Abstract
Cell-cell communication through evolutionarily conserved signaling pathways governs embryonic development and adult tissue homeostasis. Deregulation of these signaling pathways has been implicated in a wide range of human diseases including cancer. One such pathway is the Hedgehog (Hh) pathway, which was originally discovered in Drosophila and later found to play a fundamental role in human development and diseases. Abnormal Hh pathway activation is a major driver of basal cell carcinomas (BCC) and medulloblastoma. Hh exerts it biological influence through a largely conserved signal transduction pathway from the activation of the GPCR family transmembrane protein Smoothened (Smo) to the conversion of latent Zn-finger transcription factors Gli/Ci proteins from their repressor (GliR/CiR) to activator (GliA/CiA) forms. Studies from model organisms and human patients have provided deep insight into the Hh signal transduction mechanisms, revealed roles of Hh signaling in a wide range of human cancers, and suggested multiple strategies for targeting this pathway in cancer treatment.
Keywords: Hedgehog, Shh, Ptc, Smo, Gli, signaling, phosphorylation, BCC, medulloblastoma, cancer
1. Introduction
Initially discovered in Drosophila, the Hedgehog (Hh) family of secreted proteins plays critical roles in both embryonic development and adult tissue homeostasis in species ranging from insects to mammals [1–4]. Not surprisingly, deregulation of Hh signaling has been linked to a wide range of human disorders including birth defects such as holoprosencephaly, Gorlin Syndrome, Greig cephalopolysyndactyly, and Pallister-Hall syndrome, and cancer such as basal cell carcinomas and medulloblastoma [2, 5–7].
Members of the Hh family exert their biological influences through a largely conserved pathway to alter the balance between the activator and repressor forms of the Gli family of zinc finger transcription factors (Fig. 1) [2, 8, 9]. While Drosophila has only one Hh and one Gli protein, Cubitus interruptus (Ci), mammals have three Hh family members: Sonic hedgehog (Shh), Indian hedgehog (Ihh) and Desert hedgehog (Dhh), and three Gli proteins: Gli1, Gli2 and Gli3. In mammals, the Gli repressor (GliR) function is mostly derived from Gli3 whereas the Gli activator (GliA) function is mainly contributed by Gli2. Gli1 is a direct transcriptional target of Hh signaling pathway and acts in a positive feedback to reinforce GliA function.
Figure 1. The conserved Hh signal transduction pathway.
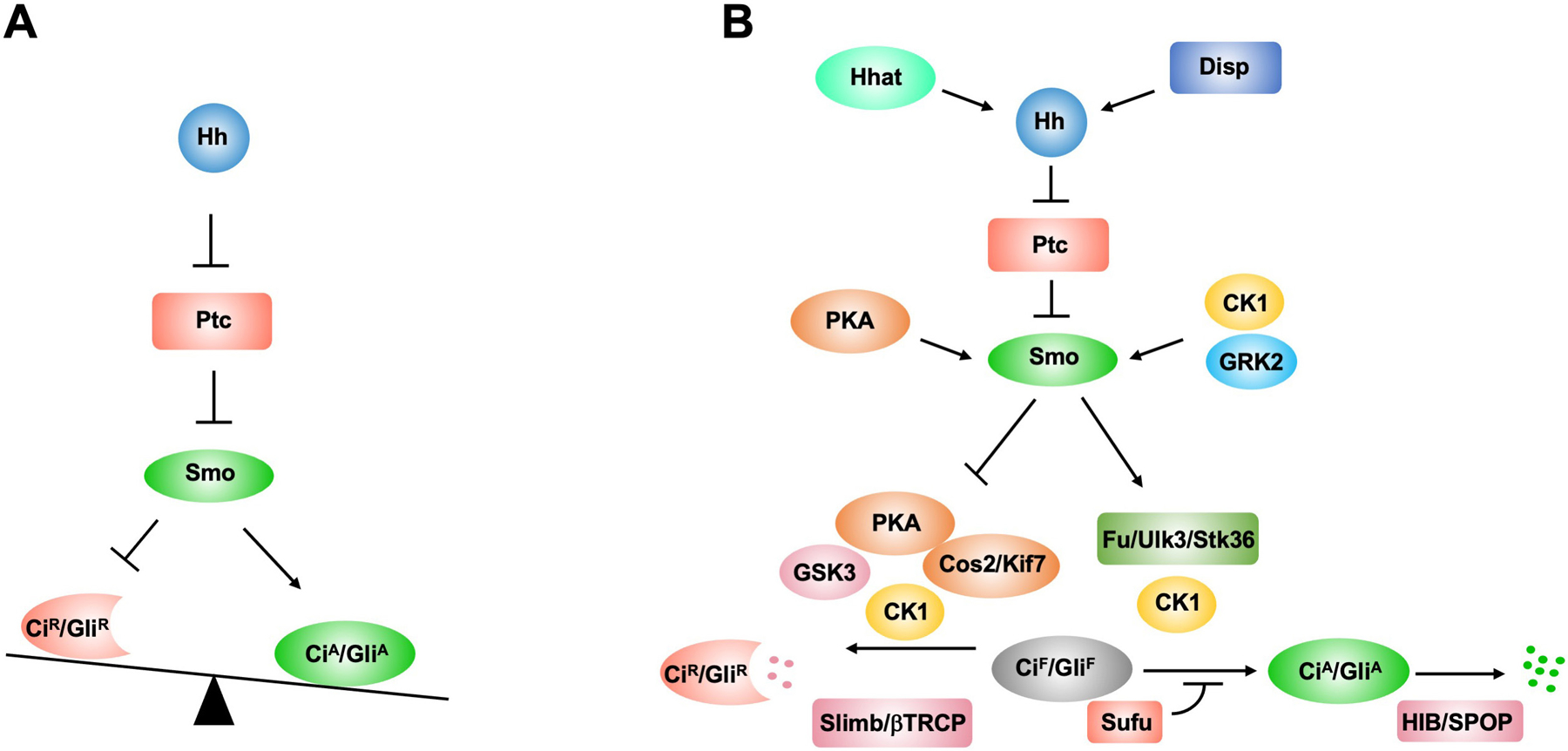
(A). A simplified scheme of Hh pathway in which Hh releases the inhibition of Smo by Ptc and activated Smo alter the balance between the activator (CiA/GliA) and repressor CiR/GliR) forms of Ci/Gli. (B) Hhat catalyzes the palmitoylation of Hh and Disp is involved in the secretion of lipidated Hh. Hh induces phosphorylation of Smo by multiple kinases including PKA (Drosophila only), CK1 and GRK2, which is required for Smo activation. In the absence of Hh, full length Ci/Gli is phosphorylated by multiple kinases including PKA, GSK3 and CK1 and subsequently targeted to ubiquitin/proteasome-mediated proteolysis through Slimb/βTRCP to generate CiR/GliR. Smo inhibits Ci/Gli phosphorylation by PKA/GSK3/CK1 to block the production of CiR/GliR. Furthermore, high levels of Hh stimulate Fu/Ulk3/Stk36-mediated phosphorylation Ci/Gli to promote the formation of CiA/GliA by antagonizing Sufu. HIB/SPOP targets CiA/GliA for degradation, which is attenuated by CK1. Cos2/Kif7 plays dual role by promoting the formation of CiR/GliR in the absence of Hh but CiA/GliA in the presence of Hh.
The Hh signal reception system consists of a twelve-span transmembrane protein Patched (Ptc) that binds directly to Hh and a GPCR family of seven-span transmembrane protein Smoothened (Smo) that transduces the Hh signal across the plasma membrane. Mammals have two Ptc proteins: Ptch1 and Ptch2, with Ptch1 as the major Hh receptor. In the absence of Hh, Ptc blocks Smo signaling activity, allowing the production of GliR/CiR that actively represses a subset of Hh target genes. Binding of Hh to Ptc alleviates its inhibition of Smo, allowing Smo to signal downstream to block GliR/CiR production and promote GliA/CiA formation (Fig. 1A). While the basic framework of Hh signal transduction pathway is similar between Drosophila and mammals (Fig. 1B), major differences exist in several regulatory steps. Most notably, vertebrate but not Drosophila Hh signal transduction depends on primary cilium, a microtube-based membrane protrusion found on most mammalian cells including cancer cells [10]. In addition, both Gli-independent noncanonical Hh signaling pathways and Hh/Smo-independent Gli activation pathways exist in mammals [11, 12]. This review focuses on the most recent understanding of the Hh-Gli signaling pathway and its role in tumorigenesis and discuss how this basic knowledge has been translated into strategies for cancer treatment.
2. Hh signal transduction
2.1. Lipid modification and dispersal of Hh ligand
Hh family members function as morphogens that act over long range with different levels pathway activity specifying distinct developmental outcomes [1]. In Hh-producing cells, full-length Hh undergoes autocleavage to release an N-terminal fragment (HhN) with a cholesterol moiety covalently linked to its C-terminus [13]. HhN is then palmitoylated near its N-terminus by the acyltransferase Skinny Hedgehog (Skn)/Hedgehog acyltransferase (Hhat) (Fig. 1B) [14–17]. While cholesterol modification increases the affinity of Hh for cell membranes and restricts its free dispersal [18, 19], dual lipid modifications facilitate the formation of large multimeric Hh complexes, allowing Hh to move over a long distance [20–26]. Dispatched (Disp), a transmembrane protein structurally related to Ptc [27, 28], is required for the secretion of lipidated Hh to the extracellular space [15, 18, 29–35]. Several Hh carriers have been implicated in long-range Hh signaling, including exosomes [36–38], Lipoprotein particles [39], cytoneme [40, 41], and the Scube family of secreted proteins [33, 42]. A recent study showed that Scube binds lipidated Shh to form a soluble signaling complex and that Hh corepressors CDON/BOC and Gas1 act cooperatively to relay Shh from Scute to Ptc [43]. In addition, many studies have uncovered the role of heparin sulfate proteoglycans (HSPGs) in modulating the release, transport, and reception of Hh [44, 45].
2.2. Hh reception: Ptc and Smo
Being the core Hh receptor, Ptc paradoxically functions as an inhibitor of Hh signaling by blocking Smo activation sub-stoichiometrically in the absence of Hh ligand [46]. How Ptc inhibits Smo and how Hh binding alleviates this inhibition have remained mysteries in the field until very recently. Several recent structural and biochemical studies suggest that Ptc functions as a cation-driven sterol transporter and that one Hh ligand binds a Ptc dimer to block the transporter activity of Ptc [47–55]. Cholesterol and its derivatives such as 24, 25-epoxycholesterol have been implicated as endogenous ligands for Smo [56–59]. Binding of these sterols to the extracellular cysteine-rich domain (CRD) and/or seven transmembrane helixes of Smo triggers conformational changes in these domains, leading to Smo activation [57, 60–64].
Ptc restricts the accessibility of sterols to Smo and thus inhibits Smo activation in the primary cilium (Fig. 2) [59, 65–67]. A recent study revealed that DHCR7 and CYP7A1, which synthesize cholesterol and oxysterol respectively, are located near the ciliary base to modulate Hh pathway activation, and that their ciliary localization is regulated by Hh [68]. Binding of Hh to Ptc inhibits its sterol transporter activity and promotes its ciliary exit in a manner depending on the Smurf family of E3 ubiquitin ligases, allowing Smo to be activated and accumulated in the primary cilium [65, 66, 69, 70]. In Drosophila, Hh induces trafficking of Ptc away and Smo toward the plasma membrane through the Smurf family of E3 ubiquitin ligases [71–74]. Although ciliary trafficking of Smo is also regulated by ubiquitin in mammalian cells, the E3 ligase(s) remains obscured [75]. Hh induces sumoylation of Smo in both Drosophila and mammalian cells and at least in Drosophila, sumoylation regulates Smo trafficking by antagonizing its ubiquitination through the recruitment of a deubiquitinating enzyme USP8/UBPY [76, 77].
Figure 2. Mammalian Hh signaling in primary cilia.
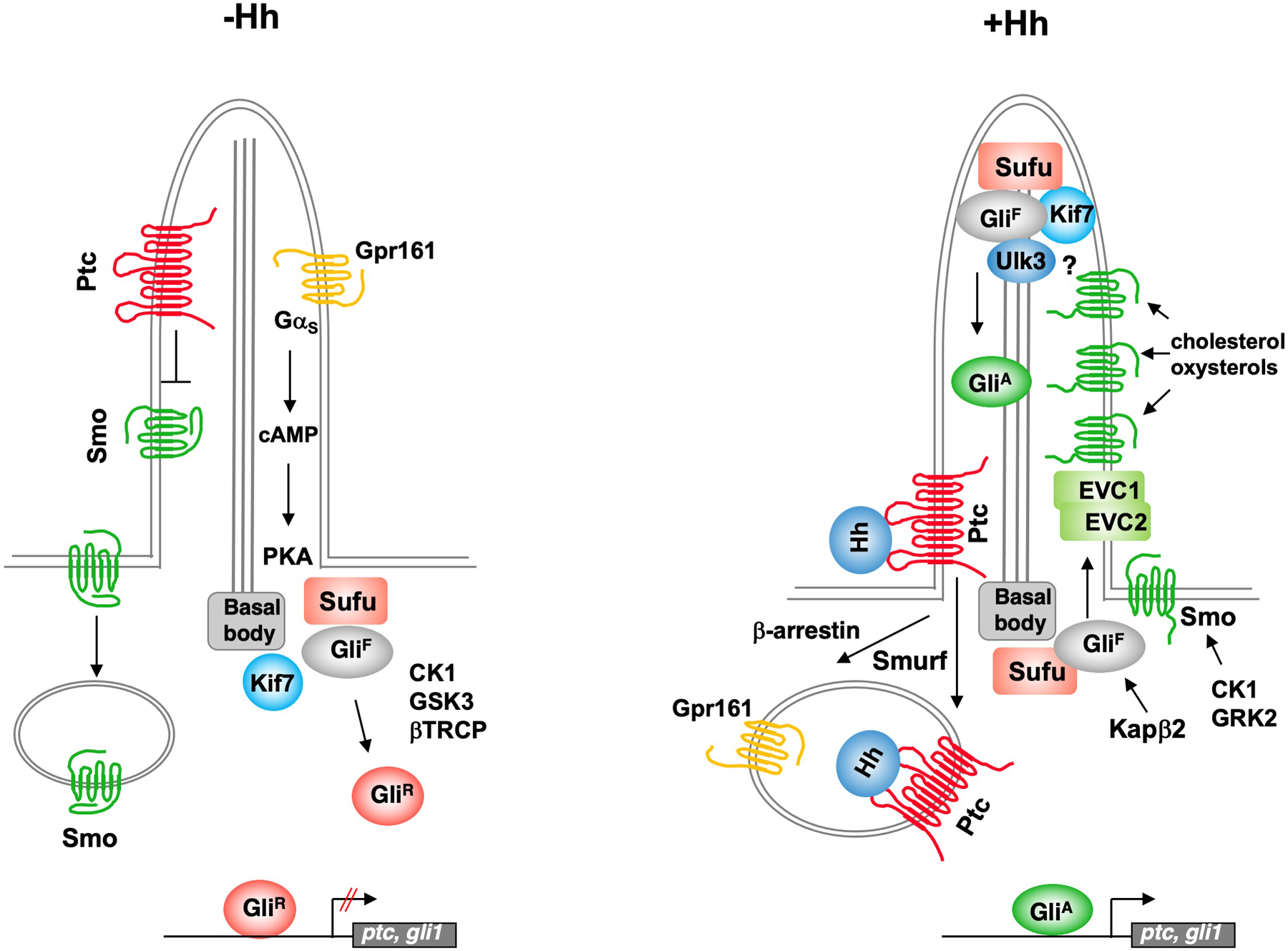
In the absence of Hh, Ptc localizes to the primary cilia and inhibits Smo ciliary accumulation. Gpr161, a ciliary localized GPCR, is required for PKA activation. Kif7 is localized at ciliary base and both Sufu and Kif 7 are required for GliR production. In the presence of Hh, Hh inhibits Ptc activity and promotes its ciliary removal depending on Smurf and, subsequently the accumulation of Smo into primary cilia depending on phosphorylation by CK1 and GRK2. Hh also promotes Gpr161 ciliary exit through Smo and β-arrestin. Smo is activated in the primary cilia by cholesterol and its derivatives oxysterols. Kif7, Gli and Sufu are localized at the ciliary tip where GliF is converted into GliA. The Evc1/Evc2 complex is localized near the ciliary base where it regulates Hh signal transduction downstream or in parallel to Smo in certain contexts. The nuclear import receptor Kapβ2 is required for Gli ciliary entry. See text for details.
Hh induces phosphorylation of Smo C-terminal intracellular tail (C-tail) by multiple kinases including PKA (Drosophila only), CK1 family kinases CK1α and CK1γ, and Gprk2/GRK2 (Fig. 1B) [78–85]. Phosphorylation of Smo promotes its cell surface/ciliary accumulation and induces conformational change and dimerization of its C-tail that adopts an open and active conformation after Hh stimulation [78, 81, 83, 86]. The mechanism by which phosphorylation promotes Smo ciliary localization remains unknown; in Drosophila however, Smo phosphorylation promotes its cell surface accumulation by inhibiting the recruitment of the Smurf family of E3 ubiquitin ligases and a Cul4-DDB1-Gβ E3 ubiquitin ligase complex [74, 87]. Hh signaling also increases the production of phosphatidylinositol 4-phosphate that binds Smo C-tail to promote Smo phosphorylation and cell surface/ciliary localization [88]. A recent study identified a membrane-tethered ubiquitination system consisting of a transmembrane protein MEGF8 and a RING superfamily E3 ligase MGRN1 that regulates mammalian Hh signaling and heart development by catalyzing the ubiquitination and degradation of Smo [89].
2.3. Hh signal transduction from Smo to Gli
Smo is a class F GPCR family member and can engage in Gαi activation in both Drosophila and mammalian cells [62, 90, 91]. Although Gαi is required for the expression of Hh target gene decapentaplegic (dpp) in Drosophila wing imaginal discs [91], whether Gαi plays a physiological role in canonical Hh signaling in mammals has remained unclear. In both Drosophila and mammals, Smo acts through its C-tail to regulate Gli/Ci. In Drosophila, Smo C-tail (SmoC) directly interacts with a multi-protein signaling complex containing Ci, the kinesin-like protein Costal 2 (Cos2) and the Ser/Thr protein kinase Fused (Fu) [92–95], and this interaction depends on the phosphorylation of SmoC by PKA, CK1 and GRK2, which triggers a conformational switch of SmoC to expose a binding pocket for Cos2-Fu [86, 96]. In addition, phosphorylation induces dimerization/oligomerization of SmoC, leading to clustering of the bound Cos2-Fu that triggers trans-autophosphorylation and activation of Fu kinase [96–98]. In the absence of Hh, Cos2-Fu serves as a molecular scaffold to bring Ci in close proximity to its kinases PKA, GSK3 and CK1, leading to efficient phosphorylation of multiple sites in its C-terminal region to generate a degron for the modular E3 ubiquitin ligase containing Slimb/βTRCP [99–104]. SCFSlimb/βTRCP catalyzes the ubiquitination of Ci, followed by proteosome-mediated partial degradation to generate a C-terminally truncated CiR (Fig. 1B) [101, 104, 105]. In the presence of Hh, activated Smo attenuates the Cos2-Ci-PKA-CK1 complex formation [103, 106], thus inhibiting Ci phosphorylation, ubiquitination, and proteolytic processing. However, blocking Ci processing is insufficient to convert accumulated full-length Ci (CiF) into CiA because the activity of CiF is blocked by additional mechanisms including cytoplasmic retention by Cos2 and proteolysis-independent inhibition by PKA phosphorylation [107–110]. Furthermore, Ci is inhibited by forming a stoichiometric complex with Sufu [111], which inhibits Ci nuclear localization and blocks the recruitment of Ci coactivator CBP even after Ci enters the nucleus [112, 113]. Previous genetic studies suggest that Fu activates Ci by antagonizing Sufu, leading to the maturation of CiF into a labile CiA [111]. A recent study demonstrated that Fu activated Ci by directly phosphorylating Ci on multiple sites, priming its further phosphorylation by CK1 on adjacent sites, and that these phosphorylation events altered Ci/Sufu interaction (Fig. 1B) [114]. CiA is short-lived and is ubiquitinated and degraded by a modular E3 ubiquitin ligase containing HIB/SPOP, leading to termination of Hh pathway activity (Fig. 1B) [115–117].
Compared to Drosophila Hh signal transduction pathway, the immediate signaling events downstream of Smo are poorly understood although phosphorylation of mammalian Smo (mSmo) C-tail by CK1 and GRK2 has been implicated in Shh signal transduction in a manner analogous to Drosophila Smo (dSmo) [83, 86, 118]. Nevertheless, the mechanism for CiR/GliR production is conserved between Drosophila and mammals and involves the phosphorylation of Gli2 and Gli3 by PKA, GSK3 and CK1, followed by SCFSlimb/βTRCP-mediated proteolytic processing [119–121]. A PDD (processing determinant domain) domain located between the Zn-finger DNA binding and Slimb/β-TRCP binding domains of Ci/Gli appears to be critical for proteasome-mediated degradation that selectively removes its C-terminal half [122]. Deletion of this domain from Ci blocks the production of CiR and renders complete degradation of Ci [123]. Gli3 is processed more efficiently than Gli2 into a truncated repressor form probably due to a more potent PDD, and Gli1 lacks a PDD and does not exhibit repressor activity [124]. The generation of GliR requires Sufu as well as Kif7, the mammalian homolog of Cos2, and Sufu recruits GSK3 for efficient Gli3 processing [125–132]. Similar to inhibition of CiA by PKA [107], there is evidence that PKA can inhibit the activity of GliA by directly phosphorylating Gli2 [133, 134]. In addition, association of Sufu with Gli inhibits GliA activity, which is reversed upon Hh stimulation [34, 114, 130, 135, 136]. A recent study revealed that an Itch/β-arrestin2 complex binds Sufu to induce its Lys63-linked polyubiquitylation, which increases its association with Gli3 and converts the full-length Gli3 into a repressor of Hh signaling, and that several Sufu mutants found in medulloblastoma patients are insensitive to Itch [137]. Another study showed that the E3 ubiquitin ligase SCFFbxl17 binds Sufu and promotes its poly-ubiquitination and degradation, leading to increased Hh pathway activity, and that Fbxl17 expression is high in Shh subgroup of medulloblastoma [138]. Sufu also has a positivity role by protecting full-length Gli from SPOP-mediated degradation [129, 139], which explains why loss of Sufu resulted in less dramatic Hh pathway activation phenotypes than loss of Ptc [140, 141].
The production of GliR depends on the primary cilia (Fig. 2)[10]. Consistent with this, the key cellular components responsible for Gli processing including PKA and proteosome are enriched at the ciliary base [133, 142–144]. In addition, Gpr161, a GPCR receptor coupled to Gαs is localized in the primary cilia, which is responsible for the local activation of PKA [145]. As a consequence, in Gpr161 mutant mice, GliR production was blocked, leading to phenotypes indicative of constitutive Hh pathway activation [145]. Hh induces the ciliary exit of Gpr161 through β-arrestin, which is recruitment to Gpr161 after it is phosphorylated by GRK2 [146]. In contrast to Gpr161, another ciliary localized orphan GPCR Gpr175 positively modulates Hh signaling by decreasing cAMP and Gli3 repressor levels via Gαi [147], suggesting that multiple ciliary localized GPCRs may regulate local cAMP and PKA activity to govern GliR production.
In response to Hh stimulation, Smo is accumulated along the entire length of primary cilia while the intracellular signaling components including Gli proteins, Sufu, and Kif7 are enriched at the tip of primary cilia where full-length Gli is thought to be converted into GliA and subsequently dissociates from Sufu and translocates to the nucleus (Fig. 2) [65, 127–129, 135, 148–150]. Like Cos2, Kif7 also interacts with Gli and has both positive and negative roles in Hh signaling [126–128]. A recent study suggested that Kif7 binds the plus end of microtubules to organize the formation of cilium tip compartment, thereby indirectly influencing Gli activation [151]. However, the mechanism by which GliF is converted into GliA at the ciliary tip has remained a mystery. In zebrafish, Fu is required for the conversion of GliF into GliA [152]; however, knockout of the Fu homolog Stk36 in mice did not cause any discernible defects Hh signaling during embryonic development [153, 154]. A recent study revealed that another Fu-related kinase Ulk3 acted semi-redundantly with Stk36 to promote Gli activation in cultured cells by directly phosphorylating a conserved site in the N-terminal region of Gli proteins [114]. In addition, phosphorylation of Gli by Ulk3/Stk36 requires Gli ciliary localization, which depends on the PY-NLS/karyopherin-beta2 nuclear import system [155]. It would be important to determine whether Ulk3 is localized to the primary cilia in response to Hh and whether Kif7 is required for the activation of Ulk3.
Additional components acting between Smo and Gli to transduce Hh signal in mammalian cells have been identified. Dlg5 was identified as a Smo interacting protein that localizes to the ciliary base where it is associated with Kif7 [156]. Dlg5 is specifically required for the formation of GliA but not for the inhibition of GliR by Hh; consistent with this, depletion of Dlg5 did not affect Gpr161 ciliary exit but reduced ciliary tip localization of Kif7 and Gli2 in response to Hh [156]. EVC and EVC2, the products of human disease genes responsible for the Ellis-van Creveld syndrome characterized by impaired Hh signaling in skeletal, cardiac, and orofacial tissues [157, 158], form a complex with Smo in response to Hh [159–161]. Interestingly, EVC2-Smo is localized in the proximal region of primary cilia, which is required for Hh pathway activation [159]. Inactivation of EVC/EVC2 did not affect Smo phosphorylation and ciliary accumulation but impaired Gli ciliary localization and activation, suggesting that EVC/EVC2 acts downstream or in parallel with Smo [159–161]. Protein purification and mass spectrometry identified two ciliary proteins EFCAB7 and IQCE that formed a complex with EVC-EVC2 to regulate Hh signaling by tethering the EVC-EVC2 complex to the base of primary cilia [162]. Centrosome-localized aPKC functions as a positive regulator of Hh signaling by phosphorylating Gli1 to increase its DNA binding activity and association with HDAC1 in basal cell carcinomas (BCCs) [163, 164]. Other kinases, including DYRK1, DYRK2, MAP3K10, Cdc2l1, Ulk3, S6K1, Plk1, and CK2 have also been identified to influence Gli activity [165–172]. Beside phosphorylation, other PTMs including ubiquitination, sumoylation, acetylation, and methylation also modulate Gli activity and Hh signaling [173–177]. A recent study revealed that a lamina-associated polypeptide 2 (LAP2) nuclear chaperoning system regulates Gli1 movement between the nuclear lamina and nucleoplasm to achieve optimal Gli1 activation in BCCs [178].
2.4. Gli action in the nucleus
Numerous studies have revealed the differential employment of GliA and GliR in various contexts during mammalian embryogenesis [2]. While GliA levels are central to cancer formation [173, 179], the involvement of GliR in repressing cancer has been implicated by studies linking primary cilia to Hh pathway-dependent tumorigenesis [180, 181]. Several genome-wide studies on Gli target genes revealed that although many target promoters contain a Gli binding consensus related to the sequence TGGGTGGTC, other target genes may not require this consensus sequence for Gli-dependent transcriptional regulation [182, 183]. Hh signaling promotes tumorigenesis by regulating genes involved in cell growth, proliferation and survival including Myc, CycD1 and D2, and Bcl2 [184]. Several cofactors have been implicated in the transcriptional regulation by Gli proteins, including the SAP18-mSin3 corepressor complex [185], the chromatin remodeling factor Brg [186], NuRD corepressor complex subunit p66b and Myc-binding protein Mycbp [187], the chromatin-associated SAFB-like transcription modulator SLTM [188], the H3K27me3-specific demethylases Jmjd3/Kdm6b [189], and BET bromodomain proteins [190]. Although Hh target genes vary depending on developmental contexts, Hh signaling universally activate Ptch1 and Gli1 in all cell types, which function as negative and positive feedback mechanisms, respectively. Therefore, Ptch1 and Gli1 upregulation serves as a signature for mammalian Hh pathway activation.
3. Hedgehog signaling in cancer
The Hh pathway has been implicated in the maintenance of stem/progenitor cells in many adult tissues, including the epithelia of many internal organs and brain [4, 191]. Not surprisingly, abnormal Hh pathway activation in many of these tissues is associated with tumorigenesis. Mutations in Hh pathway components, including Ptch1, Smo, and Sufu leading to constitutive and ligand-independent pathway activation, have been linked to basal cell carcinoma, medulloblastoma, and several other types of cancer (Fig. 3A)[192]. By contrast, ligand-dependent pathway activation has been implicated in a wide variety of cancers including gastrointestinal tumors, prostate and pancreatic cancers (Fig. 3B) [191]. These tumors generally do not harbor Hh pathway mutations and their growth can be effectively suppressed by various pathway inhibitors, such as Hh-neutralizing antibodies or Smo antagonists. These findings lead to the model that Hh ligands produced by these tumors and/or their stromal environment act on tumors to maintain stem/progenitor cells in the tumors in an undifferentiated, proliferative state. However, increasing evidence suggests that Hh signaling can also act in a paracrine fashion to promote the tumor microenvironment essential for tumor growth [193]. Thus, Hh signaling activity can influence tumor growth through both cell autonomous (acting on cancer cells) and non-autonomous (acting on stromal cells) mechanisms (Fig. 3B).
Figure 3. Hh signaling in cancer.
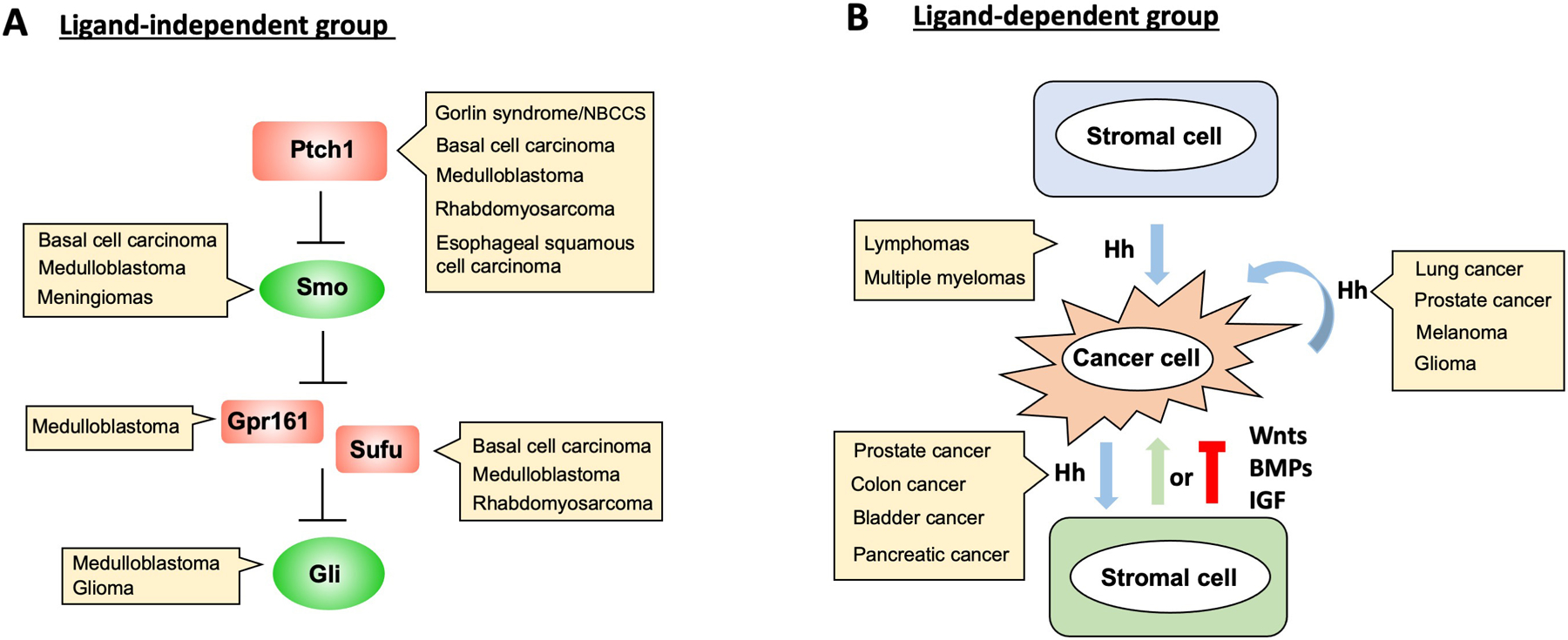
(A) In ligand-independent group of cancers, Hh pathway components are mutated or amplified (as in the case of Gli), leading constitutive pathway activation. (B) In ligand-independent group of cancers, Hh derived from cancer cells act on stromal cells or cancer cells to regulate cancer progression. Cancer cells can also be regulated by Hh derived from stromal cells as in the case of lymphomas and multiple myelomas.
3.1. Hh signaling in basal cell carcinoma
Basal cell carcinoma (BCC) is the most common skin cancer in western world. Aberrant Hh signaling was initially linked to BCC by the findings that germline mutations in Ptch1 are the culprits of Gorlin syndrome (also called nevoid BCC syndrome or NBCCS), which is characterized by predisposition of BCCs as well as other cancers [194, 195]. Subsequently, somatic mutations in Ptch1 and Smo were frequently identified in sporadic BCCs [196, 197]. Both germline and somatic mutations in Sufu were also identified in BCC albeit at much lower frequency compared to Ptch1 mutations [197]. The causal relationship of aberrant Hh signaling in BCC was established by studies showing that either loss of Ptch1 and gain of Smo drive BCC in mice [196, 198]. Lineage tracing experiments indicated that oncogenic activation of Smo in the interfollicular epidermis stem cells (IFE-SCs) but not hair follicle bulge stem cells induced BCC, identifying the interfollicular stem cells as the origin of BCC in mice [199]. Further study revealed that Smo activation in IFE-SCs drove more potent tumor growth than Smo activation in progenitor (PCs) cells because SCs have higher capability of symmetric self-renewing divisions and higher P53-dependent resistance to cell death compared with PCs [200].
3.2. Hh signaling and Medulloblastoma
Another type of cancer that Hh pathway mutations are frequently identified is medulloblastoma (MB), the most common malignant pediatric brain tumor. A major subtype of MBs (Shh group) is characterized by abnormal Hh pathway activation duo to mutations in Ptch1, Smo, and Sufu or amplification of Gli1, Gli2, and N-Myc [201–203]. Studies of Ptch1 heterozygous Gorlin syndrome patients, as well as analogous mutant mice, have strongly suggested that Hh pathway activation is critical for the transformation of granule cell precursors (GPCs) [204]. Experimental Hh pathway activation either by Ptch1 deletion or overexpression an oncogenic form of Smo in mouse neural stem cells or restricted neural progenitors revealed that medulloblastoma is derived from lineage restricted GPCs [205, 206]. Furthermore, single cell RNA-seq of developing mouse cerebellum showed that childhood SHH-MB transcriptionally mirrors the granule cell lineage, suggesting that GCPs are also the cell of origin of Shh-MB in humans [207]. Another recent single-cell transcriptome analysis of Shh-MB identified Olig2-expressing glial progenitors as transit-amplifying cells at the tumorigenic onset and showed that Olig2 promotes tumorigenesis by activating oncogenic networks including Hippo-Yap/Taz and Aurora-A/MycN pathways, implying Olig2-driven oncogenic networks as potential therapeutic targets for Shh-MB [208].
Although Sufu mutations are found in human MBs, loss of Sufu is insufficient to induce MBs in mice [209]. However, a recent study showed that deletion of SPOP together with Sufu in mice resulted in MB formation in a manner depending on Gli2 [139]. Further analysis revealed that the bHLH transcription factor Atoh1, which is a target gene of Gli2, cooperates with Gli2 to activate core Shh MB signature genes [139]. Germline mutations in Gpr161 as well as GNAS, which encodes Gαs, have been identified in infant-onset Shh-MB [210, 211]. Deletion of Gpr161 or GNAS in mice is sufficient to induce Shh-MB, highlighting the importance of the cAMP-PKA axis in restricting Shh-MB [212, 213]. A recent large-scale cancer genomic study identified frequent somatic mutations in the splicing factor U1 snRNA that resulted in inactivation of Ptch1 or activation of Gli2 and CyclinD2 in about 50% of SHHMB [214]. Another recent study identified loss-of-function mutations in the elongation factor ELP-1 leading to translation deregulation in 14% pediatric Shh-MB patients, suggesting that disruption of mRNA translation and protein homeostasis is a novel pathological mechanism for Shh-MB [215].
3.3. Hh signaling in other cancers
Beside BCC and MB, Hh pathway mutations have also been identified, albeit at much lower frequency, in several other cancers including rhabdomyosarcoma [216, 217], the most common pediatric soft tissue sarcoma, esophageal squamous cell carcinoma [218], and meningioma [219]. Furthermore, abnormal Hh pathway activation in the absence of pathway mutations has been implicated in a wide range of human malignancies including but not limited to lung cancer [220–222], prostate cancer [223–225], pancreatic [226–228], colon cancer [229], bladder cancer [230, 231], esophageal cancer [226, 232, 233], gastric cancer [226, 234, 235], liver cancer [236–238], melanomas [239, 240], gliomas [241–243], breast cancer [244–247], ovarian cancer [248], and hematological malignancies [249–251]. In most cases, elevated Hh ligand expression has been observed and transgenic expression of Hh in mice is sufficient to promote tumorigenesis [233, 252]; therefore, these cancers are classified as “ligand-dependent” group (Fig. 3B). Initial studies suggest Hh acts in an autocrine fashion to promote the growth of tumors in the ligand-dependent group [220, 226]; however, several later studies using xenograft models revealed that tumor cells did not activate Hh pathway but rather stromal cells that surround the tumor cells responded to Hh and that deletion of Smo in the stromal microenvironment inhibited tumor growth, suggesting that tumor-derived Hh acts on tumor microenvironment to promote tumor growth in a paracrine fashion [193, 253, 254].
An early study suggested that Hh-Gli signaling acts in human colon cancer cells to promote their growth, recurrence, metastasis and stem cell survival and expansion [229]. Later studies using mouse models of intestinal tumors revealed context dependent role of Hh signaling in stroma cells [255, 256]. In a mouse model of colitis-associated colon cancer, stroma-specific Hh activation markedly reduced the tumor growth and blocked progression of advanced neoplasms partly via the modulation of BMP signaling whereas attenuation of Hh signaling accelerated tumorigenesis [256]. By contrast, Hh pathway activation by Sufu reduction in gut mesenchyme promoted intestinal tumorigenesis in APCMin/− mice in part by modulating the expression of niche signals such as Wnts whereas reduction of Hh pathway activity by removing one copy of Gli2 suppressed the tumorigenesis [257]. Hence, stromal Hh signaling can either restrain or promote intestinal tumorigenesis depending on the context. Similarly, evidence for both autocrine and paracrine mechanisms for lung cancer and prostate cancer exists [193, 222, 224, 258, 259], suggesting that the mode of Hh action may vary depending on cancer subtypes and/or stages.
Tumor suppressor function of paracrine Hh signaling has also been observed in other cancers including bladder and pancreatic cancers. Shh-expressed basal cells are thought to be the cell of origin from which bladder cancer is derived [260]. Despite its initial presence in the cancer cell of origin, Shh expression is invariably lost during progression to invasive urothelial carcinoma, implying that Hh signaling could be incompatible with cancer progression [261]. In mouse models of bladder cancer, deletion of Smo from stroma dramatically accelerated cancer progression and reduced survival time due to reduced stromal expression of BMPs, which are urothelial differentiation factors, suggesting that Hh signaling restrains bladder cancer progression by stimulating stromal production of differentiation factors [261]. Whether Hh plays a similar role in human bladder cancer awaits to be determined.
Pancreatic ductal adenocarcinoma (PDA) is associated with upregulation of Shh and preclinical studies suggest that pharmacologic blockade of Hh signaling with Smo antagonists or Shh ligand-blocking antibody can reduce the growth and distant metastases of human pancreatic cancer xenografts [193, 226, 262, 263]. Activation of Gli2 can cooperate with Ras signaling to drive the formation of pancreatic intraepithelial neoplasia, the earliest stages of human PDA tumorigenesis [228]. In addition, elevated Gli2 is observed in basal-like PDA cell lines and human specimens and correlates with poor prognosis, and Gli2-mediated basal-like subtype switching can rescue PDA cell viability after KRAS ablation [264]. However, deletion of Smo in the pancreatic epithelium does not affect PDA pathogenesis in a genetically engineered mouse model of pancreatic cancer [265]. Nevertheless, Gli1, which is regulated by TGFβ and KRAS, is required both for survival and for the KRAS-mediated transformed phenotype of cultured PDA cancer cells [265]. Furthermore, Smo-independent elevation of Gli1 and Gli2 has been shown to drive chemotherapy resistance in PDA through upregulating Sox2 [266], suggesting that non-canonical and Smo-independent activation of Gli in cancer cells is essential for PDA progression and drug resistance. Modulation of Hh signaling in stroma revealed that stromal response to Hedgehog signaling restrains rather than promoting pancreatic cancer progression [267]. Consistent with this finding, high SHH levels correlate with the well-differentiated classical subtype of PDA and longer disease-free survival of PDA patients [264]. These findings may explain why clinical trial using Smo inhibitor to treat pancreatic cancer patient failed [268].
4. Targeting Hh pathway for cancer treatment
The importance of Hh signaling in a wide range of human cancers has stimulated great interest in targeting this pathway for cancer therapy. Unbiased screens using various pathway readouts as well as rational designs based on the understanding of the Hh signaling mechanism have yielded a garden-variety of pathway antagonists that block Hh signal transduction at different steps (Fig. 4), including Hh acyltransferase inhibitors [269], small molecule Hh inhibits [270–272], Smo inhibitors [273–277], and Gli inhibitors [278–280]. Multiple pathway inhibitors have entered clinical trials for many solid tumors and blood cancers and several Smo inhibitors have been approved by FDA for the treatment of advanced and metastatic BCC [281].
Figure 4. Multiple strategies for inhibiting Hh signal transduction pathway.
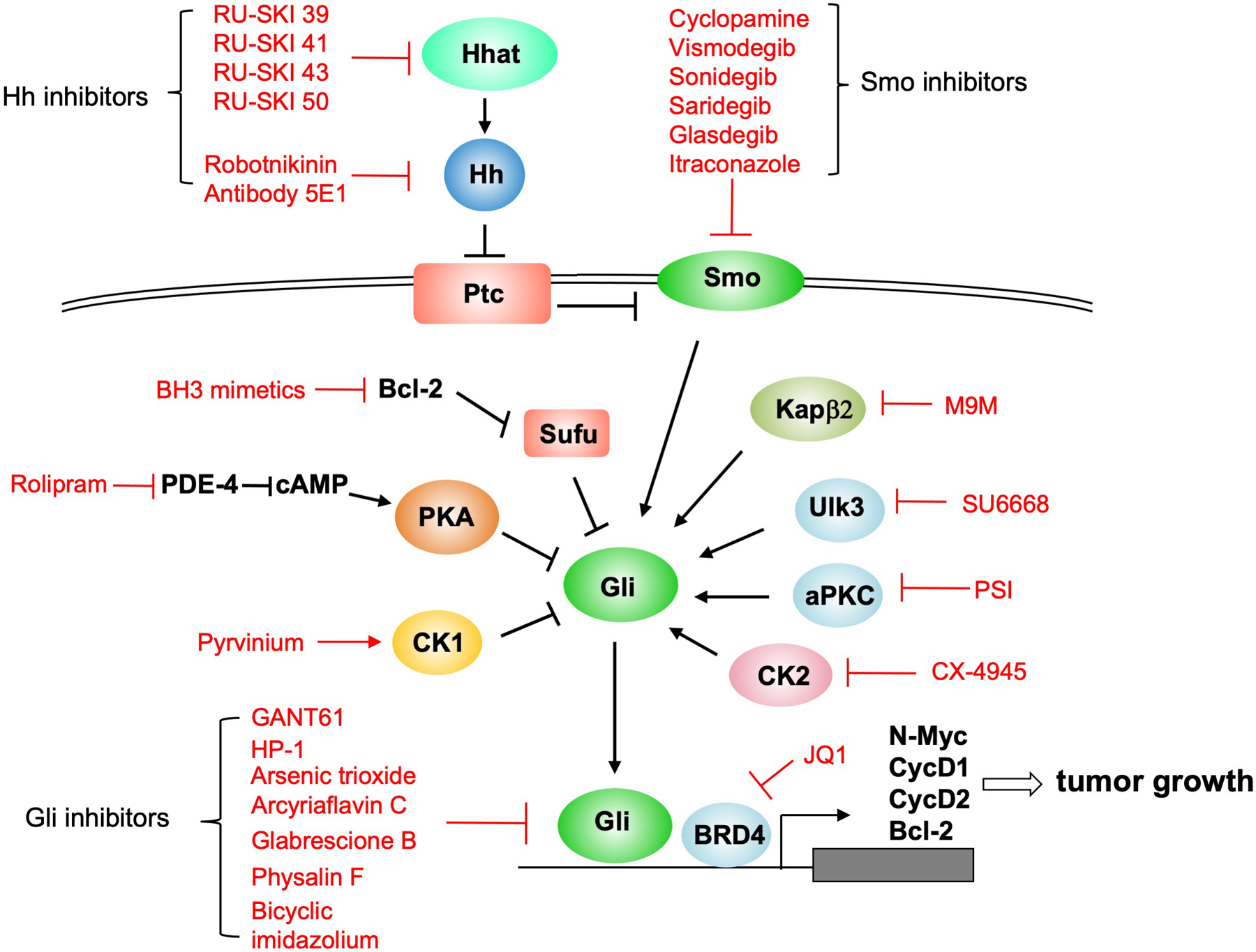
Hh signal transduction can be blocked at multiple steps including Hh inhibitors that either inhibit the activity of Hh acyltransferase (Hhat) or directly bind and inhibit Hh, Smo inhibitors, Gli inhibitors as well as pathway modulators that inhibit Gli by regulating its kinases or binding partners. See text for details.
4.1. Multiple strategies for Hh pathway inhibition
Being a GPCR family protein, Smo is the most prominent and druggable target in the Hh pathway. The first identified Smo antagonist is cyclopamine, a plant-derived steroidal alkaloid that inhibits Hh signaling by binding to Smo [273, 274]. Since its discovery, cyclopamine has been employed as a main Hh pathway blocker by numerous studies to test the requirement of Hh signaling for cancer cell growth. Nevertheless, treating cells with high dose of cyclopamine can lead to cytotoxicity that could compromise the interpretation of the results. Later, more potent and specific Smo inhibitors have been developed and entered clinic trials, which include GDC-0449/Vismodegib [282], LDE225/Sonidegib [283], IPI-926/Saridegib [284], itraconazole[285], and PF-04449913/Glasdegib [286]. However, cancer cells treated with Smo inhibitors can quickly develop resistance. In addition, tumors baring constitutive Hh pathway activity independent of Smo are insensitive to Smo inhibitors, making it necessary to develop alternative approaches to block the Hh pathway downstream of Smo.
Cell-based screens identified several compounds such as GANT61 and HP-1 as well as natural products including Arcyriaflavin C, Physalin F and Glabrescione B that selectively inhibit Gli-dependent transcription [278–280, 287]. In addition, Arsenic trioxide (ATO) can reduce the stability and ciliary accumulation of GLI2 and bind Gli1 to inhibit its transcription activity [288, 289]. A recent screen for small molecules that inhibit endogenous Gli activity in Sufu −/− MEFs identified a family of bicyclic imidazolium derivatives that can inhibit Gli-dependent transcription without affecting its ciliary trafficking or proteolytic processing [290].
Targeting the mechanisms that regulate Gli can also inhibit Gli-dependent transcription and tumorigenesis. For example, modulating the cAMP-PKA axis by rolipram, which elevates cAMP levels by selectively inhibiting phosphodiesterase-4 (PDE-4), inhibited Gli activity in granule neuron precursors [212]. Inhibitors of Gli activation kinases including aPKC and Ulk3 can attenuate Gli transcriptional activity and inhibit tumor growth [163, 291]. As CK1 acts downstream of PKA to phosphorylate Gli and promote its instability, pyrvinium, an FDA-approved anti-pinworm compound that is a potent CK1 agonist, can inhibit Hh pathway activity downstream of Smo and Sufu [292]. A phosphoproteomics study identified CK2 as critical for the stabilization and transcriptional activity of Gli2 in granule neuron precursors and demonstrated that CK2 inhibitors such as CX-4945 can decrease the viability of primary Shh-type MB patient cells in culture and block the growth of smo inhibitor-resistant MB tumors in mice [171]. A gain-of-function screen identified Bcl-2 family proteins as inhibitors of Sufu [293]. Bcl-2 proteins directly bind Sufu to disrupt its inhibition of Gli activity essential for tumor growth and small molecule BH3-mimetics can inhibit Gli by interfering with Bcl-2/Sufu interaction [293]. Another study found that BET domain proteins such as BRD4 bind Gli1 and Gli2 gene promoters to promote their transcription and that a small molecule inhibitor of BRD family proteins JQ1 can inhibit Hh-driven tumors including BCC and MB even when these tumors harbor genetic lesions rendering them resistant to Smo inhibitors [190]. Finally, ciliary localization and activation of Gli proteins are regulated by the PY-NLS/Karyopherin-β2 (Kapβ2) nuclear import machinery and inhibition of Kapβ2 by a small peptide M9M can attenuate Gli activation [155, 294, 295].
4.2. Hh pathway inhibitors in the clinics
Hh pathway inhibitors including multiple Smo antagonists such as vismodegib, sonidegib, and glasdegib have entered clinical trials for BCCs and basal cell nevus syndrome [296–299], pediatric and adult MBs [300–302], other solid cancers including small cell lung cancers [303, 304], pancreatic cancers [305–307], ovarian cancer [308], prostate cancer [309, 310], and colorectal cancer [311], as well as hematological malignancies such as acute myeloid leukemia (AML) [312–317]. The Gli inhibitor arsenic trioxide (ATO) is currently in clinical trials for treatment of Smo inhibitor-resistant BCC as well as other cancers including glioma and neuroblastoma [318]. The Smo inhibitors vismodegib and sonidegib have been approved by FDA for the treatment of locally advanced BCC in 2012 and 2015, respectively, with vismodegib also approved for metastatic BCC [281, 319]. In 2018, FDA approved glasdegib in combination with low dose cytarabine (LDAC) for the treatment of newly diagnosed AML or high-grade myelodysplastic syndrome in elderly patients or patients not suitable for intensive chemotherapy [320]. However, Hh pathway inhibitors have limited single agent activity in unselected patients with other types of cancers in early phase clinical trials [318].
4.3. Mechanisms of therapeutic resistance
Despite the high potency of Smo inhibitors in preventing BCC and MB progression in animal models and human patients, acquired drug resistance rapidly emerged, which limits long-term efficacy [321, 322]. Both genetic alterations in Smo and compensatory mechanisms downstream of Smo can contribute to the resistance to Smo inhibition. Genomic analyses of resistant tumors identified point mutations in Smo drug-binding pocket or Smo-activating mutations that account for ~ 50% of the resistant BCCs [321, 323, 324]. Genomic loss of Sufu and amplification of Gli2 or the Hh target gene Cyclin D1 have also been observed in the resistant cancer cells [324–326]. Noncanonical mechanisms that activate Gli through PI3K, aPKC, and the SRF-MLK complex have also been shown to contribute to resistance to Hh pathway inhibitors, suggesting that targeting the corresponding noncanonical pathways could be strategies to overcome cancer resistance to Smo inhibitors [163, 325, 327]. A recent study found that JNK/AP-1 and TGFβ/Smad3 pathways cooperatively activate the nuclear myocardin-related transcription factor (nMRTF), which binds SRF and acts as a coactivator for Gli1 in resistant BCCs [328], suggesting that a combinatory treatment with Smo and AP-1 inhibitors could be beneficial.
In addition to genetic and epigenetic alternations leading to persistent Gli activation that allows tumors to evade Smo inhibition, lineage plasticity and cell fate switch also contribute to drug resistance and tumor reoccurrence. In several BCC patients treated with vismodegib, invasive squamous cell carcinoma emerged from BCC [329, 330], and in one case, the recurrent lymph-node squamous cell carcinoma harbored new mutations in genes that are commonly mutated in cutaneous squamous cell carcinoma including Notch1/2 and KMT2C/MLL3, indicating that these mutations may drive BCC to SCC switch and resistance to vismodegib treatment [330]. Other studies revealed that SCCs derived from BCCs in patients treated with vismodegib activated the RAS/MAPK pathway and that loss of primary cilia appeared to be responsible for the switch from Hh to RAS/MAPK pathway in resistant tumors [331, 332]. Two recent studies using mouse models of BCC showed that Smo inhibition drove tumor evolution from Hh-dependent to Wnt-dependent tumors, suggesting that dual pathway inhibition of Hh and Wnt signaling could be a clinically relevant strategy for overcoming tumor relapse in BCC [333, 334].
Finally, Hh pathway components have been implicated in chemotherapy resistance of cancer. For example, one study suggested that high levels of Ptc in cancer cells may contribute to chemotherapy resistance by promoting multidrug efflux [335]. Another study showed that, in a phase II clinic trial of treating AML patient with ribavirin, Gli1 and the UDP glucuronosyltransferase (UGT1A) family of enzymes are elevated in resistant cells [336]. Further experiments demonstrated that Gli1 is sufficient to drive UGT1A-dependent glucuronidation of ribavirin and thus drug resistance, and that genetic or pharmacological inhibition of Gli1 can overcome drug resistance [336].
5. Conclusion
Hh signaling pathway represents one of the best examples of how basic research initiated from model organisms such as fruit fly can be translated into novel strategies for cancer treatment. Despite decades of intensive investigation, Hh signal transduction mechanism is still not fully understood. For example, there are gaps between Smo and Gli in the mammalian pathway and the precise mechanism by which primary cilia orchestrate Hh signal transduction has remained poorly understood. In addition, how Gli proteins are regulated in the nucleus and how they influence chromatin landscape or communicate with basal transcriptional machinery have largely unexplored. Understanding the detailed molecular and biochemical mechanisms of these processes may provide new avenues for targeting the Hh pathway downstream of Smo. Although many Smo antagonists have entered clinic trial and several have been proved by FDA for cancer treatment, most of the downstream pathway inhibitors including small molecule Gli inhibitors are not suitable for clinical trial due to cytotoxicity or poor bioavailability; therefore, developing clinically actionable pathway inhibitors downstream of Smo has remained a high priority. This is an urgent issue because cancer patients treated with Smo inhibitors can quickly develop drug resistance and many cancers are driven by Smo-independent Gli activation. To overcome potential bypass mechanism or drug resistance, combinatory use to Smo inhibitors with other pathway inhibitors could provide a more effective way to treat cancer. Although Smo inhibitors have been effective in treating ligand-independent cancers bearing Ptch1 or Smo mutations, early clinical trials using Smo inhibitors to treat ligand-dependent cancers where no Hh pathway mutations have been identified were disappointing. In the future clinical trials, cancer patients need to be stratified with better pathway biomarkers to ensure that their cancers are driven by activated Smo. In addition, better tumor models such as patient-derived xenograft (PDX) models and patient-derived tumor organoids can be used to test whether Hh pathway inhibitors, either alone or in combination with other pathway inhibitors, can inhibit tumor growth.
Acknowledgments
Research in J.J.’s lab is supported by grants from NIH (GM118063) and Welch foundation (I-1603). I apologize to those colleagues whose work could not be cited due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
The author declares that there are no conflicts of interest.
Reference:
- 1.Ingham PW, McMahon AP. Hedgehog signaling in animal development: paradigms and principles. Genes And Development. 2001;15(23):3059–87. doi: 10.1101/gad.938601. [DOI] [PubMed] [Google Scholar]
- 2.Jiang J, Hui CC. Hedgehog signaling in development and cancer. Dev Cell. 2008;15(6):801–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briscoe J, Therond PP. The mechanisms of Hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol. 2013;14(7):418–31. Epub 2013/05/31. doi: 10.1038/nrm3598. [DOI] [PubMed] [Google Scholar]
- 4.Petrova R, Joyner AL. Roles of Hedgehog signaling in adult organ homeostasis and repair. Development. 2014;141(18):3458–71. doi: 10.1242/dev.083691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villavicencio EH, Walterhouse DO, Iannaccone PM. The sonic hedgehog-patched-gli pathway in human development and disease. Am J Hum Genet. 2000;67(5):1047–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nieuwenhuis E, Hui CC. Hedgehog signaling and congenital malformations. Clin Genet. 2005;67(3):193–208. [DOI] [PubMed] [Google Scholar]
- 7.Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411(6835):349–54. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- 8.Wilson CW, Chuang PT. Mechanism and evolution of cytosolic Hedgehog signal transduction. Development. 2010;137(13):2079–94. Epub 2010/06/10. doi: 137/13/2079 [pii] 10.1242/dev.045021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ingham PW, Nakano Y, Seger C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat Rev Genet. 2011;12(6):393–406. Epub 2011/04/20. doi: nrg2984 [pii] 10.1038/nrg2984. [DOI] [PubMed] [Google Scholar]
- 10.Bangs F, Anderson KV. Primary Cilia and Mammalian Hedgehog Signaling. Cold Spring Harb Perspect Biol. 2017;9(5). Epub 2016/11/25. doi: 10.1101/cshperspect.a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pandit T, Ogden SK. Contributions of Noncanonical Smoothened Signaling During Embryonic Development. J Dev Biol. 2017;5(4). Epub 2018/02/06. doi: 10.3390/jdb5040011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pietrobono S, Gagliardi S, Stecca B. Non-canonical Hedgehog Signaling Pathway in Cancer: Activation of GLI Transcription Factors Beyond Smoothened. Front Genet. 2019;10:556. Epub 2019/06/28. doi: 10.3389/fgene.2019.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter JA, Ekker SC, Park WJ, von Kessler DP, Young KE, Chen CH, et al. Hedgehog patterning activity: role of a lipophilic modification mediated by the carboxy-terminal autoprocessing domain. Cell. 1996;86(1):21–34. [DOI] [PubMed] [Google Scholar]
- 14.Chamoun Z, Mann RK, Nellen D, von Kessler DP, Bellotto M, Beachy PA, et al. Skinny hedgehog, an acyltransferase required for palmitoylation and activity of the hedgehog signal. Science. 2001;293(5537):2080–4. [DOI] [PubMed] [Google Scholar]
- 15.Amanai K, Jiang J. Distinct roles of Central missing and Dispatched in sending the Hedgehog signal. Development. 2001;128(24):5119–27. [DOI] [PubMed] [Google Scholar]
- 16.Micchelli CA, The I, Selva E, Mogila V, Perrimon N. Rasp, a putative transmembrane acyltransferase, is required for Hedgehog signaling. Development. 2002;129(4):843–51. [DOI] [PubMed] [Google Scholar]
- 17.Buglino JA, Resh MD. Hhat is a palmitoylacyltransferase with specificity for N-palmitoylation of Sonic Hedgehog. J Biol Chem. 2008;283(32):22076–88. Epub 2008/06/07. doi: 10.1074/jbc.M803901200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke R, Nellen D, Bellotto M, Hafen E, Senti KA, Dickson BJ, et al. Dispatched, a novel sterol-sensing domain protein dedicated to the release of cholesterol-modified hedgehog from signaling cells. Cell. 1999;99(7):803–15. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Zhang H, Litingtung Y, Chiang C. Cholesterol modification restricts the spread of Shh gradient in the limb bud. Proc Natl Acad Sci U S A. 2006;103(17):6548–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng X, Goetz JA, Suber LM, Scott WJ Jr., Schreiner CM, Robbins DJ. A freely diffusible form of Sonic hedgehog mediates long-range signalling. Nature. 2001;411(6838):716–20. [DOI] [PubMed] [Google Scholar]
- 21.Gallet A, Rodriguez R, Ruel L, Therond PP. Cholesterol modification of hedgehog is required for trafficking and movement, revealing an asymmetric cellular response to hedgehog. Dev Cell. 2003;4(2):191–204. [DOI] [PubMed] [Google Scholar]
- 22.Lewis PM, Dunn MP, McMahon JA, Logan M, Martin JF, St-Jacques B, et al. Cholesterol modification of sonic hedgehog is required for long-range signaling activity and effective modulation of signaling by Ptc1. Cell. 2001;105(5):599–612. [DOI] [PubMed] [Google Scholar]
- 23.Chen MH, Li YJ, Kawakami T, Xu SM, Chuang PT. Palmitoylation is required for the production of a soluble multimeric Hedgehog protein complex and long-range signaling in vertebrates. Genes And Development. 2004;18(6):641–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callejo A, Torroja C, Quijada L, Guerrero I. Hedgehog lipid modifications are required for Hedgehog stabilization in the extracellular matrix. Development. 2006;133(3):471–83. [DOI] [PubMed] [Google Scholar]
- 25.Gallet A, Ruel L, Staccini-Lavenant L, Therond PP. Cholesterol modification is necessary for controlled planar long-range activity of Hedgehog in Drosophila epithelia. Development. 2006;133(3):407–18. [DOI] [PubMed] [Google Scholar]
- 26.Vyas N, Goswami D, Manonmani A, Sharma P, Ranganath HA, VijayRaghavan K, et al. Nanoscale organization of hedgehog is essential for long-range signaling. Cell. 2008;133(7):1214–27. [DOI] [PubMed] [Google Scholar]
- 27.Cannac F, Qi C, Falschlunger J, Hausmann G, Basler K, Korkhov VM. Cryo-EM structure of the Hedgehog release protein Dispatched. Sci Adv. 2020;6(16):eaay7928. Epub 2020/06/05. doi: 10.1126/sciadv.aay7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, Liu Y, Li X. Structure of human Dispatched-1 provides insights into Hedgehog ligand biogenesis. Life Sci Alliance. 2020;3(8). Epub 2020/07/11. doi: 10.26508/lsa.202000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma Y, Erkner A, Gong R, Yao S, Taipale J, Basler K, et al. Hedgehog-mediated patterning of the mammalian embryo requires transporter-like function of dispatched. Cell. 2002;111(1):63–75. [DOI] [PubMed] [Google Scholar]
- 30.Callejo A, Bilioni A, Mollica E, Gorfinkiel N, Andres G, Ibanez C, et al. Dispatched mediates Hedgehog basolateral release to form the long-range morphogenetic gradient in the Drosophila wing disk epithelium. Proc Natl Acad Sci U S A. 2011;108(31):12591–8. Epub 2011/06/22. doi: 10.1073/pnas.1106881108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caspary T, Garcia-Garcia MJ, Huangfu D, Eggenschwiler JT, Wyler MR, Rakeman AS, et al. Mouse Dispatched homolog1 is required for long-range, but not juxtacrine, Hh signaling. Curr Biol. 2002;12(18):1628–32. Epub 2002/10/10. doi: 10.1016/s0960-9822(02)01147-8. [DOI] [PubMed] [Google Scholar]
- 32.Kawakami T, Kawcak T, Li YJ, Zhang W, Hu Y, Chuang PT. Mouse dispatched mutants fail to distribute hedgehog proteins and are defective in hedgehog signaling. Development. 2002;129(24):5753–65. [DOI] [PubMed] [Google Scholar]
- 33.Tukachinsky H, Kuzmickas RP, Jao CY, Liu J, Salic A. Dispatched and scube mediate the efficient secretion of the cholesterol-modified hedgehog ligand. Cell Rep. 2012;2(2):308–20. Epub 2012/08/21. doi: 10.1016/j.celrep.2012.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D’Angelo G, Matusek T, Pizette S, Therond PP. Endocytosis of Hedgehog through dispatched regulates long-range signaling. Dev Cell. 2015;32(3):290–303. Epub 2015/01/27. doi: 10.1016/j.devcel.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 35.Hall ET, Cleverdon ER, Ogden SK. Dispatching Sonic Hedgehog: Molecular Mechanisms Controlling Deployment. Trends Cell Biol. 2019;29(5):385–95. Epub 2019/03/11. doi: 10.1016/j.tcb.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka Y, Okada Y, Hirokawa N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature. 2005;435(7039):172–7. Epub 2005/05/13. doi: 10.1038/nature03494. [DOI] [PubMed] [Google Scholar]
- 37.Matusek T, Wendler F, Poles S, Pizette S, D’Angelo G, Furthauer M, et al. The ESCRT machinery regulates the secretion and long-range activity of Hedgehog. Nature. 2014;516(7529):99–103. Epub 2014/12/05. doi: 10.1038/nature13847. [DOI] [PubMed] [Google Scholar]
- 38.Gradilla AC, Gonzalez E, Seijo I, Andres G, Bischoff M, Gonzalez-Mendez L, et al. Exosomes as Hedgehog carriers in cytoneme-mediated transport and secretion. Nat Commun. 2014;5:5649. Epub 2014/12/05. doi: 10.1038/ncomms6649. [DOI] [PubMed] [Google Scholar]
- 39.Panakova D, Sprong H, Marois E, Thiele C, Eaton S. Lipoprotein particles are required for Hedgehog and Wingless signalling. Nature. 2005;435(7038):58–65. [DOI] [PubMed] [Google Scholar]
- 40.Bischoff M, Gradilla AC, Seijo I, Andres G, Rodriguez-Navas C, Gonzalez-Mendez L, et al. Cytonemes are required for the establishment of a normal Hedgehog morphogen gradient in Drosophila epithelia. Nat Cell Biol. 2013;15(11):1269–81. Epub 2013/10/15. doi: 10.1038/ncb2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gonzalez-Mendez L, Seijo-Barandiaran I, Guerrero I. Cytoneme-mediated cell-cell contacts for Hedgehog reception. Elife. 2017;6. Epub 2017/08/22. doi: 10.7554/eLife.24045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Creanga A, Glenn TD, Mann RK, Saunders AM, Talbot WS, Beachy PA. Scube/You activity mediates release of dually lipid-modified Hedgehog signal in soluble form. Genes & development. 2012;26(12):1312–25. Epub 2012/06/09. doi: 10.1101/gad.191866.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wierbowski BM, Petrov K, Aravena L, Gu G, Xu Y, Salic A. Hedgehog Pathway Activation Requires Coreceptor-Catalyzed, Lipid-Dependent Relay of the Sonic Hedgehog Ligand. Dev Cell. 2020;55(4):450–67 e8. Epub 2020/10/11. doi: 10.1016/j.devcel.2020.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan D, Lin X. Shaping morphogen gradients by proteoglycans. Cold Spring Harbor perspectives in biology. 2009;1(3):a002493. Epub 2010/01/13. doi: 10.1101/cshperspect.a002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bandari S, Exner S, Ortmann C, Bachvarova V, Vortkamp A, Grobe K. Sweet on Hedgehogs: regulatory roles of heparan sulfate proteoglycans in Hedgehog-dependent cell proliferation and differentiation. Curr Protein Pept Sci. 2015;16(1):66–76. Epub 2015/02/19. doi: 10.2174/1389203716666150213162649. [DOI] [PubMed] [Google Scholar]
- 46.Taipale J, Cooper MK, Maiti T, Beachy PA. Patched acts catalytically to suppress the activity of Smoothened. Nature. 2002;418(6900):892–7. [DOI] [PubMed] [Google Scholar]
- 47.Qi X, Schmiege P, Coutavas E, Li X. Two Patched molecules engage distinct sites on Hedgehog yielding a signaling-competent complex. Science. 2018. Epub 2018/08/25. doi: 10.1126/science.aas8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qi X, Schmiege P, Coutavas E, Wang J, Li X. Structures of human Patched and its complex with native palmitoylated sonic hedgehog. Nature. 2018;560(7716):128–32. Epub 2018/07/12. doi: 10.1038/s41586-018-0308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gong X, Qian H, Cao P, Zhao X, Zhou Q, Lei J, et al. Structural basis for the recognition of Sonic Hedgehog by human Patched1. Science. 2018;361(6402). Epub 2018/06/30. doi: 10.1126/science.aas8935. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, Bulkley DP, Xin Y, Roberts KJ, Asarnow DE, Sharma A, et al. Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched. Cell. 2018;175(5):1352–64 e14. Epub 2018/11/13. doi: 10.1016/j.cell.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian H, Cao P, Hu M, Gao S, Yan N, Gong X. Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm. Nat Commun. 2019;10(1):2320. Epub 2019/05/28. doi: 10.1038/s41467-019-10234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Myers BR, Neahring L, Zhang Y, Roberts KJ, Beachy PA. Rapid, direct activity assays for Smoothened reveal Hedgehog pathway regulation by membrane cholesterol and extracellular sodium. Proc Natl Acad Sci U S A. 2017;114(52):E11141–E50. Epub 2017/12/13. doi: 10.1073/pnas.1717891115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudolf AF, Kinnebrew M, Kowatsch C, Ansell TB, El Omari K, Bishop B, et al. The morphogen Sonic hedgehog inhibits its receptor Patched by a pincer grasp mechanism. Nat Chem Biol. 2019;15(10):975–82. Epub 2019/09/25. doi: 10.1038/s41589-019-0370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petrov K, Wierbowski BM, Liu J, Salic A. Distinct Cation Gradients Power Cholesterol Transport at Different Key Points in the Hedgehog Signaling Pathway. Dev Cell. 2020;55(3):314–27 e7. Epub 2020/08/30. doi: 10.1016/j.devcel.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qi C, Di Minin G, Vercellino I, Wutz A, Korkhov VM. Structural basis of sterol recognition by human hedgehog receptor PTCH1. Sci Adv. 2019;5(9):eaaw6490. Epub 2019/09/27. doi: 10.1126/sciadv.aaw6490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang P, Nedelcu D, Watanabe M, Jao C, Kim Y, Liu J, et al. Cellular Cholesterol Directly Activates Smoothened in Hedgehog Signaling. Cell. 2016;166(5):1176–87 e14. Epub 2016/08/23. doi: 10.1016/j.cell.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Byrne EFX, Sircar R, Miller PS, Hedger G, Luchetti G, Nachtergaele S, et al. Structural basis of Smoothened regulation by its extracellular domains. Nature. 2016;535(7613):517–22. Epub 2016/07/21. doi: 10.1038/nature18934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao X, Tang JJ, Peng C, Wang Y, Fu L, Qiu ZP, et al. Cholesterol Modification of Smoothened Is Required for Hedgehog Signaling. Mol Cell. 2017;66(1):154–62 e10. Epub 2017/03/28. doi: 10.1016/j.molcel.2017.02.015. [DOI] [PubMed] [Google Scholar]
- 59.Raleigh DR, Sever N, Choksi PK, Sigg MA, Hines KM, Thompson BM, et al. Cilia-Associated Oxysterols Activate Smoothened. Mol Cell. 2018;72(2):316–27 e5. Epub 2018/10/20. doi: 10.1016/j.molcel.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Myers BR, Sever N, Chong YC, Kim J, Belani JD, Rychnovsky S, et al. Hedgehog pathway modulation by multiple lipid binding sites on the smoothened effector of signal response. Dev Cell. 2013;26(4):346–57. Epub 2013/08/21. doi: 10.1016/j.devcel.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang P, Zheng S, Wierbowski BM, Kim Y, Nedelcu D, Aravena L, et al. Structural Basis of Smoothened Activation in Hedgehog Signaling. Cell. 2018;174(2):312–24 e16. Epub 2018/05/29. doi: 10.1016/j.cell.2018.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi X, Liu H, Thompson B, McDonald J, Zhang C, Li X. Cryo-EM structure of oxysterol-bound human Smoothened coupled to a heterotrimeric Gi. Nature. 2019;571(7764):279–83. Epub 2019/06/07. doi: 10.1038/s41586-019-1286-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Deshpande I, Liang J, Hedeen D, Roberts KJ, Zhang Y, Ha B, et al. Smoothened stimulation by membrane sterols drives Hedgehog pathway activity. Nature. 2019;571(7764):284–8. Epub 2019/07/03. doi: 10.1038/s41586-019-1355-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi X, Friedberg L, De Bose-Boyd R, Long T, Li X. Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling. Nat Chem Biol. 2020;16(12):1368–75. Epub 2020/09/16. doi: 10.1038/s41589-020-0646-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DY, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437(7061):1018–21. [DOI] [PubMed] [Google Scholar]
- 66.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317(5836):372–6. [DOI] [PubMed] [Google Scholar]
- 67.Kinnebrew M, Iverson EJ, Patel BB, Pusapati GV, Kong JH, Johnson KA, et al. Cholesterol accessibility at the ciliary membrane controls hedgehog signaling. Elife. 2019;8. Epub 2019/10/29. doi: 10.7554/eLife.50051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Findakly S, Daggubati V, Garcia G, LaStella SA, Choudhury A, Tran C, et al. Sterol and oxysterol synthases near the ciliary base activate the Hedgehog pathway. J Cell Biol. 2021;220(1). Epub 2020/12/08. doi: 10.1083/jcb.202002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J, Hsia EY, Brigui A, Plessis A, Beachy PA, Zheng X. The role of ciliary trafficking in Hedgehog receptor signaling. Sci Signal. 2015;8(379):ra55. Epub 2015/06/04. doi: 10.1126/scisignal.aaa5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yue S, Tang LY, Tang Y, Tang Y, Shen QH, Ding J, et al. Requirement of Smurf-mediated endocytosis of Patched1 in sonic hedgehog signal reception. Elife. 2014;3. doi: 10.7554/eLife.02555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xia R, Jia H, Fan J, Liu Y, Jia J. USP8 promotes smoothened signaling by preventing its ubiquitination and changing its subcellular localization. PLoS biology. 2012;10(1):e1001238. Epub 2012/01/19. doi: 10.1371/journal.pbio.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li S, Chen Y, Shi Q, Yue T, Wang B, Jiang J. Hedgehog-regulated ubiquitination controls smoothened trafficking and cell surface expression in Drosophila. PLoS biology. 2012;10(1):e1001239. Epub 2012/01/19. doi: 10.1371/journal.pbio.1001239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang S, Zhang Z, Zhang C, Lv X, Zheng X, Chen Z, et al. Activation of Smurf E3 ligase promoted by smoothened regulates hedgehog signaling through targeting patched turnover. PLoS Biol. 2013;11(11):e1001721. Epub 2013/12/05. doi: 10.1371/journal.pbio.1001721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li S, Li S, Wang B, Jiang J. Hedgehog reciprocally controls trafficking of Smo and Ptc through the Smurf family of E3 ubiquitin ligases. Sci Signal. 2018;11(516). Epub 2018/02/14. doi: 10.1126/scisignal.aan8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Desai PB, Stuck MW, Lv B, Pazour GJ. Ubiquitin links smoothened to intraflagellar transport to regulate Hedgehog signaling. J Cell Biol. 2020;219(7). Epub 2020/05/22. doi: 10.1083/jcb.201912104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma G, Li S, Han Y, Li S, Yue T, Wang B, et al. Regulation of Smoothened Trafficking and Hedgehog Signaling by the SUMO Pathway. Dev Cell. 2016;39(4):438–51. Epub 2016/10/18. doi: 10.1016/j.devcel.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang J, Liu Y, Jiang K, Jia J. SUMO regulates the activity of Smoothened and Costal-2 in Drosophila Hedgehog signaling. Sci Rep. 2017;7:42749. doi: 10.1038/srep42749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jia J, Tong C, Wang B, Luo L, Jiang J. Hedgehog signalling activity of Smoothened requires phosphorylation by protein kinase A and casein kinase I. Nature. 2004;432(7020):1045–50. doi: 10.1038/nature03179. [DOI] [PubMed] [Google Scholar]
- 79.Zhang C, Williams EH, Guo Y, Lum L, Beachy PA. Extensive phosphorylation of Smoothened in Hedgehog pathway activation. Proc Natl Acad Sci U S A. 2004;101(52):17900–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Apionishev S, Katanayeva NM, Marks SA, Kalderon D, Tomlinson A. Drosophila Smoothened phosphorylation sites essential for Hedgehog signal transduction. Nat Cell Biol. 2005;7(1):86–92. [DOI] [PubMed] [Google Scholar]
- 81.Chen Y, Li S, Tong C, Zhao Y, Wang B, Liu Y, et al. G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes And Development. 2010;24(18):2054–67. Epub 2010/09/17. doi: 24/18/2054 [pii] 10.1101/gad.1948710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cheng S, Maier D, Neubueser D, Hipfner DR. Regulation of smoothened by Drosophila G-protein-coupled receptor kinases. Dev Biol. 2010;337(1):99–109. Epub 2009/10/24. doi: 10.1016/j.ydbio.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen Y, Sasai N, Ma G, Yue T, Jia J, Briscoe J, et al. Sonic Hedgehog dependent phosphorylation by CK1alpha and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9(6):e1001083. doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li S, Ma G, Wang B, Jiang J. Hedgehog induces formation of PKA-Smoothened complexes to promote Smoothened phosphorylation and pathway activation. Sci Signal. 2014;7(332):ra62. doi: 10.1126/scisignal.2005414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li S, Li S, Han Y, Tong C, Wang B, Chen Y, et al. Regulation of Smoothened Phosphorylation and High-Level Hedgehog Signaling Activity by a Plasma Membrane Associated Kinase. PLoS Biol. 2016;14(6):e1002481. doi: 10.1371/journal.pbio.1002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao Y, Tong C, Jiang J. Hedgehog regulates smoothened activity by inducing a conformational switch. Nature. 2007;450(7167):252–8. [DOI] [PubMed] [Google Scholar]
- 87.Li S, Cho YS, Wang B, Li S, Jiang J. Regulation of Smoothened ubiquitylation and cell surface expression through a Cul4-DDB1-Gbeta E3 ubiquitin ligase complex. J Cell Sci. 2018;131(15). Epub 2018/06/23. doi: 10.1242/jcs.218016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang K, Liu Y, Fan J, Zhang J, Li XA, Evers BM, et al. PI(4)P Promotes Phosphorylation and Conformational Change of Smoothened through Interaction with Its C-terminal Tail. PLoS Biol. 2016;14(2):e1002375. Epub 2016/02/11. doi: 10.1371/journal.pbio.1002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kong JH, Young CB, Pusapati GV, Patel CB, Ho S, Krishnan A, et al. A Membrane-Tethered Ubiquitination Pathway Regulates Hedgehog Signaling and Heart Development. Dev Cell. 2020;55(4):432–49 e12. Epub 2020/09/24. doi: 10.1016/j.devcel.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Riobo NA, Saucy B, Dilizio C, Manning DR. Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci U S A. 2006;103(33):12607–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ogden SK, Fei DL, Schilling NS, Ahmed YF, Hwa J, Robbins DJ. G protein Galphai functions immediately downstream of Smoothened in Hedgehog signalling. Nature. 2008;456(7224):967–70. Epub 2008/11/07. doi: 10.1038/nature07459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jia J, Tong C, Jiang J. Smoothened transduces Hedgehog signal by physically interacting with Costal2/Fused complex through its C-terminal tail. Genes And Development. 2003;17(21):2709–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lum L, Zhang C, Oh S, Mann RK, von Kessler DP, Taipale J, et al. Hedgehog signal transduction via Smoothened association with a cytoplasmic complex scaffolded by the atypical kinesin, Costal-2. Mol Cell. 2003;12(5):1261–74. [DOI] [PubMed] [Google Scholar]
- 94.Ruel L, Rodriguez R, Gallet A, Lavenant-Staccini L, Therond PP. Stability and association of Smoothened, Costal2 and Fused with Cubitus interruptus are regulated by Hedgehog. Nat Cell Biol. 2003;5(10):907–13. [DOI] [PubMed] [Google Scholar]
- 95.Ogden SK, Ascano M Jr., Stegman MA, Suber LM, Hooper JE, Robbins DJ. Identification of a functional interaction between the transmembrane protein Smoothened and the kinesin-related protein Costal2. Curr Biol. 2003;13(22):1998–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shi Q, Li S, Jia J, Jiang J. The Hedgehog-induced Smoothened conformational switch assembles a signaling complex that activates Fused by promoting its dimerization and phosphorylation. Development. 2011;138(19):4219–31. Epub 2011/08/20. doi: dev.067959 [pii] 10.1242/dev.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou Q, Kalderon D. Hedgehog Activates Fused through Phosphorylation to Elicit a Full Spectrum of Pathway Responses. Dev Cell. 2011;20(6):802–14. Epub 2011/06/15. doi: S1534–5807(11)00174–2 [pii] 10.1016/j.devcel.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang Y, Mao F, Lu Y, Wu W, Zhang L, Zhao Y. Transduction of the Hedgehog signal through the dimerization of Fused and the nuclear translocation of Cubitus interruptus. Cell Res. 2011;21(10):1436–51. Epub 2011/08/17. doi: 10.1038/cr.2011.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiang J, Struhl G. Protein kinase A and hedgehog signaling in Drosophila limb development. Cell. 1995;80(4):563–72. doi: Doi 10.1016/0092-8674(95)90510-3. [DOI] [PubMed] [Google Scholar]
- 100.Jia J, Amanai K, Wang G, Tang J, Wang B, Jiang J. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature. 2002;416(6880):548–52. [DOI] [PubMed] [Google Scholar]
- 101.Jia J, Zhang L, Zhang Q, Tong C, Wang B, Hou F, et al. Phosphorylation by double-time/CKIepsilon and CKIalpha targets cubitus interruptus for Slimb/beta-TRCP-mediated proteolytic processing. Dev Cell. 2005;9(6):819–30. [DOI] [PubMed] [Google Scholar]
- 102.Price MA, Kalderon D. Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell. 2002;108(6):823–35. [DOI] [PubMed] [Google Scholar]
- 103.Zhang W, Zhao Y, Tong C, Wang G, Wang B, Jia J, et al. Hedgehog-regulated costal2-kinase complexes control phosphorylation and proteolytic processing of cubitus interruptus. Dev Cell. 2005;8(2):267–78. [DOI] [PubMed] [Google Scholar]
- 104.Smelkinson MG, Kalderon D. Processing of the Drosophila hedgehog signaling effector Ci-155 to the repressor Ci-75 is mediated by direct binding to the SCF component slimb. Curr Biol. 2006;16(1):110–6. [DOI] [PubMed] [Google Scholar]
- 105.Jiang J, Struhl G. Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature. 1998;391(6666):493–6. [DOI] [PubMed] [Google Scholar]
- 106.Ruel L, Gallet A, Raisin S, Truchi A, Staccini-Lavenant L, Cervantes A, et al. Phosphorylation of the atypical kinesin Costal2 by the kinase Fused induces the partial disassembly of the Smoothened-Fused-Costal2-Cubitus interruptus complex in Hedgehog signalling. Development. 2007;134(20):3677–89. [DOI] [PubMed] [Google Scholar]
- 107.Wang G, Wang B, Jiang J. Protein kinase A antagonizes Hedgehog signaling by regulating both the activator and repressor forms of Cubitus interruptus. Genes And Development. 1999;13(21):2828–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang G, Amanai K, Wang B, Jiang J. Interactions with Costal2 and suppressor of fused regulate nuclear translocation and activity of cubitus interruptus. Genes And Development. 2000;14(22):2893–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang QT, Holmgren RA. Nuclear import of cubitus interruptus is regulated by hedgehog via a mechanism distinct from Ci stabilization and Ci activation. Development. 2000;127(14):3131–9. [DOI] [PubMed] [Google Scholar]
- 110.Wang G, Jiang J. Multiple Cos2/Ci interactions regulate Ci subcellular localization through microtubule dependent and independent mechanisms. Dev Biol. 2004;268(2):493–505 [DOI] [PubMed] [Google Scholar]
- 111.Ohlmeyer JT, Kalderon D. Hedgehog stimulates maturation of Cubitus interruptus into a labile transcriptional activator. Nature. 1998;396(6713):749–53. doi: 10.1038/25533. [DOI] [PubMed] [Google Scholar]
- 112.Shi Q, Han Y, Jiang J. Suppressor of fused impedes Ci/Gli nuclear import by opposing Trn/Kapbeta2 in Hedgehog signaling. J Cell Sci. 2014;127(Pt 5):1092–103. doi: 10.1242/jcs.142828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Han Y, Shi Q, Jiang J. Multisite interaction with Sufu regulates Ci/Gli activity through distinct mechanisms in Hh signal transduction. Proc Natl Acad Sci U S A. 2015;112(20):6383–8. Epub 2015/05/06. doi: 10.1073/pnas.1421628112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Han Y, Wang B, Cho YS, Zhu J, Wu J, Chen Y, et al. Phosphorylation of Ci/Gli by Fused Family Kinases Promotes Hedgehog Signaling. Dev Cell. 2019;50(5):610–26 e4. Epub 2019/07/08. doi: 10.1016/j.devcel.2019.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kent D, Bush EW, Hooper JE. Roadkill attenuates Hedgehog responses through degradation of Cubitus interruptus. Development. 2006;133(10):2001–10. [DOI] [PubMed] [Google Scholar]
- 116.Zhang Q, Zhang L, Wang B, Ou CY, Chien CT, Jiang J. A hedgehog-induced BTB protein modulates hedgehog signaling by degrading Ci/Gli transcription factor. Dev Cell. 2006;10(6):719–29. [DOI] [PubMed] [Google Scholar]
- 117.Zhang Q, Shi Q, Chen Y, Yue T, Li S, Wang B, et al. Multiple Ser/Thr-rich degrons mediate the degradation of Ci/Gli by the Cul3-HIB/SPOP E3 ubiquitin ligase. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(50):21191–6. Epub 2009/12/04. doi: 10.1073/pnas.0912008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen Y, Jiang J. Decoding the phosphorylation code in Hedgehog signal transduction. Cell Res. 2013;23(2):186–200. doi: 10.1038/cr.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100(4):423–34. [DOI] [PubMed] [Google Scholar]
- 120.Wang B, Li Y. Evidence for the direct involvement of {beta}TrCP in Gli3 protein processing. Proc Natl Acad Sci U S A. 2006;103(1):33–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tempe D, Casas M, Karaz S, Blanchet-Tournier MF, Concordet JP. Multisite protein kinase A and glycogen synthase kinase 3beta phosphorylation leads to Gli3 ubiquitination by SCFbetaTrCP. Mol Cell Biol. 2006;26(11):4316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Methot N, Basler K. Hedgehog controls limb development by regulating the activities of distinct transcriptional activator and repressor forms of Cubitus interruptus. Cell. 1999;96(6):819–31. [DOI] [PubMed] [Google Scholar]
- 123.Smelkinson MG, Zhou Q, Kalderon D. Regulation of Ci-SCFSlimb binding, Ci proteolysis, and hedgehog pathway activity by Ci phosphorylation. Dev Cell. 2007;13(4):481–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pan Y, Wang B. A novel protein-processing domain in Gli2 and Gli3 differentially blocks complete protein degradation by the proteasome. J Biol Chem. 2007;282(15):10846–52. [DOI] [PubMed] [Google Scholar]
- 125.Kise Y, Morinaka A, Teglund S, Miki H. Sufu recruits GSK3beta for efficient processing of Gli3. Biochemical and biophysical research communications. 2009;387(3):569–74. Epub 2009/07/23. doi: 10.1016/j.bbrc.2009.07.087. [DOI] [PubMed] [Google Scholar]
- 126.Cheung HO, Zhang X, Ribeiro A, Mo R, Makino S, Puviindran V, et al. The kinesin protein Kif7 is a critical regulator of Gli transcription factors in mammalian hedgehog signaling. Sci Signal. 2009;2(76):ra29. Epub 2009/06/25. doi: 2/76/ra29 [pii] 10.1126/scisignal.2000405. [DOI] [PubMed] [Google Scholar]
- 127.Endoh-Yamagami S, Evangelista M, Wilson D, Wen X, Theunissen JW, Phamluong K, et al. The mammalian Cos2 homolog Kif7 plays an essential role in modulating Hh signal transduction during development. Curr Biol. 2009;19(15):1320–6. Epub 2009/07/14. doi: S0960–9822(09)01323–2 [pii] 10.1016/j.cub.2009.06.046. [DOI] [PubMed] [Google Scholar]
- 128.Liem KF Jr., He M, Ocbina PJ, Anderson KV. Mouse Kif7/Costal2 is a cilia-associated protein that regulates Sonic hedgehog signaling. Proc Natl Acad Sci U S A. 2009;106(32):13377–82. Epub 2009/08/12. doi: 0906944106 [pii] 10.1073/pnas.0906944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chen MH, Wilson CW, Li YJ, Law KK, Lu CS, Gacayan R, et al. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes And Development. 2009;23(16):1910–28. Epub 2009/08/18. doi: 23/16/1910 [pii] 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes And Development. 2010;24(7):670–82. Epub 2010/04/03. doi: 24/7/670 [pii] 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang C, Pan Y, Wang B. Suppressor of fused and Spop regulate the stability, processing and function of Gli2 and Gli3 full-length activators but not their repressors. Development. 2010;137(12):2001–9. Epub 2010/05/14. doi: 10.1242/dev.052126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jia J, Kolterud A, Zeng H, Hoover A, Teglund S, Toftgard R, et al. Suppressor of Fused inhibits mammalian Hedgehog signaling in the absence of cilia. Dev Biol. 2009;330(2):452–60. doi: 10.1016/j.ydbio.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tuson M, He M, Anderson KV. Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development. 2011;138(22):4921–30. Epub 2011/10/19. doi: 10.1242/dev.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, et al. Gli protein activity is controlled by multisite phosphorylation in vertebrate Hedgehog signaling. Cell Rep. 2014;6(1):168–81. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191(2):415–28. Epub 2010/10/20. doi: jcb.201004108 [pii] 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang Y, Fu L, Qi X, Zhang Z, Xia Y, Jia J, et al. Structural insight into the mutual recognition and regulation between Suppressor of Fused and Gli/Ci. Nat Commun. 2013;4:2608. doi: 10.1038/ncomms3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Infante P, Faedda R, Bernardi F, Bufalieri F, Lospinoso Severini L, Alfonsi R, et al. Itch/beta-arrestin2-dependent non-proteolytic ubiquitylation of SuFu controls Hedgehog signalling and medulloblastoma tumorigenesis. Nat Commun. 2018;9(1):976. Epub 2018/03/09. doi: 10.1038/s41467-018-03339-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Raducu M, Fung E, Serres S, Infante P, Barberis A, Fischer R, et al. SCF (Fbxl17) ubiquitylation of Sufu regulates Hedgehog signaling and medulloblastoma development. Embo Journal. 2016;35(13):1400–16. Epub 2016/05/29. doi: 10.15252/embj.201593374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yin WC, Satkunendran T, Mo R, Morrissy S, Zhang X, Huang ES, et al. Dual Regulatory Functions of SUFU and Targetome of GLI2 in SHH Subgroup Medulloblastoma. Dev Cell. 2019;48(2):167–83 e5. Epub 2018/12/18. doi: 10.1016/j.devcel.2018.11.015. [DOI] [PubMed] [Google Scholar]
- 140.Cooper AF, Yu KP, Brueckner M, Brailey LL, Johnson L, McGrath JM, et al. Cardiac and CNS defects in a mouse with targeted disruption of suppressor of fused. Development. 2005;132(19):4407–17. [DOI] [PubMed] [Google Scholar]
- 141.Svard J, Heby-Henricson K, Persson-Lek M, Rozell B, Lauth M, Bergstrom A, et al. Genetic elimination of Suppressor of fused reveals an essential repressor function in the mammalian Hedgehog signaling pathway. Dev Cell. 2006;10(2):187–97. [DOI] [PubMed] [Google Scholar]
- 142.Wigley WC, Fabunmi RP, Lee MG, Marino CR, Muallem S, DeMartino GN, et al. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145(3):481–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Barzi M, Berenguer J, Menendez A, Alvarez-Rodriguez R, Pons S. Sonic-hedgehog-mediated proliferation requires the localization of PKA to the cilium base. J Cell Sci. 2010;123(Pt 1):62–9. doi: 10.1242/jcs.060020. [DOI] [PubMed] [Google Scholar]
- 144.Mick DU, Rodrigues RB, Leib RD, Adams CM, Chien AS, Gygi SP, et al. Proteomics of Primary Cilia by Proximity Labeling. Dev Cell. 2015;35(4):497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, et al. The ciliary G-protein-coupled receptor Gpr161 negatively regulates the Sonic hedgehog pathway via cAMP signaling. Cell. 2013;152(1–2):210–23. Epub 2013/01/22. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 146.Pal K, Hwang SH, Somatilaka B, Badgandi H, Jackson PK, DeFea K, et al. Smoothened determines beta-arrestin-mediated removal of the G protein-coupled receptor Gpr161 from the primary cilium. J Cell Biol. 2016;212(7):861–75. Epub 2016/03/24. doi: 10.1083/jcb.201506132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Singh J, Wen X, Scales SJ. The Orphan G Protein-coupled Receptor Gpr175 (Tpra40) Enhances Hedgehog Signaling by Modulating cAMP Levels. J Biol Chem. 2015;290(49):29663–75. Epub 2015/10/10. doi: 10.1074/jbc.M115.665810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haycraft CJ, Banizs B, Aydin-Son Y, Zhang Q, Michaud EJ, Yoder BK. Gli2 and gli3 localize to cilia and require the intraflagellar transport protein polaris for processing and function. PLoS Genet. 2005;1(4):e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kim J, Kato M, Beachy PA. Gli2 trafficking links Hedgehog-dependent activation of Smoothened in the primary cilium to transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 2009;106(51):21666–71. Epub 2009/12/10. doi: 10.1073/pnas.0912180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wen X, Lai CK, Evangelista M, Hongo JA, de Sauvage FJ, Scales SJ. Kinetics of hedgehog-dependent full-length Gli3 accumulation in primary cilia and subsequent degradation. Molecular and cellular biology. 2010;30(8):1910–22. Epub 2010/02/16. doi: 10.1128/MCB.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.He M, Subramanian R, Bangs F, Omelchenko T, Liem KF Jr., Kapoor TM, et al. The kinesin-4 protein Kif7 regulates mammalian Hedgehog signalling by organizing the cilium tip compartment. Nat Cell Biol. 2014;16(7):663–72. doi: 10.1038/ncb2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Wolff C, Roy S, Ingham PW. Multiple muscle cell identities induced by distinct levels and timing of hedgehog activity in the zebrafish embryo. Curr Biol. 2003;13(14):1169–81. [DOI] [PubMed] [Google Scholar]
- 153.Chen MH, Gao N, Kawakami T, Chuang PT. Mice deficient in the fused homolog do not exhibit phenotypes indicative of perturbed hedgehog signaling during embryonic development. Mol Cell Biol. 2005;25(16):7042–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Merchant M, Evangelista M, Luoh SM, Frantz GD, Chalasani S, Carano RA, et al. Loss of the serine/threonine kinase fused results in postnatal growth defects and lethality due to progressive hydrocephalus. Mol Cell Biol. 2005;25(16):7054–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Han Y, Xiong Y, Shi X, Wu J, Zhao Y, Jiang J. Regulation of Gli ciliary localization and Hedgehog signaling by the PY-NLS/karyopherin-beta2 nuclear import system. PLoS Biol. 2017;15(8):e2002063. Epub 2017/08/05. doi: 10.1371/journal.pbio.2002063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chong YC, Mann RK, Zhao C, Kato M, Beachy PA. Bifurcating action of Smoothened in Hedgehog signaling is mediated by Dlg5. Genes And Development. 2015;29(3):262–76. Epub 2015/02/04. doi: 10.1101/gad.252676.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ruiz-Perez VL, Blair HJ, Rodriguez-Andres ME, Blanco MJ, Wilson A, Liu YN, et al. Evc is a positive mediator of Ihh-regulated bone growth that localises at the base of chondrocyte cilia. Development. 2007;134(16):2903–12. Epub 2007/07/31. doi: 10.1242/dev.007542. [DOI] [PubMed] [Google Scholar]
- 158.Blair HJ, Tompson S, Liu YN, Campbell J, MacArthur K, Ponting CP, et al. Evc2 is a positive modulator of Hedgehog signalling that interacts with Evc at the cilia membrane and is also found in the nucleus. BMC Biol. 2011;9:14. Epub 2011/03/02. doi: 10.1186/1741-7007-9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dorn KV, Hughes CE, Rohatgi R. A Smoothened-Evc2 complex transduces the Hedgehog signal at primary cilia. Dev Cell. 2012;23(4):823–35. Epub 2012/09/18. doi: 10.1016/j.devcel.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yang C, Chen W, Chen Y, Jiang J. Smoothened transduces Hedgehog signal by forming a complex with Evc/Evc2. Cell Res. 2012;22(11):1593–604. doi: 10.1038/cr.2012.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Caparros-Martin JA, Valencia M, Reytor E, Pacheco M, Fernandez M, Perez-Aytes A, et al. The ciliary Evc/Evc2 complex interacts with Smo and controls Hedgehog pathway activity in chondrocytes by regulating Sufu/Gli3 dissociation and Gli3 trafficking in primary cilia. Hum Mol Genet. 2013;22(1):124–39. Epub 2012/10/03. doi: 10.1093/hmg/dds409. [DOI] [PubMed] [Google Scholar]
- 162.Pusapati GV, Hughes CE, Dorn KV, Zhang D, Sugianto P, Aravind L, et al. EFCAB7 and IQCE regulate hedgehog signaling by tethering the EVC-EVC2 complex to the base of primary cilia. Dev Cell. 2014;28(5):483–96. Epub 2014/03/04. doi: 10.1016/j.devcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Atwood SX, Li M, Lee A, Tang JY, Oro AE. GLI activation by atypical protein kinase C iota/lambda regulates the growth of basal cell carcinomas. Nature. 2013;494(7438):484–8. doi: 10.1038/nature11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mirza AN, Fry MA, Urman NM, Atwood SX, Roffey J, Ott GR, et al. Combined inhibition of atypical PKC and histone deacetylase 1 is cooperative in basal cell carcinoma treatment. JCI Insight. 2017;2(21). Epub 2017/11/03. doi: 10.1172/jci.insight.97071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Evangelista M, Lim TY, Lee J, Parker L, Ashique A, Peterson AS, et al. Kinome siRNA screen identifies regulators of ciliogenesis and hedgehog signal transduction. Sci Signal. 2008;1(39):ra7. [DOI] [PubMed] [Google Scholar]
- 166.Varjosalo M, Bjorklund M, Cheng F, Syvanen H, Kivioja T, Kilpinen S, et al. Application of active and kinase-deficient kinome collection for identification of kinases regulating hedgehog signaling. Cell. 2008;133(3):537–48. [DOI] [PubMed] [Google Scholar]
- 167.Mao J, Maye P, Kogerman P, Tejedor FJ, Toftgard R, Xie W, et al. Regulation of Gli1 transcriptional activity in the nucleus by Dyrk1. J Biol Chem. 2002;277(38):35156–61. Epub 2002/07/26. doi: 10.1074/jbc.M206743200. [DOI] [PubMed] [Google Scholar]
- 168.Wang Y, Ding Q, Yen CJ, Xia W, Izzo JG, Lang JY, et al. The crosstalk of mTOR/S6K1 and Hedgehog pathways. Cancer cell. 2012;21(3):374–87. Epub 2012/03/24. doi: 10.1016/j.ccr.2011.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Maloverjan A, Piirsoo M, Michelson P, Kogerman P, Osterlund T. Identification of a novel serine/threonine kinase ULK3 as a positive regulator of Hedgehog pathway. Exp Cell Res. 2010;316(4):627–37. Epub 2009/11/03. doi: S0014–4827(09)00447–9 [pii] 10.1016/j.yexcr.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 170.Jia H, Liu Y, Xia R, Tong C, Yue T, Jiang J, et al. Casein kinase 2 promotes Hedgehog signaling by regulating both smoothened and Cubitus interruptus. The Journal of biological chemistry. 2010;285(48):37218–26. Epub 2010/09/30. doi: 10.1074/jbc.M110.174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Purzner T, Purzner J, Buckstaff T, Cozza G, Gholamin S, Rusert JM, et al. Developmental phosphoproteomics identifies the kinase CK2 as a driver of Hedgehog signaling and a therapeutic target in medulloblastoma. Sci Signal. 2018;11(547). Epub 2018/09/13. doi: 10.1126/scisignal.aau5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Zhang T, Xin G, Jia M, Zhuang T, Zhu S, Zhang B, et al. The Plk1 kinase negatively regulates the Hedgehog signaling pathway by phosphorylating Gli1. J Cell Sci. 2019;132(2). Epub 2018/12/24. doi: 10.1242/jcs.220384. [DOI] [PubMed] [Google Scholar]
- 173.Huntzicker EG, Estay IS, Zhen H, Lokteva LA, Jackson PK, Oro AE. Dual degradation signals control Gli protein stability and tumor formation. Genes And Development. 2006;20:276–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Canettieri G, Di Marcotullio L, Greco A, Coni S, Antonucci L, Infante P, et al. Histone deacetylase and Cullin3-REN(KCTD11) ubiquitin ligase interplay regulates Hedgehog signalling through Gli acetylation. Nat Cell Biol. 2010;12(2):132–42. Epub 2010/01/19. doi: 10.1038/ncb2013. [DOI] [PubMed] [Google Scholar]
- 175.Zhou Z, Yao X, Li S, Xiong Y, Dong X, Zhao Y, et al. Deubiquitination of Ci/Gli by Usp7/HAUSP regulates Hedgehog Signaling. Developmental cell. 2015:accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Han L, Pan Y, Wang B. Small ubiquitin-like Modifier (SUMO) modification inhibits GLI2 protein transcriptional activity in vitro and in vivo. J Biol Chem. 2012;287(24):20483–9. Epub 2012/05/03. doi: 10.1074/jbc.M112.359299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Fu L, Wu H, Cheng SY, Gao D, Zhang L, Zhao Y. Set7 mediated Gli3 methylation plays a positive role in the activation of Sonic Hedgehog pathway in mammals. Elife. 2016;5. Epub 2016/05/06. doi: 10.7554/eLife.15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mirza AN, McKellar SA, Urman NM, Brown AS, Hollmig T, Aasi SZ, et al. LAP2 Proteins Chaperone GLI1 Movement between the Lamina and Chromatin to Regulate Transcription. Cell. 2019;176(1–2):198–212 e15. Epub 2018/12/07. doi: 10.1016/j.cell.2018.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ruiz i Altaba A, Mas C, Stecca B. The Gli code: an information nexus regulating cell fate, stemness and cancer. Trends Cell Biol. 2007;17(9):438–47. Epub 2007/09/12. doi: S0962–8924(07)00171–7 [pii] 10.1016/j.tcb.2007.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Han YG, Kim HJ, Dlugosz AA, Ellison DW, Gilbertson RJ, Alvarez-Buylla A. Dual and opposing roles of primary cilia in medulloblastoma development. Nature medicine. 2009;15(9):1062–5. Epub 2009/08/25. doi: 10.1038/nm.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wong SY, Seol AD, So PL, Ermilov AN, Bichakjian CK, Epstein EH Jr., et al. Primary cilia can both mediate and suppress Hedgehog pathway-dependent tumorigenesis. Nature medicine. 2009;15(9):1055–61. Epub 2009/08/25. doi: 10.1038/nm.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124(1):47–59. [DOI] [PubMed] [Google Scholar]
- 183.Vokes SA, Ji H, McCuine S, Tenzen T, Giles S, Zhong S, et al. Genomic characterization of Gli-activator targets in sonic hedgehog-mediated neural patterning. Development. 2007;134(10):1977–89. [DOI] [PubMed] [Google Scholar]
- 184.Ruiz i Altaba A Gli proteins and Hedgehog signaling: development and cancer. Trends Genet. 1999;15(10):418–25. [DOI] [PubMed] [Google Scholar]
- 185.Cheng SY, Bishop JM. Suppressor of Fused represses Gli-mediated transcription by recruiting the SAP18-mSin3 corepressor complex. Proc Natl Acad Sci U S A. 2002;99(8):5442–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhan X, Shi X, Zhang Z, Chen Y, Wu JI. Dual role of Brg chromatin remodeling factor in Sonic hedgehog signaling during neural development. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(31):12758–63. Epub 2011/07/20. doi: 10.1073/pnas.1018510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lin C, Yao E, Wang K, Nozawa Y, Shimizu H, Johnson JR, et al. Regulation of Sufu activity by p66beta and Mycbp provides new insight into vertebrate Hedgehog signaling. Genes And Development. 2014;28(22):2547–63. doi: 10.1101/gad.249425.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zhang Z, Zhan X, Kim B, Wu J. A proteomic approach identifies SAFB-like transcription modulator (SLTM) as a bidirectional regulator of GLI family zinc finger transcription factors. J Biol Chem. 2019. Epub 2019/02/21. doi: 10.1074/jbc.RA118.007018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Shi X, Zhang Z, Zhan X, Cao M, Satoh T, Akira S, et al. An epigenetic switch induced by Shh signalling regulates gene activation during development and medulloblastoma growth. Nat Commun. 2014;5:5425. doi: 10.1038/ncomms6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tang Y, Gholamin S, Schubert S, Willardson MI, Lee A, Bandopadhayay P, et al. Epigenetic targeting of Hedgehog pathway transcriptional output through BET bromodomain inhibition. Nature medicine. 2014;20(7):732–40. Epub 2014/06/30. doi: 10.1038/nm.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Beachy PA, Karhadkar SS, Berman DM. Tissue repair and stem cell renewal in carcinogenesis. Nature. 2004;432(7015):324–31. Epub 2004/11/19. doi: nature03100 [pii] 10.1038/nature03100. [DOI] [PubMed] [Google Scholar]
- 192.Wicking C, Smyth I, Bale A. The hedgehog signalling pathway in tumorigenesis and development. Oncogene. 1999;18(55):7844–51. Epub 2000/01/12. doi: 10.1038/sj.onc.1203282. [DOI] [PubMed] [Google Scholar]
- 193.Yauch RL, Gould SE, Scales SJ, Tang T, Tian H, Ahn CP, et al. A paracrine requirement for hedgehog signalling in cancer. Nature. 2008;455(7211):406–10. Epub 2008/08/30. doi: nature07275 [pii] 10.1038/nature07275. [DOI] [PubMed] [Google Scholar]
- 194.Hahn H, Wicking C, Zaphiropoulous PG, Gailani MR, Shanley S, Chidambaram A, et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell. 1996;85(6):841–51. [DOI] [PubMed] [Google Scholar]
- 195.Johnson RL, Rothman AL, Xie J, Goodrich LV, Bare JW, Bonifas JM, et al. Human Homolog of patched, a candidate gene ofr the basal cell nevus syndrome. Science. 1996;272:1668–71. [DOI] [PubMed] [Google Scholar]
- 196.Xie J, Murone M, Luoh S-M, Ryan A, Gu Q, Zhang C, et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature. 1998;391:90–2. [DOI] [PubMed] [Google Scholar]
- 197.Bonilla X, Parmentier L, King B, Bezrukov F, Kaya G, Zoete V, et al. Genomic analysis identifies new drivers and progression pathways in skin basal cell carcinoma. Nat Genet. 2016;48(4):398–406. Epub 2016/03/08. doi: 10.1038/ng.3525. [DOI] [PubMed] [Google Scholar]
- 198.Nitzki F, Becker M, Frommhold A, Schulz-Schaeffer W, Hahn H. Patched knockout mouse models of Basal cell carcinoma. J Skin Cancer. 2012;2012:907543. Epub 2012/10/02. doi: 10.1155/2012/907543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Youssef KK, Van Keymeulen A, Lapouge G, Beck B, Michaux C, Achouri Y, et al. Identification of the cell lineage at the origin of basal cell carcinoma. Nat Cell Biol. 2010;12(3):299–305. Epub 2010/02/16. doi: 10.1038/ncb2031. [DOI] [PubMed] [Google Scholar]
- 200.Sanchez-Danes A, Hannezo E, Larsimont JC, Liagre M, Youssef KK, Simons BD, et al. Defining the clonal dynamics leading to mouse skin tumour initiation. Nature. 2016;536(7616):298–303. Epub 2016/07/28. doi: 10.1038/nature19069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–9. Epub 2010/12/18. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488(7409):49–56. Epub 2012/07/27. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Garcia-Lopez J, Kumar R, Smith KS, Northcott PA. Deconstructing Sonic Hedgehog Medulloblastoma: Molecular Subtypes, Drivers, and Beyond. Trends Genet. 2020. Epub 2020/12/05. doi: 10.1016/j.tig.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 204.Goodrich L, Milenkovic L, Higgins K, Scott M. Altered neural cell fates and medulloblastoma in mouse patched mutants. Science. 1997;277(5329):1109–13. [DOI] [PubMed] [Google Scholar]
- 205.Schuller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14(2):123–34. Epub 2008/08/12. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yang ZJ, Ellis T, Markant SL, Read TA, Kessler JD, Bourboulas M, et al. Medulloblastoma can be initiated by deletion of Patched in lineage-restricted progenitors or stem cells. Cancer Cell. 2008;14(2):135–45. Epub 2008/08/12. doi: 10.1016/j.ccr.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Vladoiu MC, El-Hamamy I, Donovan LK, Farooq H, Holgado BL, Sundaravadanam Y, et al. Childhood cerebellar tumours mirror conserved fetal transcriptional programs. Nature. 2019;572(7767):67–73. Epub 2019/05/03. doi: 10.1038/s41586-019-1158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Zhang L, He X, Liu X, Zhang F, Huang LF, Potter AS, et al. Single-Cell Transcriptomics in Medulloblastoma Reveals Tumor-Initiating Progenitors and Oncogenic Cascades during Tumorigenesis and Relapse. Cancer Cell. 2019;36(3):302–18 e7. Epub 2019/09/03. doi: 10.1016/j.ccell.2019.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lee Y, Kawagoe R, Sasai K, Li Y, Russell HR, Curran T, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26(44):6442–7. Epub 2007/04/25. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 210.Begemann M, Waszak SM, Robinson GW, Jager N, Sharma T, Knopp C, et al. Germline GPR161 Mutations Predispose to Pediatric Medulloblastoma. J Clin Oncol. 2020;38(1):43–50. Epub 2019/10/15. doi: 10.1200/JCO.19.00577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Huh JY, Kwon MJ, Seo KY, Kim MK, Chae KY, Kim SH, et al. Novel nonsense GNAS mutation in a 14-month-old boy with plate-like osteoma cutis and medulloblastoma. J Dermatol. 2014;41(4):319–21. Epub 2014/02/13. doi: 10.1111/1346-8138.12284. [DOI] [PubMed] [Google Scholar]
- 212.He X, Zhang L, Chen Y, Remke M, Shih D, Lu F, et al. The G protein alpha subunit Galphas is a tumor suppressor in Sonic hedgehog-driven medulloblastoma. Nature medicine. 2014;20(9):1035–42. Epub 2014/08/26. doi: 10.1038/nm.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Shimada IS, Hwang SH, Somatilaka BN, Wang X, Skowron P, Kim J, et al. Basal Suppression of the Sonic Hedgehog Pathway by the G-Protein-Coupled Receptor Gpr161 Restricts Medulloblastoma Pathogenesis. Cell Rep. 2018;22(5):1169–84. Epub 2018/02/02. doi: 10.1016/j.celrep.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Suzuki H, Kumar SA, Shuai S, Diaz-Navarro A, Gutierrez-Fernandez A, De Antonellis P, et al. Recurrent noncoding U1 snRNA mutations drive cryptic splicing in SHH medulloblastoma. Nature. 2019;574(7780):707–11. Epub 2019/10/31. doi: 10.1038/s41586-019-1650-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Waszak SM, Robinson GW, Gudenas BL, Smith KS, Forget A, Kojic M, et al. Germline Elongator mutations in Sonic Hedgehog medulloblastoma. Nature. 2020;580(7803):396–401. Epub 2020/04/17. doi: 10.1038/s41586-020-2164-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Hahn H, Wojnowski L, Zimmer AM, Hall J, Miller G, Zimmer A. Rhabdomyosarcomas and radiation hypersensitivity in a mouse model of Gorlin syndrome. Nature medicine. 1998;4(5):619–22. Epub 1998/05/19. doi: 10.1038/nm0598-619. [DOI] [PubMed] [Google Scholar]
- 217.Tostar U, Malm CJ, Meis-Kindblom JM, Kindblom LG, Toftgard R, Unden AB. Deregulation of the hedgehog signalling pathway: a possible role for the PTCH and SUFU genes in human rhabdomyoma and rhabdomyosarcoma development. J Pathol. 2006;208(1):17–25. Epub 2005/11/19. doi: 10.1002/path.1882. [DOI] [PubMed] [Google Scholar]
- 218.Zhang L, Zhou Y, Cheng C, Cui H, Cheng L, Kong P, et al. Genomic analyses reveal mutational signatures and frequently altered genes in esophageal squamous cell carcinoma. Am J Hum Genet. 2015;96(4):597–611. Epub 2015/04/04. doi: 10.1016/j.ajhg.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Clark VE, Erson-Omay EZ, Serin A, Yin J, Cotney J, Ozduman K, et al. Genomic analysis of non-NF2 meningiomas reveals mutations in TRAF7, KLF4, AKT1, and SMO. Science. 2013;339(6123):1077–80. Epub 2013/01/26. doi: 10.1126/science.1233009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Watkins DN, Berman DM, Burkholder SG, Wang B, Beachy PA, Baylin SB. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422(6929):313–7. [DOI] [PubMed] [Google Scholar]
- 221.Yuan Z, Goetz JA, Singh S, Ogden SK, Petty WJ, Black CC, et al. Frequent requirement of hedgehog signaling in non-small cell lung carcinoma. Oncogene. 2007;26(7):1046–55. Epub 2006/08/16. doi: 10.1038/sj.onc.1209860. [DOI] [PubMed] [Google Scholar]
- 222.Park KS, Martelotto LG, Peifer M, Sos ML, Karnezis AN, Mahjoub MR, et al. A crucial requirement for Hedgehog signaling in small cell lung cancer. Nature medicine. 2011;17(11):1504–8. Epub 2011/10/11. doi: 10.1038/nm.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Karhadkar SS, Bova GS, Abdallah N, Dhara S, Gardner D, Maitra A, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431(7009):707–12. [DOI] [PubMed] [Google Scholar]
- 224.Sanchez P, Hernandez AM, Stecca B, Kahler AJ, DeGueme AM, Barrett A, et al. Inhibition of prostate cancer proliferation by interference with SONIC HEDGEHOG-GLI1 signaling. Proc Natl Acad Sci U S A. 2004;101(34):12561–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Sheng T, Li C, Zhang X, Chi S, He N, Chen K, et al. Activation of the hedgehog pathway in advanced prostate cancer. Mol Cancer. 2004;3:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Berman DM, Karhadkar SS, Maitra A, Montes De Oca R, Gerstenblith MR, Briggs K, et al. Widespread requirement for Hedgehog ligand stimulation in growth of digestive tract tumours. Nature. 2003;425(6960):846–51. [DOI] [PubMed] [Google Scholar]
- 227.Thayer SP, di Magliano MP, Heiser PW, Nielsen CM, Roberts DJ, Lauwers GY, et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature. 2003;425(6960):851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pasca di Magliano M, Sekine S, Ermilov A, Ferris J, Dlugosz AA, Hebrok M. Hedgehog/Ras interactions regulate early stages of pancreatic cancer. Genes And Development. 2006;20(22):3161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Varnat F, Duquet A, Malerba M, Zbinden M, Mas C, Gervaz P, et al. Human colon cancer epithelial cells harbour active HEDGEHOG-GLI signalling that is essential for tumour growth, recurrence, metastasis and stem cell survival and expansion. EMBO molecular medicine. 2009;1(6–7):338–51. doi: 10.1002/emmm.200900039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Fei DL, Sanchez-Mejias A, Wang Z, Flaveny C, Long J, Singh S, et al. Hedgehog signaling regulates bladder cancer growth and tumorigenicity. Cancer Res. 2012;72(17):4449–58. Epub 2012/07/21. doi: 10.1158/0008-5472.CAN-11-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Pignot G, Vieillefond A, Vacher S, Zerbib M, Debre B, Lidereau R, et al. Hedgehog pathway activation in human transitional cell carcinoma of the bladder. Br J Cancer. 2012;106(6):1177–86. Epub 2012/03/01. doi: 10.1038/bjc.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ma X, Sheng T, Zhang Y, Zhang X, He J, Huang S, et al. Hedgehog signaling is activated in subsets of esophageal cancers. International Journal Of Cancer. 2006;118(1):139–48. Epub 2005/07/09. doi: 10.1002/ijc.21295. [DOI] [PubMed] [Google Scholar]
- 233.Wang DH, Clemons NJ, Miyashita T, Dupuy AJ, Zhang W, Szczepny A, et al. Aberrant epithelial-mesenchymal Hedgehog signaling characterizes Barrett’s metaplasia. Gastroenterology. 2010;138(5):1810–22. Epub 2010/02/09. doi: 10.1053/j.gastro.2010.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ma X, Chen K, Huang S, Zhang X, Adegboyega PA, Evers BM, et al. Frequent activation of the hedgehog pathway in advanced gastric adenocarcinomas. Carcinogenesis. 2005;26(10):1698–705. Epub 2005/05/21. doi: 10.1093/carcin/bgi130. [DOI] [PubMed] [Google Scholar]
- 235.Fukaya M, Isohata N, Ohta H, Aoyagi K, Ochiya T, Saeki N, et al. Hedgehog signal activation in gastric pit cell and in diffuse-type gastric cancer. Gastroenterology. 2006;131(1):14–29. Epub 2006/07/13. doi: 10.1053/j.gastro.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 236.Huang S, He J, Zhang X, Bian Y, Yang L, Xie G, et al. Activation of the hedgehog pathway in human hepatocellular carcinomas. Carcinogenesis. 2006;27(7):1334–40. Epub 2006/02/28. doi: 10.1093/carcin/bgi378. [DOI] [PubMed] [Google Scholar]
- 237.Patil MA, Zhang J, Ho C, Cheung ST, Fan ST, Chen X. Hedgehog signaling in human hepatocellular carcinoma. Cancer Biol Ther. 2006;5(1):111–7. Epub 2006/01/07. doi: 10.4161/cbt.5.1.2379. [DOI] [PubMed] [Google Scholar]
- 238.Eichenmuller M, Gruner I, Hagl B, Haberle B, Muller-Hocker J, von Schweinitz D, et al. Blocking the hedgehog pathway inhibits hepatoblastoma growth. Hepatology. 2009;49(2):482–90. Epub 2009/01/30. doi: 10.1002/hep.22649. [DOI] [PubMed] [Google Scholar]
- 239.Stecca B, Mas C, Clement V, Zbinden M, Correa R, Piguet V, et al. Melanomas require HEDGEHOG-GLI signaling regulated by interactions between GLI1 and the RAS-MEK/AKT pathways. Proc Natl Acad Sci U S A. 2007;104(14):5895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Santini R, Vinci MC, Pandolfi S, Penachioni JY, Montagnani V, Olivito B, et al. Hedgehog-GLI signaling drives self-renewal and tumorigenicity of human melanoma-initiating cells. Stem Cells. 2012;30(9):1808–18. Epub 2012/06/26. doi: 10.1002/stem.1160. [DOI] [PubMed] [Google Scholar]
- 241.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol. 2007;17(2):165–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ehtesham M, Sarangi A, Valadez JG, Chanthaphaychith S, Becher MW, Abel TW, et al. Ligand-dependent activation of the hedgehog pathway in glioma progenitor cells. Oncogene. 2007;26(39):5752–61. Epub 2007/03/14. doi: 10.1038/sj.onc.1210359. [DOI] [PubMed] [Google Scholar]
- 243.Becher OJ, Hambardzumyan D, Fomchenko EI, Momota H, Mainwaring L, Bleau AM, et al. Gli activity correlates with tumor grade in platelet-derived growth factor-induced gliomas. Cancer Res. 2008;68(7):2241–9. Epub 2008/04/03. doi: 10.1158/0008-5472.CAN-07-6350. [DOI] [PubMed] [Google Scholar]
- 244.Kubo M, Nakamura M, Tasaki A, Yamanaka N, Nakashima H, Nomura M, et al. Hedgehog signaling pathway is a new therapeutic target for patients with breast cancer. Cancer Res. 2004;64(17):6071–4. Epub 2004/09/03. doi: 10.1158/0008-5472.CAN-04-0416. [DOI] [PubMed] [Google Scholar]
- 245.O’Toole SA, Machalek DA, Shearer RF, Millar EK, Nair R, Schofield P, et al. Hedgehog overexpression is associated with stromal interactions and predicts for poor outcome in breast cancer. Cancer Res. 2011;71(11):4002–14. Epub 2011/06/03. doi: 10.1158/0008-5472.CAN-10-3738. [DOI] [PubMed] [Google Scholar]
- 246.Neelakantan D, Zhou H, Oliphant MUJ, Zhang X, Simon LM, Henke DM, et al. EMT cells increase breast cancer metastasis via paracrine GLI activation in neighbouring tumour cells. Nat Commun. 2017;8:15773. Epub 2017/06/13. doi: 10.1038/ncomms15773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Cazet AS, Hui MN, Elsworth BL, Wu SZ, Roden D, Chan CL, et al. Targeting stromal remodeling and cancer stem cell plasticity overcomes chemoresistance in triple negative breast cancer. Nat Commun. 2018;9(1):2897. Epub 2018/07/26. doi: 10.1038/s41467-018-05220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Chen X, Horiuchi A, Kikuchi N, Osada R, Yoshida J, Shiozawa T, et al. Hedgehog signal pathway is activated in ovarian carcinomas, correlating with cell proliferation: it’s inhibition leads to growth suppression and apoptosis. Cancer Sci. 2007;98(1):68–76. Epub 2006/11/07. doi: 10.1111/j.1349-7006.2006.00353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Peacock CD, Wang Q, Gesell GS, Corcoran-Schwartz IM, Jones E, Kim J, et al. Hedgehog signaling maintains a tumor stem cell compartment in multiple myeloma. Proc Natl Acad Sci U S A. 2007;104(10):4048–53. Epub 2007/03/16. doi: 10.1073/pnas.0611682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Dierks C, Grbic J, Zirlik K, Beigi R, Englund NP, Guo GR, et al. Essential role of stromally induced hedgehog signaling in B-cell malignancies. Nature medicine. 2007;13(8):944–51. Epub 2007/07/17. doi: 10.1038/nm1614. [DOI] [PubMed] [Google Scholar]
- 251.Dierks C, Beigi R, Guo GR, Zirlik K, Stegert MR, Manley P, et al. Expansion of Bcr-Abl-positive leukemic stem cells is dependent on Hedgehog pathway activation. Cancer Cell. 2008;14(3):238–49. Epub 2008/09/06. doi: S1535–6108(08)00257–2 [pii] 10.1016/j.ccr.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 252.Wang DH, Tiwari A, Kim ME, Clemons NJ, Regmi NL, Hodges WA, et al. Hedgehog signaling regulates FOXA2 in esophageal embryogenesis and Barrett’s metaplasia. The Journal of clinical investigation. 2014;124(9):3767–80. Epub 2014/08/02. doi: 10.1172/JCI66603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fan L, Pepicelli CV, Dibble CC, Catbagan W, Zarycki JL, Laciak R, et al. Hedgehog signaling promotes prostate xenograft tumor growth. Endocrinology. 2004;145(8):3961–70. Epub 2004/05/11. doi: 10.1210/en.2004-0079. [DOI] [PubMed] [Google Scholar]
- 254.Tian H, Callahan CA, DuPree KJ, Darbonne WC, Ahn CP, Scales SJ, et al. Hedgehog signaling is restricted to the stromal compartment during pancreatic carcinogenesis. Proc Natl Acad Sci U S A. 2009;106(11):4254–9. Epub 2009/02/28. doi: 10.1073/pnas.0813203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Buller NV, Rosekrans SL, Metcalfe C, Heijmans J, van Dop WA, Fessler E, et al. Stromal Indian hedgehog signaling is required for intestinal adenoma formation in mice. Gastroenterology. 2015;148(1):170–80 e6. Epub 2014/10/14. doi: 10.1053/j.gastro.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 256.Gerling M, Buller NV, Kirn LM, Joost S, Frings O, Englert B, et al. Stromal Hedgehog signalling is downregulated in colon cancer and its restoration restrains tumour growth. Nat Commun. 2016;7:12321. Epub 2016/08/06. doi: 10.1038/ncomms12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Coquenlorge S, Yin WC, Yung T, Pan J, Zhang X, Mo R, et al. GLI2 Modulated by SUFU and SPOP Induces Intestinal Stem Cell Niche Signals in Development and Tumorigenesis. Cell Rep. 2019;27(10):3006–18 e4. Epub 2019/06/06. doi: 10.1016/j.celrep.2019.05.016. [DOI] [PubMed] [Google Scholar]
- 258.Singh S, Wang Z, Liang Fei D, Black KE, Goetz JA, Tokhunts R, et al. Hedgehog-producing cancer cells respond to and require autocrine Hedgehog activity. Cancer Res. 2011;71(13):4454–63. doi: 10.1158/0008-5472.CAN-10-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Shaw A, Gipp J, Bushman W. The Sonic Hedgehog pathway stimulates prostate tumor growth by paracrine signaling and recapitulates embryonic gene expression in tumor myofibroblasts. Oncogene. 2009;28(50):4480–90. Epub 2009/09/29. doi: 10.1038/onc.2009.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Shin K, Lim A, Odegaard JI, Honeycutt JD, Kawano S, Hsieh MH, et al. Cellular origin of bladder neoplasia and tissue dynamics of its progression to invasive carcinoma. Nat Cell Biol. 2014;16(5):469–78. Epub 2014/04/22. doi: 10.1038/ncb2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Shin K, Lim A, Zhao C, Sahoo D, Pan Y, Spiekerkoetter E, et al. Hedgehog signaling restrains bladder cancer progression by eliciting stromal production of urothelial differentiation factors. Cancer Cell. 2014;26(4):521–33. Epub 2014/10/15. doi: 10.1016/j.ccell.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Feldmann G, Dhara S, Fendrich V, Bedja D, Beaty R, Mullendore M, et al. Blockade of hedgehog signaling inhibits pancreatic cancer invasion and metastases: a new paradigm for combination therapy in solid cancers. Cancer Res. 2007;67(5):2187–96. Epub 2007/03/03. doi: 10.1158/0008-5472.CAN-06-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Feldmann G, Habbe N, Dhara S, Bisht S, Alvarez H, Fendrich V, et al. Hedgehog inhibition prolongs survival in a genetically engineered mouse model of pancreatic cancer. Gut. 2008;57(10):1420–30. Epub 2008/06/03. doi: 10.1136/gut.2007.148189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Adams CR, Htwe HH, Marsh T, Wang AL, Montoya ML, Subbaraj L, et al. Transcriptional control of subtype switching ensures adaptation and growth of pancreatic cancer. Elife. 2019;8. Epub 2019/05/29. doi: 10.7554/eLife.45313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME, et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes And Development. 2009;23(1):24–36. Epub 2009/01/13. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Jia Y, Gu D, Wan J, Yu B, Zhang X, Chiorean EG, et al. The role of GLI-SOX2 signaling axis for gemcitabine resistance in pancreatic cancer. Oncogene. 2019;38(10):1764–77. Epub 2018/11/02. doi: 10.1038/s41388-018-0553-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lee JJ, Perera RM, Wang H, Wu DC, Liu XS, Han S, et al. Stromal response to Hedgehog signaling restrains pancreatic cancer progression. Proc Natl Acad Sci U S A. 2014;111(30):E3091–100. Epub 2014/07/16. doi: 10.1073/pnas.1411679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Rosow DE, Liss AS, Strobel O, Fritz S, Bausch D, Valsangkar NP, et al. Sonic Hedgehog in pancreatic cancer: from bench to bedside, then back to the bench. Surgery. 2012;152(3 Suppl 1):S19–32. Epub 2012/07/10. doi: 10.1016/j.surg.2012.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Petrova E, Rios-Esteves J, Ouerfelli O, Glickman JF, Resh MD. Inhibitors of Hedgehog acyltransferase block Sonic Hedgehog signaling. Nat Chem Biol. 2013;9(4):247–9. Epub 2013/02/19. doi: 10.1038/nchembio.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Stanton BZ, Peng LF, Maloof N, Nakai K, Wang X, Duffner JL, et al. A small molecule that binds Hedgehog and blocks its signaling in human cells. Nat Chem Biol. 2009;5(3):154–6. Epub 2009/01/20. doi: 10.1038/nchembio.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Peng LF, Stanton BZ, Maloof N, Wang X, Schreiber SL. Syntheses of aminoalcohol-derived macrocycles leading to a small-molecule binder to and inhibitor of Sonic Hedgehog. Bioorg Med Chem Lett. 2009;19(22):6319–25. Epub 2009/10/13. doi: 10.1016/j.bmcl.2009.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wang Y, Arvanites AC, Davidow L, Blanchard J, Lam K, Yoo JW, et al. Selective identification of hedgehog pathway antagonists by direct analysis of smoothened ciliary translocation. ACS Chem Biol. 2012;7(6):1040–8. Epub 2012/05/05. doi: 10.1021/cb300028a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Taipale J, Chen JK, Cooper MK, Wang B, Mann RK, Milenkovic L, et al. Effects of oncogenic mutations in Smoothened and Patched can be reversed by cyclopamine. Nature. 2000;406(6799):1005–9. [DOI] [PubMed] [Google Scholar]
- 274.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci U S A. 2002;99(22):14071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Williams JA, Guicherit OM, Zaharian BI, Xu Y, Chai L, Wichterle H, et al. Identification of a small molecule inhibitor of the hedgehog signaling pathway: effects on basal cell carcinoma-like lesions. Proc Natl Acad Sci U S A. 2003;100(8):4616–21. Epub 2003/04/08. doi: 10.1073/pnas.0732813100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Bijlsma MF, Spek CA, Zivkovic D, van de Water S, Rezaee F, Peppelenbosch MP. Repression of smoothened by patched-dependent (pro-)vitamin D3 secretion. PLoS Biol. 2006;4(8):e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Song S, Jiang J, Zhao L, Wang Q, Lu W, Zheng C, et al. Structural optimization on a virtual screening hit of smoothened receptor. Eur J Med Chem. 2019;172:1–15. Epub 2019/04/03. doi: 10.1016/j.ejmech.2019.03.057. [DOI] [PubMed] [Google Scholar]
- 278.Lauth M, Bergstrom A, Shimokawa T, Toftgard R. Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A. 2007;104(20):8455–60. Epub 2007/05/15. doi: 10.1073/pnas.0609699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Hyman JM, Firestone AJ, Heine VM, Zhao Y, Ocasio CA, Han K, et al. Small-molecule inhibitors reveal multiple strategies for Hedgehog pathway blockade. Proc Natl Acad Sci U S A. 2009;106(33):14132–7. Epub 2009/08/12. doi: 0907134106 [pii] 10.1073/pnas.0907134106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Infante P, Mori M, Alfonsi R, Ghirga F, Aiello F, Toscano S, et al. Gli1/DNA interaction is a druggable target for Hedgehog-dependent tumors. Embo Journal. 2015;34(2):200–17. Epub 2014/12/06. doi: 10.15252/embj.201489213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Sekulic A, Von Hoff D. Hedgehog Pathway Inhibition. Cell. 2016;164(5):831. Epub 2016/02/27. doi: 10.1016/j.cell.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 282.Robarge KD, Brunton SA, Castanedo GM, Cui Y, Dina MS, Goldsmith R, et al. GDC-0449-a potent inhibitor of the hedgehog pathway. Bioorg Med Chem Lett. 2009;19(19):5576–81. Epub 2009/09/01. doi: 10.1016/j.bmcl.2009.08.049. [DOI] [PubMed] [Google Scholar]
- 283.Pan S, Wu X, Jiang J, Gao W, Wan Y, Cheng D, et al. Discovery of NVP-LDE225, a Potent and Selective Smoothened Antagonist. ACS Med Chem Lett. 2010;1(3):130–4. Epub 2010/06/10. doi: 10.1021/ml1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Tremblay MR, Lescarbeau A, Grogan MJ, Tan E, Lin G, Austad BC, et al. Discovery of a potent and orally active hedgehog pathway antagonist (IPI-926). J Med Chem. 2009;52(14):4400–18. [DOI] [PubMed] [Google Scholar]
- 285.Kim J, Tang JY, Gong R, Kim J, Lee JJ, Clemons KV, et al. Itraconazole, a commonly used antifungal that inhibits Hedgehog pathway activity and cancer growth. Cancer Cell. 2010;17(4):388–99. Epub 2010/04/14. doi: 10.1016/j.ccr.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Fukushima N, Minami Y, Kakiuchi S, Kuwatsuka Y, Hayakawa F, Jamieson C, et al. Small-molecule Hedgehog inhibitor attenuates the leukemia-initiation potential of acute myeloid leukemia cells. Cancer Sci. 2016;107(10):1422–9. Epub 2016/10/30. doi: 10.1111/cas.13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Hosoya T, Arai MA, Koyano T, Kowithayakorn T, Ishibashi M. Naturally occurring small-molecule inhibitors of hedgehog/GLI-mediated transcription. Chembiochem. 2008;9(7):1082–92. Epub 2008/03/22. doi: 10.1002/cbic.200700511. [DOI] [PubMed] [Google Scholar]
- 288.Kim J, Lee JJ, Kim J, Gardner D, Beachy PA. Arsenic antagonizes the Hedgehog pathway by preventing ciliary accumulation and reducing stability of the Gli2 transcriptional effector. Proc Natl Acad Sci U S A. 2010;107(30):13432–7. Epub 2010/07/14. doi: 10.1073/pnas.1006822107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Beauchamp EM, Ringer L, Bulut G, Sajwan KP, Hall MD, Lee YC, et al. Arsenic trioxide inhibits human cancer cell growth and tumor development in mice by blocking Hedgehog/GLI pathway. The Journal of clinical investigation. 2011;121(1):148–60. Epub 2010/12/25. doi: 10.1172/JCI42874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Hom ME, Ondrus AE, Sakata-Kato T, Rack PG, Chen JK. Bicyclic Imidazolium Inhibitors of Gli Transcription Factor Activity. ChemMedChem. 2020;15(12):1044–9. Epub 2020/04/09. doi: 10.1002/cmdc.202000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Piirsoo A, Kasak L, Kauts ML, Loog M, Tints K, Uusen P, et al. Protein kinase inhibitor SU6668 attenuates positive regulation of Gli proteins in cancer and multipotent progenitor cells. Biochim Biophys Acta. 2014;1843(4):703–14. Epub 2014/01/15. doi: 10.1016/j.bbamcr.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Li B, Fei DL, Flaveny CA, Dahmane N, Baubet V, Wang Z, et al. Pyrvinium attenuates Hedgehog signaling downstream of smoothened. Cancer Res. 2014;74(17):4811–21. Epub 2014/07/06. doi: 10.1158/0008-5472.CAN-14-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Wu X, Zhang LS, Toombs J, Kuo YC, Piazza JT, Tuladhar R, et al. Extra-mitochondrial prosurvival BCL-2 proteins regulate gene transcription by inhibiting the SUFU tumour suppressor. Nat Cell Biol. 2017;19(10):1226–36. Epub 2017/09/26. doi: 10.1038/ncb3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Torrado B, Grana M, Badano JL, Irigoin F. Ciliary Entry of the Hedgehog Transcriptional Activator Gli2 Is Mediated by the Nuclear Import Machinery but Differs from Nuclear Transport in Being Imp-alpha/beta1-Independent. PLoS One. 2016;11(8):e0162033. doi: 10.1371/journal.pone.0162033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Cansizoglu AE, Lee BJ, Zhang ZC, Fontoura BM, Chook YM. Structure-based design of a pathway-specific nuclear import inhibitor. Nat Struct Mol Biol. 2007;14(5):452–4. Epub 2007/04/17. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Von Hoff DD, LoRusso PM, Rudin CM, Reddy JC, Yauch RL, Tibes R, et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N Engl J Med. 2009;361(12):1164–72. Epub 2009/09/04. doi: 10.1056/NEJMoa0905360. [DOI] [PubMed] [Google Scholar]
- 297.Sekulic A, Migden MR, Oro AE, Dirix L, Lewis KD, Hainsworth JD, et al. Efficacy and safety of vismodegib in advanced basal-cell carcinoma. N Engl J Med. 2012;366(23):2171–9. Epub 2012/06/08. doi: 10.1056/NEJMoa1113713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Tang JY, Mackay-Wiggan JM, Aszterbaum M, Yauch RL, Lindgren J, Chang K, et al. Inhibiting the hedgehog pathway in patients with the basal-cell nevus syndrome. N Engl J Med. 2012;366(23):2180–8. Epub 2012/06/08. doi: 10.1056/NEJMoa1113538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Migden MR, Guminski A, Gutzmer R, Dirix L, Lewis KD, Combemale P, et al. Treatment with two different doses of sonidegib in patients with locally advanced or metastatic basal cell carcinoma (BOLT): a multicentre, randomised, double-blind phase 2 trial. Lancet Oncol. 2015;16(6):716–28. Epub 2015/05/20. doi: 10.1016/S1470-2045(15)70100-2. [DOI] [PubMed] [Google Scholar]
- 300.Gajjar A, Stewart CF, Ellison DW, Kaste S, Kun LE, Packer RJ, et al. Phase I study of vismodegib in children with recurrent or refractory medulloblastoma: a pediatric brain tumor consortium study. Clin Cancer Res. 2013;19(22):6305–12. Epub 2013/10/01. doi: 10.1158/1078-0432.CCR-13-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Robinson GW, Orr BA, Wu G, Gururangan S, Lin T, Qaddoumi I, et al. Vismodegib Exerts Targeted Efficacy Against Recurrent Sonic Hedgehog-Subgroup Medulloblastoma: Results From Phase II Pediatric Brain Tumor Consortium Studies PBTC-025B and PBTC-032. J Clin Oncol. 2015;33(24):2646–54. Epub 2015/07/15. doi: 10.1200/JCO.2014.60.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Kieran MW, Chisholm J, Casanova M, Brandes AA, Aerts I, Bouffet E, et al. Phase I study of oral sonidegib (LDE225) in pediatric brain and solid tumors and a phase II study in children and adults with relapsed medulloblastoma. Neuro Oncol. 2017;19(11):1542–52. Epub 2017/06/13. doi: 10.1093/neuonc/nox109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Pietanza MC, Litvak AM, Varghese AM, Krug LM, Fleisher M, Teitcher JB, et al. A phase I trial of the Hedgehog inhibitor, sonidegib (LDE225), in combination with etoposide and cisplatin for the initial treatment of extensive stage small cell lung cancer. Lung Cancer. 2016;99:23–30. Epub 2016/08/28. doi: 10.1016/j.lungcan.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Belani CP, Dahlberg SE, Rudin CM, Fleisher M, Chen HX, Takebe N, et al. Vismodegib or cixutumumab in combination with standard chemotherapy for patients with extensive-stage small cell lung cancer: A trial of the ECOG-ACRIN Cancer Research Group (E1508). Cancer. 2016;122(15):2371–8. Epub 2016/05/11. doi: 10.1002/cncr.30062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kim EJ, Sahai V, Abel EV, Griffith KA, Greenson JK, Takebe N, et al. Pilot clinical trial of hedgehog pathway inhibitor GDC-0449 (vismodegib) in combination with gemcitabine in patients with metastatic pancreatic adenocarcinoma. Clin Cancer Res. 2014;20(23):5937–45. Epub 2014/10/04. doi: 10.1158/1078-0432.CCR-14-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Catenacci DV, Junttila MR, Karrison T, Bahary N, Horiba MN, Nattam SR, et al. Randomized Phase Ib/II Study of Gemcitabine Plus Placebo or Vismodegib, a Hedgehog Pathway Inhibitor, in Patients With Metastatic Pancreatic Cancer. J Clin Oncol. 2015;33(36):4284–92. Epub 2015/11/04. doi: 10.1200/JCO.2015.62.8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ko AH, LoConte N, Tempero MA, Walker EJ, Kate Kelley R, Lewis S, et al. A Phase I Study of FOLFIRINOX Plus IPI-926, a Hedgehog Pathway Inhibitor, for Advanced Pancreatic Adenocarcinoma. Pancreas. 2016;45(3):370–5. Epub 2015/09/22. doi: 10.1097/MPA.0000000000000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Kaye SB, Fehrenbacher L, Holloway R, Amit A, Karlan B, Slomovitz B, et al. A phase II, randomized, placebo-controlled study of vismodegib as maintenance therapy in patients with ovarian cancer in second or third complete remission. Clin Cancer Res. 2012;18(23):6509–18. Epub 2012/10/04. doi: 10.1158/1078-0432.CCR-12-1796. [DOI] [PubMed] [Google Scholar]
- 309.Antonarakis ES, Heath EI, Smith DC, Rathkopf D, Blackford AL, Danila DC, et al. Repurposing itraconazole as a treatment for advanced prostate cancer: a noncomparative randomized phase II trial in men with metastatic castration-resistant prostate cancer. Oncologist. 2013;18(2):163–73. Epub 2013/01/24. doi: 10.1634/theoncologist.2012-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Maughan BL, Suzman DL, Luber B, Wang H, Glavaris S, Hughes R, et al. Pharmacodynamic study of the oral hedgehog pathway inhibitor, vismodegib, in patients with metastatic castration-resistant prostate cancer. Cancer Chemother Pharmacol. 2016;78(6):1297–304. Epub 2016/11/09. doi: 10.1007/s00280-016-3191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Berlin J, Bendell JC, Hart LL, Firdaus I, Gore I, Hermann RC, et al. A randomized phase II trial of vismodegib versus placebo with FOLFOX or FOLFIRI and bevacizumab in patients with previously untreated metastatic colorectal cancer. Clin Cancer Res. 2013;19(1):258–67. Epub 2012/10/20. doi: 10.1158/1078-0432.CCR-12-1800. [DOI] [PubMed] [Google Scholar]
- 312.Minami Y, Minami H, Miyamoto T, Yoshimoto G, Kobayashi Y, Munakata W, et al. Phase I study of glasdegib (PF-04449913), an oral smoothened inhibitor, in Japanese patients with select hematologic malignancies. Cancer Sci. 2017;108(8):1628–33. Epub 2017/05/31. doi: 10.1111/cas.13285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Martinelli G, Oehler VG, Papayannidis C, Courtney R, Shaik MN, Zhang X, et al. Treatment with PF-04449913, an oral smoothened antagonist, in patients with myeloid malignancies: a phase 1 safety and pharmacokinetics study. Lancet Haematol. 2015;2(8):e339–46. Epub 2015/12/22. doi: 10.1016/S2352-3026(15)00096-4. [DOI] [PubMed] [Google Scholar]
- 314.Savona MR, Pollyea DA, Stock W, Oehler VG, Schroeder MA, Lancet J, et al. Phase Ib Study of Glasdegib, a Hedgehog Pathway Inhibitor, in Combination with Standard Chemotherapy in Patients with AML or High-Risk MDS. Clin Cancer Res. 2018;24(10):2294–303. Epub 2018/02/22. doi: 10.1158/1078-0432.CCR-17-2824. [DOI] [PubMed] [Google Scholar]
- 315.Cortes JE, Douglas Smith B, Wang ES, Merchant A, Oehler VG, Arellano M, et al. Glasdegib in combination with cytarabine and daunorubicin in patients with AML or high-risk MDS: Phase 2 study results. Am J Hematol. 2018;93(11):1301–10. Epub 2018/08/04. doi: 10.1002/ajh.25238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Bixby D, Noppeney R, Lin TL, Cortes J, Krauter J, Yee K, et al. Safety and efficacy of vismodegib in relapsed/refractory acute myeloid leukaemia: results of a phase Ib trial. Br J Haematol. 2019;185(3):595–8. Epub 2018/09/12. doi: 10.1111/bjh.15571. [DOI] [PubMed] [Google Scholar]
- 317.Cortes JE, Dombret H, Merchant A, Tauchi T, DiRienzo CG, Sleight B, et al. Glasdegib plus intensive/nonintensive chemotherapy in untreated acute myeloid leukemia: BRIGHT AML 1019 Phase III trials. Future Oncol. 2019;15(31):3531–45. Epub 2019/09/14. doi: 10.2217/fon-2019-0373. [DOI] [PubMed] [Google Scholar]
- 318.Xie H, Paradise BD, Ma WW, Fernandez-Zapico ME. Recent Advances in the Clinical Targeting of Hedgehog/GLI Signaling in Cancer. Cells. 2019;8(5). Epub 2019/05/01. doi: 10.3390/cells8050394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Axelson M, Liu K, Jiang X, He K, Wang J, Zhao H, et al. U.S. Food and Drug Administration approval: vismodegib for recurrent, locally advanced, or metastatic basal cell carcinoma. Clin Cancer Res. 2013;19(9):2289–93. Epub 2013/03/22. doi: 10.1158/1078-0432.CCR-12-1956. [DOI] [PubMed] [Google Scholar]
- 320.Goldsmith SR, Lovell AR, Schroeder MA. Glasdegib for the treatment of adult patients with newly diagnosed acute myeloid leukemia or high-grade myelodysplastic syndrome who are elderly or otherwise unfit for standard induction chemotherapy. Drugs Today (Barc). 2019;55(9):545–62. Epub 2019/10/05. doi: 10.1358/dot.2019.55.9.3020160. [DOI] [PubMed] [Google Scholar]
- 321.Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326(5952):572–4. Epub 2009/09/04. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Chang AL, Oro AE. Initial assessment of tumor regrowth after vismodegib in advanced Basal cell carcinoma. Arch Dermatol. 2012;148(11):1324–5. Epub 2012/08/23. doi: 10.1001/archdermatol.2012.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Atwood SX, Sarin KY, Whitson RJ, Li JR, Kim G, Rezaee M, et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):342–53. Epub 2015/03/12. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Sharpe HJ, Pau G, Dijkgraaf GJ, Basset-Seguin N, Modrusan Z, Januario T, et al. Genomic analysis of smoothened inhibitor resistance in basal cell carcinoma. Cancer Cell. 2015;27(3):327–41. Epub 2015/03/12. doi: 10.1016/j.ccell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Buonamici S, Williams J, Morrissey M, Wang A, Guo R, Vattay A, et al. Interfering with resistance to smoothened antagonists by inhibition of the PI3K pathway in medulloblastoma. Sci Transl Med. 2010;2(51):51ra70. Epub 2010/10/01. doi: 10.1126/scitranslmed.3001599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Dijkgraaf GJ, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 2011;71(2):435–44. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- 327.Whitson RJ, Lee A, Urman NM, Mirza A, Yao CY, Brown AS, et al. Noncanonical hedgehog pathway activation through SRF-MKL1 promotes drug resistance in basal cell carcinomas. Nature medicine. 2018;24(3):271–81. Epub 2018/02/06. doi: 10.1038/nm.4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Yao CD, Haensel D, Gaddam S, Patel T, Atwood SX, Sarin KY, et al. AP-1 and TGFss cooperativity drives non-canonical Hedgehog signaling in resistant basal cell carcinoma. Nat Commun. 2020;11(1):5079. Epub 2020/10/10. doi: 10.1038/s41467-020-18762-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Zhu GA, Sundram U, Chang AL. Two different scenarios of squamous cell carcinoma within advanced Basal cell carcinomas: cases illustrating the importance of serial biopsy during vismodegib usage. JAMA Dermatol. 2014;150(9):970–3. Epub 2014/04/18. doi: 10.1001/jamadermatol.2014.583. [DOI] [PubMed] [Google Scholar]
- 330.Ransohoff KJ, Tang JY, Sarin KY. Squamous Change in Basal-Cell Carcinoma with Drug Resistance. N Engl J Med. 2015;373(11):1079–82. Epub 2015/09/10. doi: 10.1056/NEJMc1504261. [DOI] [PubMed] [Google Scholar]
- 331.Zhao X, Ponomaryov T, Ornell KJ, Zhou P, Dabral SK, Pak E, et al. RAS/MAPK Activation Drives Resistance to Smo Inhibition, Metastasis, and Tumor Evolution in Shh Pathway-Dependent Tumors. Cancer Res. 2015;75(17):3623–35. Epub 2015/07/02. doi: 10.1158/0008-5472.CAN-14-2999-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Kuonen F, Huskey NE, Shankar G, Jaju P, Whitson RJ, Rieger KE, et al. Loss of Primary Cilia Drives Switching from Hedgehog to Ras/MAPK Pathway in Resistant Basal Cell Carcinoma. J Invest Dermatol. 2019;139(7):1439–48. Epub 2019/02/02. doi: 10.1016/j.jid.2018.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Biehs B, Dijkgraaf GJP, Piskol R, Alicke B, Boumahdi S, Peale F, et al. A cell identity switch allows residual BCC to survive Hedgehog pathway inhibition. Nature. 2018;562(7727):429–33. Epub 2018/10/10. doi: 10.1038/s41586-018-0596-y. [DOI] [PubMed] [Google Scholar]
- 334.Sanchez-Danes A, Larsimont JC, Liagre M, Munoz-Couselo E, Lapouge G, Brisebarre A, et al. A slow-cycling LGR5 tumour population mediates basal cell carcinoma relapse after therapy. Nature. 2018;562(7727):434–8. Epub 2018/10/10. doi: 10.1038/s41586-018-0603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Bidet M, Tomico A, Martin P, Guizouarn H, Mollat P, Mus-Veteau I. The Hedgehog receptor patched functions in multidrug transport and chemotherapy resistance. Mol Cancer Res. 2012;10(11):1496–508. Epub 2012/07/04. doi: 10.1158/1541-7786.MCR-11-0578. [DOI] [PubMed] [Google Scholar]
- 336.Zahreddine HA, Culjkovic-Kraljacic B, Assouline S, Gendron P, Romeo AA, Morris SJ, et al. The sonic hedgehog factor GLI1 imparts drug resistance through inducible glucuronidation. Nature. 2014;511(7507):90–3. Epub 2014/05/30. doi: 10.1038/nature13283. [DOI] [PMC free article] [PubMed] [Google Scholar]


