Abstract
The androgen receptor is unusual among nuclear receptors in that most, if not all, of its activity is mediated via the constitutive activation function in the N terminus. Here we demonstrate that p160 coactivators such as SRC1 (steroid receptor coactivator 1) interact directly with the N terminus in a ligand-independent manner via a conserved glutamine-rich region between residues 1053 and 1123. Although SRC1 is capable of interacting with the ligand-binding domain by means of LXXLL motifs, this interaction is not essential since an SRC1 mutant with no functional LXXLL motifs retains its ability to potentiate androgen receptor activity. In contrast, mutants lacking the glutamine-rich region are inactive, indicating that this region is both necessary and sufficient for recruitment of SRC1 to the androgen receptor. This recruitment is in direct contrast to the recruitment of SRC1 to the estrogen receptor, which requires interaction with the ligand-binding domain.
The androgen receptor (AR), a member of the nuclear receptor superfamily (6, 45), is a ligand-activated transcription factor with the major ligands testosterone and dihydrotestosterone. It has the overall domain structure common to nuclear receptors, comprising an N-terminal activation domain (activation function 1 [AF1]), a central DNA-binding domain (DBD), and a C-terminal ligand-binding domain (LBD). A second, ligand-dependent activation function (AF2) in several nuclear receptors, including other steroid hormone receptors, has been well characterized (5, 15, 19, 66), but until recently there was no evidence to support such a function for the AR LBD. Deletion of the LBD results in a molecule with constitutive activity which in many reporter activation assays is equivalent to the maximum activity of the full-length receptor in the presence of ligand, implying that AF1 contributes all the activity of the receptor (35, 56, 61, 82). This finding is in contrast to what occurs with the closely related estrogen receptor (ER), in which AF2 is the major activation domain and AF1 has little independent activity (67). The situation in the AR is still more complex, in that two discrete, overlapping activation domains exist in the N-terminal domain and their usage is context dependent. While almost the entire N terminus (residues 1 to 494) is required for full activity of the full-length receptor, a core that contributes 50% of activity is located between residues 101 and 360, and this region has been termed TAU1. However, in the absence of the LBD a different region, termed TAU5 (residues 370 to 494), mediates activation (34).
Upon binding of ligand, steroid hormone receptors adopt an active conformation that facilitates the dissociation of heat shock proteins, dimerization, and binding to response elements in the promoters of responsive genes. These receptors have been shown to interact with some components of the basal transcriptional machinery (3, 9, 10, 32, 46, 59) and also to promote transcription of the gene by interacting with coactivator proteins (23, 68). Coactivators, which interact only with transcriptionally competent receptors in a ligand-dependent manner and which potentiate ligand-dependent transcriptional activation of the receptor, include the general cofactor CREB-binding protein CBP and the related protein p300 (60), the thyroid receptor-associated protein (TRAP) or vitamin D receptor-interacting protein (DRIP) complexes (33, 55), and the p160 family of coactivators. These three 160-kDa proteins, highly homologous but encoded by separate genes, are known by various acronyms and will herein be termed SRC1 (37, 53, 63) (stands for steroid receptor coactivator 1), TIF2 (73) (stands for transcription intermediary factor 2, the human homologue of mouse GRIP1 [29]), and AIB1 (2) (stands for amplified in breast cancer-1, the human homologue of mouse pCIP [69], also known as ACTR [14], RAC3 [43], or TRAM1 [64]). They may function by recruiting CBP/p300, which possesses histone acetylase activity (4, 51), to target promotors to facilitate chromatin remodeling. The TRAP and DRIP complexes may function in a manner similar to that of the mediator complex in Saccharomyces cerevisiae (21), resulting in the recruitment of RNA polymerase II. Coactivators interact with nuclear receptors via short motifs consisting of the amino acid sequence LXXLL, where L is leucine and X is any amino acid (25, 69). Other proteins which interact with receptors and contain these motifs include RIP140, originally postulated to be a coactivator but which displays some characteristics of a repressor molecule (12, 71), and ARA70 (also called ELE1) (1, 80). ARA70 was originally reported as being a coactivator specific for the AR and was demonstrated to potentiate AR activity in the prostate cancer cell line DU-145 (80).
Elucidation of the LBD crystal structures of several nuclear receptors, in the presence and absence of ligands, has revealed that they contain a series of 11 or 12 alpha-helices (reviewed in references 54 and 78). The most C terminal of these, termed helix 12, realigns in the presence of ligand, and this realignment is believed to form a hydrophobic cleft, composed of helices 3, 5, and 12, which binds the LXXLL motifs of coactivators (16, 50). The ligand-dependent AF2 function was mapped to residues in helix 12 (15), which shows a high degree of conservation between nuclear receptors. Mutations of hydrophobic residues within helices 3, 5, and 12 and of a lysine residue in helix 3 disrupt the interaction of the activated receptor with coactivators while not significantly affecting ligand binding (15, 27, 44).
While much has been published on the mechanism of action of the ER, less is known about the AR. It appears to differ from other steroid receptors in that no separable AF2 function has been shown for mammalian cells and potentiation of its activity by coactivator proteins is less pronounced than for other receptors. A ligand-dependent interaction between the two termini of the AR has also been demonstrated to be necessary for maximum activation of the full-length receptor (7, 30, 38, 39). We therefore decided to investigate whether activation by the AR occurs via the same pathways as activation by other steroid receptors.
MATERIALS AND METHODS
Plasmid construction.
Unless otherwise stated, all restriction enzymes were supplied by New England Biolabs. All constructs created by PCR amplification, with Elongase enzyme mix (Gibco BRL), were verified by sequencing.
The following plasmids have been described previously: pSVARo (8); pSG5-SRC1a, pSG5-SRC1e, pSG5-SRC1eM123, and Gal4BDBD-SRC1 fragments (37); pSG5-TIF2 (73); pEF-RIP140 (12); and pBL1 and pASV (42). The p300 expression vector pCMVβ-p300 was a gift from R. Goodman. The AR mammalian expression construct pAR123 (deletion of residues 1 to 360) and the yeast expression constructs Gal-AR.N8 and Gal-AR.N14 (which contain full-length AF1 and residues 360 to 494, respectively, fused to the Gal4 DBD) were kind gifts from A. Brinkmann and J. Trapman (7, 34).
The ARA70 expression vector was created by isolating the ARA70 sequence from a B-cell pCDNA3 plasmid library (24) with primers homologous to either end of the published sequence (80) and inserting it into pSG5-MCS (37). Sequence of the clone agreed with that published by Yeh et al. (80). The expression plasmids encoding truncated SRC-1, pSG5-SRC1(1–988), pSG5-SRC1(1–1105), and pSG5-SRC1(1–1240), were created by PCR of the relevant fragment with primers incorporating restriction sites and insertion into the expression vector pSG5-MCS. The deletion constructs pSG5-SRC1e(Δ1053–1123) and pSG5-SRC1eΔAD1 were created by the two-step recombinant-PCR method (28) to create a deleted fragment which was digested with the natural restriction sites MscI and BamHI and used to replace the corresponding wild-type fragment in pSG5-SRC1e. The AR helix 12 mutations (E893Q-E897Q, M894A, M894A-M895A, F891A, and F891A-I898T) were introduced into full-length AR in the mammalian expression vector pSVARo by recombinant PCR. For the mammalian two-hybrid study, AR residues 1 to 538 were amplified by PCR and cloned into the BglII site of the pSNATCH vector (11).
For expression in the yeast strain W303-1B, the LBD of the AR (residues 625 to 919) was amplified with primers incorporating KspI and BglII restriction sites and inserted into the expression vector pBL1 by using KspI and BamHI restriction sites. The resultant construct, pBL1-ARAF2, expressed the AR LBD in frame with the heterologous ER LBD. The helix 12 mutations were inserted into this vector by amplifying the region from the relevant AR expression construct with the same primers and inserting them into pBL1. Coactivators RIP140, TIF2(596–773), SRC1a(1240–1441), and SRC1a(1240–1441LXXAA) were amplified by PCR for insertion into the same plasmid with the same sites. Prey vectors for yeast two-hybrid assays in this system were constructed by amplification of the relevant DNA fragment [AR LBD and SRC1a(1240–1441)] and insertion into the vector pASV3, which expresses the fragment in frame with the VP16 activation domain. Full-length SRC1 expression vectors were created by inserting the amplified coding sequence into Yep20 (24a), a derivative of Yep10 (26).
The yeast vectors used in the L40 strain encoding the LexA DNA-binding site fused to AR or AR AF1 were created by amplifying amino acids 1 to 919 or 1 to 556 of the AR by PCR with primers incorporating restriction sites and by inserting them into BTM116 (74). LexA-SRC1(989–1240) fragment fusions were constructed by amplification of the region between residues 989 and 1240 with either pSG5-SRC1e or pSG5-SRC1e(Δ1053–1123) as the template and by insertion into BTM116. Vectors bearing genes encoding fusions of the Gal4 activation domain and fragments of SRC1 were created by excising the SRC1 fragments from the relevant glutathione S-transferase (GST)–SRC1 fusion constructs (37) and inserting them into pACT2 (Clontech). The Gal4-AR AF1 fusion vector was created by amplification of AR residues 1 to 556 from LexA+AR with an upstream lexA primer and a downstream AR primer and insertion into pGAD424 (Clontech). The deletion of residues 1 to 35 was achieved by cutting this fragment at a natural SmaI site.
GST fusion vectors were created by insertion of the relevant fragment [AR(1–556), SRC1(989–1240), or SRC1(989–1240Δ1053–1123)] into the EcoRI and SalI sites of pGEX-6P-1 (Pharmacia Biotech).
Cell culture and transient transfection.
COS-1 and HeLa cells were routinely maintained in E4 supplemented with 10% fetal bovine serum. Twenty-four hours before transfection, cells were plated in 24-well microtiter plates (Falcon) in phenol red-free medium supplemented with 5% dextran charcoal-stripped fetal calf serum. Transfection was performed by a modified calcium phosphate method (13), with each well receiving 25 ng of the AR expression vector, 1 μg of the androgen-responsive reporter plasmid (pG29GtkCAT [58]), and 150 ng of plasmid pCMV-βgalactosidase, together with various amounts of coactivator expression plasmid plus the empty vector to standardize the amounts of DNA. For HeLa cells the amounts used were 100 ng of the AR expression vector, 600 ng of the reporter, and 100 ng of plasmid BOS-βgalactosidase. After incubation for 16 h, the cells were washed and fresh medium containing 10−8 M mibolerone (a synthetic androgen) or vehicle was added. For dose-response assays, various concentrations of mibolerone were used. After a further 24 h, cells were washed twice with phosphate-buffered saline and lysed in 10 mM Tris-HCl (pH 8)–1 mM EDTA–150 mM NaCl–0.65% Nonidet P-40. Extracts were analyzed for chloramphenicol acetyltransferase (CAT) activity (62) or luciferase activity (15), and values were corrected for β-galactosidase activity, measured by the Galacto-Light chemiluminescence assay (Tropix). Unless otherwise stated in the figure legends, results are the averages from at least three independent experiments (except in cases where very little or no activity was seen, when the number of repetitions was occasionally two) performed in duplicate ± standard errors of the means.
For the mammalian two-hybrid assay, COS-1 cells were transfected with Fugene reagent (Roche) in accordance with the manufacturer’s instructions. Per well, the following amounts of DNA were added: 250 ng of VP16 or VP16-AR AF1, 100 ng of (Gal4)5tata-luc (20) (a gift from G. Folkers), 50 ng of cytomegalovirus lacZ, and 100 ng of the Gal4 DBD or Gal4 DBD-SRC1 fragment. The Fugene-DNA mix was left on the cells overnight, and then cells were cultured for a further 24 h before being harvested. Luciferase was assayed according to the instructions from the Promega luciferase assay kit and normalized for β-galactosidase activity.
Yeast culture and transfection.
The yeast strains W303-1B (HMLαMATαHMRa his3-11,15 trp-1 ade2-1 can1-100 leu2-3,11 ura3) and L40 [MATα trp1 his3 leu2 ade2 LYS2::(LexAop)4-HIS3 URA3::(LexAop)8-lacZ)] containing a LexA-responsive lacZ reporter were maintained as described previously (36). They were transformed by electroporation with vectors bearing genes encoding fusion proteins and transformants selected for the appropriate plasmid markers. W303-1B was first transformed with a reporter plasmid, pRLΔ21-U3ERE, which contains a lacZ gene driven by three estrogen response elements (48). To perform two-hybrid assays, transformants were grown to late log phase in 15 ml of selective medium (yeast nitrogen base containing 1% glucose and appropriate supplements), where appropriate in the presence of 10−7 M mibolerone. For dose-response curves, various concentrations of mibolerone were used. Cells were then harvested, washed, suspended in 0.1 M Tris-HCl (pH 7.5)–0.5% Triton X-100, snap frozen in a dry ice-ethanol bath, and thawed. An aliquot of this extract was assayed for β-galactosidase activity as described previously (17), and the protein content was measured by reading the optical density at 600 nm (OD600). Activity was calculated with the equation (1,000 × OD420)/(OD600 × reaction time in minutes) and expressed as β-galactosidase units. Unless otherwise stated in the figure legends, the assay was repeated with at least three independent transformants and the data are the means ± standard deviations of these readings.
GST pull-down assays.
Recombinant cDNAs in the pSG5 expression vector were transcribed and translated in vitro in the presence of [35S]methionine in reticulocyte lysate (Promega) according to the manufacturer’s protocol. GST fusion proteins were induced, purified, bound to Sepharose beads (Pharmacia), and incubated with translated proteins as previously described (37) in NETN buffer (20 nM Tris [pH 8.0], 1 nM EDTA, 0.5% NP-40, 150 mM NaCl) in the presence or absence of 10−7 mibolerone. After being washed, samples were separated on sodium dodecyl sulfate–8% polyacrylamide gels, which were fixed and dried; samples were then visualized by autoradiography. Quantitation of binding was achieved by fluorography.
RESULTS
The AR contains a ligand-dependent activation function in helix 12, which interacts with coactivators via LXXLL motifs.
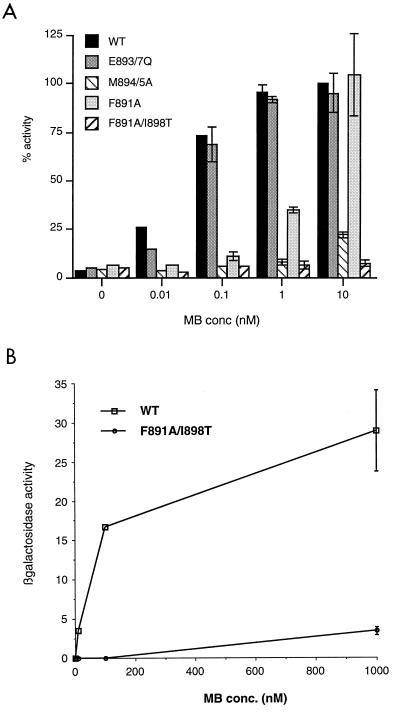
Point mutations were introduced into the full-length AR expression vector in the conserved region between residues 893 and 900, called helix 12 by analogy with those steroid receptors for which the structures of the LBDs have been solved. The mutants were cotransfected into COS-1 cells with androgen-responsive reporter vector, and the activity was measured over a range of concentrations of androgen (Fig. 1A). Elements of helix 12 which are conserved in steroid receptors are an invariant glutamic acid (residue 897 in the AR), flanked by two pairs of hydrophobic residues, and a second negatively charged residue (glutamic acid 893 in the AR). Mutation of the hydrophobic pair methionine-894 and methionine-895 abolished activity of the AR. A single substitution (I898T) in the C-terminal hydrophobic pair also abolished the activity of an otherwise active receptor (F891A). In contrast, the negatively charged residues were not essential for activity, as has been previously shown for the ER but not other receptors (5, 15, 19). N terminal of helix 12 in the ER is a tyrosine residue which appears to maintain AF2 in an inactive state in the absence of ligand, since mutation to a nonphosphorylatable alanine residue resulted in a constitutively active receptor (77). The corresponding position in the AR is occupied by a proline adjacent to a conserved phenylalanine residue. Mutation of this phenylalanine to an alanine (F891A) did not have a similar effect but merely decreased receptor activity at lower ligand concentrations.
FIG. 1.
The AR contains AF2, which is essential for function. (A) HeLa cells were plated in 24-well plates and transfected as described in Materials and Methods. Transfected cells were treated with various concentrations (conc) of the synthetic androgen mibolerone (MB) for 18 to 24 h. CAT activity was assayed and corrected for β-galactosidase activity. The activity of wild-type AR (WT) in the presence of 10 nM mibolerone was set at 100% for each experiment, and other values are expressed relative to this. (B) The LBD of the AR contains a ligand-dependent activation function. The LBD fused to a heterologous DBD was expressed in yeast strain W303-1B along with a lacZ reporter vector. Individual transformants were incubated in liquid culture overnight with various concentrations of mibolerone, and cells were pelleted and assayed for β-galactosidase activity and protein content as described in Materials and Methods. The experiment was repeated with five individual transformants for the wild type and three transformants for the mutant construct.
Transient-transfection experiments with mammalian cells and receptors lacking the N-terminal domain or consisting of the LBD fused to a heterologous Gal4 DBD with an appropriate reporter failed to demonstrate any measurable activity of the AR LBD in the presence of ligand and in the absence of any added coactivators (data not shown). However, when the AR LBD (residues 625 to 919) was fused to a heterologous DBD and expressed in yeast in the presence of an appropriate reporter vector, a ligand-dependent activity was observed (Fig. 1B). Thus, a separable, ligand-dependent AF2 function exists in this region. The activity of this function, which is much lower than that of the constitutive AF1 region in yeast (data not shown), was destroyed by the F891A and I898T mutations in helix 12, which were previously shown to abolish the activity of the full-length receptor.
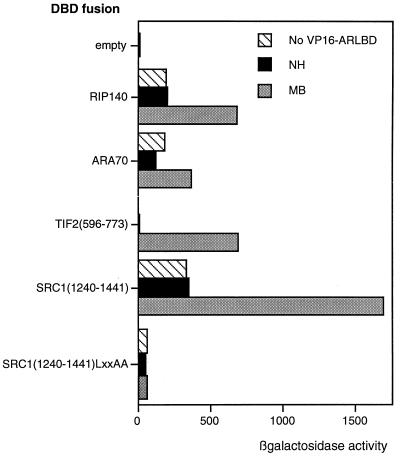
We next investigated the ability of the LBD to interact with a number of putative coactivator proteins. The AR LBD fused to a heterologous VP16 activation domain was able to interact with full-length RIP140 and ARA70 and with fragments of the coactivators SRC1 and TIF2, fused to a heterologous DBD, in a yeast two-hybrid system (Fig. 2). Interactions were ligand dependent, but in some cases activity was observed in the absence of androgen due to the presence of an independent activation function in the coactivator or coactivator fragment (hatched bars). All the putative coactivators or fragments used contain at least one receptor-interacting LXXLL motif. Using the fragment of SRC1a from residues 1240 to 1441, which contains one such motif, we showed that the interaction observed is dependent on the integrity of the leucine motif, as mutation of this region to LXXAA (where A is alanine) destroyed the ligand-dependent interaction. Further, the interaction was also destroyed by the same point mutations (F891A and I898T) in helix 12 of the AR LBD as were previously shown to destroy the activity of the full-length receptor (data not shown).
FIG. 2.
The AR interacts with coactivator proteins via the AF2 core. A two-hybrid assay was performed with yeast and coactivator fragments or full-length protein, as indicated, as bait. The activity of the lacZ reporter in the presence of the VP16 activation domain alone (hatched bars) or fused to the AR LBD without (black bars) or with (gray bars) androgen was measured, and values represent intrinsic activities of the coactivator and the ligand-dependent interaction with AR. NH, no hormone; MB, mibolerone.
AR activity is enhanced by p160 coactivators and p300.
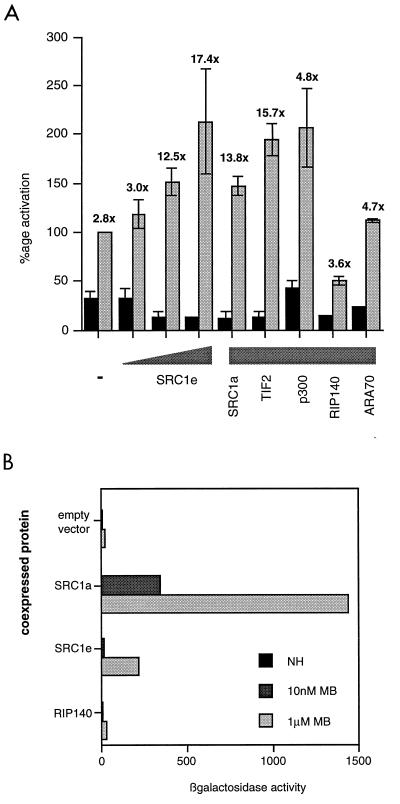
We characterized the potentiation of the ligand-dependent activity of the AR by coactivators in transient-transfection experiments. The activity of full-length AR on an androgen-responsive reporter (consisting of two consensus androgen-responsive elements in front of the thymidine kinase promoter driving the CAT gene) was enhanced by the coexpression of full-length coactivator proteins (Fig. 3A). Potentiation of AR activity was modest compared with the effect of coactivators on ER activity. However, whereas SRC1 increased the ligand-independent as well as the ligand-dependent activity of the ER, it had the effect of decreasing the ligand-independent AR activity and thus increasing the fold activation to a far greater extent. Maximum enhancement was seen with the SRC1e isoform, which enhanced the ligand-dependent fold activation in a concentration-dependent manner from 3-fold to over 17-fold. The SRC1a isoform and TIF2 also increased androgen-dependent transcription of the reporter, as did the general coactivator p300. However, ARA70, which has been described as an AR-specific coactivator, did not potentiate androgen-dependent transcription in this experimental system. Likewise, coexpression of RIP140 did not increase androgen-dependent transcription and appeared to cause a twofold decrease in CAT activity in the presence of ligand. This lack of potentiation by RIP140 and ARA70 did not reflect the inability of the AR to interact with these proteins, as a yeast two-hybrid assay showed the ligand-dependent interaction of the AR LBD with both these full-length proteins (Fig. 2). It was also possible to demonstrate potentiation of AF2 activity by SRC1 in the yeast system, where activity of the AR LBD was enhanced by coexpression of full-length SRC1 in the presence of ligand (Fig. 3B). In contrast to the situation in mammalian cells, where the 1e isoform shows greater potentiation of AR activity, in yeast cells the 1a isoform is the more potent coactivator. This may be due in part to the fact that, at least in yeast, the C-terminal LXXLL motif (motif 4), which is present only in the 1a isoform, shows far stronger affinity for the AR than any of the other three motifs (49a). Further, we observed that yeast cultures expressing full-length SRC1e exhibited a reduced growth rate; thus, it is possible that AR AF2 potentiation by SRC1e is underestimated in these experiments. In agreement with the result with the full-length receptor, RIP140 did not potentiate AF2 activity.
FIG. 3.
Coactivators potentiate the activities of the full-length AR and AF2 alone. (A) COS-1 cells were transfected as described in Materials and Methods. The coactivator was added at 200 ng per well, or various concentrations (20, 50, and 200 ng) of SRC1e were added. After transfection, cells were incubated with or without 10 nM mibolerone for 18 to 24 h before being harvested and assayed for CAT and β-galactosidase activities. Black bars show activity in the absence and gray bars show activity in the presence of hormone. The activity of AR in the absence of a coactivator and in the presence of mibolerone was set at 100%, and other values are expressed relative to this. The values above each of the gray bars show the fold induction of hormone-dependent activity relative to hormone-independent activity for each condition. (B) AR AF2 activity is potentiated by full-length SRC1 in yeast. Yeast (W3031B) was cotransfected with the AR LBD fused to a heterologous DBD, a lacZ reporter, and full-length SRC1a, SRC1e, or RIP140 and incubated overnight in the presence and absence of 100 nM mibolerone (MB). β-Galactosidase activity was measured and normalized for protein content. The experiment was repeated on several individual transformants. Results of a representative example are shown. NH, no hormone.
Coactivation of AR by SRC1 does not require leucine motifs.
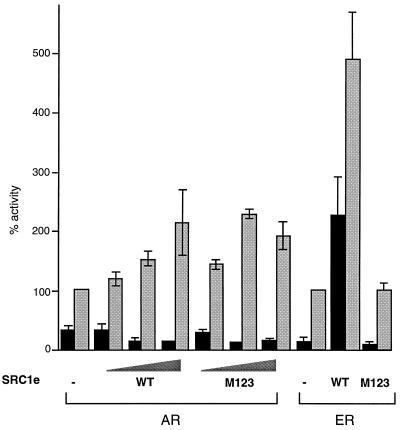
In an attempt to determine which of the three LXXLL motifs present in SRC1e was most important for its effect on AR activity, we cotransfected AR and an AR-responsive reporter vector with SRC1e containing mutations (LXXAA) in one, two, or all three of the motifs. Mutation of any two of these motifs impairs, while mutation of all three abolishes, the ability of SRC1e to potentiate ER activity (37) (Fig. 4). In contrast, mutating any one, two, or all three of the motifs has no effect on the ability of SRC1e to potentiate AR activity (data not shown and Fig. 4). This result suggests that, while the LXXLL motifs are capable of interacting with the AR LBD as shown in yeast two-hybrid assays, an additional site of interaction exists between SRC1e and the AR which is able to compensate for the lack of interaction via LXXLL motifs with the M123 mutant.
FIG. 4.
Leucine motifs are not required for potentiation of AR activity by SRC1e. COS-1 cells were transfected as described previously or with the ER expression vector pSG5-MOR and the estrogen-responsive reporter as previously described. Concentrations of coactivators used were 50, 100, and 200 ng for AR and 50 and 200 ng for ER. Activity was measured in the absence (black bars) or presence (gray bars) of 10 nM ligand. WT, wild type.
SRC1 interacts with the N terminus of the AR via a glutamine-rich domain.
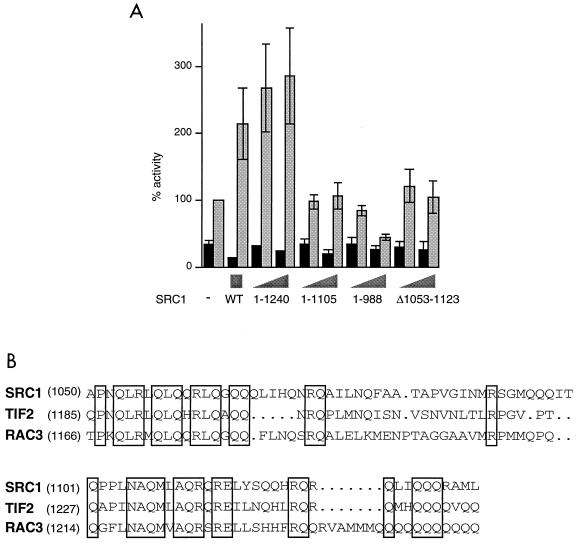
Our previous results suggested that an interaction site exists between the AR and SRC1, in addition to that between AF2 and the LXXLL motifs. To begin to map this site, we used progressive C-terminal SRC1 deletions (Fig. 5A). Residues 1240 to 1399 were found to be dispensable for SRC1 action, while deletion to residue 988 resulted in a loss of coactivation activity and a dominant-negative effect, implicating residues 989 to 1240 in interaction with the AR. SRC1 (1–1107) was also inactive on AR, implying that the C-terminal half of this region is more important for SRC1 function. The region 989 to 1240 is glutamine rich and shows a high degree of conservation between the p160 proteins (Fig. 5B). We made an internal deletion in full-length SRC1e of the most highly conserved residues, from 1053 to 1123. In transfection assays this mutant was almost inactive (Fig. 5A), indicating that this region may be necessary for interaction with the AR or potentiation of its activity.
FIG. 5.
Potentiation of the AR by SRC1 requires the glutamine-rich region, residues 1053 to 1123. (A) COS-1 cells were transfected with 50 or 200 ng of each deletion mutant of SRC1. CAT activity was measured in the absence (black bars) or presence (gray bars) of 10 nM mibolerone. (B) Alignment of the p160 coactivator proteins in the glutamine-rich region. Residues conserved across all three proteins are boxed.
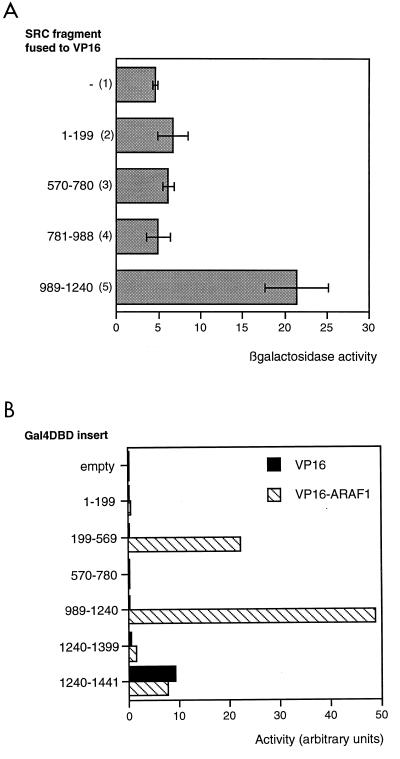
We tested the hypothesis that residues 989 to 1240 were able to interact with the AR using the yeast two-hybrid system. Full-length AR was fused to the lexA DBD, while fragments of SRC1 were fused to the Gal4 activation domain. In the absence of ligand, it was possible to detect activation of the lexA reporter gene due to ligand-independent interaction between AR and the fragment of SRC1 from residues 989 to 1240 (Fig. 6A, lane 5). In the presence of ligand, activation of the full-length AR masked any ligand-dependent interaction between the AR and SRC1 fragments (data not shown). A second two-hybrid assay was performed to determine whether AF1 was the target of SRC1(989–1240) in mammalian cells. SRC1 fragments were fused to a Gal4 DBD, and activation of a Gal4-responsive reporter was determined in the presence of an AF1-VP16 activation domain fusion (Fig. 6B). Interaction was seen between AF1 and two SRC1 fragments, one containing residues 989 to 1240 and one containing residues 199 to 569.
FIG. 6.
AR AF1 interacts with the glutamine-rich region of SRC1. (A) Interaction of full-length AR with fragments of SRC1 in the absence of ligand in yeast. Full-length AR fused to the lexA DBD was coexpressed in yeast (strain L40) with the VP16 activation domain alone (lane 1) or fused to fragments of SRC1 (lanes 2 to 5) in the presence of a lexA-responsive lacZ reporter. Yeast were incubated in liquid culture overnight, and β-galactosidase activity was assayed and corrected for protein content. (B) Interaction of AF1 with fragments of SRC1 in COS cells. SRC1 fragments fused to the Gal4 DBD were coexpressed in COS cells with a Gal4-responsive reporter and VP16 activation domain either alone (black bars) or fused to AR AF1 (hatched bars). The experiment was repeated several times, and results of a representative example are shown.
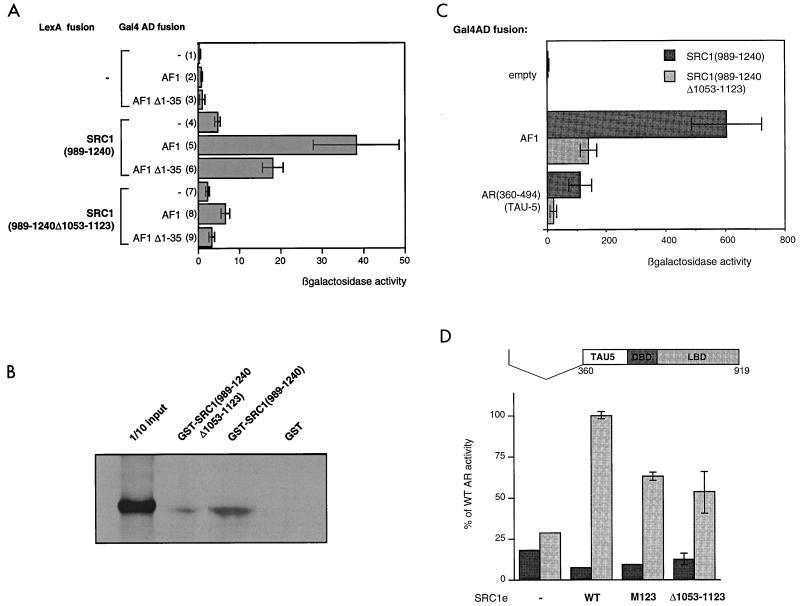
We began to characterize the interaction between AF1 and SRC1(989–1240) further. A fusion between SRC1(989–1240) and the lexA DBD showed no intrinsic activation function (Fig. 7A, lane 4). However, when AF1 fused to the Gal4 activation domain was also expressed, the reporter gene was activated, confirming interaction between AF1 and SRC1(989–1240) (Fig. 7A, lane 5). The same internal deletion, residues 1053 to 1123, which abrogated coactivation of AR by SRC1e, effectively abolished this interaction (lane 8). Thus, the glutamine-rich region between 1053 and 1123 is necessary for interaction with AR AF1.
FIG. 7.
Interaction of AF1 with SRC1 requires residues 1053 to 1123 of SRC1 and involves residues 360 to 494 of AF1 (TAU5). (A) Interaction of SRC1 residues 989 to 1240 with the AF1 region of AR requires the glutamine-rich region of SRC1 but not the N-terminal 35 amino acids of AR. The SRC1 fragment, with or without residues 1053 to 1123, was fused to the lexA DBD and expressed in yeast (L40) with the Gal4 activation domain alone (lanes 1, 4, and 7) or fused to AF1 (lanes 2, 5, and 6) or an AF1 deletion mutant (Δ1–35) (lanes 3, 6, and 9) (with the vector pGAF424). Yeast cells were grown and assayed as described in the text. (B) AR interacts in vitro with the glutamine-rich region of SRC1. GST fusion proteins, coupled to Sepharose beads, were incubated with in vitro-translated [35S]methionine-labeled full-length AR. After being washed extensively, samples were boiled and run on sodium dodecyl sulfate–8% polyacrylamide gel, which was fixed and dried, and the bound labeled protein was visualized by autoradiography. (C) TAU5 is able to interact with SRC1, dependent on the region from residues 1053 to 1123, but is not sufficient for maximal interaction. SRC1 fragments were used as bait, and the interaction with AF1 or with a fragment of AF1 comprising the TAU5 region fused to the Gal4 activation domain (in the pACT2 vector) was measured. (D) The TAU5 region of AF1 is sufficient for SRC1 potentiation of the AR. The Δ1–360 mutant (represented at the top of the figure) was cotransfected into COS cells with SRC1 mutants as shown, and activities were measured in the absence (black bars) and presence (gray bars) of hormone as described in the text. WT, wild type.
Using the GST pull-down system, we investigated the ability of the two proteins to interact in vitro. Full-length in vitro-translated AR bound to SRC1(989–1240), fused to GST, and immobilized on beads (Fig. 7B). This interaction was reduced from 3 to 0.6% of the input by deletion of residues 1052 to 1123 from the GST construct and was entirely ligand independent, as identical results were obtained in the presence of ligand (data not shown). In the converse experiment, AF1 alone fused to GST was able to bind in vitro-translated full-length SRC1e but not SRC1eΔ1053–12123 (data not shown). Thus, the interaction between AF1 and SRC1 appears to be direct and dependent on the glutamine-rich region.
An interaction between the two termini of the AR (the N-terminal domain, or AF1, and the LBD) has been shown to be necessary for full activation of the receptor. Two regions of AF1 have been implicated in this interaction, the N terminus (residues 14 to 36) and residues 370 to 494, which constitute TAU5. The AF1-LBD interaction may be direct or may be mediated via a bridging protein, for which SRC1 is a candidate. If SRC1 were acting as a bridging factor, it would be expected that regions of AF1 that are necessary for the interaction with AF2 are also necessary for interaction with SRC1. However, deletion of the first 35 residues in the yeast two-hybrid assay effected only a twofold decrease in the interaction between SRC1 and AF1 (Fig. 7A, lane 6). We investigated the second possible interaction site, residues 370 to 494. Yeast two-hybrid experiments showed that this region in isolation is able to interact with SRC1(989–1240), albeit less strongly than intact AF1 (Fig. 7C). Further, deletion of residues 1053 to 1123 greatly reduced this interaction. Thus, it is conceivable that the same regions involved in the AF1-AF2 interaction are involved in binding to SRC1. Note that it is not possible to directly compare numbers obtained for the interaction between AF1 and SRC1(989–1240) as seen in Fig. 7A and C; this is due to the use of different vectors for the AF1 fusion constructs in the two experiments.
An N-terminal deletion mutant containing the TAU5 region of AF1 (consisting of residues 360 to 919) was active in the COS cell reporter assay, showing activity up to 25% of that of full-length AR (Fig. 7D). Moreover, this was greatly enhanced (fourfold) by coexpression of SRC1e, supporting the theory that this region of AF1 (residues 360 to 494) mediates interaction with SRC1e. Surprisingly, potentiation of AR(360–919) was not completely abolished by deletion of the glutamine-rich region. This residual activity may be explained if another region of SRC1 mediates the interaction with AF1, such as residues 199 to 569, which showed interaction in the mammalian two-hybrid assay (Fig. 6B). Alternatively, in this context interactions between the LXXLL motifs and the LBD may be sufficient to promote some degree of SRC1 activity. The latter hypothesis is supported by the observation that potentiation by the M123 mutant is less than that of wild-type SRC1 on this AR mutant (Fig. 7D).
Coactivation of AR by SRC1e requires AD1 but not AD2.
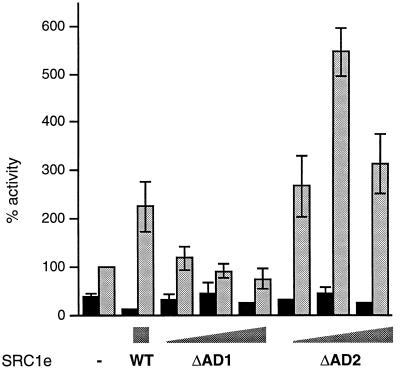
We investigated the relative importance of each of the two activation domains present in SRC1 (AD1 and AD2) for its effect on AR activity. AD1, responsible for recruitment of the general coactivator CBP/p300, maps to residues 900 to 950, while AD2 maps to the C terminus (residues 1241 to 1385). A deletion mutant lacking residues 900 to 950 was inactive, whereas the truncated SRC1(1–1240) potentiated AR activity to a greater extent than full-length SRC1e (Fig. 8). Thus, we conclude that AD1 but not AD2 is essential for potentiation of AR activity by SRC1.
FIG. 8.
Potentiation of AR activity by SRC1 requires AD1 but not AD2. COS-1 cells were transfected as described for Fig. 1 with either 200 ng of wild-type SRC1e (WT) or 50, 100, or 200 ng of the deletion mutants, in the absence (black bars) or presence (gray bars) of 10 nM mibolerone.
DISCUSSION
While it is widely accepted that the C-terminal LBDs of many nuclear receptors contain a ligand-dependent activation function, AF2 (5, 19, 65, 76), the existence of such a function in the AR has been less well established. The N terminus of the AR can activate a reporter gene in the absence of the LBD to the same extent as the full-length receptor in the presence of ligand (35, 56, 82), implying that AF1 contributes most, if not all, of the activity of the AR in many cell lines and on many promoters. Further, the C terminus alone fused to the homologous or a heterologous DBD shows little or no activity in the presence of ligand in mammalian cell lines. Residues encoding part of AF2 as determined in other receptors, which lie in a predicted amphipathic alpha-helix (helix 12) in the LBD, show a high degree of conservation across the nuclear receptor superfamily, not least in the AR. Reasoning that sequence conservation should imply functional conservation, we mutated conserved residues and examined their effects on the activity of the receptor. Our results were comparable to those obtained previously with the ER and glucocorticoid receptor (15), in which the integrity of the hydrophobic pairs is essential for the activity of the full-length receptor although the charged glutamic acids are not. Thus, although AF1 is capable of stimulating reporter genes to the maximum extent, the integrity of the AF2 core region is vital for the activity of the full-length receptor. Since the LBD alone exhibits ligand-dependent activation in S. cerevisiae (49), which we have shown is dependent on residues in helix 12, it is conceivable that a separable AF2 function exists within AR, the use of which depends on cell type and target promoter.
Initially we assumed that the interaction of coactivators with the AR was via the LBD and were indeed able to see ligand-dependent interaction of the AR LBD with several putative coactivators or coactivator fragments in a yeast two-hybrid assay. This interaction was dependent on the integrity of the LXXLL motif in the coactivator and of helix 12 in the receptor. Thus, we have demonstrated that the AR AF2 core interacts with coactivators via LXXLL motifs, as has been shown for several other receptors.
It was surprising then that an SRC1 mutant containing no functional LXXLL motifs was able to potentiate transcriptional activity of the AR, but not the ER, to the same extent as wild-type SRC1e. This result suggested an alternative method of recruitment. The region we identified as the primary site of interaction with the AR, residues 989 to 1240, contains a glutamine-rich span conserved in all p160 family members. The required residues for the interaction with AR lie between residues 1053 and 1123, a well-conserved, highly glutamine-rich region, with the C-terminal half from 1105 to 1123 perhaps being the more important. This hormone-independent interaction is with AF1 of the AR. A corresponding region in GRIP1, the mouse homologue of TIF2, has been shown by Webb et al. to promote weak interaction between AF1 of the ER and GRIP1 (75). However, this is not sufficient for SRC1 recruitment since we have demonstrated that the leucine motifs are essential for potentiating the transcriptional activity of the ER. Further evidence that this interaction, although sufficient to mediate SRC1 potentiation of AR activity, is not as important for ER activity is the fact that the mutant with a deletion of residues 1053 to 1123, which showed no activity on AR, stimulated ER activity to an even greater extent than wild-type SRC1e (our unpublished results).
It has been shown that an interaction between the N and C termini of the AR that occurs after ligand binding is vital for full activity of the AR. This interaction is implicated in receptor stabilization, reducing ligand dissociation and increasing DNA-binding affinity (7, 18, 31, 39, 40, 57, 81). Residues 14 to 36 and so-called TAU5 (residues 370 to 494) in the N-terminal domain have been implicated in the interaction (7, 40), and it has been suggested that coactivators such as SRC1 and CBP could promote this interaction, as has been shown for the ER (31, 47). Our data supports this suggestion, with SRC1 bridging AF1 and AF2 by means of interactions between the glutamine-rich region and TAU5 and between the LXXLL motifs and AF2. Clearly, the interaction of SRC1 with AF2 is also important, as evidenced by the inactivating effects of helix 12 mutations and the fact that the M123 mutant in some circumstances shows reduced potentiation of activity. However, our results suggest that the interaction with AF1 is the crucial step, since mutations in the LXXLL motifs which prevent binding to AF2 do not abolish the potentiation of wild-type AR activity by SRC1 while deletion of the AF1 interaction site does.
SRC1 contains two activation domains (37, 72), the strongest of which, AD1, has been demonstrated to recruit the general coactivator CBP/p300 (69, 72). It is postulated that SRC1 potentiates nuclear receptor activity by recruiting these coactivators to the promoter, where it may facilitate transcription by histone acetylation and may also interact with the basal transcription machinery. It is also interesting that CBP/p300 is another candidate for acting as a bridge between AF1 and AF2: this molecule promoted the interaction in yeast (31), and there is evidence that it can interact with the AR via both the N terminus and the LBD (22). The targets of AD2, which is located at the C terminus (resides 1241 to 1385), are unknown. We observed that removal of AD2 resulted in an increase, rather than a decrease, in potentiation of AR activity. A possible explanation for this lies with a weak interaction we have observed in yeast between the glutamine-rich and the C-terminal regions of SRC1 (data not shown). If this interaction occurs in vivo, then competition between AF1 and the C terminus of SRC1 for binding to the glutamine-rich region might inhibit the potentiation of AR activity by full-length SRC1 and removal of the C terminus would increase SRC1 activity. This phenomenon is not as marked for the ER, in which AF2 is more important.
Potentiation of AR activity by the p160 proteins TIF2 and SRC1, and by the general coactivator p300, was modest compared to their effects on other receptors. However, SRC1 also stimulates the ligand-independent activity of the ER such that fold activity in the presence of ligand is actually reduced by coexpression of SRC1. In contrast, androgen-independent activity is reduced by SRC1 in a reproducible, concentration-dependant manner, resulting in fold activation by androgen increasing (from 3-fold to 17.4-fold) in the presence of SRC1e. Thus, p160 coactivators appear to have the effect of increasing the androgen sensitivities of promoters. This observation is paradoxical, as we have demonstrated recruitment of SRC1 to AF1 of AR in a ligand-independent fashion and have also observed potentiation of the activity of isolated AF1 in the presence of SRC1 (data not shown). The inability of SRC1 to potentiate the transcriptional activity of the AR in the absence of ligand is consistent with the observation that the unliganded C terminus inhibits AF1 activity (82). We postulate either that this region prevents SRC1 binding to AF1 until the conformational change triggered by ligand binding has taken place or that inhibition by the unliganded C terminus cannot be overcome by the binding of SRC1 to AF1.
The putative AR-specific coactivator ARA70 had no effect in our hands, although previous reports suggest that it has modest coactivator properties in COS cells and the prostate cancer cell line DU145 (1, 79, 80). The difference may reflect the use of different promoters. The cofactor RIP140, which inhibits ER (12) and peroxisome proliferator-activated receptor (71) activities when it is overexpressed, similarly inhibited AR activity. This result is consistent with the hypothesis that RIP140 functions either to deactivate receptors (12) or as a competitive inhibitor, possibly by competing with activators such as SRC1 for binding to the liganded receptor (71).
In conclusion, we have shown that, like other nuclear receptors, the AR contains a ligand-dependent AF2 function within its LBD that interacts with coactivators via LXXLL motifs. However, unlike many other receptors, this interaction is not essential for coactivation of this receptor by p160 coactivator proteins. A second interaction occurs, between the glutamine-rich region of the coactivator and the large N-terminal region of AR that contains AF1. This interaction involves TAU5, implicated in the interaction between the N and C termini of AR, implying that one role of SRC1 may be to promote this interaction and thus allow maximum activity of the receptor. While the mechanisms of recruitment may differ, CBP/p300 is essential for the function of both the AR and the ER. Thus, transcriptional activation by the AR retains steps common to other nuclear receptors but has important differences, which may reflect the differences in relative levels of importance of AF1 and AF2 in this receptor. It is becoming increasingly evident that coactivators are recruited to AF1 of steroid receptors (41, 52, 70, 75), and this may be more important for AR activity in which AF1 plays a crucial role.
ACKNOWLEDGMENTS
We are grateful to A. Brinkmann, P. Chambon, G. Folkers, and J. Trapman for gifts of plasmids. We thank I. Goldsmith and staff for oligonucleotides; G. Clark and staff for sequencing; and Eric Kalkhoven, Ho Yi Mak, Christian Landles, Janet Valentine, and members of the Molecular Endocrinology Laboratory for plasmids, helpful discussion, and critical reading of the manuscript.
This work was supported by the Imperial Cancer Research Fund. F.C. was supported by the Belgian FWO (Fonds voor Wetenschappelijk Onderzoek), and D.M.H. was supported by the European Community TMR programme.
REFERENCES
- 1.Alen P, Claessens F, Schoenmakers E, Swinnen J V, Verhoeven G, Rombauts W, Peeters B. Interaction of the putative androgen receptor-specific coactivator ARA70/ELE1a with multiple steroid receptors and identification of an internally deleted ELE1β isoform. Mol Endocrinol. 1999;13:117–128. doi: 10.1210/mend.13.1.0214. [DOI] [PubMed] [Google Scholar]
- 2.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X-Y, Sauter G, Kallioniemi O-P, Trent J M, Meltzer P S. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 3.Baniahmad A, Ha I, Reinberg D, Tsai S, Tsai M-J, O’Malley B W. Interaction of human thyroid hormone receptor β with transcription factor TFIIB may mediate target gene derepression and activation by thyroid hormone. Proc Natl Acad Sci USA. 1993;90:8832–8836. doi: 10.1073/pnas.90.19.8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannister A J, Kouzarides T. The CBP co-activator is a histone acetyltransferase. Nature. 1996;1996:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 5.Barettino D, Vivanco Ruiz M, Stunnenburg H. Characterization of the ligand-dependent transactivation domain of thyroid hormone receptor. EMBO J. 1994;13:3039–3049. doi: 10.1002/j.1460-2075.1994.tb06603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beato M, Herrlich P, Schutz G. Steroid hormone receptors—many actors in search of a plot. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 7.Berrevoets C A, Doesburg P, Sketetee K, Trapman J, Brinkmann A O. Functional interactions of the AF-2 domain core region of the human androgen receptor with the amino-terminal domain and with the transcriptional coactivator TIF-2 (transcriptional intermediary factor-2) Mol Endocrinol. 1998;12:1172–1183. doi: 10.1210/mend.12.8.0153. [DOI] [PubMed] [Google Scholar]
- 8.Brinkmann A O, Faber P W, van Rooij H C J, Kuiper G G J M, Ris C, Klaasen P, van der Korput J A G M, Voorhorst M M, van Laar J H, Mulder E, Trapman J. The human androgen receptor: domain structure, genomic organisation and regulation of expression. J Steroid Biochem. 1989;34:307–310. doi: 10.1016/0022-4731(89)90098-8. [DOI] [PubMed] [Google Scholar]
- 9.Brou C, Chaudhary S, Davidson I, Lutz Y, Wu J, Egly J-M, Tora L, Chambon P. Distinct TFIID complexes mediate the effect of different transcriptional activators. EMBO J. 1993;12:489–499. doi: 10.1002/j.1460-2075.1993.tb05681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brou C, Wu J, Ali S, Scheer E, Lang C, Davidson I, Chambon P, Tora L. Different TBP-associated factors are required for mediating the stimulation of transcription in vitro by the acidic transactivator GAL-VP16 and the two nonacidic activation functions of the estrogen receptor. Nucleic Acids Res. 1993;21:5–12. doi: 10.1093/nar/21.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchert M, Schneider S, Adams M T, Hefti H, Moelling K, Hovens C M. Useful vectors for the two-hybrid system in mammalian cells. BioTechniques. 1997;23:396–402. doi: 10.2144/97233bm10. [DOI] [PubMed] [Google Scholar]
- 12.Cavailles V, Dauvois S, L’Horset F, Lopez G, Hoare S, Kushner P J, Parker M G. Nuclear factor RIP140 modulates transcriptional activation by the estrogen receptor. EMBO J. 1995;14:3741–3751. doi: 10.1002/j.1460-2075.1995.tb00044.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with p/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 15.Danielian P S, White R, Lees J A, Parker M G. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darimont B D, Wagner R L, Apriletti J W, Stallcup M R, Kushner P J, Baxter J D, Fletterick R J, Yamamoto K R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dodou E, Treisman R. The Saccharomyces cerevisiae MADS-box transcription factor Rlm1 is a target for the Mpk1 mitogen-activated protein kinase pathway. Mol Cell Biol. 1997;17:1848–1859. doi: 10.1128/mcb.17.4.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doesburg P, Kuil C W, Berrevoets C A, Steketee K, Faber P W, Mulder E, Brinkmann A O, Trapman J. Functional in vivo interaction between the amino-terminal, transactivating domain and the ligand binding domain of the androgen receptor. Biochemistry. 1997;36:1052–1064. doi: 10.1021/bi961775g. [DOI] [PubMed] [Google Scholar]
- 19.Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;13:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Folkers G E, van der Saag P T. Adenovirus E1A functions as a cofactor for retinoic acid receptor β (RARβ) through direct interaction with RARβ Mol. Cell Biol. 1995;15:5868–5878. doi: 10.1128/mcb.15.11.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freedman L P. Increasing the complexity of coactivation in nuclear receptor signaling. Cell. 1999;97:5–8. doi: 10.1016/s0092-8674(00)80708-4. [DOI] [PubMed] [Google Scholar]
- 22.Fronsdal K, Engedal N, Slagsvold T, Saatcioglu F. CREB binding protein is a coactivator for the androgen receptor and mediates cross-talk with AP-1. J Biol Chem. 1998;273:31853–31859. doi: 10.1074/jbc.273.48.31853. [DOI] [PubMed] [Google Scholar]
- 23.Glass C, Rose D W, Rosenfeld M G. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- 24.Glynne R, Kerr L A, Mockridge I, Beck S, Kelly A, Trowsdale J. The major histocompatibility complex-encoded proteosome component LMP7: alternative first exons and post-translational processing. Eur J Immunol. 1993;23:860–866. doi: 10.1002/eji.1830230414. [DOI] [PubMed] [Google Scholar]
- 24a.Heery, D. M. Unpublished data.
- 25.Heery D M, Kalkhoven E, Hoare S, Parker M G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 26.Heery D M, Zacharewski T, Pierrat B, Gronemeyer H, Chambon P, Losson R. Efficient transactivation by retinoic acid receptors in yeast requires retinoid X receptors. Proc Natl Acad Sci USA. 1993;90:4281–4285. doi: 10.1073/pnas.90.9.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henttu P M, Kalkhoven E, Parker M G. AF-2 activity and recruitment of steroid receptor coactivator-1 to the estrogen receptor depend on a lysine residue conserved in nuclear receptors. Mol Cell Biol. 1997;17:1832–1839. doi: 10.1128/mcb.17.4.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higuchi R. Recombinant PCR. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR protocols: a guide to methods and applications. London, United Kingdom: Academic Press Ltd.; 1990. pp. 177–183. [Google Scholar]
- 29.Hong H, Kohli K, Trivedi A, Johnson D L, Stallcup M R. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci USA. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ikonen T, Palvimo J J, Janne O A. Heterodimerization is mainly responsible for the dominant negative activity of amino-terminally truncated rat androgen receptor forms. FEBS Lett. 1998;430:393–396. doi: 10.1016/s0014-5793(98)00701-7. [DOI] [PubMed] [Google Scholar]
- 31.Ikonen T, Palvimo J J, Janne O A. Interaction between the amino- and carboxyl-terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem. 1997;272:29821–29828. doi: 10.1074/jbc.272.47.29821. [DOI] [PubMed] [Google Scholar]
- 32.Ing N, Beekman J M, Tsai S Y, Tsai M-J, O’Malley B. Members of the steroid hormone superfamily interact with TFIIB (S300-II) J Biol Chem. 1992;267:17617–17623. [PubMed] [Google Scholar]
- 33.Ito M, Yuan C-X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z-Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 34.Jenster G, van der Korput H, Trapman J, Brinkmann A O. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J Biol Chem. 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- 35.Jenster G, van der Korput H A G M, van Vroonhoven C, van der Kwast T H, Trapman J, Brinkmann A O. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–1404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- 36.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 37.Kalkhoven E, Valentine J E, Heery D M, Parker M G. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J. 1998;17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karvonen U, Kallio P J, Janne O A, Palvimo J J. Interaction of androgen receptors with androgen response element in intact cells. Roles of amino- and carboxyl-terminal regions and the ligand. J Biol Chem. 1997;272:15973–15979. doi: 10.1074/jbc.272.25.15973. [DOI] [PubMed] [Google Scholar]
- 39.Langley E, Kemppainen J A, Wilson E M. Intermolecular NH2-/carboxy-terminal interactions in androgen receptor dimerization revealed by mutations that cause androgen insensitivity. J Biol Chem. 1998;273:92–101. doi: 10.1074/jbc.273.1.92. [DOI] [PubMed] [Google Scholar]
- 40.Langley E, Zhou Z-X, Wilson E M. Evidence for anti-parallel orientation of the ligand-activated human androgen receptor dimer. J Biol Chem. 1995;270:29983–29990. doi: 10.1074/jbc.270.50.29983. [DOI] [PubMed] [Google Scholar]
- 41.Lanz R B, McKenna N J, Onate S, Albrecht U, Wong J, Tsai S Y, Tsai M-J, O’Malley B W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- 42.Le Douarin B, Pierrat B, vom Baur E, Chambon P, Losson R. A new version of the two-hybrid assay for detection of protein-protein interactions. Nucleic Acids Res. 1995;23:876–878. doi: 10.1093/nar/23.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Gomes P J, Chen J D. RAC3, a steroid/nuclear receptor-associated protein that is related to SRC-1 and TIF-2. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mak H Y, Hoare S, Henttu P M, Parker M G. Molecular determinants of the estrogen receptor-coactivator interface. Mol Cell Biol. 1999;19:3895–3903. doi: 10.1128/mcb.19.5.3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangelsdorf D J, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans R M. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McEwan I J, Gustafsson J. Interaction of the human androgen receptor transactivation function with the general transcription factor TFIIF. Proc Natl Acad Sci USA. 1997;94:8485–8490. doi: 10.1073/pnas.94.16.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McInerney E, Tsai M-J, O’Malley B, Katzenellenbogen B S. Analysis of estrogen receptor transcriptional enhancement by a nuclear hormone coactivator. Proc Natl Acad Sci USA. 1998;93:10069–10073. doi: 10.1073/pnas.93.19.10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Metzger D, Losson R, Bornet J-M, Lemoine Y, Chambon P. Promoter specificity of the two transcriptional activation functions of the human oestrogen receptor in yeast. Nucleic Acids Res. 1992;20:2813–2817. doi: 10.1093/nar/20.11.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moilanen A, Rouleau N, Ikonen T, Palvimo J J, Janne O A. The presence of a transcription activation function in the hormone-binding domain of androgen receptor is revealed by studies in yeast cells. FEBS Lett. 1998;412:355–358. doi: 10.1016/s0014-5793(97)00791-6. [DOI] [PubMed] [Google Scholar]
- 49a.Needham, M., et al. Unpublished observations.
- 50.Nolte R T, Wisely G B, Westin S, Cobb J E, Lambert M H, Kurokawa R, Rosenfeld M G, Willson T M, Glass C K, Milburn M V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 51.Ogryzko V V, Schiltz R L, Russanova V, Howard B H, Nakaiani Y. The transcriptional co-activator p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–960. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- 52.Onate S A, Boonyaratanakornkit V, Spencer T E, Tsai S Y, Tsai M-J, Edwards D P, O’Malley B W. The steroid receptor coactivator-1 contains multiple receptor interacting and activation domains that cooperatively enhance the activation function 1 (AF1) and AF2 domains of steroid receptors. J Biol Chem. 1998;273:12101–12108. doi: 10.1074/jbc.273.20.12101. [DOI] [PubMed] [Google Scholar]
- 53.Oñate S A, Tsai S Y, Tsai M-J, O’Malley B W. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 54.Parker M G, White R. Nuclear receptors spring into action. Nat Struct Biol. 1996;3:113–115. doi: 10.1038/nsb0296-113. [DOI] [PubMed] [Google Scholar]
- 55.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Näär A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 56.Rundlett S E, Wu X-P, Miesfeld R L. Functional characterizations of the androgen receptor confirm that the molecular basis of androgen action is transcriptional regulation. Mol Endocrinol. 1990;4:708–714. doi: 10.1210/mend-4-5-708. [DOI] [PubMed] [Google Scholar]
- 57.Scheller A, Hughes E, Golden K L, Robins D. Multiple receptor domains interact to permit, or restrict, androgen-specific gene activation. J Biol Chem. 1998;273:24216–24222. doi: 10.1074/jbc.273.37.24216. [DOI] [PubMed] [Google Scholar]
- 58.Schüle R, Muller M, Kaltschmidt C, Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science. 1988;242:1418–1420. doi: 10.1126/science.3201230. [DOI] [PubMed] [Google Scholar]
- 59.Schulman I G, Chakravarti D, Juguilon H, Romo A, Evans R M. Interactions between the retinoid X receptor and a conserved region of the tata-binding protein mediate hormone-dependent transactivation. Proc Natl Acad Sci USA. 1995;92:8288–8292. doi: 10.1073/pnas.92.18.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shikama N, Lyon J, La Thangue N B. The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- 61.Simental J A, Sar M, Lane M V, French F S, Wilson E M. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J Biol Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- 62.Sleigh M J. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eucaryotic cells. Anal Biochem. 1986;156:251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- 63.Spencer T E, Jenster G, Burcin M M, Allis C D, Zhou J, Mizzen C A, McKenna N J, Onate S A, Tsai S Y, Tsai M J, O’Malley B W. Steroid receptor coactivator-1 is a histone acetyltransferase. Nature. 1997;389:194–198. doi: 10.1038/38304. [DOI] [PubMed] [Google Scholar]
- 64.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. TRAM-1, a novel 160-kDa thyroid hormone receptor activator molecule, exhibits properties distinct from steroid receptor co-activator-1. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 65.Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Distinct classes of transcriptional activating domains function by different mechanisms. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- 66.Tone Y, Collingwood T N, Adams M, Chatterjee V K. Functional analysis of a transactivation domain in the thyroid hormone receptor. J Biol Chem. 1994;269:31157–31161. [PubMed] [Google Scholar]
- 67.Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human estrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:477–497. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- 68.Torchia J, Glass C, Rosenfeld M G. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- 69.Torchia J, Rose D W, Inostroza J, Kmei Y, Westin S, Glass C, Rosenfeld M. The transcriptional coactivator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;382:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 70.Tremblay A, Tremblay G B, Labrie F, Giguere V. Ligand-independent recruitment of SRC-1 to estrogen receptor β through phosphorylation of activation function AF-1. Mol Cell. 1999;3:513–9. doi: 10.1016/s1097-2765(00)80479-7. [DOI] [PubMed] [Google Scholar]
- 71.Treuter E, Albrektsen T, Johansson L, Leers J, Gustafsson J A. A regulatory role for RIP140 in nuclear receptor activation. Mol Endocrinol. 1998;12:864–881. doi: 10.1210/mend.12.6.0123. [DOI] [PubMed] [Google Scholar]
- 72.Voegel J J, Heine M J S, Tini M, Vivat V, Chambon P, Gronemeyer H. The co-activator TIF2 contains three nuclear receptor binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J. 1998;17:507–519. doi: 10.1093/emboj/17.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Voegel J J, Heine M J S, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:101–108. [PMC free article] [PubMed] [Google Scholar]
- 74.Vojtek B A, Hollenberg S M, Cooper J A. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 75.Webb P, Nguyen P, Shinsako J, Anderson C, Feng W, Nguyen M P, Chen D, Huang S-M, Subramanian S, McKinerny E, Katzenellenbogen B S, Stallcup M R, Kushner P J. Estrogen receptor activation function 1 works by binding p160 proteins. Mol Endocrinol. 1998;12:1605–1618. doi: 10.1210/mend.12.10.0185. [DOI] [PubMed] [Google Scholar]
- 76.Webster N J G, Green S, Tasset D, Ponglikitmongkol M, Chambon P. The transcriptional activation function located in the hormone-binding domain of the human oestrogen receptor is not encoded in a single exon. EMBO J. 1988;8:1441–1446. doi: 10.1002/j.1460-2075.1989.tb03526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.White R, Sjöberg M, Kalkhoven E, Parker M G. Ligand-independent activation of the oestrogen receptor by mutation of a conserved tyrosine. EMBO J. 1997;16:1427–1435. doi: 10.1093/emboj/16.6.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wurtz J-M, Bourguet W, Renaud J-P, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand-binding domain of nuclear receptors. Nat Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- 79.Yeh S, Miyamoto H, Shima H, Chang C. From estrogen to androgen receptor: a new pathway for sex hormones in prostate. Proc Natl Acad Sci USA. 1998;95:5527–5532. doi: 10.1073/pnas.95.10.5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yeh S Y, Chang C S. Cloning and characterization of a specific coactivator, ARA(70), for the androgen receptor in human prostate cells. Proc Natl Acad Sci USA. 1996;93:5517–5521. doi: 10.1073/pnas.93.11.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhou Z-X, Lane M V, Kemppainen J A, French F S, Wilson E M. Specificity of ligand-dependent androgen receptor stabilization: receptor domain interactions influence ligand dissociation and receptor stability. Mol Endocrinol. 1995;9:208–218. doi: 10.1210/mend.9.2.7776971. [DOI] [PubMed] [Google Scholar]
- 82.Zhou Z-X, Sar M, Simental J A, Lane M V, Wilson E M. A ligand-dependent bipartite nuclear targeting signal in the human androgen receptor. J Biol Chem. 1994;269:13115–13123. [PubMed] [Google Scholar]