Abstract
Transcription factor IIF (TFIIF) is a protein allosteric effector for RNA polymerase II during the initiation and elongation phases of the transcription cycle. In initiation, TFIIF induces promoter DNA to wrap almost a full turn around RNA polymerase II in a complex that includes the general transcription factors TATA-binding protein, TFIIB, and TFIIE. During elongation, TFIIF also supports a more active conformation of RNA polymerase II. This conformational model for elongation is supported by three lines of experimental evidence. First, a region within the RNA polymerase II-associating protein 74 (RAP74) subunit of TFIIF (amino acids T154 to M177), a region that is critical for isomerization of the preinitiation complex, is also critical for elongation stimulation. Amino acid substitutions within this region are shown to have very similar effects on initiation and elongation, and mutagenic analysis indicates that L155, W164, N172, I176, and M177 are the most important residues in this region for transcription. Second, TFIIF is shown to have a higher affinity for rapidly elongating RNA polymerase II than for the stalled elongation complex, indicating that RNA polymerase II alternates between active and inactive states during elongation and that TFIIF stimulates elongation by supporting the active conformational state of RNA polymerase II. The deleterious I176A substitution in the critical region of RAP74 decreases the affinity of TFIIF for the active form of the elongation complex. Third, TFIIF is shown by Arrhenius analysis to stimulate elongation by populating an activated state of RNA polymerase II.
In transcriptional initiation by multisubunit RNA polymerases, “isomerization” is a progression of conformational changes in DNA, RNA polymerase, and the general transcription factors that leads to formation of an open complex capable of forming phosphodiester bonds (8). In Escherichia coli, promoter DNA bends around RNA polymerase holoenzyme in the upstream promoter region and again around the +1 region of the promoter as the template passes through the RNA polymerase active site (28). In the vicinity of positions −5 to +15, the DNA is buried within a processivity sliding clamp made up of evolutionarily conserved domains of the RNA polymerase catalytic subunits β and β′ (26–28). E. coli RNA polymerase and promoter DNA progress through a series of conformational intermediates in the initiation mechanism (5, 36) involving DNA bending (25, 30), DNA wrapping around RNA polymerase (9, 28, 33), helix untwisting (1, 15), closing of the sliding clamp around the DNA (26–28), and separation of DNA strands for initiation (8). A mammalian homolog of E. coli RNA polymerase, RNA polymerase II, which functions in concert with the general transcription factor TATA-binding protein (TBP) and transcription factor IIB (TFIIB), TFIIF, TFIIE, and TFIIH, appears to function in initiation by a very similar mechanism, involving DNA bending and wrapping, closing of a processivity sliding clamp, and helix untwisting prior to open complex formation (8, 20, 32, 34).
TFIIF is an α2β2 heterotetramer of the RNA polymerase II-associating protein 74 (RAP74) and RAP30 subunits (6, 12, 13, 21, 42). TFIIF binds to RNA polymerase II in solution and helps deliver RNA polymerase II to the promoter (7, 13, 19). In a preinitiation complex formed on the adenovirus major late promoter, RAP74 helps to wrap DNA almost a full turn around RNA polymerase II and the general transcription factors (34). RAP74 develops or tightens specific contacts between promoter DNA and RAP30, TFIIE, and RNA polymerase II (14, 34, 35). The RAP74 subunit of TFIIF has been suggested to stimulate helix untwisting for initiation, because RAP74 mutants that are defective in preinitiation complex isomerization appear to be partially complemented in initiation by supercoiling of the DNA template (32). TFIIF, therefore, may have a role in isomerization of the preinitiation complex, causing helix untwisting prior to formation of the open complex. From a DNA template that is not negatively supercoiled, separation of DNA strands requires at least one of the helicase subunits of TFIIH (40), so helix untwisting may precede strand separation in the normal course of RNA polymerase II initiation.
In addition to its activities in RNA polymerase II recruitment, isomerization, and initiation, TFIIF stimulates the rate of elongation by RNA polymerase II (3, 4, 16, 18, 22, 31, 39). TFIIF decreases the dwell time by RNA polymerase II at pause sites or possibly at all nucleotide addition sites. TFIIF has not been reported to alter the pattern of pause sites by RNA polymerase II, indicating that TFIIF may not interact directly with the RNA polymerase catalytic center but rather may have a less direct effect on elongation rates. Both the RAP74 and RAP30 subunits are required to stimulate elongation (18, 22, 39). In this paper, we report that mutants within the N-terminal domain of the RAP74 subunit of TFIIF have remarkably similar effects on initiation and elongation. We advance a number of arguments for the conclusion that TFIIF stimulates elongation by stabilizing an activated conformation of the elongation complex.
MATERIALS AND METHODS
Initiation assays.
Initiation assays were done as previously described (32). An extract derived from the nuclei of human HeLa cells (purchased from the National Cell Culture Center, Minneapolis, Minn.) was used as the source of transcription factors. TFIIF was removed from the extract by immunoprecipitation with anti-RAP74 and anti-RAP30 antibodies. Activity was reconstituted by addition of human recombinant TFIIF or a TFIIF mutant. The buffer for transcription reactions consisted of 12 mM HEPES, (pH 7.9), 12% (wt/vol) glycerol, 0.12 mM EDTA, 0.12 mM EGTA, and 1.2 mM dithiothreitol and contained 60 mM KCl and 12 mM MgCl2. The template for transcription was plasmid pML, carrying the adenovirus major late promoter from position −258 to +196. The template was digested with endonuclease SmaI at position +217 relative to the transcription start site. Recombinant TFIIF, extract, and DNA template were combined for 60 min at 30°C. For the initiation data shown in Fig. 4A, transcription was initiated with 100 μM (each) ATP, CTP, and GTP and 5 μCi of [α-32P]UTP (0.25 μM) for 1 min. For the data shown in Fig. 4B, transcription was initiated with 100 μM (each) ATP and CTP and 0.25 μM [α-32P]UTP for 1 min. After pulse labeling, reactions were chased for 30 min with 1 mM (each) ATP, CTP, GTP, and UTP in the presence of 0.25% Sarkosyl.
FIG. 4.
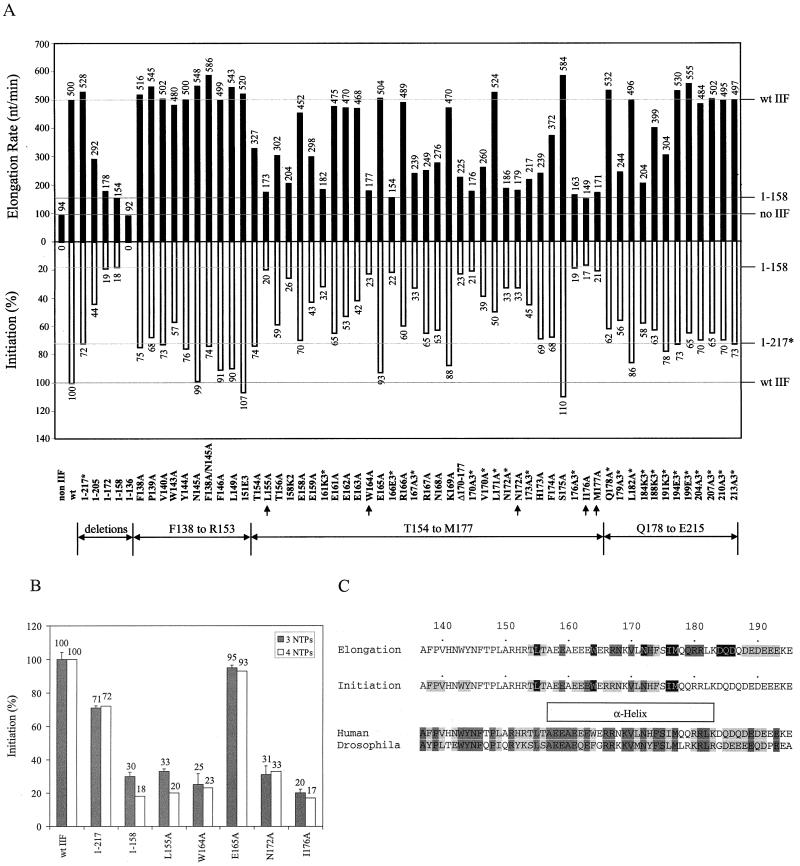
RAP74 mutants that are defective in stimulating the elongation rate of RNA polymerase II are also defective in accurate initiation. (A) Comparison of the activities of RAP74 mutants in elongation rate stimulation (solid bars) and accurate initiation (open bars). Accurate initiation was determined with an extract system depleted of TFIIF by immunoprecipitation with anti-RAP30 and anti-RAP74 antibodies. The template was the adenovirus major late promoter digested with endonuclease SmaI at position +217. Transcription was initiated with a 1-min pulse of 100 μM (each) ATP, CTP, and GTP and 0.25 μM [α-32P]UTP, followed by a 30-min chase with 1 mM (each) NTP and 0.25% Sarkosyl (32). The average of duplicate determinations is reported. The average elongation rate of mutants was estimated as described in the legend to Fig. 3. An asterisk (∗) indicates that a TFIIF mutant was constructed in RAP74(1–217) rather than in wt RAP74. (B) To confirm that the pulse-chase protocol primarily depends on initiation rather than early elongation, the procedure was repeated for select TFIIF samples with a pulse of 100 μM (each) ATP and CTP and 0.25 μM [α-32P]UTP, followed by a 30-min chase with 1 mM (each) NTP and 0.25% Sarkosyl (shaded bars). Samples were done in triplicate, and error bars represent standard deviations. The data shown for the four-NTP-pulse protocol (open bars) is from Fig. 4A. (C) Summary of mutagenic analysis, primarily considering single amino acid substitutions. Positions at which mutations showed transcriptional defects are shaded. Darker shading indicates a more severe defect, as described in the text. The predicted alpha-helix from A157 to K183 is indicated by a box below the sequence. An alignment of human and Drosophila RAP74 (17) is shown at the bottom to indicate the extent of evolutionary conservation. Darker shading indicates amino acid identity; lighter shading indicates amino acid similarity. Drosophila RAP74 can functionally replace human RAP74 in stimulating elongation and in supporting accurate initiation (18).
Immobilized DNA templates for elongation assays.
All of the templates used in these experiments contained the adenovirus major late promoter. Templates were synthesized by PCR with an upstream biotinylated primer. Biotinylated DNA was bound to paramagnetic streptavidin beads (Vector Laboratories, Inc., Burlingame, Calif.) (2, 22, 23). For the experiments represented by Fig. 1 through 4A and 5A, the upstream primer was 5′-biotin-CCCTCGAGCGGTGTTCCGCGGTCCTCCTCG-3′, the downstream primer was 5′-GTGCTCATCATTGGAAAACGTTCTT-3′, and the template for PCR was plasmid pML, containing the natural adenovirus major late promoter from position −263 to +196 relative to the transcriptional start site (+1). The DNA amplification product extends from position −263 to +1250. For the experiment represented by Fig. 5D, the upstream primer was 5′-biotin-ACGACAGGTTTCCCGACTGGAAAGC-3′, the downstream primer was 5′-GGTGTGAAATACCGCACAGATGCGT-3′, and the template was pML20-42 (the kind gift of Donal Luse) (38). Plasmid pML20-42 is useful to generate a template for synthesis of the defined U20 elongation complex, in which transcription is stalled at position +20, with a uridine base at the 3′ position. The template extended from position −300 to +300. For the experiments represented by Fig. 5C and 6, the upstream primer was 5′-biotin-ACGACAGGTTTCCCGACTGGAAAGC-3′, the downstream primer was 5′-GAGTACTCAACCAAGTCATTCTGAG-3′, and the template was pML20-42. The resulting amplification product extends from position −300 to +1000. For the experiment represented by Fig. 7, the upstream primer was 5′-biotin-ACGACAGGTTTCCCGACTGGAAAGC-3′, the downstream primer was 5′-GAGTACTCAACCAAGTCATTCTGAG-3′, and the template was pML20-49 (the kind gift of Donal Luse). Plasmid pML20-49 differs from pML20-42 in that pML20-49 contains a G-less cassette from position +24 to +120 downstream of the adenovirus major late promoter (38). The resulting amplification product extends from position −300 to +1087. This template is also convenient for synthesizing the U20 complex.
FIG. 1.
TFIIF stimulates elongation rate. Elongation complexes, initiated from the adenovirus major late promoter, were formed on immobilized templates and washed with 1% Sarkosyl and 0.5 M KCl to remove nascent elongation factors. After equilibration of complexes with transcription buffer, 20 pmol of TFIIF was added to the indicated samples (+) and elongation continued for the indicated times. Notice the similarity in the positions of pause sites in the presence and absence of TFIIF.
FIG. 5.
TFIIF can be committed to the DNA template. (A) Similar amounts of wt or mutant TFIIF are required to saturate the elongation assay mixture. TFIIF or TFIIF carrying the I176A RAP74 mutation (0, 5, 10, 20, 40, or 60 pmol) was added to elongation complexes. Elongation was for 30 s. (B) PhosphorImager quantitation of the data shown in panel A. (C) TFIIF stimulates elongation rates from bead templates after washing. TFIIF or a TFIIF mutant (20 pmol) was incubated with U20 elongation complexes. Complexes were either washed or not washed (as indicated), 1 mM (each) NTP was added, and elongation continued for 30 s. (D) Release of DNA from beads does not reduce TFIIF stimulation of elongation. U20 elongation complexes were incubated in the presence or absence of TFIIF and washed two times with 200 μl of transcription buffer. EcoRI was used to release the U20 elongation complex from the beads. F, supernatant (free) fraction; B, bead fraction. TFIIF stimulates elongation from both free and bead-associated DNA templates.
FIG. 6.
wt TFIIF has a higher affinity than mutant TFIIF for the rapid elongation complex. (A) Competition for the stalled elongation complex. The reaction scheme is shown above the gel. wt or mutant TFIIF was added to U20 elongation complexes. After 5 min, a competitor TFIIF sample was added, as indicated (5 or 50 pmol). After an additional 5 min of incubation, 1 mM (each) ATP, GTP, CTP, and UTP were added for 30 s. The order of addition of wt TFIIF and the I176A mutant did not affect competition, demonstrating that both wt and mutant TFIIF can exchange on the stalled elongation complex. Exchange was observed with a 10-fold molar excess of I176A competitor but was not apparent at molar equivalence of wt and I176A TFIIF. (B) Competition for the rapid elongation complex. The reaction scheme is shown above the gel. When RNA polymerase II elongation complexes were saturated with wt TFIIF, followed by addition of NTPs, and then the I176A mutant was added as a competitor, elongation was not noticeably inhibited (lanes 10, 11, 16, and 17; compare to control lanes 5 and 9). When RNA polymerase II elongation complexes were saturated with the I176A mutant, followed by addition of NTPs, and then wt TFIIF was added as a competitor, elongation was stimulated (lanes 12, 13, 20, and 21; compare to control lanes 6 and 8). These data show that wt TFIIF rapidly displaces the I176A mutant during elongation but that the I176A mutant is unable to noticeably displace wt TFIIF during elongation. Some control samples were chased for 45 s, because a 45-s chase provided a more reasonable comparison to a particular experimental sample. The amounts of proteins added are indicated (10 or 40 pmol).
FIG. 7.
TFIIF stimulates the elongation rate of RNA polymerase II by increasing the fraction of active elongation complexes rather than by altering the catalytic mechanism. (A) The average elongation rate of U20 complexes was determined at 23, 30, 37, and 42°C in the absence or presence of TFIIF or the strongly defective I176A mutant. (B) The data shown in panel A are displayed as an Arrhenius plot of the log average elongation rate versus the reciprocal of the absolute temperature (in kelvins). In the presence of TFIIF, the slope of the Arrhenius plot (−EA/R, where EA is the activation energy and R is the gas constant) remains constant but the intercept (log A, where A is the frequency factor) increases.
Elongation assays.
The source of transcription factors was an extract derived from human HeLa cells. Immobilized DNA template and extract were combined and incubated for 60 min at 30°C. For the experiments represented by Fig. 1 through 4A and 5A, transcription was initiated with a 1-min pulse of 100 μM (each) ATP, CTP, and GTP and 0.5 μM [α-32P]UTP. To form the U20 complex for the experiments represented by Fig. 5C, 5D, 6, and 7, transcription was initiated with a 10-min pulse of 1 mM ApC dinucleotide, 10 μM deoxy-ATP, 100 μM UTP, and 10 μCi of [α-32P]CTP, followed by a 10-min chase with 100 μM CTP (38). Complexes were then washed twice with 500 μl of transcription buffer containing 1% Sarkosyl, 0.5 M KCl, and 1 mg of bovine serum albumin per ml to remove nascent elongation and termination factors and twice with 500 μl of transcription buffer containing 60 mM KCl and 1 mg of bovine serum albumin per ml. Beads were collected with a magnetic particle separator. Complexes were resuspended in an appropriate volume of transcription buffer containing 60 mM KCl and 12 mM MgCl2 and subaliquoted into individual 20-μl samples. TFIIF or a TFIIF mutant was added to the reaction mixtures, as indicated in the figure legends. Elongation was continued by addition of 1 mM (each) ATP, CTP, UTP, and GTP. Elongation reactions were at room temperature, except for the experiment represented by Fig. 7, in which the temperature for elongation was varied. Reactions were stopped by addition of 200 μl of 0.1 M sodium acetate (pH 5.4), 0.5% sodium dodecyl sulfate, 2 mM EDTA, and 100 μg of yeast tRNA per ml. Samples were then extracted with phenol-chloroform and precipitated with ethanol. Samples were electrophoresed in a 10% polyacrylamide gel containing 50% (wt/vol) urea and Tris-borate-EDTA buffer. Gel images were analyzed with a Molecular Dynamics PhosphorImager.
RESULTS
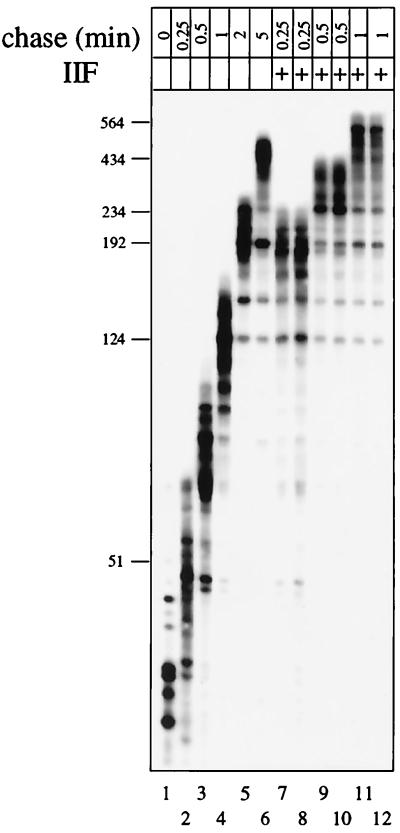
To measure the effect of TFIIF on elongation rate, RNA polymerase II elongation complexes were formed on immobilized templates (Fig. 1). In an extract derived from HeLa cell nuclei, transcription was accurately initiated from the adenovirus major late promoter with a 1-min pulse of 100 μM (each) ATP, CTP, and GTP, and 0.5 μM [α-32P]UTP. Transcription elongation complexes, generally containing RNA chains shorter than 50 nucleotides, were washed free of elongation and termination factors with buffer containing 1% Sarkosyl and 0.5 M KCl. Complexes were reequilibrated with transcription buffer, and a saturating amount of TFIIF was added to the indicated reaction mixtures. TFIIF stimulated the elongation rate about fivefold (Fig. 1; compare lane 6 with lanes 11 and 12). TFIIF has been proposed to reduce the dwell time of RNA polymerase II at pause sites. RNA polymerase II was found to pause at similar positions whether or not TFIIF was present in the reaction mixture.
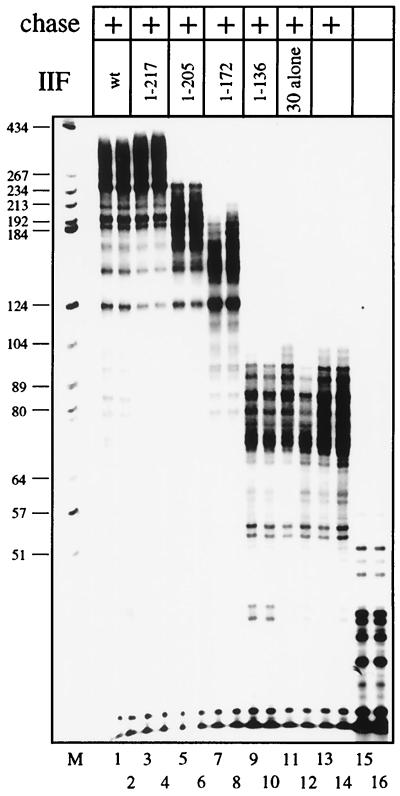
A series of deletion mutants in the RAP74 subunit of TFIIF was tested for the ability to stimulate elongation (Fig. 2). TFIIF containing the deletion mutant RAP74(1–217) (i.e., RAP74 consisting of amino acids 1 to 217) consistently stimulated elongation slightly better than did wild-type (wt) TFIIF. TFIIF containing RAP74(1–205) had reduced activity in elongation. RAP74(1–172) was further reduced in activity, and RAP74(1–136) was inactive. RAP74(1–136) is the only one of the mutants that does not form a stable complex with the RAP30 subunit of TFIIF (41, 43). RAP30 alone (Fig. 2, lanes 11 and 12) and RAP74 alone (data not shown) failed to stimulate elongation. This experiment shows that amino acids 136 to 217 of human RAP74 are very important for stimulating elongation. This is an interesting result, because the same region of RAP74 has previously been shown to be critically important for initiation (22, 32).
FIG. 2.
Identification of a region of RAP74 that is critical for elongation stimulation. TFIIF or a TFIIF mutant, containing the indicated RAP74 deletion, was added to elongation complexes as described in the legend to Fig. 1. Transcription was continued for 30 s (samples designated “chase”). The lane designated “M” contains a single-stranded 5′ 32P-labeled DNA marker.
To analyze TFIIF mutants, a quantitative elongation rate assay was developed for RNA polymerase II (Fig. 3). Sarkosyl- and salt-washed elongation complexes were prepared, a saturating amount of a TFIIF sample was added, and elongation was continued with 1 mM (each) nucleoside triphosphate (NTP) for 15, 30, or 60 s. RNA polymerase II is active in elongation in the absence of TFIIF (Fig. 3, lanes 2 to 4). In the presence of wt TFIIF (lanes 5 to 7) the rate of elongation was stimulated five- to sixfold. A PhosphorImager was used to determine the average length of transcripts in each gel lane, and this value was plotted versus the elongation time (Fig. 3B). The slope of the line is reported as the average elongation rate. Quantitation of elongation rates was precise within each experiment and consistent between independent experiments. The R2 value for the slope of the line varied between 0.99 and 1.0. Variation between experiments for a TFIIF sample was less than 10%. The calculated rate for wt TFIIF, present in every gel, was used to scale data between experiments.
FIG. 3.
Elongation rate stimulation assay. (A) Washed elongation complexes were prepared as described in the legend to Fig. 1. After addition of 20 pmol of TFIIF or the indicated TFIIF mutant, 1 mM (each) NTP was added and the reaction was stopped after 15, 30, or 60 s (chase). The size marker (M) is a 5′-end-labeled single-stranded DNA marker for estimation of RNA sizes. TFIIF mutants are named for the RAP74 mutation they contain (e.g., I176A has the isoleucine at codon 176 replaced by an alanine). (B) Quantitation of the data shown in panel A. A PhosphorImager was used to estimate the midpoint of the distribution of RNA bands. Average transcript length is plotted against elongation time. The slope is reported as the average elongation rate. Slopes vary between R2 of 0.99 and 1.0.
The elongation rate assay (Fig. 3) was used to analyze a large set of site-directed mutations constructed in the N-terminal domain of RAP74 between amino acids 136 and 217 (Fig. 4A). Construction of these mutants and characterizations of their activities in initiation were described previously (32). In Fig. 4A, the activities of these mutants in the elongation rate assay are compared to their activities in a pulse-chase assay that depends primarily on initiation. In the pulse-chase protocol shown in Fig. 4A, transcription was initiated by addition of 100 μM (each) ATP, CTP, and GTP and 0.25 μM [α-32P]UTP for 1 min. To block new initiation, Sarkosyl was added to 0.25%, and transcripts were elongated in the presence of 1 mM (each) NTP. To confirm that the pulse-chase protocol depends primarily on productive initiation, the assay was repeated for selected samples with a protocol that utilized a pulse with 100 μM ATP and CTP and 0.25 μM [α-32P]UTP for 1 min (Fig. 4B). Because GTP was omitted from the reaction mixture, elongation was blocked beyond C10 (5′-pppACUCUCUUCC-3′) prior to addition of G11. The results of the pulse-chase assays were very similar whether three or four NTPs were included in the pulse protocol (Fig. 4B), indicating that both protocols primarily measure accurate initiation rather than early elongation. Furthermore, we have shown that the RAP74 L155A and I176A mutants are defective in formation of the first phosphodiester bond from the adenovirus major late promoter (32).
Mutations throughout the region of RAP74 between amino acids 136 and 217 have startlingly similar effects on both elongation rate and initiation (Fig. 4A). In order to compare RAP74 mutants using these assays, we assigned the following descriptions of TFIIF mutants. For elongation, a “severe” defect is defined as an average elongation rate of 149 to 225 nucleotides per min, a “moderate” defect is defined as an average elongation rate of 239 to 276 nucleotides per min, and a “slight” defect is defined as an average elongation rate of 298 to 399 nucleotides per min. The average rate of elongation for wt TFIIF was determined to be 500 nucleotides per min at room temperature. The average rate of elongation for the RAP74(1–217) deletion mutant was determined to be 528 nucleotides per min. For initiation, severe, moderate, and slight defects are defined as 17 to 23%, 33 to 43%, and 50 to 76% of wt activity, respectively. The RAP74(1–217) mutant was determined to have 72% wt activity in initiation. Mutants denoted with an asterisk (see below) were constructed in RAP74(1–217), so in the initiation assay, these mutants should be compared to the RAP74(1–217) mutant. By this classification scheme, mutants with transcriptional defects were detected between amino acids F138 and E193. These data are summarized in Fig. 4C.
Substitutions between amino acids F138 and R153 had no detectable effect on elongation. The F138A, P139A, V140A, W143A, and Y144A substitutions, however, caused slight defects in initiation (57 to 76% wt activity). These residues may be involved directly or indirectly in interactions with the RAP30 subunit of TFIIF, because the inactive RAP74(1–136) deletion mutant fails to interact with the RAP30 subunit (41, 43). Substitutions between amino acids T154 and M177 affect both elongation and initiation. The overlapping sequence between amino acids A157 and K183 is predicted by PHD analysis (37) to form an alpha-helix (Fig. 4C). Of the single amino acid substitution mutants tested, the L155A, W164A, N172A, I176A, and M177A substitutions caused the greatest defects in transcription, and these positions are highlighted in Fig. 4C. The average elongation rates of these mutants varied between 149 and 179 nucleotides per min. These values should be compared to the rates for the RAP74(1–158) (154 nucleotides per min) and RAP74(1–172) (178 nucleotides per min) deletion mutants and the no-TFIIF control (94 nucleotides per min). In initiation, the L155A, W164A, N172A, I176A, and M177A mutants varied between 17 and 33% wt activity. The N172A mutant had a moderate defect in initiation (33% wt) but a severe defect in elongation (179 nucleotides per min). The L155A, W164A, I176A, and M177A substitutions at hydrophobic residues have activities very similar to the RAP74(1–158) and RAP74(1–172) deletion mutants in both elongation rate stimulation and initiation. The L155, W164, N172, I176, and M177 side chains, therefore, appear to be the most important in the F138-to-E215 region for transcriptional functions. A string of glutamic acids, 158-EEAEEE-163, appears to be important for both elongation and initiation. The 158K2 (E158K, E159K) and 161K3 (E161K, E162K, E163K) charge-reversal substitutions have severe defects in elongation (204 and 182 nucleotides per min) and initiation (26 and 32% wt). Substitutions of individual glutamates with alanines had little effect on elongation except at the E159 position, at which a moderate defect was observed (298 nucleotides per min). Single alanine substitutions at any of the five glutamates affected initiation, and the greatest effects were observed at positions E159 (43% wt) and E163 (42% wt). The negative charge in the E158-to-E163 region of RAP74, therefore, appears to contribute to transcription. A positively charged cluster (166-RRNK-169) appears to be important in transcription. 166E3* (R166E, R167E, K169E) is severely defective in elongation (154 nucleotides per min) and initiation [22% wt, 30% of RAP74(1–217)]. The 167A3* (R167A, N168A, K169A) mutation, however, which preserves positive charge at R166, has a moderate defect in elongation (239 nucleotides per min) and initiation [33% wt, 46% of RAP74(1–217)]. R166A did not show a notable defect in elongation (489 nucleotides per min) but showed a slight defect in initiation (60% wt). R167A and N168A showed moderate defects in elongation (249 and 276 nucleotides per min, respectively) and slight defects in initiation (65 and 63% wt). V170A* showed a moderate defect in both elongation (260 nucleotides per min) and initiation [39% wt, 54% of RAP74(1–217)]. L171A* showed no defect in elongation but a slight defect in initiation [50% wt, 69% of RAP74(1–217)]. H173A and F174A have slight to moderate defects in elongation (239 and 372 nucleotides per min) and slight defects in initiation (69 and 68% wt). The region from amino acid Q179 to E193 appears to be more important for elongation than for initiation. The mutants in this region are triple amino acid substitutions constructed in the RAP74(1–217) deletion. Mutants 179A3*, 184K3*, 188K3*, and 191K3* have slight to severe defects in elongation rate stimulation (244, 204, 399, and 304 nucleotides per min). None of these mutants appear to have a significant defect in initiation compared to RAP74(1–217). These triple substitutions significantly alter the charge within the Q179-to-E193 region, which may account for the defects of these mutants in elongation. Triple substitutions may disrupt the predicted helical structure between amino acids A157 and K183. This disruption may be more deleterious to an elongation reaction in which RNA polymerase II, TFIIF, DNA, and RNA are expected to be the only significant macromolecular components. The initiation system contains many additional basal and regulatory factors that could influence the activity of TFIIF. The major conclusion from this mutagenic analysis is that the region of RAP74 between amino acids T154 and M177 is very important for both elongation rate stimulation and initiation and that most mutations within this and the surrounding region have strikingly similar effects on elongation and initiation.
TFIIF (20 pmol) was added to reaction mixtures for the elongation assays represented by Fig. 1 to 4A, because this was determined to be close to a saturating level (Fig. 5A and B). By contrast, for initiation, 1 to 2 pmol of TFIIF was sufficient to saturate the assay mixture (32), indicating that interactions between general factors and TFIIF might enhance recruitment of TFIIF to the initiation complex. Addition of 60 pmol of TFIIF or more did not inhibit elongation (Fig. 5B and data not shown).
Because 20 pmol of TFIIF or more was necessary to saturate the assay, TFIIF appeared to have a low affinity for the elongation complex. Furthermore, previous experiments have indicated that TFIIF can dissociate from elongation complexes (29, 44). We therefore tested whether the stimulatory effect of TFIIF would withstand washing of the bead template (Fig. 5C). Elongation complexes were formed, stalled at position +20 downstream of a modified adenovirus major late promoter. These complexes are referred to as U20 complexes (5′-ACUCUCUUCCCCUUCUCUUU-3′), because the 3′ end of the RNA is a UMP residue. U20 complexes were incubated in the absence or presence of TFIIF or in the presence of TFIIF mutants. Complexes were washed extensively with transcription buffer containing 60 mM KCl and 1 mg of bovine serum albumin per ml, and the activity of washed complexes in elongation was compared to unwashed complexes. Some beads were lost in the washing process, but no difference was observed in TFIIF stimulation after washing (Fig. 5C; compare lanes 2 to 6 with lanes 8 to 12). Once TFIIF was incorporated into the complex, therefore, it appeared to remain associated.
We considered the possibility that the apparent commitment of TFIIF to the bead template might be due to an experimental artifact. Specifically, TFIIF bound to the magnetic beads might provide a pool of the factor for stimulating elongation throughout the reaction time course even as TFIIF was continuously exchanged on and off the elongation complex. To eliminate this possibility, U20 elongation complexes were incubated in the presence or absence of wt TFIIF. Complexes were washed free of excess TFIIF and then separated from beads by digestion with EcoRI endonuclease, which cleaves the DNA at position −130 upstream of the transcriptional start (Fig. 5D). Because the template was immobilized with an upstream biotinylated primer, digestion with EcoRI released the elongation complex from the beads. In the presence of TFIIF, about 50% of complexes were released from the beads (Fig. 5D; compare lane 6 with lanes 7 and 8). The bead-associated and free fractions of elongation complexes were separated, and NTPs were added for 30 s. TFIIF stimulated elongation even after washing of complexes with buffer and separation of complexes from the magnetic beads. TFIIF therefore remained committed to the elongation complex even when complexes were subjected to significant dilution.
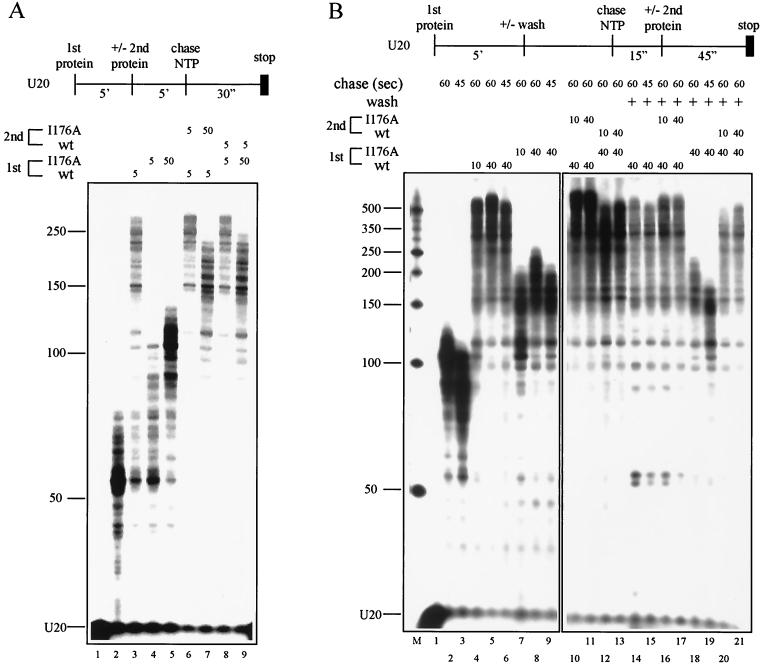
We next determined whether commitment of TFIIF to elongation complexes could be demonstrated in a competition experiment between wt TFIIF and a defective mutant (Fig. 6). We reasoned that if TFIIF were fully committed to the elongation complex, no exchange of wt and mutant TFIIF would be observed. The data in Fig. 6A, however, show that some exchange is possible between TFIIF samples bound to RNA polymerase II elongation complexes. Addition of 5 pmol of wt TFIIF before addition of 5 pmol of the I176A mutant produced the same stimulation of elongation as did addition of wt TFIIF with no competitor (Fig. 6A; compare lanes 3 and 6). However, addition of 50 pmol of I176A after 5 pmol of wt TFIIF reduced stimulation, indicating that, when added at a 10-fold molar excess, the I176A mutant could partially displace wt TFIIF (Fig. 6A; compare lanes 3, 6, and 7). Additionally, wt TFIIF can displace the I176A mutant. When the I176A mutant was added first for 5 min, followed by addition of wt TFIIF, the level of stimulation was the same as when wt TFIIF was added first (Fig. 6A; compare lanes 6 and 8 and lanes 7 and 9). On the stalled elongation complex, therefore, wt and mutant TFIIF molecules can be exchanged. wt TFIIF appears to be preferentially incorporated into the complex, at least during elongation, because even with a large excess of the I176A mutant the level of stimulation observed is more similar to that expected of wt TFIIF than of the I176A mutant (Fig. 6A; compare lanes 3, 5, 7, and 9).
Because wt TFIIF appeared to interact somewhat more strongly with the elongation complex than did the I176A mutant but exchange was possible when the mutant was added in excess, we considered the possibility that wt TFIIF might have a higher affinity for the rapid elongation complex than does the I176A mutant. Exchange might occur on the stalled elongation complex, but wt TFIIF might have an advantage in the competition for the rapid elongation complex. To test these ideas, an experiment was designed to examine exchange of wt and mutant TFIIF during elongation (Fig. 6B). The strategy was to saturate elongation complexes with either wt or mutant TFIIF. After addition of the first TFIIF sample, elongation complexes on beads were either washed with buffer to remove excess TFIIF (lanes 14 to 21) or not washed (lanes 2 to 13). At 15 s after beginning the elongation phase of the reaction, the competing TFIIF sample was added and elongation was continued for an additional 45 s. We reasoned that if wt TFIIF had a higher affinity than the I176A mutant for the rapid elongation complex, wt TFIIF would displace the mutant, resulting in an enhanced elongation rate. When wt TFIIF was added to the complex first, mutant TFIIF would not be observed to displace wt TFIIF from the elongation complex and the elongation rate would not be noticeably inhibited.
Indeed, wt TFIIF does have a higher affinity for the rapid elongation complex than does the I176A mutant. When wt TFIIF was the first protein added to the elongation complex, mutant TFIIF did not appear to displace wt TFIIF after elongation had commenced (Fig. 6B; compare lanes 10 and 11 with lanes 5 and 9). Removal of excess wt TFIIF by washing of complexes with buffer did not enhance the ability of the mutant to compete (Fig. 6B; compare lanes 16 and 17 with lanes 9 and 14). Once rapid elongation began, therefore, wt TFIIF was not noticeably displaced by the I176A mutant. Conversely, wt TFIIF displaced the I176A mutant during elongation, even when the complexes were not washed to remove excess mutant protein (Fig. 6B; compare lanes 12 and 13 with lanes 8 and 6). wt TFIIF rapidly displaced the I176A mutant and stimulated elongation, as if no I176A mutant protein had been added. These experiments demonstrate that wt TFIIF has a higher affinity than the I176A mutant for the running elongation complex, although wt and mutant TFIIF can exchange on a stalled elongation complex (Fig. 6A). In the Discussion (below), competition experiments are interpreted in terms of a model for RNA polymerase II elongation.
The average rate of elongation from U20 complexes was measured at 23, 30, 37, and 42°C (Fig. 7). Determinations were done for RNA polymerase II in the absence of TFIIF or in the presence of wt TFIIF or mutant TFIIF containing the deleterious I176A substitution. RNA synthesis was in the presence of 1 mM (each) NTP, so elongation rates were not limited by the availability of substrates. The apparent Km for NTPs is approximately 100 μM and is not affected by the presence of TFIIF in the reaction mixture (data not shown). TFIIF stimulates elongation strongly at each of the temperatures tested, and the I176A mutant has activity that is intermediate between the wt TFIIF-stimulated and unstimulated rates. In this experiment, elongation rates were also determined at 15°C (data not shown). In the presence of wt TFIIF, RNA polymerase II was weakly active at 15°C (1.6 nucleotides [nt]/s) but apparently inactive in the presence of the I176A mutant or in the absence of TFIIF.
The temperature dependence of a rate constant can be used to derive the activation energy for a reaction and the “frequency factor” (5, 10, 36). The frequency factor relates to the fraction of molecules in a population that are properly oriented with substrates to catalyze the reaction. In the context of RNA polymerase II, this is the fraction of molecules that are in an activated state for elongation. When the temperature dependence of the elongation rate is displayed as an Arrhenius plot (Fig. 7B) (log average elongation rate versus the reciprocal of the absolute temperature in kelvins), the average activation energy can be evaluated from the slope of the line (slope = −EA/R, where EA is the activation energy and R is the gas constant) and the frequency factor can be evaluated from the y-coordinate intercept (log A, where A is the frequency factor). No deviations from linearity were observed in the Arrhenius plot within the temperature range of 23 to 42°C, indicating that the rate-limiting step in catalysis did not change over this temperature interval. The slopes of the three lines in the Arrhenius plot are very similar to one another, demonstrating that the average activation energy for elongation (5.5 kcal) does not change in the presence or absence of TFIIF. The y-coordinate intercept, however, does change, showing that TFIIF increases the frequency factor. In the absence of TFIIF, the frequency factor is 2.8 × 109 s−1. In the presence of wt TFIIF, the frequency factor is 10.8 × 109 s−1. In the presence of the I176A mutant, the frequency factor is 5.3 × 109 s−1. We conclude that TFIIF stimulates elongation by maintaining RNA polymerase II in a more active conformation for elongation and that the I176A mutant is defective in maintaining the isomerized state of the elongation complex. TFIIF does not appear to stimulate elongation by affecting the catalytic steps in RNA synthesis. If TFIIF did have a direct effect on catalysis, the average activation energies in the presence and absence of TFIIF would be different.
DISCUSSION
Mutagenic analysis reveals that the region of human RAP74 between T154 and M177 is important both for stimulating the rate of elongation by RNA polymerase II and for supporting productive initiation (Fig. 4). To measure initiation, a pulse-chase protocol in which elongation was blocked beyond the first nine phosphodiester bonds from the adenovirus major late promoter was used (Fig. 4B). In other work, we have demonstrated that the L155A and I176A mutants from this region of RAP74 are defective in formation of the first phosphodiester bond from the adenovirus major late promoter (32). We have further shown that the defect of the L155A and I176A mutants in formation of short C14 and C15 transcripts can be attributed to their prior defect in first-bond formation (32). These previous abortive and productive initiation assays were done in a system of recombinant general transcription factors and purified RNA polymerase II using a supercoiled DNA template. Differences in the magnitude of mutant defects in that study and this study are probably attributable to the use of supercoiled templates in the previous study. As we argue in that paper, untwisting of the DNA helix may partially complement TFIIF function in initiation. The defect of RAP74 mutants in supporting elongation by RNA polymerase II appears to mimic their defect in formation of the first phosphodiester bond from the adenovirus major late promoter.
Although the elongation and initiation assays are very different, the similarities in the effects of RAP74 mutants are striking. Clearly, TFIIF performs very similar functions during different phases of the transcription cycle. In this study, the accurate initiation assay was done in a reconstituted human cell extract system. Transcription from the adenovirus major late promoter was dependent on ATP hydrolysis, on all of the general transcription factors (TFIIA, TFIID, TFIIB, RNA polymerase II, TFIIF, TFIIE, and TFIIH), and on the presence of the DNA binding site for the transcriptional regulator USF/MLTF (upstream sequence factor/major late transcription factor) (data not shown). Many general factors and regulatory proteins, therefore, could modulate TFIIF activity in this system. On the other hand, in the elongation stimulation assay, complexes were washed with 1% Sarkosyl and 0.5 M KCl buffer. After dissociation of elongation and termination factors and readdition of TFIIF, the crucial macromolecular components of elongation reactions are likely to be RNA polymerase II, template DNA, RNA, and TFIIF. It is therefore a surprise that results with the initiation and elongation assays mirrored one another so closely (Fig. 4A). In these assays, TFIIF activity appeared to be mediated primarily through interactions between TFIIF and RNA polymerase II and/or the DNA template, the common components of the elongation and initiation systems.
The T154 to M177 region of RAP74 is conserved in evolution, and the overlapping region between A157 and K183 is predicted to be alpha-helical (Fig. 4C). Of the single amino acid substitutions tested, L155A, W164A, N172A, I176A, and M177A caused the greatest defects in transcription, indicating that these amino acid side chains are most important for the function of this region of RAP74 in transcription. The activities of these mutants were very similar to the activity of the RAP74(1–158) deletion mutant, from which most of this critical region is removed. Many other mutations between T154 and M177 have significant effects on elongation and initiation. Even the most severely affected mutants appear to enter transcription complexes with wild-type affinity (32). Assuming that the A157-to-K183 region is alpha-helical, there is no particular relationship between the face of the helix on which side chains are displayed and the observed defects of these mutations in transcription. Notably, of the positions at which substitutions have the greatest effects, W164 would be located on the opposite side of a helix from L155, N172, I176, and M177. In the future, a molecular structure of TFIIF will assist in the full interpretation of mutations in this critical region of RAP74.
Site-specific DNA-protein photo-cross-linking studies indicated that the region of RAP74 between amino acids 172 and 205 is critically important for forming a tight wrap of adenovirus major late promoter DNA around a preinitiation complex containing TBP, TFIIB, RNA polymerase II, RAP30, and TFIIE (34). Because the RAP74(1–172) and RAP74(1–158) deletion mutants have very similar activities in elongation stimulation and accurate initiation to the L155A, W164A, N172A, I176A, and M177A substitutions (Fig. 4A), both of these deletions appear to inactivate the T154-to-M177 region of RAP74. In photo-cross-linking studies, the RAP74(1–172) mutant assembles into the preinitiation complex but fails to support a tightly wrapped structure and fails to support formation of many specific photo-cross-links to general transcription factors and RNA polymerase II within the core promoter. RAP74(1–205), on the other hand, appears to support DNA wrapping with the same efficiency as wt RAP74. RAP74(1–205) is not as active in transcription as wt RAP74, but its transcriptional defect has not yet been revealed in photo-cross-linking experiments. The interpretation of the photo-cross-linking data was that the region of RAP74 between amino acids 172 and 205 is critically important for forming the tight DNA wrap around RNA polymerase II and for isomerization of the preinitiation complex. Isomerization was hypothesized to include tight wrapping of promoter DNA around the general factor complex and bending of DNA through the RNA polymerase II active site. The region between T154 and M177, which we have identified by mutagenic analysis, appears to correspond to this region of RAP74 that is critically important for isomerization.
Because RAP74 mutants that are defective in isomerization of the preinitiation complex have such similar defects in initiation and elongation, these mutants may also fail to properly isomerize the elongation complex. In Fig. 4A, we show that a large panel of TFIIF mutants have very similar effects in initiation and elongation. E. coli RNA polymerase (11) and yeast RNA polymerase III (24) have been shown to elongate transcription by branched kinetic pathways in which the RNA polymerase converts between active and inactive conformations. Based on these previous reports for other multisubunit RNA polymerases and the data presented in this paper, a branched kinetic model for elongation by RNA polymerase II is proposed (Fig. 8). Like E. coli RNA polymerase and yeast RNA polymerase III, RNA polymerase II elongates transcription asynchronously, as if RNA polymerase II partitions between the active, isomerized form of the elongation complex (EC*) and the inactive, stalled form (EC) during elongation. In this model, the rate of phosphodiester bond synthesis for the next NTP to be added to an RNA chain (length, n bases) is represented by k*(n + 1). Conversion of EC*(n) and EC(n) occurs through rate constants ki(n) and k−i(n), respectively. In this model, transcriptional pausing occurs at positions at which k*(n + 1) is slow, k−i(n) is fast, and/or ki(n) is slow, relative to other positions in the RNA chain.
FIG. 8.
Branched kinetic model for elongation by RNA polymerase II. TFIIF is proposed to stimulate elongation by increasing the fraction of active, isomerized RNA polymerase II elongation complexes (EC*). PIC*, isomerized preinitiation complex.
TFIIF binds to the stalled elongation complex (Fig. 5C and D), but exchange of TFIIF samples on stalled complexes was observed (Fig. 6A). During elongation, however, wt TFIIF was found to be resistant to exchange with the defective I176A mutant (Fig. 6B). The I176A mutant, on the other hand, appeared to be rapidly replaced by wt TFIIF when wt TFIIF was added to elongating complexes that previously had been saturated with the I176A mutant. These results can be explained in terms of the kinetic model shown in Fig. 8. wt TFIIF is proposed to stabilize the EC* form of the elongation complex. In the presence of wt TFIIF, therefore, the elongation complex rarely exits the EC* state. We propose that in the EC* state, little or no exchange of TFIIF occurs. In the presence of the I176A mutant, however, the elongation complex is not efficiently held in the EC* state and often lapses into the EC state, in which exchange with competitor TFIIF can occur. The transcriptionally inactive EC state is proposed to be similar in conformation to the stalled elongation complex formed in the absence of added NTPs. The experiment represented by Fig. 6A indicates that wt and mutant TFIIF can be exchanged on the stalled U20 complex, at least over a period of 5 min of incubation at a 10-fold molar excess of mutant-to-wt TFIIF. The experiment represented by Fig. 6B, however, demonstrates that during elongation wt TFIIF is highly resistant to exchange with the I176A mutant. In contrast, the I176A mutant appears to be rapidly displaced by wt TFIIF during elongation. This work supports the idea that stalled elongation complexes (EC) are conformationally distinct from elongating complexes (EC*) and that wt TFIIF stimulates elongation by stabilizing the activated conformation of RNA polymerase II (EC*).
The temperature dependence of the average rate of RNA polymerase II elongation (Fig. 7B) provides further evidence for the conformational elongation model. Arrhenius analysis showed that TFIIF does not affect the average activation energy for the elongation reaction. Instead, TFIIF increases the frequency factor, which represents the population of RNA polymerase II elongation complexes that are active for elongation. According to the kinetic model in Fig. 8, this would be the population of molecules in the EC* state. The I176A mutant supports elongation by RNA polymerase II with the same activation energy as that of wt TFIIF. The mutant supports a lower frequency factor, however, indicating that the mutant stabilizes a smaller population of RNA polymerase II molecules in the EC* state. Because TFIIF appears to affect the fraction of molecules in the EC* state, TFIIF appears to stimulate elongation by accelerating ki and/or inhibiting k−i. TFIIF does not appear to affect k*(n + 1), the rate of phosphodiester bond formation. If TFIIF affected the chemical step, TFIIF would alter the activation energy for the polymerization reaction and TFIIF would also be expected to affect the selection of pause sites by RNA polymerase II. The experiment represented by Fig. 1 indicated that RNA polymerase II selects similar pause sites in the presence and absence of TFIIF. Based on these experiments, TFIIF does not appear to have a direct effect on catalysis during elongation. This result is consistent with the previous suggestion that the active site of multisubunit RNA polymerases is concealed behind a sliding processivity clamp that binds both downstream template DNA and the exiting RNA chain during elongation (26–28). TFIIF appears to accelerate elongation by stabilizing the active conformation of the RNA polymerase II elongation complex.
ACKNOWLEDGMENTS
We thank Donal Luse for clones. We gratefully acknowledge Stephan Reimers for some of the data shown in Fig. 4B and Augen Pioszak for construction of some of the RAP74 mutants used in this work.
This research was supported by a grant from the American Cancer Society. Additional support was provided by the Research Excellence Fund of Michigan State University and the Michigan State University Agricultural Experiment Station.
REFERENCES
- 1.Amouyal M, Buc H. Topological unwinding of strong and weak promoters by RNA polymerase. A comparison between the lac wild-type and the UV5 sites of Escherichia coli. J Mol Biol. 1987;195:795–808. doi: 10.1016/0022-2836(87)90485-2. [DOI] [PubMed] [Google Scholar]
- 2.Arias J A, Dynan W S. Promoter-dependent transcription by RNA polymerase II using immobilized enzyme complexes. J Biol Chem. 1989;264:3223–3229. [PubMed] [Google Scholar]
- 3.Aso T, Conaway J W, Conaway R C. The RNA polymerase II elongation complex. FASEB J. 1995;9:1419–1428. doi: 10.1096/fasebj.9.14.7589983. [DOI] [PubMed] [Google Scholar]
- 4.Bengal E, Flores O, Krauskopf A, Reinberg D, Aloni Y. Role of the mammalian transcription factors IIF, IIS, and IIX during elongation by RNA polymerase II. Mol Cell Biol. 1991;11:1195–1206. doi: 10.1128/mcb.11.3.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buc H, McClure W R. Kinetics of open complex formation between Escherichia coli RNA polymerase and the lac UV5 promoter. Evidence for a sequential mechanism involving three steps. Biochemistry. 1985;24:2712–2723. doi: 10.1021/bi00332a018. [DOI] [PubMed] [Google Scholar]
- 6.Conaway J W, Conaway R C. A multisubunit transcription factor essential for accurate initiation by RNA polymerase II. J Biol Chem. 1989;264:2357–2362. [PubMed] [Google Scholar]
- 7.Conaway R C, Garrett K P, Hanley J P, Conaway J W. Mechanism of promoter selection by RNA polymerase II: mammalian transcription factors alpha and beta gamma promote entry of polymerase into the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:6205–6209. doi: 10.1073/pnas.88.14.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulombe B, Burton Z F. DNA bending and wrapping around RNA polymerase: a “revolutionary” model to describe transcriptional mechanisms. Microbiol Mol Biol Rev. 1999;63:457–478. doi: 10.1128/mmbr.63.2.457-478.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Craig M L, Suh W C, Record M T., Jr HO· and DNase I probing of E sigma 70 RNA polymerase-lambda PR promoter open complexes: Mg2+ binding and its structural consequences at the transcription start site. Biochemistry. 1995;34:15624–15632. doi: 10.1021/bi00048a004. [DOI] [PubMed] [Google Scholar]
- 10.Eisenberg D S, Crothers D M. Physical chemistry with applications to the life sciences. Menlo Park, Calif: Benjamin/Cummings Publishing Company, Inc.; 1979. [Google Scholar]
- 11.Erie D A, Hajiseyedjavadi O, Young M C, von Hippel P H. Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription. Science. 1993;262:867–873. doi: 10.1126/science.8235608. [DOI] [PubMed] [Google Scholar]
- 12.Flores O, Ha I, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and subunit composition of transcription factor IIF. J Biol Chem. 1990;265:5629–5634. [PubMed] [Google Scholar]
- 13.Flores O, Lu H, Killeen M, Greenblatt J, Burton Z F, Reinberg D. The small subunit of transcription factor IIF recruits RNA polymerase II into the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:9999–10003. doi: 10.1073/pnas.88.22.9999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forget D, Robert F, Grondin G, Burton Z F, Greenblatt J, Coulombe B. RAP74 induces promoter contacts by RNA polymerase II upstream and downstream of a DNA bend centered on the TATA box. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamper H B, Hearst J E. A topological model for transcription based on unwinding angle analysis of E. coli RNA polymerase binary, initiation and ternary complexes. Cell. 1982;29:81–90. doi: 10.1016/0092-8674(82)90092-7. [DOI] [PubMed] [Google Scholar]
- 16.Izban M G, Luse D S. Factor-stimulated RNA polymerase II transcribes at physiological elongation rates on naked DNA but very poorly on chromatin templates. J Biol Chem. 1992;267:13647–13655. [PubMed] [Google Scholar]
- 17.Kephart D D, Price M P, Burton Z F, Finkelstein A, Greenblatt J, Price D H. Cloning of a Drosophila cDNA with sequence similarity to human transcription factor RAP74. Nucleic Acids Res. 1993;21:1319. doi: 10.1093/nar/21.5.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kephart D D, Wang B Q, Burton Z F, Price D H. Functional analysis of Drosophila factor 5 (TFIIF), a general transcription factor. J Biol Chem. 1994;269:13536–13543. [PubMed] [Google Scholar]
- 19.Killeen M, Coulombe B, Greenblatt J. Recombinant TBP, transcription factor IIB, and RAP30 are sufficient for promoter recognition by mammalian RNA polymerase II. J Biol Chem. 1992;267:9463–9466. [PubMed] [Google Scholar]
- 20.Kim T K, Lagrange T, Wang Y H, Griffith J D, Reinberg D, Ebright R H. Trajectory of DNA in the RNA polymerase II transcription preinitiation complex. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kitajima S, Tanaka Y, Kawaguchi T, Nagaoka T, Weissman S M, Yasukochi Y. A heteromeric transcription factor required for mammalian RNA polymerase II. Nucleic Acids Res. 1990;18:4843–4849. doi: 10.1093/nar/18.16.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei L, Ren D, Finkelstein A, Burton Z F. Functions of the N- and C-terminal domains of human RAP74 in transcriptional initiation, elongation, and recycling of RNA polymerase II. Mol Cell Biol. 1998;18:2130–2142. doi: 10.1128/mcb.18.4.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall N F, Price D H. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol Cell Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuzaki H, Kassavetis G A, Geiduschek E P. Analysis of RNA chain elongation and termination by Saccharomyces cerevisiae RNA polymerase III. J Mol Biol. 1994;235:1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- 25.Meyer-Almes F J, Heumann H, Porschke D. The structure of the RNA polymerase-promoter complex. DNA-bending-angle by quantitative electrooptics. J Mol Biol. 1994;236:1–6. doi: 10.1006/jmbi.1994.1112. [DOI] [PubMed] [Google Scholar]
- 26.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science. 1996;273:211–217. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 27.Nudler E, Gusarov I, Avetissova E, Kozlov M, Goldfarb A. Spatial organization of transcription elongation complex in Escherichia coli. Science. 1998;281:424–428. doi: 10.1126/science.281.5375.424. [DOI] [PubMed] [Google Scholar]
- 28.Polyakov A, Severinova E, Darst S A. Three-dimensional structure of E. coli core RNA polymerase: promoter binding and elongation conformations of the enzyme. Cell. 1995;83:365–373. doi: 10.1016/0092-8674(95)90114-0. [DOI] [PubMed] [Google Scholar]
- 29.Price D H, Sluder A E, Greenleaf A L. Dynamic interaction between a Drosophila transcription factor and RNA polymerase II. Mol Cell Biol. 1989;9:1465–1475. doi: 10.1128/mcb.9.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rees W A, Keller R W, Vesenka J P, Yang G, Bustamante C. Evidence of DNA bending in transcription complexes imaged by scanning force microscopy. Science. 1993;260:1646–1649. doi: 10.1126/science.8503010. [DOI] [PubMed] [Google Scholar]
- 31.Reines D, Conaway J W, Conaway R C. The RNA polymerase II general elongation factors. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- 32.Ren D, Lei L, Burton Z F. A region within the RAP74 subunit of human transcription factor IIF is critical for initiation but dispensable for complex assembly. Mol Cell Biol. 1999;19:7377–7387. doi: 10.1128/mcb.19.11.7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivetti C, Guthold M, Bustamante C. Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO J. 1999;18:4464–4475. doi: 10.1093/emboj/18.16.4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robert F, Douziech M, Forget D, Egly J-M, Greenblatt J, Burton Z F, Coulombe B. Wrapping of promoter DNA around the RNA polymerase II initiation complex induced by TFIIF. Mol Cell. 1998;2:341–351. doi: 10.1016/s1097-2765(00)80278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. Localization of subunits of transcription factors IIE and IIF immediately upstream of the transcriptional initiation site of the adenovirus major late promoter. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 36.Roe J H, Burgess R R, Record M T., Jr Temperature dependence of the rate constants of the Escherichia coli RNA polymerase-lambda PR promoter interaction. Assignment of the kinetic steps corresponding to protein conformational change and DNA opening. J Mol Biol. 1985;184:441–453. doi: 10.1016/0022-2836(85)90293-1. [DOI] [PubMed] [Google Scholar]
- 37.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 38.Samkurashvili I, Luse D S. Structural changes in the RNA polymerase II transcription complex during transition from initiation to elongation. Mol Cell Biol. 1998;18:5343–5354. doi: 10.1128/mcb.18.9.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan S, Aso T, Conaway R C, Conaway J W. Roles for both the RAP30 and RAP74 subunits of transcription factor IIF in transcription initiation and elongation by RNA polymerase II. J Biol Chem. 1994;269:25684–25691. [PubMed] [Google Scholar]
- 40.Tirode F, Busso D, Coin F, Egly J M. Reconstitution of the transcription factor TFIIH: assignment of functions for the three enzymatic subunits, XPB, XPD, and cdk7. Mol Cell. 1999;3:87–95. doi: 10.1016/s1097-2765(00)80177-x. [DOI] [PubMed] [Google Scholar]
- 41.Wang B Q, Burton Z F. Functional domains of human RAP74 including a masked polymerase binding domain. J Biol Chem. 1995;270:27035–27044. doi: 10.1074/jbc.270.45.27035. [DOI] [PubMed] [Google Scholar]
- 42.Wang B Q, Lei L, Burton Z F. Importance of codon preference for production of human RAP74 and reconstitution of the RAP30/74 complex. Protein Expr Purif. 1994;5:476–485. doi: 10.1006/prep.1994.1067. [DOI] [PubMed] [Google Scholar]
- 43.Yonaha M, Aso T, Kobayashi Y, Vasavada H, Yasukochi Y, Weissman S M, Kitajima S. Domain structure of a human general transcription initiation factor, TFIIF. Nucleic Acids Res. 1993;21:273–279. doi: 10.1093/nar/21.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zawel L, Kumar K P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]