Abstract
Pancreatic ductal adenocarcinoma (PDAC) is known for its poor prognosis with few long-term survivors. This study aimed to establish a prognostic score using unique transcriptomic profiles of long-term survivors to be used as a patient selection tool for meaningful clinical intervention in PDAC. In TCGA PDAC cohort, 16 genes were significantly upregulated in the long-term survivor tumors. A prognostic score was established using these 16 genes by LASSO Cox regression, and PHKG1, HOXA4, ISL2, DMRT3 and TRA2A gene expressions were included in the score. The prognostic value was confirmed in both testing and validation cohorts. The characteristics of the high score tumor was investigated by bioinformatical approach. The high score tumor was associated with TP53 mutation but not with other commonly enhanced signaling pathways in PDAC. The high score tumor was associated with higher tumor mutational burden and unfavorable tumor microenvironment (TME), such as lower infiltration of CD8-positive T cells and dendritic cells, and less cell composition of mature blood vessels and fibroblasts. The high score tumor was also associated with enhanced cell proliferation and margin positivity after surgery. The impact of score component genes on the cell proliferation was investigated by in vitro experiments. Silencing of the score component genes promoted cell proliferation. In conclusion, the prognostic score predicted PDAC patient survival and was associated with cancer aggressiveness such as unfavorable TME and enhanced cell proliferation.
Keywords: Pancreatic ductal adenocarcinoma, long-term survivor, transcriptome, score, survival cancer, HOX
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive and lethal cancers, ranking third in cancer-related cause of death in the USA, with an increasing incidence of 57,600 estimated new cases in 2020 [1]. More than half of PDAC patients have metastatic disease at the time of diagnosis resulting in a dismal prognosis, with median survival of 3-6 months and 5-year survival rate of approximately 10% [2]. Surgical resection is the only curative therapeutic option currently available. However, due to the anatomical location and function of the pancreas, surgery is accompanied with a relatively higher rate of severe complication. Gemcitabine monotherapy has been the standard of care in unresectable or recurrent tumors for decades. Recently FOLFIRINOX and nab-paclitaxel together with gemcitabine became an alternative treatment option. Unfortunately, multiple agent combination therapy has shown severe toxicity limiting its use. Given severe adverse effects and complications of the available medical and surgical treatments, as well as poor prognosis, identifying a biomarker that predicts patient prognosis may be useful for appropriate patient selection to guide treatment.
Known clinical factors that are associated with poor prognosis in PDAC include AJCC TNM staging, surgical margin positivity, perineural and lymphovascular invasion, and elevated tumor markers [3,4]. Some of these clinical factors are also associated with long-term survival of PDAC [5]. PDAC has a complex tumor microenvironment (TME) that includes extracellular matrix, immune cells, blood and lymphatic vessels and fibroblasts. TME plays a crucial role in cancer biology and affects patient prognosis in a myriad of cancers, including PDAC [6,7]. Thus, some TME signatures, assessed by pathology or genetic profiles, can predict patient prognosis [8-11]. We, and others, have reported specific molecules and signaling pathways associated with patient prognosis in PDAC [3,12,13]. Additionally, prognostic scoring systems have been proposed, by combining specific characteristics, such as proliferation and TME [14]. However, these characteristics are only a part of many biological factors, and any unknown factor is not considered when making these scoring systems. Thus, an unbiased strategy to identify prognostic factors may be more straightforward in predicting patient prognosis. Some of the previous studies that used such strategies identified candidate genes by comparing normal and cancerous tissues [15-19]. We argue that differentially expressed genes between normal and cancer tissues may reflect carcinogenesis, generation of cancer from a normal cell, instead of cancer progression, which is worsening of an already existing cancer. Previous studies identified these genes by comparing PDAC patient’s with short survival (defined as less than 1 year) to those who lived longer than a year [20,21]. However, many recent surgical cases live longer than a year, making this comparison less clinically relevant. To this end, we believe comparing long-term survival PDAC patient’s (more than 5-year survival) to common clinical course (less than 3-year survival) is more clinically meaningful. Additionally, patients who undergo surgical resection show better prognosis compared to patients with non-resectable tumors. Therefore, they should be separately analyzed in order to refine the population. In this study, we aimed to establish a prognostic score for surgically resectable PDAC using long-term survivor unique transcriptomic signatures, identified by unbiased strategy, and to investigate underlying PDAC biology associated with patient prognosis utilizing large patient cohorts.
Materials and methods
Data acquisition and pre-processing
There are 186 patients in The Cancer Genome Atlas (TCGA) pancreatic cancer cohort. The clinical data and the gene expression data from RNA-sequence were downloaded through cBioportal (TCGA provisional dataset) [22,23]. Out of 185, 154 patients were pathologically diagnosed as PDAC. Of those, 147 patients have gene expression data from RNA sequence. The patients were divided into long-term survivors who survived more than 5 years and common clinical course patients who died within 3 years after surgery. As a validation cohort, we obtained gene expression profiles together with survival data of 102 PDAC patients from the Gene Expression Omnibus (GEO) database, Gene Series Expression (GSE) 21501 [24]. Since TCGA and GEO are de-identified publicly available cohorts, institutional review board approval was waived, which was the case with previous publications [6].
Gene set enrichment analysis (GSEA)
Gene set enrichment analysis (GSEA) was carried out comparing the high and low prognostic score PDACs using software provided by the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp) using hallmark gene sets, as we described previously [12,25-31]. For each gene expression, the patients were divided by median cutoff. In the circumstance with more than half patient’s expression was lower limit, patients were divided into lower limit and above limit groups. False discovery rate (FDR) <0.25 was considered as significant.
Cell culture and reagents
Human PDAC cell line, MiaPACA-2 was obtained from ATCC (Manassas, VA) and cultured in DMEM (Gibco, Gaithersburg, MD) with 10% fetal bovine serum (FBS) (Gibco) in a humidified incubator at 37°C in 5% CO2. All cell lines were used within 20 passages after revival and were shown to be mycoplasma free using the PlasmoTest kit (InVivoGen, San Diego, CA). Short-hairpin RNA (shRNA) was used for knockdown experiments. MiaPACA-2 cells were transduced with pGIPZ-shNT (non-targeting), pGIPZ-shPHKG1 (clone ID: V2LHS_58678), pGIPZ-shHOXA4 (clone ID: V2LHS_133124), pGIPZ-shISL2 (clone ID: V2LHS_57876), pGIPZ-shTRA2A (clone ID: V2LHS_70113), and selected by puromycin (Gibco) to establish stable knockdown cells.
qPCR
Total RNA was extracted using RNeasy Mini Kit (Qiagen, Velno, Netherlands). Reverse transcription was carried out using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Waltham, MA) according to manufacturer’s instructions. β-actin was used as an internal control. Sequences of primers were shown in Table S1. Data were analyzed using ΔΔCt method and normalized by shNT cells.
Cell proliferation and drug sensitivity assay
5,000 cells were seeded per well of 96-well plate. Cell proliferation was measured at the indicated time point using Cell Counting Kit-8 (Dojindo, Japan), according to the manufacturer’s protocol.
Statistical analysis
Heatmap was generated by Morpheus (https://software.broadinstitute.org/morpheus). The prognostic score was shown to be higher in worse prognosis patients by LASSO Cox regression. Survival analysis was conducted by Kaplan-Meier curve with log-rank test. The patients were divided into the high and low score groups using median cutoff. Clinical demographics were compared by Fisher’s exact test. Continuous value was compared by Student’s t-test. Cell composition fraction of the tumor was estimated by xCell algorithm [32] and compared by Wilcoxon test. Correlation analyses were conducted using Pearson’s correlation coefficiency. Two-sided P<0.05 was considered as statistically significant for all tests. All statistical analyses were performed using R software (http:///www.r-project.org/), Bioconductor (http://bioconductor.org/) and GraphPad Prism 9.0 (GraphPad Software).
Results
Multiple genes are upregulated in PDAC long-term survivors
We investigated transcriptomic profiles using TCGA PDAC cohort. Median overall survival (OS) of the entire PDAC cohort in TCGA was 18.7 months (Figure 1A). Since 3-year OS rate was approximately 20%, we defined the patients who died within 3 years after surgery as common clinical course PDAC (n=76). Conversely, there were 4 patients who survived more than 5 years after surgery (Figure 1B). Surprisingly, we found that all the patients in the long-term survivor group were Stage II, whereas all stages were represented in the common clinical course PDAC group (6, 67, 1 and 2 patients of Stage I, II, III and IV, respectively), suggesting that clinical staging based upon anatomical morphology alone is not enough to predict long-term survival. There was no significant difference in patient demographics investigated between long-term survivor and common clinical course PDAC groups (Table S2).
Figure 1.
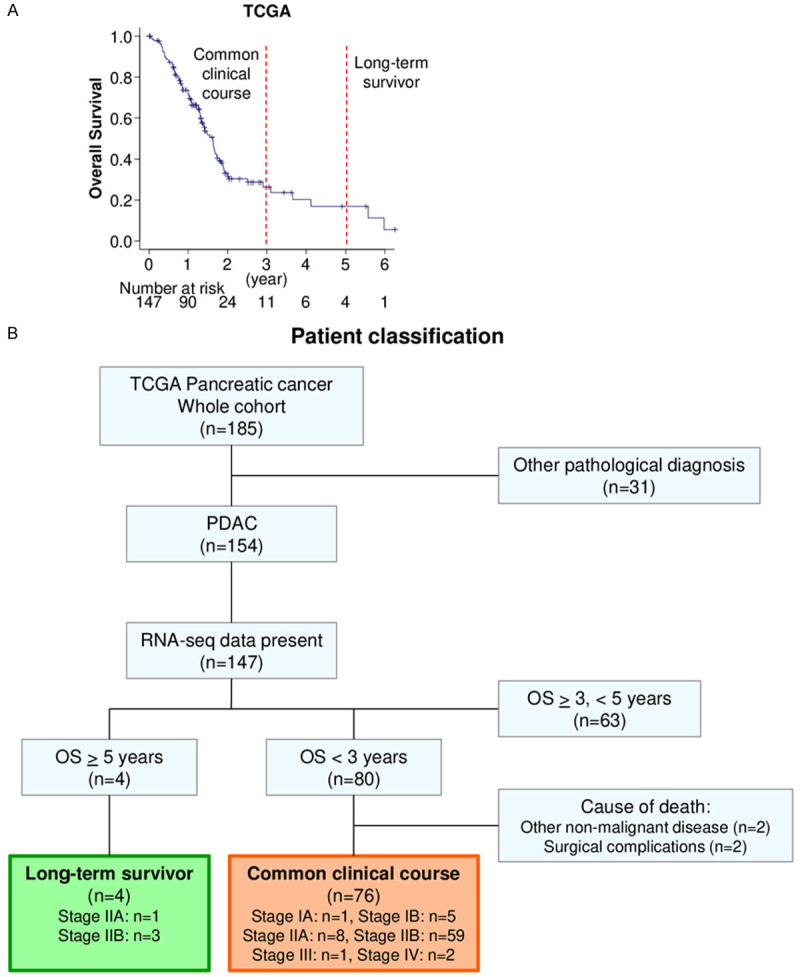
Pancreatic ductal adenocarcinoma (PDAC) cohort of TCGA. A. Overall survival of TCGA PDAC cohort. B. Patient classification of long-term survivor and common clinical course groups of TCGA pancreatic cancer cohort.
In order to investigate long-term survivor unique transcriptomic signatures, gene expression profiles were compared between the long-term survivor and the common clinical course groups. 16 genes were identified as significantly differentially expressed genes with Bonferroni adjusted P<0.05 (Figure 2A). Interestingly, 6 out of 16 genes were members of HOXA family, which were reported as oncogenes [33]. Nevertheless, they were upregulated in the tumor of the long-term survivor. There were 4 clusters by correlation analysis, and 4 of HOXA genes were in the same cluster with high correlation (Figure 2B). All of them were upregulated in the long-term survivor group (Figure 2C).
Figure 2.
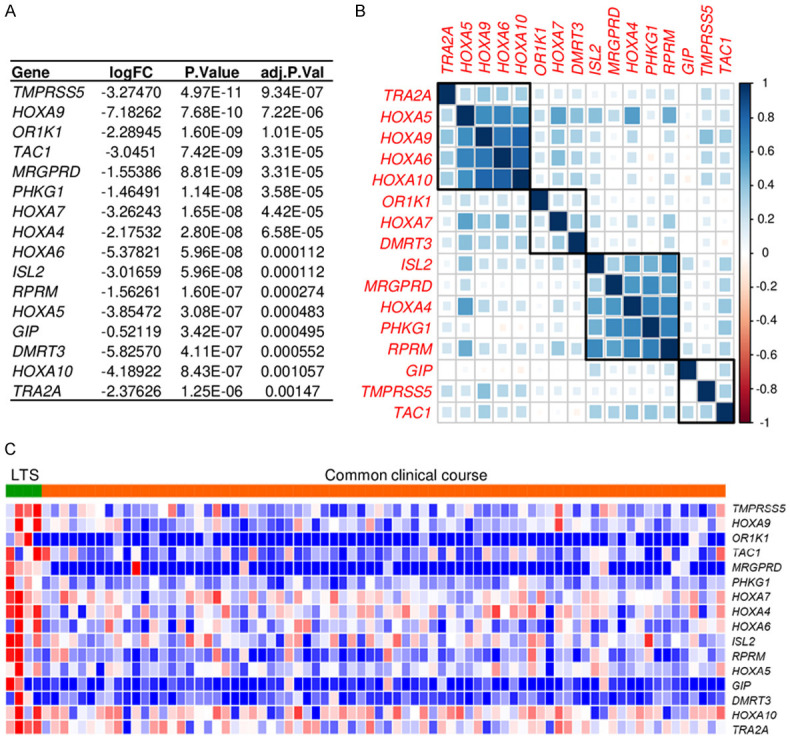
Transcriptomic differences between long-term survivors (LTSs) and common clinical course patients in pancreatic ductal adenocarcinoma (PDAC). A. Genes significantly differentially expressed comparing the tumors of long-term survivor and common clinical course patients with Bonferroni P<0.05 in TCGA. B. Correlation matrix of significantly differentially expressed genes in the whole TCGA PDAC cohort. C. Heatmap of significantly differentially expressed genes in LTSs and common clinical course course patients in TCGA.
We have explored the mechanism by which these 16 genes are highly expressed in long-term survivor by GSEA using TCGA cohort. Interestingly, although these genes were highly expressed in the long-term survivor, 4 of them were significantly associated with epithelial-mesenchymal transition (EMT) (Figure S1). In addition, 3 of them were significantly associated with interferon (IFN)-α response (Figure S1). Contrarily, 4 of them were negatively associated with glycolysis and 3 of them were associated with mTORC1 signaling and protein secretion (Figure S1). Taken together, these genes may associate with high anti-cancer immunity and with less glycolysis and mTOR signaling, resulting in long-term survival, but also associate with EMT.
Establishment of prognostic score in PDAC
In order to establish a prognostic score for PDAC, we applied LASSO Cox regression to the whole PDAC cohort of TCGA utilizing the identified 16 genes for OS. Utilizing cross-validation in the LASSO Cox regression model, the partial likelihood deviance is plotted in log(λ) in which vertical lines are shown at the optimal values by minimum criteria and 1-SE criteria (Figure 3A). Among the identified 16 genes, 5 genes were selected for use in the prognostic score, including PHKG1, HOXA4, ISL2, DMRT3 and TRA2A. Coefficient profile graph was shown in Figure 3B. The prognostic score was generated using these identified 5 genes, correlating with higher scores and worse prognosis patients as shown in Figure 3C.
Figure 3.
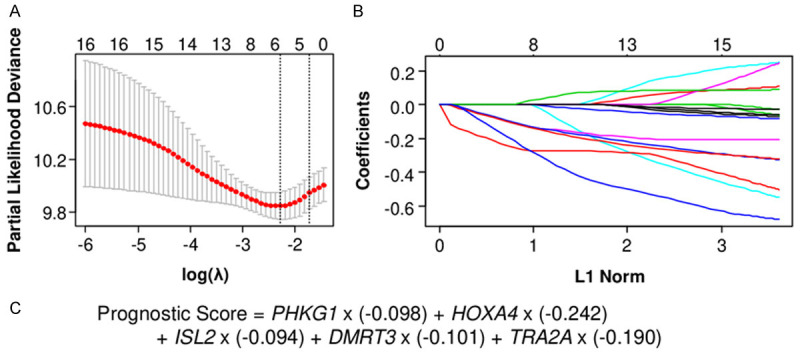
Establishment of the prognostic score in pancreatic ductal carcinoma (PDAC). A. Cross-validation for turning parameter selection in the LASSO Cox regression model. λ is the turning parameter. The partial likelihood deviance is plotted in log(λ), in which vertical lines are shown at the optimal values by minimum criteria and 1-SE criteria. B. Coefficient profiles of 16 identified genes in the LASSO Cox regression model. C. Formula of the prognostic score.
To confirm the prognostic value of the score, whole PDAC patients in TCGA cohort were divided into the high and the low score groups by median cutoff. As expected, the patients with the high score tumor showed significantly worse prognosis in both disease-free survival (DFS) (median survival time: 10.1 vs 25.1 months, P<0.001) and OS (median survival time: 15.6 vs 30.0 months, P<0.001) in the testing TCGA cohort (Figure 4A, 4B). The high prognostic score patients were confirmed to have significantly worse OS in the validation cohort, GSE21501 with 102 PDAC patients (P=0.009) (Figure 4C). On the other hand, the previously reported factors, including AJCC pT, pN and stage did not show significant difference in OS (Figure S2). This is most likely due to patient distribution since most of the patients were diagnosed as pT3 (84%) and Stage II (88%) surgically resected PDACs.
Figure 4.
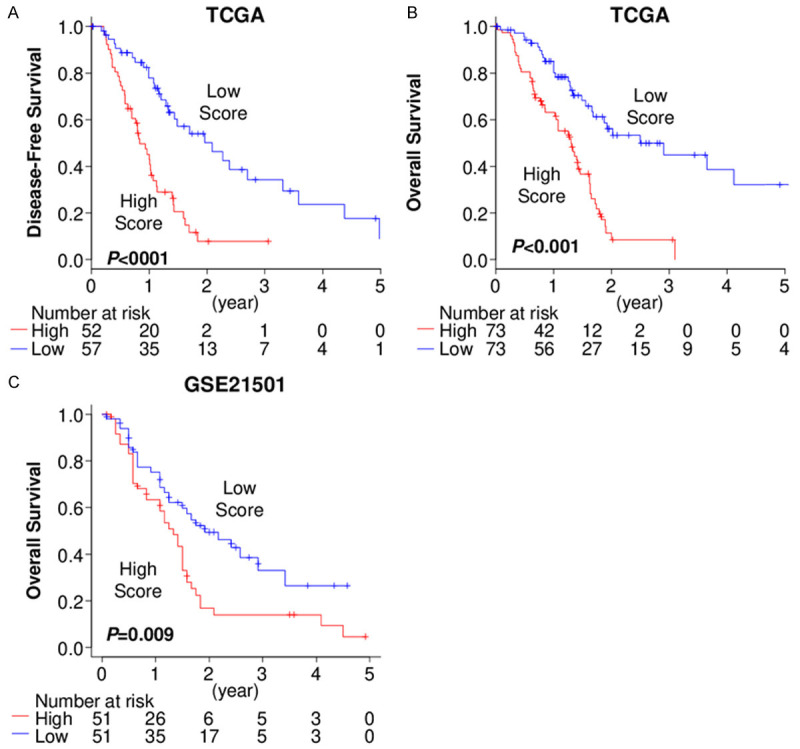
The prognostic score predicts pancreatic ductal adenocarcinoma (PDAC) patient prognosis. A. Disease-free survival comparing the patients with the high and low prognostic score tumors in TCGA. B. Overall survival comparing the patients with the high and low prognostic score tumors in TCGA. C. Overall survival comparing the patients with the high and low prognostic score tumors in GSE21501.
No association of prognostic score with specific signaling pathway
We investigated if the prognostic score is associated with specific gene mutations or known enhanced signaling pathways in PDAC. We found that TP53, KRAS, SMAD4, and TITIN (TTN) were the top 4 highly mutated genes in TCGA PDAC cohort. TP53 mutated tumors have a higher prognostic score than non-mutated tumors (P=0.012) (Figure 5A). However, there was no significant difference in the prognostic score between non-mutated and mutated tumors among KRAS, SMAD4, or TTN (Figure 5A). We further investigated the association of the prognostic score with signaling pathways, including K-ras, TGF-β, Wnt/β-catenin, and Hedgehog signaling, which are known to be enhanced in PDAC [34] and p53 pathway. Unexpectedly, GSEA revealed that none of these gene sets were associated with the prognostic score (Figure 5B).
Figure 5.
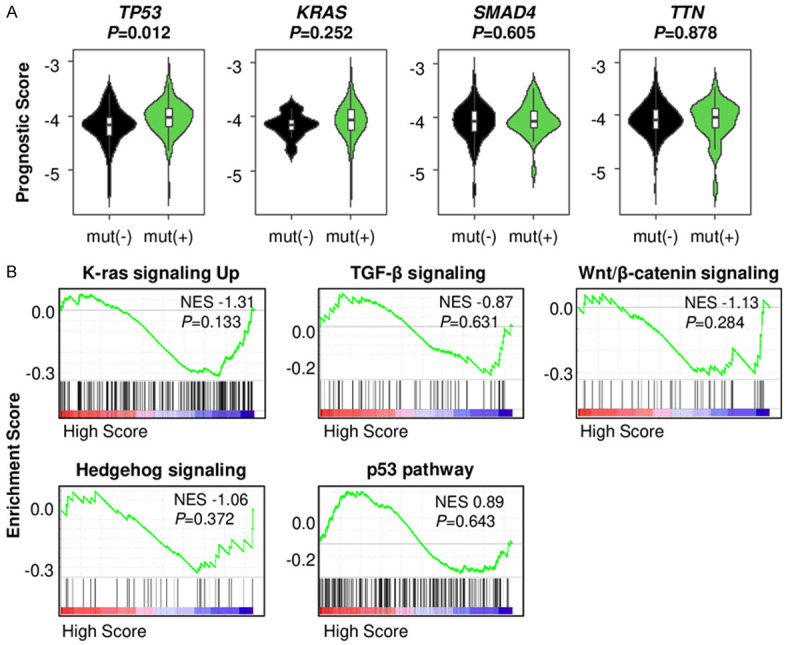
The association of the prognostic score with mutation of genes and signaling pathways in pancreatic ductal adenocarcinoma (PDAC). A. The prognostic score comparison in tumors between non-mutated and mutated TP53, KRAS, SMAD4 and TTN in TCGA PDAC cohort. B. Gene set enrichment analysis (GSEA) of signaling pathways comparing the low and high prognostic score PDACs in TCGA.
High score PDACs are associated with unfavorable tumor immune microenvironment (TME)
It is well known that tumors with an enhanced anti-cancer immune microenvironment show better prognosis [35]. To this end, we investigated whether the high prognostic score tumors have attenuated tumor immune microenvironments. High tumor mutational burden (TMB) is not only one of the aggressive characteristics of cancer [36], but also known as an attractant of immune cells by production of neoantigens. We found that the high score tumors were associated with higher TMB as compared to the low score tumors (P=0.029) (Figure 6A). The high score tumors were associated with significantly lower infiltration of CD8-positive T cells, which play a main role in anti-cancer immunity (P=0.004) (Figure 6B). Furthermore, dendritic cells, which are antigen-presenting cells that act as messengers between the innate and the adaptive immune systems, were also significantly lower in the high score tumors (P<0.001) (Figure 6B).
Figure 6.
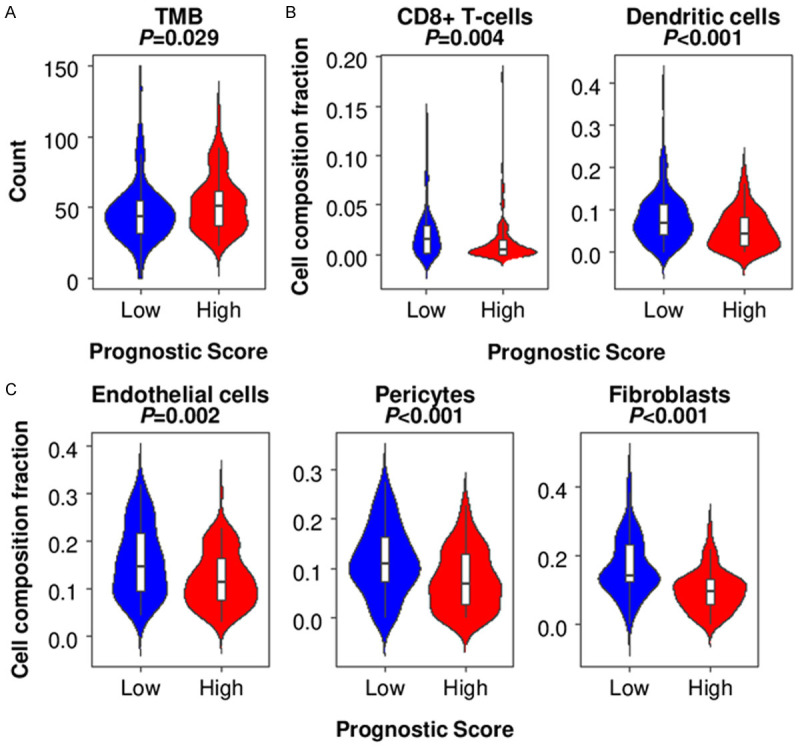
The association of the prognostic score and tumor microenvironment (TME) in pancreatic ductal adenocarcinoma (PDAC) in TCGA. A. The tumor mutational burden (TMB) comparison between the low and high prognostic score PDACs in TCGA. B. Infiltrated immune cell comparison between the low and high prognostic score tumors. C. Other component comparison between the low and high prognostic score tumors.
Recently we reported that PDAC with more mature blood vessels, formed by endothelial cells and pericytes, have improved survival with enhanced anti-cancer immune response [6]. In accordance with this previous finding, high score tumors were associated with significantly lower amounts of endothelial cells (P=0.002) and pericyte (P<0.001) (Figure 6C). We also previously found that PDAC with high amounts of fibroblasts showed higher curative resection rates, leading to better prognosis [37]. As expected, high score tumors showed lower fibroblasts (P<0.001) (Figure 6C). These findings suggest that high prognostic score PDAC is associated with high TMB and unfavorable TME with less anti-cancer immune cell infiltration, less mature blood vessels and fibroblasts.
High score tumor is associated with enhanced cell proliferation and margin positivity after resection
Given that high score tumor is associated with high TMB and unfavorable TME, we hypothesized that high score tumor is associated with cancer progression and aggressiveness. Indeed, MKI67 expression level, which encodes proliferation marker Ki-67, was significantly higher in the high score tumors (P=0.010) (Figure 7A). Furthermore, the fraction of epithelial cells, which mainly reflect cancer cells, was higher in the high score tumors (P=0.022) (Figure 7A). Along these lines, tumors with positive margins, after resection (R1/2 resection), reflecting cancer invasiveness rather than surgical technique, showed significantly higher prognostic scores (P=0.010) (Figure 7B). Unexpectedly, prognostic score was not associated with histological grade or AJCC cancer staging (Figure 7B). However, GSEA revealed that high prognostic score tumors significantly enriched cell proliferation related gene sets, including G2/M checkpoint (NES 1.86, P=0.023), E2F targets (NES 1.81, P=0.026), and MYC targets (NES 1.66, P=0.036) (Figure 7C). Taken together, these findings suggest that high prognostic score tumor, reflecting short survival PDAC, is associated with enhanced cell proliferation.
Figure 7.
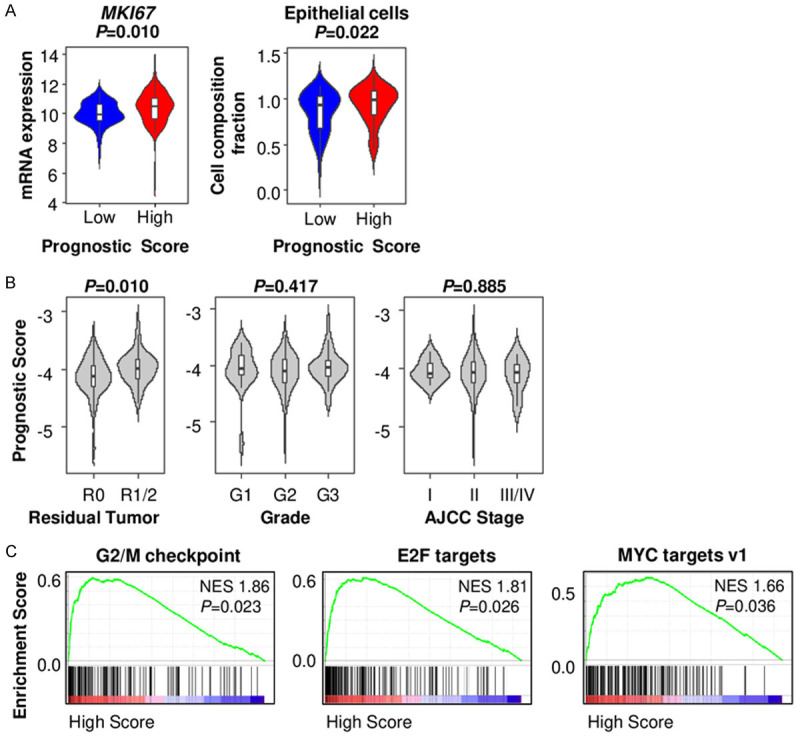
The association of prognostic score and cancer aggressiveness in pancreatic ductal adenocarcinoma (PDAC). A. MKI67 expression and epithelial cell composition comparison between the low and high prognostic score PDACs in TCGA. B. Prognostic score comparison by residual tumor status, histological grade, and AJCC stage in TCGA PDACs. C. Gene set enrichment analysis (GSEA) of cell proliferation related genes comparing between the low and high prognostic score PDACs in TCGA.
To investigate whether the genes included in the score relate to the promotion of cell proliferation, we used human PDAC cell line, MiaPACA-2. Each gene was silenced by transducing shRNA. PHKG1, HOXA4, ISL2 and TRA2A knockdown was confirmed by qPCR in shRNA transduced cells (Figure 8A). DMRT3 expression was undetectable in the control cells, thus excluded from further evaluation. We found that silencing of each of the 4 genes promoted cell proliferation as compared to control cells (shNT) (Figure 8B).
Figure 8.
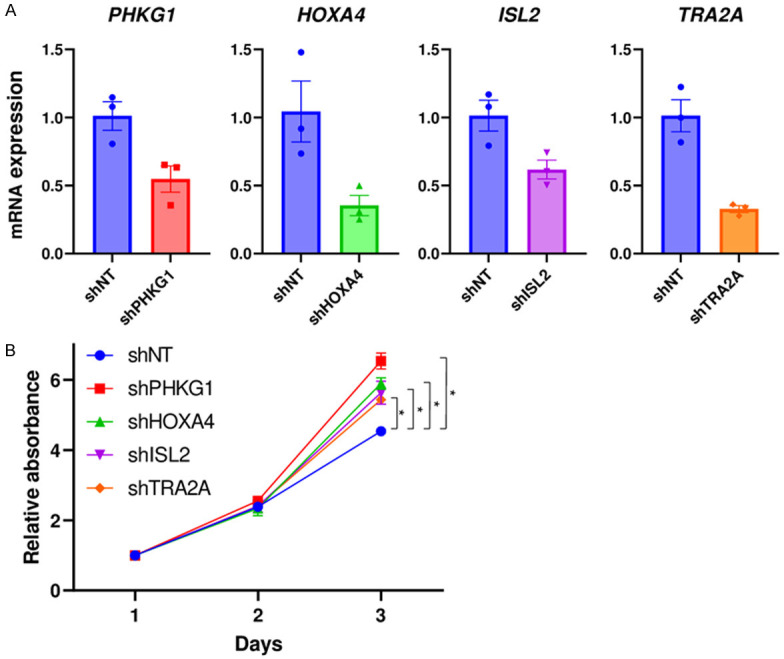
The association of prognostic score component genes and cell proliferation in pancreatic ductal adenocarcinoma (PDAC). A. mRNA expression by qPCR in shRNA transduced cells. B. Cell proliferation assay quantified by CCK-8 in shRNA transduced cells. n=6, each.
Discussion
In this study we established a prognostic score for PDAC patients with surgically resectable tumor utilizing long-term survivor unique transcriptome profiles, identified by comparing gene expression profiles of patients with greater than 5 years survival and patients with common clinical course who died within 3 years after surgery. Our method was unbiased and straightforward with higher scores able to predict patient prognosis. The score identified the PDAC patients with poor prognosis in the testing TCGA cohort and confirmed in the validation GSE21501 cohort. High score tumor was associated with TP53 mutation but no other commonly enhanced signaling pathways in PDAC. Additionally, high score tumor was associated with high TMB and unfavorable TME, including less anti-cancer immune cells, less mature blood vessels and fibroblasts. High prognostic score PDAC was associated with enhanced cell proliferation and positive surgical margin. Each score component gene promoted cell proliferation.
We found that 6 of HOXA genes were upregulated in tumors of long-term survivors, including HOXA4, HOXA5, HOXA6, HOXA7, HOXA9 and HOXA10. HOX genes are highly conserved genes encoding transcriptional factors [33]. There are 4 HOX families, HOXA, HOXB, HOXC, and HOXD, and each family has 9 to 11 members [33]. Some of HOX genes have been reported as oncogenes in cancers. HOXA4 and HOXA9 play roles in stem cell differentiation, and promote self-renewal and proliferation of cancer stem cells in colorectal cancer [38,39]. High expression of HOXA4 is associated with worse prognosis in colorectal cancer [33], and high expression of HOXA9 is associated with poor prognosis in acute myeloid leukemia [33]. Lymphoblastic leukemia with high expression of HOX genes, including HOXA4, HOXA5, HOXA7 and HOXA9 is associated with worse prognosis [40,41]. Conversely, HOXA5 has been reported to induce apoptosis in breast cancer [42,43]. Interestingly, we identified 6 HOXA genes upregulated in the favorable prognosis PDAC patients, which is contradictory to a majority of previous reports.
In addition to HOXA gene, PHKG1, ISL2, DMRT3 and TRA2A genes were utilized in the prognostic score. PHKG1 plays roles in angiogenesis and glycolysis in cancer [44,45]. ISL2 has never been reported to have any role in cancer, thus this is the first report to demonstrate its association with a better prognosis in cancer. DMRT3 is also a transcription factor that associates with heterogeneity of lung squamous cell carcinoma, and regulates genes involved in epithelial cell differentiation together with TP63/SOX2, which are known as oncogenic transcription factors [46]. TRA2A plays a role in the regulation of pre-mRNA splicing. TRA2A promotes chemo-resistance in breast cancer [47], and aggressive cancer characteristics, such as proliferation, migration, invasion and epithelial mesenchymal transition in glioblastoma [48]. Our finding that PHKG1, DMRT3 and TRA2A were upregulated in the PDACs of favorable prognosis patients is also contradictory to previous reports. PDAC may have unique transcriptome profiles compared to other cancers.
TMB reflects neoantigen production, resulting in high anti-cancer immunity and improved prognosis [35], yet we found that high TMB is also associated with cancer aggressiveness [36]. Studies have shown that PDAC is less immunogenic and less TMB compared to other tumors [49], and high TMB in PDAC is inversely correlated with CD8-positive T cells, leading it to be used as a poor prognostic indicator in PDAC [50]. In the current study, we observed that high prognostic score patient with worse prognosis was associated with the high TMB and attenuated anti-cancer immunity, including less CD8-positive T cells. This finding implies that the clinical significance of TMB varies among different types of cancer.
We recently found that PDAC tumors with high amounts of mature blood vessels have enhanced anti-cancer immunity with better prognosis [6]. We also found that higher fibroblast composition in PDAC is associated with higher rates of negative surgical margins, along with high mature blood density and less cancer cells in the tumor [37]. The fraction of cancer cells within a tumor is thought to reflect cancer cell proliferation [9]. This current study is consistent with these findings in that high prognostic score tumors with worse survival were associated with enhanced cell proliferation, shown by high number of epithelial cells, high MKI67 expression, as well as less mature blood vessel cells and fibroblasts. Furthermore, high prognostic score tumor was associated with cell proliferation related gene sets, G2/M checkpoint, E2F targets and MYC targets, of which we have recently reported to be enriched in aggressive breast cancer [51,52]. Given these results, our newly established prognostic score enables us to detect PDAC with high TMB, unfavorable TME, enhanced cell proliferation, and poor survival.
In this study, we established the prognostic score by analyzing resectable PDAC patients. Our results demonstrated that the knockdown of the genes that constitute the score, which were highly expressed in the tumor of long-term survivor, promoted cell proliferation. Furthermore, the score was associated with residual tumor status. These findings suggest that long-term survival may be achieved by a less invasive phenotype due to suppressed cell proliferation, resulting in R0 resection.
There are limitations in this study. First, there were only 4 patients who survived more than 5 years after diagnosis of pancreatic cancer in TCGA cohort resulting in a less than ideal sample size for long-term survivor group statistical analyses. This was because 1) PDAC is not the most common type of cancer, 2) PDAC has very poor prognosis with 5 year survival of approximately 5% that severely limits availability of tumor samples with both transcriptome and clinical parameters from patients that survived for more than 5 years. As a result, the current study is the first, to the best of our knowledge, which was able to generate a score using transcriptomics of pancreatic cancer patients who survived more than 5 years. Future prospective study should be conducted to confirm our findings using large cohorts with additional long-term survivors. Second, there are some important analyses missing because we could not obtain those data within TCGA PDAC cohort. We are unable to show the superiority of the score compared to previously reported factors, including anatomical factors (resectability according to the NCCN guideline), biological factors (positivity of lymph node metastasis, tumor markers) and conditional factors (performance status, immune-nutritional status). Optimal treatment selection is essential to achieve long-term survival in PDAC. Unfortunately, we were unable to access any PDAC cohort with both transcriptome and treatment response on long-term survivor’s data large enough to conduct bioinformatic analyses despite an exhaustive, comprehensive search. TCGA cohort does not include detailed treatment information, therefore prohibiting us from analyzing the relationship between treatment selection and patient survival. We were also unable to analyze the relationship between tumor markers, such as CEA and CA19-9, and patient prognosis, including score superiority compared to tumor markers, despite their role as predictors of patient survival in PDAC. In addition, TCGA cohort includes only surgically resected PDACs, accounting for only 20% of PDAC cases. Therefore, the score utility in non-resectable PDAC is unknown. Further, the mechanistic analysis was conducted using only bioinformatic analysis and a few in vitro experiments, thus, the exact roles of genes in the prognostic score are also not elucidated. To investigate it, further experimental approach is needed.
In conclusion, we established a prognostic score for PDAC patients who underwent surgical resection using long-term survivor unique transcriptomic profiles. We found that poor prognosis patients with a high score were associated with aggressive characteristics such as unfavorable TME and enhanced cell proliferation. This score may be useful in clinical practice to decide treatment strategy by predicting patient prognosis.
Acknowledgements
This work was supported by NIH grant R01CA160688, R01CA250412, R37CA248018, DOD grant W81XWH-19-1-0674 (BC180510) as well as the Edward K. Duch Foundation and Paul & Helen Ellis Charitable Trust to KT, and National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Cancer Comprehensive Cancer Center Shared Resources. KT is the Alfiero Foundation Chair of Breast Oncology at Roswell Park Comprehensive Cancer Center. shRNA transduction was conducted by Renae Holtz at the Gene Modulation Shared Resource of Roswell Park Comprehensive Cancer Center.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Ilic M, Ilic I. Epidemiology of pancreatic cancer. World J Gastroenterol. 2016;22:9694–9705. doi: 10.3748/wjg.v22.i44.9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilici A. Prognostic factors related with survival in patients with pancreatic adenocarcinoma. World J Gastroenterol. 2014;20:10802–10812. doi: 10.3748/wjg.v20.i31.10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi H, Katsuta E, Yan L, Tokumaru Y, Katz MHG, Takabe K. Transcriptomic profile of lymphovascular invasion, a known risk factor of pancreatic ductal adenocarcinoma metastasis. Cancers (Basel) 2020;12:2033. doi: 10.3390/cancers12082033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rochefort P, Lardy-Cleaud A, Sarabi M, Desseigne F, Cattey-Javouhey A, de la Fouchardière C. Long-term survivors in metastatic pancreatic ductal adenocarcinoma: a retrospective and matched pair analysis. Oncologist. 2019;24:1543–1548. doi: 10.1634/theoncologist.2018-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katsuta E, Qi Q, Peng X, Hochwald SN, Yan L, Takabe K. Pancreatic adenocarcinomas with mature blood vessels have better overall survival. Sci Rep. 2019;9:1310. doi: 10.1038/s41598-018-37909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takahashi K, Ehata S, Koinuma D, Morishita Y, Soda M, Mano H, Miyazono K. Pancreatic tumor microenvironment confers highly malignant properties on pancreatic cancer cells. Oncogene. 2018;37:2757–2772. doi: 10.1038/s41388-018-0144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, Rashid NU, Williams LA, Eaton SC, Chung AH, Smyla JK, Anderson JM, Kim HJ, Bentrem DJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Yeh JJ. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–1178. doi: 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, Laklai H, Sugimoto H, Kahlert C, Novitskiy SV, De Jesus-Acosta A, Sharma P, Heidari P, Mahmood U, Chin L, Moses HL, Weaver VM, Maitra A, Allison JP, LeBleu VS, Kalluri R. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer Cell. 2015;28:831–833. doi: 10.1016/j.ccell.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 10.Takahashi K, Hisaka T, Horiuchi H, Ishikawa H, Nakayama M, Nakashima O, Kawahara A, Kage M, Yano H, Akagi Y, Yonemoto K, Kinoshita H, Shirouzu K. Neoplastic spindle cells are an independent prognostic factor in pancreatic cancer. Pancreas. 2015;44:742–749. doi: 10.1097/MPA.0000000000000337. [DOI] [PubMed] [Google Scholar]
- 11.Wartenberg M, Cibin S, Zlobec I, Vassella E, Eppenberger-Castori S, Terracciano L, Eichmann MD, Worni M, Gloor B, Perren A, Karamitopoulou E. Integrated genomic and immunophenotypic classification of pancreatic cancer reveals three distinct subtypes with prognostic/predictive significance. Clin Cancer Res. 2018;24:4444–4454. doi: 10.1158/1078-0432.CCR-17-3401. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi H, Katsuta E, Yan L, Dasgupta S, Takabe K. High expression of Annexin A2 is associated with DNA repair, metabolic alteration, and worse survival in pancreatic ductal adenocarcinoma. Surgery. 2019;166:150–156. doi: 10.1016/j.surg.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee J, Lee J, Kim JH. Identification of matrix metalloproteinase 11 as a prognostic biomarker in pancreatic cancer. Anticancer Res. 2019;39:5963–5971. doi: 10.21873/anticanres.13801. [DOI] [PubMed] [Google Scholar]
- 14.Kandimalla R, Tomihara H, Banwait JK, Yamamura K, Singh G, Baba H, Goel A. A 15-gene immune, stromal, and proliferation gene signature that significantly associates with poor survival in patients with pancreatic ductal adenocarcinoma. Clin Cancer Res. 2020;26:3641–3648. doi: 10.1158/1078-0432.CCR-19-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou YY, Chen LP, Zhang Y, Hu SK, Dong ZJ, Wu M, Chen QX, Zhuang ZZ, Du XJ. Integrated transcriptomic analysis reveals hub genes involved in diagnosis and prognosis of pancreatic cancer. Mol Med. 2019;25:47. doi: 10.1186/s10020-019-0113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang J, Jiang Y, He R, Liu W, Yang M, Tao L, Fu X, Shen Y, Li J, Liu D, Huo Y, Zhang J, Hua R, Zhang Z, Sun Y. DKK2 impairs tumor immunity infiltration and correlates with poor prognosis in pancreatic ductal adenocarcinoma. J Immunol Res. 2019;2019:8656282. doi: 10.1155/2019/8656282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song J, Xu Q, Zhang H, Yin X, Zhu C, Zhao K, Zhu J. Five key lncRNAs considered as prognostic targets for predicting pancreatic ductal adenocarcinoma. J Cell Biochem. 2018;119:4559–4569. doi: 10.1002/jcb.26598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan X, Wan H, Hao X, Lan T, Li W, Xu L, Yuan K, Wu H. Importance of gene expression signatures in pancreatic cancer prognosis and the establishment of a prediction model. Cancer Manag Res. 2019;11:273–283. doi: 10.2147/CMAR.S185205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou C, Zhao Y, Yin Y, Hu Z, Atyah M, Chen W, Meng Z, Mao H, Zhou Q, Tang W, Wang P, Li Z, Weng J, Bruns C, Popp M, Popp F, Dong Q, Ren N. A robust 6-mRNA signature for prognosis prediction of pancreatic ductal adenocarcinoma. Int J Biol Sci. 2019;15:2282–2295. doi: 10.7150/ijbs.32899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Zhu D, Xing H, Hou Y, Sun Y. A 6-gene risk score system constructed for predicting the clinical prognosis of pancreatic adenocarcinoma patients. Oncol Rep. 2019;41:1521–1530. doi: 10.3892/or.2019.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontana A, Copetti M, Di Gangi IM, Mazza T, Tavano F, Gioffreda D, Mattivi F, Andriulli A, Vrhovsek U, Pazienza V. Development of a metabolites risk score for one-year mortality risk prediction in pancreatic adenocarcinoma patients. Oncotarget. 2016;7:8968–8978. doi: 10.18632/oncotarget.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stratford JK, Bentrem DJ, Anderson JM, Fan C, Volmar KA, Marron JS, Routh ED, Caskey LS, Samuel JC, Der CJ, Thorne LB, Calvo BF, Kim HJ, Talamonti MS, Iacobuzio-Donahue CA, Hollingsworth MA, Perou CM, Yeh JJ. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 2010;7:e1000307. doi: 10.1371/journal.pmed.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katsuta E, Maawy AA, Yan L, Takabe K. High expression of bone morphogenetic protein (BMP) 6 and BMP7 are associated with higher immune cell infiltration and better survival in estrogen receptorpositive breast cancer. Oncol Rep. 2019;42:1413–1421. doi: 10.3892/or.2019.7275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katsuta E, Yan L, Nagahashi M, Raza A, Sturgill JL, Lyon DE, Rashid OM, Hait NC, Takabe K. Doxorubicin effect is enhanced by sphingosine-1-phosphate signaling antagonist in breast cancer. J Surg Res. 2017;219:202–213. doi: 10.1016/j.jss.2017.05.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okano M, Oshi M, Butash AL, Asaoka M, Katsuta E, Peng X, Qi Q, Yan L, Takabe K. Estrogen receptor positive breast cancer with high expression of androgen receptor has less cytolytic activity and worse response to neoadjuvant chemotherapy but better survival. Int J Mol Sci. 2019;20:2665. doi: 10.3390/ijms20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okano M, Oshi M, Butash AL, Katsuta E, Tachibana K, Saito K, Okayama H, Peng X, Yan L, Kono K, Ohtake T, Takabe K. Triple-negative breast cancer with high levels of annexin A1 expression is associated with mast cell infiltration, inflammation, and angiogenesis. Int J Mol Sci. 2019;20:4197. doi: 10.3390/ijms20174197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sporn JC, Katsuta E, Yan L, Takabe K. Expression of MicroRNA-9 is associated with overall survival in breast cancer patients. J Surg Res. 2019;233:426–435. doi: 10.1016/j.jss.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 30.Terakawa T, Katsuta E, Yan L, Turaga N, McDonald KA, Fujisawa M, Guru KA, Takabe K. High expression of SLCO2B1 is associated with prostate cancer recurrence after radical prostatectomy. Oncotarget. 2018;9:14207–14218. doi: 10.18632/oncotarget.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katsuta E, Yan L, Takeshita T, McDonald K, Dasgupta S, Opyrchal M, Takabe K. High MYC mRNA expression is more clinically relevant than MYC DNA amplification in triple-negative breast cancer. Int J Mol Sci. 2020;21:217. doi: 10.3390/ijms21010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhatlekar S, Fields JZ, Boman BM. Role of HOX genes in stem cell differentiation and cancer. Stem Cells Int. 2018;2018:3569493. doi: 10.1155/2018/3569493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katsuta E, Rashid OM, Takabe K. Clinical relevance of tumor microenvironment: immune cells, vessels, and mouse models. Hum Cell. 2020;33:930–937. doi: 10.1007/s13577-020-00380-4. [DOI] [PubMed] [Google Scholar]
- 36.Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10:1852. doi: 10.1038/s41598-020-58995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katsuta E, Rashid OM, Takabe K. Fibroblasts as a biological marker for curative resection in pancreatic ductal adenocarcinoma. Int J Mol Sci. 2020;21:3890. doi: 10.3390/ijms21113890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhatlekar S, Addya S, Salunek M, Orr CR, Surrey S, McKenzie S, Fields JZ, Boman BM. Identification of a developmental gene expression signature, including HOX genes, for the normal human colonic crypt stem cell niche: overexpression of the signature parallels stem cell overpopulation during colon tumorigenesis. Stem Cells Dev. 2014;23:167–179. doi: 10.1089/scd.2013.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhatlekar S, Viswanathan V, Fields JZ, Boman BM. Overexpression of HOXA4 and HOXA9 genes promotes self-renewal and contributes to colon cancer stem cell overpopulation. J Cell Physiol. 2018;233:727–735. doi: 10.1002/jcp.25981. [DOI] [PubMed] [Google Scholar]
- 40.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 41.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 42.Chen H, Chung S, Sukumar S. HOXA5-induced apoptosis in breast cancer cells is mediated by caspases 2 and 8. Mol Cell Biol. 2004;24:924–935. doi: 10.1128/MCB.24.2.924-935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raman V, Martensen SA, Reisman D, Evron E, Odenwald WF, Jaffee E, Marks J, Sukumar S. Compromised HOXA5 function can limit p53 expression in human breast tumours. Nature. 2000;405:974–978. doi: 10.1038/35016125. [DOI] [PubMed] [Google Scholar]
- 44.Camus S, Quevedo C, Menendez S, Paramonov I, Stouten PF, Janssen RA, Rueb S, He S, Snaar-Jagalska BE, Laricchia-Robbio L, Izpisua Belmonte JC. Identification of phosphorylase kinase as a novel therapeutic target through high-throughput screening for anti-angiogenesis compounds in zebrafish. Oncogene. 2012;31:4333–4342. doi: 10.1038/onc.2011.594. [DOI] [PubMed] [Google Scholar]
- 45.Jarrar Y, Zihlif M, Al Bawab AQ, Sharab A. Effects of intermittent hypoxia on expression of glucose metabolism genes in MCF7 breast cancer cell line. Curr Cancer Drug Targets. 2020;20:216–222. doi: 10.2174/1568009619666191116095847. [DOI] [PubMed] [Google Scholar]
- 46.Zhang S, Li M, Ji H, Fang Z. Landscape of transcriptional deregulation in lung cancer. BMC Genomics. 2018;19:435. doi: 10.1186/s12864-018-4828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu T, Sun H, Zhu D, Dong X, Liu F, Liang X, Chen C, Shao B, Wang M, Wang Y, Sun B. TRA2A promoted paclitaxel resistance and tumor progression in triple-negative breast cancers via regulating alternative splicing. Mol Cancer Ther. 2017;16:1377–1388. doi: 10.1158/1535-7163.MCT-17-0026. [DOI] [PubMed] [Google Scholar]
- 48.Tan Y, Hu X, Deng Y, Yuan P, Xie Y, Wang J. TRA2A promotes proliferation, migration, invasion and epithelial mesenchymal transition of glioma cells. Brain Res Bull. 2018;143:138–144. doi: 10.1016/j.brainresbull.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Martinez-Bosch N, Vinaixa J, Navarro P. Immune evasion in pancreatic cancer: from mechanisms to therapy. Cancers (Basel) 2018;10:6. doi: 10.3390/cancers10010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen H, Yang G, Xiao J, Zheng L, You L, Zhang T. Neoantigen-based immunotherapy in pancreatic ductal adenocarcinoma (PDAC) Cancer Lett. 2020;490:12–19. doi: 10.1016/j.canlet.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, Endo I, Takabe K. G2M cell cycle pathway score as a prognostic biomarker of metastasis in Estrogen Receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21:2921. doi: 10.3390/ijms21082921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, Endo I, Takabe K. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9:1643. doi: 10.3390/cells9071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


