Abstract
The influence of microbiological species has gained increased visibility and traction in the medical domain with major revelations about the role of bacteria on symbiosis and dysbiosis. A large reason for these revelations can be attributed to advances in deep-sequencing technologies. However, the research on the role of fungi has lagged. With the continued utilization of sequencing technologies in conjunction with traditional culture assays, we have the opportunity to shed light on the complex interplay between the bacteriome and the mycobiome as they relate to human health. In this review, we aim to offer a comprehensive overview of the human mycobiome in healthy and diseased states in a systematic way. The authors hope that the reader will utilize this review as a scaffolding to formulate their understanding of the mycobiome and pursue further research.
Keywords: dysbiosis, mycobiome, health, disease, microbiome, commensals
Introduction
The role of commensal and pathologic bacteria in influencing human health has been studied for over a century with numerous direct correlations to disease states as well as symbiotic conditions (Ubeda et al., 2017; Ranjan and Dongari-Bagtzoglou, 2018; Libertucci and Young, 2019). While we have made great strides in our understanding of bacteria, our understanding of fungal commensalism remains limited. Not until recently were fungi thought to be a crucial aspect of the human microbiome, now individually referred to as the mycobiome. Furthermore, due to increasing utilization of antimicrobials and the resultant damage they cause to our microbiome, we are just starting to appreciate the beneficial roles of microbes. With increased understanding of the impact that microbes have in the human physiology and pathology, and advancing technology, there has been an emphasis on exploring the interplay between bacterial and fungal communities as it relates to human health (Ghannoum, 2016).
While bacteria comprise the overwhelming majority of biodiversity (>99%) in humans, the shear sizes of the fungal cells, relative to their bacterial counterparts, compensate for this difference (Seed, 2014). Previously, fungi were considered to be irrelevant when they were co-isolated with bacteria, or were considered to be environmental contaminants (Brown et al., 2012; Huffnagle and Noverr, 2013). Now we understand that fungal flora is crucial in maintenance, metabolism, and immunity of the microbiota and the host (Qin et al., 2010). Healthy state is an interplay between an intact immune system, genetic polymorphisms, and microbiota (Iliev et al., 2012). This interplay between bacteria, fungi, and the host are integral in the symbiosis and dysbiosis seen in human health (Figure 1). In this comprehensive review, we will explore the mycobiome present in humans that contribute in a symbiotic manner, as well as the alterations in diversity that may lead to dysbiosis and pathologic states.
FIGURE 1.
Mycobiome affects a wide spectrum of diseases across various niches in the human body. The cooperative evolution of fungi and bacteria is a complex interplay, where it may be mutualistic in some conditions, while antagonistic in others.
Advances in Technology to Evaluate Mycobiome
Traditionally, fungi have been evaluated using culture-based protocols; however, with advances in deep-sequencing technologies, investigators are able to study fungi that were previously unculturable in a lab. These developments have exponentially accelerated our understanding, and have led to the “human microbiome project,” which focused on the bacterial community, an undertaking that was initiated over a decade ago (Turnbaugh et al., 2007). With increased knowledge, we come to appreciate the complexity of fungi in a more detailed and comprehensive manner. Furthermore, we are able to evaluate the inter-kingdom interactions to improve our understanding of the microbial diversity and the complex network of species that yield the healthy and pathologic states.
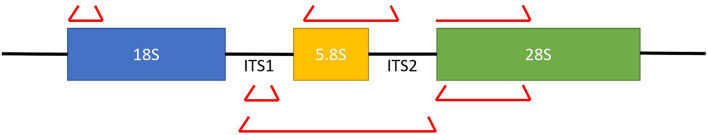
In comparison to the Sanger sequencing method, next generation sequencing allows us to perform community-level analyses in a more efficient and cost-effective manner (Ackerman and Underhill, 2017). Multiple gene loci have been targeted for fungal gene profiling including 18S ribosomal rRNA (small subunit) and 28S rRNA region (large subunit), along with the two internal transcribed spacer units 1 and 2 (ITS1/ITS2) (Dollive et al., 2012). The small subunit is highly conserved and lacks discriminatory power, while the larger subunit is more hypervariable in comparison to the 18s ribosomal rRNA. Overall, the ITS1 and ITS2 are used most commonly to detect fungi as it has the best combination of variability to identify species while also conserving targets that can be readily targeted (Figure 2; Dollive et al., 2012; Findley et al., 2013). The ITS sequences are aligned to the operational taxonomic units (OTUs) in fungal databases to identify species. A standing challenge with further examination of the mycobiome remains the abundant fungal diversity in the environment; however, with concerted efforts we are able to identify approximately 80% of the mycobiome found in fecal and mucosal samples (Findley et al., 2013; Underhill and Iliev, 2014; Tang et al., 2015; Motooka et al., 2017). Despite this, our understanding of the fungal world in limited due to their vast diversity with limited effort from the scientific community, variations in the DNA extraction techniques, and lack of well-maintained and comprehensive database for sequence comparison.
FIGURE 2.
A diagrammatic representation of the various fungal ribosomal genes that can be targeted for sequencing. The ITS sequences provide the best balance of conservation which facilitates targeting, while also providing diversity to differentiate between organisms. The red brackets represent primer sequences that can be utilized to sequence the desired region.
Significance of Mycobiome in Cross-Kingdom Interactions
As mentioned previously, there is increased appreciation for inter-kingdom interactions between bacteria and fungi as they relate to human health. For example: there is increasing evidence that infections may frequently be co-inhabited by fungi and bacteria. This is certainly true in cases of wound infections, but also in cases of “isolated” fungal infections. Various studies have found reported cases of candidemia to have mixed microbial infections up to 38% of the time (Hermann et al., 1999; Klotz et al., 2007; Kim et al., 2013). In particular, our experiences with chronic wound patients have suggested that bacteria and fungi co-inhabit the wound bed to increase pathogenicity and tolerance of antibiotics (Ghannoum, 2016).
In a 2016 study by Kalan et al. (2016), the authors utilized an in vitro model to demonstrate a close interaction between bacterial and fungal cells, with yeast cells forming the biofilm core, and bacteria forming the biofilm periphery. In this model, the fungi benefit by obtaining virulence factors (i.e., the ability to form hyphae and the ability to secrete extracellular enzymes), while the bacteria gain antibacterial tolerance provided by the protective fungal matrix (Hoarau et al., 2016; Kalan et al., 2016). Similar interplay has been reported with Escherichia coli/Staphylococcus aureus and Candida albicans (De Brucker et al., 2015; Kong et al., 2016). Our group has demonstrated a cooperative survival technique utilizing mixed biofilms between the bacteriome and the mycobiome in Crohn’s disease (CD) patients, where we see inter-kingdom association between Candida tropicalis and Serratia marcescens/E. coli (Hoarau et al., 2016). Figure 3 provides an in vitro and in vivo representation of this finding. These findings have also been demonstrated by Sokol et al. (2017), who show positive correlation between Saccharomyces and Malassezia with various bacterial species in CD. Similarly, patients with cystic fibrosis (CF) who suffer from Psuedomonas aeruginosa infection are frequently found to have persistent infections from Aspergillus fumigatus or C. albicans compared to patients without these fungi (Amin et al., 2010; Chotirmall et al., 2010). These findings further reinforce our appreciation of the inter-kingdom interplay that may need to be addressed in treating complex chronic wounds, patients with severe CD, or patients with CF who suffer recurrent pulmonary infections.
FIGURE 3.
(A) A microscopic image of an in vitro biofilm. (B) A medial section of a mouse colon demonstrati ng microorganisms present on the villi under a protective barrier formed by the biofilm matrix.
In contrast to synergistic interactions highlighted above, microbes may also interact in an antagonistic manner, where one organism may inhibit the growth of another organism (Brogden et al., 2005; Rall and Knoll, 2016). To add another layer of complexity, the host and the environment may further impact the proliferation or inhibition of microbial growth. These factors dynamically influence the synergistic or antagonistic influence of various microbes (Wargo and Hogan, 2006; Kapitan et al., 2018). The prevalence of certain organisms and the variations in biodiversity may lead to a state of dysbiosis in the host, which may make the host vulnerable to various pathologic conditions. Kruger et al. (2019) aptly describe the interactions between fungi and bacteria as one of five modes: (1) physical interaction, (2) environmental interaction, (3) biofilm formation, (4) chemical and metabolic interaction, and (5) competitive interaction.
A unique challenge with studying the bacterio- and the myco-biome simultaneously is the difficulty with extracting nucleic acids from these species without a biased protocol. The rigid cell walls of fungi, and the mechanical, chemical, and enzymatic steps needed to effectively retrieve nucleic acids from bacteria can inherently introduce bias into the protocol (Wesolowska-Andersen et al., 2014; Vesty et al., 2017). Consequently, most protocols are targeted at identifying specific species, which may not permit other species to be properly amplified using nucleic acid based assays (Underhill and Iliev, 2014). This challenge leads to variations in the micro- and mycobiome analysis between individuals and protocols. To date there is no established gold standard protocol to evaluate the inter-kingdom interactions between bacteria and fungi (Tang et al., 2015; Enaud et al., 2018). To complicate matters further, there are often transient colonizers in various niches, which further skew genetic analyses or culture results (Chen et al., 2011). All of these factors combined, lead to a consistent challenge and require a disciplined approach to studying the microbial flora in human physiology and disease states. While there remains variability in findings in this domain, there is little debate that the interplay between bacteria and fungi are crucial to a comprehensive understanding of the human health (Brogden et al., 2005; Wargo and Hogan, 2006; Shirtliff et al., 2009; Leclair and Hogan, 2010; Morales and Hogan, 2010; Peleg et al., 2010; Peters et al., 2012; Allison et al., 2016; Forster et al., 2016; Rall and Knoll, 2016; Kapitan et al., 2018).
Mycobiome in Healthy and Diseased States
With the stated limitations in mind, we will explore various niches within human health and characterize the normal fungal flora. Furthermore, we aim to comment on the fungal and bacterial interactions in these domains during a healthy state, and the alterations occurring in dysbiotic states that may lead to pathologic conditions.
Gastrointestinal Mycobiome
Oral Mycobiome
The oral cavity is the initial contact of the human gastrointestinal flora to the environment (Dewhirst et al., 2010). This cavity is a unique reservoir for microbial growth due to the various surfaces present. For example: the microbes on the tongue, a mucosal surface with constant shedding, is drastically different than the teeth, a surface that represents a prime estate for biofilm formation (Arweiler and Netuschil, 2016). As mentioned previously, biofilm are three dimensional structures comprised of bacterial and fungal biomass that increases microbial pathogenicity and tolerance to antibiotics (Costerton et al., 1999). This may lead to pathologic states of gingival plaques and cavities (Zhang Y. et al., 2018).
Compared to the bacteriome, the mycobiome of the oral cavity is vastly understudied. The most common fungal organisms identified in the human oral cavity are Candida (with C. albicans as the most dominant species), Cladosporium, Aureobasidium, Saccharomycetales, Aspergillus, Fusarium, Cryptococcus, and Malassezia (Ghannoum et al., 2010; Dupuy et al., 2014). Once again, the protocols utilized to characterize the biodiversity in the oral cavity leads to inherent variations in the species. In fact, some studies suggest that not only is there significant microbial variability between individuals, but there can be intrapersonal variability in biodiversity through the course of a day (Costello et al., 2009; Caporaso et al., 2011; Stahringer et al., 2012; David et al., 2014a; Ding and Schloss, 2014; Flores et al., 2014; Cameron et al., 2015; Belstrom et al., 2016; Utter et al., 2016). This was further demonstrated within the mycobiome between individuals; however, the intrapersonal fungal diversity remained relatively stable over the course of a 30 day period (Monteiro-da-Silva et al., 2014).
In states of dysbiosis, the fungal and the bacterial biodiversity may be compromised (Selwitz et al., 2007; Takahashi and Nyvad, 2011; Zhang Y. et al., 2018). Once again, the interplay between the bacteriome and mycobiome is critical in these scenarios. For example: a frequent intake of carbohydrates combined with poor oral care may lead to acidification of the niche due to sugar-fermentation and acidification from the bacteria. This change in pH may provide a selective pressure in which certain microbes may thrive while others may perish (Takahashi and Nyvad, 2011). Similarly, exposures to antibiotics or states of immunodeficiency may provide a selective pressure that leads to a decrease in biodiversity while allowing organisms such as Candida to overgrow and proliferate (Costa et al., 2013; Chanda et al., 2017). This state may lead to individuals suffering from oral candidiasis or thrush (Millsop and Fazel, 2016). Interestingly, many newborns who have not achieved adequate biodiversity suffer from thrush, which further implicates the need for microbes in maintaining a competent immune system to keep these organisms under control (Dickstein, 1964; Hoepelman and Dupont, 1996; Jain et al., 2010; Sampaio-Maia and Monteiro-Silva, 2014; Dzidic et al., 2018).
Gingivitis is another condition associated with oral dysbiosis, which may further progress to periodontitis after a period of chronic inflammation (Zhang Y. et al., 2018). Various fungal organisms have been identified in patients suffering from this condition (Hajishengallis, 2014). Specifically, Candida and Rhodoturula, were found in many such patients; however, only C. albicans was found in all patients (Canabarro et al., 2013). Furthermore, C. albicans was most frequently found in patients with bacterial species that operate in mutualistic relationship to increase pathogenicity and lead to virulent plaque formation and increase disease severity (Gross et al., 2012). The two most common bacterial species identified were Streptococcus mutans and Streptococcus viridans. Focusing on S. mutans, this species of bacteria is commonly found in biofilms formed on teeth (Gross et al., 2012; Metwalli et al., 2013). The pathogenic potential of this organism is believed to arise from the formation of extracellular polysaccharides (EPS) by utilizing an exoenzyme called glucosyltransferase (Falsetta et al., 2012, 2014; Metwalli et al., 2013). This enzyme forms biofilms by adhering to surfaces, and ultimately creating an acidic environment, which leads to the formation of dental plaques (Forssten et al., 2010; Metwalli et al., 2013). An in vitro study demonstrated that presence of C. albicans leads to increased expression of glucosyltransferase B, which in turn binds to the surface of C. albicans and leads to formation of extracellular matrix (Falsetta et al., 2014). Furthermore, EPS also sequesters antifungal drugs such as fluconazole, which renders the C. albicans tolerant to the medication (Kim et al., 2018). Lastly, fungal products such as farnesol lead to S. mutans proliferation in high concentration (Kim et al., 2017).
This mutualistic relationship is challenged by Willems et al. (2016), who argue that C. albicans actually increases the pH, which would inhibit S. mutans proliferation. This has been further supported by Barbosa et al. (2016), who found reduced hyphal formation in C. albicans in larval tissue when co-injected with S. mutans.
Despite the conflicting conclusions of the interplay between C. albicans and Streptococcus mutans, there are various other organisms that are known to operate synergistically with C. albicans in infections, and oral candidiasis (Diaz et al., 2012). Streptococcus oralis, Streptococcus sanguinis, and Streptococcus gordonii have been associated with potentiating C. albicans infections and leading to a deep organ dissemination of this pathogenic yeast (Diaz et al., 2012; Xu et al., 2016). In addition, C. albicans has been found to potentiate the growth of anaerobic bacteria in aerobic conditions to exacerbate gingivitis (Karkowska-Kuleta et al., 2018). Furthermore a simultaneous C. albicans infection in an individual with oral lichen planus can lead to exacerbation of symptoms in these individuals who suffer from a chronic inflammatory condition. The diagnosis and treatment can be challenging in these patients, as the typical treatment for lichen planus is steroids, but it may promote further confluence of the C. albicans (Karkowska-Kuleta et al., 2018).
Overall, the impact of oral mycobiome in oral pathologic conditions is incompletely understood, but there is mounting evidence that fungal organisms play a critical role in potentiating disease (Janus et al., 2017). In addition, most studies are performed with limited number of organisms, which is another shortcoming of these studies, as the full complexity of a biodiverse environment cannot be entirely evaluated (Janus et al., 2017).
Intestines
The microbiome of the gut is well characterized compared to others organs due to the ability to readily evaluate this flora via fecal samples (Durban et al., 2011; Carstens et al., 2018; Altomare et al., 2019). However, this methodology is more indicative of the intraluminal organisms rather than the mucosal or the tissue specific microbiota (Zoetendal et al., 2008; Nash et al., 2017). Despite the stated limitation, the mucosal microbiome has been studied extensively in patients with inflammatory bowel disease (IBD) in whom tissue from biopsies and surgeries are routinely collected (Lepage et al., 2005; Sepehri et al., 2007; Ott et al., 2008; Altomare et al., 2019). Similar to the oral microbiome, there is interpersonal variations in the gut flora, which can be influenced by various factors such as diet, gender, age, concurrent comorbidities, medications, and environment (Wu et al., 2011; David et al., 2014b; Theriot et al., 2014; Strati et al., 2016). In addition, characteristics inherent to the intestinal environment such as pH, oxygen level, and the presence of macro- and micronutrients may impact the biodiversity and floral density reffered to as abundance (Qin et al., 2010; Donaldson et al., 2016).
It is estimated that 0.1% of the total gut microbiome is comprised of fungi (Qin et al., 2010; Ianiro et al., 2016). Some of the major fungal taxa identified in healthy individuals include Ascomycota and Basidiomycota (Ott et al., 2008; Hoffmann et al., 2013; Nash et al., 2017). Other genera may include Saccharomyces, Candida, Malassezia, and Cladosporium (Ott et al., 2008; Hoffmann et al., 2013; Nash et al., 2017). Once again, the interplay between bacteria and fungi are integral to maintaining a healthy state. Multiple studies have demonstrated dysbiosis in pathologic states, especially IBD (Scupham et al., 2006; Dollive et al., 2013; Hoarau et al., 2016; Wheeler et al., 2016; Sokol et al., 2017; Angebault et al., 2018; Kalyana Chakravarthy et al., 2018; Skalski et al., 2018; Sovran et al., 2018; El Jurdi et al., 2019). In a study of patients with CD, there was a close association between pathogenic bacteria and the fungus C. tropicalis, with a lower prevalence of beneficial bacteria (Hoarau et al., 2016). Sokol et al. (2017) demonstrated an imbalance of Basidiomycota/Ascomycota ratio in patients with IBD. In fact, dysbiotic states in the intestines have been correlated to the onset of CD(Ott et al., 2008; Hoarau et al., 2016; Sokol et al., 2017).
The study of the intestinal microbiome serves as a great opportunity to discuss the concept of co-occurrence/co-exclusion. Patterns of co-occurrence have traditionally been explained by species that prefer similar environmental attributes and exhibit similar tolerances such as pH, temperature, habitat, and biogeographic distribution. This may include interactions that may be mutualistic or parasitic. Identifying co-occurrence may shed light on essential interactions that impact the ecosystem and the biodiversity. These patterns have been studied both at the macro- and the microbiome level. The human genome project has shed light on the interactions within the human gut and oral flora (Ott et al., 2008; Durban et al., 2011; Hoarau et al., 2016). As the reader progresses through the rest of this manuscript, they should attempt to contextualize these interactions to improve their understanding of the human mycobiome.
Fungal prevalence in the gut can be affected by administration of antifungal medications. In a study by Wheeler et al. (2016), it was demonstrated that Malassezia and C. albicans have a regulatory role in preventing infections from opportunistic organisms such as Aspergillus amstelodami, Epicoccoum nigrum, and Wallemia sebi. These opportunistic pathogens may thrive in a state of dysbiosis initiated by the administration of fluconazole, as demonstrated in a murine model (Wheeler et al., 2016). Ultimately, this imbalance led to acute or chronic colitis in the murine model used. This beneficial effect of fungi was further illustrated by Jiang et al. (2017), where they demonstrated that commensal fungi such as Saccharomyces cerevisiae, C. albicans, or fungal cell wall mannans may recapitulate the protective benefits of intestinal bacteria.
In a study evaluating the beneficial effects of fungi, S. cerevisiae (baker’s yeast), a commonly studied fungi, was used as a probiotic to prophylactically prevent Clostridium difficile infections (Massot et al., 1984). In another porcine study, S. cerevisiae was able to reduce the translocation of enterotoxigenic E. coli (ETEC) to confer mucosal immunity (Lessard et al., 2009). The competitive inhibition towards C. difficile demonstrated by S. cerevisiae does not hold true for other fungal species. Studies examining the protective effects of candidal species on preventing C. difficile related colitis, and mortality have yielded conflicting conclusions. Zuo et al. found that high abundance of C. albicans was correlated with more severe C. difficile infection in a murine model (Zuo et al., 2018). Meanwhile, Markey et al. found C. albicans to serve a protective role in their mouse model (Markey et al., 2020). They concluded that C. albicans co-inhabitation led to an increased diversity in the overall microbiota, which limited the severity of disease cause by C. difficile.
Another notable organism present in the gut that interacts with C. albicans is Enterococcus faecalis (Allison et al., 2016). In previous studies, E. faecalis has been cited to have potentiating effect on C. albicans and thought to promote hyphal formation in endotracheal biofilms (Adair et al., 1999). However, in the gut, E. faecalis is an inhibitory organism to C. albicans (Cruz et al., 2013; Graham et al., 2017). These findings emphasize the variability in inter-kingdom interactions among these organisms based on their niche.
Similar to the dysbiotic effects of antifungal usage, indiscriminate use of antibacterial medications may lead to increased fungal abundance and disseminated candidiasis (Kennedy and Volz, 1985; Mason et al., 2012). In a murine study, the administration of antibiotics led to overwhelming colonization by Candida species, while depleting the abundance of Bacteroidetes and Synergistetes (Erb Downward et al., 2013). It appears that the presence of Bacteroides and Firmicutes are critical to regulating candidal confluence (Erb Downward et al., 2013). Overall, Candida colonization in the gut can be both beneficial or detrimental. It is beneficial in circumstances when it prevents the overcolonization of other organisms and aids in breakdown of food in collaboration with bacteria. Specifically, Candida can breakdown complex carbohydrates leading to by-products that can be metabolized by Prevotella (Kraneveld et al., 2012). Which in turn produce by-products that can be used by Candida. However, it can be detrimental in states where Candida can become overwhelmingly abundant and limit biodiversity.
Lungs
The advancements in culturing techniques and development of next generation sequencing has debunked the previously held idea that the lungs were sterile (Mitchell and Glanville, 2018). Recent studies have demonstrated that much like the gut, the lungs provide a unique habitat for select organisms to thrive. However, compared to the gut, the overall bioburden in the lungs is significantly lower (Budden et al., 2017). Environmental factors that impact organisms in the lungs are pH level, temperature, and oxygen concentration (Ingenito et al., 1987). Similar to other organ systems, there is variability within varying parts of the lungs themselves based on the variations in the factors listed above (West, 1978).
The most common fungi identified in the lungs of healthy subjects are Aspergillus, Candida, Penicillium, Clavispora, and Davidiellaceae (Charlson et al., 2012; van Woerden et al., 2013). In addition, many healthy individuals are colonized by Pneumocystis which is of low pathogenic potential in immunocompetent patients; however, it may lead to pneumonia in immunocompromised individuals such as HIV infected patients (Peterson and Cushion, 2005; Montes-Cano et al., 2007; Sethi and Murphy, 2008; Chabe et al., 2011; Huang et al., 2011).
The study of pulmonary microbiome is nascent; however, there is increasing evidence that the interplay between fungal and bacterial species is crucial in dysbiosis. Majority of literature in this domain is based on findings in patients with CF (Cui et al., 2014; Marsland and Gollwitzer, 2014; Mitchell and Glanville, 2018). Due to a defective CF transmembrane conductance regulator protein, these patients form copious amounts of thick mucus in their lungs (along with pathologic manifestations in other organs) which promotes microbial growth and colonization. Furthermore, poor penetration into the viscous mucus prevents adequate treatment with antimicrobials (Robinson et al., 1999). Co-colonization by multiple species has been documented and leads to higher morbidity and mortality in patients with CF (Peleg et al., 2010). Specifically, bacteria such as P. aeruginosa and S. aureus, along with fungi such as A. fumigatus and C. albicans are commonly isolated in these patients (Delhaes et al., 2012).
Interestingly, the lungs were one of the first niches where the co-inhabitation of fungi and bacteria was investigated (Kerr et al., 1999). The Gram-negative bacteria, P. aeruginosa has been extensively studied as a common cause of infection in patients with CF (Gordee and Matthews, 1969; Kerr, 1994; Hogan et al., 2004; Mowat et al., 2010). Furthermore, the interaction between P. aeruginosa and A. fumigatus and C. albicans have been evaluated in both in vitro and in vivo models (Kerr et al., 1999; Briard et al., 2016; Chen et al., 2018). The findings from these studies further highlight our previous point of the infancy of microbial interactions as they affect human health and disease.
Despite a dedicated effort in evaluating the impact of P. aeruginosa on human health, the in vitro and in vivo models conclude opposite interactions between the listed organisms. The in vitro studies concluded that P. aeruginosa inhibits the growth of A. fumigatus and C. albicans by decreasing filamentation, biofilm formation, and conidia biomass (Mowat et al., 2010). This inhibitory effect is also seen with other fungi that are commonly isolated in the lungs of CF patients (Gordee and Matthews, 1969; Hogan et al., 2004; Kaur et al., 2015; Sedlacek et al., 2015). In return, A. fumigatus has the ability to produce gliotoxin which prevents bacterial biofilm formation by P. aeruginosa along with other organisms (Reece et al., 2018). The exact mechanism of this antagonism is not fully understood (Chen et al., 2018).
In contrast to the in vitro findings, studies using in vivo murine models show a synergistic relationship between P. aeruginosa and C. albicans (Kerr et al., 1999; Hogan et al., 2004; Morales and Hogan, 2010; Roux et al., 2013). This is validated clinically in patients with CF, who have significantly worse clinical course when colonized by two organisms rather than one (Leclair and Hogan, 2010). This points to the major limitation in our understanding of the microbiome as the complexity of host, pathogen, and resources cannot be adequately simulated in an in vitro model. It is hypothesized that these organisms synergistically facilitate each other’s growth by inhibiting the action of the alveolar macrophages (Ader et al., 2011; Roux et al., 2013). This was further supported by a clinical study demonstrating prolonged hospitalization in patients with polymicrobial pneumonia, as compared to an infection caused by a single organism (Azoulay et al., 2006).
Another well-investigated model evaluating the interaction between bacteria and fungi involve Klebsiella species (Kollef et al., 2005). Klebsiella pneumoniae are routinely identified in ventilator-associated pneumonia (Kollef et al., 2005). These species are also found in the gut and lungs of healthy individuals. Nogueira et al. (2019) studied the interaction of Klebsiella with Aspergillus species and found that the bacteria prevent hyphal formation and spore germination in the fungal organism. The more important finding in the study was identifying the importance of physical contact between the two species that leads to the inhibitory effect (Nogueira et al., 2019). Klebsiella also prevents biofilm formation by the Aspergillus species, which further inhibits fungal growth (Fox et al., 2014). But similar to previous examples, there is contradictory evidence when the organisms in the model are changed. For example: Frases et al. (2006) demonstrated a synergistic relationship between Cryptococcus neoformans and Klebsiella. Therefore, it is not possible to generalize the interactions between species, and isolated models may not fully capture the complexity of inter-kingdom interactions. Increased understanding in this domain has clinical implications as immunocompromised patients may suffer from pathologic infections from species such as Mucorales, while the ability to decrease fungal virulence by utilizing another competitive species may offer a therapeutic modality (Ibrahim et al., 2008; Kobayashi and Crouch, 2009; Hover et al., 2016).
Skin
The human skin is colonized by bacteria, fungi, viruses, and parasites. In context of this review, the majority of studies evaluating cutaneous mycobiome have looked at patients with atopic dermatitis, seborrheic dermatitis, dandruff, or dermatophytosis (Gupta et al., 2004; Havlickova et al., 2008; Seebacher et al., 2008; Gaitanis et al., 2012; Clavaud et al., 2013). Overall, most of the fungi found on the skin are commensal organisms; however, some may harbor pathologic potential (Leung et al., 2016). The pathologic manifestations from these organisms mostly occur in patients who are immunocompromised.
While culture based studies have been used in the past to characterize the skin flora, these studies are unable to fully capture the complexity of the microbiome as well as the interactions that occur between the numerous organisms co-colonizing the skin (Seebacher et al., 2008). The predominant organisms identified on the skin of healthy individuals are members of the Malassezia genus, with less common species including Aspergillus, Candida, Saccharomyces, and Epicoccum (Seebacher et al., 2008; Clavaud et al., 2013). The fungal skin mycobiome differs with age as younger individuals have greater fungal diversity and less Malassezia cultured compared to their adult counterparts (Seebacher et al., 2008). The increase in Malassezia occurs sharply around adolescence which may be linked to the change in sebaceous activity. Consequently, availability of lipids provides a needed nutrient for favorable growth of this yeast as most species lack genes for fatty acid synthesis (Jo et al., 2017).
The identification of Malassezia has provided insight into various dermatologic conditions such as tinea versicolor, atopic dermatitis, seborrheic dermatitis (Gupta et al., 2004; Gaitanis et al., 2012). Seborrheic dermatitis is a great example to illustrate the possible implications of fungi in cutaneous manifestations and subsequent therapeutic intervention.
Seborrheic dermatitis is a common skin condition that is associated with the scalp and with Malassezia as the etiologic agent (Shuster, 1984; Pierard et al., 1997; Nakabayashi et al., 2000). Decreased immune status is associated with the disease, suggesting that immunity may play a significant role in predisposing individuals to seborrheic dermatitis. Antifungal shampoos have a profound impact on the condition, implicating fungi as an important player in the progression of the disease. Because of its connection to sebaceous-dependent organisms, it often starts with adolescents (White et al., 2014). In this population, the disease is linked to dandruff and may have severe manifestations (White et al., 2014). Moreover, Malassezia globosa and Malassezia restricta are often isolated from patients (Pedrosa et al., 2014; White et al., 2014). M. globosa and M. restricta are both common components of the skin mycobiome, indicating they are not necessarily the initial cause of the infection. Mycobiome analysis showed a decrease in fungal diversity, with an increase in the Basidiomycota phylum (Park et al., 2012). Additionally, there is a significant decrease in Cryptococcus and Didymella, accompanied by a significant increase in Filobasidium floriforme, Malassezia, and Penicillium (Park et al., 2012).
As the largest organ and the most readily accessible organ in the human body, the skin will be a centerpiece of mycobiome analysis as we move forward. The advancing technology will certainly offer us greater insight into the complex interaction amongst the various organisms that are present on the skin. Moreover, unpublished data from our team shows that interactions between Alternaria and S. aureus occurs in the setting of atopic dermatitis (data not shown).
Genitourinary Tract
Vagina
The microbiome of the vagina is influenced by multiple host and environmental factors such as age, menstruation, pregnancy, health status, hygienic practices, and medications. For example: during reproductive years there is a high supply of glycogen and nutrients which leads to a high confluence of anaerobic organisms, which create an acidic niche (Bradford and Ravel, 2017; Fuochi et al., 2017; Vaneechoutte, 2017; Greenbaum et al., 2019). Candida is the most commonly identified vaginal fungus, followed by Saccharomycetales, Davidiellaceae, Cladosporium, and Pichia in young women (Drell et al., 2013).
One of the most well-studied inter-kingdom interactions in the human body is between Candida and Lactobacilli (Sobel et al., 1998; De Bernardis et al., 2018). Vulvovaginal candidiasis is a common manifestation of dysbiosis that affects 75% of women at some point through their lifetime (Sobel et al., 1998). Most frequently this condition is caused by C. albicans, but may also result from Candida glabrata, Candida tropicalis, and Candida parapsilosis (Sobel et al., 1998; De Bernardis et al., 2018). Alterations in the vaginal microbiome by changes in one of the factors listed above may result in the loss of beneficial bacteria such as Lactobacilli, which may lead to dysbiosis and subsequent candidiasis. In a healthy female, Lactobacillus inhibits candidal growth by competing for nutrients, preventing adhesion, and excreting fungicidal compounds (Basson, 2000; Kaewsrichan et al., 2006; Forster et al., 2016).
Lactobacilli have also been shown to inhibit hyphal formation, which decreases the virulence of Candida (Noverr and Huffnagle, 2004; Nguyen et al., 2011). The excretion of butyric acid by the bacteria has been shown to inhibit germ-tube formation (Noverr and Huffnagle, 2004). This finding has been tested in vivo as well, where pretreatment of Galleria mellonella larvae resulted in improved survival after C. albicans infection (de Oliveira et al., 2017; Ribeiro et al., 2017; Rossoni et al., 2017). Lactobacilli also have an anti-inflammatory effect which decreases production of cytokines such as IL-1β, IL-6, and IL-8 (de Oliveira Perrucini et al., 2019). Interestingly, studies have shown that when Candida is in a state of overgrowth, it keeps the abundance of Lactobacillus under control so that it maintains its dominance (de Oliveira et al., 2017).
With increased understanding of the interplay between bacteria and fungi, Lactobacilli probiotic supplements are widely utilized in women who suffer from recurrent vaginal candidiasis (Tachedjian et al., 2017; Zhang Z. et al., 2018). The supplements can be administered orally or topically, and have been shown to increase the efficacy of antifungal treatments along with preventing relapse (Kovachev and Vatcheva-Dobrevska, 2015; Pendharkar et al., 2015; Tachedjian et al., 2017).
Not all bacteria interact antagonistically with C. albicans. Streptococci (specifically Group B) and E. coli interact synergistically with Candida and can lead to a variety of pregnancy related complications such as preterm birth, low birth weight, and sepsis (Cools et al., 2016). For this reason, pregnant women in the US are routinely screened for Group B Streptococci (GBS) and treated appropriately prior to delivery. Multiple studies have demonstrated the potentiating effect of GBS on Candida which leads to increased virulence of this pathogenic yeast by increasing the adhesion of Candida to the host cells, along with increasing inflammation (Cools et al., 2016; Pidwill et al., 2018; Yu et al., 2018).
Genitourinary Tract
Till recently, urine in healthy individuals without an infection was considered to be sterile (Proctor, 2011; Magistro and Stief, 2019). However, this idea was based primarily on culture based studies, which mainly evaluated aerobic, fast-growing uropathogens (Proctor, 2011). Due to this early perception, the evaluation of urine was not included in the Human Microbiome Project (Turnbaugh et al., 2007). However, recent advancement in microbiome analyses fueled by high-throughput DNA based techniques have not only proven that microbes exist in healthy human urine, but have also shown that multiple urologic conditions may be associated with dysbiotic microbial states (Siddiqui et al., 2011; Khasriya et al., 2013; Hilt et al., 2014; Pearce et al., 2014; Thomas-White et al., 2016).
Compared to some of the other niches described in this review, the urologic investigation of microbes lags behind. There is significant interpersonal variability in urinary microbiome based on various factors such as age, sex, hygiene, diet, and hydration status. Furthermore, the advancement in the domain has been hampered by the lack of standardized sampling methods along with variable protocol for genetic analysis (Aragon et al., 2018). In addition, the relatively low abundance of microbial species, specifically fungi, in urine poses a challenge in DNA extraction and amplification (Ackerman and Underhill, 2017). The majority of investigations primarily focus on the bacterial alterations in the urinary microbiome that has been correlated with conditions such as overactive bladder, urinary incontinence, interstitial cystitis, neuropathic bladder, sexually transmitted infections, and chronic prostatitis/chronic pelvic pain syndrome (Ackerman and Underhill, 2017). Dysbiotic states have also been demonstrated in urologic malignancies such as bladder and prostate cancer (Bucevic Popovic et al., 2018).
The only two fungi that are consistently identified in urine belong to the Saccharomycetes class, which are Candida and Saccharomyces (Pearce et al., 2014; Nickel et al., 2016). Some of the other fungal species that have been identified to be pathogenic to humans include Cryptococcus, Aspergillus, Coccidioides, Histoplasma, and Blastomyces (Enoch et al., 2006; Sanglard, 2016).
The dearth of literature related to mycobiome in the urologic realm underscores the importance of continued efforts in understanding the complex interplay between host, bacteria, and fungi. While patients and practitioners are utilizing probiotics to reduce risk of infection in patients with recurrent urinary tract infections, the literature lags behind the practice. With improved understanding, we may be able to identify ideal candidates who may benefit from probiotic treatments or those who may need microbiome guided therapeutic intervention that may restore a symbiotic state to prevent future recurrences.
Immune System
It is essential that the host immune system not only defends against invasive fungal growth, but also permits colonization of fungal commensals (Iliev and Underhill, 2013). The host’s response to endogenous and exogenous insults is directly determined by the microbiome. Host cells contain pattern receptors that allow them to detect the pathogen-associated molecular patterns, which are fungal cell wall components. In doing so, immune response is activated, turning on downstream proinflammatory and antimicrobial signaling pathways (Westerberg and Voyack, 2013; Tipton et al., 2017). In microbiome dysbiosis, this mechanism is altered and leads to inhibition of physiological immune responses and progression of disease. As such, the crosstalk between the microbiome and the host’s immune system is vital for the continued maintenance of homeostasis (Oever and Netea, 2014).
There are several mechanisms by which the microbiome mediates host immune defense. One important mechanism is via competitive exclusion by residing bacteria in mucosal niches (Oever and Netea, 2014). By doing so, the microbiota is protecting the body against invasive microorganisms, such as fungal pathogens. This response is particularly crucial against the overgrowth of Candida in invasive candidiasis as well as in gastrointestinal candidiasis since the severity of this disease is dependent on the fungal abundance and load colonizing the mucosa (Oever and Netea, 2014). Modulation of host antifungal immune response via bacterial commensals has been demonstrated with Lactobacillus. Lactobacillus is positively correlated with protection of the vaginal mucosa and the maintenance of a healthy microenvironment (Bradford and Ravel, 2017). Studies have shown that the lack of Lactobacillus in the vagina is strongly correlated with urogenital infections and HIV (Drell et al., 2013; Brown et al., 2015; Bradford and Ravel, 2017). In such situations, use of probiotics to increase concentrations of Lactobacillus are beginning to show promise (Oever and Netea, 2014; Bradford and Ravel, 2017).
A pathologic condition where the lack of immune system can be most readily studied is HIV. Mycobiome status is severely shifted in immunocompromised patients, specifically HIV patients. In fact, many complications exhibited in HIV patients are a product of mycobiome disruptions. For example, oral candidiasis is the most common oral complication in HIV patients (Mukherjee et al., 2014; Brown et al., 2015; Chandra et al., 2015). A study conducted by Brown et al. (2015) analyzed and compared the fungal and bacterial composition of HIV subjects against controls. They found that Pichia was absent from all HIV samples but present in control samples. Pichia is known to inhibit Candida and the formation of biofilms via the release of secretory factors and nutrient competition (Mukherjee et al., 2014; Brown et al., 2015; Chandra et al., 2015). The authors therefore proposed that the lack of Pichia in the HIV cohort probably contributes to the fungal dysfunction that allows for oral candidiasis to develop (Mukherjee et al., 2014; Brown et al., 2015). Mukherjee et al. (2014) conducted in vivo testing in which mice treated with Pichia spent medium (PSM) exhibited significantly decreased infection score and fungal burden compared to untreated mice. This study identified one such treatment that has the potential to develop into a novel antifungal in treating oral candidiasis in HIV patients.
Despite availability of antiretroviral therapy, pulmonary diseases pose as significant comorbidities in HIV patients (Cui et al., 2015). Specifically, COPD has been linked to HIV though the cause is poorly understood (Cui et al., 2015). It is possible that increased susceptibility to fungal infection in HIV manifests as pulmonary disease in these patients. The most common fungus in HIV patients, with or without COPD, is Pneumocystis and it is the leading cause of pneumonia in HIV-infected and immunocompromised patients (Cui et al., 2015). Cui et al. (2015) found that P. jirovecii and Candida lacerata are two overrepresented fungal species in the lungs of HIV patients; both of these species are known lung pathogens.
The immunocompromised status of HIV patients predisposes them to complications and comorbidities like oral candidiasis, COPD, and pneumonia. The mycobiome is inherently involved in this process and manipulating the fungal abundance in such patients to restore mycological balance could potentially alleviate the side effects exhibited in HIV.
Allergy
With the advances in next-gen sequencing, investigators have been able to evaluate the fungal bioburden found in patients who suffer from conditions that have been correlated to allergic reactions. One of these conditions is allergic rhinitis, which refers to the chronic inflammation of the nasal cavity. In their 2015 study, Jung et al. (2015) evaluated the mycobiome profile of individuals with allergic rhinitis compared to healthy controls. They found that both groups had a high level of Malassezia species. However, the abundance of specific Malassezia species (e.g., M. globosa, M. restricta) varied from person to person. Additionally, there was a trend toward greater fungal diversity in allergic rhinitis individuals compared to healthy controls albeit this was not statistically significant (Jung et al., 2015). These findings may be significant in offering therapeutic interventions to this commonly encountered condition, which leads to significant compromise in quality of life.
Conclusion
It is clear that the mycobiome plays a critical role in health and disease. However, the crosstalk between fungi and bacteria may be just as important as the fungi present. The current limited interest in the crosstalk between fungi and bacteria is a direct consequence of an incomplete understanding of the mycobiome, and the complexities associated with studying these polymicrobial interactions. It is therefore imperative to fully analyze and establish the fungal niches within the body to gain a broader understanding of the functionality of the microbiome, as a whole. This is evident in multiple body sites.
More research is needed to fully understand the interplay between fungus, bacteria, and the host, but it is evident that fungus can have a profound impact on health and disease. With the advancement in our sequencing technologies, and further elucidation of the organisms present in various niches, it is important to place a concerted effort in identifying approaches that may yield clinical efficacy in reestablishing a microbial diversity that prevents diseases and restores a healthy balance. These may include probiotics or dietary products based on our developing understanding of the inter-kingdom relationships between the microbiome and the mycobiome.
Author Contributions
All authors involved equally in the conceiving of this project, writing, and editing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
- ITS
Internal transcribed spacer
- OUT
Operational taxonomic units
- CD
Crohn’s disease
- CF
Cystic fibrosis
- EPS
extracellular polysaccharide
- GBS
Group B Streptococci.
Funding
The payment and cost for this manuscript were funded by departmental funds.
References
- Ackerman A. L., Underhill D. M. (2017). The mycobiome of the human urinary tract: potential roles for fungi in urology. Ann. Transl. Med. 5:31. 10.21037/atm.2016.12.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adair C. G., Gorman S. P., Feron B. M., Byers L. M., Jones D. S., Goldsmith C. E. (1999). Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med. 25 1072–1076. [DOI] [PubMed] [Google Scholar]
- Ader F., Jawhara S., Nseir S., Kipnis E., Faure K., Vuotto F. (2011). Short term Candida albicans colonization reduces Pseudomonas aeruginosa-related lung injury and bacterial burden in a murine model. Crit. Care 15:R150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison D. L., Willems H. M., Jayatilake J. A., Bruno V. M., Peters B. M., Shirtliff M. E. (2016). Candida-bacteria interactions: their impact on human disease. Microbiol. Spectr. 4. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Altomare A., Putignani L., Del Chierico F., Cocca S., Angeletti S., Ciccozzi M., et al. (2019). Gut mucosal-associated microbiota better discloses inflammatory bowel disease differential patterns than faecal microbiota. Dig. Liver Dis. 51 648–656. 10.1016/j.dld.2018.11.021 [DOI] [PubMed] [Google Scholar]
- Amin R., Dupuis A., Aaron S. D., Ratjen F. (2010). The effect of chronic infection with Aspergillus fumigatus on lung function and hospitalization in patients with cystic fibrosis. Chest 137 171–176. 10.1378/chest.09-1103 [DOI] [PubMed] [Google Scholar]
- Angebault C., Ghozlane A., Volant S., Botterel F., d’Enfert C., Bougnoux M. E. (2018). Combined bacterial and fungal intestinal microbiota analyses: impact of storage conditions and DNA extraction protocols. PLoS One 13:e0201174. 10.1371/journal.pone.0201174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon I. M., Herrera-Imbroda B., Queipo-Ortuno M. I., Castillo E., Del Moral J. S., Gomez-Millan J. (2018). The urinary tract microbiome in health and disease. Eur. Urol. Focus 4 128–138. [DOI] [PubMed] [Google Scholar]
- Arweiler N. B., Netuschil L. (2016). The oral microbiota. Adv. Exp. Med. Biol. 902 45–60. [DOI] [PubMed] [Google Scholar]
- Azoulay E., Timsit J. F., Tafflet M., de Lassence A., Darmon M., Zahar J. R. (2006). Candida colonization of the respiratory tract and subsequent pseudomonas ventilator-associated pneumonia. Chest 129 110–117. 10.1378/chest.129.1.110 [DOI] [PubMed] [Google Scholar]
- Barbosa J. O., Rossoni R. D., Vilela S. F., de Alvarenga J. A., Velloso Mdos S., Prata M. C. (2016). Streptococcus mutans can modulate biofilm formation and attenuate the virulence of Candida albicans. PLoS One 11:e0150457. 10.1371/journal.pone.0150457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson N. J. (2000). Competition for glucose between Candida albicans and oral bacteria grown in mixed culture in a chemostat. J. Med. Microbiol. 49 969–975. 10.1099/0022-1317-49-11-969 [DOI] [PubMed] [Google Scholar]
- Belstrom D., Holmstrup P., Bardow A., Kokaras A., Fiehn N. E., Paster B. J. (2016). Temporal stability of the salivary microbiota in oral health. PLoS One 11:e0147472. 10.1371/journal.pone.0147472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford L. L., Ravel J. (2017). The vaginal mycobiome: a contemporary perspective on fungi in women’s health and diseases. Virulence 8 342–351. 10.1080/21505594.2016.1237332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briard B., Heddergott C., Latge J. P. (2016). Volatile compounds emitted by Pseudomonas aeruginosa stimulate growth of the fungal pathogen Aspergillus fumigatus. mBio 7:e00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogden K. A., Guthmiller J. M., Taylor C. E. (2005). Human polymicrobial infections. Lancet 365 253–255. 10.1016/s0140-6736(05)70155-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown G. D., Denning D. W., Gow N. A., Levitz S. M., Netea M. G., White T. C. (2012). Hidden killers: human fungal infections. Sci. Transl. Med. 4:165rv13. 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- Brown R. E., Ghannoum M. A., Mukherjee P. K., Gillevet P. M., Sikaroodi M. (2015). Quorum-sensing dysbiotic shifts in the HIV-infected oral metabiome. PLoS One 10:e0123880. 10.1371/journal.pone.0123880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucevic Popovic V., Situm M., Chow C. T., Chan L. S., Roje B., Terzic J. (2018). The urinary microbiome associated with bladder cancer. Sci. Rep. 8:12157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budden K. F., Gellatly S. L., Wood D. L., Cooper M. A., Morrison M., Hugenholtz P. (2017). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15 55–63. 10.1038/nrmicro.2016.142 [DOI] [PubMed] [Google Scholar]
- Cameron S. J., Huws S. A., Hegarty M. J., Smith D. P., Mur L. A. (2015). The human salivary microbiome exhibits temporal stability in bacterial diversity. FEMS Microbiol. Ecol. 91:fiv091. 10.1093/femsec/fiv091 [DOI] [PubMed] [Google Scholar]
- Canabarro A., Valle C., Farias M. R., Santos F. B., Lazera M., Wanke B. (2013). Association of subgingival colonization of Candida albicans and other yeasts with severity of chronic periodontitis. J. Periodontal. Res. 48 428–432. 10.1111/jre.12022 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Lauber C. L., Costello E. K., Berg-Lyons D., Gonzalez A., Stombaugh J. (2011). Moving pictures of the human microbiome. Genome Biol. 12:R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens A., Roos A., Andreasson A., Magnuson A., Agreus L., Halfvarson J. (2018). Differential clustering of fecal and mucosa-associated microbiota in ‘healthy’ individuals. J. Dig. Dis. 19 745–752. 10.1111/1751-2980.12688 [DOI] [PubMed] [Google Scholar]
- Chabe M., Aliouat-Denis C. M., Delhaes L., Aliouat el M., Viscogliosi E., Dei-Cas E. (2011). Pneumocystis: from a doubtful unique entity to a group of highly diversified fungal species. FEMS Yeast Res. 11 2–17. 10.1111/j.1567-1364.2010.00698.x [DOI] [PubMed] [Google Scholar]
- Chanda W., Joseph T. P., Wang W., Padhiar A. A., Zhong M. (2017). The potential management of oral candidiasis using anti-biofilm therapies. Med. Hypotheses 106 15–18. 10.1016/j.mehy.2017.06.029 [DOI] [PubMed] [Google Scholar]
- Chandra J., Retuerto M., Mukherjee P. K., Ghannoum M. (2015). The fungal biome of the oral cavity. Methods Mol. Biol. 1356 107–135. 10.1007/978-1-4939-3052-4_9 [DOI] [PubMed] [Google Scholar]
- Charlson E. S., Diamond J. M., Bittinger K., Fitzgerald A. S., Yadav A., Haas A. R. (2012). Lung-enriched organisms and aberrant bacterial and fungal respiratory microbiota after lung transplant. Am. J. Respir. Crit. Care Med. 186 536–545. 10.1164/rccm.201204-0693oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. C., Patel S., Meyer W., Chapman B., Yu H., Byth K. (2018). Pseudomonas aeruginosa inhibits the growth of scedosporium and lomentospora in vitro. Mycopathologia 183 251–261. 10.1007/s11046-017-0140-x [DOI] [PubMed] [Google Scholar]
- Chen Y., Chen Z., Guo R., Chen N., Lu H., Huang S. (2011). Correlation between gastrointestinal fungi and varying degrees of chronic hepatitis B virus infection. Diagn. Microbiol. Infect. Dis. 70 492–498. 10.1016/j.diagmicrobio.2010.04.005 [DOI] [PubMed] [Google Scholar]
- Chotirmall S. H., Greene C. M., McElvaney N. G. (2010). Candida species in cystic fibrosis: a road less travelled. Med. Mycol. 48 Suppl 1 S114–S124. [DOI] [PubMed] [Google Scholar]
- Clavaud C., Jourdain R., Bar-Hen A., Tichit M., Bouchier C., Pouradier F. (2013). Dandruff is associated with disequilibrium in the proportion of the major bacterial and fungal populations colonizing the scalp. PLoS One 8:e58203. 10.1371/journal.pone.0058203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools P., Jespers V., Hardy L., Crucitti T., Delany-Moretlwe S., Mwaura M. (2016). A multi-country cross-sectional study of vaginal carriage of group B streptococci (GBS) and Escherichia coli in resource-poor settings: prevalences and risk factors. PLoS One 11:e0148052. 10.1371/journal.pone.0148052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A. C., Pereira C. A., Junqueira J. C., Jorge A. O. (2013). Recent mouse and rat methods for the study of experimental oral candidiasis. Virulence 4 391–399. 10.4161/viru.25199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello E. K., Lauber C. L., Hamady M., Fierer N., Gordon J. I., Knight R. (2009). Bacterial community variation in human body habitats across space and time. Science 326 1694–1697. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J. W., Stewart P. S., Greenberg E. P. (1999). Bacterial biofilms: a common cause of persistent infections. Science 284 1318–1322. 10.1126/science.284.5418.1318 [DOI] [PubMed] [Google Scholar]
- Cruz M. R., Graham C. E., Gagliano B. C., Lorenz M. C., Garsin D. A. (2013). Enterococcus faecalis inhibits hyphal morphogenesis and virulence of Candida albicans. Infect. Immun. 81 189–200. 10.1128/iai.00914-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Lucht L., Tipton L., Rogers M. B., Itch A., Kessinger C., et al. (2015). Topographic diversity of the respiratory tract mycobiome and alteration in HIV and lung disease. Am. J. Respir. Crit. Care Med. 191 932–942. 10.1164/rccm.201409-1583oc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Morris A., Huang L., Beck J. M., Twigg H. L., III, von Mutius E. (2014). The microbiome and the lung. Ann. Am. Thorac. Soc. 11 Suppl 4 S227–S232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A., Materna A. C., Friedman J., Campos-Baptista M. I., Blackburn M. C., Perrotta A. (2014a). Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15:R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L. A., Maurice C. F., Carmody R. N., Gootenberg D. B., Button J. E., Wolfe B. E. (2014b). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505 559–563. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bernardis F., Graziani S., Tirelli F., Antonopoulou S. (2018). Candida vaginitis: virulence, host response and vaccine prospects. Med. Mycol. 56(suppl_1) 26–31. [DOI] [PubMed] [Google Scholar]
- De Brucker K., Tan Y., Vints K., De Cremer K., Braem A., Verstraeten N. (2015). Fungal beta-1,3-glucan increases ofloxacin tolerance of Escherichia coli in a polymicrobial E. coli/Candida albicans biofilm. Antimicrob. Agents Chemother. 59 3052–3058. 10.1128/aac.04650-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Perrucini P. D., Poli-Frederico R. C., de Almeida Pires-Oliveira D. A., Bertin L. D., Pires F. B., Shimoya-Bittencourt W., et al. (2019). Anti-inflammatory and healing effects of pulsed ultrasound therapy on fibroblasts. Am. J. Phys. Med. Rehabil. 99 19–25. 10.1097/phm.0000000000001265 [DOI] [PubMed] [Google Scholar]
- de Oliveira F. E., Rossoni R. D., de Barros P. P., Begnini B. E., Junqueira J. C., Jorge A. O. C. (2017). Immunomodulatory effects and anti-Candida activity of lactobacilli in macrophages and in invertebrate model of Galleria mellonella. Microb. Pathog. 110 603–611. 10.1016/j.micpath.2017.08.006 [DOI] [PubMed] [Google Scholar]
- Delhaes L., Monchy S., Frealle E., Hubans C., Salleron J., Leroy S. (2012). The airway microbiota in cystic fibrosis: a complex fungal and bacterial community–implications for therapeutic management. PLoS One 7:e36313. 10.1371/journal.pone.0036313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst F. E., Chen T., Izard J., Paster B. J., Tanner A. C., Yu W. H. (2010). The human oral microbiome. J. Bacteriol. 192 5002–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz P. I., Xie Z., Sobue T., Thompson A., Biyikoglu B., Ricker A. (2012). Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect. Immun. 80 620–632. 10.1128/iai.05896-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickstein B. (1964). Neonatal oral candidiasis: evaluation of a new chemotherapeutic agent. Clin. Pediatr. (Phila) 3 485–488. 10.1177/000992286400300811 [DOI] [PubMed] [Google Scholar]
- Ding T., Schloss P. D. (2014). Dynamics and associations of microbial community types across the human body. Nature 509 357–360. 10.1038/nature13178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollive S., Peterfreund G. L., Sherrill-Mix S., Bittinger K., Sinha R., Hoffmann C., et al. (2012). A tool kit for quantifying eukaryotic rRNA gene sequences from human microbiome samples. Genome Biol. 13:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dollive S., Peterfreund G. L., Sherrill-Mix S., Bittinger K., Sinha R., Hoffmann C. (2013). Fungi of the murine gut: episodic variation and proliferation during antibiotic treatment. PLoS One 8:e71806. 10.1371/journal.pone.0071806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G. P., Lee S. M., Mazmanian S. K. (2016). Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 14 20–32. 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drell T., Lillsaar T., Tummeleht L., Simm J., Aaspollu A., Vain E. (2013). Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS One 8:e54379. 10.1371/journal.pone.0054379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy A. K., David M. S., Li L., Heider T. N., Peterson J. D., Montano E. A. (2014). Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One 9:e90899. 10.1371/journal.pone.0090899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durban A., Abellan J. J., Jimenez-Hernandez N., Ponce M., Ponce J., Sala T. (2011). Assessing gut microbial diversity from feces and rectal mucosa. Microb. Ecol. 61 123–133. 10.1007/s00248-010-9738-y [DOI] [PubMed] [Google Scholar]
- Dzidic M., Collado M. C., Abrahamsson T., Artacho A., Stensson M., Jenmalm M. C. (2018). Oral microbiome development during childhood: an ecological succession influenced by postnatal factors and associated with tooth decay. ISME J. 12 2292–2306. 10.1038/s41396-018-0204-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Jurdi N., Filali-Mouhim A., Salem I., Retuerto M., Dambrosio N. M., Baer L., et al. (2019). Gastrointestinal microbiome and mycobiome changes during autologous transplantation for multiple myeloma: results of a prospective pilot study. Biol. Blood Marrow Transplant 25 1511–1519. 10.1016/j.bbmt.2019.04.007 [DOI] [PubMed] [Google Scholar]
- Enaud R., Vandenborght L. E., Coron N., Bazin T., Prevel R., Schaeverbeke T. (2018). The mycobiome: a neglected component in the microbiota-gut-brain axis. Microorganisms 6:22. 10.3390/microorganisms6010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch D. A., Ludlam H. A., Brown N. M. (2006). Invasive fungal infections: a review of epidemiology and management options. J. Med. Microbiol. 55(Pt 7) 809–818. 10.1099/jmm.0.46548-0 [DOI] [PubMed] [Google Scholar]
- Erb Downward J. R., Falkowski N. R., Mason K. L., Muraglia R., Huffnagle G. B. (2013). Modulation of post-antibiotic bacterial community reassembly and host response by Candida albicans. Sci. Rep. 3:2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta M. L., Klein M. I., Lemos J. A., Silva B. B., Agidi S., Scott-Anne K. K., et al. (2012). Novel antibiofilm chemotherapy targets exopolysaccharide synthesis and stress tolerance in Streptococcus mutans to modulate virulence expression in vivo. Antimicrob. Agents Chemother. 56 6201–6211. 10.1128/aac.01381-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsetta M. L., Klein M. I., Lemos J. A., Silva B. B., Agidi S., Scott-Anne K. K. (2014). Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 82 1968–1981. 10.1128/iai.00087-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K., Oh J., Yang J., Conlan S., Deming C., Meyer J. A. (2013). Topographic diversity of fungal and bacterial communities in human skin. Nature 498 367–370. 10.1038/nature12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores G. E., Caporaso J. G., Henley J. B., Rideout J. R., Domogala D., Chase J. (2014). Temporal variability is a personalized feature of the human microbiome. Genome Biol. 15:531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forssten S. D., Bjorklund M., Ouwehand A. C. (2010). Streptococcus mutans, caries and simulation models. Nutrients 2 290–298. 10.3390/nu2030290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster T. M., Bjorklund M., Ouwehand A. C. (2016). Enemies and brothers in arms: Candida albicans and gram-positive bacteria. Cell Microbiol. 18 1709–1715. 10.1111/cmi.12657 [DOI] [PubMed] [Google Scholar]
- Fox E. P., Cowley E. S., Nobile C. J., Hartooni N., Newman D. K., Johnson A. D. (2014). Anaerobic bacteria grow within Candida albicans biofilms and induce biofilm formation in suspension cultures. Curr. Biol. 24 2411–2416. 10.1016/j.cub.2014.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frases S., Chaskes S., Dadachova E., Casadevall A. (2006). Induction by Klebsiella aerogenes of a melanin-like pigment in Cryptococcus neoformans. Appl. Environ. Microbiol. 72 1542–1550. 10.1128/aem.72.2.1542-1550.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuochi V., Li Volti G., Furneri P. M. (2017). Commentary: lactobacilli dominance and vaginal pH: why is the human vaginal microbiome unique? Front. Microbiol. 8:1815. 10.3389/fmicb.2017.01815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanis G., Magiatis P., Hantschke M., Bassukas I. D., Velegraki A. (2012). The Malassezia genus in skin and systemic diseases. Clin. Microbiol. Rev. 25 106–141. 10.1128/cmr.00021-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum M. (2016). Cooperative evolutionary strategy between the bacteriome and mycobiome. mBio 7:e01951–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghannoum M. A., Jurevic R. J., Mukherjee P. K., Cui F., Sikaroodi M., Naqvi A. (2010). Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6:e1000713. 10.1371/journal.ppat.1000713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordee R. S., Matthews T. R. (1969). Systemic antifungal activity of pyrrolnitrin. Appl. Microbiol. 17 690–694. 10.1128/am.17.5.690-694.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham C. E., Cruz M. R., Garsin D. A., Lorenz M. C. (2017). Enterococcus faecalis bacteriocin EntV inhibits hyphal morphogenesis, biofilm formation, and virulence of Candida albicans. Proc. Natl. Acad. Sci. U.S.A. 114 4507–4512. 10.1073/pnas.1620432114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum S., Greenbaum G., Moran-Gilad J., Weintraub A. Y. (2019). Ecological dynamics of the vaginal microbiome in relation to health and disease. Am. J. Obstet. Gynecol. 220 324–335. 10.1016/j.ajog.2018.11.1089 [DOI] [PubMed] [Google Scholar]
- Gross E. L., Beall C. J., Kutsch S. R., Firestone N. D., Leys E. J., Griffen A. L. (2012). Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLoS One 7:e47722. 10.1371/journal.pone.0047722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. K., Batra R., Bluhm R., Boekhout T., Dawson T. L., Jr. (2004). Skin diseases associated with Malassezia species. J. Am. Acad. Dermatol. 51 785–798. 10.1016/j.jaad.2003.12.034 [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. (2014). The inflammophilic character of the periodontitis-associated microbiota. Mol. Oral. Microbiol. 29 248–257. 10.1111/omi.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havlickova B., Czaika V. A., Friedrich M. (2008). Epidemiological trends in skin mycoses worldwide. Mycoses 51 Suppl 4 2–15. 10.1111/j.1439-0507.2008.01606.x [DOI] [PubMed] [Google Scholar]
- Hermann C., Hermann J., Munzel U., Ruchel R. (1999). Bacterial flora accompanying Candida yeasts in clinical specimens. Mycoses 42 619–627. 10.1046/j.1439-0507.1999.00519.x [DOI] [PubMed] [Google Scholar]
- Hilt E. E., McKinley K., Pearce M. M., Rosenfeld A. B., Zilliox M. J., Mueller E. R. (2014). Urine is not sterile: use of enhanced urine culture techniques to detect resident bacterial flora in the adult female bladder. J. Clin. Microbiol. 52 871–876. 10.1128/jcm.02876-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoarau G., Mukherjee P. K., Gower-Rousseau C., Hager C., Chandra J., Retuerto M. A. (2016). Bacteriome and mycobiome interactions underscore microbial dysbiosis in familial crohn’s disease. mBio 7:e01250–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepelman I. M., Dupont B. (1996). Oral candidiasis: the clinical challenge of resistance and management. Int. J. Antimicrob. Agents 6 155–159. 10.1016/0924-8579(95)00050-x [DOI] [PubMed] [Google Scholar]
- Hoffmann C., Dollive S., Grunberg S., Chen J., Li H., Wu G. D. (2013). Archaea and fungi of the human gut microbiome: correlations with diet and bacterial residents. PLoS One 8:e66019. 10.1371/journal.pone.0066019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan D. A., Vik A., Kolter R. (2004). A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54 1212–1223. 10.1111/j.1365-2958.2004.04349.x [DOI] [PubMed] [Google Scholar]
- Hover T., Maya T., Ron S., Sandovsky H., Shadkchan Y., Kijner N. (2016). Mechanisms of bacterial (Serratia marcescens) attachment to, migration along, and killing of fungal hyphae. Appl. Environ. Microbiol. 82 2585–2594. 10.1128/aem.04070-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L., Cattamanchi A., Davis J. L., den Boon S., Kovacs J., Meshnick S. (2011). HIV-associated Pneumocystis pneumonia. Proc. Am. Thorac. Soc. 8 294–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle G. B., Noverr M. C. (2013). The emerging world of the fungal microbiome. Trends Microbiol. 21 334–341. 10.1016/j.tim.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianiro G., Tilg H., Gasbarrini A. (2016). Antibiotics as deep modulators of gut microbiota: between good and evil. Gut 65 1906–1915. 10.1136/gutjnl-2016-312297 [DOI] [PubMed] [Google Scholar]
- Ibrahim A. S., Gebremariam T., Liu M., Chamilos G., Kontoyiannis D., Mink R. (2008). Bacterial endosymbiosis is widely present among zygomycetes but does not contribute to the pathogenesis of mucormycosis. J. Infect. Dis. 198 1083–1090. 10.1086/591461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev I. D., Underhill D. M. (2013). Striking a balance: fungal commensalism versus pathogenesis. Curr. Opin. Microbiol. 16 366–373. 10.1016/j.mib.2013.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev I. D., Funari V. A., Taylor K. D., Nguyen Q., Reyes C. N., Strom S. P. (2012). Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science 336 1314–1317. 10.1126/science.1221789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingenito E. P., Solway J., McFadden E. R., Jr., Pichurko B., Bowman H. F., Michaels D. (1987). Indirect assessment of mucosal surface temperatures in the airways: theory and tests. J. Appl. Physiol. (1985) 63 2075–2083. 10.1152/jappl.1987.63.5.2075 [DOI] [PubMed] [Google Scholar]
- Jain A., Jain S., Rawat S. (2010). Emerging fungal infections among children: a review on its clinical manifestations, diagnosis, and prevention. J. Pharm. Bioallied. Sci. 2 314–320. 10.4103/0975-7406.72131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janus M. M., Crielaard W., Volgenant C. M., van der Veen M. H., Brandt B. W., Krom B. P. (2017). Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J. Oral Microbiol. 9:1270613. 10.1080/20002297.2016.1270613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T. T., Shao T. Y., Ang W. X. G., Kinder J. M., Turner L. H., Pham G. (2017). Commensal fungi recapitulate the protective benefits of intestinal bacteria. Cell Host Microbe 22 809–816e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo J. H., Kennedy E. A., Kong H. H. (2017). Topographical and physiological differences of the skin mycobiome in health and disease. Virulence 8 324–333. 10.1080/21505594.2016.1249093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W. H., Croll D., Cho J. H., Kim Y. R., Lee Y. W. (2015). Analysis of the nasal vestibule mycobiome in patients with allergic rhinitis. Mycoses 58 167–172. 10.1111/myc.12296 [DOI] [PubMed] [Google Scholar]
- Kaewsrichan J., Peeyananjarassri K., Kongprasertkit J. (2006). Selection and identification of anaerobic lactobacilli producing inhibitory compounds against vaginal pathogens. FEMS Immunol. Med. Microbiol. 48 75–83. 10.1111/j.1574-695x.2006.00124.x [DOI] [PubMed] [Google Scholar]
- Kalan L., Loesche M., Hodkinson B. P., Heilmann K., Ruthel G., Gardner S. E., et al. (2016). Redefining the chronic-wound microbiome: fungal communities are prevalent, dynamic, and associated with delayed healing. mBio 7 e1058–e1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyana Chakravarthy S., Jayasudha R., Ranjith K., Dutta A., Pinna N. K., Mande S. S. (2018). Alterations in the gut bacterial microbiome in fungal Keratitis patients. PLoS One 13:e0199640. 10.1371/journal.pone.0199640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitan M., Niemiec M. J., Steimle A., Frick J. S., Jacobsen I. D. (2018). Fungi as part of the microbiota and interactions with intestinal bacteria. Curr. Top. Microbiol. Immunol. 422 265–301. 10.1007/82_2018_117 [DOI] [PubMed] [Google Scholar]
- Karkowska-Kuleta J., Bartnicka D., Zawrotniak M., Zielinska G., Kieronska A., Bochenska O. (2018). The activity of bacterial peptidylarginine deiminase is important during formation of dual-species biofilm by periodontal pathogen Porphyromonas gingivalis and opportunistic fungus Candida albicans. Pathog. Dis. 76:fty033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur J., Pethani B. P., Kumar S., Kim M., Sunna A., Kautto L. (2015). Pseudomonas aeruginosa inhibits the growth of Scedosporium aurantiacum, an opportunistic fungal pathogen isolated from the lungs of cystic fibrosis patients. Front. Microbiol. 6:866. 10.3389/fmicb.2015.00866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. J., Volz P. A. (1985). Effect of various antibiotics on gastrointestinal colonization and dissemination by Candida albicans. Sabouraudia 23 265–273. 10.1080/00362178585380391 [DOI] [PubMed] [Google Scholar]
- Kerr J. (1994). Inhibition of fungal growth by Pseudomonas aeruginosa and Pseudomonas cepacia isolated from patients with cystic fibrosis. J. Infect. 28 305–310. 10.1016/s0163-4453(94)91943-7 [DOI] [PubMed] [Google Scholar]
- Kerr J. R., Taylor G. W., Rutman A., Hoiby N., Cole P. J., Wilson R. (1999). Pseudomonas aeruginosa pyocyanin and 1-hydroxyphenazine inhibit fungal growth. J. Clin. Pathol. 52 385–387. 10.1136/jcp.52.5.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khasriya R., Sathiananthamoorthy S., Ismail S., Kelsey M., Wilson M., Rohn J. L. (2013). Spectrum of bacterial colonization associated with urothelial cells from patients with chronic lower urinary tract symptoms. J. Clin. Microbiol. 51 2054–2062. 10.1128/jcm.03314-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Liu Y., Benhamou R. I., Sanchez H., Simon-Soro A., Li Y. (2018). Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J. 12 1427–1442. 10.1038/s41396-018-0113-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Sengupta A., Niepa T. H., Lee B. H., Weljie A., Freitas-Blanco V. S. (2017). Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep. 7:41332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Yoon Y. K., Kim M. J., Sohn J. W. (2013). Risk factors for and clinical implications of mixed Candida/bacterial bloodstream infections. Clin. Microbiol. Infect. 19 62–68. 10.1111/j.1469-0691.2012.03906.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz S. A., Chasin B. S., Powell B., Gaur N. K., Lipke P. N. (2007). Polymicrobial bloodstream infections involving Candida species: analysis of patients and review of the literature. Diagn. Microbiol. Infect. Dis. 59 401–406. 10.1016/j.diagmicrobio.2007.07.001 [DOI] [PubMed] [Google Scholar]
- Kobayashi D. Y., Crouch J. A. (2009). Bacterial/Fungal interactions: from pathogens to mutualistic endosymbionts. Annu. Rev. Phytopathol. 47 63–82. 10.1146/annurev-phyto-080508-081729 [DOI] [PubMed] [Google Scholar]
- Kollef M. H., Shorr A., Tabak Y. P., Gupta V., Liu L. Z., Johannes R. S. (2005). Epidemiology and outcomes of health-care-associated pneumonia: results from a large US database of culture-positive pneumonia. Chest 128 3854–3862. [DOI] [PubMed] [Google Scholar]
- Kong E. F., Tsui C., Kucharikova S., Andes D., Van Dijck P., Jabra-Rizk M. A. (2016). Commensal protection of staphylococcus aureus against antimicrobials by candida albicans biofilm matrix. mBio 7:e01365–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovachev S. M., Vatcheva-Dobrevska R. S. (2015). Local probiotic therapy for vaginal Candida albicans infections. Probiotics Antimicrob. Proteins 7 38–44. 10.1007/s12602-014-9176-0 [DOI] [PubMed] [Google Scholar]
- Kraneveld E. A., Buijs M. J., Bonder M. J., Visser M., Keijser B. J., Crielaard W. (2012). The relation between 10.1371/journal.pone.0042770 oral Candida load and bacterial microbiome profiles in Dutch older adults. PLoS One 7:e42770. 10.1371/journal.pone.0042770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger W., Vielreicher S., Kapitan M., Jacobsen I. D., Niemiec M. J. (2019). Fungal-bacterial interactions in health and disease. Pathogens 8:70. 10.3390/pathogens8020070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclair L. W., Hogan D. A. (2010). Mixed bacterial-fungal infections in the CF respiratory tract. Med. Mycol. 48 Suppl 1 S125–S132. [DOI] [PubMed] [Google Scholar]
- Lepage P., Seksik P., Sutren M., de la Cochetiere M. F., Jian R., Marteau P. (2005). Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 11 473–480. 10.1097/01.mib.0000159662.62651.06 [DOI] [PubMed] [Google Scholar]
- Lessard M., Dupuis M., Gagnon N., Nadeau E., Matte J. J., Goulet J. (2009). Administration of Pediococcus acidilactici or Saccharomyces cerevisiae boulardii modulates development of porcine mucosal immunity and reduces intestinal bacterial translocation after Escherichia coli challenge. J. Anim. Sci. 87 922–934. 10.2527/jas.2008-0919 [DOI] [PubMed] [Google Scholar]
- Leung M. H., Chan K. C., Lee P. K. (2016). Skin fungal community and its correlation with bacterial community of urban Chinese individuals. Microbiome 4:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libertucci J., Young V. B. (2019). The role of the microbiota in infectious diseases. Nat. Microbiol. 4 35–45. 10.1038/s41564-018-0278-4 [DOI] [PubMed] [Google Scholar]
- Magistro G., Stief C. G. (2019). The urinary tract microbiome: the answer to all our open questions? Eur. Urol. Focus 5 36–38. 10.1016/j.euf.2018.06.011 [DOI] [PubMed] [Google Scholar]
- Markey L., Hooper A., Melon L. C., Baglot S., Hill M. N., Maguire J., et al. (2020). Colonization with the commensal fungus Candida albicans perturbs the gut-brain axis through dysregulation of endocannabinoid signaling. Psychoneuroendocrinology. 121:104808. 10.1016/j.psyneuen.2020.104808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsland B. J., Gollwitzer E. S. (2014). Host-microorganism interactions in lung diseases. Nat. Rev. Immunol. 14 827–835. 10.1038/nri3769 [DOI] [PubMed] [Google Scholar]
- Mason K. L., Erb Downward J. R., Mason K. D., Falkowski N. R., Eaton K. A., Kao J. Y. (2012). Candida albicans and bacterial microbiota interactions in the cecum during recolonization following broad-spectrum antibiotic therapy. Infect. Immun. 80 3371–3380. 10.1128/iai.00449-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massot J., Sanchez O., Couchy R., Astoin J., Parodi A. L. (1984). Bacterio-pharmacological activity of Saccharomyces boulardii in clindamycin-induced colitis in the hamster. Arzneimittelforschung 34 794–797. [PubMed] [Google Scholar]
- Metwalli K. H., Khan S. A., Krom B. P., Jabra-Rizk M. A. (2013). Streptococcus mutans, Candida albicans, and the human mouth: a sticky situation. PLoS Pathog. 9:e1003616. 10.1371/journal.ppat.1003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millsop J. W., Fazel N. (2016). Oral candidiasis. Clin. Dermatol. 34 487–494. [DOI] [PubMed] [Google Scholar]
- Mitchell A. B., Glanville A. R. (2018). The human respiratory microbiome: implications and impact. Semin. Respir. Crit. Care Med. 39 199–212. 10.1055/s-0037-1617441 [DOI] [PubMed] [Google Scholar]
- Monteiro-da-Silva F., Araujo R., Sampaio-Maia B. (2014). Interindividual variability and intraindividual stability of oral fungal microbiota over time. Med. Mycol. 52 498–505. 10.1093/mmy/myu027 [DOI] [PubMed] [Google Scholar]
- Montes-Cano M. A., de la Horra C., Dapena F. J., Mateos I., Friaza V., Respaldiza N., et al. (2007). Dynamic colonisation by different Pneumocystis jirovecii genotypes in cystic fibrosis patients. Clin. Microbiol. Infect. 13 1008–1011. 10.1111/j.1469-0691.2007.01789.x [DOI] [PubMed] [Google Scholar]
- Morales D. K., Hogan D. A. (2010). Candida albicans interactions with bacteria in the context of human health and disease. PLoS Pathog. 6:e1000886. 10.1371/journal.ppat.1000886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motooka D., Fujimoto K., Tanaka R., Yaguchi T., Gotoh K., Maeda Y. (2017). Fungal ITS1 deep-sequencing strategies to reconstruct the composition of a 26-species community and evaluation of the gut mycobiota of healthy japanese individuals. Front. Microbiol. 8:238. 10.3389/fmicb.2017.00238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowat E., Rajendran R., Williams C., McCulloch E., Jones B., Lang S. (2010). Pseudomonas aeruginosa and their small diffusible extracellular molecules inhibit Aspergillus fumigatus biofilm formation. FEMS Microbiol. Lett. 313 96–102. 10.1111/j.1574-6968.2010.02130.x [DOI] [PubMed] [Google Scholar]
- Mukherjee P. K., Chandra J., Retuerto M., Sikaroodi M., Brown R. E., Jurevic R. (2014). Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog. 10:e1003996. 10.1371/journal.ppat.1003996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi A., Sei Y., Guillot J. (2000). Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med. Mycol. 38 337–341. 10.1080/714030958 [DOI] [PubMed] [Google Scholar]
- Nash A. K., Auchtung T. A., Wong M. C., Smith D. P., Gesell J. R., Ross M. C. (2017). The gut mycobiome of the human microbiome project healthy cohort. Microbiome 5:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L. N., Lopes L. C., Cordero R. J., Nosanchuk J. D. (2011). Sodium butyrate inhibits pathogenic yeast growth and enhances the functions of macrophages. J. Antimicrob. Chemother. 66 2573–2580. 10.1093/jac/dkr358 [DOI] [PubMed] [Google Scholar]
- Nickel J. C., Stephens A., Landis J. R., Mullins C., van Bokhoven A., Lucia M. S. (2016). Assessment of the lower urinary tract microbiota during symptom flare in women with urologic chronic pelvic pain syndrome: a MAPP network study. J. Urol. 195 356–362. 10.1016/j.juro.2015.09.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira M. F., Pereira L., Jenull S., Kuchler K., Lion T. (2019). Klebsiella pneumoniae prevents spore germination and hyphal development of Aspergillus species. Sci. Rep. 9:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noverr M. C., Huffnagle G. B. (2004). Regulation of Candida albicans morphogenesis by fatty acid metabolites. Infect. Immun. 72 6206–6210. 10.1128/iai.72.11.6206-6210.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oever J. T., Netea M. G. (2014). The bacteriome-mycobiome interaction and antifungal host defense. Eur. J. Immunol. 44 3182–3191. 10.1002/eji.201344405 [DOI] [PubMed] [Google Scholar]
- Ott S. J., Kuhbacher T., Musfeldt M., Rosenstiel P., Hellmig S., Rehman A. (2008). Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand. J. Gastroenterol. 43 831–841. 10.1080/00365520801935434 [DOI] [PubMed] [Google Scholar]
- Park H. K., Ha M. H., Park S. G., Kim M. N., Kim B. J., Kim W. (2012). Characterization of the fungal microbiota (mycobiome) in healthy and dandruff-afflicted human scalps. PLoS One 7:e32847. 10.1371/journal.pone.0032847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce M. M., Hilt E. E., Rosenfeld A. B., Zilliox M. J., Thomas-White K., Fok C. (2014). The female urinary microbiome: a comparison of women with and without urgency urinary incontinence. mBio 5:e01283–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrosa A. F., Lisboa C., Goncalves Rodrigues A. (2014). Malassezia infections: a medical conundrum. J. Am. Acad. Dermatol. 71 170–176. 10.1016/j.jaad.2013.12.022 [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Hogan D. A., Mylonakis E. (2010). Medically important bacterial-fungal interactions. Nat. Rev. Microbiol. 8 340–349. 10.1038/nrmicro2313 [DOI] [PubMed] [Google Scholar]
- Pendharkar S., Brandsborg E., Hammarstrom L., Marcotte H., Larsson P. G. (2015). Vaginal colonisation by probiotic lactobacilli and clinical outcome in women conventionally treated for bacterial vaginosis and yeast infection. BMC Infect. Dis. 15:255. 10.1186/s12879-015-0971-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B. M., Jabra-Rizk M. A., O’May G. A., Costerton J. W., Shirtliff M. E. (2012). Polymicrobial interactions: impact on pathogenesis and human disease. Clin. Microbiol. Rev. 25 193–213. 10.1128/cmr.00013-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. C., Cushion M. T. (2005). Pneumocystis: not just pneumonia. Curr. Opin. Microbiol. 8 393–398. 10.1201/b14162-21 [DOI] [PubMed] [Google Scholar]
- Pidwill G. R., Rego S., Jenkinson H. F., Lamont R. J., Nobbs A. H. (2018). Coassociation between Group B streptococcus and Candida albicans promotes interactions with vaginal epithelium. Infect. Immun. 86:e00669–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierard G. E., Arrese J. E., Pierard-Franchimont C., Doncker D. E. (1997). Prolonged effects of antidandruff shampoos – time to recurrence of Malassezia ovalis colonization of skin. Int. J. Cosmet. Sci. 19 111–117. 10.1046/j.1467-2494.1997.171706.x [DOI] [PubMed] [Google Scholar]
- Proctor L. M. (2011). The human microbiome project in 2011 and beyond. Cell Host Microbe 10 287–291. 10.1016/j.chom.2011.10.001 [DOI] [PubMed] [Google Scholar]
- Qin J., Li R., Raes J., Arumugam M., Burgdorf K. S., Manichanh C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rall G., Knoll L. J. (2016). Development of complex models to study co- and polymicrobial infections and diseases. PLoS Pathog. 12:e1005858. 10.1371/journal.ppat.1005858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A., Dongari-Bagtzoglou A. (2018). Tipping the balance: C. albicans adaptation in polymicrobial environments. J. Fungi (Basel) 4:112. 10.3390/jof4030112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece E., Doyle S., Greally P., Renwick J., McClean S. (2018). Aspergillus fumigatus inhibits Pseudomonas aeruginosa in Co-culture: implications of a mutually antagonistic relationship on virulence and inflammation in the cf airway. Front Microbiol 9:1205. 10.3389/fmicb.2018.01205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro F. C., de Barros P. P., Rossoni R. D., Junqueira J. C., Jorge A. O. (2017). Lactobacillus rhamnosus inhibits Candida albicans virulence factors in vitro and modulates immune system in Galleria mellonella. J. Appl. Microbiol. 122 201–211. [DOI] [PubMed] [Google Scholar]
- Robinson M., Daviskas E., Eberl S., Baker J., Chan H. K., Anderson S. D. (1999). The effect of inhaled mannitol on bronchial mucus clearance in cystic fibrosis patients: a pilot study. Eur. Respir. J. 14 678–685. 10.1034/j.1399-3003.1999.14c30.x [DOI] [PubMed] [Google Scholar]
- Rossoni R. D., Fuchs B. B., de Barros P. P., Velloso M. D., Jorge A. O., Junqueira J. C. (2017). Lactobacillus paracasei modulates the immune system of Galleria mellonella and protects against Candida albicans infection. PLoS One 12:e0173332. 10.1371/journal.pone.0173332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux D., Gaudry S., Khoy-Ear L., Aloulou M., Phillips-Houlbracq M., Bex J. (2013). Airway fungal colonization compromises the immune system allowing bacterial pneumonia to prevail. Crit. Care Med. 41 e191–e199. [DOI] [PubMed] [Google Scholar]
- Sampaio-Maia B., Monteiro-Silva F. (2014). Acquisition and maturation of oral microbiome throughout childhood: an update. Dent. Res. J. (Isfahan) 11 291–301. [PMC free article] [PubMed] [Google Scholar]
- Sanglard D. (2016). Emerging threats in antifungal-resistant fungal pathogens. Front. Med. (Lausanne) 3:11. 10.3389/fmed.2016.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scupham A. J., Presley L. L., Wei B., Bent E., Griffith N., McPherson M. (2006). Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 72 793–801. 10.1128/aem.72.1.793-801.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlacek L., Graf B., Schwarz C., Albert F., Peter S., Wurstl B. (2015). Prevalence of Scedosporium species and Lomentospora prolificans in patients with cystic fibrosis in a multicenter trial by use of a selective medium. J. Cyst. Fibros 14 237–241. 10.1016/j.jcf.2014.12.014 [DOI] [PubMed] [Google Scholar]
- Seebacher C., Bouchara J. P., Mignon B. (2008). Updates on the epidemiology of dermatophyte infections. Mycopathologia 166 335–352. 10.1007/s11046-008-9100-9 [DOI] [PubMed] [Google Scholar]
- Seed P. C. (2014). The human mycobiome. Cold Spring Harb. Perspect. Med. 5:a019810. 10.1101/cshperspect.a019810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwitz R. H., Ismail A. I., Pitts N. B. (2007). Dental caries. Lancet 369 51–59. [DOI] [PubMed] [Google Scholar]
- Sepehri S., Kotlowski R., Bernstein C. N., Krause D. O. (2007). Microbial diversity of inflamed and noninflamed gut biopsy tissues in inflammatory bowel disease. Inflamm. Bowel Dis. 13 675–683. 10.1002/ibd.20101 [DOI] [PubMed] [Google Scholar]
- Sethi S., Murphy T. F. (2008). Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med. 359 2355–2365. [DOI] [PubMed] [Google Scholar]
- Shirtliff M. E., Peters B. M., Jabra-Rizk M. A. (2009). Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol. Lett. 299 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuster S. (1984). The aetiology of dandruff and the mode of action of therapeutic agents. Br. J. Dermatol. 111 235–242. 10.1111/j.1365-2133.1984.tb04050.x [DOI] [PubMed] [Google Scholar]
- Siddiqui H., Nederbragt A. J., Lagesen K., Jeansson S. L., Jakobsen K. S. (2011). Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol. 11:244. 10.1186/1471-2180-11-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skalski J. H., Limon J. J., Sharma P., Gargus M. D., Nguyen C., Tang J. (2018). Expansion of commensal fungus Wallemia mellicola in the gastrointestinal mycobiota enhances the severity of allergic airway disease in mice. PLoS Pathog. 14:e1007260. 10.1371/journal.ppat.1007260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel J. D., Faro S., Force R. W., Foxman B., Ledger W. J., Nyirjesy P. R. (1998). Vulvovaginal candidiasis: epidemiologic, diagnostic, and therapeutic considerations. Am. J. Obstet. Gynecol. 178 203–211. 10.1016/s0002-9378(98)80001-x [DOI] [PubMed] [Google Scholar]
- Sokol H., Leducq V., Aschard H., Pham H. P., Jegou S., Landman C. (2017). Fungal microbiota dysbiosis in IBD. Gut 66 1039–1048. 10.1136/gutjnl-2015-310746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovran B., Planchais J., Jegou S., Straube M., Lamas B., Natividad J. M. (2018). Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 6:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahringer S. S., Clemente J. C., Corley R. P., Hewitt J., Knights D., Walters W. A. (2012). Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood. Genome Res. 22 2146–2152. 10.1101/gr.140608.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strati F., Di Paola M., Stefanini I., Albanese D., Rizzetto L., Lionetti P. (2016). Age and gender affect the composition of fungal population of the human gastrointestinal tract. Front. Microbiol. 7:1227. 10.3389/fmicb.2016.01227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachedjian G., Aldunate M., Bradshaw C. S., Cone R. A. (2017). The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res. Microbiol. 168 782–792. 10.1016/j.resmic.2017.04.001 [DOI] [PubMed] [Google Scholar]
- Takahashi N., Nyvad B. (2011). The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 90 294–303. 10.1177/0022034510379602 [DOI] [PubMed] [Google Scholar]
- Tang J., Iliev I. D., Brown J., Underhill D. M., Funari V. A. (2015). Mycobiome: approaches to analysis of intestinal fungi. J. Immunol. Methods 421 112–121. 10.1016/j.jim.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot C. M., Koenigsknecht M. J., Carlson P. E., Jr., Hatton G. E., Nelson A. M., Li B. (2014). Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5:3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-White K. J., Hilt E. E., Fok C., Pearce M. M., Mueller E. R., Kliethermes S. (2016). Incontinence medication response relates to the female urinary microbiota. Int. Urogynecol. J. 27 723–733. 10.1007/s00192-015-2847-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipton L., Ghedin E., Morris A. (2017). The lung mycobiome in the next-generation sequencing era. Virulence 8 334–341. 10.1080/21505594.2016.1235671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Hamady M., Fraser-Liggett C. M., Knight R., Gordon J. I. (2007). The human microbiome project. Nature 449 804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda C., Djukovic A., Isaac S. (2017). Roles of the intestinal microbiota in pathogen protection. Clin. Transl. Immunol. 6:e128. 10.1038/cti.2017.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill D. M., Iliev I. D. (2014). The mycobiota: interactions between commensal fungi and the host immune system. Nat. Rev. Immunol. 14 405–416. 10.1038/nri3684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utter D. R., Mark Welch J. L., Borisy G. G. (2016). Individuality, stability, and variability of the plaque microbiome. Front. Microbiol. 7:564. 10.3389/fmicb.2016.00564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Woerden H. C., Gregory C., Brown R., Marchesi J. R., Hoogendoorn B., Matthews I. P. (2013). Differences in fungi present in induced sputum samples from asthma patients and non-atopic controls: a community based case control study. BMC Infect Dis. 13:69. 10.1186/1471-2334-13-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaneechoutte M. (2017). The human vaginal microbial community. Res. Microbiol. 168 811–825. 10.1016/j.resmic.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Vesty A., Biswas K., Taylor M. W., Gear K., Douglas R. G. (2017). Evaluating the impact of DNA extraction method on the representation of human oral bacterial and fungal communities. PLoS One 12:e0169877. 10.1371/journal.pone.0169877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wargo M. J., Hogan D. A. (2006). Fungal–bacterial interactions: a mixed bag of mingling microbes. Curr. Opin. Microbiol. 9 359–364. 10.1016/j.mib.2006.06.001 [DOI] [PubMed] [Google Scholar]
- Wesolowska-Andersen A., Bahl M. I., Carvalho V., Kristiansen K., Sicheritz-Ponten T., Gupta R. (2014). Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2:19. 10.1186/2049-2618-2-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J. B. (1978). Regional differences in the lung. Chest 74 426–437. 10.1016/s0012-3692(15)37392-x [DOI] [PubMed] [Google Scholar]
- Westerberg D. P., Voyack M. J. (2013). Onychomycosis: current trends in diagnosis and treatment. Am. Fam. Phys. 88 762–770. [PubMed] [Google Scholar]
- Wheeler M. L., Limon J. J., Bar A. S., Leal C. A., Gargus M., Tang J. (2016). Immunological consequences of intestinal fungal dysbiosis. Cell Host Microbe 19 865–873. 10.1016/j.chom.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White T. C., Findley K., Dawson T. L., Jr., Scheynius A., Boekhout T., Cuomo C. A., et al. (2014). Fungi on the skin: dermatophytes and Malassezia. Cold Spring Harb. Perspect. Med. 4:a019802. 10.1101/cshperspect.a019802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems H. M., Kos K., Jabra-Rizk M. A., Krom B. P. (2016). Candida albicans in oral biofilms could prevent caries. Pathog. Dis. 74:ftw039. 10.1093/femspd/ftw039 [DOI] [PubMed] [Google Scholar]
- Wu G. D., Chen J., Hoffmann C., Bittinger K., Chen Y. Y., Keilbaugh S. A. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334 105–108. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Sobue T., Bertolini M., Thompson A., Dongari-Bagtzoglou A. (2016). Streptococcus oralis and Candida albicans synergistically activate mu-calpain to degrade e-cadherin from oral epithelial junctions. J. Infect. Dis. 214 925–934. 10.1093/infdis/jiw201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. Y., Fu F., Kong W. N., Xuan Q. K., Wen D. H., Chen X. Q. (2018). Streptococcus agalactiae inhibits candida albicans hyphal development and diminishes host vaginal mucosal TH17 response. Front. Microbiol. 9:198. 10.3389/fmicb.2018.00198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wang X., Li H., Ni C., Du Z., Yan F. (2018). Human oral microbiota and its modulation for oral health. Biomed. Pharmacother. 99 883–893. 10.1016/j.biopha.2018.01.146 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Lv J., Pan L., Zhang Y. (2018). Roles and applications of probiotic Lactobacillus strains. Appl. Microbiol. Biotechnol. 102 8135–8143. 10.1007/s00253-018-9217-9 [DOI] [PubMed] [Google Scholar]
- Zoetendal E. G., Rajilic-Stojanovic M., de Vos W. M. (2008). High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57 1605–1615. 10.1136/gut.2007.133603 [DOI] [PubMed] [Google Scholar]
- Zuo T., Wong S. H., Cheung C. P., Lam K., Lui R., Cheung K., et al. (2018). Gut fungal dysbiosis correlates with reduced efficacy of fecal microbiota transplantation in Clostridium difficile infection. Nat. Commun. 9:3663. 10.1038/s41467-018-06103-6 [DOI] [PMC free article] [PubMed] [Google Scholar]