Editor—We recently demonstrated an association between intraoperative opioid exposure and oncological outcomes in a retrospective study of 740 patients with early-stage lung adenocarcinoma who underwent primary tumour resection. This cohort was unique in that all patients also underwent next-generation sequencing1 of the tumour samples, permitting investigation into the interaction of tumour genomics with opioid dose to modify outcomes. In particular, we found that alterations in two tumour genomic factors (high fraction genome altered and CDKN2A gene deep deletions) were associated with worse overall survival with increased opioid exposure, compared with patients without these genetic alterations. By contrast, increasing intraoperative dose in patients with alteration in either the Wnt or the Hippo oncogenic signalling pathway reversed the pro-tumour association, resulting in improved recurrence-specific survival (RSS) with increased opioid exposure, compared with patients with unaltered pathways.2
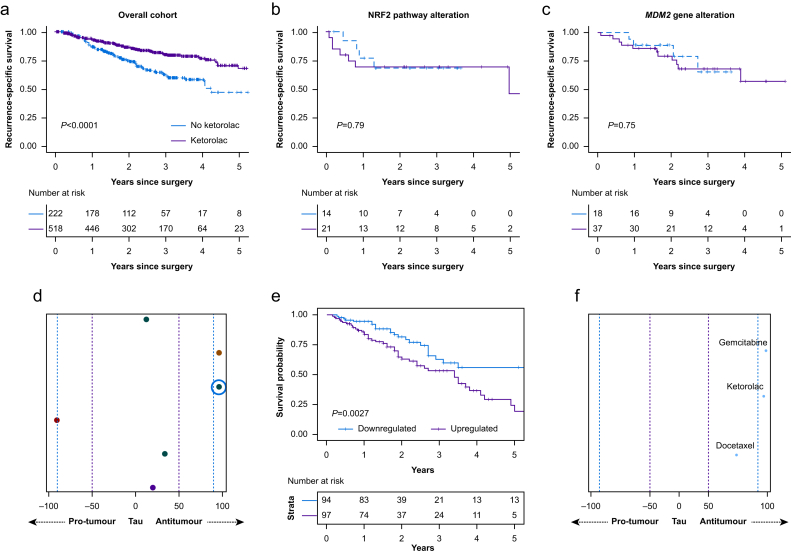
Motivated by the extensive literature suggesting that NSAIDs may have antitumour effects,3 we decided to look at intraoperative ketorolac exposure in this cohort (IRB# 18–391, Memorial Sloan Kettering Cancer Center, approval date September 7, 2018). Five hundred and eighteen of the 740 patients in our cohort received ketorolac intraoperatively (either 15 or 30 mg i.v.). Ketorolac administration generally occurred at the conclusion of the surgery after tumour removal, and most of these patients (415/518) received additional doses by the end of the first postoperative day. This exposure was associated with improved RSS on univariable analysis (hazard ratio [HR] 0.48; 95% confidence interval [CI]: 0.35–0.66; P<0.001) (Fig. 1a). Consequently, we repeated the original multivariable analysis, this time including ketorolac. The association remained after covariate adjustment (HR 0.64; 95% CI: 0.46–0.89; P=0.007). Of note, the inclusion of ketorolac did not change (and was independent of) the previously reported association of opioids with worse overall survival and the association of ketamine with improved RSS (Supplementary Table 1). Patient demographics for this cohort have been previously described in detail.2
Fig 1.
Kaplan–Meier curves for recurrence-specific survival for patients who did (purple) or did not (blue) receive ketorolac, for (a) all patients in the cohort, (b) patients with nuclear factor erythroid 2-related factor 2 (NRF2) oncogenic pathway alteration, and (c) patients with MDM2 gene alteration. (d) Predicted pro-vs antitumour regulation by ketorolac of the six survival-associated gene networks in The Cancer Genome Atlas Lung Adenocarcinoma (TCGA-LUAD) external validation cohort; each row corresponds to one survival-associated gene network, and the coloured circle represents ketorolac's strength and direction (pro-vs antitumour) of regulation of that network in comparison with all 8558 other drugs and small molecules in the Connectivity Map database, given as a percentile rank (tau), where the purple dotted line represents the 50th percentile and the blue dotted line represents the 90th percentile. (e) Survival curves for patients in the TCGA-LUAD cohort with up-vs downregulation of the survival-associated gene network highlighted by the blue circle in (d). (f) Predicted antitumour regulation by ketorolac of the highlighted gene network (x-axis tau, as described previously), with other drugs commonly used in the treatment of lung adenocarcinoma highlighted for comparison.
Next, we probed whether ketorolac interacted with any of the tumour genomic factors to modify this association. Of the oncogenic pathways and high-frequency gene alterations previously studied, two factors were found to have statistically significant interactions with ketorolac to modify the association with RSS (Supplementary Table 2). Alteration in either the nuclear factor erythroid 2-related factor 2 (NRF2) oncogenic pathway (P=0.052) or the MDM2 gene (part of the TP53 oncogenic pathway) (P=0.009) neutralised the improved RSS seen in the overall cohort, possibly reversing the association such that ketorolac was associated with worse RSS (Fig. 1b and c), although the relatively low frequency of these alterations limited the ability to statistically differentiate between these two possibilities in this analysis. Of note, the NRF2 pathway was altered in 4.1% of patients who did and 6.3% of patients who did not receive ketorolac; the MDM2 gene was altered in 7.1% who did and 8.1% who did not receive ketorolac. Both genomic factors have been related to the effects of NSAIDs on cancer in preclinical data. Increased levels of MDM2, a proto-oncogene ubiquitin ligase and negative regulator of TP53, inactivate cellular apoptosis.4 Ketorolac has been shown to block DDX3, an ATP-dependent RNA helicase that regulates cell cycle activity, which results in downregulation of MDM2.5 Additionally, NSAIDs have been shown to activate the NRF2 pathway, an antioxidant response regulatory pathway.6 Note that the number of pathways altered, a global biomarker of genomic mutability across the patient cohort, also had an effect on the association between ketorolac exposure and recurrence.
We sought to further validate this finding in an external surgical cohort (The Cancer Genome Atlas Lung Adenocarcinoma [TCGA-LUAD] cohort)7 through the use of a computational methodology we developed to predict the effects of drugs on oncological outcomes.8 The method inputs a patient cohort with both outcome and RNA sequencing data, such as TCGA, and a priori knowledge of the effects of a drug on gene expression, generally from the cancer-derived Connectivity Map database.9 The method then calculates gene co-expression networks associated with patient survival in a data-driven manner, and projects the effects of a drug on gene expression onto these networks. The result is a prediction of the drug's regulation of the survival-associated networks and, by extension, the survival outcome itself. For example, a drug that upregulates a network associated with improved survival would be expected to itself improve survival.
Application of this method to ketorolac for the TCGA-LUAD cohort of 392 surgical patients predicts a net antitumour effect of ketorolac on lung adenocarcinoma; ketorolac regulates five of six survival-associated gene networks in an antitumour direction Fig. 1d. Survival curves for one of the strongest regulated networks (highlighted by the blue circle in Fig. 1d) are shown in Figure 1e, where it is seen that downregulation is associated with improved survival. (Note that, because of greater reliability and validity of this endpoint in TCGA, overall survival is used in the TCGA analysis; see Supplementary material.) The predicted antitumour regulation of this highlighted network by ketorolac is shown in Figure 1f, where ketorolac is seen to be in the >90th percentile, compared with all 8558 other drugs in the Connectivity Map database, in terms of strength of network regulation, with specific drugs commonly used for the treatment of lung adenocarcinoma highlighted as well for comparison. We note that, whereas this method provides external validation of the antitumour effect of ketorolac seen in the overall cohort, it is more limited in addressing the specific tumour genomic alterations found in our clinical cohort (i.e. NRF2 and TP53 pathways), as gene co-expression networks are fundamentally different from mutational signatures and transcend individual functional genomic pathways. That said, the method predicts that ketorolac does regulate one of the six survival-associated co-expression networks in a pro-tumour direction, which is consistent with the existence of smaller genomic subgroups in which ketorolac results in worse patient survival.
We recognise the exploratory nature of this study and the need for further investigation to definitively determine associations uncovered here, in which a single intraoperative ketorolac dose potentially impacts oncological outcomes. Nonetheless, this is, to our knowledge, the first time that tumour genomics have been demonstrated to modify the ketorolac–recurrence association. The specific genomic factors identified here (NRF2 pathway and MDM2 gene alteration), although exploratory and hypothesis generating, could be of significant clinical importance in that there may exist a subgroup of patients for whom ketorolac negates or reverses the RSS benefit observed more generally. Perioperative use of ketorolac has increased with the adoption of enhanced recovery after surgery protocols.10 It is therefore important before surgery to identify particular patients for whom ketorolac could negatively affect survival because of underlying tumour genomics. This information can be derived from next-generation sequencing of a tumour biopsy specimen (or even plasma-derived cell-free DNA) obtained preoperatively, enabling patient-based precision ketorolac dosing in the perioperative period.
Acknowledgements
JSM and HVG acknowledge John Chodera of the Sloan Kettering Institute for his support and insightful discussions. JSM also acknowledges Sahrena London for helpful conversations.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bja.2021.05.032.
Contributor Information
David R. Jones, Email: jonesd2@mskcc.org.
Joshua S. Mincer, Email: mincerj@mskcc.org.
Declarations of interest
PJM's spouse has an equity interest in Johnson & Johnson. PSA has received research funding from Atara Biotherapeutics and ACEA Biosciences; has served on the Scientific Advisory Board or as consultant to Atara Biotherapeutics, Bayer, CARISMA Therapeutics, Imugene, and Takeda Therapeutics; and has patents, royalties, and intellectual property on mesothelin-targeted CARs and other T-cell therapies, method for detection of cancer cells using virus, and pending patent applications on T-cell therapies. GR has financial relationships with Scanlan. JMI is a consultant for Genentech and has an equity interest in LumaCyte. MJB serves as a consultant for AstraZeneca. GWF is on the speaker's bureau and serves as a consultant for Edwards Lifesciences. DRJ serves as a consultant for Merck and AstraZeneca. All other authors have no potential conflicts to disclose.
Funding
National Cancer Institute (R01CA217169 and R01CA240472) to DRJ, (R01CA236615) to PSA, (T32CA009501) to JGC, (P30 CA008748 to Memorial Sloan Kettering Cancer Center); Hamilton Family Foundation to DRJ; Department of Defense (LC160212) to PSA; Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cheng D.T., Mitchell T.N., Zehir A. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Connolly J., Tan K.S., Mastrogiacomo B. Associations between intraoperative opioid exposure and tumor genomic alterations linked to survival differences in lung adenocarcinoma. Br J Anaesth. 2021 Jul;127:75–84. doi: 10.1016/j.bja.2021.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cata J.P., Guerra C.E., Chang G.J., Gottumukkala V., Joshi G.P. Non-steroidal anti-inflammatory drugs in the oncological surgical population: beneficial or harmful? A systematic review of the literature. Br J Anaesth. 2017;119:750–764. doi: 10.1093/bja/aex225. [DOI] [PubMed] [Google Scholar]
- 4.Gupta A., Shah K., Oza M.J., Behl T. Reactivation of p53 gene by MDM2 inhibitors: a novel therapy for cancer treatment. Biomed Pharmacother. 2019;109:484–492. doi: 10.1016/j.biopha.2018.10.155. [DOI] [PubMed] [Google Scholar]
- 5.He Y., Zhang D., Yang Y. A double-edged function of DDX3, as an oncogene or tumor suppressor, in cancer progression (review) Oncol Rep. 2018;39:883–892. doi: 10.3892/or.2018.6203. [DOI] [PubMed] [Google Scholar]
- 6.Staurengo-Ferrari L., Badaro-Garcia S., Hohmann M.S.N. Contribution of Nrf2 modulation to the mechanism of action of analgesic and anti-inflammatory drugs in pre-clinical and clinical stages. Front Pharmacol. 2018;9:1536. doi: 10.3389/fphar.2018.01536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scarpa J.R., DiNatale R.G., Mano R. Identifying clear cell renal cell carcinoma coexpression networks associated with opioid signaling and survival. Cancer Res. 2021;81:1101–1110. doi: 10.1158/0008-5472.CAN-20-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subramanian A., Narayan R., Corsello S.M. A next generation Connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452. doi: 10.1016/j.cell.2017.10.049. e1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mayor M.A., Khandhar S.J., Chandy J., Fernando H.C. Implementing a thoracic enhanced recovery with ambulation after surgery program: key aspects and challenges. J Thorac Dis. 2018;10:S3809–S3814. doi: 10.21037/jtd.2018.10.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.