Abstract
Hepatocellular carcinoma (HCC) is still one of the most common malignancies worldwide. The accuracy of biomarkers for predicting the prognosis of HCC and the therapeutic effect is not satisfactory. N6-methyladenosine (m6A) methylation regulators play a crucial role in various tumours. Our research aims further to determine the predictive value of m6A methylation regulators and establish a prognostic model for HCC. In this study, the data of HCC from The Cancer Genome Atlas (TCGA) database was obtained, and the expression level of 15 genes and survival was examined. Then we identified two clusters of HCC with different clinical factors, constructed prognostic markers and analysed gene set enrichment, proteins’ interaction and gene co-expression. Three subgroups by consensus clustering according to the expression of the 13 genes were identified. The risk score generated by five genes divided HCC patients into high-risk and low-risk groups. In addition, we developed a prognostic marker that can identify high-risk HCC. Finally, a novel prognostic nomogram was developed to accurately predict HCC patients’ prognosis. The expression levels of 13 m6A RNA methylation regulators were significantly upregulated in HCC samples. The prognosis of cluster 1 and cluster 3 was worse. Patients in the high-risk group show a poor prognosis. Moreover, the risk score was an independent prognostic factor for HCC patients. In conclusion, we reveal the critical role of m6A RNA methylation modification in HCC and develop a predictive model based on the m6A RNA methylation regulators, which can accurately predict HCC patients’ prognosis and provide meaningful guidance for clinical treatment.
Introduction
Hepatocellular carcinoma (HCC) has become the fourth most common cancer with a second highest mortality rate and a poor survival outcome worldwide (1,2). Most HCC patients are in an advanced stage at initial diagnosis (3–5). Recently, although immune checkpoint inhibitors and targeted therapy have been shown to improve the survival rate of advanced HCC patients, the overall survival (OS) rate is still low (6–10). Therefore, it is still an urgent task to predict the prognosis of HCC with high accuracy and find new biomarkers and therapeutic targets for the diagnosis and treatment of HCC. Currently, the tumour staging system (TNM) is still the most extensively used prognostic indicator for monitoring HCC progression (11,12). Some prognostic models were recently based on the differentially expressed genes between HCC tissues and non-tumour tissues. Unfortunately, these prognostic models’ predictive sensitivity and specificity are not high (13,14).
N6-methyladenosine (m6A) has reversible modification in messenger RNA (mRNA), regulating the processing, translation and degradation of mRNA (15,16). Recent evidence has revealed that the expression of m6A regulators is closely related to pathological processes such as stem cell differentiation, tumorigenesis and metastasis (17–19). Moreover, the dysregulation of m6A genes will affect the expression of oncogenes (20–22). m6A modification is also associated with the proliferation, differentiation, invasion and metastasis of malignancy (23–25).
The m6A-related genes have been widely studied in various tumours (15). The methylation modification of m6A includes three enzymes: ‘writers’ (methyltransferase, WTAP, METTL3, METTL14, KIAA1429, ZC3H13 and RBM15), ‘reader’ (YTH domain-containing RNA binding proteins, YTHDF1, YTHDF22, YTHDF3, YTHDC1, YTHDC2, HNPNPA2B1 and HNPNPC) and ‘eraser’ (demethylase, FTO and ALKBH5) (26,27). However, the relationship between m6A-related regulators and HCC remains unclear.
Therefore, our research aims to evaluate gene expression characteristics, analyse associations between m6A methylation regulators and clinicopathological features in patients with HCC and generate a novel predictive model based on the m6A genes HCC.
Material and Methods
Data collection
The RNA-seq transcriptome data and corresponding clinical data of HCC samples were obtained from the public database, The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). mRNA expression data and clinical parameters including age, gender, tumour grade, tumour stage, TNM stage and vascular invasion were downloaded. A total of 50 non-tumour tissues and 374 tumour tissues were included for subsequent analysis.
Data processing and differential expression analysis of m6A RNA methylation regulators
R software (Version 3.5.3) was performed for evaluating differentially expressed genes between HCC tissues and non-tumour tissues. The differential expression of 15 currently known m6A RNA methylation-related genes (HNRNPC, HNRNPA2B1, YTHDF1, YTHDC1, RBM15, YTHDF2, YTHDC2, FTO, KIAA1429, ALKBH5, METTL14, WTAP, METTL3, YTHDF3 and ZC3H13) in tumour samples and non-tumour samples were analysed. The adjusted P < 0.05 was considered significant in the test. For validation, the research compared expression levels of these 15 genes in HCC tissues and non-tumour tissues in the Gene Expression Profiling Interactive Analysis 2 (GEPIA2) database (http://gepia2.cancer-pku.cn/#index).
Protein–protein interaction network and gene co-expression network analysis
STRING (http://string-db.org; version 11.0) is an online biological database used to analyse the protein–protein interaction among m6A RNA methylation regulators. The Coexpedia database (http://www.coexpedia.org/) was used to analyse the co-expression of m6A RNA methylation-related genes in HCC samples.
Consensus clustering analysis
Consensus clustering is a kind of class discovery technique used to detect unknown possible clusters composed of items with similar intrinsic characteristics. Based on the comprehensive expression of the 15 genes, we identified distinct subgroups of 374 HCC samples with ‘ConsensusClusterPlus’ in R, using principal component analysis to verify the grouping results. The OS rate among different clusters was evaluated by the Kaplan–Meier method and log-rank test. The distribution of age, gender, tumour grade, tumour stage and vascular invasion between various clusters was analysed by the Chi-square test.
Prognostic signatures generation and prediction
To evaluate the association between 15-gene expression and patients’ survival, Cox regression analysis and least absolute shrinkage and selection operator (LASSO) were performed by the ‘glment’ package in R software. High-risk and low-risk group patients were grouped by the median risk scores and Kaplan–Meier method was used for further survival analysis. The receiver operating characteristic curve was used to detect the predictive efficiency of the prediction model. Independent risk factors for HCC were defined by univariate and multivariate Cox regression analysis.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA; version 3.0) was performed to explore the m6A RNA methylation regulators related to signalling pathways in HCC. Gene expression enrichment analysis was performed between datasets with low or high mRNA expression. The normalised enrichment score, nominal P-value and false discovery rate q-value were assessed.
Statistical analysis
A two‑sided P < 0.05 was considered to indicate a statistically significant difference. IBM SPSS (version 24.0) and R software (version 3.5.3) were used for statistical analysis.
Results
Profiling of m6A RNA methylation regulators in HCC
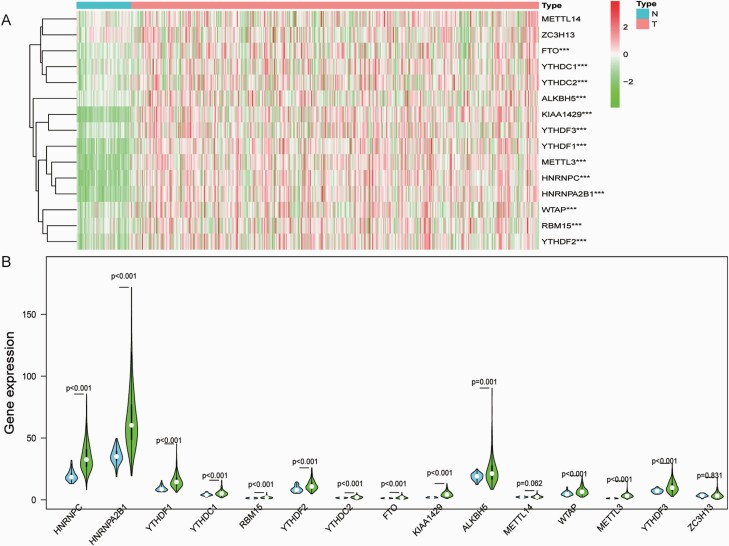
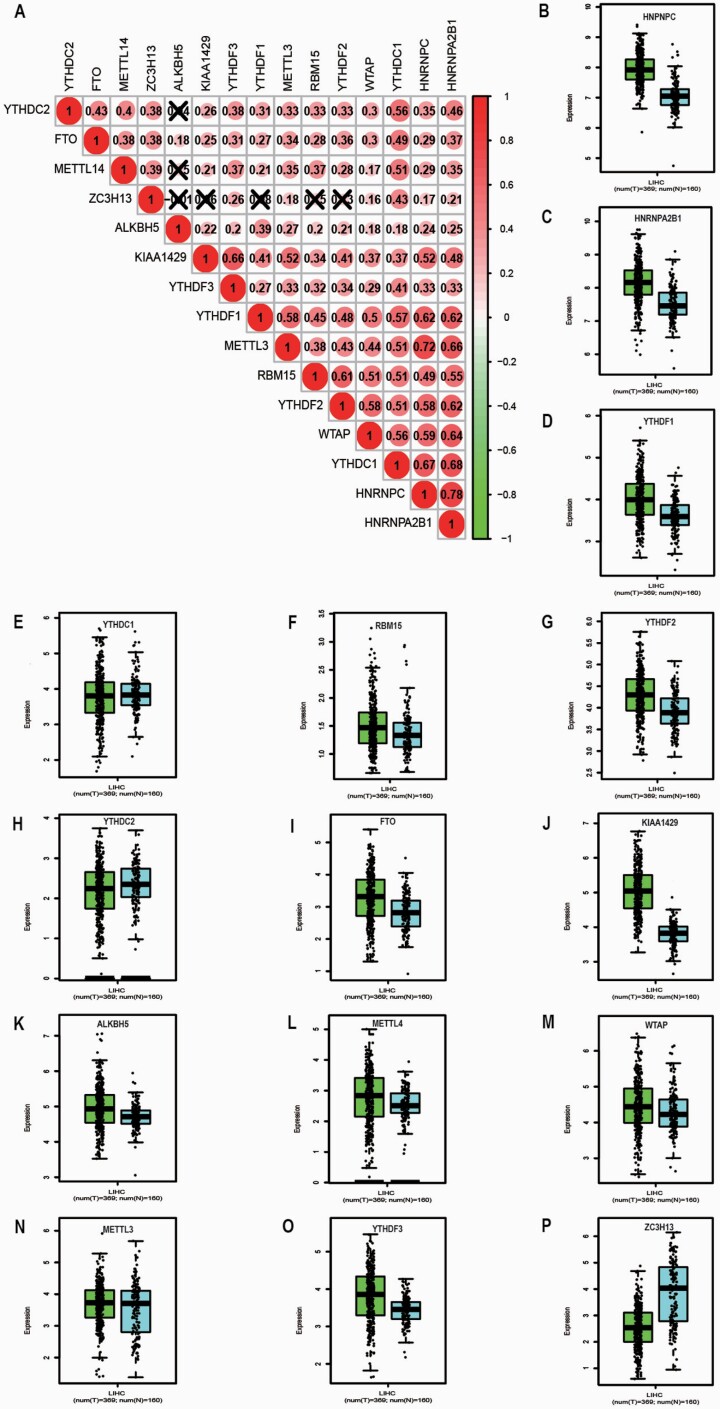
Heatmap and violin plot were generated to visualise the specially expressed m6A RNA methylation regulators between HCC samples and non-tumour samples (Figure 1A and B). Red or green colour in the plots represented relatively high or low expression, respectively. The expression levels of FTO (P < 0.001), YTHDC1 (P < 0.001), YTHDC2 (P < 0.001), ALKBH5 (P < 0.001), KIAA1429 (P < 0.001), YTHDF3 (P < 0.001), YTHDF1 (P < 0.001), METTL3 (P < 0.001), HNRNPC (P < 0.001), HNRNPA2B1 (P < 0.001), WTAP (P < 0.001), RBM15 (P < 0.001) and YTHDF2 (P < 0.001) were significantly upregulated in HCC tissues compared to non-tumour tissues. METTL14 (P = 0.062) and ZC3H13 (P = 0.831) showed no significant difference. As shown in Figure 2A, Pearson correlation analysis demonstrated that all 15 genes showed a positive correlation with each other. HNRNPC and HNRNPA2B1 have the strongest correlation (r = 0.78). METTL3 and HNRNPC (r = 0.72), YTHDC1 and HNRNPA2B1 (r = 0.68), YTHDC1 and HNRNPC (r = 0.67), KIAA1429 and YTHDF3 (r = 0.66), METTL3 and HNRNPA2B1 (r = 0.66), WTAP and HNRNPA2B1 (r = 0.64), YTHDF1 and HNRNPA2B1 (r = 0.62), YTHDF1 and HNRNPC (r = 0.62), YTHDF2 and HNRNPA2B1 (r = 0.62), as well as RBM15 and YTHDF2 (r = 0.61) were also correlated. For validation, we performed an analysis of the expression of m6A RNA methylation-related genes in HCC samples with a threshold set as P-value ≤ 0.05, fold change ≥ 1 in the GEPIA2 database. FTO, ALKBH5, KIAA1429, YTHDF3, YTHDF1, HNRNPC, HNRNPA2B1, WTAP, RBM15 and YTHDF2 were significantly upregulated in HCC tissues compared with non-tumour tissues (All P < 0.05). However, ZC3H13 was significantly upregulated in non-tumour than tumour (P < 0.05). METTL14, YTHDC1, YTHDC2 and METTL3 showed no significant difference (Figure 2B–P).
Fig. 1.
The profiling of m6A RNA methylation regulators in HCC tissues and non-tumour tissues. (A) The heatmap of 15 m6A RNA methylation regulators in tumour samples and non-tumour samples (red is upregulated and green is downregulated; *P < 0.05, **P < 0.01 and *** P < 0.001); (B) Vioplot visualising the differentially m6A RNA methylation regulators in HCC (blue is non-tumour and green is tumour).
Fig. 2.
(A) Correlation matrix of interaction in m6A methylation-related genes. Correlation coefficients are plotted with negative correlation (green) and positive correlation (red); (B–P) Expression of 15 m6A RNA methylation regulators in tumour samples and non-tumour samples from GEPIA2 database (blue is non-tumour and green is tumour).
Protein–protein interaction network and gene co-expression network analysis
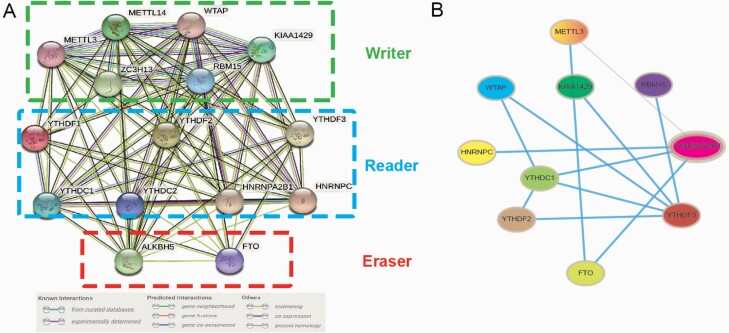
The interaction network further supported the result of the correlation analysis. The STRING database analysed the interactions among the 15 m6A RNA methylation regulators (Figure 3A). WTAP, YTHDF2 and HNRNPA2B1 seemed to be the interaction network’s hub genes, and their interactions or co-expressions with FTO, YTHDC1, METTL3, YTHDF3, HNRNPC, KIAA1429 and RBM15 were supported both by the Coexpedia database (Figure 3B).
Fig. 3.
(A) Protein–protein interaction network was constructed to evaluate the interaction among m6A RNA methylation regulators; (B) Construction of gene co-expression networks among m6A RNA methylation regulators.
Consensus clustering of 15 m6A RNA methylation regulators
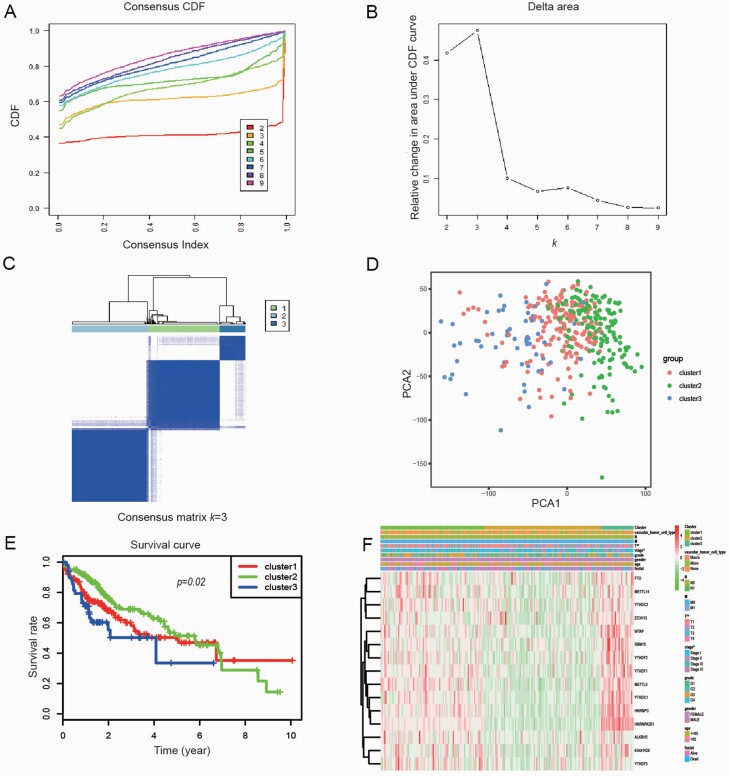
Based on m6A methylation-related gene expression levels, we identified distinct subgroups of 374 HCC samples by ‘ConsensusClusterPlus package’ in R and found that k = 3 achieved adequate selection to divide these samples into three clusters, namely cluster 1, cluster 2 and cluster 3 (Figure 4A–C). We further proved the correctness of our grouping by principal component analysis. The significant variance between the three subgroups was shown in the principal component analysis modal (Figure 4D). HCC patients in cluster 2 and cluster 3 had a poor OS than cluster 2 (P = 0.02; Figure 4E). Then the associations between the clustering and clinicopathological parameters were examined. The significant difference was found between cluster 2 and cluster 1, cluster 3 for the tumour stage (P < 0.05) and T classification (P < 0.01). In contrast, other parameters such as age, gender, tumour grade, N classification, M classification and vascular invasion were no significant different (Figure 4F).
Fig. 4.
Identification of consensus clusters by m6A RNA methylation regulators. (A) Consensus clustering cumulative distribution function (CDF) for k = 2–9; (B) Relative change in area under CDF curve for k = 2–9; (C) Consensus clustering matrix for k = 3; (D) Principal component analysis of the three subgroups; (E) Kaplan–Meier survival plots of the three subgroups; (F) Heatmap and clinicopathologic features of the three clusters defined by the m6A RNA methylation regulators consensus expression (red is upregulated and green is downregulated; *P < 0.05).
Identification of prognostic signature
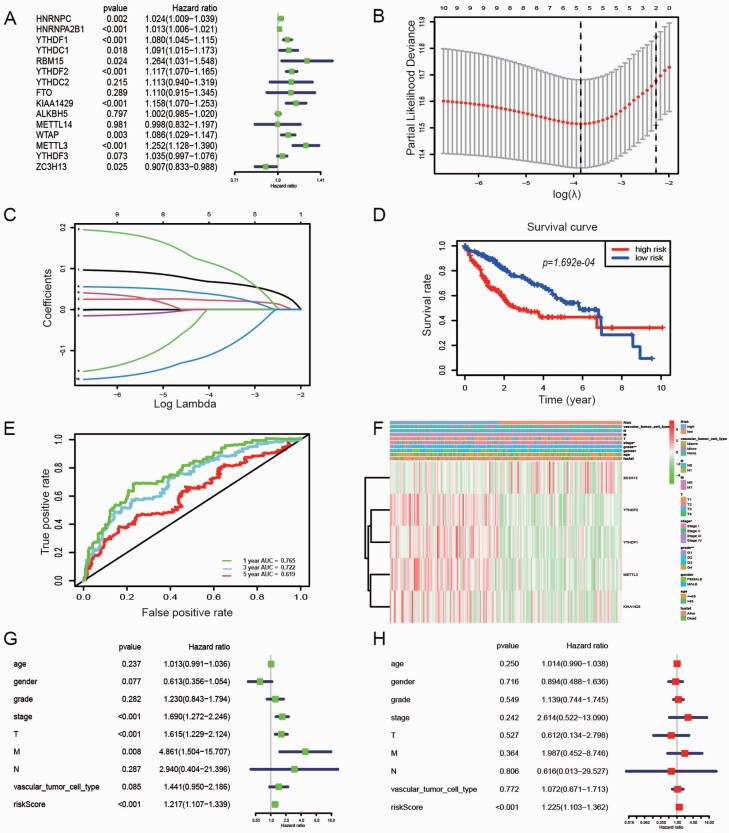
We used univariate Cox regression to analyse 15 genes, and 13 candidate genes were selected with P < 0.05 as a screening condition (Figure 5A). Besides, this research screened predictive genes as prognostic indicators through the LASSO Cox regression model. The value of λ was selected according to the minimum residual sum of squares. Five potential predictors (Figure 5B and C). ZC3H13, YTHDF2, YTHDF1, METTL3 and KIAA1429 were identified as prognostic factors for HCC. The risk score of five genes was also calculated for further univariate and multivariate Cox regression analyses. The risk score formula to predict OS was developed as follows: risk score = (0.068 × YTHDF2) + (0.023 × YTHDF1)+ (0.113 × METTL3) + (0.038 × KIAA1429) + (−0.109 × ZC3H13). In addition, HCC patients were grouped into the high-risk and low-risk groups based on the median risk score. The survival analysis showed that HCC patients in the high-risk group had a significantly poorer OS than those in the low-risk group (P = 1.692e−04; Figure 5D). We also evaluated the prognostic efficiency of risk factors through receiver operating characteristic curves, which showed that the area under the receiver operating characteristic curve (AUC) for 1-, 3- and 5-year OS was 0.765, 0.722 and 0.619, respectively (Figure 5E). Figure 5F revealed that the expression of ZC3H13, YTHDF2, YTHDF1, METTL3 and KIAA1429 in the high-risk and low-risk groups. Significant differences were found between the high- and low-risk groups concerning tumour stage (P < 0.05) and tumour grade (P < 0.01). Univariable and multivariable Cox regression analyses were performed to investigate whether the prognostic signature-based risk score was an independent prognostic indicator. After deleting cases with incomplete values, such as age, gender, tumour grade, tumour stage, vascular invasion, etc., a total of 208 patients were used for subsequent analysis. The univariate analysis showed that the tumour stage (P < 0.001, HR = 1.690, 95% CI = 1.272–2.246), T classification (P < 0.001, HR = 1.615, 95% CI = 1.229–2.124), M classification (P = 0.008, HR = 4.861, 95% CI = 1.504–15.707) and risk score (P < 0.001, HR = 1.217, 95% CI = 1.107–1.339) were significantly correlated with the OS (Figure 5G). Moreover, multivariate Cox regression analysis further identified that risk score (P < 0.001, HR = 1.225, 95% CI = 1.103–1.362 as shown in Figure 5H), was an independent prognostic factor in HCC.
Fig. 5.
The effect of m6A RNA methylation regulators, the risk score and clinicopathological variables on the prognosis of HCC. (A) Cox univariate analysis of m6A RNA methylation regulators; (B) Partial likelihood deviance versus log (λ) was drawn using LASSO Cox regression model; (C) Coefficients of selected features are shown by lambda parameter. (D) The Kaplan–Meier OS curves for HCC patients assigned to high- and low-risk groups based on the risk score; (E) Time‑dependent risk receiver operating characteristic curves. The 1‑, 3‑ and 5‑year risk AUC were 0.765, 0.722 and 0.619, respectively; (F) The heatmap shows the expression of 5 m6A RNA methylation regulators and the distribution of clinicopathological variables between the high- and low-risk groups (red is upregulated and green is downregulated; *P < 0.05, **P < 0.01); (G) Forest plot of univariate Cox regression analysis in HCC; (H) Forest plot of multivariate Cox regression analysis in HCC.
GSEA biological process enrichment
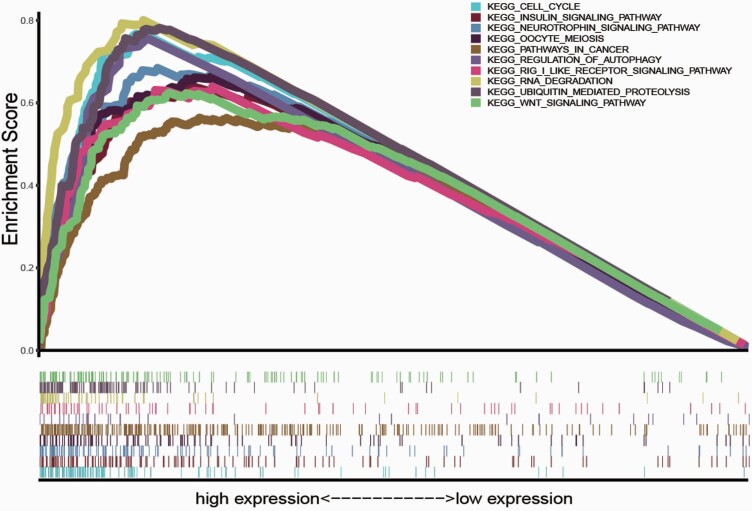
Based on the TCGA data, we explored mRNAs’ function associated with the m6A RNA methylation regulators and their related signal transduction pathways through GSEA. Based on normalised enrichment score, false discovery rate q-value and nominal P-value, significantly enriched signalling pathways were selected. In this study, the top enrichments involved in the cell cycle, insulin signalling pathway, neurotrophin signalling pathway, oocyte meiosis, pathways in cancer, regulation of autophagy, RIG I-like receptor signalling pathway, RNA degradation, ubiquitin-mediated proteolysis and Wnt signalling pathway (Figure 6).
Fig. 6.
GSEA of the established m6A RNA methylation regulators.
Establishment of a prognostic nomogram for HCC
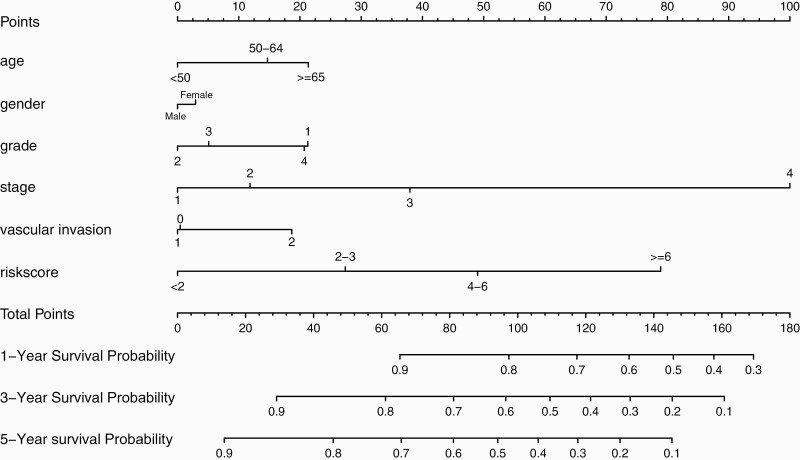
We developed a new prognostic nomogram based on age, gender, tumour grade, tumour stage, vascular invasion and risk score to provide a quantitative method for predicting individuals’ survival (Figure 7). The results revealed that the nomogram could systematically predict HCC patients’ OS at 1-, 3- and 5-years.
Fig. 7.
Prognostic nomogram was established by combining clinicopathological parameters and risk score.
Discussion
HCC is still one of the most common cancer globally, and its incidence rate is increasing year by year. The initiation and development of HCC is a multistep process that involves the interaction of biological factors, environmental factors and genetic factors (28). The efficacy of current treatment strategies is not so good that the OS of HCC patients is far from satisfactory (1). Therefore, it is necessary to study cancer development’s molecular mechanism and find novel predictive biomarkers and valuable therapeutic targets. Recent studies have confirmed that N6-methyladenosine (m6A), the most abundant RNA modification, is closely related to various human diseases, including cancer (29–32). Aberrant methylation of m6A RNA can regulate the carcinogenesis of many tumour types (19,25). However, the prognostic value of m6A genes in HCC has not been well confirmed. This study aimed to analyse the differential expression of m6A genes, establish a prognosis model of HCC, and find potential therapeutic targets.
In this study, we compared the expression of 15 m6A RNA methylation regulators (including KIAA1429, RBM15, ZC3H13, METTL3, METTL14, WTAP, YTHDF1, YTHDF2, YTHDF3, HNRNPC, HNRNPA2B1, YTHDC1, YTHDC2, ALKBH5 and FTO) in non-tumour tissues and HCC tissues based on TCGA data. We found that these regulatory factors were upregulated in tumour samples. According to the risk score of m6A regulatory factors, we divided HCC patients into high-risk and low-risk groups. The results showed significant differences in OS and some clinicopathological features between high-risk and low-risk groups. Finally, we established a prognosis model consisting of the m6A RNA methylation regulators. It is encouraging that the risk score can independently predict the prognosis of patients with HCC. Thus, the risk characteristics in this study can more accurately predict the individualised survival of HCC patients.
As shown in our predictive model, 5 m6A RNA methylation-related genes, including ZC3H13, YTHDF1, YTHDF2, METTL3 and KIAA1429, were identified as prognostic factors for HCC. It has been confirmed that ZC3H13 has different roles in distinct human cancers (33,34). Zhu et al. and Kim et al. reported that ZC3H13 had a tumour-inhibitory effect in colorectal cancer (33,35). In contrast, Gewurz et al. revealed that ZC3H13 might play a role in tumour progression as an oncogene (36). Therefore, the specific function of ZC3H13 in HCC remains unclear, which requires in-depth study.
Both YTHDF1 and YTHDF2 are members of the YTH domain family. YTHDF1 is an m6A ‘reader’, which recognises and binds to m6A modified mRNA, thus improving their target RNAs’ translation efficiency (27). It is reported that YTHDF1 is overexpressed in HCC with a poor prognosis (37). Zhao’s study yielded similar results (38). YTHDF2 has also been reported to be associated with various kinds of tumours (39). Recent studies have shown that the high expression of YTHDF2 can inhibit the proliferation and the growth of HCC. In the mechanism, YTHDF2 can promote the degradation of EGFR mRNA by binding to the m6A modified site of EGFR 3′-UTR, thus inhibiting the proliferation and growth of HCC cells (40). On the contrary, Hou et al. proved that the expression of YTHDF2 was downregulated in HCC YTHDF2 might inhibit tumour progression by promoting the decay of IL11 and SERPINE2 mRNA, suggesting that YTHDF2 is a suppressor of HCC. Their research also demonstrated that reducing YTHDF2 in HCC cells promotes inflammation, vascular reconstruction and metastasis (41). Therefore, the specific role of YTHDF2 in HCC is also controversial. The difference in these results may be related to the high heterogeneity of liver cancer.
METTL3, an RNA methyltransferase, is involved in mRNA degradation, translation and biogenesis control through m6A modification (42,43). METTL3 has been proved to be involved in tumorigenesis and the progression of some cancers (42). For example, Chen et al. reported that METTL3 is engaged in HCC and lung metastasis progress (44). Besides, high-level expression of METTL3 promotes cancer metastasis and poor prognosis in HCC patients (45). Aravalli et al. demonstrated that METTL3 gene knockout could significantly inhibit the occurrence and progression of HCC (46). The above results suggest that METTL3 may be an oncogene of HCC (44). The role of METTL3 in other gastrointestinal tumours, such as gastric cancer (47–49), colorectal cancer (50,51) and pancreatic cancer (52), has been confirmed to be similar to HCC.
Overexpression of KIAA1429 was also associated with a poor prognosis of HCC (53). LAN et al. found that KIAA1429 is an essential driving factor of hepatocarcinogenesis and metastasis. Their research showed that silencing KIAA1429 can inhibit the proliferation and metastasis of cancer cells, which indicates that KIAA1429 can be used as a new gene for the treatment of HCC (53). At the mechanism level, KIAA1429 may inhibit ID2 by upregulating its m6A level, thus promoting the progress of HCC (54). In other tumours, Qian et al. revealed that KIAA1429 was highly expressed in breast cancer tissues, but it was often downregulated in non-tumour tissues (55), similar to the results of the HCC study. KIAA1429, as a component of m6A ‘writers’, can significantly reduce the mRNA of m6A after being knocked out, which indicates that KIAA1429 plays a vital role in cancer progression and can be used as a novel gene for the treatment of cancer patients.
Our study indicates that ZC3H13, YTHDF1, YTHDF2, METTL3 and KIAA1429 are all associated with HCC patients’ prognosis. In addition, the risk score and clinical features of Cox regression analysis confirmed that these five genes could be used as prognostic markers and even targets for new therapies for HCC. Moreover, the combination of m6A methylation-related genes and clinical parameters may have better predictive ability than a single biomarker for HCC patients. Recently, m6A methylation-related genes have shown great potential in predicting the survival of cancer patients. M6A methylation has been reported to be involved in many essential signalling pathways of malignancies, which affect tumour invasion, metastasis, immune cell infiltration and immune escape (56–61). We explored mRNAs’ related signal pathways through GSEA. In this study, the top enrichments involved in the pathways in cancer, cell cycle, oocyte meiosis, regulation of autophagy, RIG I-like receptor signalling pathway, RNA degradation, Wnt signalling pathway and so on. The enrichment analysis of m6A and signal pathway can provide more personalised treatments for HCC patients, including targeted therapy and immunotherapy. Our study preliminarily proved that m6A methylation-related genes play a significant role in the occurrence and development of HCC, suggesting that this marker can be regarded as a prognostic factor for the disease provide meaningful guidance for the selection of treatment strategies. Genes related to the methylation of m6A RNA may be potential therapeutic targets. Our research comprehensively analysed 13 m6A regulatory factors and established a valuable prognostic model based on 5 regulatory factors of m6A RNA methylation to predict the prognosis of HCC patients, which provides meaningful guidance for clinical treatment and will be helpful for future research. In addition, a prognostic nomogram generated based on risk score and clinicopathological features is easy to use and interpret and has a satisfactory accuracy in predicting HCC patients’ prognosis. However, we have to admit that this study has limitations. Firstly, since our data are from the TCGA database, further experimental evidence is needed to confirm our findings. Secondly, the number of HCC samples is significantly higher than that of non-tumour samples, which may affect the results’ reliability and accuracy. Finally, extending the results to other races needs further verification because the TCGA database population mainly comes from white and black people.
In conclusion, we found that the expression of m6A RNA methylation regulators is closely related to the prognosis of HCC. And the prognosis model generated by 5 m6A methylation-related genes can predict the prognosis of HCC patients and point out the direction for further study of the pathogenesis of HCC. Further functional experiments are needed to confirm these results in the future.
Supplementary Material
Acknowledgements
We acknowledge the TCGA database for providing its platform and contributors for uploading their meaningful datasets.
Conflict of interest statement: The authors declare that they have no conflicts of interest.
Funding
This study was supported by the Natural Science Foundation of Guangdong Province (No. 2018A0303130098); Science and Technology Planning Project of Guangdong Province (No. 2017A020215112); Science and Technology Planning Project of Guangzhou (No. 201903010017 and No. 201904010479); and Medical Innovation Project of the First Affiliated Hospital of Guangzhou University of Chinese Medicine (2019IIT18).
Data availability statement
Publicly available datasets were analysed in this study. These can be found in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/); The GEPIA2 database (http://gepia2.cancer-pku.cn/#index). The original data, named ‘m6Aexp.xlsx’ and ‘clinical.xls’, have been uploaded as supplementary data.
References
- 1.Bray, F., Ferlay, J., Soerjomataram, I., Siegel, R. L., Torre, L. A. and Jemal, A. (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin., 68, 394–424. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay, J., Colombet, M., Soerjomataram, I., Mathers, C., Parkin, D. M., Piñeros, M., Znaor, A. and Bray, F. (2019) Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer, 144, 1941–1953. [DOI] [PubMed] [Google Scholar]
- 3.Llovet, J. M., Zucman-Rossi, J., Pikarsky, E., Sangro, B., Schwartz, M., Sherman, M. and Gores, G. (2016) Hepatocellular carcinoma. Nat. Rev. Dis. Primers, 2, 16018. [DOI] [PubMed] [Google Scholar]
- 4.European Association for the Study of the Liver. Electronic address: easloffice@easloffice.eu and European Association for the Study of the Liver. (2018) EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J. Hepatol., 69, 182–236. [DOI] [PubMed] [Google Scholar]
- 5.Marrero, J. A., Kulik, L. M., Sirlin, C. B., Zhu, A. X., Finn, R. S., Abecassis, M. M., Roberts, L. R. and Heimbach, J. K. (2018) Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology, 68, 723–750. [DOI] [PubMed] [Google Scholar]
- 6.Arellano, L. M. and Arora, S. P. (2018) Systemic treatment of advanced hepatocellular carcinoma in older adults. J. Nat. Sci., 4. [PMC free article] [PubMed] [Google Scholar]
- 7.Kudo, M., Finn, R. S., Qin, S., et al. (2018) Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: a randomised phase 3 non-inferiority trial. Lancet, 391, 1163–1173. [DOI] [PubMed] [Google Scholar]
- 8.Jindal, A., Thadi, A. and Shailubhai, K. (2019) Hepatocellular carcinoma: etiology and current and future drugs. J. Clin. Exp. Hepatol., 9, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roderburg, C., Özdirik, B., Wree, A., Demir, M. and Tacke, F. (2020) Systemic treatment of hepatocellular carcinoma: from sorafenib to combination therapies. Hepat. Oncol., 7, HEP20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zongyi, Y. and Xiaowu, L. (2020) Immunotherapy for hepatocellular carcinoma. Cancer Lett., 470, 8–17. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, Y., Chen, S. W., Liu, L. L., Yang, X., Cai, S. H. and Yun, J. P. (2018) A model combining TNM stage and tumor size shows utility in predicting recurrence among patients with hepatocellular carcinoma after resection. Cancer Manag. Res., 10, 3707–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elshaarawy, O., Alkhatib, A., Elhelbawy, M., Gomaa, A., Allam, N., Alsebaey, A., Rewisha, E. and Waked, I. (2019) Validation of modified albumin-bilirubin-TNM score as a prognostic model to evaluate patients with hepatocellular carcinoma. World J. Hepatol., 11, 542–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Long, J., Zhang, L., Wan, X., Lin, J., Bai, Y., Xu, W., Xiong, J. and Zhao, H. (2018) A four-gene-based prognostic model predicts overall survival in patients with hepatocellular carcinoma. J. Cell. Mol. Med., 22, 5928–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li, W., Lu, J., Ma, Z., Zhao, J. and Liu, J. (2019) An integrated model based on a six-gene signature predicts overall survival in patients with hepatocellular carcinoma. Front. Genet., 10, 1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, X. Y., Zhang, J. and Zhu, J. S. (2019) The role of m6A RNA methylation in human cancer. Mol. Cancer, 18, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He, L., Li, H., Wu, A., Peng, Y., Shu, G. and Yin, G. (2019) Functions of N6-methyladenosine and its role in cancer. Mol. Cancer, 18, 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tong, J., Flavell, R. A. and Li, H. B. (2018) RNA m6A modification and its function in diseases. Front. Med., 12, 481–489. [DOI] [PubMed] [Google Scholar]
- 18.Yang, C., Hu, Y., Zhou, B., Bao, Y., Li, Z., Gong, C., Yang, H., Wang, S. and Xiao, Y. (2020) The role of m6A modification in physiology and disease. Cell Death Dis., 11, 960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang, J., Chen, J., Fei, X., Wang, X. and Wang, K. (2020) N6-methyladenine RNA modification and cancer. Oncol. Lett., 20, 1504–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Z., Weng, H., Su, R., et al. (2017) FTO plays an oncogenic role in acute myeloid leukemia as a N6-methyladenosine RNA demethylase. Cancer Cell, 31, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vu, L. P., Pickering, B. F., Cheng, Y., et al. (2017) The N6-methyladenosine (m6A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med., 23, 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, J., Eckert, M. A., Harada, B. T., et al. (2018) m6A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nat. Cell Biol., 20, 1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cui, Q., Shi, H., Ye, P., et al. (2017) m6A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep., 18, 2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwok, C. T., Marshall, A. D., Rasko, J. E. and Wong, J. J. (2017) Genetic alterations of m6A regulators predict poorer survival in acute myeloid leukemia. J. Hematol. Oncol., 10, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huo, F. C., Zhu, Z. M. and Pei, D. S. (2020) N6-methyladenosine (m6A) RNA modification in human cancer. Cell Prolif., 53, e12921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu, Y., Dominissini, D., Rechavi, G. and He, C. (2014) Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet., 15, 293–306. [DOI] [PubMed] [Google Scholar]
- 27.Wang, X., Zhao, B. S., Roundtree, I. A., et al. (2015) N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell, 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levrero, M. and Zucman-Rossi, J. (2016) Mechanisms of HBV-induced hepatocellular carcinoma. J. Hepatol., 64, S84–S101. [DOI] [PubMed] [Google Scholar]
- 29.Gokhale, N. S., McIntyre, A. B. R., McFadden, M. J., et al. (2016) N6-methyladenosine in Flaviviridae viral RNA genomes regulates infection. Cell Host Microbe, 20, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, B., Li, Y., Song, R., Xue, C. and Xu, F. (2019) Functions of RNA N6-methyladenosine modification in cancer progression. Mol. Biol. Rep., 46, 2567–2575. [DOI] [PubMed] [Google Scholar]
- 31.Mathiyalagan, P., Adamiak, M., Mayourian, J., et al. (2019) FTO-dependent N6-Methyladenosine regulates cardiac function during remodeling and repair. Circulation, 139, 518–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, Y., Shen, F., Huang, W., Qin, S., Huang, J. T., Sergi, C., Yuan, B. F. and Liu, S. M. (2019) Glucose is involved in the dynamic regulation of m6A in patients with type 2 diabetes. J. Clin. Endocrinol. Metab., 104, 665–673. [DOI] [PubMed] [Google Scholar]
- 33.Zhu, D., Zhou, J., Zhao, J., Jiang, G., Zhang, X., Zhang, Y. and Dong, M. (2019) ZC3H13 suppresses colorectal cancer proliferation and invasion via inactivating Ras-ERK signaling. J. Cell. Physiol., 234, 8899–8907. [DOI] [PubMed] [Google Scholar]
- 34.Wang, T., Kong, S., Tao, M. and Ju, S. (2020) The potential role of RNA N6-methyladenosine in cancer progression. Mol. Cancer, 19, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim, Y. R., Chung, N. G., Kang, M. R., Yoo, N. J. and Lee, S. H. (2010) Novel somatic frameshift mutations of genes related to cell cycle and DNA damage response in gastric and colorectal cancers with microsatellite instability. Tumori, 96, 1004–1009. [PubMed] [Google Scholar]
- 36.Gewurz, B. E., Towfic, F., Mar, J. C., et al. (2012) Genome-wide siRNA screen for mediators of NF-κB activation. Proc. Natl. Acad. Sci. U. S. A., 109, 2467–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou, Y., Yin, Z., Hou, B., Yu, M., Chen, R., Jin, H. and Jian, Z. (2019) Expression profiles and prognostic significance of RNA N6-methyladenosine-related genes in patients with hepatocellular carcinoma: evidence from independent datasets. Cancer Manag. Res., 11, 3921–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao, X., Chen, Y., Mao, Q., Jiang, X., Jiang, W., Chen, J., Xu, W., Zhong, L. and Sun, X. (2018) Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark., 21, 859–868. [DOI] [PubMed] [Google Scholar]
- 39.Tanabe, A., Tanikawa, K., Tsunetomi, M., et al. (2016) RNA helicase YTHDC2 promotes cancer metastasis via the enhancement of the efficiency by which HIF-1α mRNA is translated. Cancer Lett., 376, 34–42. [DOI] [PubMed] [Google Scholar]
- 40.Zhong, L., Liao, D., Zhang, M., Zeng, C., Li, X., Zhang, R., Ma, H. and Kang, T. (2019) YTHDF2 suppresses cell proliferation and growth via destabilizing the EGFR mRNA in hepatocellular carcinoma. Cancer Lett., 442, 252–261. [DOI] [PubMed] [Google Scholar]
- 41.Hou, J., Zhang, H., Liu, J., et al. (2019) YTHDF2 reduction fuels inflammation and vascular abnormalization in hepatocellular carcinoma. Mol. Cancer, 18, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin, S., Choe, J., Du, P., Triboulet, R. and Gregory, R. I. (2016) The m(6)A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell, 62, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barbieri, I., Tzelepis, K., Pandolfini, L., et al. (2017) Promoter-bound METTL3 maintains myeloid leukaemia by m6A-dependent translation control. Nature, 552, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen, M., Wei, L., Law, C. T., et al. (2018) RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology, 67, 2254–2270. [DOI] [PubMed] [Google Scholar]
- 45.Ma, J. Z., Yang, F., Zhou, C. C., et al. (2017) METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N6-methyladenosine-dependent primary MicroRNA processing. Hepatology, 65, 529–543. [DOI] [PubMed] [Google Scholar]
- 46.Aravalli, R. N., Cressman, E. N. and Steer, C. J. (2013) Cellular and molecular mechanisms of hepatocellular carcinoma: an update. Arch. Toxicol., 87, 227–247. [DOI] [PubMed] [Google Scholar]
- 47.Yue, B., Song, C., Yang, L., Cui, R., Cheng, X., Zhang, Z. and Zhao, G. (2019) METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol. Cancer, 18, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang, C., Zhang, M., Ge, S., Huang, W., Lin, X., Gao, J., Gong, J. and Shen, L. (2019) Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med., 8, 4766–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Q., Chen, C., Ding, Q., et al. (2020) METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut, 69, 1193–1205. [DOI] [PubMed] [Google Scholar]
- 50.Deng, R., Cheng, Y., Ye, S., et al. (2019) m6A methyltransferase METTL3 suppresses colorectal cancer proliferation and migration through p38/ERK pathways. Onco. Targets. Ther., 12, 4391–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, T., Hu, P. S., Zuo, Z., et al. (2019) METTL3 facilitates tumor progression via an m6A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol. Cancer, 18, 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taketo, K., Konno, M., Asai, A., et al. (2018) The epitranscriptome m6A writer METTL3 promotes chemo- and radioresistance in pancreatic cancer cells. Int. J. Oncol., 52, 621–629. [DOI] [PubMed] [Google Scholar]
- 53.Lan, T., Li, H., Zhang, D., et al. (2019) KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol. Cancer, 18, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng, X., Li, M., Rao, X., Zhang, W., Li, X., Wang, L. and Huang, G. (2019) KIAA1429 regulates the migration and invasion of hepatocellular carcinoma by altering m6A modification of ID2 mRNA. Onco. Targets. Ther., 12, 3421–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qian, J. Y., Gao, J., Sun, X., et al. (2019) KIAA1429 acts as an oncogenic factor in breast cancer by regulating CDK1 in an N6-methyladenosine-independent manner. Oncogene, 38, 6123–6141. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, S. Y., Zhang, S. W., Fan, X. N., Zhang, T., Meng, J. and Huang, Y. (2019) FunDMDeep-m6A: identification and prioritization of functional differential m6A methylation genes. Bioinformatics, 35, i90–i98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen, Y., Zhou, C., Sun, Y., He, X. and Xue, D. (2020) m6A RNA modification modulates gene expression and cancer-related pathways in clear cell renal cell carcinoma. Epigenomics, 12, 87–99. [DOI] [PubMed] [Google Scholar]
- 58.Fang, J., Hu, M., Sun, Y., Zhou, S. and Li, H. (2020) Expression profile analysis of m6A RNA methylation regulators indicates they are immune signature associated and can predict survival in Kidney Renal Cell Carcinoma. DNA Cell Biol. doi: 10.1089/dna.2020.5767 [DOI] [PubMed] [Google Scholar]
- 59.Wang, J., Zhang, C., He, W. and Gou, X. (2020) Effect of m6A RNA methylation regulators on malignant progression and prognosis in renal clear cell Carcinoma. Front. Oncol., 10, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yi, D., Wang, R., Shi, X., Xu, L., Yilihamu, Y. and Sang, J. (2020) METTL14 promotes the migration and invasion of breast cancer cells by modulating N6‑methyladenosine and hsa‑miR‑146a‑5p expression. Oncol. Rep., 43, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang, Q., Zhang, Q., Li, Q., Zhang, J. and Zhang, J. (2021) Clinicopathological and immunological characterization of RNA m6 A methylation regulators in ovarian cancer. Mol. Genet. Genomic Med., 9, e1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analysed in this study. These can be found in The Cancer Genome Atlas (https://portal.gdc.cancer.gov/); The GEPIA2 database (http://gepia2.cancer-pku.cn/#index). The original data, named ‘m6Aexp.xlsx’ and ‘clinical.xls’, have been uploaded as supplementary data.