeneity and differential susceptibility as possible may help clarify requisite population immunity levels to attenuate viral transmission [9] and inform development of early interventions for those who exhibit characteristics that are predictive of symptom severity.
We propose that the gut microbiome contributes to differential susceptibility or immunological dark matter underlying SARS-CoV-2 infection. Indeed, the potential of the gut microbiome to confer risk or resilience against diseases has already been investigated in the context of other pathogens. For example, gut microbes have been shown to modulate susceptibility to Salmonella infection (caused by a bacterium) [10] and malaria (caused by a plasmodium) [11]. We suggest here that the gut microbiome may confer upon hosts both risk and resilience to SARS-CoV-2, depending on the baseline microbial profile of the host. Researchers have already proposed important ways in which pandemic-related conditions (e.g., lockdowns and distancing measures) affect microbial communities [12] and here we discuss the converse, focusing on the gut microbiome as a biomarker and a contributing factor to COVID-19 outcomes. We first synthesise recent research connecting the gut microbiome to SARS-CoV-2 and briefly evaluate associations between the gut microbiome and relevant aspects of host physiology in the context of immune responses to SARS-CoV-2. We then analyse the gut microbiome as it relates to prominent risk factors for COVID-19 symptom severity. Finally, we explore the implications of the gut microbiome and differential susceptibility for management of the COVID-19 pandemic. At present, the gut microbiome→risk factor→COVID-19 pathway is largely speculative and has not yet been clearly established. Nevertheless, we hope that discussion of these associations and potential underlying mechanisms may guide future research into these connections.
The gut microbiome and host immunity in the context of COVID-19
The gut microbiome, host antiviral responses, and inflammation
The gut microbiome plays critical roles in training and regulating the immune system of the host [13., 14., 15.]. Gut microbes confer colonisation resistance, reducing the probability of pathogens successfully establishing themselves in the local gut ecosystem [16]. Gut microbes also regulate early antiviral responses. For instance, germ-free mice possess mononuclear phagocytes that show impaired cytokine gene expression, especially with respect to type I interferons, which are critical for effective host antiviral response [17]. Similarly, antibiotic-induced ablation of gut microbial populations reduces the capacity of the host to launch robust antiviral responses, and thus, influenza virus infection triggers more severe symptoms in antibiotic-treated mice compared with untreated controls [18]. These findings suggest that a healthy gut microbiome is necessary for appropriate antiviral defence.
Gut microbial imbalances have been shown to affect acute inflammation well beyond the gut, including the lung. There is now extensive research elucidating the influence of gut microbes on pulmonary immunity through what is commonly referred to as the microbiome–gut–lung axis [19., 20., 21.]. Gut dysbiosis has been associated with infection by various respiratory viruses, including influenza A virus (IAV) and respiratory syncytial virus (RSV) [22,23]. Moreover, gut dysbiosis during influenza infection has been shown to contribute to pneumococcal superinfection in the lung [24]. The mechanisms underlying microbiome–gut–lung associations continue to be explored, but disturbances in the gut microbiome can increase the permeability of the intestinal barrier, permitting translocation of gut bacteria to the lung, which may trigger local immune responses [25]. Microbial metabolites, such as short-chain fatty acids (SCFAs) derived from gut microbial fermentation of dietary fibre, have also been shown to modulate immunological reactivity in the lung [21,26] and have been detected in the lung compartment itself [27]. For instance, mice with high circulating SCFA levels were protected against allergic lung inflammation after exposure to dust mite extract [21]. This reduced reactivity was attributable to SCFA-induced alteration of bone marrow haematopoiesis, leading to the lungs having dendritic cells with an impaired capacity to activate effector TH2 cells, which potentiate proinflammatory responses. Overall, such results indicate roles for the gut microbiome in altering host responses to at least some respiratory infections.
SARS-CoV-2 entry into susceptible cells is promoted by virus–cell fusion, which occurs when the SARS-CoV-2 spike protein binds to the cellular receptor angiotensin-converting enzyme 2 (ACE2) and is cleaved by transmembrane protease serine 2 (TMPRSS2) and other host proteases, triggering conformational changes that enable membrane fusion [28,29]. ACE2 and TMPRSS2 are abundant in alveolar tissue [28,30], but are also widely distributed across other host tissues, including the gut [31., 32., 33., 34.]. Therefore, any effects of gut microbes on ACE2 and TMPRSS2 might be expected to affect the risk of SARS-CoV-2 infection.
Research connecting the microbiome, gastrointestinal function, and COVID-19
COVID-19 can present with gastrointestinal symptoms and inflammation [35,36], which has prompted research into gut microbial contributions to COVID-19 outcomes [37]. In Table 1 , we summarise an emerging body of empirical research on the associations between SARS-CoV-2, COVID-19 outcomes, and microbial communities. Although we focus mainly on the gut microbiome in Table 1, we also report data on host-associated microbiomes at other body sites. Collectively, these studies report differences in the microbial composition of patients with COVID-19 compared with those of healthy controls or patients with non-COVID-19 respiratory tract infections [38., 39., 40., 41., 42., 43.]. Available studies suggest that the gut microbial communities of patients with COVID-19 exhibit lower taxonomic diversity [42., 43., 44., 45.] and an increase in opportunistic pathogens [42., 43., 44.], although the extent to which these differences are attributable to concomitant antibiotic treatment in the hospital setting remains unclear. Moreover, the composition of the gut microbiota has been found to covary with disease severity [44,46,47] and dysfunctional immune responses [46]. Compared with healthy controls, researchers have observed higher relative abundances in patients with COVID-19 of gut bacterial taxa including Ruminococcus gnavus, Clostridium ramosum, Coprobacillus, Akkermansia muciniphila, and Eggerthella lenta, and lower relative abundances of Alistipes shahii and several butyrate producers, such as Roseburia intestinalis, Eubacterium hallii, Ruminococcus bromii, and Faecalibacterium prausnitzii [43,44]. The depletion of butyrate producers may be relevant to disease aetiology because butyrate is the primary metabolic fuel for colonocytes and important for maintaining colonic epithelial integrity [48]. Additionally, some gut bacterial taxa, including Bacteroides spp. found to be inversely correlated with faecal SARS-CoV-2 load in hospitalised patients [43], are capable of modulating the expression and function of cell surface receptors that regulate viral entry into cells, including ACE2 in the gut [43]. Relatedly, Bacteroides has been associated with the modification of lung heparan sulfate, which may in turn alter virion adhesion to host cells [49]. Heparan sulfate is a glycosaminoglycan that regulates structural and functional processes in lung tissue, with implications for lung pathophysiology [50] (Table 1). Such research supports an association between the abundance of particular microbial species and the capacity of the virus to successfully infect the host.
Table 1.
Correlations between host-associated microbial features and SARS-CoV-2 features and outcomes
| Study summary | Sample characteristics |
Samples and microbial analysis | Key findings | Refs | |||
|---|---|---|---|---|---|---|---|
| Groups | Sample size | Female/male | Reported age in years | ||||
| Examined sputum and airway microbial features of patients with COVID-19 | Patients with COVID-19; patients with non-COVID-19 pneumonia | 62; 125 | 22/40; 55/70 | 45 (25–82)a; 48 (15–88)a | Metatranscriptome sequencing of sputum or nasopharyngeal swabs | Airway microbiome in patients with COVID-19 showed reduced α-diversity compared with patients with non-COVID-19 pneumonia | [38] |
| Examined lung microbial features of patients with COVID-19 | Patients with COVID-19; uninfected controls | 19; 23 | NA | NA | Metatranscriptome sequencing of bronchoalveolar lavage fluid samples | Pathogens (e.g., pneumonia-causing Klebsiella oxytoca), immunomodulatory taxa (e.g., lactic acid bacteria and Faecalibacterium prausnitzii), and tobacco mosaic virus were enriched in the COVID-19 group, suggesting microbiota dysbiosis. Analysis of microbial α- and β-diversity also showed large differences between patients and controls | [39] |
| Examined lung microbial features in deceased patients with COVID-19 | Deceased patients with COVID-19 | 20 | 6/14 | 66 (60.75–77.0)b | 16S rRNA sequencing used to profile lung tissue acquired via postmortem needle core biopsies immediately after death | Significant enrichment of pathogenic microbes in lungs. Most prevalent bacterial genera were Acinetobacter (80.70% of total sequences), Chryseobacterium (2.68%), Burkholderia (2.00%), Brevundimonas (1.18%), Sphingobium (0.93%), and Enterobacteriaceae (0.68%), together comprising 92.32% of total sequences and regularly detected in all subjects. Biopsies also revealed that most patients had mixed bacterial and fungal infections | [40] |
| Examined lung microbial features of patients with COVID-19 | Patients with COVID-19; patients with non-COVID-19 pneumonia; uninfected controls | 8; 25; 20 | 3/5; NA; NA | 49.0 (±7.9)a; NA; NA | Metatranscriptome sequencing of bronchoalveolar lavage fluid samples | Relative to healthy controls, sequenced bronchoalveolar lavage fluid samples from patients with COVID-19 were similar to those with community-acquired pneumonia, and were either dominated by pathogens or displayed elevated levels of oral and upper respiratory commensal bacteria | [41] |
| Examined gut microbial features of hospitalised patients with COVID-19 or H1N1 | Patients with COVID-19; patients with H1N1; uninfected controls | 30; 24; 30 | 13/17; 9/15; 13/17 | 55.0 (48.0–62.0)b; 48.5 (33.3–66.8)b; 53.5 (43.8–60.3)b | 16S rRNA (V3-V4) gene sequencing of faecal samples | COVID-19 was associated with significantly reduced bacterial α-diversity, higher relative abundance of opportunistic pathogens (e.g., Streptococcus, Rothia, Veillonella, and Actinomyces). Patients with H1N1 displayed lower diversity and different overall microbial composition compared with patients with COVID-19, with seven microbial biomarkers distinguishing the two cohorts | [42] |
| Examined gut microbial features of COVID-19 patients over the course of hospitalisation | Patients with COVID-19; patients with non-COVID-19 pneumonia; uninfected controls | 15; 6; 15 | 8/7; 2/4; 6/9 | 55 (44–67.5)b; 50 (44–65)b; 48 (45–48)b | Shotgun metagenomic sequencing of faecal samples | COVID-19 was associated with significantly higher relative abundance of opportunistic pathogens (including Clostridium hathewayi, Actinomyces viscosus, and Bacteroides nordii) and lower relative abundance of commensal symbionts (including Eubacterium, Faecalibacterium prausnitzii, Roseburia, and Lachnospiraceae). Changes in abundance of of Bacteroides spp. over the course of hospitalisation were inversely correlated with SARS-CoV-2 load in faecal samples. Several of these bacteria (e.g., Bacteroides massiliensis, Bacteroides dorei, and Bacteroides thetaiotaomicron) have been linked with downregulation of ACE2 in the murine gut, and were inversely correlated with SARS-CoV-2 load in human faecal samples | [43] |
| Examined whether COVID-19 alters nasal, throat and gut microbial features in children | Children with COVID-19; uninfected controls | 9; 14 | NA; NA | 7-139 months; age-matched | 16S rRNA (V4) gene sequencing of faecal samples, nasal swabs, and throat swabs | Relative to healthy controls, sustained enrichment of Pseudomonas veronii in both respiratory and gut microbiomes observed in children with COVID-19, with relative abundance of this taxon exceeding 20% in most of the children with COVID-19 | [179] |
| Examined whether a co-receptor of SARS-CoV-2, heparan sulfate, was sensitive to modification via microbes | NA | NA | NA | NA | No direct measure of microbial composition | In human cell cultures, heparan sulfate was highly sensitive to modification by Bacteroides species. Specifically, Bacteroides ovatus and Bacteroides thetaiotaomicron could catabolise cell-surface heparan sulfate and block SARS-CoV-2 from binding to human lung cells. Across two large human microbiome data sets, two of the established risk factors for COVID-19, older age and male sex, were associated with significant reductions in microbes capable of modifying heparan sulfate | [49] |
| Examined whether gut microbial features could be predictive of blood proteomic biomarkers of severe COVID-19 disease | Patients with severe COVID-19; patients with non-severe COVID-19; uninfected controls | 13; 18; 990 | NA; NA; 668/322 | NA; NA; 58.79 (±5.6)a | Quantitative proteomics analysis of serum samples; 16S rRNA (V3-V4) gene sequencing of fecal samples | Identified set of serum proteomic inflammatory markers capable of predicting progression to severe COVID-19, and established that these serum markers could in turn be accurately predicted by gut microbiota composition. These inflammatory markers were positively associated with the genera Ruminococcus, Blautia, and Lactobacillus, and negatively associated with the genera Bacteroides and Streptococcus and the order Clostridiales | [47] |
| Investigated whether gut microbial features are linked to disease severity in patients with COVID-19 | Patients with COVID-19; uninfected controls | 100; 78 | 47/53; 45/33 | 36.4 (±18.7)a; 45.5 (±13.3)a | Shotgun metagenomic sequencing of faecal samples | Gut microbiota composition covaried with disease severity and dysfunctional immune responses in patients with COVID-19. Microbial composition in recovered patients remained altered compared with individuals not infected with SARS-CoV-2 | [46] |
| Analysed oropharyngeal microbial features in patients with mild, moderate, and severe COVID-19, in patients with non-SARS-CoV-2 infections, and in healthy controls | Patients with mild COVID-19; patients with moderate COVID-19; patients with severe COVID-19; patients with upper respiratory tract infections; uninfected controls | 36; 27; 66; 112; 74 | 24/12; 10/17; 14/46; 75/37; 48/26 | 50 (36.75–55.5)b; 57 (46.25–72.75)b; 64 (53.25–72.75)b; 46 (31.0–57.0)b; 36 (29.75–53.25)b | Shotgun metagenomic sequencing of oropharyngeal swabs | Significantly diminished oropharyngeal microbial diversity and high dysbiosis in hospitalised patients with severe COVID-19, which was further linked to a loss of microbial genes and metabolic pathways. Random forest machine learning revealed that oropharyngeal microbiota abundances of Haemophilus or Streptococcus spp. were most important microbial features for segregating clinical outcomes in patients hospitalised with COVID-19 | [180] |
| Investigated associations between oropharyngeal microbial features and fatality in patients with COVID-19 | Patients with COVID-19; uninfected controls | 192; 95 | 78/114; 57/38 | 58 (49–68)b; 47 (33–61)b | Metatranscriptome sequencing of longitudinal oropharyngeal swabs. Swabs were obtained on days 1, 5, 10, 14, 21, and 28 after admission when patient's condition allowed | Abundance of Streptococcus on admission, particularly that of Streptococcus parasanguinis, was identified as a strong predictor of fatality (39/192 cases of COVID-19 were fatal) | [181] |
| Investigated changes in gut microbial features from acute COVID-19 through postconvalescence | Patients with COVID-19; uninfected controls | 30; 30 | 11/19; 11/19 | 53.5 (39.75–59)b; 53.5 (45.25–58)b | 16S rRNA (V3-V4) gene sequencing of longitudinal faecal samples, acquired during acute phase of infection (from onset of illness to host clearing of virus), convalescence (from host clearing of virus to 2 weeks post discharge from hospital), and postconvalescence (6 months post discharge from hospital) | Significant differences in gut microbial communities between patients with COVID-19 and uninfected controls. Microbial richness was lower among patients with COVID-19 than healthy controls at all three time points. Trend toward increasing richness following infection, but richness remained substantially lower even 6 months after host had cleared the virus | [45] |
| Compared throat and gut microbiomes of patients with COVID-19 with samples from uninfected individuals obtained for an earlier study; human observations then extended to vaccinated and unvaccinated murine models | Patients with COVID-19; uninfected controls | 13; 5 | 7/6; NA | 48 (15–85)b; NA | Metagenomic and metatranscriptomic sequencing of throat swabs and faecal samples from patients with COVID-19, and intestinal and faecal samples from infected vaccinated or infected unvaccinated mice | Gut bacterial diversity lower in patients with COVID-19 relative to uninfected controls. Relative to mild and moderate cases of COVID-19, severe COVID-19 was associated with increase in abundance of opportunistic pathogens (e.g., Corynebacterium, Enterococcus, Campylobacter, Citrobacter, Enterobacter) and a loss of butyrate-producing gut bacteria (e.g., Eubacterium and species such as Faecalibacterium prausnitzii). Relative abundances of both Akkermansia muciniphila and Odoribacter higher in both patients with COVID-19 and infected unvaccinated mice related to infected vaccinated mice, with A. muciniphila also being more transcriptionally active in infected unvaccinated mice, suggesting these taxa are associated with disease processes. As a result of antibiotic treatment in several patients, some bacterial changes were difficult to attribute to effects of SARS-CoV-2 versus concomitant antibiotic exposure | [44] |
Abbreviations: NA, not available
Age presented as mean (±S.D.).
Age presented as median (interquartile range).
Recent research raises the possibility that the gut microbiome impacts SARS-CoV-2 infection and COVID-19 severity by modulating T and B cell function. There are two principal subtypes of T cell: CD4+ T cells and CD8+ T cells. CD4+ T cells release cytokines that activate other immune cells, thereby potentiating an immune response, whereas CD8+ T cells kill virus-infected cells. B cells produce neutralising antibodies that bind to peptides in the viral spike and other viral proteins, inhibiting the capacity of the virion to infect host cells. Through these actions, T cells and B cells help clear acute infection with SARS-CoV-2 [51], modulate COVID-19 disease severity [52,53], and play an important role in immunity against reinfection [54,55], including against emerging variants of concern [56].
While the studies summarised in Table 1 are mostly correlational, some of the findings do appear to reconcile with the role of gut bacteria in training and regulating the immune system and the particularly well-established link between gut microbes and the shaping of adaptive T and B cell responses, both locally and systemically [57., 58., 59.]. Gnotobiotic studies have shown that both T cells and B cells are regulated by the microbiome. For instance, T cell differentiation and proliferation depend on the presence of gut microbes [15]. Gut microbes have also been shown to play a crucial role in activating mucosal-associated invariant T (MAIT) cells [60], which were recently found to be associated with COVID-19 disease severity [61,62]. Similarly, with respect to B cells, experiments in mice indicate that B cell differentiation is promoted indirectly by the gut microbiota through upregulation of interleukin (IL) production, specifically IL-1β and IL-6 [63].
Gut microbial regulation of host inflammatory status
Some of the dominant risk factors for severe COVID-19 outcomes are inflammation-linked metabolic and cardiovascular comorbidities, such as diabetes and hypertension, and demographic characteristics, such as older age (Box 1 ) and male biological sex (Box 2 ) [1,2,64]. Each of these characteristics is associated with variations in gut microbial composition and function.
Box 1. The gut microbiome and host age.
Greater age is an important risk factor for outcome severity in SARS-CoV-2 infection [2,114., 115., 116., 117., 118.]. Ageing also affects gut microbial structure and function, an association thought to be connected to senescence of the host immune system [119., 120., 121.]. Covariation between host age, immunity, and microbial composition likely exerts joint effects on inflammatory status and risk of disease. Although much human variation remains to be characterised, ageing appears to result in generally lower α-diversity coupled with increased relative abundance of pathobionts [121., 122., 123., 124.]. A recent study investigated the causal role of gut microbes in regulating ageing-related inflammation and immunopathology by transplanting microbes from young or aged mice into young, germ-free recipients [124]. The germ-free recipients of microbial transplants from aged mice displayed a range of immunological changes relative to recipients with young donors, including elevated inflammation and increased gut barrier permeability, as evidenced by increased translocation of microbial products (e.g., lipopolysaccharide) into systemic circulation [124]. Taken together, these general patterns suggest that the microbiome may contribute to elevated inflammation in older individuals, which may then act as a risk factor for more severe outcomes upon SARS-CoV-2 infection.
Alt-text: Box 1
Box 2. The gut microbiome and host biological sex.
Researchers have found sex differences in immune responses to SARS-CoV-2 [125]. Biological males are at increased risk of experiencing more virulent SARS-CoV-2 infection and greater rates of mortality [126,187]. However, biological females appear more likely to display persistent, post-infection symptoms, including fatigue, anosmia, dyspnoea, and cognitive dysfunction [128., 129., 130.]. These findings suggest that female sex does not confer general protection, but rather that biological sex confers differential risks and protection for different SARS-CoV-2 infection courses. Furthermore, males and females respond differently to vaccines [130], including vaccines against SARS-CoV-2. For instance, recent human findings suggest that vaccination triggers greater levels of immunoglobulin G (IgG) in females relative to males [131].
Potential mechanisms
The X chromosome is enriched in genes regulating host immunity [132], and this chromosomal difference may be one of the causes underlying sex differences in host responses to SARS-CoV-2 [126]. Another mechanism underlying observed sex differences in COVID-19 outcomes may be the action of sex steroids. Sex steroids regulate the availability of both ACE2 and TMPRSS2, the cell surface molecular features exploited by SARS-CoV-2 virions. For instance, it appears that androgens (predominant in males) upregulate both ACE2 and TMPRSS2 [134., 135., 136., 137., 138.], whereas oestrogens (predominant in females) appear to downregulate these molecular targets [139., 140., 141.]. Notably, castration of mice or androgen deprivation in mouse and human cells reduced ACE2 and TMPRSS2 expression and protein levels, and androgen deprivation or anti-androgen treatment diminished entry of SARS-CoV-2 into lung cells [133]. Correspondingly, human males undergoing androgen-deprivation treatment for prostate cancer experience less severe COVID-19 outcomes [134], and blocking androgenic signalling is associated with reduced ACE2 levels and reduced risk of severe COVID-19 outcomes [135].
Moreover, sex hormones also regulate host inflammatory tone. Intriguingly, androgens have been reported to exert mostly anti-inflammatory effects [141], despite their association with more severe COVID-19 outcomes. By contrast, oestrogens may exert either anti-inflammatory or proinflammatory effects, depending on the receptor [141]. In general, hormone levels vary across individuals of a given biological sex. This variation may influence the extent to which host inflammation is a risk factor, and may dampen associations between biological sex and COVID-19 outcomes, potentially explaining some variation in susceptibility among individuals of a given biological sex.
Potential microbial involvement
There is some evidence of sex differences in the mammalian gut microbiome [119., 120., 121.,143., 144., 145., 146., 147., 148., 149.]. Crucially, the gut microbiome regulates the bioavailability of sex steroids [107,144,149]. For example, germ-free male mice show markedly lower testosterone concentrations relative to typically colonised controls, suggesting that the presence of microbial populations increases the quantity of testosterone in males [144]. Moreover, transferring gut microbes from adult males into preadolescent female mice increases testosterone concentrations in recipients [144], adding inductive support to the causal role of the microbiome in androgen regulation. Gut microbes also regulate oestradiol bioavailability. For instance, numerous gut bacterial taxa secrete β-glucuronidase, which deconjugates oestradiol that has undergone conjugation by the liver and excretion into the gut lumen via bile. Deconjugation returns luminal oestradiol to its active form and enables re-entry into host circulation, thus increasing the bioavailable oestrogen in the host [151., 152., 153.]. Correspondingly, antibiotic administration increased quantities of conjugated oestrogens excreted in host faeces in both males and females [154., 155., 156.]. Moreover, gut microbes can increase oestrogen activity via biotransformation of diet-derived phytoestrogens [156,157]. Overall, therefore, gut microbes play important roles in sex steroid bioavailability, and sex steroids in turn regulate inflammation and the availability of the cell surface features exploited by SARS-CoV-2.
Alt-text: Box 2
Various gut microbiome–immune interactions modulate systemic and acute inflammation in the host. In some cases, the inflammatory landscape may in turn increase risk for virulent SARS-CoV-2 infection, culminating in cytokine storms characterised by very high concentrations of circulating cytokines [e.g., IL-1β, IL-6, tumour necrosis factor-α (TNF-α), monocyte chemoattractant protein-1 (MCP-1), and macrophage inflammatory protein-1α (MIP-1α/CCL3)], hyperinflammation (measured by biomarkers such as C-reactive protein) and, frequently, multiorgan failure and death [1]. Given the relationships between gut microbes, host immunity, and host inflammatory status, it is conceivable that microbiome composition and function may confer risk for, or resilience against, the immune dysregulation and excessive inflammation characterising severe COVID-19 (Figure 1 ).
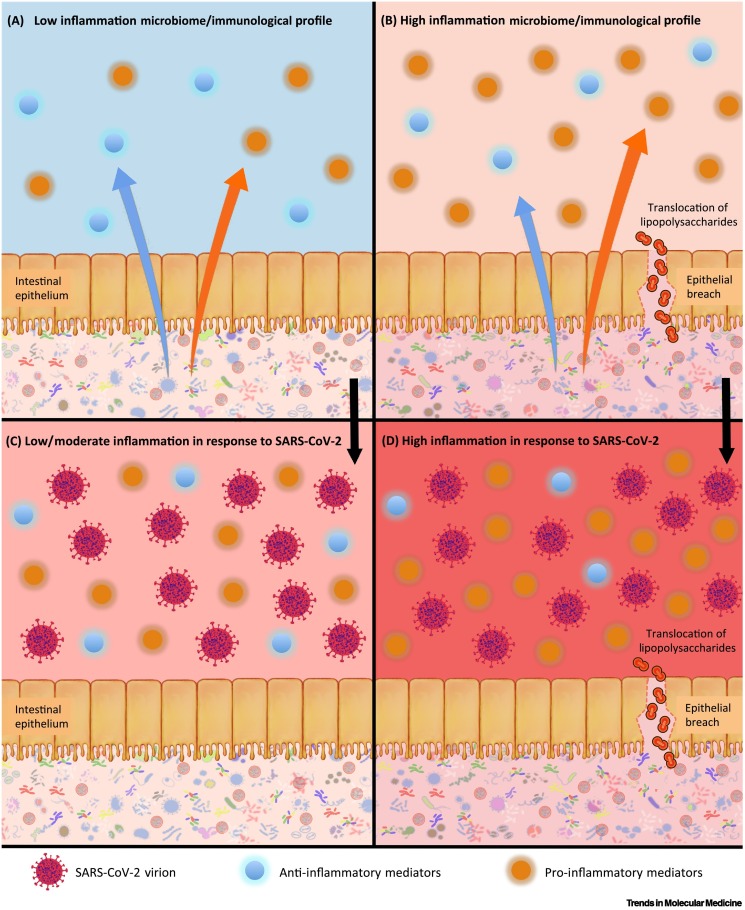
Figure 1.
Microbiome-associated inflammation profiles and potential reactions to severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2).
The microbiome influences host inflammation through the regulation of host immunological processes (proinflammatory effects are indicated by the orange arrow, and anti-inflammatory effects by the blue arrow). These include the production of host pro- and anti-inflammatory mediators, and the migration of bacteria or bacterial products, such as lipopolysaccharide, through the gut lining. (A) A microbiome that contributes to a balanced or low-inflammation state (low-inflammation properties are indicated by the blue systemic background and the pale-pink microbial background) with a relatively higher level of anti-inflammatory cytokines. (B) A microbiome that contributes to a proinflammatory state (high-inflammation properties are indicated by the pale-orange systemic background and the darker-pink gut background), with a relatively higher number of proinflammatory mediators, and potential translocation of bacterial products into systemic circulation (the proinflammatory state is indicated by the relatively larger orange arrow). The bottom two images show the putative reactions of these systems to SARS-CoV-2 infection, denoted by the presence of virions. (C) A relatively muted proinflammatory response to viral infection, represented by the pink systemic background behind the immune cells. (D) A strong proinflammatory response to viral infection, represented by the red systemic background behind the immune cells.
Murine research has confirmed a role for microbially regulated host immunity and inflammation as a risk factor for cytokine storms [65,66]. Cohousing specific pathogen-free laboratory mice with pathogen-harbouring pet-store mice increased quantities of circulating monocytes and neutrophils expressing toll-like receptor 2 (TLR2) and TLR4 in the laboratory animals. These changes affected outcomes in a subsequent immune challenge involving Listeria monocytogenes, with cohoused laboratory mice exhibiting enhanced bacterial clearance but heightened sensitivity to cytokine storms and sepsis [65]. Similarly, exposure of laboratory mice to the microbial communities characterising wild-living mice led to elevated immune responses and increased probability of cytokine storms relative to unexposed laboratory controls [66]. While these studies reported immune responses to novel pathogens (and thus strong immune reactions are predicted a priori), these results do demonstrate that differences in baseline microbial composition among hosts are sufficient to affect the probability of cytokine storms.
The vertebrate immune system has evolved to simultaneously defend against potential pathogens while developing tolerance for, and actively curating (to the extent possible), beneficial and commensal microbes colonising the host [67,68]. These contribute to the development and training of host immunity during early life, following which molecular dialogues between the microbiome and the host regulate immunity throughout the lifespan [15,69,70]. Indeed, a recent study demonstrated that human gut microbes are closely associated with circulating immune cells [71]. This study found strong associations between the genera Faecalibacterium, Ruminococcus, and Akkermansia and blood concentrations of neutrophils, lymphocytes, and monocytes. Moreover, gut microbes are implicated in the aetiopathogenesis of other diseases that are characterised by hyperinflammation, such as the autoimmune diseases rheumatoid arthritis and systemic lupus erythematosus. In these diseases, the gut microbiome can trigger symptoms in genetically susceptible individuals by producing orthologues of autoantigens, which can lead to microbiota-directed immune responses targeting host tissues instead [72].
Impaired gut barrier function may also exacerbate inflammation in both autoimmune diseases [72] and COVID-19 [73]. While the gut microbiome in most healthy individuals facilitates immune homeostasis, compositional imbalances induced by host-driven or environmental factors can affect the integrity and selective permeability of the intestinal barrier, with implications for inflammation [74] and cytokine storms [75]. Elevated intestinal barrier permeability can permit translocation of commensal and pathogenic microbes and their products into systemic circulation, which may in turn trigger heightened systemic inflammatory reactions, increased tissue damage, and ultimately chronic inflammation [76]. Unlike acute inflammatory responses, chronic inflammation is characterised by a dense infiltration of primary immune cells (e.g., lymphocytes and macrophages) in tissues. These cells produce proinflammatory cytokines, growth factors, and enzymes (e.g., nitric oxide synthase and matrix metalloproteinase), which lead to sustained inflammation and persistent cycles of tissue damage and repair [77,78]. Chronic low-grade inflammation may increase the risk of harmful hyperinflammation in COVID-19 [1], suggesting links between gut microbiota-mediated changes in gut barrier integrity and the risk of cytokine storms.
Microbial translocation into peripheral circulation, which occurs when gut barrier integrity is compromised, has been described as a characteristic feature of systemic inflammation among children with SARS-CoV-2 complicated by multisystem inflammatory syndrome (MIS-C) [79]. Compared with children with acute COVID-19 and either seropositive children or controls, patients with MIS-C displayed elevated serum lipopolysaccharide (LPS) [79]. Similarly, children with acute COVID-19 showed higher serum LPS levels compared with seropositive individuals, and seropositive individuals showed higher serum LPS levels compared with controls, suggesting that the quantity of LPS entering circulation due to enhanced gut permeability is correlated with the severity of COVID-19 infection [79]. These correlations could be driven, in part, by interactions between LPS and the SARS-CoV-2 spike protein. The SARS-CoV-2 spike protein has been shown to bind directly with LPS, affect the function of LPS-binding protein, and modulate the aggregation state of LPS, thereby boosting proinflammatory activity [80]. Combinations of LPS and the SARS-CoV-2 spike protein increased TLR4-mediated cytokine responses in human blood and peripheral blood mononuclear cells, an effect that was also confirmed in vivo using NF-κB reporter mice [80]. Cytokine storms typically result in intestinal damage [81,82], suggesting that impaired barrier function could anchor a vicious cycle of escalating inflammation.
Apart from altering probabilities of hyperinflammation, variation in the baseline gut microbial profile and the resilience of the gut microbiota in response to SARS-CoV-2 infection may also modulate inflammation associated with metabolic diseases now known to increase the risk of severe COVID-19 outcomes, including diabetes mellitus [1]. For instance, several studies found that antibiotic administration reduced inflammation and insulin resistance in high-fat diet-fed mice or mice genetically susceptible to metabolic disease [83,84]. Moreover, germ-free mice receiving microbial transfers from insulin-resistant mice exhibited more inflammation than counterparts receiving microbial transfers from controls [84]. These results demonstrate a causal role for gut microbes in the generation of an inflammatory phenotype that could then serve as a risk factor in SARS-CoV-2 infection.
Animal models to investigate microbial associations with SARS-CoV-2 outcomes
Associations between the microbiome and risk factors for COVID-19 such as host age and biological sex (see Box 1, Box 2, respectively) can be investigated experimentally using animal models. Although mice are most commonly used in studies of both immune function and the microbiome, the existing SARS-CoV-2 virus now in circulation does not readily infect mice due to inefficient interactions with the mouse orthologue of ACE2 [85]. This limitation could be overcome through the use of ACE2 transgenic mice [86,87], mice transduced with adenoviruses encoding human ACE2 [88,89], or alternatively, by exposing common murine strains such as C57BL/6 or Swiss Webster to the recently developed mouse-adapted SARS-CoV-2 MA virus [90]. Such techniques have already delivered interesting results pertaining to interactions between the host microbiome and SARS-CoV-2 infection. For instance, researchers have recently examined the actions of SARS-CoV-2 in vaccinated and unvaccinated ACE2 transgenic mice [44]. Infected unvaccinated mice showed reduced gut bacterial diversity relative to vaccinated mice exposed to SARS-CoV-2. Similar to changes previously observed in humans with COVID-19 relative to healthy controls (see Table 1), infected unvaccinated mice showed elevations in Odoribacter and A. muciniphila, as well as reductions in Lactobacillus reuteri and Bacteroides uniformis, relative to vaccinated animals.
Primate models that are susceptible to SARS-CoV-2 infection, such as macaques [91], will doubtless also prove useful, given their closer phylogenetic relatedness and physiological similarity to humans. For instance, investigators have recently examined the microbial effects of SARS-CoV-2 infection in rhesus macaques and cynomolgus macaques [91]. SARS-CoV-2 infection triggered changes and ecological perturbations in the host gut microbiome that persisted over the course of the infection. The abundances of several bacterial taxa were found to be correlated with infection-related parameters, including nasal and rectal viral loads, C-reactive protein levels, and circulating levels of several proinflammatory cytokines. The concentrations of several SCFAs were observed to decline in faeces during infection, although it remains unclear whether this reduction was attributable to reduced bacterially mediated SCFA production or increased SCFA utilisation by host cells [91].
Animal models can similarly be used to examine microbial associations with host differential susceptibility to SARS-CoV-2. For instance, consider the possibility that an aged microbiome confers increased risk for severe infection. This could be tested via experimental colonisation studies in gnotobiotic mice susceptible to SARS-CoV-2 or treated with the mouse-adapted SARS-CoV-2 MA virus [90]. Germ-free animals could be colonised with faecal microbial communities from young and old mice, or young and old humans, and then be exposed to the virus. If harbouring an aged microbiome is a risk factor for COVID-19, then we might expect faecal microbiota transplantation to trigger more severe symptoms in recipients of microbiotas from older individuals, relative to recipients with younger donors. In general, this type of research demonstrates that animal models may be able to generate useful and fine-grained information that can inform our understanding of the role of the microbiome in COVID-19.
Impacts of differential susceptibility to SARS-CoV-2 on pandemic modelling
It has become increasingly apparent that the enormous heterogeneity of exposure, susceptibility, virulence, transmission, and vaccination status play key roles in shaping the time course of the pandemic and variations between individuals, groups, communities, and countries [6,7,93., 94., 95.]. Accounting for this heterogeneity suggests that the requisite levels of herd immunity may be lower than assumed under conventional models, which often do not consider heterogeneity and differential susceptibility in detail. Differential susceptibility, although biologically realistic, is often difficult to incorporate into epidemiological models. To explain quantitative dissociations between the incidence of infection, time-varying infection:fatality ratios, and fluctuations in seroprevalence, it is necessary to model populations as mixtures of individuals who are: (i) more or less exposed to the virus, due to vaccination status, geospatial factors, population density, population-level behavioural factors (such as mask-wearing, hand washing, self-isolation, time spent outdoors, and vaccination), and cultural factors (such as the value placed on personal space); (ii) differentially capable of transmitting the virus, due to differences in vaccination status or viral shedding [96., 97., 98.]; and (iii) more or less susceptible to virulent infection, due to vaccination status, potential cross-reactive immunity with other β-coronaviruses, individual differences in the expression of ACE2 or TMPRSS2 [7,99., 100., 101.], or other host factors that may constitute immunological dark matter.
In this context of differential susceptibility, the gut microbiome is a host factor that varies within and between individuals and, as described here, is broadly related to several major risk factors for severe COVID-19. Furthermore, variations in host genetics may also contribute to microbiome-associated differential susceptibility and protection, although these immunogenetic relationships await further study (Box 3 ). Our suggestion is not that the microbiome is responsible for most of the variance associated with SARS-CoV-2 infection, but rather that the microbiome, being meaningfully correlated with several host factors relating to host immunity, may shed light on some of the immunological dark matter underlying differential susceptibility. Including information about the gut microbiome, and its impact on host immunity, into quantitative assessments of variations in risk and resilience, as summarised in Figure 2 (Key figure), could contribute nuance to the development of public health interventions and messaging. For example, the identification of specific bacterial taxa associated with host susceptibility, combined with the application of rapid and low-cost methods for profiling gut microbiomes, such as taxon-specific quantitative polymerase chain reaction (qPCR) targeting the 16S rRNA gene, could enable pre-emptive population surveys or individual tests capable of estimating relative risk. These efforts could be furthered through large-scale prospective studies to assess relationships between microbiome composition and infection outcomes. In addition to collecting microbiome data, prospective studies could leverage data pertaining to host inflammatory status, sex, age, and metabolic and cardiovascular health, each of which is known to interact with the microbiome. Notably, because gut microbial interactions with inflammation and host factors such as age and sex may influence susceptibility and outcomes with respect to multiple viral pathogens, these efforts may yield insights relevant to SARS-CoV-2, other circulating viruses, and viruses that may become problematic in the future.
Box 3. The microbiome, host genetics, and COVID-19.
Immunogenetic variation has been associated with variation in COVID-19 susceptibility and outcomes [159., 160., 161.]. Host genes also contribute to variation in gut microbial composition, as evidenced by twin studies [162., 163., 164., 165.], genome-wide association studies in humans [166., 167., 168., 169., 170.], and large-scale, long-term studies in nonhuman primates [170]. Although global genetic factors explain less variation than do environmental factors [171], specific gene variants have been shown to meaningfully affect gut microbial community composition, including LCT, VDR, NOD2, ABO, and FUT2 [161,165,169,172]. Researchers have also been interested in the interactions between host genes and the gut microbiome in the context of disease [161]. Although associations are generally small and many mechanisms remain unclear, genome–microbiome–disease interactions raise the possibility that, in addition to modulating COVID-19 resistance and susceptibility through the immune system, host genes may also affect disease resistance and susceptibility through effects on the gut microbiome.
A further interesting immunogenetic connection of potential relevance to COVID-19 outcomes is the blood group of the host. Importantly, natural blood group antibodies can recognise members of the gut microbiome [173] and also shape gut microbial composition and ecology [172,174]. These associations have been studied in the context of inflammatory conditions, such as inflammatory bowel disease [172], but have not yet been extended to COVID-19. This connection is particularly relevant because blood groups have also been associated with COVID-19 outcomes, with O and Rh– blood groups appearing to confer protective effects in the context of SARS-CoV-2 infection [175]. These findings provide a further potential link between the gut microbiota and COVID-19 outcomes.
Alt-text: Box 3
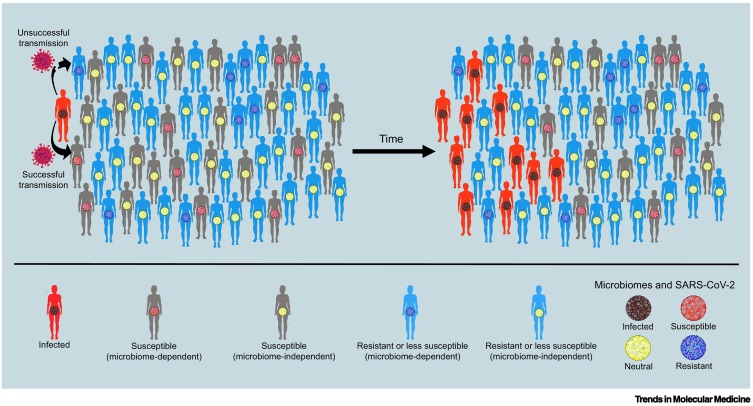
Figure 2.
Key figure. Microbial contributions to differential susceptibility and immunological dark matter.
Viral transmission in a population in which individuals are infected (red silhouettes), susceptible (grey silhouettes), or resistant (blue silhouettes) to severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2) infection. While differential susceptibility may be mediated by many factors, this figure emphasises hypothetical effects of the gut microbiome. The microbiome may enhance susceptibility (red microbiomes) or resilience (blue microbiomes), or may be unrelated to risk or resilience (yellow microbiomes). Thus, grey individuals with red microbiomes represent the subpopulation for which the microbiome is a hypothetical source of risk for SARS-CoV-2 infection. Grey individuals with yellow microbiomes are susceptible to SARS-CoV-2 for reasons unrelated to the microbiome. Blue individuals with blue microbiomes represent the subpopulation for which the microbiome is the hypothetical source of resistance to SARS-CoV-2. Blue individuals with yellow microbiomes represent the subpopulation that resists or is less susceptible to SARS-CoV-2 for reasons unrelated to the microbiome. Infected individuals (red silhouettes) have microbiomes that carry a signature of the infection (black microbiomes). Differential susceptibility increases the variability in the propensity of individuals to spread the virus, a phenomenon known as ‘overdispersion’ in epidemiological modelling (put simply: ‘the few infect the many’). Overdispersion can exert profound effects on viral spread at the population level, making it a potentially important source of immunological dark matter. Such dynamics are an important explanation for why an epidemic does not unfold as expected, under often implausible assumptions of homogenous and well-mixed populations.
It is also generally thought that differential susceptibility and transmission reduce effective herd immunity thresholds [7,101]. Differential susceptibility to SARS-CoV-2 may arise from diverse sources. For example, there is interest in the role of variations in vitamin D levels in COVID-19 risk [102]. If differences in the gut microbiome also contribute to differential susceptibility, then consistent population-level differences in the gut microbiome (such as those seen between industrialised and non-industrialised populations and across lifestyle gradients [104., 105., 106.]) may meaningfully contribute to modelling population-level disease risks and herd immunity thresholds.
Of course, these situations are highly simplified. There is no single ‘resilient’ microbiome. There is also no strictly ‘neutral’ microbiome. Furthermore, the sources of differential susceptibility (e.g., vaccination status, prior immunity, comorbidities, age, sex, and microbial composition) do not confer their effects in isolation. Rather, they interact with one another to generate complex profiles of risk and resilience. Moreover, individual resilience and symptom severity are dynamic and continuous, as opposed to categorical, and are known to depend on the extent and duration of exposure to the virus (i.e., contact rates and mean periods of infectiousness). Accordingly, susceptible individuals may avoid infection, and resistant or less-susceptible individuals may still contract the virus, although the latter would be expected to suffer fewer symptoms, recover more quickly, and be less infectious due to faster recovery, lower viral loads, or lower viral shedding rates.
Concluding remarks
We have highlighted how explicit considerations of the gut microbiome may elucidate aspects of COVID-19 risk and resilience. To our knowledge, the microbiome is not presently systematically considered in COVID-19 research, despite its potential to affect infection response and supplement disease management. In this vein, there remain numerous important issues in need of investigation (see Outstanding questions). There are several areas in which research can readily be conducted. For example, faecal samples can be acquired easily and non-invasively and used to assess interactions between the gut microbiome, COVID-19 risk factors, therapeutic interventions, and disease outcomes. Moreover, as discussed earlier, there are now several animal models being used to study COVID-19 [87., 88., 89., 90., 91., 92.,106], and these too provide ready sources of microbial data if researchers choose to collect faecal samples and profile the associated microbial communities through sequencing or targeted qPCR. Experiments with gnotobiotic animal models may prove especially useful because these offer unique opportunities to isolate causal effects of the gut microbiome on host phenotypes. There are also several areas in which clinicians may benefit and to which they could contribute (see Clinician’s corner).
Outstanding questions.
To what extent does baseline gut microbiome profile modulate the risks of SARS-CoV-2 infection and COVID-19 severity given infection?
Given abundant virus × bacteria interactions within the gut microbiome [182], do SARS-CoV-2 virions interact directly with host microbes? If so, how do these interactions influence local microbial ecology and disease severity?
Researchers across biological and medical science have raised concerns about the overuse of antibiotics and the rise of antibiotic resistance. Notably, a recent meta-analysis suggests that patients with COVID-19 are prescribed antibiotics at rates much higher than warranted for bacterial co-infection [176]. Could the overuse of antibiotics be affecting gut microbial composition in ways that affect host immune function and the response to SARS-CoV-2 virions?
Are the observed increases in gut permeability with worsening COVID-19 causes or outcomes of symptom exacerbation?
Can prebiotics or probiotics be used as complementary or adjuvant methods in the treatment of COVID-19? Notably, the endogenous populations of at least some probiotic candidates have been associated with poorer COVID-19 outcomes. For instance, A. muciniphila, a probiotic under investigation for its metabolic benefits [184,185], has also been found to be elevated in SARS-CoV-2 infection in human and rodent studies [44]. As such, the use of probiotics should be carefully considered against evidence of microbial changes occuring in COVID-19.
How can the human challenge studies for COVID-19 that are now being initiated be used to elucidate gut microbiome-mediated risk and resilience? For example, patient monitoring in these trials could be modified to include assessment of gut microbial composition before, during, and after the period of infection.
Alt-text: Outstanding questions
Clinician’s corner.
Working to characterise the gut microbiome in infected and non-infected cohorts, and longitudinally in individuals before, during, and after infection, could shed light on the immunological dark matter underlying differential susceptibility to SARS-CoV-2 infection and COVID-19 outcomes. Obtaining faecal samples from patients with COVID-19 for downstream analysis of gut microbial structure and function is technically straightforward, non-invasive, and relatively easy to add on to current patient care and monitoring. Analysis of these samples in conjunction with patient records could help us understand how the gut microbiome contributes to individual and population-level risk and resilience.
COVID-19 is primarily investigated as a disease of the respiratory tract. However, given the widespread systemic effects of the virus, it may be fruitful for gastroenterologists to be further involved in researching and treating SARS-CoV-2 infection.
During treatment, clinicians could seek to obtain information about ongoing and previous antibiotic use by patients with COVID-19. This will enable inferences about the patient’s baseline level of gut dysbiosis, and will help to contextualise the results of any microbial profiling studies carried out.
A recent meta-analysis suggests that physicians prescribe antibiotics to patients with COVID-19 at rates substantially higher than estimated rates of bacterial co-infection in COVID-19 cases, suggesting some degree of antibiotic overuse [176]. If antibiotics are used to treat bacterial infections associated with COVID-19, clinicians should consider whether it may be appropriate to replenish beneficial and commensal microbes via the use of prebiotics or probiotics. In addition, if the circumstances permit, it may be feasible to use narrow-spectrum antibiotics, which could help to preserve broader gut microbial integrity.
During treatment, clinicians could consider assessing circulating markers of compromised gut barrier function, such as serum LPS. This will help researchers understand the extent to which gut permeability is increased in COVID-19 and perhaps whether gut permeability predicts disease severity.
Given that many therapeutic drugs are chemically modified by the gut microbiome and that such activities can profoundly alter their effectiveness [177,178], it would be valuable to assess the relationships between the gut microbiome and patient outcomes in the context of current COVID-19 treatments. Advancing such knowledge could ultimately enable clinicians to one day use the microbiome as a diagnostic tool to contribute to decisions about the best course of treatment.
Alt-text: Clinician’s corner
We note that many inconsistencies and contradictions exist across studies reporting interactions between the gut microbiome and human health [108,109,183,186]. Part of the difficulty arises from day-to-day gut microbial plasticity in response to variable environmental factors such as diet [110,111], and the high degree of interindividual gut microbial dissimilarity across humans, which often necessitates longitudinal sampling or very large data sets to derive reliable patterns [112]. The COVID-19 pandemic provides precisely such an opportunity on a global scale. Accordingly, we recommend routine collection and analysis of faecal samples alongside standard patient data. We also emphasise the need for prospective approaches characterising longitudinal microbiome change as individuals progress from healthy to infected to recovered, as well as before and after vaccination. Overall, investigating the microbiome in the context of COVID-19 could deepen our understanding of the current pandemic, facilitate responses to future outbreaks, and advance our basic appreciation of the microbiome as a fundamental component, and regulator, of host immunity.
Acknowledgments
Acknowledgments
We thank Cary Allen-Blevins, Katia Chadaideh, Benjamin Ho, Laura Schell, and Emily Venable for helpful feedback and discussion. A.H.M. is supported by the National Institutes of Health (R35 GM138284). S.L.K. is supported by the Johns Hopkins Serological Center of Excellence (U54CA260492), the Johns Hopkins Specialized Center of Research Excellence (U54AG062333), and the Johns Hopkins Center of Excellence in Influenza Research and Surveillance (HHSN272201400007C). S.E.E. is supported by the National Institutes of Health (P30 ES02109, U01 CA164337, R01CA108854), the Theron Randolph Foundation, the Angiogenesis Foundation, and the John Templeton Foundation. K.J.F. is supported by funding for the Wellcome Centre for Human Neuroimaging (205103/Z/16/Z). R.N.C. is supported by the National Science Foundation (BCS-1919892), National Institutes of Health (R01AG049395), and the Harvard Dean’s Competitive Fund for Promising Scholarship. A.S. and S.H. declare no research funding.
Declaration of interests
None declared by authors.
Glossary
- Cytokine
any of several proteins secreted by various immune cells (e.g., chemokines, interleukins, interferons, and tumour necrosis factors). Cytokines facilitate cell–cell signalling, and can exert proinflammatory or anti-inflammatory effects.
- Cytokine storm
state of systemic hyperinflammation involving dramatically elevated concentrations of proinflammatory cytokines and other immune cells. Cytokine storms can lead to dysfunction of secondary organs, multiorgan failure, and death.
- Differential susceptibility
general phenomenon that susceptibility to a disease is not uniform in a population but varies among individuals. In COVID-19, differential susceptibility is relevant both at the level of clinical susceptibility (i.e., being at higher or lower risk of developing COVID-19) and the level of transmission (i.e., some individuals may experience more virulent replication of SARS-CoV-2 and are more likely to infect others).
- Dynamic causal modelling
analytical method that exploits Bayesian model comparisons (a model selection technique based on variational Bayes) to best explain empirical timeseries, enabling tests of various hypotheses. Dynamic causal modelling was originally developed for the analysis of neuroimaging timeseries, but could be used to test many types of quantitative hypothesis, including epidemiological hypotheses.
- Dysbiosis
state in which the homeostatic integrity of a host-associated microbial community is impaired. Gut dysbiosis can entail changes in microbial composition, diversity, abundance, or function.
- Germ-free
biological state entailing complete microbial exclusion. Germ-free animals are born and raised under strict laboratory conditions that ensure sterility. Given that germ-free animals do not experience microbial colonisation, comparisons between germ-free animals and conventional (wild-type) animals enable assessments of causal gut microbial contributions to host physiological development and function.
- Gnotobiotic
a state in which all microbial exposures are known. This term encompasses germ-free animals and formerly germ-free animals that have been intentionally colonised with specific microbial taxa or predetermined microbial communities.
- Immunological dark matter
term intended to convey the existence of latent causes underlying clinical and epidemiological data, such as infection rates, notification rates, recovery rates, and fatality rates. These latent causes are hidden or ‘dark’ in that they must be inferred through modelling, rather than by direct observation. Key examples include heterogeneity in exposure, transmission (i.e., overdispersion), and susceptibility within a population.
- Lipopolysaccharide (LPS)
canonical proinflammatory compound derived from the outer membrane of Gram-negative bacteria.
- Microbiome
total collection of microbes, microbial genes, and microbial products inhabiting a particular environment or body site. Human-associated microbiomes differ across populations, individuals, and body sites within a given individual, and over time for a given individual body site.
- Microbiota
community of microbes, including bacteria, archaea, viruses, and eukaryotes (e.g., protists and fungi), inhabiting a particular environment or body site. The gastrointestinal tract is home to the largest, densest, and most diverse human-associated microbiota, with the large intestine alone hosting trillions of organisms.
References
- 1.Apicella M., et al. COVID-19 in people with diabetes: understanding the reasons for worse outcomes. Lancet Diabetes Endocrinol. 2020;8:782–792. doi: 10.1016/S2213-8587(20)30238-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williamson E.J., et al. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friston K.J., et al. Dynamic causal modelling. NeuroImage. 2003;19:1273–1302. doi: 10.1016/s1053-8119(03)00202-7. [DOI] [PubMed] [Google Scholar]
- 4.Lloyd-Smith J.O., et al. Superspreading and the effect of individual variation on disease emergence. Nature. 2005;438:355–359. doi: 10.1038/nature04153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Endo A., et al. Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. Wellcome Open Res. 2020;5:67. doi: 10.12688/wellcomeopenres.15842.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friston K., et al. ‘Dark matter’, second waves and epidemiological modelling. BMJ Glob. Health. 2020;5 doi: 10.1136/bmjgh-2020-003978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gomes M.G.M., et al. Individual variation in susceptibility or exposure to SARS-CoV-2 lowers the herd immunity threshold. medRxiv. 2020 doi: 10.1101/2020.04.27.20081893. Published online May 24, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parr T., et al. Dynamic causal modelling of immune heterogeneity. Sci. Rep. 2021;11:11400. doi: 10.1038/s41598-021-91011-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lourenço J., et al. The impact of host resistance on cumulative mortality and the threshold of herd immunity for SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.07.15.20154294. Published online October 1 2020. [DOI] [Google Scholar]
- 10.Velazquez E.M., et al. Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat. Microbiol. 2019;4:1057–1064. doi: 10.1038/s41564-019-0407-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villarino N.F., et al. Composition of the gut microbiota modulates the severity of malaria. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2235–2240. doi: 10.1073/pnas.1504887113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Finlay B.B., et al. The hygiene hypothesis, the COVID pandemic, and consequences for the human microbiome. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2010217118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooper L.V., et al. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung H., et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buffie C.G., Pamer E.G. Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganal S.C., et al. Priming of natural killer cells by nonmucosal mononuclear phagocytes requires instructive signals from commensal microbiota. Immunity. 2012;37:171–186. doi: 10.1016/j.immuni.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 18.Abt M.C., et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Budden K.F., et al. Emerging pathogenic links between microbiota and the gut–lung axis. Nat. Rev. Microbiol. 2017;15:55–63. doi: 10.1038/nrmicro.2016.142. [DOI] [PubMed] [Google Scholar]
- 20.Marsland B.J., et al. The gut-lung axis in respiratory disease. Ann. Am. Thorac. Soc. 2015;12:S150–S156. doi: 10.1513/AnnalsATS.201503-133AW. [DOI] [PubMed] [Google Scholar]
- 21.Trompette A., et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 22.Harding J.N., et al. Altered gut microbiota in infants is associated with respiratory syncytial virus disease severity. BMC Microbiol. 2020;20:140. doi: 10.1186/s12866-020-01816-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yildiz S., et al. Influenza A virus infection impacts systemic microbiota dynamics and causes quantitative enteric dysbiosis. Microbiome. 2018;6:9. doi: 10.1186/s40168-017-0386-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sencio V., et al. Gut dysbiosis during influenza contributes to pulmonary pneumococcal superinfection through altered short-chain fatty acid production. Cell Rep. 2020;30:2934–2947. doi: 10.1016/j.celrep.2020.02.013. [DOI] [PubMed] [Google Scholar]
- 25.Fanos V., et al. Lung microbiota in the acute respiratory disease: from coronavirus to metabolomics. J. Pediatr. Neonatal Individ. Med. 2020;9 [Google Scholar]
- 26.Tian X., et al. Elevated gut microbiome-derived propionate levels are associated with reduced sterile lung inflammation and bacterial immunity in mice. Front. Microbiol. 2019;10:159. doi: 10.3389/fmicb.2019.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang A.T., Marsland B.J. Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 2019;12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walls A.C., et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shang J., et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamming I., et al. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qi F., et al. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burgueño J.F., et al. Expression of SARS-CoV-2 entry molecules ACE2 and TMPRSS2 in the gut of patients with IBD. Inflamm. Bowel Dis. 2020;26:797–808. doi: 10.1093/ibd/izaa085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zang R., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Effenberger M., et al. Faecal calprotectin indicates intestinal inflammation in COVID-19. Gut. 2020;69:1543–1544. doi: 10.1136/gutjnl-2020-321388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y., et al. COVID-19 and gastrointestinal symptoms. Br. J. Surg. 2020;107:e382–e383. doi: 10.1002/bjs.11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto S., et al. The human microbiome and COVID-19: a systematic review. PLoS ONE. 2021;16 doi: 10.1371/journal.pone.0253293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang H., et al. Metatranscriptomic characterization of Coronavirus Disease 2019 identified a host transcriptional classifier associated with immune signaling. Clin. Infect. Dis. 2021;73:376–385. doi: 10.1093/cid/ciaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han Y., et al. The active lung microbiota landscape of COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.08.20.20144014. Published online August 23, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fan J., et al. The lung tissue microbiota features of 20 deceased patients with COVID-19. J. Infect. 2020;81:e64–e67. doi: 10.1016/j.jinf.2020.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen Z., et al. Genomic diversity of Severe Acute Respiratory Syndrome–Coronavirus 2 in patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2020;71:713–720. doi: 10.1093/cid/ciaa203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu S., et al. Alterations of the gut microbiota in patients with Coronavirus Disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020;71:2669–2678. doi: 10.1093/cid/ciaa709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuo T., et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159:944–955. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cao J., et al. Integrated gut virome and bacteriome dynamics in COVID-19 patients. Gut Microbes. 2021;13:1887722. doi: 10.1080/19490976.2021.1887722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., et al. Six-month follow-up of gut microbiota richness in patients with COVID-19. Gut. 2021 doi: 10.1136/gutjnl-2021-324090. Published online April 8, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeoh Y.K., et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gou W., et al. Gut microbiota, inflammation, and molecular signatures of host response to infection. J. Genet. Genomics. 2021;48:792–802. doi: 10.1016/j.jgg.2021.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Geirnaert A., et al. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 2017;7:11450. doi: 10.1038/s41598-017-11734-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martino C., et al. Bacterial modification of the host glycosaminoglycan heparan sulfate modulates SARS-CoV-2 infectivity. bioRxiv. 2020 doi: 10.1101/2020.08.17.238444. Published online August 18, 2020. [DOI] [Google Scholar]
- 50.Haeger S.M., et al. Heparan sulfate in the developing, healthy, and injured lung. Am. J. Respir. Cell Mol. Biol. 2016;55:5–11. doi: 10.1165/rcmb.2016-0043TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rydyznski Moderbacher C., et al. Antigen-specific adaptive immunity to SARS-CoV-2 in acute COVID-19 and associations with age and disease severity. Cell. 2020;183:996–1012. doi: 10.1016/j.cell.2020.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tan A.T., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo X., et al. Long-term persistence of IgG antibodies in SARS-CoV infected healthcare workers. medRxiv. 2020 doi: 10.1101/2020.02.12.20021386. Published online February 14, 2020. [DOI] [Google Scholar]
- 55.Li C.K., et al. T cell responses to whole SARS coronavirus in humans. J. Immunol. 2008;181:5490–5500. doi: 10.4049/jimmunol.181.8.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarke A., et al. Impact of SARS-CoV-2 variants on the total CD4+ and CD8+ T cell reactivity in infected or vaccinated individuals. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chattopadhyay I., Shankar E.M. SARS-CoV-2-indigenous microbiota nexus: does gut microbiota contribute to inflammation and disease severity in COVID-19? Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.590874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCoy K.D., et al. The microbiome and immune memory formation. Immunol. Cell Biol. 2019;97:625–635. doi: 10.1111/imcb.12273. [DOI] [PubMed] [Google Scholar]
- 59.Saeidi A., et al. Functional role of mucosal-associated invariant T cells in HIV infection. J. Leukoc. Biol. 2016;100:305–314. doi: 10.1189/jlb.4RU0216-084R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Constantinides M.G., et al. MAIT cells are imprinted by the microbiota in early life and promote tissue repair. Science. 2019;366 doi: 10.1126/science.aax6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jouan Y., et al. Phenotypical and functional alteration of unconventional T cells in severe COVID-19 patients. J. Exp. Med. 2020;217 doi: 10.1084/jem.20200872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parrot T., et al. MAIT cell activation and dynamics associated with COVID-19 disease severity. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abe1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rosser E.C., et al. Regulatory B cells are induced by gut microbiota–driven interleukin-1β and interleukin-6 production. Nat. Med. 2014;20:1334–1339. doi: 10.1038/nm.3680. [DOI] [PubMed] [Google Scholar]
- 64.Chen T., et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: retrospective study. BMJ. 2020;368 doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huggins M.A., et al. Microbial exposure enhances immunity to pathogens recognized by TLR2 but increases susceptibility to cytokine storm through TLR4 sensitization. Cell Rep. 2019;28:1729–1743. doi: 10.1016/j.celrep.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosshart S.P., et al. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 2019;365 doi: 10.1126/science.aaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Foster K.R., et al. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McFall-Ngai M. Care for the community. Nature. 2007;445:153. doi: 10.1038/445153a. [DOI] [PubMed] [Google Scholar]
- 69.Gomez de Aguero M., et al. The maternal microbiota drives early postnatal innate immune development. Science. 2016;351:1296–1302. doi: 10.1126/science.aad2571. [DOI] [PubMed] [Google Scholar]
- 70.Thaiss C.A., et al. The microbiome and innate immunity. Nature. 2016;535:65–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 71.Schluter J., et al. The gut microbiota is associated with immune cell dynamics in humans. Nature. 2020;588:303–307. doi: 10.1038/s41586-020-2971-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dehner C., et al. The microbiome in systemic autoimmune disease: mechanistic insights from recent studies. Curr. Opin. Rheumatol. 2019;31:201–207. doi: 10.1097/BOR.0000000000000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim H.S. Do an altered gut microbiota and an associated leaky gut affect COVID-19 severity? mBio. 2021;12 doi: 10.1128/mBio.03022-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cani P.D., et al. Microbial regulation of organismal energy homeostasis. Nat. Metab. 2019;1:34–46. doi: 10.1038/s42255-018-0017-4. [DOI] [PubMed] [Google Scholar]
- 75.Santos-Oliveira J.R., et al. Evidence that lipopolysaccharide may contribute to the cytokine storm and cellular activation in patients with visceral leishmaniasis. PLoS Negl. Trop. Dis. 2011;5 doi: 10.1371/journal.pntd.0001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tilg H., Moschen A.R. Microbiota and diabetes: an evolving relationship. Gut. 2014;63:1513–1521. doi: 10.1136/gutjnl-2014-306928. [DOI] [PubMed] [Google Scholar]
- 77.Jaiswal M., et al. Nitric oxide in gastrointestinal epithelial cell carcinogenesis: linking inflammation to oncogenesis. Am. J. Physiol.-Gastrointest. Liver Physiol. 2001;281:G626–G634. doi: 10.1152/ajpgi.2001.281.3.G626. [DOI] [PubMed] [Google Scholar]
- 78.Ulisse S., et al. Expression of cytokines, inducible nitric oxide synthase, and matrix metalloproteinases in pouchitis: effects of probiotic treatment. Am. J. Gastroenterol. 2001;96:2691–2699. doi: 10.1111/j.1572-0241.2001.04139.x. [DOI] [PubMed] [Google Scholar]
- 79.Kumar N.P., et al. Systemic inflammation and microbial translocation are characteristic features of SARS-CoV-2-related multisystem inflammatory syndrome in children. Open Forum Infect. Dis. 2021;8 doi: 10.1093/ofid/ofab279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Petruk G., et al. SARS-CoV-2 spike protein binds to bacterial lipopolysaccharide and boosts proinflammatory activity. J. Mol. Cell Biol. 2021;12:916–932. doi: 10.1093/jmcb/mjaa067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vignesh R., et al. Could perturbation of gut microbiota possibly exacerbate the severity of COVID-19 via cytokine storm? Front. Immunol. 2021;11:607734. doi: 10.3389/fimmu.2020.607734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gu J., Korteweg C. Pathology and pathogenesis of severe acute respiratory syndrome. Am. J. Pathol. 2007;170:1136–1147. doi: 10.2353/ajpath.2007.061088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cani P.D., et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 84.Vijay-Kumar M., et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiang R-D., et al. Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell. 2020;182:50–58. doi: 10.1016/j.cell.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bao L., et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature. 2020;583:830–833. doi: 10.1038/s41586-020-2312-y. [DOI] [PubMed] [Google Scholar]
- 89.Hassan A.O., et al. A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell. 2020;182:744–753. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun J., et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182:734–743. doi: 10.1016/j.cell.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dinnon K.H., et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sokol H., et al. SARS-CoV-2 infection in nonhuman primates alters the composition and functional activity of the gut microbiota. Gut Microbes. 2021;13:1–19. doi: 10.1080/19490976.2021.1893113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Friston K.J., et al. How vaccination and contact isolation might interact to suppress transmission of Covid-19: a DCM study. medRxiv. 2021 doi: 10.1101/2021.01.03.20248972. Published online January 4, 2021. [DOI] [Google Scholar]
- 94.Friston K.J., et al. Dynamic causal modelling of COVID-19. Wellcome Open Res. 2020;5:89. doi: 10.12688/wellcomeopenres.15881.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Britton T., et al. A mathematical model reveals the influence of population heterogeneity on herd immunity to SARS-CoV-2. Science. 2020;369:846–849. doi: 10.1126/science.abc6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Long Q-X., et al. Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 2020;26:1200–1204. doi: 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 97.van Kampen J.J.A., et al. Duration and key determinants of infectious virus shedding in hospitalized patients with coronavirus disease-2019 (COVID-19) Nat. Commun. 2021;12:267. doi: 10.1038/s41467-020-20568-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wölfel R., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 99.Bunyavanich S., et al. Nasal gene expression of angiotensin-converting enzyme 2 in children and adults. JAMA. 2020;323:2427. doi: 10.1001/jama.2020.8707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Le Bert N., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 101.Seo G-Y., et al. The role of innate lymphoid cells in response to microbes at mucosal surfaces. Mucosal Immunol. 2020;13:399–412. doi: 10.1038/s41385-020-0265-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ng K.W., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370:1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Razdan K., et al. Vitamin D levels and COVID-19 susceptibility: is there any correlation? Med. Drug Discov. 2020;7 doi: 10.1016/j.medidd.2020.100051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jha A.R., et al. Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Smits S.A., et al. Seasonal cycling in the gut microbiome of the Hadza hunter-gatherers of Tanzania. Science. 2017;357:802–806. doi: 10.1126/science.aan4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vangay P., et al. US immigration westernizes the human gut microbiome. Cell. 2018;175:962–972. doi: 10.1016/j.cell.2018.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Muñoz-Fontela C., et al. Animal models for COVID-19. Nature. 2020;586:509–515. doi: 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sarkar A., et al. The role of the microbiome in the neurobiology of social behaviour. Biol. Rev. 2020;95:1131–1166. doi: 10.1111/brv.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hanage W.P. Microbiology: microbiome science needs a healthy dose of scepticism. Nature. 2014;512:247–248. doi: 10.1038/512247a. [DOI] [PubMed] [Google Scholar]
- 110.Walter J., et al. Establishing or exaggerating causality for the gut microbiome: lessons from human microbiota-associated rodents. Cell. 2020;180:221–232. doi: 10.1016/j.cell.2019.12.025. [DOI] [PubMed] [Google Scholar]
- 111.Carmody R.N., et al. Cooking shapes the structure and function of the gut microbiome. Nat. Microbiol. 2019;4:2052–2063. doi: 10.1038/s41564-019-0569-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.David L.A., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wilkinson J.E., et al. A framework for microbiome science in public health. Nat. Med. 2021;27:766–774. doi: 10.1038/s41591-021-01258-0. [DOI] [PubMed] [Google Scholar]
- 114.Davies N.G., et al. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat. Med. 2020;26:1205–1211. doi: 10.1038/s41591-020-0962-9. [DOI] [PubMed] [Google Scholar]
- 115.Jordan R.E., et al. Covid-19: risk factors for severe disease and death. BMJ. 2020;368 doi: 10.1136/bmj.m1198. [DOI] [PubMed] [Google Scholar]
- 116.Laxminarayan R., et al. Epidemiology and transmission dynamics of COVID-19 in two Indian states. Science. 2020;370:691–697. doi: 10.1126/science.abd7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu K., et al. Clinical features of COVID-19 in elderly patients: a comparison with young and middle-aged patients. J. Infect. 2020;80:e14–e18. doi: 10.1016/j.jinf.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou F., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jašarević E., et al. Sex differences in the gut microbiome–brain axis across the lifespan. Philos. Trans. R. Soc. B Biol. Sci. 2016;371:20150122. doi: 10.1098/rstb.2015.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kundu P., et al. Our gut microbiome: the evolving inner self. Cell. 2017;171:1481–1493. doi: 10.1016/j.cell.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 121.Yatsunenko T., et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Biagi E., et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Claesson M.J., et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zwielehner J., et al. Combined PCR-DGGE fingerprinting and quantitative-PCR indicates shifts in fecal population sizes and diversity of Bacteroides, bifidobacteria and Clostridium cluster IV in institutionalized elderly. Exp. Gerontol. 2009;44:440–446. doi: 10.1016/j.exger.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 125.Fransen F., et al. Aged gut microbiota contributes to systemical inflammaging after transfer to germ-free mice. Front. Immunol. 2017;8:1385. doi: 10.3389/fimmu.2017.01385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Takahashi T., et al. Sex differences in immune responses that underlie COVID-19 disease outcomes. Nature. 2020;588:315–320. doi: 10.1038/s41586-020-2700-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Scully E.P., et al. Considering how biological sex impacts immune responses and COVID-19 outcomes. Nat. Rev. Immunol. 2020;20:442–447. doi: 10.1038/s41577-020-0348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bliddal S., et al. Acute and persistent symptoms in non-hospitalized PCR-confirmed COVID-19 patients. Sci. Rep. 2021;11:13153. doi: 10.1038/s41598-021-92045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Peghin M., et al. Post-COVID-19 symptoms 6 months after acute infection among hospitalized and non-hospitalized patients. Clin. Microbiol. Infect. 2021;27:1507–1513. doi: 10.1016/j.cmi.2021.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sudre C.H., et al. Attributes and predictors of long COVID. Nat. Med. 2021;27:626–631. doi: 10.1038/s41591-021-01292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Flanagan K.L., et al. Sex and gender differences in the outcomes of vaccination over the life course. Annu. Rev. Cell Dev. Biol. 2017;33:577–599. doi: 10.1146/annurev-cellbio-100616-060718. [DOI] [PubMed] [Google Scholar]
- 132.Demonbreun A.R., et al. COVID-19 mRNA vaccination generates greater immunoglobulin G levels in women compared to men. J. Infect. Dis. 2021;224:793–797. doi: 10.1093/infdis/jiab314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 134.Deng Q., et al. Targeting androgen regulation of TMPRSS2 and ACE2 as a therapeutic strategy to combat COVID-19. iScience. 2021;24:102254. doi: 10.1016/j.isci.2021.102254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Montopoli M., et al. Androgen-deprivation therapies for prostate cancer and risk of infection by SARS-CoV-2: a population-based study (N = 4532) Ann. Oncol. 2020;31:1040–1045. doi: 10.1016/j.annonc.2020.04.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Samuel R.M., et al. Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. Cell Stem Cell. 2020;27:876–889. doi: 10.1016/j.stem.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stopsack K.H., et al. TMPRSS2 and COVID-19: serendipity or opportunity for intervention? Cancer Discov. 2020;10:779–782. doi: 10.1158/2159-8290.CD-20-0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Qiao Y., et al. Targeting transcriptional regulation of SARS-CoV-2 entry factors ACE2 and TMPRSS2. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2021450118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bukowska A., et al. Protective regulation of the ACE2/ACE gene expression by estrogen in human atrial tissue from elderly men. Exp. Biol. Med. 2017;242:1412–1423. doi: 10.1177/1535370217718808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu J., et al. Sex differences in renal angiotensin converting enzyme 2 (ACE2) activity are 17β-oestradiol-dependent and sex chromosome-independent. Biol. Sex Differ. 2010;1:6. doi: 10.1186/2042-6410-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stelzig K.E., et al. Estrogen regulates the expression of SARS-CoV-2 receptor ACE2 in differentiated airway epithelial cells. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2020;318:L1280–L1281. doi: 10.1152/ajplung.00153.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bereshchenko O., et al. Glucocorticoids, sex hormones, and immunity. Front. Immunol. 2018;9:1332. doi: 10.3389/fimmu.2018.01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.He M., et al. Host gender and androgen levels regulate gut bacterial taxa in pigs leading to sex-biased serum metabolite profiles. Front. Microbiol. 2019;10:1359. doi: 10.3389/fmicb.2019.01359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kaliannan K., et al. Estrogen-mediated gut microbiome alterations influence sexual dimorphism in metabolic syndrome in mice. Microbiome. 2018;6:205. doi: 10.1186/s40168-018-0587-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Markle J.G.M., et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 146.Org E., et al. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016;7:313–322. doi: 10.1080/19490976.2016.1203502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xiao L., et al. A reference gene catalogue of the pig gut microbiome. Nat. Microbiol. 2016;1:16161. doi: 10.1038/nmicrobiol.2016.161. [DOI] [PubMed] [Google Scholar]
- 148.Yurkovetskiy L., et al. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang X., et al. Sex- and age-related trajectories of the adult human gut microbiota shared across populations of different ethnicities. Nat. Aging. 2021;1:87–100. doi: 10.1038/s43587-020-00014-2. [DOI] [PubMed] [Google Scholar]
- 150.Jaggar M., et al. You’ve got male: sex and the microbiota-gut-brain axis across the lifespan. Front. Neuroendocrinol. 2020;56:100815. doi: 10.1016/j.yfrne.2019.100815. [DOI] [PubMed] [Google Scholar]
- 151.Dabek M., et al. Distribution of β-glucosidase and β-glucuronidase activity and of β-glucuronidase gene gus in human colonic bacteria: β-glycosidase activity in human gut bacteria. FEMS Microbiol. Ecol. 2008;66:487–495. doi: 10.1111/j.1574-6941.2008.00520.x. [DOI] [PubMed] [Google Scholar]
- 152.Kwa M., et al. The intestinal microbiome and estrogen receptor-positive female breast cancer. J. Natl. Cancer Inst. 2016;108:djw029. doi: 10.1093/jnci/djw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Plottel C.S., Blaser M.J. Microbiome and malignancy. Cell Host Microbe. 2011;10:324–335. doi: 10.1016/j.chom.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Adlercreutz H., et al. Effect of ampicillin administration on the excretion of twelve oestrogens in pregnancy urine. Acta Endocrinol. 1975;80:551–557. doi: 10.1530/acta.0.0800551. [DOI] [PubMed] [Google Scholar]
- 155.Hamalainen E., et al. Effect of oxytetracycline administration on intestinal metabolism of oestrogens and on plasma sex hormones in healthy men. Gut. 1987;28:439–445. doi: 10.1136/gut.28.4.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Martin F., et al. Excretion of progesterone metabolites and estriol in faeces from pregnant women during ampicillin administration. J. Steroid Biochem. 1975;6:1339–1346. doi: 10.1016/0022-4731(75)90363-5. [DOI] [PubMed] [Google Scholar]
- 157.Bess E.N., et al. Genetic basis for the cooperative bioactivation of plant lignans by Eggerthella lenta and other human gut bacteria. Nat. Microbiol. 2020;5:56–66. doi: 10.1038/s41564-019-0596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mabrok H.B., et al. Lignan transformation by gut bacteria lowers tumor burden in a gnotobiotic rat model of breast cancer. Carcinogenesis. 2012;33:203–208. doi: 10.1093/carcin/bgr256. [DOI] [PubMed] [Google Scholar]
- 159.The Severe Covid-19 GWAS Group Genomewide association study of severe Covid-19 with respiratory failure. N. Engl. J. Med. 2020;383:1522–1534. doi: 10.1056/NEJMoa2020283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.COVID-19 Host Genetics Initiative Mapping the human genetic architecture of COVID-19. Nature. 2021 doi: 10.1038/s41586-021-03767-x. Published online July 8, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zeberg H., Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587:610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- 162.Hall A.B., et al. Human genetic variation and the gut microbiome in disease. Nat. Rev. Genet. 2017;18:690–699. doi: 10.1038/nrg.2017.63. [DOI] [PubMed] [Google Scholar]
- 163.Goodrich J.K., et al. Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Goodrich J.K., et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xie H., et al. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst. 2016;3:572–584. doi: 10.1016/j.cels.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang J., et al. Genome-wide association analysis identifies variation in vitamin D receptor and other host factors influencing the gut microbiota. Nat. Genet. 2016;48:1396–1406. doi: 10.1038/ng.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Turpin W., et al. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat. Genet. 2016;48:1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- 168.Bonder M.J., et al. The effect of host genetics on the gut microbiome. Nat. Genet. 2016;48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 169.Blekhman R., et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kolde R., et al. Host genetic variation and its microbiome interactions within the Human Microbiome Project. Genome Med. 2018;10:6. doi: 10.1186/s13073-018-0515-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Grieneisen L., et al. Gut microbiome heritability is nearly universal but environmentally contingent. Science. 2021;373:181–186. doi: 10.1126/science.aba5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rothschild D., et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 173.Rühlemann M.C., et al. Genome-wide association study in 8,956 German individuals identifies influence of ABO histo-blood groups on gut microbiome. Nat. Genet. 2021;53:147–155. doi: 10.1038/s41588-020-00747-1. [DOI] [PubMed] [Google Scholar]
- 174.Stowell S.R., et al. Innate immune lectins kill bacteria expressing blood group antigen. Nat. Med. 2010;16:295–301. doi: 10.1038/nm.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Arnolds K.L., et al. Blood type and the microbiome- untangling a complex relationship with lessons from pathogens. Curr. Opin. Microbiol. 2020;56:59–66. doi: 10.1016/j.mib.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ray J.G., et al. Association between ABO and Rh blood groups and SARS-CoV-2 infection or severe COVID-19 illness: a population-based cohort study. Ann. Intern. Med. 2021;174:308–315. doi: 10.7326/M20-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Langford B.J., et al. Antibiotic prescribing in patients with COVID-19: rapid review and meta-analysis. Clin. Microbiol. Infect. 2021;27:520–531. doi: 10.1016/j.cmi.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Zimmermann M., et al. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570:462–467. doi: 10.1038/s41586-019-1291-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Spanogiannopoulos P., et al. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 2016;14:273–287. doi: 10.1038/nrmicro.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xu R., et al. Progressive worsening of the respiratory and gut microbiome in children during the first two months of COVID-19. medRxiv. 2020 doi: 10.1101/2020.07.13.20152181. Published online July 17, 2020. [DOI] [Google Scholar]
- 181.de Castilhos J., et al. COVID-19 severity and complications associated with low diversity, dysbiosis and predictive metagenome features of the oropharyngeal microbiome. Res. Sq. 2021 doi: 10.21203/rs.3.rs-127621/v1. Published online January 6, 2021. [DOI] [Google Scholar]
- 182.Ren L., et al. Dynamics of the upper respiratory tract microbiota and its association with mortality in COVID-19. Am. J. Respir. Crit. Care Med. 2021 doi: 10.1164/rccm.202103-0814OC. Published online September 17, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Li N., et al. The commensal microbiota and viral infection: a comprehensive review. Front. Immunol. 2019;10:1551. doi: 10.3389/fimmu.2019.01551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Sarkar A., et al. The microbiome in psychology and cognitive neuroscience. Trends Cogn. Sci. 2018;22:611–636. doi: 10.1016/j.tics.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 185.Depommier C., et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 2019;25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Everard A. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Carmody R.N., et al. Gut microbiota through an evolutionary lens. Science. 2021;372:462–463. doi: 10.1126/science.abf0590. [DOI] [PubMed] [Google Scholar]
- 188.Peckham H. Male sex identified by global COVID-19 meta-analysis as a risk factor for death and ITU admission. Nat. Commun. 2020;11:6317. doi: 10.1038/s41467-020-19741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]