Abstract
The reverse transcriptase (RT) assay is a simple, relatively inexpensive, widely used assay that can detect all retroviruses (known and novel retroviruses as well as infectious and defective retroviruses) on the basis of the divalent cation requirement of their RT enzyme, i.e., Mg2+ or Mn2+. Descriptions of various RT assays have been published; however, they cannot be directly applied to the analysis of biological products or clinical samples without further standardization to determine the lower limit of virus detection (sensitivity), assay variability (reproducibility), or ability to detect different retroviruses (specificity). We describe the detection of type E and type D primate retroviruses, which may be pathogenic for humans, by a new 32P-based, Mg2+-containing RT assay. The results show that the sensitivity of detection is <3.2 50% tissue culture infective doses (TCID50s) for human immunodeficiency virus type 1 (HIV-1) and <1 TCID50 for simian immunodeficiency virus isolated from a rhesus macaque (SIVmac). Analysis of recombinant HIV-1 RT enzyme indicated that 10−5 U, which is equivalent to 4.25 × 104 virions, could be detected. Additionally, genetically distinct type D retroviruses such as simian AIDS retrovirus and squirrel monkey retrovirus were also detected in the assay with similar sensitivities. Thus, the improved RT assay can be used to detect genetically divergent Mg2+-dependent retroviruses of human and simian origin that can infect human cells and that therefore pose a potential health risk to humans.
All retroviruses can be divided into two categories on the basis of the presence of an Mg2+- or Mn2+-requiring RNA-dependent DNA polymerase that is termed reverse transcriptase (RT) (1, 22) and that is critical in the retroviral life cycle (23, 24). Each group includes retroviruses of diverse origins that are structurally distinct and genetically divergent but that share similar cation requirements for their RT activity. For example, different retrovirus types (21) such as avian type C retroviruses, primate type D retroviruses, and primate type E lentiviruses, which includes human immunodeficiency virus (HIV) type 1 (HIV-1), can be grouped together on the basis of the presence of an Mg2+-requiring RT in these viruses. Thus, the detection of RT activity can generally indicate the presence of a retrovirus in the absence of specific information regarding its genome or protein content. Although RT assays generally detect about 105 to 106 virus particles and are not as sensitive as infectivity or PCR assays, they are broadly reactive and have been used for the detection and isolation of different types of novel retroviruses including HIV-1 (2, 6). In addition, RT assays are routinely used in infectivity studies for the rapid and easy monitoring of retrovirus infection and replication. The detection of small amounts of retrovirus by the RT assay may be made possible by virus amplification in a susceptible cell line or by increasing the virus concentration in a sample so that it is above the detection limit of the assay, e.g., by centrifugation. The RT assay is also widely used for analysis of potential retroviral contaminants in biological products, which may be introduced during passage through animals, during propagation in cell substrates, or from biological raw materials used in production (12).
Several Mg2+-based RT assays have been developed; however, the viruses used in most of the studies have been avian myeloblastosis virus or HIV-1. Furthermore, the previous studies describe details regarding assay development; however, there is little information about assay standardization, including sensitivity of virus detection, ability to detect different retroviruses, or assay variability. Such information is especially important when an RT assay is used to demonstrate the absence of retroviral contaminants in biological products and in analyses of clinical samples from potentially infected individuals. Current HIV-1 RT assays have been used in infectivity studies, neutralization assays, and assessments of antiviral effects. The original assays used [3H]deoxynucleoside triphosphates to extend the oligonucleotide primer to produce the cDNA copy of the homopolymer template (2, 6, 10). Modifications of the RT assays have been made to increase the sensitivity of virus detection which include the use of 32P- and 125I-radiolabeled nucleotide substrates (7, 25). Additionally, RT assays with increased sensitivities have been developed with nonisotopically labeled nucleotides; however, this was achieved after a prolonged incubation, i.e., 15 to 24 h (3, 5, 20). In this paper we describe the standardization of a new 32P-based RT cocktail, with a 2-h incubation period, for the general detection of retroviruses that contain Mg2+-requiring RT, including type E lentiviruses (e.g., HIV-1 and simian immunodeficiency virus [SIV]) as well as two distinct type D retroviruses, i.e., simian AIDS retrovirus (SRV) (4, 16) and squirrel monkey retrovirus (SMRV) (8).
MATERIALS AND METHODS
RT assays.
The new RT cocktail contains the following: 6 mM MgCl2, 3 μg of poly(A) (P-L Biochemicals Inc. Milwaukee, Wis.) per ml, 0.021 μg of p(dT)12–18 (Pharmacia Biotech, Piscataway, N.J.) per ml, 0.12% Nonidet P-40, 24 mM triethanolamine (TEA; U.S. Biochemical Corp., Cleveland, Ohio), and 28.8 mM EGTA (U.S. Biochemical Corp.). TEA-EGTA was formulated separately by mixing 3.93 ml of TEA and 13.146 g of EGTA in a final volume of 300 ml (pH 8.0). This cocktail was aliquoted and stored at −20°C such that it would undergo only two freeze-thaws prior to its use. Additionally, the cocktail was thawed on ice or quickly at 37°C and was immediately put on ice. Under these handling conditions the cocktail was found to remain stable for at least a year, as determined by the RT activity of a standard control virus. Just prior to use of the cocktail, the following were added per milliliter of chilled cocktail: 4 μl of 1 M dithiothreitol, 3 μl of [α-32P]dTTP (1.5 μCi; >400 Ci/mmol; Amersham Corp., Arlington Heights, Ill.), and 1 μl of 10−4 M dTTP. The dithiothreitol and nonradioactive dTTP solutions were also aliquoted individually and stored at −20°C to avoid additional freezing and thawing. Ten microliters of test sample was incubated with 50 μl of the RT cocktail in a screw-cap, 1.5-ml conical polypropylene tube (Starstedt, Arlington, Tex.) for 2 h in a 37°C water bath. Five microliters of the reaction mixture was spotted, in duplicate, onto DE81 paper (Whatman, Maidstone, United Kingdom), air dried, and washed at room temperature on a rocker in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) four times each for 5 min and two times each for 1 min with 95% ethanol. The paper was air dried and exposed overnight at −80°C to X-OMAT AR film (Kodak, Rochester, N.Y.). The next day the spots were cut out and counted in a scintillation counter.
In all the experiments described in this paper, the radioisotope was used prior to or on the manufacturer’s reference date. Dilutions of viruses and enzyme were prepared as described below and were immediately used for RT analyses. The negative control was complete medium (described below).
Viruses.
HIV-1 was prepared in human peripheral blood mononuclear cells (PBMCs) by using strain LAI, which had been grown in CEM clone 12D7 cells (obtained as strain LAV.04 from M. A. Martin, National Institute of Allergy and Infectious Diseases). For virus titration, human PBMCs (106 cells/ml) were stimulated with phytohemagglutinin (PHA; Murex Diagnostics, Dartford, United Kingdom), which was used at a final concentration of 250 ng/ml in complete RPMI 1640 medium containing 10% fetal bovine serum (Hyclone, Logan, Utah), 10 mM HEPES (Biofluids, Rockville Md.), 250 U of penicillin per ml, 250 μg of streptomycin per ml, and 2 mM l-glutamine (Life Technologies, Gibco BRL, Grand Island, N.Y.). After 3 days, the PHA-containing medium was replaced with medium containing 10% interleukin-2 (Hemagen Diagnostics, Columbia, Md.), and the cells were resuspended at a final concentration of 106 cells/0.1 ml. Virus infection was set up in a 24-well plate: 0.1 ml of 10−1 to 10−6 serial dilutions of virus were initially incubated for 1 h at 37°C with 0.1 ml of 106 PBMCs. After 1 h, 1.8 ml of medium was added and the cells were cultured in a 5% CO2 atmosphere at 37°C. After 3 to 4 days, half of the medium was removed and was replaced with fresh medium. On day 7 postinfection, the supernatant was filtered through a 0.45-μm-pore-size filter unit (Spin-X Centrifuge Tube Filters; Costar, Cambridge, Mass.) and collected for the RT assay. The virus infections were set up in quadruplicate. The virus titer was calculated as described by Reed and Muench (18) and was expressed as the 50% tissue culture infectious dose (TCID50) per milliliter. The titer of the HIV-1 stock was 104.5 TCID50s/ml in human PBMCs.
SIVmac was obtained by ligation of cloned 5′ and 3′ DNA fragments of SIVmac-mm239 (11) and transfection in 174× CEM cells. The virus obtained was propagated in rhesus monkey PBMCs to produce a large-scale virus stock (6a). The titer of the monkey-grown SIVmac stock was determined as described above for HIV-1, except that the monkey PBMCs were stimulated with 500 ng of PHA per ml. The titer of SIVmac239 at passage 1 in autologous monkey PBMCs was determined as 104.0 TCID50s/ml.
For RT assay analysis of HIV-1 and SIV, each virus was serially diluted in complete RPMI 1640 medium. At least two independently prepared dilutions were assayed in at least two separate RT assays.
SRV type 1 (SRV-1) and SMRV were purchased from Advanced Biotechnologies Inc. (Columbia, Md.) and were at 8.26 × 107 and 1.92 × 1011 virus particles/ml, respectively, on the basis of electron microscopy. Serial dilutions of the viruses were made in complete RPMI 1640 medium. For each virus, one dilution series was used and the RT assay was done in duplicate.
RT enzyme.
Recombinant HIV-1 RT was purchased from Worthington Biochemical Corporation (Freehold, N.J.; supplied as 48.7 U/μl). Serial dilutions of the enzyme were made in complete medium and were used immediately in RT assays.
RESULTS
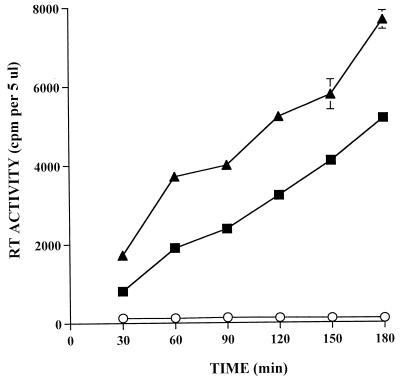
The linear range of the RT reaction was determined by assaying the RT activity at various times ranging from 30 to 180 min (Fig. 1). Independent RT reactions with 10 μl of undiluted HIV-1 and SIVmac were set up for each time point as described in Materials and Methods. Each reaction was terminated at the times indicated in Fig. 1 by spotting in duplicate 5 μl of the reaction mixture onto DE81 filter paper. All the reaction mixtures were spotted on a single filter paper until the reaction mixture from last time point was spotted. The paper was then washed as described in Materials and Methods. The results indicated that the RT activities for HIV and SIV were directly proportional to the reaction times up to 180 min. The rate of dTTP incorporation was found to be directly proportional to the virus concentration as well as to the amount of [32P]dTTP used in the assay (data not shown). The RT activity increased with the amount of [32P]dTTP with 1 to 10 μl (0.5 to 5 μCi) of the radiolabeled deoxynucleoside triphosphate.
FIG. 1.
Linear range of the RT assay. The linearity of the RT assay was determined by HIV-1LAI and SIVmac-mm239. Ten microliters of undiluted virus was assayed at the indicated time intervals. The RT activity is indicated. The mean ± standard deviation from one experiment is indicated. ▴, HIV-1; ■, SIV; ○, medium.
The sensitivity of detection of type E lentiviruses in the RT assay was determined with HIV-1 and SIV. The results are presented in Table 1. Ten microliters of undiluted virus (first sample) and serial dilutions were assayed for RT activity. The sensitivity of detection of HIV-1 was <3.2 TCID50s. A similar sensitivity of detection was seen with a different, independently prepared HIV-1 stock (13). The sensitivity of detection of SIVmac was <1 TCID50. Furthermore, in repeated, parallel experiments, the sensitivity of HIV-1 and SIV detection was 10-fold greater by the new RT assay than by a previously published assay (25). In a comparison of the new RT assay with another HIV-1 RT assay published by Hoffman et al. (10), sevenfold greater RT activity was detected when two HIV-1 concentrations were tested with the new cocktail (18a). To further assess assay sensitivity, RT analysis was done with serial dilutions of recombinant HIV-1 RT. Similar results were obtained with two independently prepared dilutions. The results of RT analysis of one dilution series, which was assayed in duplicate, are presented in Fig. 2. The sensitivity of detection was 10−5 U of HIV-1 RT enzyme.
TABLE 1.
Detection of HIV-1 and SIV in RT assay
| HIV-1
|
SIV
|
||||
|---|---|---|---|---|---|
| No. of infectious particlesa | RT activityb | Autorad resultc | No. of infectious particlesa | RT activityb | Autorad resultc |
| 316.2 | 4,253 ± 1,705 | + | 100 | 3,545 ± 1,184 | + |
| 63.2 | 1,347 ± 410 | + | 20 | 969 ± 123 | + |
| 31.6 | 735 ± 188 | + | 10 | 505 ± 116 | + |
| 6.3 | 258 ± 18 | + | 2 | 183 ± 40 | + |
| 3.2 | 183 ± 21 | + | 1 | 147 ± 45 | + |
| 0.63 | 115 ± 27 | − | 0.2 | 108 ± 31 | − |
| 0.32 | 109 ± 39 | − | 0.1 | 108 ± 48 | − |
| 0.063 | 98 ± 32 | − | 0.02 | 108 ± 37 | − |
| 0.032 | 107 ± 33 | − | 0.01 | 100 ± 36 | − |
| Medium | 102 ± 44 | − | Medium | 102 ± 44 | − |
Number of infectious virions in 10 μl of undiluted (first) sample and serially diluted samples on the basis of an HIV-1 stock titer of 104.5 TCID50s per ml in human PBMCs and an SIV stock titer of 104.0 TCID50s per ml in rhesus monkey PBMCs.
RT activity is counts per minute per 5 μl of spotted reaction mixture. In the case of HIV-1 the mean ± standard deviation was calculated for two independent RT assays in which each sample was spotted twice, and in the case of SIV the mean ± standard deviation was calculated for four spots (two per sample) from two independent RT assays. Each assay was performed with an independently prepared virus dilution series.
Autorad, autoradiography. An autoradiogram was exposed overnight. The results are indicated as positive and negative and do not reflect differences in the intensities of the positive signals.
FIG. 2.
Detection of recombinant HIV-1 RT enzyme. Serial dilutions of HIV-1 RT enzyme were analyzed by the RT assay. The RT activity is indicated. The mean ± standard deviation was calculated for four spots obtained from two RT assays which were done in duplicate with one dilution series.
The ability of the new RT assay to detect other retroviruses containing an Mg2+-requiring RT enzyme, such as type D retroviruses, was assessed with SRV and SMRV. In this case the sensitivity of detection was based upon the use of virus stocks with known numbers of particles, as determined by electron microscopy, and therefore may be less accurate than when the HIV-1 and SIV stocks were used, in which case the number of infectious particles was determined on the basis of an infectivity assay. The results presented in Table 2 indicate that about 2 × 105 virions of SMRV were detected and that about 8 × 104 virions of SRV were detected.
TABLE 2.
Detection of type D retroviruses in RT assays
| Virus and no. of virus particlesa | RT activityb | Autorad resultc |
|---|---|---|
| SMRV | ||
| 1.92 × 107 | 5,163 ± 228 | + |
| 1.92 × 106 | 574 ± 24 | + |
| 1.92 × 105 | 114 ± 10 | + |
| 1.92 × 104 | 79 ± 5 | − |
| 1.92 × 103 | 77 ± 5 | − |
| SRV | ||
| 8.25 × 105 | 547 ± 146 | + |
| 8.25 × 104 | 85 ± 9 | + |
| 8.25 × 103 | 65 ± 15 | − |
The number of virus particles in 10 μl is indicated on the basis of the particle count obtained by electron microscopy by Advanced Biotechnologies Inc. The first sample in the case of SMRV represents a 1:100 dilution of the original sample, and the rest of the samples are 10-fold serial dilutions. In the case of SRV the first sample is undiluted virus and the rest of the samples are 10-fold serial dilutions.
The RT activity is counts per minute per 5 μl and is reported as the mean ± standard deviation for two RT assays which were done in duplicate, with each sample being spotted twice.
Autorad, autoradiography. Results are based upon visual examination of an overnight exposure of the autoradiogram. The results are indicated as positive and negative and do not reflect the differences in the intensities of the positive signals.
It should be noted that because 32P was used in the assay, the results could be monitored by both scintillation counting and autoradiography. A parallel analysis was done in all cases; it was found that a weakly positive signal was easier to visualize from the autoradiogram. On the basis of autoradiography, a result was positive if the counts were 50% above the background RT activity.
DISCUSSION
The RT assay is widely used for the general detection of known and novel retroviruses. This is primarily because it is easy, quick, and relatively inexpensive to perform. Recently, highly sensitive PCR-based RT assays which can detect 3 to 100 virions have been developed (9, 17, 19); however, their use at this time is limited because the assay is technically demanding and expensive. Thus, parallel efforts have continued to increase the sensitivity of retrovirus detection by the conventional RT assays.
In this paper, we describe the standardization of an improved Mg2+-based RT assay which can detect different types of primate retroviruses. The sensitivity of the assay was determined to be <1 TCID50 of SIVmac and <3.2 TCID50s of HIV-1. Analysis of the recombinant HIV-1 RT enzyme indicated the detection of 10−5 U, which, on the basis of the molecular weight (117,000), was equivalent to 3.4 × 106 molecules of RT. Since it is reported that 80 molecules of RT are present per HIV-1 particle (15), the sensitivity of detection was calculated to be <4.25 × 104 virions. These results thus indicate a possible 1:10,000 ratio of infectious virus to total particles, which is consistent with previously reported results (14). In addition to primate lentiviruses, type D retroviruses of genetically diverse origins were detected at about 8 × 104 virions of SRV, which was isolated from an Old World monkey, and about 2 × 105 virions of SMRV, which was isolated from a New World monkey.
The sensitivity of detection was achieved with a relatively short incubation time (2 h), in contrast to some other RT assays that require prolonged incubation times (i.e., 15 to 24 h) for increased detection (5, 20). The sensitivity of the new RT assay for HIV and SIV detection can be further improved by increasing the incubation time to up to 3 h and/or by increasing the amount of [32P]dTTP. Conversely, with samples containing adequate amounts of virus, a minimum amount of radiolabeled dTTP and/or a reduced reaction time can be used, since the counts obtained are directly proportional to the incubation time and to the [32P]dTTP concentration.
The results obtained with the new RT assay were highly reproducible; similar results were obtained with regard to the sensitivity of detection of HIV and SIV when independently prepared virus stocks or dilutions were used. However, some variability in the counts per minute incorporated can occur between different assays, especially due to handling, e.g., when the sample or the radioisotope is pipetted or when the final reaction mixture is spotted onto the filter paper. We have reduced interassay variability by using designated and accurately calibrated pipetting devices for the different handling procedures. One caveat of the new RT assay is that if the reaction is done in a CO2 incubator, screw-cap tubes must be used to avoid lowering of the pH in the reaction, which suppresses the RT activity (18b).
The RT assay has been used for the detection of known and novel retroviruses from infected cells both in assessing the replication of viruses and for evaluating antiretroviral treatments (e.g., in the case of treatments for HIV-1 infection). We have used the RT assay described in this paper as a general detection strategy in a multicombinational analysis with specific detection strategies such as DNA and RNA PCR assays to demonstrate the absence of detectable HIV or SIV in several monovalent lots of oral, poliovirus vaccine (13). The new RT assay was especially useful because of its increased sensitivity for the detection of retroviral RT compared to those of other, similar assays and because of its low background signal.
ACKNOWLEDGMENTS
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases: p239SpE3′ and p239SpSp5′ cloned DNAs (from Ronald Desrosiers). We thank Teresa A. Galvin for preparing the titered SIVmac-mm239 stock, S. Tabriz Ali for preparing the titered HIV-1 stock, and Theodore Bryan for technical assistance. We also acknowledge Malcolm A. Martin for support in formulation of the RT cocktail and Keith Peden, Hana Golding, and Muhammad Shahabuddin for review of the manuscript.
REFERENCES
- 1.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 2.Barre-Sinoussi F, Chermann J C, Rey F, Nugeyre M T, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rosenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 3.Cook R F, Cook S J, Issel C J. A nonradioactive micro-assay for released reverse transcriptase activity of a lentivirus. BioTechniques. 1991;13:380–386. [PubMed] [Google Scholar]
- 4.Daniel M D, King N W, Letvin N L, Hunt R D, Sehgal P K, Desrosiers R C. A new type D retrovirus isolated from macaques with an immunodeficiency syndrome. Science. 1984;223:602–605. doi: 10.1126/science.6695172. [DOI] [PubMed] [Google Scholar]
- 5.Eberle J, Seibl R. A new method for measuring reverse transcriptase activity by ELISA. J Virol Methods. 1992;40:347–356. doi: 10.1016/0166-0934(92)90092-r. [DOI] [PubMed] [Google Scholar]
- 6.Gallo R C, Salahuddin S Z, Popovic M, Shearer G M, Kaplan M, Haynes B F, Palker T J, Redfield R, Oleske J, Safai B, White G, Foster P, Markham P D. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science. 1984;224:500–503. doi: 10.1126/science.6200936. [DOI] [PubMed] [Google Scholar]
- 6a.Galvin, T. A., and A. S. Khan. Unpublished data.
- 7.Gronowitz J S, Neumuller M, Lennerstrand J, Bhikhabhai R, Unge T, Weltman H, Kallander C F R. Carrier bound templates for single tube reverse transcriptase assays and for combined purification and activity analyses, with special reference to HIV. Biotechnol Appl Biochem. 1991;13:127–142. [PubMed] [Google Scholar]
- 8.Heberling R L, Barker S T, Kalter S S, Smith G C, Helmke R J. Oncornavirus: isolation from a squirrel monkey (Saimiri sciureus) lung culture. Science. 1977;195:289–292. doi: 10.1126/science.63993. [DOI] [PubMed] [Google Scholar]
- 9.Heneine W, Yamamoto S, Switzer W M, Spira T J, Folks T M. Detection of reverse transcriptase by a highly sensitive assay in sera from persons infected with human immunodeficiency virus type 1. J Infect Dis. 1995;171:1210–1216. doi: 10.1093/infdis/171.5.1210. [DOI] [PubMed] [Google Scholar]
- 10.Hoffman A D, Banapour B, Levy J A. Characterization of the AIDS-associated retrovirus reverse transcriptase and optimal conditions for its detection in virions. Virology. 1985;147:326–335. doi: 10.1016/0042-6822(85)90135-7. [DOI] [PubMed] [Google Scholar]
- 11.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science. 1990;248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 12.Khan A S. Retrovirus screening of vaccine cell substrates. In: Brown F, Lubiniecki A S, editors. Developments in biological standardization: viral safety and evaluation of viral clearance from biopharmaceutical products. Vol. 88. Basel, Switzerland: Karger; 1996. pp. 155–160. [PubMed] [Google Scholar]
- 13.Khan A S, Shahabuddin M, Bryan T, Joshi B H, Lee S, Hewlett I K. Analysis of live, oral poliovirus vaccine monopools for human immunodeficiency virus type 1 and simian immunodeficiency virus. J Infect Dis. 1996;174:1185–1190. doi: 10.1093/infdis/174.6.1185. [DOI] [PubMed] [Google Scholar]
- 14.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Layne S P, Merges M J, Dembo M, Spouge J L, Conley S R, Moore J P, Raina J L, Renz H, Gelderblom H R, Nara P L. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology. 1992;189:695–714. doi: 10.1016/0042-6822(92)90593-e. [DOI] [PubMed] [Google Scholar]
- 16.Marx P A, Maul D H, Osborn K G, Lerche N W, Moody P, Lowenstine L J, Henrickson R V, Arthur L O, Gravell M, London W T, Sever J L, Levy J A, Munn R B, Gardner M B. Simian AIDS: isolation of a type D virus and disease transmission. Science. 1984;223:1083–1086. doi: 10.1126/science.6695196. [DOI] [PubMed] [Google Scholar]
- 17.Pyra H, Boni J, Schupach J. Ultrasensitive retrovirus detection by a reverse transcriptase assay based on product enhancement. Proc Natl Acad Sci USA. 1994;91:1544–1548. doi: 10.1073/pnas.91.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reed L J, Muench H A. Simple method of estimating fifty per cent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 18a.Repaske, R. Data not shown.
- 18b.Repaske, R. Unpublished data.
- 19.Silver J, Maudru T, Fujita K, Repaske R. An RT-PCR assay for the enzyme activity of reverse transcriptase capable of detecting single virions. Nucleic Acids Res. 1993;21:3593–3594. doi: 10.1093/nar/21.15.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K, Saito T, Kondo M, Osanai M, Watanabe S, Kano T, Kano K, Imai M. Poly A-linked non-isotopic microtiter plate reverse transcriptase assay for sensitive detection of clinical human immunodeficiency virus isolates. J Virol Methods. 1995;55:347–356. doi: 10.1016/0166-0934(95)00073-5. [DOI] [PubMed] [Google Scholar]
- 21.Teich N. Taxonomy of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses, molecular biology of tumor viruses. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 25–207. [Google Scholar]
- 22.Temin H M, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature (London) 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 23.Varmus H, Swanstrom R. Replication of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses, molecular biology of tumor viruses. 2nd ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. 369–512. [Google Scholar]
- 24.Varmus H, Swanstrom R. Replication of retroviruses. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses, molecular biology of tumor viruses. 2nd ed. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1985. pp. 75–134. [Google Scholar]
- 25.Willey R L, Smith D H, Lasky L A, Theodore T S, Earl P L, Moss B, Capon D J, Martin M A. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62:139–147. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]