Abstract
In this prospective study, nasal swab samples from patients with acute respiratory infections were evaluated for the presence of Mycoplasma pneumoniae. This PCR-plus-hybridization-based detection was associated with the detection of other viral agents. During the five winter surveillance periods, 3,897 samples were collected by 75 medical practitioners participating in the Groupe Régional d’Observation de la Grippe surveillance network in Rhône-Alpes (France). M. pneumoniae was detected in 283 samples (7.3%); its rate of detection ranged from 10.1 to 2.0% over the five periods, and it was the second most frequently isolated pathogen during the survey, after influenza A. Three high-prevalence winters were observed, yielding an early winter peak of M. pneumoniae infection which was observed in all age groups. No statistically significant difference was detected between rates of infections in the different age groups, but M. pneumoniae infection was significantly related to lower respiratory tract infection during periods of high prevalence. This study defined the frequency of M. pneumoniae detection from nasal swab specimens in patients with acute respiratory infections, confirming its high prevalence and the presence of large outbreaks due to this pathogen.
Mycoplasma pneumoniae is a common respiratory pathogen, found mainly during fall and winter (9), that is responsible for mild acute respiratory infections (ARIs) such as sore throats, pharyngitis, rhinitis, and tracheobronchitis (2). Apart from the seasonal variation, epidemic peaks of M. pneumoniae infections have been observed in some countries about every 4 to 7 years (6, 14).
Besides mild infections, M. pneumoniae is also a causative agent of atypical pneumonia resistant to β-lactam therapy. Rapid diagnosis procedures are required to diagnose severe pneumonia due to this pathogen in order to implement appropriate therapy. This diagnosis used to rely on culture, a time-consuming and relatively insensitive technique, and serological methods, which are also insensitive and give only a retrospective diagnosis (11, 20). The lack of convenient classical methods led to the development of alternative rapid tests such as antigen detection, hybridization with DNA probes, and genomic amplification by PCR (3). Among these, PCR has shown to be promising because of its sensitivity and specificity (3, 7, 13, 25). Several procedures for sample preparation and different primer sets have already been described (4, 21, 26).
The development of these rapid and accurate techniques means that they can also be of help in estimating the frequency of M. pneumoniae mild upper respiratory tract infections occurring in outpatients, as well as for the monitoring of possible M. pneumoniae epidemics (1, 13).
In France, detection and monitoring of influenza virus outbreaks are performed each year by the Groupe Régional d’Observation de la Grippe (GROG) surveillance network. This network was implemented in 1987 in different regions of France in accordance with World Health Organization guidelines. Every year, virological surveillance of influenza outbreaks is performed by nasal swab specimen collection by volunteer general practitioners and pediatricians from outpatients presenting with clinical signs and symptoms suggestive of viral ARI (17, 19). The swab samples that are sent to the laboratory and analyzed for the presence of influenza viruses are also processed for the detection of other microorganisms (19). Since winter 1992, M. pneumoniae has been systematically sought by PCR in each swab received through the GROG network of Rhône-Alpes (France).
Here we present the results of a five-winter surveillance (i) to analyze the overall incidence of respiratory M. pneumoniae infections occurring in outpatients presenting with ARI, (ii) to characterize the form and frequency of M. pneumoniae epidemics among years, and (iii) to analyze the frequency of M. pneumoniae infections in different age groups and compare the infection rates with those due to respiratory viral pathogens.
MATERIALS AND METHODS
Nasal swab samples from patients.
Volunteer general practitioners and pediatricians (55 and 20, respectively) located in different areas of the Rhône-Alpes region of France (5.4 million inhabitants) were included in the network and performed the sampling. This network represents approximately 1% of the medical practitioners in the region. The study periods were (i) between week 40 of 1992 and week 13 of 1993, (ii) between week 42 of 1993 and week 16 of 1994, (iii) between week 40 of 1994 and week 11 of 1995, between week 41 of 1995 and week 12 of 1996, and between week 40 of 1996 and week 13 of 1997.
During these five winters, a total of 3,897 nasal swabs were sent to the National Influenza Center in the Reference Laboratory of Virology in Lyon (average, 37.8 per week; range, 1 to 157 per week). Some samples were hand carried to the laboratory, and some were sent to the laboratory by mail. Mailed samples were Virocult swabs kept in transport medium to ensure the viability of the viruses (15).
The only criterion for the enrollment of a patient was the presence of an ARI with clinical signs and symptoms suggestive of influenza (fever of over 38°C plus one respiratory symptom including cough, sore throat, or nasal symptoms plus one constitutional symptom including headache, myalgias, chills, or fatigue). The specimens were systematically sent in with a completed, standardized questionnaire including patient demographics, time of onset of symptoms, temperature, clinical symptoms, physical findings, influenza vaccination status, and the presence of other similar cases of illness in the family or in contact persons.
Sample preparation for M. pneumoniae detection by PCR.
Detection of M. pneumoniae by PCR was systematically performed for each swab. Briefly, at arrival in the laboratory, the swab was removed from the transport tube and its content were expressed in a sterile glass tube containing 2.5 ml of Eagle’s minimum essential medium (Biowhittaker, Verviers, Belgium); the transport medium was also added. The tube was vigorously agitated with a vortex mixer, and 300 μl of this mixture was subsequently used for PCR. The remaining volume was used for virus detection.
The 300-μl aliquot was centrifuged at 14,000 × g for 20 min. The supernatant was discarded, and the pellet was resuspended in 100 μl of lysis buffer (10 mM Tris HCl [pH 8], 1 mM EDTA, 0.1% Triton X-100 [Sigma], 100 μg of proteinase K/ml [Boehringer Mannheim]). The mixture was incubated for 30 min at 55°C and then for 10 min at 95°C. The sample was then ready for PCR or could be frozen and stored for later use.
PCR amplification.
For M. pneumoniae-specific amplification, we used primers MPP11 (5′TGCCATCAACCCGCGCTTAAC) and MPP12 (5′CCTTTGCAACTGCTATAGTA) (purchased from Eurogentec) (7). The target sequence for amplification was a 466-bp segment of the gene coding for the P1 cytadhesin protein. The final volume of the PCR mixture was 50 μl, and it contained 10 μl of the extracted solution described above; 0.5 μM each primer; 200 μM each dATP, dCTP, dTTP (Eurogentec), and dGTP, 50 mM KCl, 10 mM Tris HCl (pH 9); 1.5 mM MgCl2; 0.1% (vol/vol) Triton X-100; and 1 U of Taq DNA polymerase (Promega, Madison, Wis.). The reaction was performed in a 96-well microplate (Thermowell-H; Costar, Cambridge, Mass.) placed in a thermocycler (Omnigene; Hybaid). PCR runs consisted of a denaturation step of 95°C for 3 min, followed by 36 cycles of amplification, each consisting of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min. A final elongation step of 5 min at 72°C was done to ensure that the polymerization of every amplified fragment was completed. A positive control (M. pneumoniae FH) and negative controls (water, DNA-free extraction mixture, and DNA-free PCR mixture) were systematically run in parallel.
Analysis of amplified samples.
The PCR products were analyzed on a 1.5% (wt/vol) agarose gel (Seakem GTG; FMC, Rockland, Maine). Electrophoresis was run in 1× Tris-borate-EDTA (Sigma) and followed by ethidium bromide staining. Colored gels were photographed under UV light by using an MP4+ Polaroid camera.
Dot blot hybridization in the winters of 1992-1993 and 1993-1994.
The dot blot hybridization assay is considered a reference method for the detection of amplified M. pneumoniae DNA (12). Briefly, 10 μl of the amplified product was applied to a prewetted (10× SSC [1× SSC is 0.15 M NaCl plus 0.015 sodium citrate]), positively charged nylon membrane (Hybond N+; Amersham) by using a dot blot manifold (Bio-Rad). The samples were immobilized and denatured on the nylon membrane by UV irradiation. The membrane was prehybridized for 30 min at 53°C with 1-ml/cm2 1% blocking buffer (Amersham) in 10 mM maleic acid–15 mM NaCl (pH 7.5). Fluorescein-labeled probe MP-I (5′ CAA ACC GGG CAG ATC ACC TTT; Eurogentec) was added to a concentration of 500 pM to the hybridization solution (5× SSC, 0.1% N-lauroylsarcosine [Sigma], 0.02% sodium dodecyl sulfate, 5% blocking reagent), and hybridization was performed at 53°C. After 45 min of incubation, three washes were done in 1× SSC–1% sodium dodecyl sulfate for 5 min each at 53°C, followed by one wash with 1× SSC. The fluorescein-labeled hybrid was detected immunologically. The detection system used a chemiluminescence assay (Boehringer Mannheim). The bound DNA probe was visualized after incubation for 15 min to overnight at 37°C in the dark.
Detection of amplified DNA by DEIA since the winter of 1994-1995.
The Gen-Eti-K-DNA enzyme immunoassay (DEIA; DiaSorin) is based on the immobilization of a capture probe-amplification product hybrid on a solid phase by using a biotin-avidin bridge (23). DNA duplexes are detected by an anti-DNA mouse monoclonal antibody that specifically reacts with double-stranded DNA (dsDNA) but not with single-stranded DNA. The specific ds-DNA–anti-DNA antibody complexes are visualized by a peroxidase-conjugated anti-mouse antibody and a chromogen-substrate mixture. The detection of amplified M. pneumoniae DNA with the DEIA was carried out in accordance with the manufacturer’s recommendations. Avidin-coated plates were incubated overnight at 4°C with 50 ng of biotinylated oligonucleotide probe MP-I per well. Wells were washed five times with 300 μl of washing solution. Twenty microliters of the sample was denatured by incubation for 10 min at 96°C and then quickly cooled on ice. The sample was incubated with 100 μl of hybridization buffer at 50°C for 60 min on a heating block (Microelisa System; Organon Teknika). After five washes done as described above, 100 μl of the anti-dsDNA antibody diluted 1:50 was added and the mixture was incubated for 60 min at room temperature. After a new washing procedure, bound anti-dsDNA antibody was detected by adding 100 μl of horseradish peroxidase linked to rabbit anti-mouse immunoglobulin G. Following 60 min of incubation at room temperature and five washes, 100 μl of chromogen substrate mixture was added. The colorimetric reaction was allowed to develop for 30 min in the dark at room temperature prior to being stopped with 200 μl of 1 N sulfuric acid. A450 and A630 were determined with a microtiter plate reader. The A630 was subtracted from the A450. The DEIA cutoff value was calculated as the mean value of all negative controls plus 0.2 absorbance unit, providing a cutoff value of 0.25 to 0.30 absorbance unit.
Detection of other infectious agents.
As the samples were collected by the GROG network, implemented for the detection and monitoring of influenza epidemics, detection and/or inoculation onto tissue cell culture were systematically performed for the diagnosis of influenza A and B viruses as previously described (18). The swabs were also systematically processed to detect other viruses, such as respiratory syncytial virus (RSV), coronavirus, rhinovirus, and parainfluenza virus, as previously described (19).
Statistical analysis.
Statistical analysis was performed by using a chi-square test and multivariate analysis for determination of odds ratios (ORs) and relative risks (RRs). Estimation of impact was based on the facts that GROG practitioners represent 1% of the practitioners in the region and that they sample approximately 2% of their patients presenting with ARI.
RESULTS
Frequency of detection of M. pneumoniae and other pathogens.
Practitioners in the network have collected swabs from an average of 1.8% (1 of 55) of their patients presenting with influenza-like illnesses (ILIs) (range, 1.2 to 2.5%). Among the 3,897 nasal swabs collected, 283 (7.6%) were positive for M. pneumoniae by PCR. Over the five periods of surveillance, different rates of detection were recorded (97 of 959 [10.1%] in 1992-1993, 67 of 690 [9.7%] in 1993-1994, 81 of 933 [8.7%] in 1994-1995, 15 of 766 [2.0%] in 1995-1996, and 23 of 549 [4.2%] in 1996-1997.
Apart from M. pneumoniae, other etiologic agents responsible for respiratory tract infections were also identified. The frequency of detection of respiratory pathogens varied from year to year as shown in Table 1. Mixed infections combining M. pneumoniae and a viral agent (influenza A or B virus or RSV) were also recorded every year (Table 2).
TABLE 1.
Number of samples positive for different respiratory pathogens detected in enrolled patients
| Winter | Number (%) of samples positive
|
Total no. (%) of positive samples | Total no. of samples | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| M. pneumoniae | Influenza A virus | Influenza B virus | RSV | Coronavirus | Rhinovirus | Adenovirus | Parainfluenzavirus | |||
| 1992–1993 | 97 (10.1) | 15 (1.6) | 143 (15) | 65 (6.8) | 10 (1) | 15 (1.6) | 11 (1.2) | 2 (0.3) | 358 (35.7) | 959 |
| 1993–1994 | 67 (9.7) | 176 (43.8) | 8 (1.1) | 29 (4.2) | 96 (13.9) | 11 (1.6) | 10 (1.5) | 4 (0.6) | 401 (58) | 690 |
| 1994–1995 | 81 (8.7) | 64 (6.8) | 47 (5) | 108 (11.6) | 64 (6.8) | 35 (3.7) | 22 (2.3) | 3 (0.3) | 424 (45.4) | 933 |
| 1995–1996 | 15 (2.0) | 150 (19.6) | 19 (2.5) | 36 (4.7) | NDa | 3 (0.4) | 8 (1) | 8 (1) | 239 (31.2) | 766 |
| 1996–1997 | 23 (4.2) | 110 (20) | 76 (13.8) | 17 (3) | ND | 4 (0.7) | 8 (1.4) | 8 (1.4) | 254 (46.2) | 549 |
| Total | 283 (7.3) | 515 (13.2) | 293 (7.5) | 255 (6.5) | 170 (4.3) | 68 (1.7) | 59 (1.5) | 25 (0.5) | 1,668 (42.8) | 3,897 |
ND, not determined.
TABLE 2.
Number of mixed infections detected during the 5-year surveillance
| Winter | No. of samples positive
|
Total no. of samples | |||
|---|---|---|---|---|---|
| Influenza A virus + M. pneumoniae | Influenza B virus + M. pneumoniae | RSV + M. pneumoniae | M. pneumoniae totala | ||
| 1992–1993 | 0 | 22 | 8 | 97 | 959 |
| 1993–1994 | 21 | 0 | 2 | 67 | 690 |
| 1994–1995 | 2 | 0 | 20 | 81 | 933 |
| 1995–1996 | 4 | 0 | 1 | 15 | 766 |
| 1996–1997 | 11 | 0 | 1 | 23 | 549 |
Including mixed infections.
The distribution of the pathogens changed during the 5-year surveillance; M. pneumoniae ranked first to fifth in frequency of detection among samples. After influenza A virus, M. pneumoniae was the second most frequent pathogen detected during this 5-year survey. The annual impact of M. pneumoniae measured during the 6-month surveillance ranged from 1,234 cases/100,000 inhabitants during the 1994-1995 surveillance to 190 cases/100,000 inhabitants during the 1995-1996 surveillance.
Seasonal distribution.
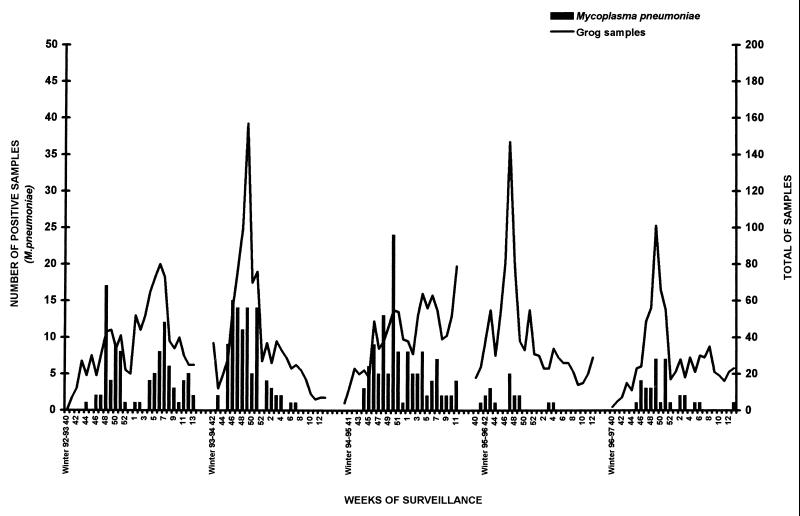
Every year, at least one peak of M. pneumoniae infection was observed during late autumn (October to December), and it was variable in duration and intensity, as summarized in Fig. 1 and 2. During the winter 1992-1993 surveillance, a second peak was observed. During the first three winters of the study, M. pneumoniae was frequently detected, accounting for approximately 25% of the pathogens detected (Table 1) and up to 50% of the positive nasal swabs processed during M. pneumoniae peaks. During these peaks, no other viral pathogen was detected in more than 10% of the samples. On the other hand, during the winter of 1995-1996, only 18 swabs were positive, reflecting a very low rate of diffusion.
FIG. 1.
Seasonal distribution of collected specimens (curves) and samples positive for M. pneumoniae (bars). The surveillance was stopped between weeks 14 and 40 of each year.
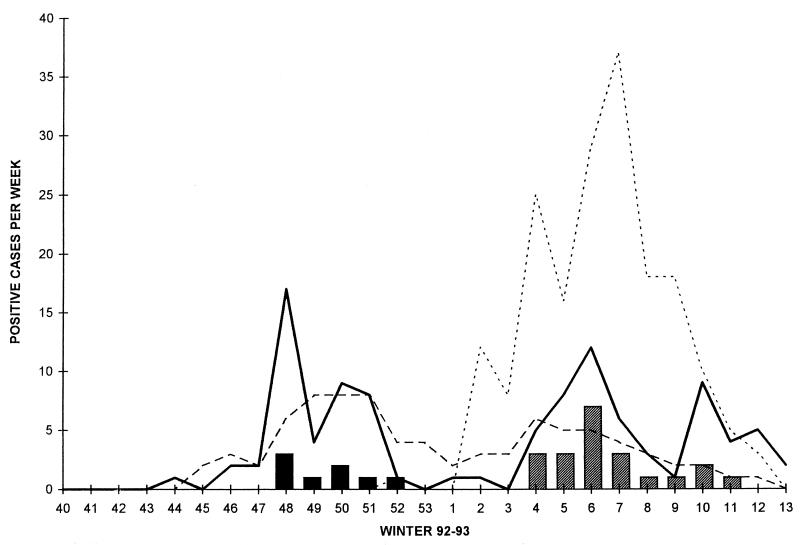
FIG. 2.
Seasonal distribution of samples positive for M. pneumoniae (——), RSV (––––), influenza B virus (… …), and mixed infections due to M. pneumoniae and RSV (■) and M. pneumoniae and influenza B virus (▨) during the winter 1992-1993 surveillance.
As a global viral surveillance was performed at the same time, various viruses were detected or isolated from GROG swabs. Every year, the influenza epidemic was monitored and other virus-related epidemics were recorded (data not shown).
Age distribution.
The age distribution of M. pneumoniae infections is presented in Table 3. These infections were detected in all age groups. No statistically significant difference was observed between rates of detection in the different age groups, including the <4-year-old group that represented a large number of samples (P > 0.05; OR < 0.5).
TABLE 3.
Distribution of samples collected and samples positive for M. pneumoniae over the five periods of surveillance and among age groups
| Age of patients (yr) | Winter 1992–1993
|
Winter 1993–1994
|
Winter 1994–1995
|
Winter 1995–1996
|
Winter 1996–1997
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of samples (% of total) | No. of samples positive for M. pneumoniae (% of total) | No. of samples (% of total) | No. of samples positive for M. pneumoniae (% of total) | No. of samples (% of total) | No. of samples positive for M. pneumoniae (% of total) | No. of samples (% of total) | No. of samples positive for M. pneumoniae (% of total) | No. of samples (% of total) | No. of samples positive for M. pneumoniae (% of total) | |
| <4 | 283 (29.5) | 32 (11.3) | 242 (35.1) | 21 (31.3) | 377 (40.4) | 41 (50.6) | 278 (36.3) | 7 (46.7) | 137 (25.0) | 4 (17.4) |
| 5–14 | 220 (22.9) | 24 (10.9) | 118 (17.1) | 8 (6.8) | 155 (16.6) | 14 (9.0) | 177 (23.1) | 3 (1.7) | 91 (16.6) | 4 (4.4) |
| 15–29 | 147 (15.3) | 15 (10.2) | 122 (17.7) | 11 (9.0) | 128 (13.7) | 10 (7.8) | 103 (13.4) | 1 (1.0) | 98 (17.9) | 5 (5.1) |
| 30–44 | 135 (14.1) | 12 (8.9) | 106 (15.4) | 14 (13.2) | 134 (14.4) | 6 (4.5) | 113 (14.8) | 1 (0.9) | 86 (15.7) | 2 (2.3) |
| 45–59 | 55 (5.7) | 7 (12.7) | 55 (8.0) | 9 (16.4) | 78 (8.4) | 6 (7.7) | 48 (6.3) | 1 (2.1) | 61 (11.1) | 3 (4.9) |
| 60–65 | 23 (2.4) | 3 (13.0) | 10 (1.4) | 1 (10.0) | 16 (1.7) | 0 | 10 (1.3) | 0 | 21 (3.8) | 0 |
| >65 | 18 (1.9) | 0 | 22 (3.2) | 2 (9.1) | 32 (3.4) | 3 (9.4) | 16 (2.1) | 1 (6.3) | 36 (6.6) | 4 (11.1) |
| ? | 78 (8.1) | 4 (5.1) | 15 (2.2) | 1 (6.7) | 13 (1.4) | 1 (7.7) | 21 (2.7) | 1 (4.8) | 19 (3.5) | 1 (5.3) |
| Total | 959 | 97 (10.1) | 690 | 67 (9.7) | 933 | 81 (8.7) | 766 | 15 (2.0) | 549 | 23 (4.2) |
Clinical presentation.
During the three winters with a high prevalence of M. pneumoniae (1992 to 1994) and whatever the age group, the lower respiratory tract infection (LRTI) syndrome was significantly associated with M. pneumoniae (P < 0.0001; OR > 3, RR > 2). However, during the winters with a low prevalence of M. pneumoniae (1995 to 1997), no difference was observed between clinical signs of LRTI recorded from M. pneumoniae-infected patients and patients infected with other agents (P > 0.05; OR = 0.58; Table 4).
TABLE 4.
Percentage of clinical syndromes associated with M. pneumoniae diagnosis
| Winters | No. of samples positive for M. pneumoniae/total no. of samples | % of samples associated with M. pneumoniae (other agents) and:
|
||||
|---|---|---|---|---|---|---|
| URTIa | ILI | LRTI | Otitis | Other diagnoses | ||
| 1992–1994 | 245/2,582 | 27.1 (34.6) | 24.5 (33.3) | 31.9b (13.4) | 6.7 (8.1) | 9.6 (5.1) |
| 1995–1997 | 38/1,315 | 29.5 (43) | 29 (34) | 15.7c (14) | 7.2 (7) | 18 (2) |
URTI, upper respiratory tract infection.
Statistically significant difference (P < 0.00001; OR = 3.02).
No statistically significant difference (OR = 0.58).
DISCUSSION
In our study, M. pneumoniae was the second most frequent cause of respiratory tract infection in all age groups of outpatients, with a calculated impact reaching 1,234 cases per 100,000 inhabitants during the winter of 1994-1995. This result is in agreement with previously reported studies (2, 10, 11). This high prevalence was certainly due to the use of a reliable, sensitive detection technique; PCR has proved its usefulness for accurate diagnosis and is now accepted as a clinically useful technique (18). Our study also confirmed that M. pneumoniae can be responsible for epidemics. During our survey, it was detected in up to 50% of the swabs sampled from patients presenting with ARI in a given period (Fig. 1). Even if M. pneumoniae infection provoked various clinical presentations, LRTIs were more frequently observed in patients infected with M. pneumoniae (Table 4). This was previously observed in a study conducted in a pediatric practice (5). The various presentations of M. pneumoniae infection might be a consequence of a bias due to the recruitment of patients. These were included since they presented with ARI, a nonspecific syndrome that suggests an influenza ARI, as well as those due to other respiratory pathogens (16). On the other hand, since the major aim of the GROG network is to detect the onsets of influenza epidemics, practitioners are keen to collect samples from patients with an identical presentation, regardless of specific ILI criteria (one symptom may be lacking). This behavior encouraged detection of epidemics due to any specific pathogen and facilitated the surveillance for respiratory tract infections, as we have shown in our previously published study (19).
The high rate of detection of M. pneumoniae during the first three winters raised the question of the pathogenicity of the strains detected. The dramatic drop in its detection observed during the last 2 years of the surveillance (Table 1) suggests that the detection of M. pneumoniae observed in previous years was directly correlated with its epidemiological pattern among positive patients, i.e., sporadic circulation versus epidemic events (1). Moreover, the low incidence correlated with the lack of specific association with LRTI as observed during high-incidence periods (Table 4).
According to Jacobs (14), M. pneumoniae strains can be divided into two groups (groups 1 and 2) with different adherence abilities; the immune response lacks adherence-inhibiting antibodies during M. pneumoniae group 2 infection. Jacobs suggested that group 1 M. pneumoniae strains are more likely to develop epidemics. Since our PCR cannot type the strains, we are unable to confirm this finding.
Possible M. pneumoniae carriage has been suggested by several authors (4, 9, 11, 18). Among them, Foy (9) reported that M. pneumoniae was recovered by culture from throat samples up to 4 months after illness. It has been suggested that a quantitative PCR assay technique would be useful in determining the number of DNA copies (or the equivalent number of CFU per milliliter) of M. pneumoniae in positive patients; the determination of a threshold in this quantitative assay could then differentiate between putative carriers and patients with M. pneumoniae-related respiratory tract infections (4, 18). This approach has been developed by Williamson et al. (27). They suggested that the threshold is 104 DNA copies per ml (equivalent to 103 CFU/ml) of throat washing (2 to 5 ml). Beyond this value, all patients were unequivocally infected with M. pneumoniae. Gnarpe et al. (11) detected M. pneumoniae in up to 13.5% of the throat samples from a healthy population. They concluded that M. pneumoniae is a common finding in the general population and that the rate of detection varies greatly from one period to another. As the sensitivity of our PCR assay is approximately 102 CFU/ml (unpublished data), we may have detected nonpathogenic strains collected from carriers. This detection of nonpathogenic M. pneumoniae strains may have occurred during low-prevalence years, explaining the lack of association with LRTI, as observed during years of high prevalence. According to our results, it is likely that M. pneumoniae may be isolated or detected in patients with no evidence of disease. The use of a quantitative PCR assay will help us to learn more about the carrier state (9). In studies recruiting patients presenting with mild infections such as the common cold (22), the prevalence of M. pneumoniae was low (2%). This is in accordance with our results and suggests that M. pneumoniae is not a causative agent of such mild diseases. The 2% detection rate might just reflect the percentage of healthy carriers.
In some of the cases in our study, M. pneumoniae coinfected with viral strains (influenza A or B virus or RSV). These mixed infections were not severe but were responsible for specific clinical presentations such as otitis in children (13). During the 1992-1993 surveillance, two peaks of M. pneumoniae infection were recorded. These coincided with two viral epidemics (RSV and influenza B virus). The mixed infections were recorded during the coincident peaks, indicating cocirculation of the strains (Fig. 2). This strongly suggests that M. pneumoniae can superinfect patients presenting with viral infections, either in the early stage of infection (the first 3 days) or in the late stage during the recovery of respiratory cells. During the surveillance, the rate of infection varied with the year. This variation was related neither to the rate of detection of other pathogens nor to any specific climatic changes. Such dramatic changes, which have already been observed by others, suggest cycles of M. pneumoniae epidemics (8, 14, 24). The high rate observed during the winter of 1992-1993 was also reported in Poland by Rastawicki et al. (24).
This analysis of outpatients infected with M. pneumoniae emphasizes its pathogenic role but revealed no specific sign or symptoms. M. pneumoniae was responsible for rhinopharyngitis, bronchitis, and otitis. These symptoms were observed in the different age groups, unlike in previous studies (8). Interestingly, no specific increase in severe atypical pneumonia was observed during winters with a high prevalence of M. pneumoniae infection.
M. pneumoniae is a frequent pathogen in upper respiratory tract infection, and since it can be cured by appropriate therapy, its detection should be systematically performed in patients presenting with ARI.
ACKNOWLEDGMENTS
We are grateful to all of the general practitioners and pediatricians participating in the GROG Rhône-Alpes network; without them, this work would have been impossible. Thanks to François Chappuis for his help with the statistical analysis.
REFERENCES
- 1.Abele-Horn M, Busch U, Nitschiko H, Jacobs E, Bax R, Pfaff F, Schaffer B, Heesemann J. Molecular approaches to diagnosis of pulmonary diseases due to Mycoplasma pneumoniae. J Clin Microbiol. 1998;36:548–551. doi: 10.1128/jcm.36.2.548-551.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atmar R L, Greenberg S B. Pneumonia caused by Mycoplasma pneumoniae and the TWAR agent. Semin Respir Infect. 1989;4:19–31. [PubMed] [Google Scholar]
- 3.Bernet C, Garret M, de Barbeyrac B, Bebear C, Bonnet J. Detection of Mycoplasma pneumoniae by using the polymerase chain reaction. J Clin Microbiol. 1989;27:2492–2496. doi: 10.1128/jcm.27.11.2492-2496.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buck G E, O’Hara L C, Summersgill J T. Rapid, sensitive detection of Mycoplasma pneumoniae in simulated clinical specimens by DNA amplification. J Clin Microbiol. 1992;30:3280–3283. doi: 10.1128/jcm.30.12.3280-3283.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chapman R S, Henderson F W, Clyde W A, Jr, Collier A M, Denny F W. The epidemiology of tracheobronchitis in pediatric practice. Am J Epidemiol. 1981;114:786–797. doi: 10.1093/oxfordjournals.aje.a113249. [DOI] [PubMed] [Google Scholar]
- 6.Clyde W A., Jr Clinical overview of typical Mycoplasma pneumoniae infections. Clin Infect Dis. 1993;17(Suppl. 1):32–36. [PubMed] [Google Scholar]
- 7.De Barbeyrac B, Bernet-Poggi C, Febrer F, Renaudin H, Dupon M, Bebear C. Detection of Mycoplasma pneumoniae and Mycoplasma genitalium in clinical samples by polymerase chain reaction. Clin Infect Dis. 1993;17:83–89. doi: 10.1093/clinids/17.supplement_1.s83. [DOI] [PubMed] [Google Scholar]
- 8.Dominguez A, Minguell S, Torres J, Serrano A, Vidal J, Salleras L. Community outbreak of acute respiratory infection by Mycoplasma pneumoniae. Eur J Epidemiol. 1996;12:131–134. doi: 10.1007/BF00145497. [DOI] [PubMed] [Google Scholar]
- 9.Foy H M. Infections caused by Mycoplasma pneumoniae and possible carrier state in different populations of patients. Clin Infect Dis. 1993;17(Suppl. 1):37–46. doi: 10.1093/clinids/17.supplement_1.s37. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh K, Clements G B. Surveillance of Mycoplasma pneumoniae infections in Scotland 1986–1991. J Infect. 1992;25:221–227. doi: 10.1016/0163-4453(92)94196-5. [DOI] [PubMed] [Google Scholar]
- 11.Gnarpe J, Lundbäck A, Sündelöf B, Gnarpe H. Prevalence of M. pneumoniae in subjectively healthy individuals. Scand J Infect Dis. 1992;24:161–164. doi: 10.3109/00365549209052607. [DOI] [PubMed] [Google Scholar]
- 12.Göbel V B, Geiser A, Stanbridge E J. Oligonucleotide probes complementary to variable regions of ribosomal RNA discriminate between Mycoplasma species. J Gen Microbiol. 1987;133:1969–1974. doi: 10.1099/00221287-133-7-1969. [DOI] [PubMed] [Google Scholar]
- 13.Ieven M, Ursi D, Van Bever H, Quint W, Niesters H G M, Goossens H. Detection of Mycoplasma pneumoniae by two polymerase chain reactions and role of M. pneumoniae in acute respiratory tract infections in pediatric patients. J Infect Dis. 1996;173:1445–1452. doi: 10.1093/infdis/173.6.1445. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs E. Mycoplasma infections of the human respiratory tract. Wien Klin Wochenschr. 1997;109:574–577. [PubMed] [Google Scholar]
- 15.Jensen C, Johnson F B. Comparison of various transport media for viability maintenance of herpes simplex virus, respiratory syncytial virus and adenovirus. Diagn Microbiol Infect Dis. 1994;19:137–142. doi: 10.1016/0732-8893(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 16.Kok T, Higgins G. Prevalence of respiratory viruses and Mycoplasma pneumoniae in sputum samples from unselected adult patients. Pathology. 1997;29:300–302. doi: 10.1080/00313029700169135. [DOI] [PubMed] [Google Scholar]
- 17.Layani M P, Gras I, Allard J P, Aymard M. Program and abstracts of the 10th International Congress of the International Organization for Mycoplasmology (IOM), 19–26 July, Bordeaux, France. 1994. Detection of Mycoplasma pneumoniae in clinical samples from the GROG during winter 92–93 by polymerase chain reaction; pp. 472–473. [Google Scholar]
- 18.Leng Z, Kenny G E, Roberts M C. Evaluation of the detection limits of PCR for identification of Mycoplasma pneumoniae in clinical samples. Mol Cell Probes. 1994;8:125–130. doi: 10.1006/mcpr.1994.1017. [DOI] [PubMed] [Google Scholar]
- 19.Lina B, Valette M, Foray S, Luciani J, Stagnara J, See D M, Aymard M. Surveillance of community-acquired viral infections due to respiratory viruses in Rhône-Alpes (France) during winter 1994 to 1995. J Clin Microbiol. 1996;34:3007–3011. doi: 10.1128/jcm.34.12.3007-3011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lind K, Bentzon M W. Ten and a half years seroepidemiology of Mycoplasma pneumoniae infection in Denmark. Epidemiol Infect. 1991;107:189–199. doi: 10.1017/s0950268800048810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lüneberg E, Skov Jensen J, Frosch M. Detection of Mycoplasma pneumoniae by polymerase chain reaction and nonradioactive hybridization in microtiter plates. J Clin Microbiol. 1993;31:1088–1094. doi: 10.1128/jcm.31.5.1088-1094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mäkelä M J, Puhakka T, Ruuskanen O, Leinonen M, Saikku P, Kimpimäki M, Blomqvist S, Hyypiä T, Arstila P. Viruses and bacteria in etiology of the common cold. J Clin Microbiol. 1998;36:539–542. doi: 10.1128/jcm.36.2.539-542.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mantero G, Antonella Z, Albertini A. DNA enzyme immunoassay: general method for detecting products of polymerase chain reaction. Clin Chem. 1991;37:422–429. [PubMed] [Google Scholar]
- 24.Rastawicki W, Kaluzewski S, Jagielski M. Occurrence of serologically verified Mycoplasma pneumoniae infections in Poland in 1971–1995. Eur J Epidemiol. 1998;14:37–40. doi: 10.1023/a:1007431932087. [DOI] [PubMed] [Google Scholar]
- 25.Razin S. DNA probes and PCR in diagnosis of Mycoplasma infections. Mol Cell Probes. 1994;8:497–511. doi: 10.1006/mcpr.1994.1071. [DOI] [PubMed] [Google Scholar]
- 26.Talkington D F, Thacker W L, Keller D W, Jensen J S. Diagnosis of Mycoplasma pneumoniae infection in autopsy and open-lung biopsy tissues by nested PCR. J Clin Microbiol. 1998;36:1151–1153. doi: 10.1128/jcm.36.4.1151-1153.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williamson J, Marmion B P, Worswick D A, Kok T-W, Tannock G, Herd R, Harris R J. Laboratory diagnosis of Mycoplasma pneumoniae infection. 4. Antigen capture and PCR-gene amplification for detection of the mycoplasma: problems of clinical correlation. Epidemiol Infect. 1992;109:519–537. doi: 10.1017/s0950268800050512. [DOI] [PMC free article] [PubMed] [Google Scholar]