Abstract
This review evaluates the pediatric evidence for pharmacogenetic associations for drugs that are commonly prescribed by or encountered by pediatric clinicians across multiple subspecialties, organized from most to least pediatric evidence. We begin with the pharmacogenetic research that led to warning of increased risk of death in certain pediatric populations (“ultrarapid metabolizers”) who are prescribed codeine after tonsillectomy or adenoidectomy. We review the evidence for genetic testing for thiopurine metabolism, which has become routine in multiple pediatric subspecialties. We discuss the pharmacogenetic research in proton pump inhibitors, for which clinical guidelines have recently been made available. With an increase in the prevalence of behavioral health disorders including attention deficit hyperactivity disorder (ADHD), we review the pharmacogenetic literature on selective serotonin reuptake inhibitors, selective -norepinephrine reuptake inhibitors, and ADHD medications. We will conclude this section on the current pharmacogenetic data on ondansetron.
We also provide our perspective on how to integrate the current research on pharmacogenetics into clinical care and what further research is needed. We discuss how institutions are managing pharmacogenetic test results and implementing them clinically, and how the electronic health record can be leveraged to ensure testing results are available and taken into consideration when prescribing medications.
Introduction
Pharmacogenetics is an emerging component of precision medicine. Many medications prescribed by pediatricians and pediatric subspecialists are influenced by pharmacogenetic variants. Yet, few pediatricians have training to incorporate pharmacogenetic results into practice. This review will provide an overview of pharmacogenetics, review pharmacogenetic research for exemplary gene-drug pairs, discuss clinical implementation, and provide a general perspective for the field.
Definition of Key Terms and Resources
Discussion of pharmacogenetic literature is facilitated by knowledge of key terms and resources (Table 1) (1–5). While the terms pharmacogenetics and pharmacogenomics are often used interchangeably, pharmacogenetics is the study of single genes and their effect on drug response, and pharmacogenomics is focused on how the entire genome influences drug response. Some pharmacogenes affect drug absorption, distribution, metabolism or excretion (“what the body does to the drug,” or pharmacokinetics). Others influence the therapeutic effects or risk of adverse drug events (ADEs) (“what the drug does to the body,” or pharmacodynamics). Precision medicine incorporates pharmacogenetic, clinical, environmental, and lifestyle factors into prescribing decisions.
Table 1:
Definitions of Key Terms and Resources in the Field of Pharmacogenetics
| Pharmacogenetics | Study of individual or a few genes and their effect on the variability of drug responses |
| Pharmacogenomics | Field that combines pharmacology and genomics to study how all genes in the genome affect the drug response |
| Pharmacokinetics | What the body does to the drug, including absorption, distribution, metabolism and elimination |
| Adverse drug event (ADE) | Harm caused by the use of a drug |
| Pharmacodynamics | What the drug does to the body, including therapeutic response and adverse effects |
| Precision Medicine | The provision of treatments and preventions based on an individual’s genetic, environmental and lifestyle factors |
| Alleles/Gene Variants | Forms of the same gene with small differences in the DNA sequence/Alterations of the most common DNA sequence of a gene |
| Metabolizing Enzymes | Enzymes that are responsible for the breakdown of molecules and chemicals, including drugs, to metabolites. During the process of metabolism, drugs can be activated (prodrug → active drug) or fully or partially inactivated (active drug → partially active or inactive metabolites). |
| Cytochrome P450 Genes and Enzymes | Enzymes encoded by the cytochrome P450 genes and expressed primarily in the liver are involved in the synthesis and metabolism of various molecules and chemicals, including drugs. Each cytochrome gene is named with CYP, followed by the gene’s subgroup and a number of the gene within the subgroup (e.g. CYP2D6). |
| Star (*) Alleles | Combinations of genetic variants (haplotypes) in drug metabolism genes are designated using “star nomenclature”. Star alleles are reported to cause no, decreased, normal, or increased function of the enzyme. |
| Gene-drug pair/interaction | Change in the effect of a drug due to differences in a gene |
| Metabolizer Phenotypes | Description of clinical phenotypes based on the combined impact of both alleles Normal metabolizer: Fully functional metabolizing enzyme activity Rapid metabolizer: Increased enzyme activity compared to normal metabolizers but less than ultrarapid metabolizers Ultrarapid metabolizers: Increased enzyme activity compared to rapid metabolizers Intermediate metabolizer: Decreased enzyme activity compared to normal metabolizer but more than poor metabolizers Poor metabolizer: Little to no enzyme activity |
| Drug-Drug Interaction (DDI) | Change in the effect of a drug due to the presence of another drug in the body |
| PharmVar (Pharmacogene Variation Consortium) | Repository for pharmacogene allelic variation to facilitate the interpretation of pharmacogenetic test results for precision medicine |
| Pharmacogenomics Knowledge Base (PharmGKB) | Resource sponsored by the National Institutes of Health that curates information on human genetic variation and drug responses |
| Clinical Pharmacogenetics Implementation Consortium (CPIC) | International group interested in facilitating the clinical use of pharmacogenetics testing through the creation of peer-reviewed and evidence-based gene/drug clinical practice guidelines |
Individual differences in drug metabolism can be partially attributed to variants (alterations of the DNA sequence) in genes that code for metabolizing enzymes responsible for drug breakdown. Cytochrome P450 (CYP450) is a superfamily of enzymes expressed in the liver, intestinal tissue, and elsewhere that are involved in drug metabolism. Alleles (versions of the same gene with one or more variants) of genes coding for CYP450 and other metabolizing enzymes are identified using the “star (*) nomenclature,” where *1 is the reference sequence to which all alleles are compared and generally the functional enzyme (6). The activity of each allele is assessed and determined based on in vitro activity and in vivo evidence for associated drugs (gene-drug pair/interaction) (4) (Table 2). The functional phenotype or metabolizer status resulting from the combined effect of both alleles is categorized using standard nomenclature: poor, intermediate, normal, rapid, and ultrarapid. This results in a spectrum of possible enzyme activity ranging from no function in poor metabolizers to increased function in ultrarapid metabolizers. Medications may be administered as inactive prodrugs or active drugs. Prodrugs are activated by the process of metabolism as they are transformed into active molecules. In contrast, active drugs can be inactivated by metabolism by being broken down into partially active or inactive metabolites (Figure 1A). Knowing whether a prescribed medication is activated or inactivated by metabolism influences the clinical effects of altered enzyme function, and the risk of ADEs or drug-drug interactions (Figure 1B).
Table 2:
Example drug-gene pairs for medications prescribed to children with significant evidence for recommendations in prescribing actions as defined by the Clinical Pharmacogenetics Implementation Consortium
| Drug | Gene |
|---|---|
| allopurinol | HLA-B |
| aminoglycoside antibacterials | MT-RNR1 |
| amitriptyline | CYP2C19 |
| CYP2D6 | |
| atomoxetine | CYP2D6 |
| azathioprine | TPMT |
| NUDT15 | |
| carbamazepine | HLA-A |
| HLA-B | |
| citalopram | CYP2C19 |
| clopidogrel | CYP2C19 |
| codeine | CYP2D6 |
| escitalopram | CYP2C19 |
| fluvoxamine | CYP2D6 |
| ibuprofen | CYP2C9 |
| lansoprazole | CYP2C19 |
| mercaptopurine | TPMT |
| NUDT15 | |
| nortriptyline | CYP2D6 |
| omeprazole | CYP2C19 |
| ondansetron | CYP2D6 |
| pantoprazole | CYP2C19 |
| paroxetine | CYP2D6 |
| phenytoin | HLA-B |
| CYP2C9 | |
| sertraline | CYP2C19 |
| simvastatin | SLCO1B1 |
| succinylcholine | RYR1 |
| CACNA1S | |
| tacrolimus | CYP3A5 |
| tramadol | CYP2D6 |
| voriconazole | CYP2C19 |
| warfarin | CYP2C9 |
| CYP4F2 | |
| VKORC1 |
Figure 1:
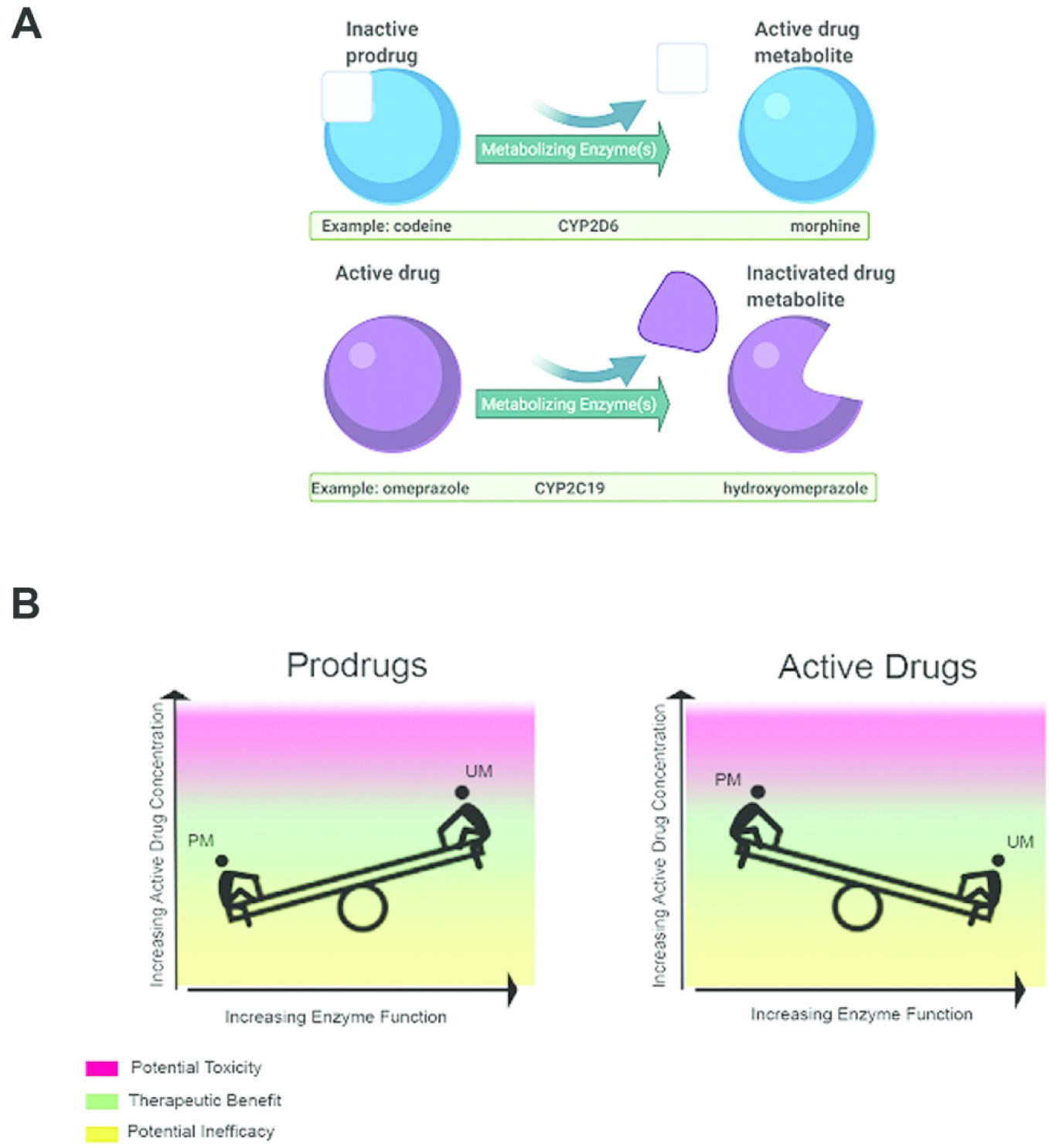
The relationship between prodrugs, active drugs and inactivated drug metabolites, and metabolizer status. (A) Inactive prodrugs (top) need to be metabolized into an active drug metabolite to have therapeutic effect. In contrast, when an active drug is administered, metabolism of the drug can result in an inactivated drug metabolite. (B) Since prodrugs (left) are metabolized to an active form of the drug to achieve therapeutic effect, poor metabolizers who have low enzyme function will have low active drug concentrations and are at risk for inefficacy. Ultrarapid metabolizers who have high enzyme function are at risk of high active drug concentrations and potential toxicity. Active drugs (right) are metabolized into inactive drugs that have minimal or no therapeutic effect. Poor metabolizers are therefore at risk of high concentrations of active drugs and thus potential toxicity, while ultrarapid metabolizers will have low concentrations of active drug and may not achieve therapeutic efficacy.
Pharmacogenetic knowledge is rapidly changing with new evidence being produced at exponential rates due to active research. PharmVar, the Pharmacogene Variation Consortium, and PharmGKB, resources supported by the National Institutes of Health, curate and centralize pharmacogenetic information to facilitate the interpretation of pharmacogenetic testing (2,7). The Clinical Pharmacogenetic Implementation Consortium (CPIC) produces evidence-based clinical guidelines on gene-drug pairs and interactions (5), including statements regarding how recommendations apply to pediatric patients. Finally, the Food and Drug Administration (FDA) has provided lists of pharmacogenetic associations for which data support management recommendations and how drug labels have incorporated the data (8,9).
Examples of Pharmacogenomic Research Supporting Clinical Implementation
Codeine and Morphine
One of the most well-known gene-drug interactions in children associated with life threatening ADEs and drug label changes is CYP2D6 and codeine. While the prodrug codeine has minimal analgesic effects, its most active metabolite, morphine, has a 200-fold greater affinity for opioid receptors, provides potent analgesic effects, and has the potential to cause somnolence, respiratory depression, and death (10). Low production of morphine from codeine may result in inadequate pain control.
Codeine is metabolized to morphine by CYP2D6, the enzyme encoded by the CYP2D6 gene. In vitro studies demonstrated variable morphine production by human liver tissue and purified enzymes based on CYP2D6 genotypes (11–15). Liver enzymes isolated from ultrarapid metabolizers have higher morphine production when compared to enzymes from normal metabolizers, while enzymes from poor metabolizers produce little morphine. In vivo, poor metabolizers (prevalence of 0.4–5.4% (16)) have lower morphine production, less analgesia, and fewer ADEs compared to normal and ultrarapid metabolizers (17–27). These findings have been confirmed in children (28).
Given the low potency, codeine was considered safe for outpatient use, but reports of severe ADEs in infants of breastfeeding mothers who were taking codeine raised concern. High morphine concentrations were found in symptomatic infants of breastfeeding mothers taking codeine (29,30). In most cases the high morphine concentration was due to the dose of codeine, though some were also identified as CYP2D6 ultrarapid metabolizers. The FDA advised caution when prescribing codeine to a breastfeeding mother who is an ultrarapid metabolizer (31). The prevalence of ultrarapid metabolizer status varies depending on the biogeographic population (32), but can be above 10% in certain groups including Oceanian, Ashkenazi Jewish, and Middle Eastern populations (16).
In 2009, a 2 year-old child with sleep apnea was reported to have died after taking codeine following an uncomplicated adenotonsillectomy (33). Postmortem analysis revealed high morphine levels and functional duplication of CYP2D6 leading to ultrarapid metabolizer phenotype. After review of ADEs reported in children taking codeine after tonsillectomy and/or adenoidectomy (T&A) and who had altered CYP2D6 metabolism (34,35), the FDA issued a new boxed warning stating codeine was contraindicated in children following a T&A (36). In 2014, CPIC guidelines strongly recommended avoidance of codeine in ultrarapid metabolizers due to potential for toxicity and in poor metabolizers due to lack of efficacy (36,37). While codeine prescriptions for children have decreased since the initial FDA warning in 2013, use has persisted in some pediatric practices (38,39).
After reviewing ADEs reported over a 45-year period in children under the age of 18, the FDA identified over 60 cases of severe breathing problems and 24 deaths following codeine administration (40). In 2017, the FDA updated codeine drug labels to strengthen their warnings, including a contraindication for codeine to treat pain or cough in children under 12 years of age. A new warning was also added, recommending against codeine use in adolescents ages 12 to 18 who have underlying breathing issues, such as obstructive sleep apnea, and in breastfeeding mothers.
Some have advocated for an exception to the FDA contraindication if pharmacogenetic testing is done, which could enable safe codeine use (41). By eliminating codeine as an option to treat pain, clinicians have resorted to alternative analgesics, including more potent opioids, with higher risk of serious ADEs and which also have gene-drug interactions. Patients with sickle cell disease often require opioids when they are in crises, and codeine was frequently used as it is the only Schedule III opioid analgesic available in the United States. Incorporation of CYP2D6 testing into clinical practice with a clinical decision support tool within the electronic health record (EHR) has been demonstrated to be a safe and effective way to prescribe codeine to children with sickle cell disease while preventing codeine use after T&A and in patients with CYP2D6 ultrarapid and poor metabolizer genotypes (42). Further research is required to identify other patient populations and indications in which the benefits of pharmacogenetic-guided codeine use outweighs the risks.
With the removal of codeine from many pediatric formularies, tramadol may be used more frequently for pain management. However, tramadol is also a substrate of CYP2D6 metabolism and requires transformation into an active metabolite to provide pain relief (43). Similar to codeine, poor metabolizers are at risk of inadequate pain control and ultrarapid metabolizers have higher risk of ADEs (44).
Thiopurines
Azathioprine (a prodrug of mercaptopurine), mercaptopurine, and thioguanine are thiopurine immunosuppressants prescribed for pediatric dermatologic, gastrointestinal, oncologic, and rheumatologic diseases. Their ADEs include myelosuppression, pancreatitis, and hepatotoxicity, and their use may carry an increased risk of lymphoma (45). All three drugs are considered prodrugs and metabolized to active thioguanine nucleotides. Thiopurines have significant interactions with two pharmacogenes, thiopurine methyltransferase (TPMT) and nudix (nucleoside diphopshate linked moiety X)-type motif 15 (NUDT15). Each of these genes strongly influences clinical response and development of ADEs (46). Preemptive (pre-prescription) genetic testing for one or both of these genes has become routine for some pediatric subspecialists.
The interactions between TPMT and mercaptopurine, to which azathioprine is converted, are complex (46,47). TPMT inactivates mercaptopurine through methylation, converting the drug to methylmercaptopurine base and thus decreasing the amount of parent drug available to produce active thioguanine nucleotides (TGNs). However, TPMT also acts on a secondary metabolite of mercaptopurine, thioinosine monophoshate (TIMP), and converts it to methyl-TIMP which has immunosuppressive effects and contributes to hepatotoxicity. In vitro studies have shown that mercaptopurine’s conversion to methylmercaptopurine was lower in cells and organ tissues isolated from intermediate metabolizers and absent in poor metabolizers, when compared to normal metabolizers (48–51). In poor metabolizers, proteosomal degradation of TPMT was responsible for enzyme deficiency (52–54).
Clinically, TPMT poor metabolizers are at high risk for severe life threatening myelosuppression due to toxic TGN levels (47,55). While intermediate metabolizers have higher TGNs than normal metabolizers, many are able to tolerate full doses of mercaptopurine or azathioprine because of reduced methyl-TIMP levels (46). Individualized mercaptopurine and azathioprine dosing based on TPMT phenotype and measurement of thiopurine metabolites can successfully treat acute lymphoblastic leukemia (56) and inflammatory bowel disease (57–59), while reducing ADEs.
TPMT testing identifies only a portion of patients at risk of myelosuppression. More recently, NUDT15 variants have been demonstrated to affect thiopurine tolerability, especially in Asian and Hispanic patients. Thiopurines are metabolized into cytotoxic thioguanine triphosphate (TGTP), the primary anti-leukemic metabolite and significant contributor to myelosuppression (46). NUDT15 converts TGTP to thioguanine monophosphate, a less toxic metabolite. In vitro, NUDT15 decreases thiopurine cytotoxicity by inactivating thiopurine metabolites (60). In children, NUDT15 deficient alleles led to increased levels of active thiopurine metabolites and cytotoxicity, requiring lower mercaptopurine doses (60–63).
CPIC updated guidelines in 2018 on thiopurine dosing based on TPMT and/or NUDT15 genotypes (46). Recommendations on initial doses depend on the indication and the phenotype for each enzyme. While genotyping errors can occur and phenotypes can vary within metabolizer status (e.g. TPMT intermediate metabolizers), evaluation of markers of disease progression, myelosuppression or other toxicities, and even metabolites, will allow clinicians to adjust thiopurine doses from the genotype-guided starting doses.
Proton Pump Inhibitors
Proton pump inhibitors (PPIs) are commonly prescribed to children (64–66). FDA approved indications (with variable age ranges) include short term therapy for gastroesophageal reflux disease, erosive esophagitis, peptic ulcer disease, and H. pylori eradication (67–69). PPIs are often used off label in younger children, and for indications including eosinophilic esophagitis (for which PPIs are considered standard of care) and some upper respiratory tract inflammatory conditions, with conflicting data to support efficacy (70–72). These drugs act at the gastric cells by inactivating the proton acid pump which suppresses acid secretion (73). Common ADEs of PPIs include headache and gastrointestinal distress, and children are more prone to respiratory infections (67). Prior studies have suggested that the main route of metabolism is via sulfoxidation and hydroxylation through CYP450 enzymes with correlation of intrinsic clearance of PPIs to enzyme function (74). Omeprazole is considered a CYP2C19 clinical index inhibitor and has been used in drug-drug interaction studies (75). All first-generation PPIs (omeprazole, lansoprazole and pantoprazole) and the second-generation PPI dexlansoprazole are primarily metabolized by CYP2C19, with CYP3A4 playing a minor role (73). Metabolism of the second-generation PPIs esomeprazole and rabeprazole are less CYP2C19 dependent (73).
Multiple adult studies have suggested that individuals with reduced CYP2C19 metabolism have increased exposure to first-generation PPIs compared to normal metabolizers (76). In adults, poor metabolizers have been shown to have a greater response and less acidic gastric pH compared to intermediate and normal metabolizers (77). Taken together, these studies suggest that CYP2C19 plays a clinically relevant role in PPI efficacy. CPIC guidelines are available to guide use of CYP2C19 metabolizer status for PPI selection and dose (78).
Prior studies have suggested increased CYP2C19 function in children compared to adults, thus the association of CYP2C19 function and clinical outcomes of PPI use should be carefully considered in the pediatric population (79). For pediatric patients, pantoprazole and lansoprazole are the two most commonly investigated PPIs with respect to CYP2C19 effects. These studies have shown that poor metabolizers have higher exposure compared to normal metabolizers with delayed clearance and longer drug half-life (80,81). Clinical studies of children taking lansoprazole have associated ADEs and efficacy with CYP2C19 metabolizer status (82). The data for omeprazole are less clear and include some studies that show no association of outcomes with CYP2C19 genotype (83,84). However, a recent study showed reduced PPI efficacy in CYP2C19 ultrarapid metabolizers vs. those with reduced or normal CYP2C19 function (85). Data for infants are lacking and would be of interest given the very low CYP2C19 function observed in fetal and neonatal periods, followed by PPI clearance approximating adult values around 6 months of age (86,87). In a cohort of children under 3 years of age (median age 7 months), increased upper respiratory infections were observed in normal metabolizers compared to those with increased CYP2C19 function (88). Taken together, these data suggest that CYP2C19 genotype can predict PPI plasma concentrations, efficacy, and toxicity in children, and support the use of CYP2C19 data to guide PPI dosing, particularly after the neonatal period.
Selective Serotonin Reuptake Inhibitors
Selective serotonin reuptake inhibitors (SSRIs) increase serotonergic activity by decreasing presynaptic serotonin reuptake (89). SSRIs are the most common antidepressant class used in pediatric patients (90). Some SSRIs are FDA approved for some pediatric indications such as depression and obsessive compulsive disorder, but are commonly prescribed off label in pediatric and adolescent patients for indications that have adult FDA approval such as anxiety disorders, post-traumatic stress disorder, and premenstrual dysphoric disorder (91). Common pediatric SSRI ADEs include activation, gastrointestinal upset, and sleep disturbance (92).
Some SSRIs, including sertraline, citalopram, and escitalopram, are extensively metabolized by CYP2C19, with other CYP450 enzymes contributing to a lesser extent (79). Paroxetine and fluvoxamine are primarily metabolized by CYP2D6, while both CYP2D6 and CYP2C19 contribute significantly to fluoxetine metabolism (79). In vitro studies correlate the effect of CYP variants and SSRI concentrations, and active drug and metabolite concentrations have been observed to correlate with the functional status of the CYP enzyme primarily responsible for drug metabolism (79). Poor and intermediate metabolizers have higher plasma concentrations than normal, rapid or ultrarapid metabolizers (93). There are CPIC guidelines pertaining to five of most commonly used SSRIs to assist in using CYP2D6 metabolic status for dosing of paroxetine and fluvoxamine and CYP2C19 status for sertraline, escitalopram, and citalopram. These guidelines generally suggest reduced starting dose or alternative therapy in poor metabolizers to avoid ADEs and alternative therapies in ultrarapid metabolizers to avoid inefficacy (79).
For children and adolescents, the level of evidence for pharmacogenomic effects of CYPs varies across SSRIs. The data regarding the association of citalopram and escitalopram show mixed results. One small pediatric study did not show an association of citalopram and escitalopram concentrations or clinical symptoms to CYP2C19 genotype (94). Another study suggested that ultrarapid metabolizers had slower escitalopram dose escalation over time compared to other metabolizers, although there was no association with the clinical endpoint point of irritability (94,95). In contrast, one pediatric study showed poor and intermediate CYP2C19 metabolizers have higher citalopram/escitalopram plasma levels although clinical effects were not reported in this study (96). Also, one study showed a higher rate of discontinuation of citalopram/escitalopram in poor metabolizers compared to faster metabolizers, likely related to increased ADEs (97). There are three prior pediatric studies that do not suggest an association of CYP2C19 function and pediatric response or toxicity to sertraline (97–99), although another study found that children with reduced CYP2C19 metabolism had fewer ADEs than normal metabolizers, opposite of findings in adults (92). Fluoxetine, which is metabolized by CYP2D6 into an active metabolite, does not have CPIC guidelines. Pediatric studies investigating CYP2D6 metabolizer status and fluoxetine response did not show a difference in active metabolites or clinical outcomes (100,101). Thus, taken together the pediatric evidence for using CYP450 genetic testing for SSRI dosage guidance in youth is mixed and requires further study.
Some studies have examined associations of SSRI pharmacodynamic targets to attempt to improve pediatric efficacy. Two studies showed a correlation of the variants that encode the serotonin transporter and receptor, SLC6A4 and HTR2A, on the dose and response of sertraline (98) and fluoxetine (101). Another study showed an association of the serotonin transporter SLC6A4 and citalopram related agitation (102). Although candidate genes are emerging to predict pediatric response to SSRIs, further work in this area is needed prior to clinical implementation.
Attention Deficit-Hyperactive Disorder Medications
ADHD is a common pediatric disorder with evidenced based guidelines suggesting medication as a first line treatment for children older than 6 years of age (103). Stimulants are most commonly used as first line agents, but non-stimulants (atomoxetine, clonidine, and guanfacine) are commonly used as second line treatments or for certain populations (103). Most methylphenidate and amphetamine-containing medications are FDA approved to treat ADHD in children 6 years and older, as are atomoxetine, clonidine, and guanfacine (104). Despite these options, there is often heterogeneity in response to ADHD medications, which might be related to genetic factors (105).
Methylphenidate is not metabolized significantly by any CYP450 enzymes, while some amphetamine medications (e.g. dextroamphetamine) utilize CYP2D6 as a primary metabolic pathway (106). There are no guidelines or adult data to suggest dosing of stimulants based on genotype. Overall, many reports propose multiple candidate genes for pediatric response to stimulants, although these often have inconsistent evidence (105). Catechol-o-methyltransferase (COMT), an enzyme that inactivates dopamine and norepinephrine, the targets of methylphenidate, is the most extensively studied. Results indicate that gene variants resulted in decreased enzyme activity (107) and decreased medication response (108,109), although conflicting data are published (110). Thus, at this time there is insufficient evidence for using pharmacogenomics to guide stimulant dosing.
With regard to the non-stimulant medications used in ADHD treatment, atomoxetine, a selective norepinephrine reuptake inhibitor (110,111), is metabolized by CYP2D6 and to a lesser extent CYP2C19 (112). Pediatric studies suggest that CYP2D6 poor metabolizers had better response to atomoxetine than normal metabolizers, yet the poor metabolizers also experienced increased ADEs (112,113). Atomoxetine has guidance for dosing based on CYP2D6 from the FDA label and CPIC. The FDA label for atomoxetine suggests CYP2D6 poor metabolizers should reach a maximum dose of 1.2 mg/kg/day while others can go to a maximum dose of 1.4 mg/kg/day (114). CPIC guidelines recommend the appropriate timing to titrate atomoxetine dose based on CYP2D6 genotype and suggest that CYP2D6 ultrarapid metabolizers will likely not achieve appropriate efficacy from this drug at standard dosing regimens (112). If a clinician is prescribing atomoxetine, CYP2D6 testing may aid in determining the titration schedule and target dose.
Ondansetron
Ondansetron is a 5-HT3 antagonist commonly prescribed to reduce acute nausea and vomiting in pediatric patients. Metabolism of ondansetron is through several CYP enzymes: CYP3A4, CYP1A2, CYP2D6 and CYP1A1 (115). Though it only accounts for ~30% of the metabolism of ondansetron (116), CYP2D6 variation can influence exposure and efficacy of the drug. Adult CYP2D6 ultrarapid metabolizers are more likely to continue to experience nausea and vomiting than normal metabolizers because of reduced exposure (117,118). One study in pediatric stem cell transplant patients confirmed the association between CYP2D6 ultrarapid metabolizers and increased episodes of emesis due to chemotherapy (119). The CPIC guideline for ondansetron suggests that an alternative antiemetic agent (e.g. granisetron) be used in CYP2D6 ultrarapid metabolizers (120).
Ondansetron efficacy also depends on pharmacodynamics. The drug prevents serotonin from binding to vagal afferent nerves after release from the intestinal enterochromaffin cells, which decreases vagus nerve signaling, reducing serotonin release in the brainstem (120,121). There are large interindividual differences in binding of ondansetron to the 5-HT3 receptors, and efficacy partially correlates with receptor occupancy (122,123). Variants in HTR3B have been associated with early efficacy of ondansetron (124,125), but more research is needed, especially in pediatric patients.
Clinical Pharmacogenetic Testing
There are several potential approaches to clinical pharmacogenetic testing. Depending on the drug, single-gene or panel-based tests are available. Clinicians must also decide on the timing of testing. “Reactive” testing is pursued at the time of prescribing the associated drug or after an ADE has been experienced (126–128) (Figure 2). Clinical utility of the results could be limited by the turnaround time for the test, particularly if a drug needs to be prescribed immediately. This obstacle could be overcome with point-of-care, rapid turnaround testing (128). Alternatively, preemptive testing is performed in advance of prescribing decisions so that turnaround time is not an issue (Figure 2). Results are available (e.g. in the EHR) for all prescribers. Given readily available technology to test multiple variants across multiple genes at low cost, many have advocated for a pre-emptive, panel-based pharmacogenetic testing strategy (126,127). Large studies have demonstrated that >90% of patients who undergo pharmacogenetic testing harbor at least one actionable variant, and, further, that most patients are exposed to at least one of the associated drugs (129,130). As costs for panel-based testing decrease, these data support a preemptive panel-based strategy (129,130). However, reimbursement by insurance companies has been identified as a barrier to implementation (131,132).
Figure 2.
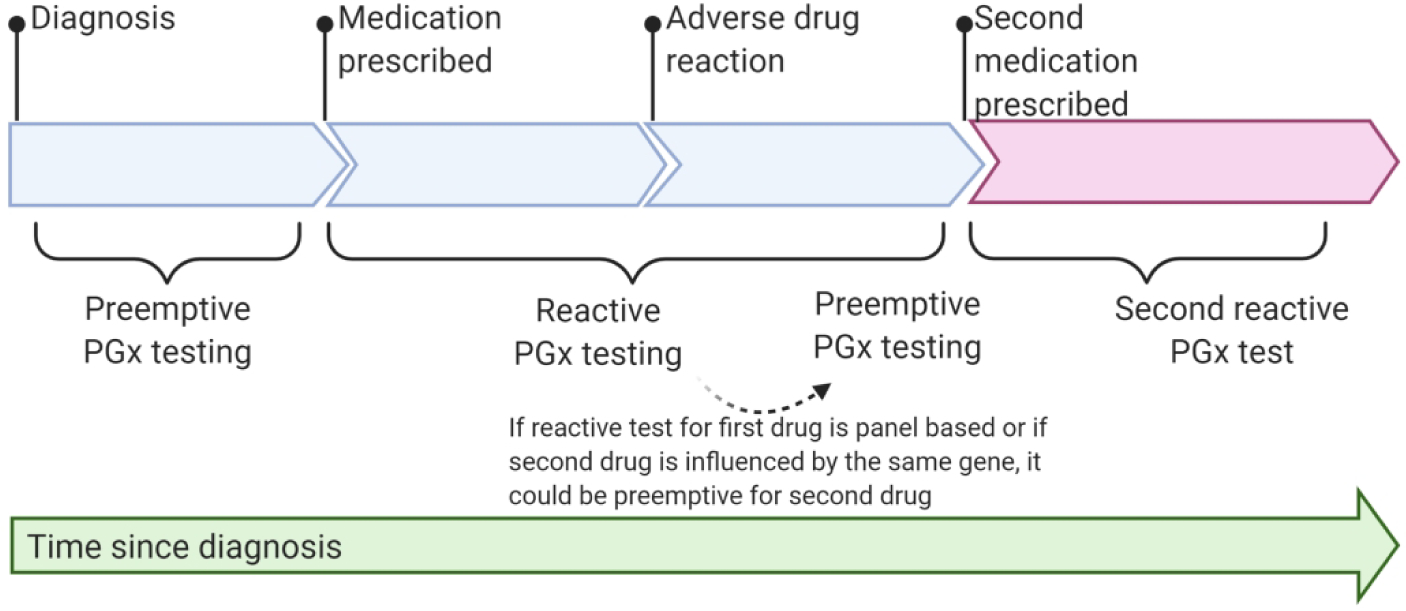
A timeline of pharmacogenetic testing to demonstrate preemptive vs reactive testing. Pharmacogenetic (PGx) testing is preemptive if it occurs prior to prescription of a medication affected by the gene tested. If a panel of genes is tested or if a second medication influenced by the same gene is prescribed, a reactive test can be preemptive for the second medication, indicated by the dotted arrow. Clinical events are indicated by the pins along the timeline and brackets indicate when a test would be considered preemptive or reactive.
For pediatricians to use pharmacogenetic data in routine clinical practice, they must have access to a valid, relevant testing assay with clear, interpretable, actionable results with appropriate turnaround time (126). Since most physicians have not received formal pharmacogenomics education and do not feel adequately informed about pharmacogenetics (133,134), pediatricians should be aware of resources that provide guidance in the interpretation of clinical pharmacogenetic tests, such as CPIC and PharmGKB. Several pharmacogenetics conferences offer continuing education credits, and for those interested in a more thorough understanding, there are certificate programs in pharmacogenetics for clinicians.
Ensuring that pharmacogenetic results are easily accessible in the EHR to all healthcare team members, including those outside the institution or the laboratory that performed the test, is an area of opportunity for implementation research. The EHR is the logical place to record results and create decision supports for current and future prescribers. Further, EHRs can be linked to pharmacogenomic records or biobanked specimens to provide research resources to further advance pharmacogenetics (135,136). Utilizing the EHR in this way requires collaboration of prescribers, pharmacists, laboratories, pharmacologists, and bioinformaticians.
Many institutions, health care systems, and countries are implementing pharmacogenomics via various strategies (137). In the US, some institutions are moving beyond recording pharmacogenomic data in the EHR by providing clinical decision support; these alerts are activated when a drug is being ordered for which a patient has an actionable genotype (137,138). Thailand has implemented wallet sized plastic cards to record pharmacogenomic results that patients carry and can present to medical providers (139). Ubiquitous Pharmacogenomics is a European effort to pre-emptively obtain pharmacogenomic testing, embed this data in the clinical record, provide patients with wallet sized cards with pharmacogenomic data, and utilize an electronic clinical decision support system to notify prescribers when a patient has a risk genotype (137).
Perspective: Recommendations for Pediatricians
For pediatric researchers, the examples provided herein demonstrate the spectrum of the maturity of pharmacogenetic research for various drugs. The association of variable CYP2D6 function, predicted by CYP2D6 genetic variants, to codeine activation to morphine was one of the sentinel discoveries in the field of pharmacogenetics. Further research exploring the clinical impact of this gene-drug interaction revealed potential toxicity from therapeutic doses of codeine affecting neonates, infants, and children. The accumulation of evidence led to regulatory and practice changes. On the other hand, for ondansetron, the evidence for the drug-gene interaction in adults is robust, but validation in pediatric cohorts is lacking. While some advocate for extrapolation of adult data to adolescents and children, there may be unique drug-gene interactions in younger patients. For SSRIs, one pediatric study indicated that ADEs are more common among CYP2C19 normal metabolizers than poor metabolizers, opposite of what is reported in adults (92). While not discussed in our earlier examples due to infrequent prescription in pediatrics, for simvastatin, an increased effect size of SLCO1B1 variation is seen in children and adolescents vs. adults (140). These examples motivate validation of pharmacogenetic associations using data from pediatric populations prior to clinical implementation. Furthermore, the tremendous physiologic and metabolic changes that occur during childhood (and particularly in infancy and adolescence) provide the opportunity for discovery of novel, pediatric-specific drug-gene interactions.
It is important to generate high quality evidence to support clinical implementation of pharmacogenomics into pediatric practice, the hallmarks of which are: 1) Adequate sample size. Many pediatric pharmacogenetic studies are of inadequate sample size to provide power to detect a difference between groups, leading to inappropriate reporting of negative findings (Type II error). Collection of large cohorts of drug-exposed children representing the full spectrum of genetic variants can be difficult, particularly for drugs that are infrequently used in children. Large biobanks are one potential solution to gathering sufficient sample size. 2) Clinically meaningful endpoints. The progression of the codeine-CYP2D6 association from research to regulatory action was facilitated by the documentation of severe, life threatening events and deaths attributed to the drug-gene interaction. Likewise, clinical implementation of TPMT and NUDT15 testing to guide thiopurine dosing was supported by the observation of potentially life threatening white blood cell suppression. While not all pharmacogenetic associations will be associated with such dramatic outcomes, studies must include clinically relevant outcomes (e.g. for ondansetron, nausea and vomiting impacting quality of life and length of hospital stay). For drugs with well-known therapeutic and toxic concentrations (e.g. voriconazole), pharmacokinetic studies may suffice, but for many others, pharmacodynamic data will be required. 3) Inclusion of participants representing those exposed to the drug with respect to age, indication, and ancestry. Drugs often have pediatric- and age-specific indications (e.g. NSAIDs for closure of patent ductus arteriosus), which may have specific pharmacogenetic associations. Neonates, infants, children, and adolescents may have developmentally regulated expression of drug transporters, metabolic enzymes, and drug response targets, leading to age-specific associations. Many genetic and pharmacogenetic studies have focused on those of European ancestry. Since the spectrum and prevalence of genetic variants often vary across ancestral populations, it is imperative to study diverse cohorts to make the findings relevant to clinical care. 4) Robust assessment of the target gene. While full sequencing for each gene of interest may be cost prohibitive, researchers must ensure that the most important and frequent variants for the gene(s) of interest, which may vary depending on the population, are assessed.
Clinical implementation of pharmacogenetics in pediatrics is challenging (132,134,136,137,141,142) and further discussion of clinical implementation is beyond the scope of this review. However, additional high quality studies with these four hallmarks will propel the field of pediatric pharmacogenetics towards clinical implementation.
Pediatric clinicians can expect increasing evidence supporting incorporation of pharmacogenetics into their care of children. However, care must be taken to critically evaluate the evidence. It has been common for some commercial pharmacogenetic testing companies to take a “more is better” approach to their recommendations, including genetic associations with very little or conflicting evidence into their pharmacogenetic test results (143). Recent FDA actions have encouraged laboratories performing pharmacogenetic testing to provide only the results of the testing, without interpretation or guidance for the prescriber. While this may avoid the inappropriate reliance on poor quality studies to guide care, it also shifts the burden of interpretation to clinicians. It is important for pediatricians to have access to high quality education about pharmacogenetics as more drug-gene interactions are demonstrated to have clinical validity for children.
Impact Statement:
While many reviews of pharmacogenetics literature are available, there are few focused on pediatrics.
Pediatricians across subspecialties will become more comfortable with pharmacogenetics terminology, know resources they can use to help inform their prescribing habits for drugs with known pharmacogenetic associations, and understand the limitations of testing and where further research is needed.
Acknowledgments
We thank Brendan (Tex) Armstreet for assistance with designing Figure 1, and Drs. Catherine Forster and Todd Florin for critical feedback on this manuscript. Figures were created with Biorender.com.
Dr. Tang Girdwood was supported by the National Institute of Child Health and Development Cincinnati Pediatric Clinical Pharmacology Postdoctoral Training Program [5T32HD069054-09] and Dr. Rossow was supported by the National Institute of Child Health and Development & National Institute of General Medical Sciences Vanderbilt Pediatric Clinical Pharmacology Postdoctoral Training Program [5T32GM007569-43].
Footnotes
Disclosure Statement: The authors have no disclosures to report.
References
- 1.Genetics Home Reference. What is pharmacogenomics? [Internet]. Genetics Home Reference. [cited 2020 Jul 18];Available from: https://ghr.nlm.nih.gov/primer/genomicresearch/pharmacogenomics
- 2.PharmGKB [Internet]. PharmGKB. [cited 2020 Jul 18];Available from: www.pharmgkb.org
- 3.Pirmohamed M. Pharmacogenetics and pharmacogenomics. Br J Clin Pharmacol 2001;52:345–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caudle KE, et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 2017;19:215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical Pharmacogenetics Implementation Consortium (CPIC®) [Internet]. [cited 2020 Jul 18];Available from: https://cpicpgx.org/
- 6.Robarge JD, Li L, Desta Z, Nguyen A, Flockhart DA. The Star-Allele Nomenclature: Retooling for Translational Genomics [Internet]. Clinical Pharmacology & Therapeutics. 2007;82:244–8. Available from: 10.1038/sj.clpt.6100284 [DOI] [PubMed] [Google Scholar]
- 7.PharmVar [Internet]. [cited 2020 Jul 18];Available from: www.pharmvar.org
- 8.Center for Devices, Radiological Health. Table of Pharmacogenetic Associations [Internet]. U.S. Food and Drug Administration. 2020. [cited 2020 Jul 18];Available from: https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations [Google Scholar]
- 9.Center for Drug Evaluation, Research. Table of Pharmacogenomic Biomarkers [Internet]. U.S. Food and Drug Administration. 2020. [cited 2020 Jul 18];Available from: https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling [Google Scholar]
- 10.Thorn CF, Klein TE, Altman RB. Codeine and morphine pathway [Internet]. Pharmacogenetics and Genomics. 2009;19:556–8. Available from: 10.1097/fpc.0b013e32832e0eac [DOI] [PubMed] [Google Scholar]
- 11.Dayer P, Desmeules J, Leemann T, Striberni R. Bioactivation of the narcotic drug codeine in human liver is mediated by the polymorphic monooxygenase catalyzing debrisoquine 4-hydroxylation (cytochrome P-450 dbl/bufI). Biochem Biophys Res Commun 1988;152:411–6. [DOI] [PubMed] [Google Scholar]
- 12.Mortimer Ö, et al. Polymorphic formation of morphine from codeine in poor and extensive metabolizers of dextromethorphan: Relationship to the presence of immunoidentified cytochrome P-450IID1 [Internet]. Clinical Pharmacology and Therapeutics. 1990;47:27–35. Available from: 10.1038/clpt.1990.4 [DOI] [PubMed] [Google Scholar]
- 13.Yu A, Kneller BM, Rettie AE, Haining RL. Expression, purification, biochemical characterization, and comparative function of human cytochrome P450 2D6.1, 2D6.2, 2D6.10, and 2D6.17 allelic isoforms. J Pharmacol Exp Ther 2002;303:1291–300. [DOI] [PubMed] [Google Scholar]
- 14.Shen H, et al. Comparative metabolic capabilities and inhibitory profiles of CYP2D6.1, CYP2D6.10, and CYP2D6.17. Drug Metab Dispos 2007;35:1292–300. [DOI] [PubMed] [Google Scholar]
- 15.Zhang W-Y, Tu Y-B, Haining RL, Yu A-M. Expression and functional analysis of CYP2D6.24, CYP2D6.26, CYP2D6.27, and CYP2D7 isozymes. Drug Metab Dispos 2009;37:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaedigk A, Sangkuhl K, Whirl-Carrillo M, Klein T, Leeder JS. Prediction of CYP2D6 phenotype from genotype across world populations. Genet Med 2017;19:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yue QY, Svensson JO, Alm C, Sjoqvist F, Sawe J. Codeine O-demethylation co-segregates with polymorphic debrisoquine hydroxylation [Internet]. British Journal of Clinical Pharmacology. 1989;28:639–45. Available from: 10.1111/j.1365-2125.1989.tb03556.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sindrup SH, et al. Codeine increases pain thresholds to copper vapor laser stimuli in extensive but not poor metabolizers of sparteine [Internet]. Clinical Pharmacology and Therapeutics. 1990;48:686–93. Available from: 10.1038/clpt.1990.212 [DOI] [PubMed] [Google Scholar]
- 19.Chen ZR, Somogyi AA, Reynolds G, Bochner F. Disposition and metabolism of codeine after single and chronic doses in one poor and seven extensive metabolisers. Br J Clin Pharmacol 1991;31:381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Desmeules J, Gascon MP, Dayer P, Magistris M. Impact of environmental and genetic factors on codeine analgesia. Eur J Clin Pharmacol 1991;41:23–6. [DOI] [PubMed] [Google Scholar]
- 21.Caraco Y, Sheller J, Wood AJ. Pharmacogenetic determination of the effects of codeine and prediction of drug interactions. J Pharmacol Exp Ther 1996;278:1165–74. [PubMed] [Google Scholar]
- 22.Poulsen L, et al. Codeine and morphine in extensive and poor metabolizers of sparteine: pharmacokinetics, analgesic effect and side effects. Eur J Clin Pharmacol 1996;51:289–95. [DOI] [PubMed] [Google Scholar]
- 23.Hedenmalm K, Sundgren M, Granberg K, Spigset O, Dahlqvist R. Urinary excretion of codeine, ethylmorphine, and their metabolites: relation to the CYP2D6 activity. Ther Drug Monit 1997;19:643–9. [DOI] [PubMed] [Google Scholar]
- 24.Mikus G, et al. Effect of codeine on gastrointestinal motility in relation to CYP2D6 phenotype* [Internet]. Clinical Pharmacology & Therapeutics. 1997;61:459–66. Available from: 10.1016/s0009-9236(97)90196-x [DOI] [PubMed] [Google Scholar]
- 25.Poulsen L, Riishede L, Brøsen K, Clemensen S, Sindrup SH. Codeine in post-operative pain. Study of the influence of sparteine phenotype and serum concentrations of morphine and morphine-6-glucuronide. Eur J Clin Pharmacol 1998;54:451–4. [DOI] [PubMed] [Google Scholar]
- 26.Lötsch J, et al. Evidence for morphine-independent central nervous opioid effects after administration of codeine: contribution of other codeine metabolites. Clin Pharmacol Ther 2006;79:35–48. [DOI] [PubMed] [Google Scholar]
- 27.Tseng CY, Wang SL, Lai MD, Lai ML, Huang JD. Formation of morphine from codeine in Chinese subjects of different CYP2D6 genotypes. Clin Pharmacol Ther 1996;60:177–82. [DOI] [PubMed] [Google Scholar]
- 28.Williams DG, Patel A, Howard RF. Pharmacogenetics of codeine metabolism in an urban population of children and its implications for analgesic reliability. Br J Anaesth 2002;89:839–45. [DOI] [PubMed] [Google Scholar]
- 29.Madadi P, et al. Safety of codeine during breastfeeding: fatal morphine poisoning in the breastfed neonate of a mother prescribed codeine. Can Fam Physician 2007;53:33–5. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Madadi P, et al. Pharmacogenetics of neonatal opioid toxicity following maternal use of codeine during breastfeeding: a case-control study. Clin Pharmacol Ther 2009;85:31–5. [DOI] [PubMed] [Google Scholar]
- 31.Graphix Group, LLC. Promethazine HCl and Codeine Phosphate Oral Solution Drug label.. [cited 2020 Jul 18];Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2008/008306s030lbl.pdf
- 32.Huddart R, et al. Standardized Biogeographic Grouping System for Annotating Populations in Pharmacogenetic Research [Internet]. Clinical Pharmacology & Therapeutics. 2019;105:1256–62. Available from: 10.1002/cpt.1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciszkowski C, Madadi P, Phillips MS, Lauwers AE, Koren G. Codeine, ultrarapid-metabolism genotype, and postoperative death. N Engl J Med 2009;361:827–8. [DOI] [PubMed] [Google Scholar]
- 34.Voronov P, Przybylo HJ, Jagannathan N. Apnea in a child after oral codeine: a genetic variant - an ultra-rapid metabolizer. Paediatr Anaesth 2007;17:684–7. [DOI] [PubMed] [Google Scholar]
- 35.Kelly LE, et al. More codeine fatalities after tonsillectomy in North American children. Pediatrics 2012;129:e1343–7. [DOI] [PubMed] [Google Scholar]
- 36.Food and Drug Administration. Safety Review Update of codeine use in children; new Boxed Warning and Contraindication on use after tonsillectomy and/or adenoidectomy. [cited 2020 Jul 18];Available from: https://www.fda.gov/media/85072/download
- 37.Crews KR, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 2014;95:376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chua K-P, Shrime MG, Conti RM. Effect of FDA Investigation on Opioid Prescribing to Children After Tonsillectomy/Adenoidectomy. Pediatrics [Internet] 2017;140. Available from: 10.1542/peds.2017-1765 [DOI] [PubMed] [Google Scholar]
- 39.Townsend JA, Sebastião YV, Cooper JN. Effect of FDA Warning on Codeine and Alternate Opioid Prescribing After Pediatric Dental Procedures in Ohio. Pediatr Dent 2019;41:439–45. [PMC free article] [PubMed] [Google Scholar]
- 40.Center for Drug Evaluation, Research. FDA Drug Safety Communication: FDA restricts use of prescription codeine pain and cough medicines and tramadol pain medicines in children; recommends against use in breastfeeding women | FDA [Internet]. U.S. Food and Drug Administration. 2019. [cited 2020 Jul 18];Available from: https://www.fda.gov/drugs/drug-safety-and-availability/fda-drug-safety-communication-fda-restricts-use-prescription-codeine-pain-and-cough-medicines-and [Google Scholar]
- 41.Gammal RS, et al. The Case for Pharmacogenetics-Guided Prescribing of Codeine in Children. Clin Pharmacol Ther 2019;105:1300–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gammal RS, et al. Pharmacogenetics for Safe Codeine Use in Sickle Cell Disease. Pediatrics [Internet] 2016;138. Available from: 10.1542/peds.2015-3479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lassen D, Damkier P, Brøsen K. The Pharmacogenetics of Tramadol. Clin Pharmacokinet 2015;54:825–36. [DOI] [PubMed] [Google Scholar]
- 44.Crews KR, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6, OPRM1, and COMT genotype and select opioid therapy. Clin Pharmacol Ther [Internet] 2021;Available from: 10.1002/cpt.2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ashworth LA, Billett A, Mitchell P, Nuti F, Siegel C, Bousvaros A. Lymphoma risk in children and young adults with inflammatory bowel disease: Analysis of a large single-center cohort [Internet]. Inflammatory Bowel Diseases. 2012;18:838–43. Available from: 10.1002/ibd.21844 [DOI] [PubMed] [Google Scholar]
- 46.Relling MV, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT 15 Genotypes: 2018 Update [Internet]. Clinical Pharmacology & Therapeutics. 2019;105:1095–105. Available from: 10.1002/cpt.1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans WE. Pharmacogenetics of thiopurine S-methyltransferase and thiopurine therapy. Ther Drug Monit 2004;26:186–91. [DOI] [PubMed] [Google Scholar]
- 48.Weinshilboum RM, Sladek SL. Mercaptopurine pharmacogenetics: monogenic inheritance of erythrocyte thiopurine methyltransferase activity. Am J Hum Genet 1980;32:651–62. [PMC free article] [PubMed] [Google Scholar]
- 49.Van Loon JA, Weinshilboum RM. Thiopurine methyltransferase biochemical genetics: Human lymphocyte activity [Internet]. Biochemical Genetics. 1982;20:637–58. Available from: 10.1007/bf00483962 [DOI] [PubMed] [Google Scholar]
- 50.Van Loon JA, Weinshilboum RM. Thiopurine methyltransferase isozymes in human renal tissue. Drug Metab Dispos 1990;18:632–8. [PubMed] [Google Scholar]
- 51.Szumlanski CL, Honchel R, Scott MC, Weinshilboum RM. Human liver thiopurine methyltransferase pharmacogenetics: biochemical properties, liver-erythrocyte correlation and presence of isozymes. Pharmacogenetics 1992;2:148–59. [PubMed] [Google Scholar]
- 52.Tai H-L, Krynetski EY, Schuetz EG, Yanishevski Y, Evans WE. Enhanced proteolysis of thiopurine S-methyltransferase (TPMT) encoded by mutant alleles in humans (TPMT*3A, TPMT*2): Mechanisms for the genetic polymorphism of TPMT activity [Internet]. Proceedings of the National Academy of Sciences. 1997;94:6444–9. Available from: 10.1073/pnas.94.12.6444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tai HL, et al. Enhanced proteasomal degradation of mutant human thiopurine S-methyltransferase (TPMT) in mammalian cells: mechanism for TPMT protein deficiency inherited by TPMT*2, TPMT*3A, TPMT*3B or TPMT*3C. Pharmacogenetics 1999;9:641–50. [DOI] [PubMed] [Google Scholar]
- 54.Wang L, Sullivan W, Toft D, Weinshilboum R. Thiopurine S-methyltransferase pharmacogenetics: chaperone protein association and allozyme degradation. Pharmacogenetics 2003;13:555–64. [DOI] [PubMed] [Google Scholar]
- 55.Relling MV, et al. Mercaptopurine therapy intolerance and heterozygosity at the thiopurine S-methyltransferase gene locus. J Natl Cancer Inst 1999;91:2001–8. [DOI] [PubMed] [Google Scholar]
- 56.Relling MV, Pui C-H, Cheng C, Evans WE. Thiopurine methyltransferase in acute lymphoblastic leukemia. Blood 2006;107:843–4. [DOI] [PubMed] [Google Scholar]
- 57.Kaskas BA, et al. Safe treatment of thiopurine S-methyltransferase deficient Crohn’s disease patients with azathioprine. Gut 2003;52:140–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee M-N, et al. Successful azathioprine treatment with metabolite monitoring in a pediatric inflammatory bowel disease patient homozygous for TPMT*3C. Yonsei Med J 2013;54:1545–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benkov K, et al. Role of thiopurine metabolite testing and thiopurine methyltransferase determination in pediatric IBD. J Pediatr Gastroenterol Nutr 2013;56:333–40. [DOI] [PubMed] [Google Scholar]
- 60.Moriyama T, et al. NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet 2016;48:367–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang JJ, et al. Inherited NUDT15 variant is a genetic determinant of mercaptopurine intolerance in children with acute lymphoblastic leukemia. J Clin Oncol 2015;33:1235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka Y, et al. Susceptibility to 6-MP toxicity conferred by aNUDT15variant in Japanese children with acute lymphoblastic leukaemia [Internet]. British Journal of Haematology. 2015;171:109–15. Available from: 10.1111/bjh.13518 [DOI] [PubMed] [Google Scholar]
- 63.Zgheib NK, et al. NUDT15 and TPMT genetic polymorphisms are related to 6-mercaptopurine intolerance in children treated for acute lymphoblastic leukemia at the Children’s Cancer Center of Lebanon. Pediatr Blood Cancer 2017;64:146–50. [DOI] [PubMed] [Google Scholar]
- 64.Illueca M, Alemayehu B, Shoetan N, Yang H. Proton pump inhibitor prescribing patterns in newborns and infants. J Pediatr Pharmacol Ther 2014;19:283–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barron JJ, Tan H, Spalding J, Bakst AW, Singer J. Proton pump inhibitor utilization patterns in infants. J Pediatr Gastroenterol Nutr 2007;45:421–7. [DOI] [PubMed] [Google Scholar]
- 66.Blank M-L, Parkin L. National Study of Off-label Proton Pump Inhibitor Use Among New Zealand Infants in the First Year of Life (2005–2012). J Pediatr Gastroenterol Nutr 2017;65:179–84. [DOI] [PubMed] [Google Scholar]
- 67.Rosen R, et al. Pediatric Gastroesophageal Reflux Clinical Practice Guidelines: Joint Recommendations of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the European Society for Pediatric Gastroenterology, Hepatology, and Nutrition. J Pediatr Gastroenterol Nutr 2018;66:516–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibbons TE, Gold BD. The use of proton pump inhibitors in children: a comprehensive review. Paediatr Drugs 2003;5:25–40. [DOI] [PubMed] [Google Scholar]
- 69.Jones NL, et al. Joint ESPGHAN/NASPGHAN Guidelines for the Management of Helicobacter pylori in Children and Adolescents (Update 2016). J Pediatr Gastroenterol Nutr 2017;64:991–1003. [DOI] [PubMed] [Google Scholar]
- 70.Dellon ES, et al. ACG clinical guideline: Evidenced based approach to the diagnosis and management of esophageal eosinophilia and eosinophilic esophagitis (EoE). Am J Gastroenterol 2013;108:679–92; quiz 693. [DOI] [PubMed] [Google Scholar]
- 71.Ummarino D, et al. Impact of antisecretory treatment on respiratory symptoms of gastroesophageal reflux disease in children. Dis Esophagus 2012;25:671–7. [DOI] [PubMed] [Google Scholar]
- 72.Wasilewska J, et al. Respiratory response to proton pump inhibitor treatment in children with obstructive sleep apnea syndrome and gastroesophageal reflux disease. Sleep Med 2012;13:824–30. [DOI] [PubMed] [Google Scholar]
- 73.Ward RM, Kearns GL. Proton Pump Inhibitors in Pediatrics. Pediatric Drugs 2013;15:119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Abelo A, et al. STEREOSELECTIVE METABOLISM OF OMEPRAZOLE BY HUMAN CYTOCHROME P450 ENZYMES. DMD 2000;28:966–72. [PubMed] [Google Scholar]
- 75.Center for Drug Evaluation, Research. Table of Substrates, Inhibitors and Inducers [Internet]. U.S. Food and Drug Administration. 2020. [cited 2020 Jul 19];Available from: https://www.fda.gov/drugs/drug-interactions-labeling/drug-development-and-drug-interactions-table-substrates-inhibitors-and-inducers [Google Scholar]
- 76.Gardiner SJ, Begg EJ. Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacol Rev 2006;58:521–90. [DOI] [PubMed] [Google Scholar]
- 77.Shirai N, et al. Effects of CYP2C19 genotypic differences in the metabolism of omeprazole and rabeprazole on intragastric pH. Aliment Pharmacol Ther 2001;15:1929–37. [DOI] [PubMed] [Google Scholar]
- 78.Lima JJ, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2C19 and Proton Pump Inhibitor Dosing. Clin Pharmacol Ther [Internet] 2020;Available from: 10.1002/cpt.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hicks JK, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther 2015;98:127–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kearns GL, et al. Single-dose pharmacokinetics of oral and intravenous pantoprazole in children and adolescents. J Clin Pharmacol 2008;48:1356–65. [DOI] [PubMed] [Google Scholar]
- 81.Ward RM, et al. Single-dose, multiple-dose, and population pharmacokinetics of pantoprazole in neonates and preterm infants with a clinical diagnosis of gastroesophageal reflux disease (GERD). Eur J Clin Pharmacol 2010;66:555–61. [DOI] [PubMed] [Google Scholar]
- 82.Gumus E, et al. Evaluation of lansoprazole as a probe for assessing cytochrome P450 2C19 activity and genotype-phenotype correlation in childhood. Eur J Clin Pharmacol 2012;68:629–36. [DOI] [PubMed] [Google Scholar]
- 83.Kearns GL, Leeder JS, Gaedigk A. Impact of the CYP2C19*17 allele on the pharmacokinetics of omeprazole and pantoprazole in children: evidence for a differential effect. Drug Metab Dispos 2010;38:894–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lima JJ, et al. Association of CYP2C19 polymorphisms and lansoprazole-associated respiratory adverse effects in children. J Pediatr 2013;163:686–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Franciosi JP, et al. Association Between CYP2C19*17 Alleles and pH Probe Testing Outcomes in Children With Symptomatic Gastroesophageal Reflux. J Clin Pharmacol 2018;58:89–96. [DOI] [PubMed] [Google Scholar]
- 86.Marier J-F, et al. Pharmacokinetics of omeprazole in healthy adults and in children with gastroesophageal reflux disease. Ther Drug Monit 2004;26:3–8. [DOI] [PubMed] [Google Scholar]
- 87.Tran A, et al. Pharmacokinetic-pharmacodynamic study of oral lansoprazole in children. Clin Pharmacol Ther 2002;71:359–67. [DOI] [PubMed] [Google Scholar]
- 88.Bernal CJ, et al. CYP2C19 Phenotype and Risk of Proton Pump Inhibitor-Associated Infections. Pediatrics [Internet] 2019;144. Available from: 10.1542/peds.2019-0857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lochmann D, Richardson T. Selective Serotonin Reuptake Inhibitors. Handb Exp Pharmacol 2019;250:135–44. [DOI] [PubMed] [Google Scholar]
- 90.Olfson M, He J-P, Merikangas KR. Psychotropic Medication Treatment of Adolescents: Results From the National Comorbidity Survey–Adolescent Supplement. J Am Acad Child Adolesc Psychiatry 2013;52:378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Aka I, et al. Clinical Pharmacogenetics of Cytochrome P450-Associated Drugs in Children. J Pers Med [Internet] 2017;7. Available from: 10.3390/jpm7040014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rossow KM, Aka IT, Maxwell-Horn AC, Roden DM, Van Driest SL. Pharmacogenetics to Predict Adverse Events Associated With Antidepressants [Internet]. Pediatrics. 2020;146:e20200957. Available from: 10.1542/peds.2020-0957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chang M, Tybring G, Dahl M-L, Lindh JD. Impact of cytochrome P450 2C19 polymorphisms on citalopram/escitalopram exposure: a systematic review and meta-analysis. Clin Pharmacokinet 2014;53:801–11. [DOI] [PubMed] [Google Scholar]
- 94.Carlsson B, et al. Enantioselective analysis of citalopram and metabolites in adolescents. Ther Drug Monit 2001;23:658–64. [DOI] [PubMed] [Google Scholar]
- 95.Bishop JR, et al. Escitalopram pharmacogenetics: CYP2C19 relationships with dosing and clinical outcomes in autism spectrum disorder. Pharmacogenet Genomics 2015;25:548–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rudberg I, Hendset M, Uthus LH, Molden E, Refsum H. Heterozygous mutation in CYP2C19 significantly increases the concentration/dose ratio of racemic citalopram and escitalopram (S-citalopram). Ther Drug Monit 2006;28:102–5. [DOI] [PubMed] [Google Scholar]
- 97.Aldrich SL, et al. Influence of CYP2C19 Metabolizer Status on Escitalopram/Citalopram Tolerability and Response in Youth With Anxiety and Depressive Disorders. Front Pharmacol 2019;10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poweleit EA, et al. Pharmacogenetics of Sertraline Tolerability and Response in Pediatric Anxiety and Depressive Disorders. J Child Adolesc Psychopharmacol 2019;29:348–61. [DOI] [PubMed] [Google Scholar]
- 99.AlOlaby RR, et al. Molecular biomarkers predictive of sertraline treatment response in young children with fragile X syndrome. Brain Dev 2017;39:483–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ramsey LB, Bishop JR, Strawn JR. Pharmacogenetics of treating pediatric anxiety and depression. Pharmacogenomics 2019;20:867–70. [DOI] [PubMed] [Google Scholar]
- 101.Troy TF, Poweleit EA, Strawn JR, Martin LJ, Ramsey LB. The Influence of Pharmacodynamic Genes on Fluoxetine Response in Pediatric Anxiety and Depressive Disorders. J Child Adolesc Psychopharmacol 2020;30:276–7. [DOI] [PubMed] [Google Scholar]
- 102.Amitai M, et al. Pharmacogenetics of citalopram-related side effects in children with depression and/or anxiety disorders. J Neural Transm 2016;123:1347–54. [DOI] [PubMed] [Google Scholar]
- 103.Wolraich ML, et al. ; Subcommittee on Children and Adolescents with Attention-Deficit/Hyperactive Disorder. Clinical Practice Guideline for the Diagnosis, Evaluation, and Treatment of Attention-Deficit/Hyperactivity Disorder in Children and Adolescents. 2019;144(4):e20192528. Pediatrics [Internet] 2020;145. Available from: 10.1542/peds.2019-3997 [DOI] [PubMed] [Google Scholar]
- 104.Bruxel EM, et al. ADHD pharmacogenetics across the life cycle: New findings and perspectives. Am J Med Genet B Neuropsychiatr Genet 2014;165B:263–82. [DOI] [PubMed] [Google Scholar]
- 105.Kambeitz J, Romanos M, Ettinger U. Meta-analysis of the association between dopamine transporter genotype and response to methylphenidate treatment in ADHD. Pharmacogenomics J 2014;14:77–84. [DOI] [PubMed] [Google Scholar]
- 106.Joshua FD, Elizabeth T, Talia P. The Child Medication Fact Book for Psychiatric Practice. 2018.
- 107.Lachman HM, et al. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics 1996;6:243–50. [DOI] [PubMed] [Google Scholar]
- 108.Cheon K-A, Jun J-Y, Cho D-Y. Association of the catechol-O-methyltransferase polymorphism with methylphenidate response in a classroom setting in children with attention-deficit hyperactivity disorder. Int Clin Psychopharmacol 2008;23:291–8. [DOI] [PubMed] [Google Scholar]
- 109.Kereszturi E, et al. Catechol-O-methyltransferase Val158Met polymorphism is associated with methylphenidate response in ADHD children. Am J Med Genet B Neuropsychiatr Genet 2008;147B:1431–5. [DOI] [PubMed] [Google Scholar]
- 110.Mills S, et al. No evidence of association between Catechol-O-Methyltransferase (COMT) Val158Met genotype and performance on neuropsychological tasks in children with ADHD: a case-control study. BMC Psychiatry 2004;4:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Michelson D, et al. CYP2D6 and clinical response to atomoxetine in children and adolescents with ADHD. J Am Acad Child Adolesc Psychiatry 2007;46:242–51. [DOI] [PubMed] [Google Scholar]
- 112.Brown JT, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Cytochrome P450 (CYP)2D6 Genotype and Atomoxetine Therapy. Clin Pharmacol Ther 2019;106:94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Trzepacz PT, et al. CYP2D6 metabolizer status and atomoxetine dosing in children and adolescents with ADHD. Eur Neuropsychopharmacol 2008;18:79–86. [DOI] [PubMed] [Google Scholar]
- 114.Eli Lily and Company. Atomoxetine Food and Drug Administration Drug Label [Internet]. June/2009. [cited 08/2020];Available from: https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/021411s029s030lbl.pdf
- 115.Huddart R, Altman RB, Klein TE. PharmGKB summary: Ondansetron and tropisetron pathways, pharmacokinetics and pharmacodynamics. Pharmacogenet Genomics 2019;29:91–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dixon CM, et al. Multiple forms of cytochrome P450 are involved in the metabolism of ondansetron in humans. Drug Metab Dispos 1995;23:1225–30. [PubMed] [Google Scholar]
- 117.Kaiser R, et al. Patient-tailored antiemetic treatment with 5-hydroxytryptamine type 3 receptor antagonists according to cytochrome P-450 2D6 genotypes. J Clin Oncol 2002;20:2805–11. [DOI] [PubMed] [Google Scholar]
- 118.Candiotti KA, et al. The impact of pharmacogenomics on postoperative nausea and vomiting: do CYP2D6 allele copy number and polymorphisms affect the success or failure of ondansetron prophylaxis? Anesthesiology 2005;102:543–9. [DOI] [PubMed] [Google Scholar]
- 119.Edwards A, Ramsey L. The influence of CYP2D6 metabolizer status on the control of emesis by ondansetron in pediatric patients undergoing hematopoietic stem cell transplantation.University of Cincinnati Master’s Thesis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bell GC, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther 2017;102:213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Endo T, et al. Neurochemistry and neuropharmacology of emesis - the role of serotonin. Toxicology 2000;153:189–201. [DOI] [PubMed] [Google Scholar]
- 122.Yamada Y, et al. Receptor occupancy theory-based analysis of antiemetic effects and standard doses of 5-HT3 receptor antagonists in cancer patients. Cancer Chemother Pharmacol 2004;54:185–90. [DOI] [PubMed] [Google Scholar]
- 123.Ayuhara H, et al. Receptor occupancy theory-based analysis of interindividual differences in antiemetic effects of 5-HT3 receptor antagonists. Int J Clin Oncol 2009;14:518–24. [DOI] [PubMed] [Google Scholar]
- 124.Tremblay P-B, et al. Variations in the 5-hydroxytryptamine type 3B receptor gene as predictors of the efficacy of antiemetic treatment in cancer patients. J Clin Oncol 2003;21:2147–55. [DOI] [PubMed] [Google Scholar]
- 125.Lehmann AS, et al. Pharmacogenetic predictors of nausea and vomiting of pregnancy severity and response to antiemetic therapy: a pilot study. BMC Pregnancy Childbirth 2013;13:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weitzel KW, Cavallari LH, Lesko LJ. Preemptive Panel-Based Pharmacogenetic Testing: The Time is Now. Pharm Res 2017;34:1551–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Krebs K, Milani L. Translating pharmacogenomics into clinical decisions: do not let the perfect be the enemy of the good. Hum Genomics 2019;13:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Roden DM, et al. Pharmacogenomics. Lancet 2019;394:521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Van Driest SL, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther 2014;95:423–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature 2015;526:343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dunnenberger HM, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol 2015;55:89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Keeling NJ, et al. Preemptive pharmacogenetic testing: exploring the knowledge and perspectives of US payers. Genet Med 2019;21:1224–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Stanek EJ, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clin Pharmacol Ther 2012;91:450–8. [DOI] [PubMed] [Google Scholar]
- 134.Owusu Obeng A, et al. Physician-Reported Benefits and Barriers to Clinical Implementation of Genomic Medicine: A Multi-Site IGNITE-Network Survey. J Pers Med [Internet] 2018;8. Available from: 10.3390/jpm8030024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Roden DM, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hoffman JM, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet 2014;166C:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.van der Wouden CH, et al. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin Pharmacol Ther 2017;101:341–58. [DOI] [PubMed] [Google Scholar]
- 138.Pulley JM, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clin Pharmacol Ther 2012;92:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sukasem C, Chantratita W. A success story in pharmacogenomics: genetic ID card for SJS/TEN. Pharmacogenomics 2016;17:455–8. [DOI] [PubMed] [Google Scholar]
- 140.Wagner JB, et al. Impact of SLCO1B1 Genotype on Pediatric Simvastatin Acid Pharmacokinetics. J Clin Pharmacol 2018;58:823–33. [DOI] [PubMed] [Google Scholar]
- 141.Liu M, et al. A Tutorial for Pharmacogenomics Implementation Through End-to-End Clinical Decision Support Based on Ten Years of Experience from PREDICT. Clin Pharmacol Ther [Internet] 2020;Available from: 10.1002/cpt.2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ramsey LB, et al. Implementation of Pharmacogenetics at Cincinnati Children’s Hospital Medical Center: Lessons Learned Over 14 Years of Personalizing Medicine. Clin Pharmacol Ther 2019;105:49–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bousman CA, Dunlop BW. Genotype, phenotype, and medication recommendation agreement among commercial pharmacogenetic-based decision support tools. Pharmacogenomics J 2018;18:613–22. [DOI] [PubMed] [Google Scholar]


