Abstract
Autosomal Dominant Polycystic Kidney Disease (ADPKD) is the most common inherited renal disorder, characterized by renal cyst development leading to end-stage renal disease. Although the appropriate choice of suitable reference is critical for quantitative RNA analysis, no comparison of frequently used “housekeeping” genes is available. Here, we determined the validity of 7 candidate housekeeping genes (Actb, Actg1, B2m, Gapdh, Hprt, Pgam1 and Ppia) in kidney tissues from mouse models orthologous to ADPKD, including a cystic mice (CY) 10–12 weeks old (Pkd1flox/flox:Nestincre/Pkd1flox/−:Nestincre, n = 10) and non-cystic (NC) controls (Pkd1flox/flox/Pkd1flox/-, n = 10), Pkd1-haploinsufficient (HT) mice (Pkd1+/−, n = 6) and wild-type (WT) controls (Pkd1+/+, n = 6) and a severely cystic (SC) mice 15 days old (Pkd1V/V, n = 7) and their controls (CO, n = 5). Gene expression data were analyzed using six distinct statistical softwares. The estimation of the ideal number of genes suggested the use of Ppia alone as sufficient, although not ideal, to analyze groups altogether. Actb, Hprt and Ppia expression profiles were correlated in all samples. Ppia was identified as the most stable housekeeping gene, while Gapdh was the least stable for all kidney samples. Stat3 expression level was consistent with upregulation in SC compared to CO when normalized by Ppia expression. In conclusion, present findings identified Ppia as the best housekeeping gene for CY + NC and SC + CO groups, while Hprt was the best for the HT + WT group.
Subject terms: Computational biology and bioinformatics, Genetics, Molecular biology, Biomarkers, Diseases, Nephrology
Introduction
Autosomal dominant polycystic kidney disease (ADPKD) is manifested by bilateral development of multiple fluid-filled epithelial-derived cysts1,2. It is the most common Mendelian disorder of the kidneys, affecting 3–5:10,000 people worldwide3 and the leading monogenic cause of end-stage kidney disease4. Mutations in the PKD1 gene account for ~ 78% of the affected families while mutations in PKD2 are detected in ~ 15% of them5,6, with the remaining ones being genetically unresolved or associated with rare mutations in the GANAB and DNAJB11 loci7.
The elucidation of PKD1/PKD2-related biology has allowed major steps toward the understanding of ADPKD pathogenesis and the development of diagnostic tools and biomarkers8. The significant number of abnormally functioning pathways involved in the disease pathophysiology, in turn, opened a number of tracks to develop potentially specific therapies. In addition to eGFR, the development of progression and prognostic markers has been also essential to guide clinical decisions in ADPKD. The classification based on total kidney volume growth9 has been widely used as well as the multiple criteria developed by Cornec-Le Gal et al.10 A combination of distinct biomarkers in a classifier including new biomarkers should further increase sensitivity and specificity11. The characterization and elucidation of specific strategic transcriptional profiles in turn, are expected to expand the options of therapeutic targets and the number of useful biomarkers for the disease.
Gene expression analysis plays a central role on identifying and characterizing pathways involved in specific phenotypes and diseases, potentially allowing elucidation of pathogenetic aspects and biomarker discovery. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) is one of the most sensitive and reproducible means of quantifying RNA expression12,13. Nonetheless, to provide accurate expression analysis this method requires expression normalization of the genes of interest to a reference gene that is stable and not affected by experimental conditions. In this scenario, expression stability is a major criterion for housekeeping gene selection14. Reference genes are generally selected among housekeeping genes ubiquitously expressed and not transcriptionally affected by experimental conditions13. While the expression of some housekeeping genes is constant under certain conditions, it can significantly change in some circumstances15 such as developmental stages, cell types and experimental conditions13,14,16,17. It is therefore essential to characterize the suitability of candidate housekeeping genes to serve as appropriate internal mRNA expression controls under a given experimental condition where transcriptional effects are being investigated.
Glyceraldehyde-3-phosphate dehydrogenase (Gapdh), actin beta (Actb) and 18 s ribosomal RNA (18 s rRNA) are the most common reference genes. In kidney tissues of Pkd1-deficient mouse models, Gapdh has been the most often used endogenous control for gene expression studies18. However, its reliability in this context has not been demonstrated yet. Moreover, the indiscriminate use of these genes may be inappropriate, as they have been implicated in disease processes19 including ADPKD20. Therefore, whether the expression of housekeeping genes is or is not stable in kidney tissues of Pkd1-deficient mouse models remains unknown. Determining the stability of genes known to be involved in ADPKD and used as housekeeping is particularly important.
To elucidate this issue and bring appropriate information to PKD experimental studies involving kidney transcriptional profiles, we aimed to identify the most stable housekeeping controls from a panel, namely seven candidate genes commonly used as endogenous controls in kidney disease, not limited to ADPKD: Actb, Actg1, B2m, Gapdh, Hprt, Pgam1, and Ppia., to be considered in kidney RT-qPCR obtained from Pkd1-deficient mouse models.
Results
Quantitative gene expression analyses: the first step to identify optimal housekeeping genes for RT-qPCR studies of renal tissue in Pkd1-deficient mouse models
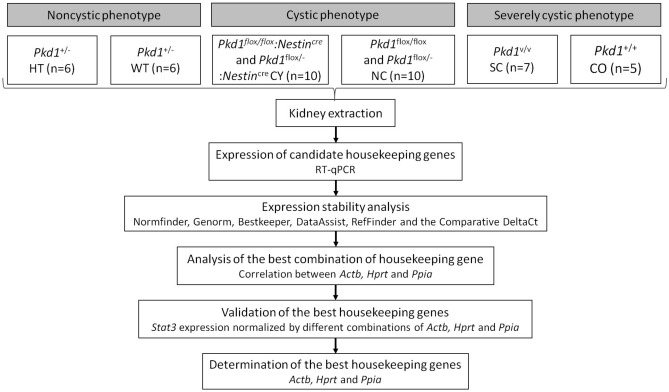
We applied a stepwise strategy to select and evaluate candidate housekeeping genes with the aim of identifying optimal housekeeping genes for RT-qPCR analyses of kidney tissue in three Pkd1-deficient mouse models: Pkd1+/− [haploinsufficient (HT), noncystic] and its control Pkd1+/+ (wild type, WT); Pkd1flox/flox:Nestincre and Pkd1flox/−:Nestincre (cystic, CY) and their controls Pkd1flox/flox and Pkd1flox/− (noncystic, NC); and Pkd1V/V [severely cystic, SC) and its control Pkd1+/+ (CO, noncystic). Such steps are outlined in Fig. 1.
Figure 1.
Workflow diagram illustrating the strategy used to identify the best housekeeping normalizer for RT-qPCR studies in Pkd1-deficient kidneys. To proceed with appropriate comparisons, we included housekeeping genes selected from the available literature (Actb, Actg1, B2m, Gapdh, Hprt, Pgam1 and Ppia). Pkd1, polycystic kidney disease 1 gene.
Raw RT-qPCR data in renal tissue were obtained for the Actb, Actg1, B2m, Gapdh, Hprt, Pgam1 and Ppia candidate housekeeping genes for each of the Pkd1-deficient mouse models. The corresponding median Ct values are shown in Fig. 2. High expression levels were obtained for Actg1 [16.60 (1.61)], B2m [19.94 (0.88)], Gapdh [19.11 (1.63)], Pgam1 [19.52 (1.10)] and Ppia [16.60 (0.75)], with median Cts lying between 15 and 20. Hprt [22.37 (0.69)] and Actb [25.38 (1.32)], in turn, displayed moderate (Ct between 20 and 25) and relatively low expression (Ct above 25), respectively (Fig. 2). This process was preceded by assessment of RNA quality, which showed that RIN (RNA Integration Numbers) ranged from 5.9 to 8.8 among the analyzed samples (Supplementary Table S4).
Figure 2.
Renal expression profiles of the seven selected candidate housekeeping genes in Pkd1-deficient mouse models. Lower threshold values (Ct) indicate higher expression levels. The median values are expressed as horizontal lines while the error bars represent the corresponding interquartile ranges. The Actb Ct values were the highest ones, reflecting the lowest expression levels whereas the Actg1 Ct values were the lowest, indicating the highest expression.
Separate assessments and comparisons involving the renal cystic mice and their corresponding controls were performed, including only the Pkd1flox/flox:Nestincre and Pkd1flox/flox genotypes as well as both cystic genotypes and corresponding controls: Pkd1flox/flox:Nestincre, Pkd1flox/−:Nestincre, Pkd1flox/flox and Pkd1flox/- (Supplementary Tables S2 and S3). The results yielded by the two analyses did not significantly differ, showing similarity and stability of genetic background between both model pairs. Of note, this pattern was also observed between such model pairs in a previous study of ours directed to identifying housekeeping genes for microRNA expression analysis21. In this context, in the current study we analyzed this cystic model merging Pkd1flox/flox:Nestincre and Pkd1flox/−:Nestincre mice in the CY group and Pkd1flox/flox and Pkd1flox/- in the NC group.
Analysis of expression stability of the candidate housekeeping genes
The following step in the process of selecting the best housekeeping genes was the assessment of expression stability for each of the candidates, taking into account the related genetic backgrounds. This task was accomplished using the NormFinder, GeNorm, RefFinder, ΔCt method, Bestkeeper and DataAssist software packages (Fig. 2), which allowed determining their relative expression stability and generating a ranking among the best ones. The most stably expressed genes in the kidney samples of the three Pkd1-deficient mouse models were defined by the lowest variability values observed in each condition of interest. It must be pointed out that the results for the CY-NC model did not differ between the two and four-genotype analyses (Supplementary Tables S2 and S3).
All candidate housekeeping mRNAs presented M values below 1.5, the GeNorm set threshold, findings that are consistent with stability22. The Bestkeeper software points out inconsistency when SD is higher than 1.0, a pattern detected only for Hprt and Pgam1 in the SC sample groups (Supplementary Table S2). NormFinder evaluates expression stability by analyzing the intra- and intergroup transcriptional variation of the candidate reference genes. An SD value lower than 0.5 is recommended to consider a gene a suitable housekeeping. Ppia, Hprt, Pgam1 and B2m had SD values below 0.5, while the Actb, Actg1 and Gapdh values lied above 0.5 in all sample groups. Moreover, Pgam1 and B2m displayed SD higher than 0.5 in the SC group, B2m in CO and SC + CO, and Gapdh in CY, NC, HT, WT, SC e SC + CO groups. These results suggest that these candidate genes should not be considered suitable housekeepings in the mentioned groups and settings.
Based on the different used algorithms and considering all ranks yielded by the referred analyses, Ppia appeared as the most stably expressed housekeeping gene for analyses including all groups, as well as for the CY, SC, CY + NC and SC + CO groups. The data that supported this conclusion were yielded by NormFinder (SD = 0.22), GeNorm (M value = 0.53), RefFinder (Geomean = 1.00), ΔCt method (Mean SD = 0.80), Bestkeeper (CV = 3.24 and SD = 0.54) and DataAssist (Score = 0.68) (Supplementary Table S2). Pgam1, on the other hand, was the most stably expressed housekeeping gene for analyses involving the WT + CO groups, Actb for the NC group, and Hprt for analyses including the HT + WT groups as well as only HT animals (Table 1).
Table 1.
Best housekeeping gene for each group of samples, each model and respective control, and all groups yielded by software analyses.
| Groups | Best housekeeping genes identified by software analyses | Best housekeeping gene | |||||
|---|---|---|---|---|---|---|---|
| NormFinder | GeNorm | RefFinder | ΔCt method | Bestkeeper | DataAssist | ||
| All | Ppia* | Actb + Actg1 | Ppia | Ppia | Hprt | Ppia | Ppia |
| HT | Pgam1 | Hprt + Ppia | Pgam1 | Pgam1 | Hprt | Hprt | Hprt/Pgam1 |
| WT | Ppia | B2m + Pgam1 | Pgam1 | Pgam1 | Hprt | Ppia | Pgam1 |
| CY | Ppia | Actg1 + Ppia | Ppia | Ppia | Actb | Ppia | Ppia |
| NC | Pgam1 | Actb + Pgam1 | Hprt | Hprt | Actb | Ppia | Actb/Hprt/Pgam1 |
| SC | Ppia | Actg1 + Hprt | Ppia | Ppia | Gapdh | Ppia | Ppia |
| CO | Pgam1 | Actg1 + Ppia | Pgam1 | Pgam1 | Hprt | Actg1 | Pgam1 |
| HT + WT | Pgam1* | Hprt + Ppia | Actg1 | Actg1 | Hprt | Hprt | Hprt |
| CY + NC | Pgam1* | Actg1 + Ppia | Ppia | Ppia | Actb | Ppia | Ppia |
| SC + CO | Ppia* | Actg1 + Hprt | Ppia | Ppia | Actb | Ppia | Ppia |
*Best reference genes considering intra- and intergroup variations.
All, all samples; HT, haploinsufficient; WT, wild type; CY, cystic; NC, noncystic; SC, severely cystic phenotype; CO, control for the severely cystic phenotype. NormFinder (version 0.953; https://moma.dk/normfinder-software), GeNorm (version 2.2; https://genorm.cmgg.be/), BestKeeper (version 1.0; https://www.gene-quantification.de/bestkeeper.html), DataAssist (version 3.01; https://www.thermofisher.com/br/en/home/technical-resources/software-downloads/dataassist-software.html), the comparative ΔCt method and RefFinder (https://www.heartcure.com.au/reffinder/).
Genes in bold: higher frequency of appearance in softwares.
Taken together, our data ranked Ppia and Hprt as the most stable candidate housekeeping genes. Gapdh, on the other hand, was deemed as the least stable one, being associated with the following values: NormFinder (SD = 0.83), GeNorm (M value = 085), RefFinder (Geomean = 7.00), ΔCt method (Mean SD = 1.38), Bestkeeper (CV = 4.32 and SD = 0.82), and DataAssist (Score = 0.95) (Supplementary Table S2).
Since the selection of the most suitable housekeeping gene depends on the Pkd1 deficiency model, it is important to conclude that Ppia was identified as the best housekeeping gene for the CY-NC and SC-CO pairs of samples while Hprt was the most suitable for HT-WT. Therefore, and interestingly, Ppia seems to be the best housekeeping in analysis involving models with cystic phenotypes (CY and SC), being associated with NormFinder (SD = 0.09), GeNorm (M value = 0.18), RefFinder (Geomean = 1.41), ΔCt method (Mean SD = 0.63), Bestkeeper (CV = 3.25 and SD = 0.84), and DataAssist (Score = 0.50) (Supplementary Table S2). Pgam1, in turn, appears as the most appropriate housekeeping gene when comparing noncystic controls (WT, NC and CO) (Table 1).
Analysis of the best combination of housekeeping genes
The use of combined housekeeping genes is a strategy often employed to improve comparisons of relative target gene expression among groups. A comprehensive analysis of potential combinations of evaluated candidates, therefore, should be performed to establish the best anchors to quantify and compare gene expression. Ultimately, this approach constitutes a way of minimizing possible outliers and inappropriate differences between measurements.
The software packages recommend at least two genes to be used together as housekeeping genes. The best combination of candidate housekeepings for each of our group of samples, based on analyses yielded by different software packages, is shown in Table 2. Such analyses revealed that the Actb + Hprt and Hprt + Ppia pairs are the combinations most often identified as the best housekeeping gene options for comparisons including all groups, with the same statistical weight; Actg1 + Ppia was the most frequent pair for the CY group; Actb + Pgam1 for NC and WT; Hprt + Pgam1 for HT; Actb + Ppia or Actg1 + Ppia for SC; and Actg1 + Pgam1 for the CO group.
Table 2.
Best combination of housekeeping genes for each group of samples, each model and respective control, and all groups yielded by software analyses.
| Groups | Best pair of housekeeping genes identified by software analyses | Best pair of housekeeping genes | Best trio of housekeeping genes | |||||
|---|---|---|---|---|---|---|---|---|
| NormFinder | GeNorm | RefFinder | ΔCt method | Bestkeeper | DataAssist | |||
| All | Actb + Pgam1* | Actb + Actg1 | Hprt + Ppia | Hprt + Ppia | Actb + Hprt | Hprt + Ppia | Actb + Hprt/Hprt + Ppia | Actb + Hprt + Ppia |
| HT | Hprt + Pgam1 | Hprt + Ppia | Actg1 + Pgam1 | Actg1 + Pgam1 | Hprt + Ppia | B2m + Hprt | Hprt + Pgam1 | Actg1 + Hprt + Pgam1/Hprt + Pgam1 + Ppia |
| WT | Pgam1 + Ppia | B2m + Pgam1 | Actb + Pgam1 | Actb + Pgam1 | Actb + Hprt | Pgam1 + Ppia | Actb + Pgam1 | Actb + Pgam1 + Ppia |
| CY | Pgam1 + Ppia | Actg1 + Ppia | Actg1 + Ppia | Actg1 + Ppia | Actb + B2m | Pgam1 + Ppia | Actg1 + Ppia | Actb + Actg1 + Ppia/Actg1 + Pgam1 + Ppia/Actg1 + B2m + Ppia |
| NC | Pgam1 + Ppia | Actb + Pgam1 | Actb + Hprt | Actb + Hprt | Actb + Actg1 | Pgam1 + Ppia | Actb + Pgam1 | Actb + Hprt + Pgam1/Actb + Pgam1 + Ppia |
| SC | Actg1 + Ppia | Actg1 + Hprt | Actb + Ppia | Actb + Ppia | Actb + Gapdh | Actg1 + Ppia | Actb + Ppia/Actg1 + Ppia | Actb + Hprt + Ppia/Actg1 + Hprt + Ppia |
| CO | Actg1 + Pgam1 | Actg1 + Ppia | Gapdh + Pgam1 | Actg1 + Pgam1 | Gapdh + Hprt | Actg1 + Pgam1 | Actg1 + Pgam1 | Actg1 + Gapdh + Pgam1 |
| HT + WT | Hprt + Pgam1* | Hprt + Ppia | Actg1 + B2m | Actg1 + B2m | Actb + Hprt | B2m + Hprt | B2m + Hprt | Actg1 + B2m + Hprt |
| CY + NC | Pgam1 + Ppia* | Actg1 + Ppia | Actg1 + Ppia | Pgam1 + Ppia | Actb + Hprt | Pgam1 + Ppia | Pgam1 + Ppia | Actg1 + Pgam1 + Ppia |
| SC + CO | Actg1 + Ppia* | Actg1 + Hprt | Actb + Ppia | Actg1 + Ppia | Actb + Hprt | Actg1 + Ppia | Actg1 + Ppia | Actb + Actg1 + Ppia/Actg1 + Hprt + Ppia |
*Best reference genes considering intra- and intergroup variations.
All, all samples; HT, haploinsufficient; WT, wild type; CY, cystic; NC, noncystic; SC, severely cystic phenotype; CO, control for the severely cystic phenotype. NormFinder (version 0.953; https://moma.dk/normfinder-software), GeNorm (version 2.2; https://genorm.cmgg.be/), BestKeeper (version 1.0; https://www.gene-quantification.de/bestkeeper.html), DataAssist (version 3.01; https://www.thermofisher.com/br/en/home/technical-resources/software-downloads/dataassist-software.html), the comparative ΔCt method and RefFinder (https://www.heartcure.com.au/reffinder/).
Genes in bold: higher frequency of appearance in softwares.
Comparisons of the different Pkd1-deficient kidney tissues with their respective controls identified Pgam1 + Ppia as the best pair of housekeeping genes for CY and NC, B2m + Hprt for HT and WT, and Actg1 + Ppia for SC and CO. Additionally, Actb + Hprt + Ppia was the most frequent best trio identified for analyses including all groups; Actg1 + Pgam1 + Ppia for CY and NC, Actg1 + B2m + Hprt for HT and WT, and Actb + Actg1 + Ppia or Actg1 + Hprt + Ppia for SC and CO (Table 2).
Notably, the identified best pair and trio combinations of housekeeping genes provided further support to the association of Ppia and Pgam1 with the cystic and non-cystic phenotypes, respectively. Moreover, considering all samples together Ppia remains the most stable candidate housekeeping gene.
Determination of the optimal number of housekeeping genes
The use of an optimal number of reference genes is important to save samples and primers, select and validate housekeeping genes, classify samples, group genes and monitor time-dependent processes. While the selection and use of stable housekeeping genes is often limited to only one, in most of such cases the stability of gene expression analysis could improve with the inclusion of one or more additional housekeeping controls. In this scenario, in the current study we established the minimum number of housekeeping genes that should be used in gene expression analysis involving each of our groups, each model with its respective control, and all groups.
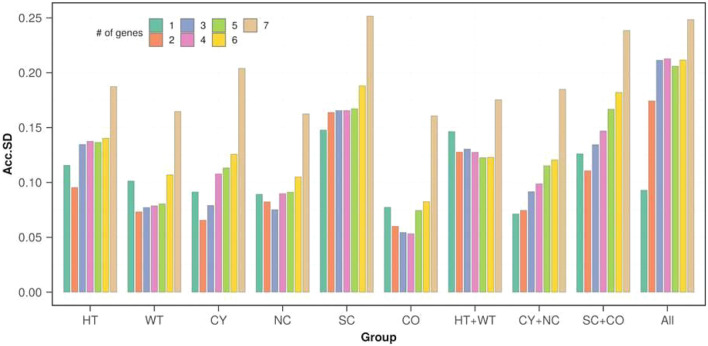
The GenEx software package was used to calculate the Acc.SD for the seven candidate housekeeping genes and to determine the optimal number of reference genes to be used for each dataset (Fig. 3). Based on the 0.15 Acc.SD cut-off, Ppia was identified as the most stable candidate gene for normalization when considering all sample groups (All; Fig. 4). The addition of another gene led to a cut-off greater than 0.15, demonstrating that the insertion of a second normalizer in the gene expression analysis would increase the Acc.SD. Moreover, the addition of more than one gene led to a cut-off greater than 0.2, indicating that the insertion of a third gene would increase even more the Acc.SD. Applying a global analysis and based on the lowest Acc.SD value, the best normalizers for each sample group were found to be: two (Actg1 + Ppia) was the optimal number of housekeeping genes to be considered for the CY group, three (Actb + Hprt + Pgam1) for the NC group, two (Hprt + Pgam1) for HT, two (Actb + Pgam1) for the WT group, one (Ppia) for SC, and three (Actg1 + Gapdh + Pgam1) for the CO group. Importantly, we observed that different numbers of housekeeping genes should be used when comparing the renal tissue of each Pkd1-deficient model with its respective control: one (Ppia) for CY + NC, two (B2m + Hprt) for HT + WT, and two (Actg1 + Ppia) for SC + CO.
Figure 3.
Optimal number of reference genes for each dataset according to GenEx analyses. Accumulated standard deviation (Acc.SD) for the five candidate housekeeping genes in all groups of samples allows estimating the ideal number of genes for normalization. Acc.SD was calculated with the GenEx software package (version 6; http://www.multid.se). Lower values of Acc.SD indicate the optimal number of reference genes. HT, Pkd1-haploinsufficient; WT, wild type; CY, cystic; NC, noncystic; SC, severely cystic; CO, control for the severely cystic phenotype.
Figure 4.
The ΔCt values of the three best candidate housekeeping genes: Actb (A–C), Hprt (D–F) and Ppia (G–I). A lower threshold value (Ct) indicates a higher gene expression level. The median values are expressed as horizontal lines while the error bars represent interquartile ranges. CY, cystic; NC, noncystic; HT, haploinsufficient; WT, wild type; SC, severely cystic phenotype; CO, control of severely cystic phenotype. Actb, target expression normalized by Actb; Hprt, target expression normalized by Hprt; Ppia, target expression normalized by Ppia; Actb + Hprt, target expression normalized by Actb + Hprt; Actb + Ppia, target expression normalized by Actb + Ppia; Hprt + Ppia, target expression normalized by Hprt + Ppia. Comparisons between the Pkd1-deficient model and its respective control were performed with the Mann–Whitney U test. P < 0.05 was not detected for any of the comparisons.
Intergene expression normalization among the top three candidate housekeeping genes
An optimal housekeeping gene should present expression stability, displaying low expression variability with respect to other housekeeping genes. This property can be evaluated by normalizing their expression by each other’s. To evaluate this aspect for the best candidate housekeeping genes, we used as the third strategy different algorithms to normalize the expression levels (Ct) of the top three candidates (Actb + Hprt + Ppia) to each other’s expression (Fig. 4). Their expression levels did not differ between any two-sample groups. These results suggest that any of these three genes are suitable to be employed as housekeeping genes among the analyzed samples. Interestingly, the presence of Ppia as a housekeeping gene decreased the gene expression dispersion of both Actb and Hprt (Fig. 4).
Correlation of mRNAs expression between pairs of the top three candidate housekeeping genes
Pair correlation analysis between the best housekeeping genes can provide options to choose relevant housekeeping genes for gene expression studies. This approach was applied to our models orthologous to ADPKD associated with different profiles of Pkd1 deficiency, displaying or not a cystic phenotype. Such correlation analyses were performed using the mRNAs expression data yielded by all evaluated kidney samples. The expression levels (Ct) of the three best candidate housekeeping genes revealed a very strong correlation between Hprt and Ppia (ρ = 0.89, p < 0.05, Fig. 5) and a strong correlation between Actb and Hprt (ρ = 0.84, p < 0.05, Fig. 5). In addition, a moderate correlation was observed between Actb and Ppia (ρ = 0.81, p < 0.05, Fig. 5). These results showed that the Actb, Hprt and Ppia expression profiles are correlated in all the samples herein evaluated and can be used together as suitable housekeeping genes.
Figure 5.
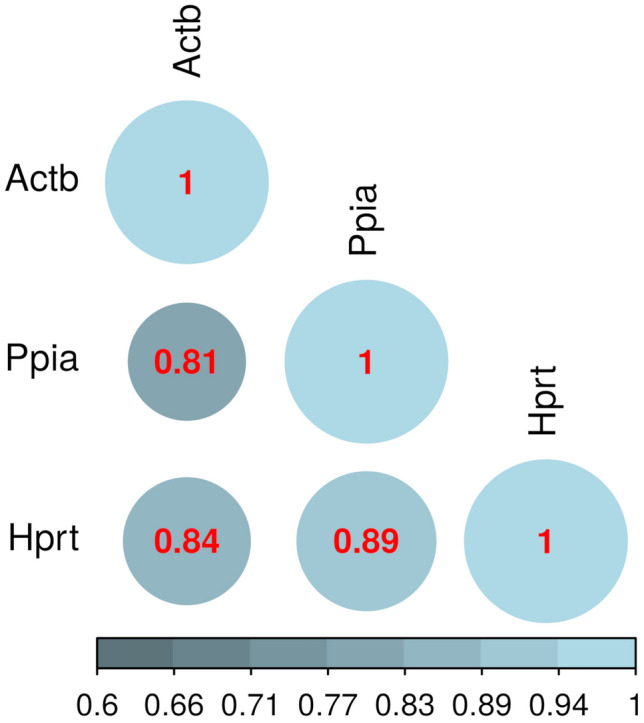
Pair correlation matrix between the number of amplification cycles of the best housekeeping candidate genes Actb, Hprt and Ppia. Circle color and radius are proportional to the Spearman’s rank correlation coefficient (significance level: p ≤ 0.05).
Validation of the best candidate housekeeping genes by normalizing expression of Stat3 target gene
In order to validate the expression stability of the three best candidate housekeeping genes, the relative expression of the Stat3 target gene was assessed using different combinations of Actb, Hprt and Ppia normalizers (Fig. 6). Stat3 was selected for this purpose because it is a well-defined gene with increased expression in ADPKD and in murine polycystic kidney disease models.
Figure 6.
The ΔCt values of Stat3 expression target gene normalized by different combinations of the three best candidate housekeeping genes (Actb, Hprt and Ppia) (A) Stat3/Actb, target expression normalized by Actb; (B) Stat3/Hprt, target expression normalized by Hprt; (C) Stat3/Ppia, target expression normalized by Ppia; (D) Stat3/Actb + Hprt, target expression normalized by Actb + Hprt; (E) Stat3/Actb + Ppia, target expression normalized by Actb + Ppia; (F) Stat3/Hprt + Ppia, target expression normalized by Hprt + Ppia, (G) Stat3/Actb + Hprt + Ppia target expression normalized by Actb + Hprt + Ppia. The median values are represented as horizontal lines and the error bars represent interquartile ranges. CY, cystic; NC, noncystic; HT, haploinsufficient; WT, wild type; SC, severely cystic phenotype; CO, control for the severely cystic phenotype. *p < 0.05 by Kruskal–Wallis with Dunn’s post-hoc test, followed by FDR correction.
The Stat3 expression level was consistent with upregulation in SC kidneys compared to its control CO when normalized by Ppia expression (Fig. 6C). Trends of upregulation were also observed in SC kidneys compared to CO when normalized by other combinations of the best candidate housekeeping genes (Figs. 6B, 6D, 6E, 6F and 6G). Trends of Stat3 upregulation in CY compared to their control kidneys were also observed when its expression was normalized by different combinations of Actb, Hprt and Ppia (Figs. 6A, 6B, 6D, 6E, 6F and 6G). In contrast, Stat3 expression was not statistically different between HT and WT kidneys regardless the housekeeping gene combination used for normalization (Fig. 6). Our results also suggest that the SC kidneys, and likely their controls CO, have expression profiles slightly different from the other analyzed mouse model kidneys. Taken together, the use of Ppia showed to be the most suitable housekeeping gene among all considered sample groups.
Many studies with other animal models, including kidney tissues, have used Gapdh as the housekeeping gene, assuming unaltered expression20,23–25. Given the weakness of Gapdh as a housekeeping gene observed in our models, it is essential that this gene be analyzed and compared to other potential housekeeping gene normalizers. Indeed, when we analyzed Stat3 expression using Gapdh as the housekeeping normalizer, we did not find different expression levels between CY and NC, HT and WT, and SC and CO (Supplementary Figure S2). These finding are in disagreement with the trends of Stat3 upregulation detected in CY kidneys compared to NC and in SC compared to CO evidenced when our housekeeping gene combinations were applied.
Expression of the best candidate housekeeping genes does not correlate with the kidney weight/body weight ratio in CY mice
Since analyses involving mouse models orthologous to human ADPKD often require assessment of severity of the cystic phenotype or cyst growth, we reasoned that the identified housekeeping genes should have their stability evaluated with respect to the renal cyst burden, reflected in kidney size/weight. This approach could allow the investigation whether the disease stage, progression and severity significantly modify the expression patterns of potential housekeeping genes. To accomplish this task, we sought for potential correlations between the expression levels of Actb, Hprt and Ppia and kidney weight/body weight ratio (KW/BW) in the CY group. The expression levels of Actb, Hprt and Ppia, however, did not correlate with KW/BW (ρ = 0.05, p < 0.871, Supplementary Figure S3A; ρ = −0.22, p < 0.42, Supplementary Figure S3B; and ρ = 0.44, p = 0.09, Supplementary Figure S3C, respectively). This lack of correlations strongly suggests that the cystic burden does not significantly influence the expression level of the analyzed housekeeping genes, further supporting their use as controls in studies involving animals with different severities of renal cystic phenotypes.
Discussion
PKD1/PKD2 pathogenic mutations deregulate a number of downstream pathways that aberrantly affect major cellular properties and contribute to the ADPKD phenotype. Such pathways encompass mTOR, cMYC, STATs, cAMP and intracellular calcium homeostasis, promoting cyst epithelial cell proliferation, apoptosis, epithelial secretion, and cellular metabolic rewiring26 which in turn constitute logical targets for the development of therapeutic interventions. Given there is no widely accepted single model of ADPKD for therapeutic testing at present, most agents are tested in multiple preclinical models27. Since orthologous animal models with slowly progressive renal cystic disease reproduce more closely the human disease, they are more appropriate to unravel mechanisms underlying ADPKD pathogenesis and generate useful biomarkers28.
To systematically evaluate the complexity of target gene expression analyses in in vivo models orthologous to ADPKD and propose the best standards for such studies, we aimed to establish the most appropriate housekeeping genes to be employed in three mouse models with distinct profiles of Pkd1 gene deficiency, in different experimental scenarios. While HT mice show almost exclusively Pkd1+/− renal cells but do not display cysts, reproducing the background cell environment found in ADPKD type 1 patients, CY mice have renal cysts presumably formed by Pkd1−/− cells, reproducing the ADPKD type 1 cystic phenotype and its expected consequences. Notably, CY mice have preserved GFR at the evaluated ages (12 weeks). To complete the diverse phenotypes associated with Pkd1 deficiency, we included in our systematic assessment, an early, severely cystic animal model, the Pkd1V/V mouse (SC).
One reason justified the use of the SC (15 days) mouse as the model of highly cystic burden and another reason supported/validated its use as such a model. The first reason is because SC mice have a very short survival due to their fast progression to renal failure29. We had to choose an age, therefore, at which the significant majority of these animals would be still alive; so the chosen age was 15 days. In contrast, CY mice had to be analyzed at a significantly higher age, to have enough time to present an adequate cystic phenotype. In this context, we chose to work with 10–12-week-old CY and NC mice, as well as 10–12-week-old HT and WT animals; CY animals display a mild to moderate renal cystic phenotype which has been shown to be associated with preserved renal function even at ages of a few more weeks than the age range used in the current study30,31. The other reason, which supported/validated our decision, was the finding that the housekeeping gene expression profiles detected in CO mice (control wild-type animals with 15 days of life) did not significantly differ from the other controls WT and NC (PPIA with median Cts lying between 16 and 17, HPRT between 21–24 and ACTB between 24–27). Moreover, Stat3 expression did not significantly differ among CO, WT and NC kidneys in all but one normalization. Finally, we assessed the correlation between the best candidate housekeeping genes and KW/BW ratio in the CY group of samples. The detected absence of correlation between the expression levels of the best ones and the KW/BW ratio in the CY group suggests that the level of cystic involvement does not lead to significant changes in housekeeping gene expression.
While the use of non-validated endogenous control genes in gene expression studies results in unreliable data, a universal, invariably expressed gene is unlikely to exist32. This hypothetical control gene, in fact, may not even exist within individual tissues and cell types. In this context, a more adequate purpose is to identify the most reliable gene or set of genes to be applied in each experimental setting. Although several studies have evaluated gene expression profiles to identify new biomarkers and therapeutic targets for ADPKD26,33, to the best of our knowledge, no study to date has addressed the appropriateness of housekeeping gene usage in animal models orthologous to ADPKD. The present study identified Ppia as the best housekeeping gene for CY + NC and SC + CO groups, while Hprt was the best for the HT + WT group.
Cui et al. (2009)34 have previously analyzed endogenous genes as potentially useful housekeeping genes for analyses of target gene expression in kidney tissue samples of cpk mice, a well-characterized recessive cystic kidney disease model34. They studied the expression of 16 commonly used housekeeping genes in seven mildly and seven severely affected whole kidney tissue samples using TaqMan RT-qPCR assays and Affymetrix GeneChip arrays, normalized and tested for overall variance and equivalence of the means. Both statistical approaches and both TaqMan- and GeneChip-based methods converged on 3 out of the 4 top-ranked genes (Ppia, Gapdh and Pgk1) that displayed the most constant expression levels across the assessed phenotypes. Such results led to the conclusion that a combination of the top-ranked genes would provide suitable endogenous internal control for gene expression studies in cpk kidney tissues across a wide range of disease severity. The different disease etiology, however, precludes the application of these data to gene expression studies performed in in vivo models orthologous to ADPKD. The rationale of the current study was to run the first assessment of candidate housekeeping genes for normalization of mRNA expression by RT-qPCR in kidney tissue of mouse models orthologous to ADPKD with distinct patterns of Pkd1 deficiency.
Leal et al. (2016)35 has successfully evaluated the suitability of reference genes in meniscus samples in pathological and control conditions using the software packages applied in our current analysis35. These investigators revealed HPRT1 as the best single reference gene but as expected, showed that two or more reference genes should be used for gene expression normalization instead. According to the samples tested and proposed comparisons, the appropriate housekeeping gene combination included HPRT1 + TBP, HPRT1 + GAPDH, or HPRT1 + TBP + GAPDH. Other authors suggested that PPIA and RPS13, especially in combination, were the best suitable references to normalize gene expression in ccRCC tissues as compared to classical reference genes such as beta-Actin, GAPDH, 18S or B2M36. Therefore, such reports served as anchors for our present investigation about the stability of seven candidate genes selected from previous studies (Actb, Actg1, B2m, Gapdh, Hprt, Pgam1 and Ppia)24,34,37–41 in kidneys samples of Pkd1-deficient noncystic and cystic mouse models. The stability of gene expression was analyzed using distinct statistical models, including a pairwise comparison model, geNorm, and an ANOVA-based model, NormFinder.
Each of the applied algorithms ranked the best candidate reference genes, identifying Ppia as the most stable and reliable housekeeping gene, while Gapdh was least stable for all kidney samples. Gapdh is the most commonly used gene as endogenous control in RT-qPCR analyses, since its expression is usually constant23–25,41,42. Using a similar approach, a recent report evaluated six commonly used reference genes (Actb, B2m, Gapdh, Hmbs, Hprt and Ppia) to identify the most constantly expressed gene under the influence of testosterone in rat43. This study showed that Hmbs and Ppia were the most stably expressed genes in the hypothalamus while Hmbs and Gapdh appeared to be the most stable genes in kidneys, indicating that in this setting the Gapdh expression profile was more stable than in our case. Concentrations of GAPDH, however, may vary among individuals44, during pregnancy45, according to developmental stage46,47 and during the cell cycle48. Other reports also documented such a limitation49–53. Therefore, the recognition that the expression of Gapdh may exhibit tissue-specific regulationis consistent with our results and emphasizes the need for validation of commercially available control assays.
In line with the aforementioned raised concerns, our findings revealed that the selection of appropriate housekeeping genes and consequent use of Ppia as the reference, allowed the detection of differences between groups regarding Stat3 expression, thought to be upregulated in kidney cysts and associated with PKD progression in mouse models8,54–60. On the other hand, the normalization using Gapdh showed no difference. Interestingly, despite the different ages between the CY and SC groups, normalization using Ppia showed very close expression profiles in these two groups, decreasing the expression dispersion observed with Actb and Hprt. These data suggest that Ppia is a good expression normalizer for models associated with renal cystic phenotypes, a piece of information that may be highly relevant to investigate and compare different cystic disease stages, progression and severity. On the other hand, Pgam1 was associated with the Pkd1-haploinsufficient non-cystic model, suggesting that this might be a more appropriate normalizer to be used in studies involving Pkd1-deficient non-cystic renal phenotypes.
One limitation of the present study relies on the selection of only seven candidate housekeeping genes to be tested for validation, a process that was based on data available in previous studies. We believe, therefore, that further research is still required is this field, since the evaluation of additional candidate housekeeping genes in additional Pkd1-orthologous mouse models and experimental conditions are likely to improve the specificies of gene normalization in this biological scenario. Moreover, the pipeline adopted in the present study should be further tested using other models as well.
In conclusion, our findings established Ppia as the most appropriate housekeeping gene for comparisons involving a cystic model and its respective control, Hprt for non-cystic Pkd1-haploinsufficient and wild-type mice, and Ppia for a severely cystic model and its corresponding wild-type control. Overall, our analysis identified Ppia as the best and the most stable housekeeping gene in the comprehensive Pkd1-deficiency scenario, while Gapdh was the least stable in the three evaluated mouse models. Such data will allow more robust and reliable analyses of target gene expression in kidney tissue of Pkd1-deficient mouse models, a reality that will contribute to the elucidation of the role of different genes in different scenarios related to ADPKD or Pkd1 biology. Normalizing to a suitable housekeeping gene or sets of them can not only remove artifactual differences due to sampling and quality of mRNA but also identify real changes in gene expression levels.
Methods
Ethical statement
The authors confirm that all experiments were carried out in accordance with ARRIVE guidelines and regulations (https://arriveguidelines.org). "The ethical approval for all animal care and procedures were given by the Ethic Committee on Animal Use of the Federal University of Sao Paulo—Brazil (CEUA/UNIFESP), protocol number CEUA 4558140219."
ADPKD mouse models
The three mouse models were maintained on the C57BL/6 strain background, an important requirement for the performance of our study. Two models were evaluated at 10–12 weeks of age: a renal cystic mouse (Pkd1flox/flox:Nestincre/Pkd1flox/−:Nestincre, CY, n = 10; and their corresponding noncystic controls Pkd1flox/flox/Pkd1flox/-, NC, n = 10) and a Pkd1-haploinsufficient noncystic mouse (Pkd1+/−, HT, n = 6) and its respective wild-type control (Pkd1+/+, WT, n = 6). The third model was assessed at 15 days of life due to its severely renal cystic phenotype (Pkd1V/V, SC, n = 7) along with its same-age wild-type control (CO, n = 5). It must be noted that the WT and CO controls harbor the same genotype (Pkd1+/+) but were analyzed at different ages. Only kidneys from male animals were analyzed in order to avoid potential gender-related experimental variability. The mice were genotyped using specific PCR reactions21.
The CY mice, homozygous for a Pkd1-floxed allele (Pkd1flox) or compound heterozygous Pkd1flox/−, display a mosaic pattern of full gene inactivation induced by a Nestin-Cre transgene through excision of exons 2–4 (Pkd1flox/flox:Nestincre)30,41,61–63. The HT model is heterozygous for a Pkd1 null allele, characterized by early transcriptional interruption30,64, and develops no renal cysts by 12 weeks of age. The SC model is homozygous for the Pkd1 knockin T3041V allele, which prevents the autoproteolytic cleavage of PC1 at the GPS site29,30. Pkd1V/V animals have no gross phenotype by postnatal day (P) 6 but develop rapid and progressive distal nephron cysts thereafter. This severe renal phenotype, which eventually leads to renal failure, is responsible for the early mortality that occurs between the 2nd and 6th week29.
The mice were fed ad libitum and housed at constant ambient temperature in a 12-h light cycle. Animal procedures were approved by the Internal Biosafety Commission of Genetically Modified Organisms of the University of São Paulo School of Medicine and by the Universidade Federal de São Paulo (UNIFESP) Research Ethics Committees. SC (Pkd1V/V) and its wild-type control (CO) animals were euthanized by cervical dislocation, whereas the other animal groups were euthanized with intraperitoneal thiopental (0.4 mg/g of body weight). Their kidneys were appropriately harvested for RT-qPCR analyses. All experiments were conducted in accordance with international standards of animal care and experimentation. Both kidneys were collected and stored at − 80 °C for further use21.
Housekeeping genes
Using the terms “genes” and “polycystic and reverse transcriptase polymerase chain reaction” in the PubMed search, we found seven articles employing tissue samples from human ADPKD and animal ADPKD-orthologous kidneys. We also included housekeeping genes selected from other tissues in the literature24,34,37–41, namely Actb, Actg1, B2m, Gapdh, Hprt, Pgam1 and Ppia. All of these seven genes are constitutively expressed in kidney tissue of mouse models orthologous to ADPKD, have independent cellular functions, and are assumed not to be co-regulated.
Primers design
The oligonucleotide primers used to detect the gene-related products were designed based on the corresponding mRNA sequences (http://www.ncbi.nlm.nih.gov/nucleotide), using the Primer-Blast program (http://www.ncbi.nlm.nih.gov/tools/primer-blast/). This tool allows the design of primers taking into account alignments, even if partial, with other sequences potentially amplifiable by the primers, an evaluation that assesses all relevant sequences together. The Oligoanalyzer program (http://www.idtdna.com/analyzer/applications/oligoanalyzer) was also used to predict possible formation of stable secondary structures (hairpins) and primer dimers (homo and heterodimers, in the case of stable 3' structure), virtually guaranteeing the amplification of single products (no primer dimers and other products). To avoid amplification of genomic DNA due to potential contamination, the designed primers flanked exon–intron and intron–exon junctions expected to amplify products > 1000 bp from genomic DNA in all analyzed cases. To warrant high amplification efficiency, a maximum product size of 150 bp was established.
RNA extraction
Renal tissue lysis was performed using zirconia beads (Interprise, USA) and the Precellys (BioAmerica, USA) homogenizer. TRIzol (Life Technologies, USA) was employed for total RNA extraction according to the manufacturer’s protocol. The RNA quantity and purity were determined using the NanoVue spectrophotometer (GE Healthcare Life Sciences, USA) and analyzed with the Agilent 2100 Bioanalyzer 6000 Nanochip (Agilent Technologies Inc., Waldbronn, BW, Germany). We treated 2 µg of total RNA with DNase (RQ1 RNase-free DNase; Promega, USA), to avoid genomic DNA contamination. Total RNA was stored at -80 °C until further use21.
cDNA preparation and RT-qPCR
Complementary DNA (cDNA) was synthesized from 2 µg of total RNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer`s instructions.
Gene expression was performed in triplicate using SYBR Green (Applied Biosystems) in the QuantStudio7 qPCR system according to the manufacturer`s instructions. Primer sequences for the 7 candidate housekeeping genes and the target gene are shown in Supplementary Table S1. Gene expression quantification was performed in the same run for each sample to eliminate technical variation. Previously, we validated the amplification efficiencies of each primer, which were within the 90%-110% range.
Analysis of housekeeping gene expression stability
The tissue samples were classified into 7 distinct groups: (1) CY, including the cystic kidney samples; (2) NC, non-cystic kidney samples; (3) HT, Pkd1-haploinsuficient kidney samples; (4) WT, wild-type kidney samples; (5) SC, severely cystic kidney samples; (6) CO, early-life, wild-type kidney samples; and (7) All, including all kidney samples. The three mouse models with distinct patterns of Pkd1 deficiency were compared with their respective control tissues: CY vs NC, HT vs WT and SC vs CO.
RT-qPCR Cts were manually settled as 0.02 while the mean Ct values of the three technical replicates were imported to six algorithms: NormFinder (version 0.953; https://moma.dk/normfinder-software),65 GeNorm (version 2.2; https://genorm.cmgg.be/),22 BestKeeper (version 1.0; https://www.gene-quantification.de/bestkeeper.html),66 DataAssist (version 3.01; https://www.thermofisher.com/br/en/home/technical-resources/software-downloads/dataassist-software.html), the comparative ΔCt method12 and RefFinder (https://www.heartcure.com.au/reffinder/)67 following the authors’ recommendations. These software packages were used to determine the relative expression stability of the candidate housekeeping genes and to generate a ranking for the best ones.
The NormFinder software is a Microsoft Excel-based application that evaluates the expression stability of candidate reference genes. The stability value is calculated by analyzing their intra- and intergroup transcriptional variation. A lower variation in the expression levels indicates more stable gene expression (low stability value)65. The GeNorm program determines the gene expression stability value M according to the average pairwise variation between one particular gene and all other candidate genes. The most stable gene expression yields the lowest M value22. The BestKeeper program calculates the standard deviation (SD) and the coefficient of variance (CV) of the Ct levels, and applies the Pearson correlation coefficient to estimate the inter-gene relations of all possible candidate gene pairs66. The DataAssist software, in turn, determines a score for the best reference genes based on the GeNorm algorithm. It uses the RQ to calculate the stability value of each gene. A lower score indicates more stable expression (Thermo Fisher, USA). The comparative ΔCt method is based on a comparison of the relative expression of possible gene pairs within each sample. Gene stability is ranked according to the reproducibility of the gene expression differences in the analyzed samples12. Lastly, the RefFinder software integrates the NormFinder, GeNorm, BestKeeper, and the comparative ΔCt method assigning an appropriate weight to each gene, calculating the geometric mean of these weights, and generating a rank of the best candidate reference genes67. The optimal number of reference genes was selected using the GenEx software package (version 6; http://www.multid.se).
Statistical analysis
The Shapiro–Wilk test showed that the Ct values of the candidate housekeeping genes were not normally distributed, so that the results are expressed as median and interquartile range (IQ). The ΔCt values were determined by: Ct(target gene)−Ct(housekeeping gene) or Ct(target gene)−mean[Ct(housekeeping gene 1);Ct(housekeeping gene 2)].
To evaluate potential expression differences of Actb, Actg1, B2m, Gapdh, Hprt, Pgam1, Ppia and Stat3 among the sample groups, the Kruskal–Wallis with Dunn’s post-hoc test, followed by FDR correction using the Benjamini–Hochberg method, was used. Comparisons between the Pkd1-deficient model model and its respective control were performed with the Mann–Whitney U test.
Lastly, the Spearman correlation test was employed to verify potential correlations among the expression levels of Actb, Hprt and Ppia, and between the expression levels of such genes and kidney weight in CY cystic kidney samples. A value between 0.30–0.50 was determined as a weak correlation, 0.50–0.70 as moderate, 0.70–0.90 as strong, and 0.90–1.00 as a very strong correlation68.
Supplementary Information
Acknowledgements
This study received funding from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) 309045/2018-5 (IPH); and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) 2018/09135-0 (JJM), 2011/21593-4 (RM) 2015/23345-9 (MAB and IPH), 2015/17152-3 (LFO).
Author contributions
J.J.M., A.C.A. and I.P.H. designed the study, conceptualization, methodology, validation, formal analysis and investigation. A.G.A., F.M.F. and R.G.M. were responsible for sample collection. J.J.M., A.C.A. and MSO performed the experiments. J.J.M., A.C.A., F.M.F., E.H.W., L.F.O. and I.P.H. analyzed the data. J.J.M., A.C.A., L.F.O. and I.P.H. wrote the first draft of the manuscript. M.A.B. provided study material. All authors gave intellectual input and reviewed the last version of the manuscript.
Data availability
All data including supporting datasets are made available as main figures or supplementary information files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Juan J. Muñoz, Ana C. Anauate, Luiz F. Onuchic and Ita P. Heilberg.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-99366-x.
References
- 1.Mason SB, et al. The biomarker enriched proteome of autosomal dominant polycystic kidney disease cyst fluid. Proteomics Clin. Appl. 2009;3:1247–1250. doi: 10.1002/prca.200800163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grantham JJ, Mulamalla S, Swenson-Fields KI. Why kidneys fail in autosomal dominant polycystic kidney disease. Nat. Rev. Nephrol. 2011;7:556–566. doi: 10.1038/nrneph.2011.109. [DOI] [PubMed] [Google Scholar]
- 3.Lanktree MB, et al. Prevalence estimates of polycystic kidney and liver disease by population sequencing. J. Am. Soc. Nephrol. 2018;29:2593–2600. doi: 10.1681/asn.2018050493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cadnapaphornchai MA. Autosomal dominant polycystic kidney disease in children. Curr. Opin. Pediatr. 2015;27:193–200. doi: 10.1097/mop.0000000000000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastos AP, Onuchic LF. Molecular and cellular pathogenesis of autosomal dominant polycystic kidney disease. Braz. J. Med. Biol. Res. 2011;44:606–617. doi: 10.1590/s0100-879x2011007500068. [DOI] [PubMed] [Google Scholar]
- 6.Cornec-Le Gall E, Torres VE, Harris PC. Genetic complexity of autosomal dominant polycystic kidney and liver diseases. J. Am. Soc. Nephrol. 2018;29:13–23. doi: 10.1681/asn.2017050483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergmann C, et al. Polycystic kidney disease. Nat. Rev. Dis. Primers. 2018;4:50. doi: 10.1038/s41572-018-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viau A, et al. Tubular STAT3 limits renal inflammation in autosomal dominant polycystic kidney disease. J. Am. Soc. Nephrol. 2020;31:1035–1049. doi: 10.1681/asn.2019090959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Irazabal MV, et al. Imaging classification of autosomal dominant polycystic kidney disease: A simple model for selecting patients for clinical trials. J. Am. Soc. Nephrol. 2015;26:160–172. doi: 10.1681/asn.2013101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornec-Le Gall E, et al. Can we further enrich autosomal dominant polycystic kidney disease clinical trials for rapidly progressive patients? Application of the PROPKD score in the TEMPO trial. Nephrol. Dial Transpl. 2018;33:645–652. doi: 10.1093/ndt/gfx188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kistler AD, et al. Urinary proteomic biomarkers for diagnosis and risk stratification of autosomal dominant polycystic kidney disease: A multicentric study. PLoS ONE. 2013;8:e53016. doi: 10.1371/journal.pone.0053016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakao R, Yamamoto S, Yasumoto Y, Kadota K, Oishi K. Impact of denervation-induced muscle atrophy on housekeeping gene expression in mice. Muscle Nerve. 2015;51:276–281. doi: 10.1002/mus.24310. [DOI] [PubMed] [Google Scholar]
- 14.Sellayah D, Sek K, Anthony FW, Hanson MA, Cagampang FR. Sensitivity of housekeeping genes in the hypothalamus to mismatch in diets between pre- and postnatal periods in mice. Neurosci. Lett. 2008;447:54–57. doi: 10.1016/j.neulet.2008.09.060. [DOI] [PubMed] [Google Scholar]
- 15.Radonić A, et al. Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2004;313:856–862. doi: 10.1016/j.bbrc.2003.11.177. [DOI] [PubMed] [Google Scholar]
- 16.Wang K, et al. Molecular engineering of DNA: Molecular beacons. Angew. Chem. Int. Ed. Engl. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manzano R, et al. Housekeeping gene expression in myogenic cell cultures from neurodegeneration and denervation animal models. Biochem. Biophys. Res. Commun. 2011;407:758–763. doi: 10.1016/j.bbrc.2011.03.096. [DOI] [PubMed] [Google Scholar]
- 18.Liu D, et al. Identification of key genes and candidated pathways in human autosomal dominant polycystic kidney disease by bioinformatics analysis. Kidney Blood Press. Res. 2019;44:533–552. doi: 10.1159/000500458. [DOI] [PubMed] [Google Scholar]
- 19.Lu S, Gu X, Hoestje S, Epner DE. Identification of an additional hypoxia responsive element in the glyceraldehyde-3-phosphate dehydrogenase gene promoter. Biochim. Biophys. Acta. 2002;1574:152–156. doi: 10.1016/s0167-4781(01)00359-1. [DOI] [PubMed] [Google Scholar]
- 20.Dioudis C, Dimitrios G, Thomas TH, West IC. Abnormal glyceraldehyde-3-phosphate dehydrogenase binding and glycolytic flux in Autosomal Dominant Polycystic Kidney Disease after a mild oxidative stress. Hippokratia. 2008;12:162–167. [PMC free article] [PubMed] [Google Scholar]
- 21.Muñoz JJ, et al. Identification of housekeeping genes for microRNA expression analysis in kidney tissues of Pkd1 deficient mouse models. Sci. Rep. 2020;10:231. doi: 10.1038/s41598-019-57112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen WC, Tzeng YS, Li H. Gene expression in early and progression phases of autosomal dominant polycystic kidney disease. BMC Res. Notes. 2008;1:131. doi: 10.1186/1756-0500-1-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hama T, et al. Aberrant Smad3 phosphoisoforms in cyst-lining epithelial cells in the. Am. J. Physiol. Renal. Physiol. 2017;313:F1223–F1231. doi: 10.1152/ajprenal.00697.2016. [DOI] [PubMed] [Google Scholar]
- 25.Sato Y, Yamamura M, Sasaki M, Harada K. Blockade of hedgehog signaling attenuates biliary cystogenesis in the polycystic kidney (PCK) rat. Am. J. Pathol. 2018;188:2251–2263. doi: 10.1016/j.ajpath.2018.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Harris PC, Torres VE. Genetic mechanisms and signaling pathways in autosomal dominant polycystic kidney disease. J. Clin. Invest. 2014;124:2315–2324. doi: 10.1172/jci72272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menezes LF, Germino GG. Murine models of polycystic kidney disease. Drug Discov. Today Dis. Mech. 2013;10:e153–e158. doi: 10.1016/j.ddmec.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers KA, et al. Differences in the timing and magnitude of Pkd1 gene deletion determine the severity of polycystic kidney disease in an orthologous mouse model of ADPKD. Physiol. Rep. 2016 doi: 10.14814/phy2.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu S, et al. Essential role of cleavage of Polycystin-1 at G protein-coupled receptor proteolytic site for kidney tubular structure. Proc. Natl. Acad. Sci. USA. 2007;104:18688–18693. doi: 10.1073/pnas.0708217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balbo BE, et al. Cardiac dysfunction in Pkd1-deficient mice with phenotype rescue by galectin-3 knockout. Kidney Int. 2016;90:580–597. doi: 10.1016/j.kint.2016.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sousa MV, et al. Smoking accelerates renal cystic disease and worsens cardiac phenotype in Pkd1-deficient mice. Sci. Rep. 2021;11:14443. doi: 10.1038/s41598-021-93633-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Wang B, He H, Sun A, Guo C. A new set of reference housekeeping genes for the normalization RT-qPCR data from the intestine of piglets during weaning. PLoS ONE. 2018;13:e0204583. doi: 10.1371/journal.pone.0204583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Breyer MD, Susztak K. The next generation of therapeutics for chronic kidney disease. Nat. Rev. Drug Discov. 2016;15:568–588. doi: 10.1038/nrd.2016.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cui X, Zhou J, Qiu J, Johnson MR, Mrug M. Validation of endogenous internal real-time PCR controls in renal tissues. Am. J. Nephrol. 2009;30:413–417. doi: 10.1159/000235993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leal MF, et al. Comprehensive selection of reference genes for expression studies in meniscus injury using quantitative real-time PCR. Gene. 2016;584:60–68. doi: 10.1016/j.gene.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 36.Dupasquier S, et al. Validation of housekeeping gene and impact on normalized gene expression in clear cell renal cell carcinoma: Critical reassessment of YBX3/ZONAB/CSDA expression. BMC Mol. Biol. 2014;15:9. doi: 10.1186/1471-2199-15-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Olsan EE, et al. Signal transducer and activator of transcription-6 (STAT6) inhibition suppresses renal cyst growth in polycystic kidney disease. Proc. Natl. Acad. Sci. USA. 2011;108:18067–18072. doi: 10.1073/pnas.1111966108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma Y, Dai H, Kong X, Wang L. Impact of thawing on reference gene expression stability in renal cell carcinoma samples. Diagn. Mol. Pathol. 2012;21:157–163. doi: 10.1097/PDM.0b013e31824d3435. [DOI] [PubMed] [Google Scholar]
- 39.Caracausi M, et al. Systematic identification of human housekeeping genes possibly useful as references in gene expression studies. Mol. Med. Rep. 2017;16:2397–2410. doi: 10.3892/mmr.2017.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li C, et al. Expression of PGAM1 in renal clear cell carcinoma and its clinical significance. Int. J. Clin. Exp. Pathol. 2015;8:9410–9415. [PMC free article] [PubMed] [Google Scholar]
- 41.Fonseca JM, et al. Renal cyst growth is the main determinant for hypertension and concentrating deficit in Pkd1-deficient mice. Kidney Int. 2014;85:1137–1150. doi: 10.1038/ki.2013.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmid H, et al. Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies. Kidney Int. 2003;64:356–360. doi: 10.1046/j.1523-1755.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- 43.Gholami K, Loh SY, Salleh N, Lam SK, Hoe SZ. Selection of suitable endogenous reference genes for qPCR in kidney and hypothalamus of rats under testosterone influence. PLoS ONE. 2017;12:e0176368. doi: 10.1371/journal.pone.0176368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bustin SA, McKay IA. The product of the primary response gene BRF1 inhibits the interaction between 14-3-3 proteins and cRaf-1 in the yeast trihybrid system. DNA Cell Biol. 1999;18:653–661. doi: 10.1089/104454999315060. [DOI] [PubMed] [Google Scholar]
- 45.Cale JM, Millican DS, Itoh H, Magness RR, Bird IM. Pregnancy induces an increase in the expression of glyceraldehyde-3-phosphate dehydrogenase in uterine artery endothelial cells. J. Soc. Gynecol. Investig. 1997;4:284–292. doi: 10.1016/S1071-5576(97)00067-1. [DOI] [PubMed] [Google Scholar]
- 46.Puissant C, Bayat-Sarmadi M, Devinoy E, Houdebine LM. Variation of transferrin mRNA concentration in the rabbit mammary gland during the pregnancy-lactation-weaning cycle and in cultured mammary cells. A comparison with the other major milk protein mRNAs. Eur. J. Endocrinol. 1994;130:522–529. doi: 10.1530/eje.0.1300522. [DOI] [PubMed] [Google Scholar]
- 47.Calvo EL, Boucher C, Coulombe Z, Morisset J. Pancreatic GAPDH gene expression during ontogeny and acute pancreatitis induced by caerulein. Biochem. Biophys. Res. Commun. 1997;235:636–640. doi: 10.1006/bbrc.1997.6716. [DOI] [PubMed] [Google Scholar]
- 48.Mansur NR, Meyer-Siegler K, Wurzer JC, Sirover MA. Cell cycle regulation of the glyceraldehyde-3-phosphate dehydrogenase/uracil DNA glycosylase gene in normal human cells. Nucleic Acids Res. 1993;21:993–998. doi: 10.1093/nar/21.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sikand K, Singh J, Ebron JS, Shukla GC. Housekeeping gene selection advisory: Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin are targets of miR-644a. PLoS ONE. 2012;7:e47510. doi: 10.1371/journal.pone.0047510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glare EM, Divjak M, Bailey MJ, Walters EH. beta-Actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax. 2002;57:765–770. doi: 10.1136/thorax.57.9.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nazari F, Parham A, Maleki AF. GAPDH, β-actin and β2-microglobulin, as three common reference genes, are not reliable for gene expression studies in equine adipose- and marrow-derived mesenchymal stem cells. J. Anim. Sci. Technol. 2015;57:18. doi: 10.1186/s40781-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Araujo LCC, Bordin S, Carvalho CRO. Reference gene and protein expression levels in two different NAFLD mouse models. Gastroenterol. Res. Pract. 2020;2020:1093235. doi: 10.1155/2020/1093235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou SM, et al. β-actin gene expression is variable among individuals and not suitable for normalizing mRNA levels in Portunus trituberculatus. Fish Shellfish Immunol. 2018;81:338–342. doi: 10.1016/j.fsi.2018.07.021. [DOI] [PubMed] [Google Scholar]
- 54.Weimbs T, Shillingford JM, Torres J, Kruger SL, Bourgeois BC. Emerging targeted strategies for the treatment of autosomal dominant polycystic kidney disease. Clin. Kidney J. 2018;11:i27–i38. doi: 10.1093/ckj/sfy089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Talbot JJ, et al. Polycystin-1 regulates STAT activity by a dual mechanism. Proc. Natl. Acad. Sci. USA. 2011;108:7985–7990. doi: 10.1073/pnas.1103816108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weimbs T, Talbot JJ. STAT3 signaling in polycystic kidney disease. Drug Discov. Today Dis. Mech. 2013;10:e113–e118. doi: 10.1016/j.ddmec.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takakura A, et al. Pyrimethamine inhibits adult polycystic kidney disease by modulating STAT signaling pathways. Hum. Mol. Genet. 2011;20:4143–4154. doi: 10.1093/hmg/ddr338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leonhard WN, et al. Curcumin inhibits cystogenesis by simultaneous interference of multiple signaling pathways: In vivo evidence from a Pkd1-deletion model. Am. J. Physiol. Renal. Physiol. 2011;300:F1193–1202. doi: 10.1152/ajprenal.00419.2010. [DOI] [PubMed] [Google Scholar]
- 59.Nowak KL, Hopp K. Metabolic reprogramming in autosomal dominant polycystic kidney disease: Evidence and therapeutic potential. Clin. J. Am. Soc. Nephrol. 2020;15:577–584. doi: 10.2215/cjn.13291019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malas TB, et al. Prioritization of novel ADPKD drug candidates from disease-stage specific gene expression profiles. EBioMedicine. 2020;51:102585. doi: 10.1016/j.ebiom.2019.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Piontek KB, et al. A functional floxed allele of Pkd1 that can be conditionally inactivated in vivo. J. Am. Soc. Nephrol. 2004;15:3035–3043. doi: 10.1097/01.Asn.0000144204.01352.86. [DOI] [PubMed] [Google Scholar]
- 62.Shillingford JM, Piontek KB, Germino GG, Weimbs T. Rapamycin ameliorates PKD resulting from conditional inactivation of Pkd1. J. Am. Soc. Nephrol. 2010;21:489–497. doi: 10.1681/asn.2009040421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meca R, et al. Caffeine accelerates cystic kidney disease in a Pkd1-deficient mouse model. Cell Physiol. Biochem. 2019;52:1061–1074. doi: 10.33594/000000072. [DOI] [PubMed] [Google Scholar]
- 64.Bastos AP, et al. Pkd1 haploinsufficiency increases renal damage and induces microcyst formation following ischemia/reperfusion. J. Am. Soc. Nephrol. 2009;20:2389–2402. doi: 10.1681/asn.2008040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.Can-04-0496. [DOI] [PubMed] [Google Scholar]
- 66.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 67.Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 2012 doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- 68.Mukaka MM. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012;24:69–71. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data including supporting datasets are made available as main figures or supplementary information files.