Abstract
For the differentiation and identification of mycobacterial species, the rpoB gene, encoding the β subunit of RNA polymerase, was investigated. rpoB DNAs (342 bp) were amplified from 44 reference strains of mycobacteria and clinical isolates (107 strains) by PCR. The nucleotide sequences were directly determined (306 bp) and aligned by using the multiple alignment algorithm in the MegAlign package (DNASTAR) and the MEGA program. A phylogenetic tree was constructed by the neighbor-joining method. Comparative sequence analysis of rpoB DNAs provided the basis for species differentiation within the genus Mycobacterium. Slowly and rapidly growing groups of mycobacteria were clearly separated, and each mycobacterial species was differentiated as a distinct entity in the phylogenetic tree. Pathogenic Mycobacterium kansasii was easily differentiated from nonpathogenic M. gastri; this differentiation cannot be achieved by using 16S rRNA gene (rDNA) sequences. By being grouped into species-specific clusters with low-level sequence divergence among strains of the same species, all of the clinical isolates could be easily identified. These results suggest that comparative sequence analysis of amplified rpoB DNAs can be used efficiently to identify clinical isolates of mycobacteria in parallel with traditional culture methods and as a supplement to 16S rDNA gene analysis. Furthermore, in the case of M. tuberculosis, rifampin resistance can be simultaneously determined.
The genus Mycobacterium comprises a wide range of organisms, including obligate parasites causing serious human and animal diseases, opportunistic pathogens, and saprophytic species found in nature. Human infections are caused mainly by slowly growing mycobacteria that need more than 7 days to form visible colonies on solid media. Traditionally, the definitive diagnosis of mycobacterial infections has been dependent on the isolation and identification of causative agents and requires a series of specialized physiological and biochemical tests. The procedures for these tests are complex, laborious, and usually impeded by the slow growth of mycobacteria in clinical laboratories. In particular, Mycobacterium leprae has not been cultivated in vitro. There have been increasing numbers of reports of infections caused by mycobacteria other than M. tuberculosis (MOTT), especially in association with human immunodeficiency virus infection. These are rarely disease associated, previously unknown or newly recognized mycobacteria that are not easy to identify; moreover, due to their phenotypic similarity to certain species, they cannot be easily characterized by the conventional methods of identification. However, mycobacterial systematics may help in the differentiation and identification of these phenotypically similar mycobacterial species.
Descriptive taxonomic analyses have been used to classify mycobacterial species. For a clearer definition of species boundaries, macromolecular comparisons, particularly of 16S rRNA, which is highly conserved throughout organisms, have been used to determine phylogenetic relationships. Mycobacterial phylogenetic analysis based on sequences of 16S rRNA (25, 31) or the 16S rRNA gene (16S rDNA) (27) has helped to define mycobacterial species. This analysis demonstrated the usefulness of genotypic studies, especially when conventional procedures are inapplicable, particularly for the differentiation and identification of novel and uncultivable mycobacteria (19). It has been suggested, however, that for the delineation of species boundaries, 16S rRNA-based phylogenetic analysis has its limitations (11). Ambiguous results due to the presence of two different 16S rRNA genes in an organism would also limit the use of 16S rDNA sequencing in the identification of mycobacterial species (23, 26). Doubts about its usefulness were raised because M. kansasii, a pathogenic mycobacterium, could not be distinguished from nonpathogenic M. gastri by this means (35). A similar result was observed in 23S rRNA sequence analysis; the sequence for M. kansasii was identical to that for M. celatum (32).
rpoB encodes the β subunit of RNA polymerase. The rpoB nucleotide sequences of three mycobacterial species were previously known (15, 16, 22). Missense mutations within rpoB’s limited region are known to be related to rifampin resistance in M. tuberculosis (34). Recently, the rpoB gene was used as an alternative tool to identify mycobacteria (14). However, only a limited number of reference species (five slowly growing and five rapidly growing species) in the genus Mycobacterium were used.
In the present study, rpoB DNAs (342 bp) comprising a highly conserved region throughout the eubacteria (5) were amplified from the 44 reference strains of mycobacteria. Their nucleotide sequences (306 bp) were directly determined and compared to study their phylogenetic relationships. To demonstrate the feasibility of using this method in which rpoB sequences are compared and a phylogenetic tree with reference species is inferred, this procedure was applied to clinical isolates. We suggest that this procedure is a useful identification method that can be completed within two working days.
MATERIALS AND METHODS
Mycobacterial strains and clinical isolates.
Forty-four reference strains of the genus Mycobacterium (Table 1) and clinical isolates used in this study were provided by the Korean Institute of Tuberculosis, the Korean National Tuberculosis Association (KNTA), and the World Health Organization/International Union Against Tuberculosis and Lung Disease-designated Supranational Reference Laboratory for Global Drug Resistance Surveillance. M. leprae (Thai 53 strain) was provided by the Institute of Hansen’s Disease, Catholic University Medical College, Seoul, Korea. Clinical isolates of M. tuberculosis (46 strains), M. avium complex (18 strains), M. kansasii (32 strains), M. fortuitum (5 strains), and M. szulgai (6 strains) were identified by conventional methods and provided for the blinded rpoB gene analysis. For the clinical samples of M. leprae, six punch biopsy specimens were obtained from active lesions of patients diagnosed on the basis of histological findings, acid-fast bacterium staining, and amplification of DNA encoding an 18-kDa protein (36) by the Institute of Hansens’ Disease.
TABLE 1.
Mycobacteria and other microorganisms used for the rpoB sequence analysis
| Species | Accession no.a | Strain | Source |
|---|---|---|---|
| M. abscessus | AF057449 | CAP97E-03 | SNUHb |
| M. africanum | AF057450 | ATCC 25420 | KNTA |
| M. asiaticum | AF057455 | ATCC 25276 | KNTA |
| M. aurum | AF057456 | ATCC 23366 | KNTA |
| M. avium | AF057457 | ATCC 25291 | KNTA |
| M. bovis | AF057451 | ATCC 19210 | KNTA |
| M. bovis BCG | AF057452 | French strain 1173P2 | KNTA |
| M. celatum | |||
| Type 1 | AF057458 | ATCC 51131 | KNTA |
| Type 2 | AF057459 | ATCC 51130 | KNTA |
| M. chelonae | AF057460 | ATCC 35749 | KNTA |
| M. chitae | AF057461 | ATCC 19627 | KNTA |
| M. fallax | AF057462 | ATCC 35219 | KNTA |
| M. flavescens | AF057463 | ATCC 14474 | KNTA |
| M. fortuitum | AF057464 | ATCC 6841 | KNTA |
| M. fortuitum 49403 | AF057465 | ATCC 49403 | KNTA |
| M. gastri | AF057466 | ATCC 15754 | KNTA |
| M. genavense | AF057467 | ATCC 51233 | KNTA |
| M. gordonae | AF057468 | ATCC 14470 | KNTA |
| M. haemophilum | AF057469 | ATCC 29548 | KNTA |
| M. interjectum | AF057470 | ATCC 51457 | KNTA |
| M. intermedium | AF057471 | ATCC 51848 | KNTA |
| M. intracellulare | AF057472 | ATCC 13950 | KNTA |
| M. kansasii | AF057473 | ATCC 12478 | KNTA |
| M. leprae | AF057474 | Thai 53 strain | IHDc |
| M. malmoense | AF057475 | ATCC 29571 | KNTA |
| M. marinum | AF057476 | ATCC 927 | KNTA |
| M. neoaurum | AF057477 | ATCC 25795 | KNTA |
| M. nonchromogenicum | AF057478 | ATCC 19530 | KNTA |
| M. paratuberculosis | AF057479 | ATCC 19698 | KNTA |
| M. phlei | AF057480 | ATCC 11758 | KNTA |
| M. peregrinum | AF057481 | ATCC 14467 | KNTA |
| M. scrofulaceum | AF057482 | ATCC 19981 | KNTA |
| M. senegalense | AF057483 | ATCC 35796 | KNTA |
| M. shimoidei | AF057486 | ATCC 27962 | KNTA |
| M. simiae | AF057484 | ATCC 25275 | KNTA |
| M. smegmatis | AF057485 | ATCC 19420 | KNTA |
| M. szulgai | AF057487 | ATCC 35799 | KNTA |
| M. terrae | AF057488 | ATCC 15755 | KNTA |
| M. thermoresistibile | AF057489 | ATCC 19527 | KNTA |
| M. triviale | AF057490 | ATCC 23292 | KNTA |
| M. tuberculosis H37Rv | AF057454 | ATCC 27294 | KNTA |
| M. ulcerans | AF057491 | ATCC 19423 | KNTA |
| M. vaccae | AF057492 | ATCC 15483 | KNTA |
| M. xenopi | AF057493 | ATCC 19250 | KNTA |
| R. equi | AF057494 | ATCC 10146 | IMSNUd |
| N. nova | AF057495 | ATCC 21197 | IMSNU |
| C. diphtheriae | AF057496 | SNUH | SNUH |
GenBank accession numbers for the rpoB sequences of the reference strains.
SNUH, Department of Clinical Pathology, Seoul National University Hospital.
IHD, Institute of Hansen’s Disease, The Catholic University Medical College.
IMSNU, Culture Collection Center, Institute of Microbiology, Seoul National University.
Preparation of DNA and PCR.
Mycobacterial DNAs were prepared by the bead beater–phenol extraction method. A loopful of culture of each isolate was suspended in 200 μl of TEN buffer (10 mM Tris-HCl, 1 mM EDTA, 100 mM NaCl; pH 8.0), placed in a 2.0-ml screw-cap microcentrifuge tube filled with 100 μl (packed volume) of glass beads (diameter, 0.1 mm; Biospec Products, Bartlesville, Okla.) and 100 μl of phenol-chloroform-isopropyl alcohol (50:49:1). To disrupt the bacteria, the tube was oscillated on a Mini-Bead Beater (Biospec Products) for 1 min, and to separate the phases, the tube was centrifuged (12,000 × g, 5 min). After the aqueous phase was transferred into another clean tube, 10 μl of 3 M sodium acetate and 250 μl of ice-cold ethanol were added; to enable the DNA to precipitate, the mixture was kept at −20°C for 10 min. The DNA pellet was washed with 70% ethanol, dissolved in 60 μl of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0), and used as a template for PCR. M. leprae (Thai 53 strain) was prepared from the footpads of nude mice (BALB/c nu/nu; B & K Universal Ltd., North Humberside, United Kingdom) that had been inoculated at the Institute of Hansen’s Disease and maintained there for 18 months. The resected swollen footpads and biopsy specimens were homogenized in 2 ml of phosphate-buffered saline, using a Mickle homogenizer (Mickle Laboratory Engineering, Surrey, United Kingdom). The supernatant was collected after tissue debris had settled (1 × g, 5 min), and M. leprae DNA was prepared as previously described (36).
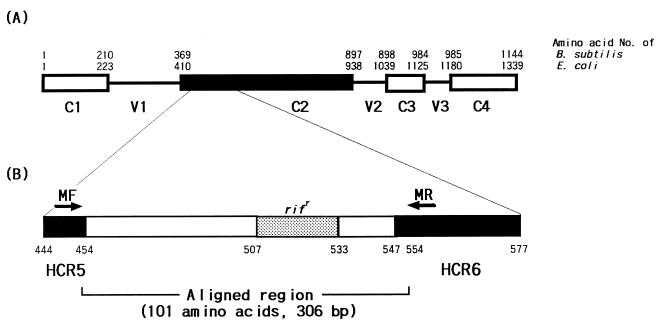
A set of primers (MF, 5′CGACCACTTCGGCAACCG3′; MR, 5′TCGATCGGGCACATCCGG3′) was used to amplify rpoB DNA (342 bp) encompassing the Rifr region, which is associated with rifampin resistance in M. tuberculosis (Fig. 1). The primers were selected from the highly conserved regions (HCR5 and HCR6) on the basis of known rpoB sequences of Bacillus subtilis (5). The amplified region (306 bp; from R454 to H554 using the codon numbering system for Escherichia coli) lies within the C2 region, one of four conserved domains (C1 to C4). The nucleotide sequence of the forward primer was identical to the corresponding sequences of M. tuberculosis, M. leprae, and M. smegmatis (GenBank accession no. L27989, Z14314, and U24474, respectively). However, one nucleotide of the reverse primer was different from the corresponding M. smegmatis sequence. Template DNA (50 ng) and 20 pmol of each primer were added to a PCR mixture tube (AccuPower PCR PreMix; Bioneer, Chungbuk, Korea) which contained 1 U of Taq DNA polymerase, 250 μM each deoxynucleoside triphosphate, 50 mM Tris-HCl (pH 8.3), 40 mM KCl, 1.5 mM HgCl2, and gel loading dye, and the volume was adjusted with distilled water to 20 μl. The reaction mixture was subjected to 30 cycles of amplification (30 s at 95°C, 30 s at 60°C, and 45 s at 72°C) followed by a 5-min extension at 72°C (model 9600 Thermocycler; Perkin-Elmer Cetus). The PCR products were electrophoresed on a 3% agarose gel and purified by the use of a QIAEX II gel extraction kit (Qiagen, Hilden, Germany).
FIG. 1.
A set of primers (MF-MR) that can amplify mycobacterial rpoB DNA (342 bp) was selected from the rpoB sequences of M. tuberculosis, M. leprae, and M. smegmatis (GenBank accession no. L27989, Z14314, and U24474, respectively) corresponding to highly conserved regions (HCR5 and HCR6) of B. subtilis and E. coli. (A) Primary sequences of the RNA polymerase β subunits of B. subtilis and E. coli are composed of four conserved domains (C1 to C4) and three variable domains (V1 to V3). (B) The rif region (nucleotides 507 to 533 [E. coli numbering]), associated with rifampin resistance in M. tuberculosis, is flanked by HCR5 (nucleotides 444 to 454) and HCR6 (nucleotides 547 to 577).
Nucleotide sequencing.
The nucleotide sequences (306 bp) of the purified PCR products (342 bp) were directly determined with forward and reverse primers, using an Applied Biosystems model 373A automatic sequencer and a BigDye Terminator Cycle Sequencing kit (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom). For the sequencing reaction, 60 ng of PCR-amplified DNA, 3.2 pmol of either the forward or the reverse primer, and 8 μl of BigDye Terminator RR mix (Perkin-Elmer Applied Biosystems; part no. 4303153) were mixed, and the contents were adjusted to a final volume of 20 μl by addition of distilled water. The reaction was run with 5% (vol/vol) dimethyl sulfoxide for 30 cycles of 15 s at 95°C, 10 s at 50°C, and 4 min at 60°C. Both strands were sequenced as a cross-check.
Sequence analysis.
Sequences were aligned by using the multiple alignment algorithm in the MegAlign package (Windows version 3.12e; DNASTAR, Madison, Wis.). The rpoB sequences of Rhodococcus equi, Nocardia nova, and Corynebacterium diphtheriae were simultaneously determined, and those of B. subtilis, Staphylococcus aureus, and E. coli from GenBank (accession no. L24376, X64172, and V0040, respectively) were also used. A phylogenetic tree of the mycobacteria was constructed by using the MEGA program (21). A bootstrap analysis (100 repeats) using R. equi as the outgroup was performed to evaluate the topology of the phylogenetic tree.
Identification of clinical isolates.
The clinical isolates (107 strains) of mycobacteria, except for six specimens of M. leprae, were identified by blind testing. They had been isolated and identified by conventional methods at KNTA and provided without information.
Nucleotide sequence accession numbers.
The rpoB gene sequences determined for the mycobacterial reference strains and other microorganisms have been deposited in GenBank (accession no. AF057449 to AF057496).
RESULTS
rpoB sequences of reference strains.
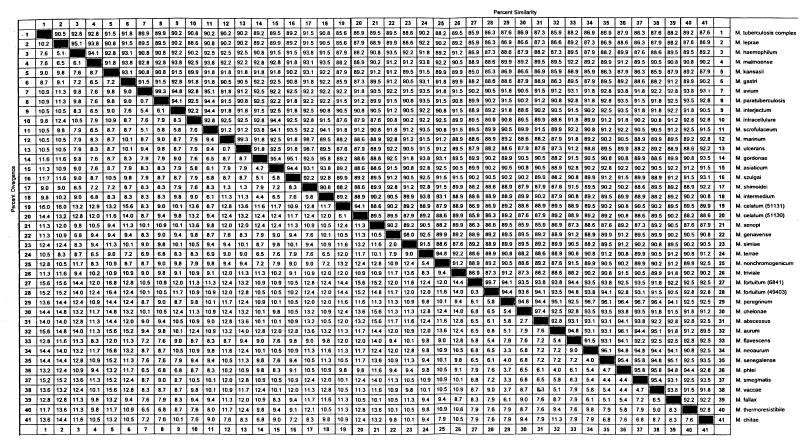
rpoB DNAs (342 bp) were amplified from 44 reference strains of mycobacteria and species of closely related genera such as R. equi and N. nova, while no amplifications from nonmycobacteria that can usually be isolated from the human body were observed (data not shown). The nucleotide sequences (306 bp) of amplified DNAs were determined and compared. The G+C contents of these amplified DNAs were 63 to 69%, reflecting the G+C contents of the total DNAs of the genus Mycobacterium (62 to 70%). No insertions or deletions were observed. The determined nucleotide sequences were compared pairwise for similarity; the results showed that the 44 mycobacterial strains were closely related to each other and were distinct from other genera. In general, 85 to 100% similarity (interspecies divergence, 0 to 15%) was observed among mycobacterial species (Fig. 2). Interestingly, the members of the M. tuberculosis complex had identical sequences. Pathogenic M. kansasii were easily differentiated from nonpathogenic M. gastri (93.1% similarity).
FIG. 2.
Sequence pair distances of 44 reference species of mycobacteria determined by using the Clustal program with weighted residue weight table (MegAlign package [Windows version 3.12e]; DNASTAR, Madison, Wis.). M. tuberculosis complex consists of M. tuberculosis, M. bovis, M. bovis BCG, and M. africanum.
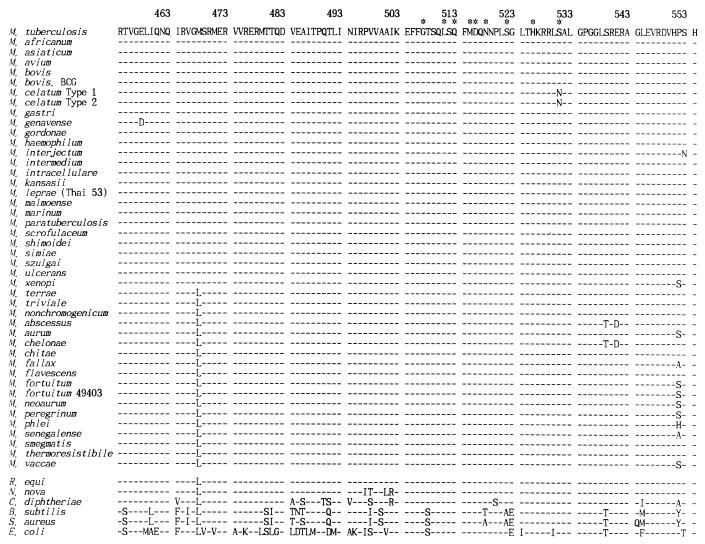
Amino acid sequences of reference strains.
The deduced amino acid sequences of amplified rpoB DNAs comprised 101 amino acid residues (R454 to H554 [E. coli numbering]) (Fig. 3). Because the variations in the nucleotide sequence were usually observed in the last nucleotide of a codon, amino acid sequences among mycobacterial species were highly conserved (97 to 100% similarity). Interestingly, instead of an M468 (ATG) residue, which was found in most of the slowly growing mycobacteria, rapidly growing mycobacteria and the M. terrae complex had an L468 (CTG, TTG, or CTC), as did nonmycobacteria. Among the investigated mycobacteria, only M. celatum, which has been reported to be completely rifampin resistant (6) upon susceptibility testing, had an N531 residue (AAC). This position is one of the most frequent sites of mutation associated with rifampin resistance in M. tuberculosis (S531→L [TCG→TTG]) (18, 34).
FIG. 3.
Deduced amino acid sequences (R454 to H554 [E. coli numbering]) of rpoB DNAs from 44 reference strains of mycobacteria and 6 nonmycobacterial species. Nucleotide sequences of B. subtilis, S. aureus, E. coli (GenBank accession no. L24376, X64172, and V0040, respectively, were used for comparisons). Asterisks indicate amino acids that are frequently changed in rifampin-resistant M. tuberculosis.
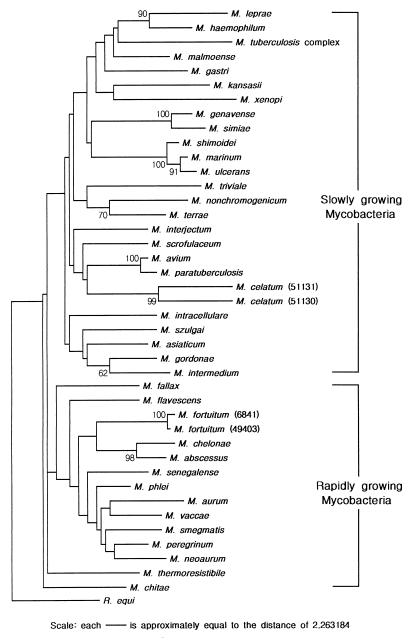
Phylogenetic tree.
A phylogenetic tree, which provided the basis for species differentiation in the genus Mycobacterium, was constructed by the neighbor-joining method (Fig. 4). All tested species showed good separation. Rapidly growing species were united, to the exclusion of the slowly growing line of descent, as in the phylogenetic tree based on the 16S rRNA or rDNA sequences (25, 27, 31). Clustering of pathogenic and potentially pathogenic species was another characteristic. M. fortuitum, M. chelonae, and M. abscessus, which are included in the taxonomic group of pathogenic, rapidly growing mycobacteria, formed a distinct cluster. M. haemophilum was the species most similar to M. leprae, in accordance with the 23S rRNA sequence analysis (32). Slowly growing, pathogenic M. kansasii and nonpathogenic M. gastri were clearly separated, though the two were not distinguished by the 16S rRNA sequence analysis (27). Also, M. szulgai was separated from M. malmoense in the tree. M. intracellulare, long regarded as being closely related to M. avium, was separated distantly from the latter species, which was clustered with M. paratuberculosis, M. celatum, and M. scrofulaceum. The reliability of the phylogenetic tree inferred was verified by the bootstrap method, using R. equi as the outgroup.
FIG. 4.
Phylogenetic tree based on rpoB gene sequences (GenBank accession no. AF057449 to AF057496) shows the relationships of the 44 reference strains of mycobacteria, including M. tuberculosis complex (M. tuberculosis, M. bovis, M. bovis BCG, and M. africanum). ATCC numbers of M. celatum and M. fortuitum strains are shown in parentheses. This tree was constructed by the neighbor-joining method. Topology was also evaluated by bootstrap analysis (MEGA program, 100 repeats, with R. equi as the outgroup). The numerical values in the tree represent bootstrap results. The distance between two strains is the sum of the branch lengths between them.
Identification of clinical isolates.
Nucleotide sequence variations of rpoB among the clinical isolates were observed. The range of variation among the strains in each species was narrow (intraspecies divergence, <1%). In general, only 1 or 2 nucleotide variations were observed among the clinical isolates (99.0 to 100% similarity) (Table 2). Variants of M. tuberculosis were found only among the rifampin-resistant strains. M. leprae, which has not yet been cultivated in vitro, was successfully identified by PCR-rpoB sequence analysis of punch biopsy specimens. The clinical isolates of M. avium complex were divided into two groups, M. avium and M. intracellulare. Separately, these strains were also identified by DT1-DT6 PCR (9) (data not shown). Interestingly, the rpoB sequence of one strain (Kan 2-13) among the 32 clinical isolates of M. kansasii exhibited a very low level of similarity (94.8%) to that of the reference strain (ATCC 12478), while those of the 31 others were identical. However, the strain was again confirmed as M. kansasii by the conventional method. By PCR-restriction fragment length polymorphism analysis (PRA) of the hsp65 gene (8) it was identified as M. kansasii group II (data not shown). By referring to the phylogenetic tree and by using reference strains, we applied this procedure to the clinical isolates and thus could identify various mycobacterial species, which had been confirmed in the conventional way. The one strain that showed a low level of similarity to the M. kansasii reference strain was distinctively separated (data not shown).
TABLE 2.
Sequence pair distances observed among the clinical isolates
| Species | Range of % similarity | No. of strains |
|---|---|---|
| M. tuberculosis | 99.0–100 | 46 |
| Rifampin susceptible | 100 | 22 |
| Rifampin resistant | 99.0–100 | 24 |
| M. avium complex | 99.3–100 | 18 |
| M. avium | 99.7–100 | 5 |
| M. intracellulare | 99.3–100 | 13 |
| M. kansasiia | 100 | 31 |
| M. leprae | 100 | 6 |
| M. fortuitum | 99.7–100 | 5 |
| M. szulgai | 99.0 | 6 |
One strain among 32 clinical isolates of M. kansasii showed 94.8% similarity.
Our results show that the rpoB sequences from the 44 reference strains of mycobacteria provide another basis for determining systematic phylogenetic relationships that can be used to identify clinically isolated mycobacteria which are pathogenic or potentially pathogenic. The PCR-mediated sequence analysis of rpoB DNA can thus be regarded as a feasible method for the identification of mycobacteria.
DISCUSSION
Because of the frequent reports and significance of MOTT infections, there is an increasing need for rapid characterization of clinically isolated mycobacteria. In cases involving mycobacterial infection, definitive diagnosis is dependent on the isolation and identification of causative agents. However, it is not easy to identify these mycobacteria by conventional methods. An alternative, when identification by conventional methods is difficult or fails, is the use of mycobacterial systematics based on genetic analysis to distinguish mycobacteria at the species level. Phylogenetic relationships of a causative agent can be inferred from the nucleotide sequences of amplified DNA. Determining phylogenetic relationships is useful, especially when conventional bacteriologic tests are inapplicable, e.g., when fastidious or uncultivable mycobacteria are to be identified.
Through an extensive sequence analysis of 16S rRNA or its coding rDNA (25, 27, 31), 23S rRNA (32), the 16S–23S rDNA internal transcribed spacer (12, 29), and the dnaJ gene (33), phylogenetic relationships among mycobacterial species have been defined. The 16S rRNA sequence analysis suggested a way to identify mycobacteria by characterizing species-specific nucleotide sequences (4, 17, 28). Clinical isolates of Mycobacterium spp. were identified by a direct sequence determination of amplified 16S rRNA gene fragments (20, 30); in addition, prompt recognition of previously undescribed species was possible. However, 16S rRNA sequence-based phylogenetic analysis has its limitations (7, 11). Despite major differences in clinical importance and different phenotypic traits, M. gastri and M. kansasii have been shown to be identical by 16S rRNA-based analysis (27). This inability to distinguish these two species by this technique has been debated, and it was suggested that additional comparative analyses were required to determine the relationship between these two organisms (35). Problems due to the rRNA gene copy number also occurred. Though it may be a rare occurrence, variations in the sequence of two copies of a specific 16S rDNA gene have been described (3, 23, 26), despite the suggestion that mycobacteria have only a single copy of the rpoB gene per genome (10). Ambiguous results that could be attributed to multiple copies of the target gene were not observed in the rpoB chromatogram obtained by automatic sequencing.
Recently, while we were preparing the manuscript for this article, a study using the rpoB gene and a DNA chip for genotyping and mycobacterial species identification (14), in which the rpoB sequences of a limited number of mycobacterial species (GenBank accession no. AF060279 to AF060367) comprising codons 482 to 715 were posted, was reported. The nucleotide sequence of their forward primer corresponded to the sequence of the M. tuberculosis rpoB gene. The nucleotide sequences were determined after the amplified DNAs were cloned. Thus, even the rpoB genes of M. avium (ATCC 25291; GenBank accession no. AF060366) and its clinical isolates have the same sequences as the M. tuberculosis gene. This ambiguity may have resulted from their sequencing method (cloning-sequencing). However, according to our results, MOTT genes showed different sequences in the corresponding primer region. For example, the M. avium gene had 3 nucleotides that differed from those of the M. tuberculosis gene. We could determine all of the sequences by the PCR-direct sequencing method.
It is difficult to believe that the partial DNA sequences (306 bp) of a single gene can reflect the phylogenetic relationships of many mycobacteria, since only a small potion of the whole rpoB gene was used in this study. As the hypervariable region A in 16S rDNA (∼350 bp), which is widely used, the use of an rpoB DNA fragment may not be sufficient to differentiate mycobacterial species under certain circumstances. Analysis of the rest of the rpoB gene may be desirable when the analysis of a fragment is not sufficient. Thus, when additional comparative analysis of mycobacterial species is required, the use of rpoB sequence analysis is strongly recommended. This technique is a simple and feasible method that nicely complements the 16S rRNA sequence analysis. To reflect more comprehensive phylogenetic relationships among the currently recognized mycobacteria, we tried to include as many species as possible. Our results reflected the classical distinction depending on the growth rate and characteristic clustering of pathogenic species obtained by other methods, which supported the usefulness of the rpoB sequence analysis.
The level of divergence of rpoB among the characterized species was usually less than 1.0% (i.e., more than 99.0% similarity was generally exhibited). However, one strain among the M. kansasii isolates showed only 94.8% similarity to the reference strain. One possible and reasonable explanation is the M. kansasii heterogeneity that has been recently revealed by molecular genotyping studies of the hsp65 PRA (8, 24) and 16S–23S rRNA gene internal transcribed spacer sequences (1, 29). By the use of PRA of the hsp65 gene (8), this strain was identified as M. kansasii group II, which again provided another good example of the advantage of rpoB sequence analysis.
Compared with 16S rRNA gene sequence analysis, rpoB sequence analysis offers several advantages. First, a target DNA, i.e., a single site without a deletion or an insertion, is sufficiently small enough to be sequenced directly in both directions at once and contains enough information to distinguish most of the currently recognized mycobacteria. Thus, there is no need to analyze several hypervariable regions or to sequence the nearly 1.5 kb of 16S rDNA. Second, problems due to the 16S rRNA (or rDNA) sequences can be eliminated. The inferred phylogenetic tree distinguished M. kansasii from M. gastri and M. szulgai from M. malmoense. Third, to be a good marker for species differentiation, a target gene should be stable and, at the same time, sequence variations should occur randomly. However, for the differentiation of species, an extremely conserved or highly variable gene may not be adequate. In other words, the high similarity value or narrow range of 16S rRNA sequences (94.3 to 100% similarity) may preclude discrimination. On the other hand, the sequence variation of rpoB DNA among mycobacteria was observed to be moderate (85 to 100% similarity). Fourth, further useful information relating to the rifampin susceptibility of a particular species (or strain) of mycobacterium is contained in rpoB sequences. As shown in our results, M. celatum, which was known to be completely rifampin resistant (6), had an N531 residue (AAC). Similar findings on natural resistance to rifampin due to the primary amino acid sequence of the β subunit of RNA polymerase have been found for Borrelia burgdorferi (S531→N) and Spiroplasma citri (S531→T) (2, 13). That is one of the most frequent sites of mutation rendering rifampin resistance in M. tuberculosis (S531→L) (18, 34).
We have demonstrated that the comparative analysis of rpoB sequences is an efficient procedure which, through the use of PCR-automated DNA sequencing, permits the identification of clinical isolates to the species level. This procedure provides a molecular tool for the diagnosis of mycobacterial infections. Because of its relative simplicity and rapidity, the method can be completed within two working days.
ACKNOWLEDGMENTS
This work was supported by grant 97-N1-02-01-A-08 from the National Project for Medical Research, funded by the Korean Ministry of Science and Technology (MOST), and in part by the Academic Research Fund (GE 97-000032) of the Korean Ministry of Education.
REFERENCES
- 1.Abed Y, Bollet C, de Micco P. Demonstration of Mycobacterium kansasii species heterogeneity by the amplification of the 16S–23S spacer region. J Med Microbiol. 1995;43:156–158. doi: 10.1099/00222615-43-2-156. [DOI] [PubMed] [Google Scholar]
- 2.Alekshun M, Kashlev M, Schwartz I. Molecular cloning and characterization of Borrelia burgdorferi rpoB. Gene. 1997;186:227–235. doi: 10.1016/s0378-1119(96)00714-7. [DOI] [PubMed] [Google Scholar]
- 3.Bercovier H, Kafri O, Sela S. Mycobacteria possess a surprisingly small number of ribosomal RNA genes in relation to the size of their genome. Biochem Biophys Res Commun. 1986;136:1136–1141. doi: 10.1016/0006-291x(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 4.Böddinghaus B, Rogall T, Flohr T, Blöcker H, Böttger E C. Detection and identification of mycobacteria by amplification of rRNA. J Clin Microbiol. 1990;28:1751–1759. doi: 10.1128/jcm.28.8.1751-1759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boor K J, Dunkan M L, Price C W. Genetic and transcriptional organization of the region encoding the β subunit of Bacillus subtilis RNA polymerase. J Biol Chem. 1995;270:20329–20336. doi: 10.1074/jbc.270.35.20329. [DOI] [PubMed] [Google Scholar]
- 6.Butler W R, O’Connor S P, Yakrus M A, Smithwick R W, Plikaytis B B, Moss C W, Floyd M M, Woodley C L, Kilburn J O, Vadney F S, Gross W M. Mycobacterium celatum sp. nov. Int J Syst Bacteriol. 1993;43:539–548. doi: 10.1099/00207713-43-3-539. [DOI] [PubMed] [Google Scholar]
- 7.Clayton R A, Sutton G, Hinkle P S, Jr, Bult C, Fields C. Intraspecific variation in small-subunit rRNA sequences in GenBank: why single sequences may not adequately represent prokaryotic taxa. Int J Syst Bacteriol. 1995;45:595–599. doi: 10.1099/00207713-45-3-595. [DOI] [PubMed] [Google Scholar]
- 8.Devallois A, Goh K S, Rastogi N. Rapid identification of mycobacteria to species level by PCR-restriction fragment length polymorphism analysis of the hsp65 gene and proposition of an algorithm to differentiate 34 mycobacterial species. J Clin Microbiol. 1997;35:2969–2973. doi: 10.1128/jcm.35.11.2969-2973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devallois A, Picardeau M, Paramasivan C N, Vincent V, Rastogi N. Molecular characterization of Mycobacterium avium complex isolates giving discordant results in AccuProbe tests by PCR-restriction enzyme analysis, 16S rRNA gene sequencing, and DT1-DT6 PCR. J Clin Microbiol. 1997;35:2767–2772. doi: 10.1128/jcm.35.11.2767-2772.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnabella V, Martiniuk F, Kinney D, Bacerdo M, Bonk S, Hanna B, Rom W N. Isolation of the gene for the beta subunit of RNA polymerase from rifampicin-resistant Mycobacterium tuberculosis and identification of new mutations. Am J Respir Cell Mol Biol. 1994;11:639–643. doi: 10.1165/ajrcmb.11.6.7946393. [DOI] [PubMed] [Google Scholar]
- 11.Fox G E, Wisotzkey J D, Jurtshunk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 12.Frothingham R, Wilson K H. Molecular phylogeny of the Mycobacterium avium complex demonstrates clinically meaningful divisions. J Infect Dis. 1994;169:305–312. doi: 10.1093/infdis/169.2.305. [DOI] [PubMed] [Google Scholar]
- 13.Gaurivaud P, Laigret F, Bove J-M. Insusceptibility of members of the class Mollicutes to rifampin: studies of the Spiroplasma citri RNA polymerase β-subunit gene. Antimicrob Agents Chemother. 1996;40:858–862. doi: 10.1128/aac.40.4.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingeras T R, Ghandour G, Wang E, Berno A, Small P M, Drobniewski F, Alland D, Desmond E, Holodniy M, Drenkow J. Simultaneous genotyping and species identification using hybridization pattern recognition analysis of generic Mycobacterium DNA arrays. Genome Res. 1998;8:435–448. doi: 10.1101/gr.8.5.435. [DOI] [PubMed] [Google Scholar]
- 15.Hetherington S V, Watson A S, Patrick C C. Sequence and analysis of the rpoB gene of Mycobacterium smegmatis. Antimicrob Agents Chemother. 1995;39:2164–2166. doi: 10.1128/aac.39.9.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honore N T, Bergh S, Chanteau S, Doucet-Populaire F, Eiglmeier K, Garnier T, Georges C, Launois P, Limpaiboon T, Newton S, Niang K, Del Portillo P, Ramesh G R, Reddi P, Ridel P R, Sittisombut N, Wu-Hunter S, Cole S T. Nucleotide sequence of the first cosmid from the Mycobacterium leprae genome project: structure and function of the Rif-Str regions. Mol Microbiol. 1993;7:207–214. doi: 10.1111/j.1365-2958.1993.tb01112.x. [DOI] [PubMed] [Google Scholar]
- 17.Hughes M S, Skuce R A, Beck L-A, Neill S D. Identification of mycobacteria from animals by restriction enzyme analysis and direct DNA cycle sequencing of polymerase chain reaction-amplified 16S rRNA gene sequences. J Clin Microbiol. 1993;31:3216–3222. doi: 10.1128/jcm.31.12.3216-3222.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim B-J, Kim S-Y, Park B-H, Lyu M-A, Park I-K, Bai G-H, Kim S-J, Cha C-Y, Kook Y-H. Mutations in the rpoB gene of Mycobacterium tuberculosis that interfere with PCR–single-strand conformation polymorphism analysis for rifampin susceptibility testing. J Clin Microbiol. 1997;35:492–494. doi: 10.1128/jcm.35.2.492-494.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirschner P, Meier A, Böttger E C. Genotypic identification and detection of mycobacteria—facing novel and uncultured pathogens. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 173–190. [Google Scholar]
- 20.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F-C, Böttger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Tamura K, Masatoshi N. MEGA: molecular evolutionary genetics analysis, version 1.01. University Park: The Pennsylvania State University; 1993. [Google Scholar]
- 22.Miller L P, Crawford J T, Shinnick T M. The rpoB gene of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:805–811. doi: 10.1128/aac.38.4.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninet B, Monod M, Emler S, Pawlowski J, Metral C, Rohner P, Auckenthaler R, Hirschel B. Two different 16S rRNA genes in a mycobacterial strain. J Clin Microbiol. 1996;34:2531–2536. doi: 10.1128/jcm.34.10.2531-2536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Picardeau M, Prod’hom G, Raskine L, LePennec M P, Vincent V. Genotypic characterization of five subspecies of Mycobacterium kansasii. J Clin Microbiol. 1997;35:25–32. doi: 10.1128/jcm.35.1.25-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitulle C, Dorsch M, Kazda J, Wolters J, Stackebrandt E. Phylogeny of rapidly growing members of the genus Mycobacterium. Int J Syst Bacteriol. 1992;42:337–343. doi: 10.1099/00207713-42-3-337. [DOI] [PubMed] [Google Scholar]
- 26.Reischl U, Feldmann K, Naumann L, Gaugler B J M, Ninet B, Hirschel B, Emler S. 16S rRNA sequence diversity in Mycobacterium celatum strains caused by presence of two different copies of 16S rRNA gene. J Clin Microbiol. 1998;36:1761–1764. doi: 10.1128/jcm.36.6.1761-1764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogall T, Wolters J, Flohr T, Böttger E C. Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int J Syst Bacteriol. 1990;40:323–330. doi: 10.1099/00207713-40-4-323. [DOI] [PubMed] [Google Scholar]
- 28.Rogall T, Flohr T, Böttger E C. Differentiation of Mycobacterium species by direct sequencing of amplified DNA. J Gen Microbiol. 1990;136:1915–1920. doi: 10.1099/00221287-136-9-1915. [DOI] [PubMed] [Google Scholar]
- 29.Roth A, Fischer M, Hamid M E, Michalke S, Ludwig W, Mauch H. Differentiation of phylogenetically related slowly growing mycobacteria based on 16S–23S rRNA gene internal transcribed spacer sequences. J Clin Microbiol. 1998;36:139–147. doi: 10.1128/jcm.36.1.139-147.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Springer B, Stockman L, Teschner K, Roberts G D, Böttger E C. Two-laboratory collaborative study on identification of mycobacteria: molecular versus phenotypic methods. J Clin Microbiol. 1996;34:296–303. doi: 10.1128/jcm.34.2.296-303.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stahl D A, Urbance J W. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J Bacteriol. 1990;172:116–124. doi: 10.1128/jb.172.1.116-124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stone B B, Nietupski R M, Breton G L, Weisburg W G. Comparison of Mycobacterium 23S rRNA sequences by high-temperature reverse transcription and PCR. Int J Syst Bacteriol. 1995;45:811–819. doi: 10.1099/00207713-45-4-811. [DOI] [PubMed] [Google Scholar]
- 33.Takewaki S-I, Okuzumi K, Manabe I, Tanimura M, Miyamura K, Nakahara K-I, Yazaki Y, Ohkubo A, Nagai R. Nucleotide sequence comparison of the mycobacterial dnaJ gene and PCR-restriction fragment length polymorphism analysis for identification of mycobacterial species. Int J Syst Bacteriol. 1994;44:159–166. doi: 10.1099/00207713-44-1-159. [DOI] [PubMed] [Google Scholar]
- 34.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 35.Wayne L G, Sramek H A. Agents of newly recognized or infrequently encountered mycobacterial diseases. Clin Microbiol Rev. 1992;5:1–25. doi: 10.1128/cmr.5.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams D L, Gillis T P, Booth R J, Looker D, Watson J D. The use of a specific DNA probe and polymerase chain reaction for the detection of Mycobacterium leprae. J Infect Dis. 1990;162:193–200. doi: 10.1093/infdis/162.1.193. [DOI] [PubMed] [Google Scholar]