Abstract
Sphingosine-1-Phosphate (S1P) is produced by Sphingosine Kinase 1 (SphK1) in the cell and is transported out of the cells by ABCC1 transporter. S1P induces inflammation, angiogenesis and modulates tumor immune microenvironment (TIME) in autocrine and paracrine manner. We hypothesized that high S1P export is associated with hepatocellular carcinoma (HCC) progression and worse survival. Transcriptome linked with clinical data were obtained from a total of 533 patients from TCGA (The Cancer Genome Atlas)-HCC (n = 350), GSE6764 (n = 75), and GSE89377 (n = 108) cohorts. Both SphK1 and ABCC1 were expressed higher in aggressive HCC than normal liver or cirrhosis and correlated with MKi67 expression. High S1P export by high expression of both SphK1 and ABCC1 enriched gene sets related with cell proliferation (E2F targets, G2M checkpoint, MYC targets), inflammation (Inflammatory response, TNFα, IL6), angiogenesis, metastasis (TGF-β, epithelial-mesenchymal transition), and immune response (allograft rejection, complement, interferon-gamma) in gene set enrichment analysis. High S1P export was associated with elevation of HGF, HSP90AA1, TRAF2, and AKR1B10. It was also associated with high intratumor heterogeneity, leucocyte fraction, macrophage regulation and lymphocyte infiltration, as well as T helper type2 cells, macrophages, dendritic cells, CD4+ T memory activated cells, B-cells and cytolytic activity score in TIME. High S1P export was associated with significantly worse disease specific survival (P = 0.034) and overall survival (P = 0.004) compared to low S1P export group. In conclusion, simultaneous high expression of SphK1 and ABCC1 that reflect S1P export is associated with enhancement of both HCC progression and immune response. Given that S1P export was also associated with worse survival, we cannot help but speculate that pro-cancer pathways activated by S1P may overwhelm the anti-cancer immune response mediated by S1P.
Keywords: Sphingosine-1-Phosphate, sphingosine kinase, hepatocellular carcinoma, S1P, HCC, liver
Introduction
Hepatocellular carcinoma (HCC) accounts for > 90% of all primary liver cancers, making it the most common liver malignancy and is second only to pancreatic cancer in terms of lethality with only 18% 5-year survival [1]. Globally, HCC is fourth most lethal cancer and sixth most common. Majority of patients with HCC have underlying cirrhosis from non-alcoholic fatty liver disease (NAFLD), alcohol abuse and viral hepatitis (Hepatitis B and C). NAFLD related HCC has increasing incidence in Western countries [2], with underlying pathophysiology of insulin resistance, oxidative stress and inflammation playing an important role in cancer initiation and progression [3].
Sphingolipids, consisting of various head groups attached to ceramide are found in cell membranes of mammals as an essential component [4]. Sphingosine, which has been associated with cell signaling is derived from deacylation of Ceramide [5]. Sphingosine kinase 1 (SphK1) and Sphingosine kinase 2 (SphK2) phosphorylate Sphingosine to form Sphingosine-1-Phosphate (S1P) [6,7]. Various physiological and pathological proceeses involved in cancer progression are regulated by the powerful pleotropic molecule involved in cell signalling-S1P [8-10]. Various hormones, cytokines and growth factors increase S1P production in cytosol, which is exported out of the cell by a subset of ATP-binding cassette (ABC) transporters, including ABCC1 and ABCG2 [11,12]. Thus, higher levels of SphK1 and ABCC1 would imply higher levels of exported S1P [13]. There are five G protein coupled S1P receptors, to which the extracellular S1P (mainly a product of SphK1) binds to S1PR1-5. This then in an autocrine or paracrine manner mediate multiple known actions of S1P, called ‘inside-out signalling’ [14]. Extracellular S1P signaling is associated with activation of TNF receptor-associated factor 2 (TRAF2), heat shock proteins, NF-κβ, increased telomerase activity, increase in cancer cell growth, regulation of immune cell trafficking implicated in tumor immunology and metastasis [15].
SphK1 and S1P are overexpressed in several HCC cell lines, and tumor samples [16,17]. Immunohistochemistry (IHC) has shown increased levels of both SphK1 and S1P in HCC [18]. There was a positive correlation between tumor stage, size and histological differentiation and SphK1 expression [19] and negative correlation with overall survival [17]. All the above studies were performed on HCC cell lines or IHC of the tumor tissues. This is one of first studies which utilized comprehensive gene expression data from the publicly available cohorts to study the role of S1P in HCC progression, to the best of our knowledge. We hypothesize that high levels of S1P export determined by high expressions of both SphK1 and ABCC1, is related to HCC progression and worse clinical outcomes.
Materials and methods
Clinical data acquisition
We retrieved gene expression levels, clinical and pathological data of patients with HCC from The Cancer Genome Atlas (TCGA) [20] via cBioPortal as described previously [20-29]. A total of 350 patients with HCC and 50 control patients were included in the analysis. Gene Expression Omnibus (GEO) data sets GSE6764 [30] and GSE89377 were used to analyze gene expression profiles at various stages of liver fibrosis, cirrhosis and cancer. GSE6764 contained 75 patient samples with 13 samples from cirrhotic tissue, 17 dysplastic nodules, and 35 HCCs. GSE89377 dataset (https://www.ncbi.nlm.nih.gov/geo/geo2r/?acc = GSE89377) was downloaded from the GEO website, and the cohort contains 108 cases in total, including 13 healthy people, 9 patients with low-grade chronic hepatitis, 12 with high-grade chronic hepatitis, 12 with cirrhosis, 11 with low-graded dysplastic nodules, 11 with high-grade dysplastic nodules, 5 with early HCC, 9 with Stage I HCC, 12 with Stage II HCC and 14 with Stage III HCC. The institutional review board was waived as TCGA and GEO datasets are publicly available and de-identified.
Gene set enrichment analysis (GSEA)
Broad Institute provided the software for Gene set enrichment analysis (GSEA) (http://software.broadinstitute.org/gsea/index.j False discovery rate (FDR) of 0.25 was deemed to be statistically significant, based on the recommendation of Broad institute.
Immune activity related scores and immune cell composition
As we previously reported [39-43], xCell computational algorithm was used to calculate immune cell composition in a tumor per guidelines stipulated in the journal, Genome Biology in 2017 by Aran D et al. [44]. Thorsson et al. previously reported data on TGF-β Response, Leukocyte Fraction, IFN-γ Response, Lymphocyte Infiltration Signature Score and regulation of TIL. [45]. We calculated Cytolytic activity (CYT) using the geometric mean of granzyme A and Perforin 1 expression values as described by Rooney et al. [46-54].
Statistical analysis
All the statistical analyses were performed using R software (http:///www.r-project.org/). Kaplan-Meier survival analysis was performed in R for the survival analysis. A p value < 0.05 was considered statistically significant. One-way ANOVA was used to determine the significance of difference in various groups. We used Mann Whitney U test (two group comparison) and Kruskal Test for multiple group comparison.
Results
Gene expression levels of SphK1 and ABCC1 were higher in tumor tissue than in normal liver
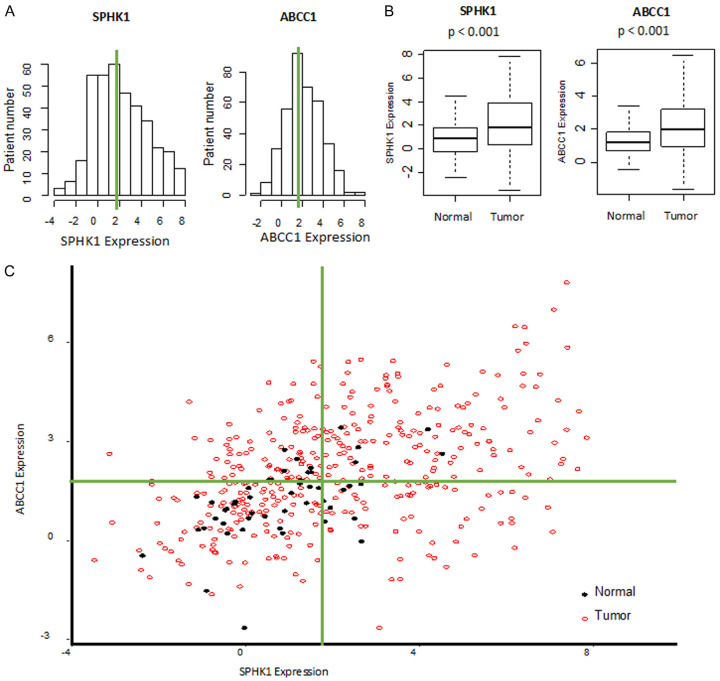
Increased gene expression levels of SphK1 and ABCC1 would indicate the higher production and extracellular export of S1P [11,13]. We found that SphK1 and ABCC1 expression in the TCGA cohort had normal distributions (Figure 1A) with higher expression in tumor tissue versus normal liver (both p < 0.001, Figure 1B). The scatterplot shows median of each gene expression in normal and HCC tumor tissues (Figure 1C), with normal liver clustered around SphK1-low expression and ABCC1-low expression quadrant. This is the first report to the best of our knowledge that there are increases in gene expression levels of SphK1/ABCC1 that suggest export of S1P in large cohort of HCC human samples, which agrees with the mechanisms reported in the previous studies.
Figure 1.
SphK1 and ABCC1 expressions in the tumor tissue is higher than normal liver in TCGA cohort. A. Histogram of gene expression of SphK1 and ABCC1 in TCGA cohort. B. Gene expressions of SphK1 and ABCC1 in tumor tissue versus normal liver samples. C. Scatter plot showing gene expressions of SphK1 and ABCC1 in tumor tissue (red open circle) and normal liver tissue (closed black circle).
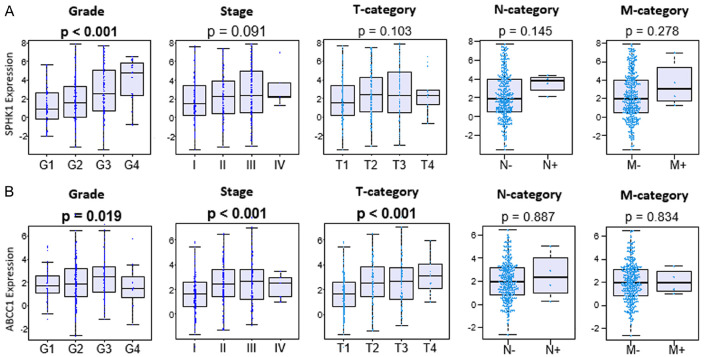
Higher S1P export was seen in advanced grade and stage of HCC
Prior study has shown increased SphK1 expression, which indicates increased production of S1P, in larger tumor size and advanced stage and grade by immunohistochemistry in HCC [17]. In agreement, there was a significant increase in SphK1 gene expression by advanced grade, but not statistically significant by stage, although there was a trend in TCGA (P < 0.001 and P = 0.091, respectively. Figure 2A). ABCC1 gene expressions were significantly higher in advanced grade and stage, particularly in the T-category, which is the size of a tumor in TCGA (P = 0.019, P < 0.001, and P < 0.001, respectively. Figure 2B). Our results are in agreement with the notion that not only the production of S1P, which is reflected by SphK1 expression but also S1P export that is reflected by both SphK1 and ABCC1 expressions are elevated in advanced grade and stage in HCC.
Figure 2.
Gene expressions of SphK1 and ABCC1 with respect to grade and stage of HCC in TCGA cohort. A. SphK1 gene expression with respect to grade and stage of HCC in TCGA. B. ABCC1 gene expression with regards to grade and stage of HCC in TCGA.
SphK1 and ABCC1 gene expressions that reflect S1P export correlate with carcinogenesis and progression of HCC
Given that gene expressions of both SphK1 and ABCC1, which implicate S1P export, were elevated in advanced grade and stage of HCC, we hypothesized that the level of S1P export increased by the stepwise carcinogenic process of HCC. To test this hypothesis, we investigated the gene expression levels of MKI67, the most commonly used cell proliferation marker, SphK1 and ABCC1 in various stages of cancer progression, which are normal liver tissue, dysplasia (DL), cirrhosis, very-early to very-advanced HCC, in GSE6764 cohort. As expected, MKI67 expression increased as carcinogenesis progressed (P < 0.001, Figure 3A). There was a significant increase in the gene expression levels of SphK1 with progression of carcinogenesis (P = 0.026), with level of ABCC1 trended up (P = 0.032). The gene expression levels of MKI67, SphK1, demonstrated significant increase in validation cohort, GSE89377 (both P < 0.001, Figure 3B) with expression of ABCC1 not significantly altered (P = 0.061, Figure 3B). The set includes dysplasia, cirrhosis, low grade and high-grade chronic hepatitis, early HCC, and low to high-grade HCC defined by the GSE98377. These results show that S1P export is elevated as carcinogenic progression advances in multiple cohorts of HCC.
Figure 3.
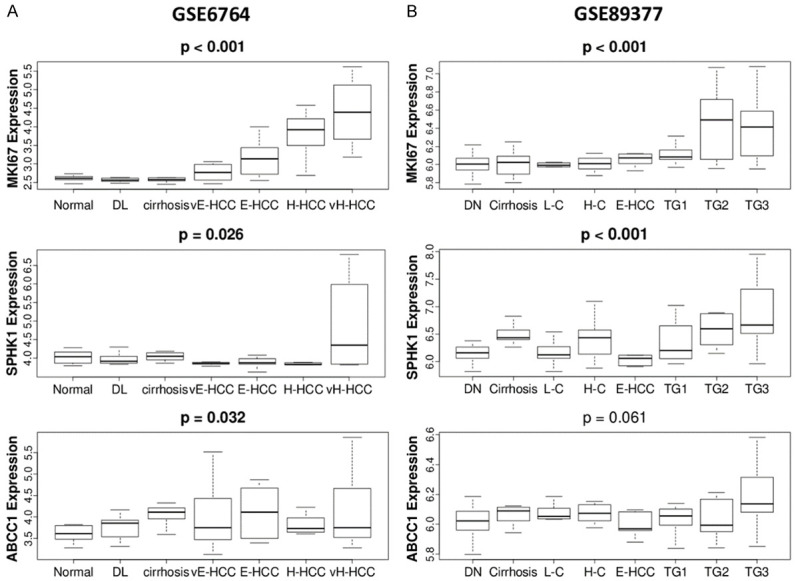
Correlation of MKI67, SphK1 and ABCC1 gene expression with carcinogenic progression of HCC in two cohorts, (A) GSE6764 (n = 75) that include normal liver tissue (n = 8), dysplasia (n = 17), cirrhosis (n = 13), very early hepatocellular carcinoma (vE-HCC) (n = 8), early HCC (E-HCC) (n = 10), advanced HCC (H-HCC) (n = 7), very advanced HCC (VH-HCC) (n = 10). (B) GSE89377 (n = 107) that include dysplastic nodule (DN) (n = 35), cirrhosis (n = 12), low grade cirrhosis (L-C) (n = 8) and high grade cirrhosis (H-C) (n = 12), early HCC (n = 5), and tumor grades (TG) 1-3 (n = 9, 12, 14, respectively) of HCC.
High levels of S1P export is associated with HCC aggravating factors
HGF (Hepatocyte Growth factor) promotes production of SphK1 and motility and invasion of several HCC cell lines, promoting HCC metastases [55]. S1P increases TRAF2 (TNF receptor-associated factor 2), NF-κβ activation [56] and with Heat shock protein association (HSP90A1), preventing apoptosis [57]. We have found that tumors with higher expressions of both SphK1 and ABCC1 had significantly increased levels of HGF (P < 0.001), HSP90A1 (P < 0.001), TRAF2 (P < 0.001) and AKR1B10 (P < 0.001) expressions when compared to tumors with low expression of both SphK1 and ABCC1 (Figure 4). These results are in agreement with the higher grade and stage of HCC associated in tumors with higher exported S1P.
Figure 4.
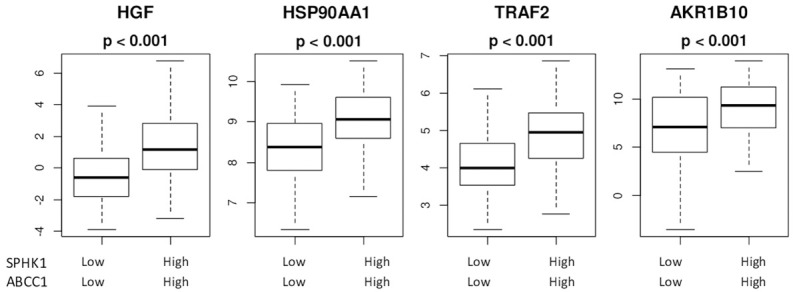
High levels of S1P export leads to factors promoting HCC progression. HGF: hepatocyte growth factor; HSP90AA1: Heat Shock protein90AA1; TRAF2: TNF receptor associated factor 2; AKR1B10-Aldo-keto reductase family1 member B10 gene.
High levels of exported S1P promotes cell proliferation, inflammation, cancer metastases, and immune response
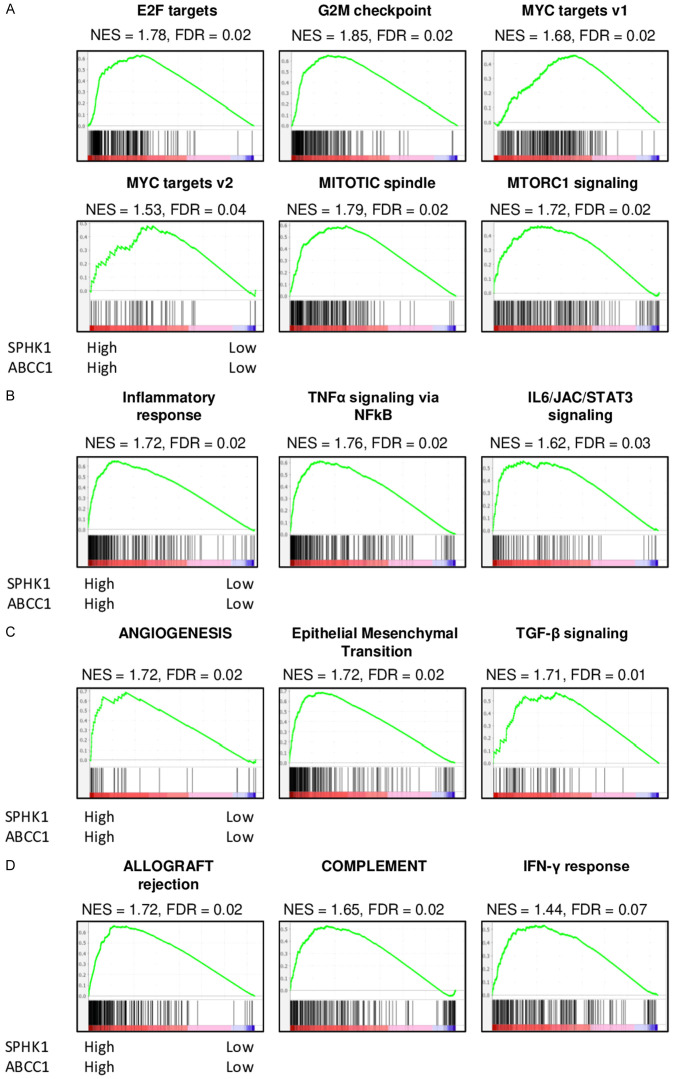
Based on our previous basic research findings that cancer with high S1P export had increased cell proliferation [13,58], evoke inflammation [59,60], promote metastasis [61,62], and enhance immune response [7,35,63], we hypothesized that HCC with high S1P export has similar features. To test this hypothesis, we conducted GSEA comparing both SphK1 and ABCC1 expression high group to both low expression group in TCGA cohort. There was significant enrichment of five cell proliferation-related gene sets; E2F targets, G2M checkpoint, MYC targets V1, MYC targets V2, Mitotic Spindle and MTORC1 signaling in HCC with high S1P export (Figure 5A). S1P export high HCC also enriched inflammation-related gene sets like Inflammatory response, TNF-α signaling via NF-κβ, and IL6/JAK/STAT3 signaling (Figure 5B). Further, Angiogenesis, Epithelial-mesenchymal transition and TGF-β signaling, which are related to metastases, were also found to be significantly enriched to S1P export high HCC (Figure 5C). Finally, statistically significant enrichment of immune response-related gene sets; Allograft rejection, Complement, and Interferon (IFN)-γ response was found in HCC with high levels of exported S1P (Figure 5D). These results demonstrate that the high levels of S1P export were significantly associated with many pathways of HCC progression like cell proliferation, inflammation, and metastases, but these tumors also promoted immune activity in the TCGA cohort, implying the complex relationship of high S1P to HCC progression.
Figure 5.
Comparison of tumors with High expression of both SphK1 and ABCC1 versus Low expression of SphK1 and ABCC1 with Gene Set Enrichment Analysis (A) Cell Proliferation gene sets (B) Inflammation related gene sets (C) metastasis related gene sets (D) Immune response related gene sets. NES: normalized enrichment score; FDR: false discovery rate.
Tumors with high levels of S1P export were linked to favorable and unfavorable tumor immune microenvironment (TIME)
We further explored the TIME to help explain the aggressiveness of HCCs with high levels of S1P export in TCGA cohort. Immune cell activity and function scores as previously described by Thorrson et al. were analyzed. Tumors with high S1P export had significantly increased intratumor heterogeneity, MKI67 expression (P < 0.001 for both) indicating aggressive tumors with higher proliferation (Figure 6A). We also found that the silent and non-silent mutation rate was not significantly higher (P = 0.149 and 0.046, respectively), in contrary to what is often found in highly proliferating tumors. The Leucocyte fraction, macrophage regulation and lymphocyte infiltration scores were found to be significantly lower (P < 0.001 or all) in tumors with high exported S1P, which could explain the aggressive nature of these tumors (Figure 6B). We used xCell, the computational algorithm to analyze the immune cell composition. There was significantly higher levels of Th2 Cells (P < 0.001), but M2 macrophage levels were not significantly increased (P = 0.964) (Figure 6C) indicating an unfavorable TIME, at the same time, there was increased M1 macrophages, dendritic cells, CD4+ memory cells and B cells (P < 0.001, P < 0.001, P = 0.063, P < 0.001, respectively) which indicate higher anti-tumor activity (favorable TIME), in TCGA cohort (Figure 6D). The CYT (Cytolytic activity) score was significantly higher in high S1P export tumors indicating a favorable TIME (Figure 6E).
Figure 6.
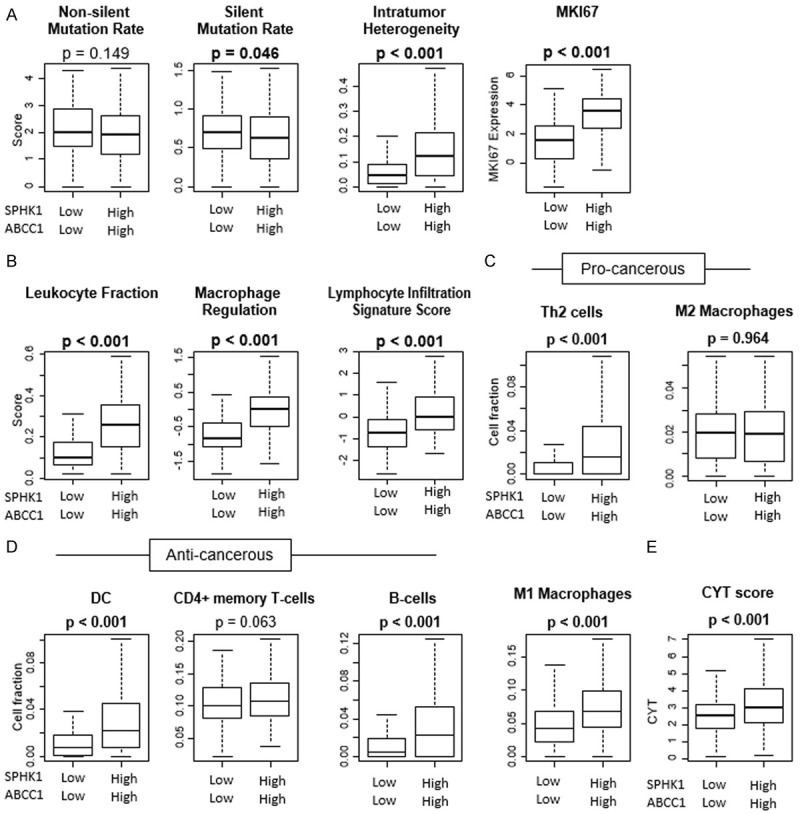
Comparison of tumors with High versus low export of S1P with respect to Tumor Immune Microenvironment in TCGA. (A) Mutation rates and intratumor heterogeneity and proliferation. (B) Leucocyte fraction, macrophage regulation and lymphocyte immune signature score. Immune cell composition in TCGA cohort using Xcell (C) Cells with unfavorable TIME (D) cells with favorable TIME (E) Cytolytic activity score (CYT).
Low levels of S1P export is associated with significantly better progression-free and overall survival
Given the role of SphK1 and ABCC1 in cancer, we investigated whether their expressions relate with disease specific (DSS) and/or overall (OS) survival in TCGA cohort. Although high expression of either of these genes had a trend to have worse survival, there was no statistical difference in the survival curves except for ABCC1 expression in OS (P = 0.002). On the other hand, we found that the DSS of patients with low S1P export (SphK1 low and ABCC1 low) was significantly better than any other combination of SphK1 and ABCC1 expressions (P = 0.034). The OS was also better in low S1P export group with p = 0.004 (Figure 7). Together with the fact that ABCC1 is not the only transporter to export S1P, this result further supports the notion that S1P export promotes HCC progression and contributes to patient survival.
Figure 7.
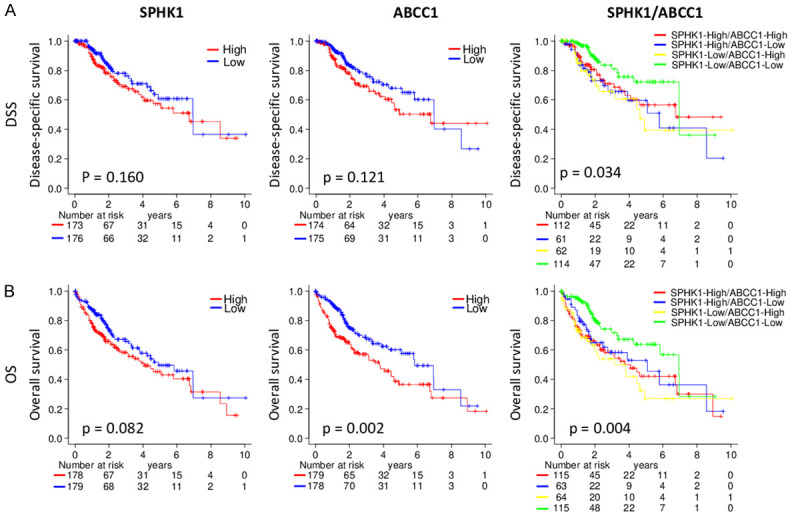
Tumors with high expression of SphK1 and ABCC1 (high S1P export) have worse prognosis. (A) Kaplan-Meier analysis of disease specific survival (B) Kaplan-Meier analysis of overall survival.
Discussion
We hypothesized that higher levels of S1P export is associated with worse prognosis and cancer biology which is aggressive in HCC patients. S1P export was reflected as simultaneous high expression of SphK1 and ABCC1, where both follow a normal distribution curve in TCGA cohort. The expressions of SphK1 and ABCC1 were consistently higher in higher grades, higher MKI67 expression, and in later stages of HCC compared from normal liver and cirrhosis, supporting the hypothesis of higher S1P export is associated with more aggressive HCCs. There were significantly higher levels of HGF, TRAF2, NF-κβ activation with heat shock protein association and AKR1B10 production, with higher levels of S1P export, all of which could explain the higher grade and stage of HCC in TCGA cohort. High S1P export HCC enriched cell proliferation, inflammation, and metastasis related gene sets in GSEA. Interestingly it also enriched anti-cancer immune response gene sets (Complement, Interferon Gamma, Allograft rejection). With xCell, higher exported S1P showed increased levels of Th2 cells (unfavorable TIME) but also found higher levels of dendritic cells, CD4+ memory cells, M1 macrophages (favorable TIME), and B cells. Cytolytic activity (CYT), was significantly elevated in high S1P export group, indicating higher immune cell activity as a whole. We found that there was a significantly better overall survival as well as disease-specific survival in HCCs with low levels of S1P export.
Several studies have shown that SphK1/S1P axis is activated in HCC cell lines [16,18]; however, this is the first study to our knowledge to report the clinical relevance of increased S1P export in HCC patients in a large cohort. Our study agrees with other studies [17] showing higher levels of S1P export in higher grade and later stage tumors, linking higher S1P export levels to aggressive HCCs. We found no significant correlation between lymph node metastases and exported S1P levels in HCC, as opposed to prior reports in breast cancer [13,64]. This could probably be explained by different pathways for metastases in these two types of cancer.
Based on this, we hypothesized that more aggressive the HCC, higher the expression of genes that contribute to S1P export, SphK1, and ABCC1. We demonstrated in two independent GEO datasets GSE6764 and GSE89377, that there were significantly increased levels of S1P export as the HCC grade increases when compared to normal tissue. We then investigated the mechanisms by which exported S1P could promote HCC progression. We found significantly higher HGF levels, known to promote motility and invasion of HCC cell lines, and promote metastases [55]. Higher TRAF2 levels, NF-κβ activation and with association of HSP90A1, prevent apoptosis [57]. AKR1B10 is increased in HCC, this is a protein associated with lipid metabolism and is related to negative outcomes [65]. High S1P export was significantly associated with increased levels of HGF, TRAF2, NF-κβ, HSP90A1 as well as AKR1B10 expressions, all of which will promote HCC progression.
Inflammation has a significant role in progression of cirrhosis to HCC, as well as promote cancer progression and metastasis [66]. We previously reported that higher levels of S1P export promotes inflammation in tumors like breast cancer, aiding cancer progression [60]. In current study we have demonstrated that exported S1P evokes inflammation, influences tumor microenvironment, promotes cell cycle progression as well as metastasis in HCC patients. GSEA demonstrates an increased ex pression of genes related to cell proliferation such as E2F targets, G2M checkpoint, MYC targets V1 and V2, and mitotic spindle. Genes related to inflammation Inflammatory response, TNFα signaling via NF-κβ, IL6/JAK/STAT3 signaling, genes promoting HCC metastases like Angiogenesis, Epithelial mesenchymal transition, TGF-β signaling, in high S1P tumors, which could make these tumors more aggressive.
The tumor immune microenvironment showed significantly higher levels of Th2 cells and macrophages indicating an unfavorable TIME as well as increased leucocyte fraction, macrophage regulation, intratumor heterogeneity, and MKI67 expression in high S1P tumors, all of which promote HCC progression, aggressiveness and metastasis.
We also found a significantly higher level of anti-cancer cells like dendritic cells, CD4+ T cells, B cells, which prevent cancer progression. GSEA also showed significant increase in anti-tumor activity genes like Complement, Interferon gamma, Allograft rejection and interferon gamma response. CYT-high HCCs lead to enhanced immunity and better survival [48], in our study we have a significantly higher CYT score in high S1P tumors. This would suggest a higher immune activity in the S1P high HCCs. We can see the complex relationship with high S1P export and HCC. We see that high S1P promotes inflammation which brings in both pro and anti-tumor cells, increased immune cell activity as evidenced by high CYT score.
Given all these results, we cannot help but speculate high S1P export promotes HCC progression and HCC metastasis. S1P also promotes anti-tumor activity in several ways, but we feel that the protumor activity probably overwhelms the anti-tumor activity in HCC, which ultimately promotes HCC growth and progression.
The current study has limitations. First, this is a retrospective study conducted using databases, TCGA which are assessable to public. Second, the clinical information of all subjects in the database is not complete. Lastly, we do need prospective studies, in-vitro and in vivo experiments, to confirm and validate the underlying mechanism of our clinical findings.
In conclusion, simultaneous high expression of SphK1 and ABCC1 that reflect S1P export is associated with enhancement of both HCC progression and immune response. Given that S1P export was also associated with worse survival, we cannot help but speculate that pro-cancer pathways activated by S1P may overwhelm the anti-cancer immune response mediated by S1P.
Acknowledgements
This work is supported by US NCI/NIH grant R01CA160688, R01CA250412, R37CA248018, as well as Department of Defense-BCRP grant W81XWH-19-1-0674. Roswell Park Comprehensive Cancer Center is supported by NCI/NIH grant P30-CA016056.
Disclosure of conflict of interest
None.
References
- 1.Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson B, Mariotto A, Lake AJ, Wilson R, Sherman RL, Anderson RN, Henley SJ, Kohler BA, Penberthy L, Feuer EJ, Weir HK. Annual report to the nation on the status of cancer, 1975-2014, featuring survival. J Natl Cancer Inst. 2017;109:djx030. doi: 10.1093/jnci/djx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Younossi Z, Stepanova M, Ong JP, Jacobson IM, Bugianesi E, Duseja A, Eguchi Y, Wong VW, Negro F, Yilmaz Y, Romero-Gomez M, George J, Ahmed A, Wong R, Younossi I, Ziayee M, Afendy A. Nonalcoholic steatohepatitis is the fastest growing cause of hepatocellular carcinoma in liver transplant candidates. Clin Gastroenterol Hepatol. 2019;17:748–755. e743. doi: 10.1016/j.cgh.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 3.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moro K, Kawaguchi T, Tsuchida J, Gabriel E, Qi Q, Yan L, Wakai T, Takabe K, Nagahashi M. Ceramide species are elevated in human breast cancer and are associated with less aggressiveness. Oncotarget. 2018;9:19874–19890. doi: 10.18632/oncotarget.24903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moro K, Nagahashi M, Gabriel E, Takabe K, Wakai T. Clinical application of ceramide in cancer treatment. Breast Cancer. 2019;26:407–415. doi: 10.1007/s12282-019-00953-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirose Y, Nagahashi M, Katsuta E, Yuza K, Miura K, Sakata J, Kobayashi T, Ichikawa H, Shimada Y, Kameyama H, McDonald KA, Takabe K, Wakai T. Generation of sphingosine-1-phosphate is enhanced in biliary tract cancer patients and is associated with lymphatic metastasis. Sci Rep. 2018;8:10814. doi: 10.1038/s41598-018-29144-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsuchida J, Nagahashi M, Nakajima M, Katsuta E, Rashid OM, Qi Q, Yan L, Okuda S, Takabe K, Wakai T. Sphingosine Kinase 1 is associated with immune cell-related gene expressions in human breast cancer. J Surg Res. 2020;256:645–656. doi: 10.1016/j.jss.2020.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. doi: 10.1038/nature13475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogretmen B. Sphingolipid metabolism in cancer signalling and therapy. Nat Rev Cancer. 2018;18:33–50. doi: 10.1038/nrc.2017.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nagahashi M, Abe M, Sakimura K, Takabe K, Wakai T. The role of sphingosine-1-phosphate in inflammation and cancer progression. Cancer Sci. 2018;109:3671–3678. doi: 10.1111/cas.13802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takabe K, Kim RH, Allegood JC, Mitra P, Ramachandran S, Nagahashi M, Harikumar KB, Hait NC, Milstien S, Spiegel S. Estradiol induces export of sphingosine 1-phosphate from breast cancer cells via ABCC1 and ABCG2. J Biol Chem. 2010;285:10477–10486. doi: 10.1074/jbc.M109.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takabe K, Spiegel S. Export of sphingosine-1-phosphate and cancer progression. J Lipid Res. 2014;55:1839–1846. doi: 10.1194/jlr.R046656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada A, Nagahashi M, Aoyagi T, Huang WC, Lima S, Hait NC, Maiti A, Kida K, Terracina KP, Miyazaki H, Ishikawa T, Endo I, Waters MR, Qi Q, Yan L, Milstien S, Spiegel S, Takabe K. ABCC1-exported sphingosine-1-phosphate, produced by sphingosine kinase 1, shortens survival of mice and patients with breast cancer. Mol Cancer Res. 2018;16:1059–1070. doi: 10.1158/1541-7786.MCR-17-0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takabe K, Paugh SW, Milstien S, Spiegel S. “Inside-out” signaling of sphingosine-1-phosphate: therapeutic targets. Pharmacol Rev. 2008;60:181–195. doi: 10.1124/pr.107.07113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maceyka M, Rohrbach T, Milstien S, Spiegel S. Role of sphingosine kinase 1 and sphingosine-1-phosphate axis in hepatocellular carcinoma. Handb Exp Pharmacol. 2020;259:3–17. doi: 10.1007/164_2019_217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, He YY, Sun JX, Guo WX, Li N, Xue J, Cheng SQ. The impact of sphingosine kinase 1 on the prognosis of hepatocellular carcinoma patients with portal vein tumor thrombus. Ann Hepatol. 2015;14:198–206. [PubMed] [Google Scholar]
- 17.Cai H, Xie X, Ji L, Ruan X, Zheng Z. Sphingosine kinase 1: a novel independent prognosis biomarker in hepatocellular carcinoma. Oncol Lett. 2017;13:2316–2322. doi: 10.3892/ol.2017.5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reynolds GM, Visentin B, Sabbadini R. Immunohistochemical detection of sphingosine-1-phosphate and sphingosine kinase-1 in human tissue samples and cell lines. Methods Mol Biol. 2018;1697:43–56. doi: 10.1007/7651_2017_44. [DOI] [PubMed] [Google Scholar]
- 19.Wang F, Wu Z. Sphingosine kinase 1 overexpression is associated with poor prognosis and oxaliplatin resistance in hepatocellular carcinoma. Exp Ther Med. 2018;15:5371–5376. doi: 10.3892/etm.2018.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, Endo I, Takabe K. A novel 4-Gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel) 2020;12:1148. doi: 10.3390/cancers12051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi H, Oshi M, Asaoka M, Ishikawa T, Endo I, Takabe K. ASO author reflections: transitioning from morphology to transcriptomics in capturing tumor biology. Ann Surg Oncol. 2020;27:4486–4487. doi: 10.1245/s10434-020-08680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi H, Oshi M, Asaoka M, Yan L, Endo I, Takabe K. Molecular biological features of nottingham histological grade 3 breast cancers. Ann Surg Oncol. 2020;27:4475–4485. doi: 10.1245/s10434-020-08608-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandhi S, Elkhanany A, Oshi M, Dai T, Opyrchal M, Mohammadpour H, Repasky EA, Takabe K. Contribution of immune cells to glucocorticoid receptor expression in breast cancer. Int J Mol Sci. 2020;21:4635. doi: 10.3390/ijms21134635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takahashi H, Katsuta E, Yan L, Tokumaru Y, Katz MHG, Takabe K. Transcriptomic profile of lymphovascular invasion, a known risk factor of pancreatic ductal adenocarcinoma metastasis. Cancers (Basel) 2020;12:2033. doi: 10.3390/cancers12082033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeshita T, Yan L, Peng X, Kimbung S, Hatschek T, Hedenfalk IA, Rashid OM, Takabe K. Transcriptomic and functional pathway features were associated with survival after pathological complete response to neoadjuvant chemotherapy in breast cancer. Am J Cancer Res. 2020;10:2555–2569. [PMC free article] [PubMed] [Google Scholar]
- 27.Oshi M, Angarita FA, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. High expression of NRF2 is associated with increased tumor-infiltrating lymphocytes and cancer immunity in ER-positive/HER2-negative breast cancer. Cancers (Basel) 2020;12:3856. doi: 10.3390/cancers12123856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takeshita T, Torigoe T, Yan L, Huang JL, Yamashita H, Takabe K. The impact of immunofunctional phenotyping on the malfunction of the cancer immunity cycle in breast cancer. Cancers (Basel) 2020;13:110. doi: 10.3390/cancers13010110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshi M, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Cherkassky L, Takabe K. Enhanced DNA repair pathway is associated with cell proliferation and worse survival in hepatocellular carcinoma (HCC) Cancers (Basel) 2021;13:323. doi: 10.3390/cancers13020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wurmbach E, Chen YB, Khitrov G, Zhang W, Roayaie S, Schwartz M, Fiel I, Thung S, Mazzaferro V, Bruix J, Bottinger E, Friedman S, Waxman S, Llovet JM. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10:1852. doi: 10.1038/s41598-020-58995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tokumaru Y, Oshi M, Katsuta E, Yan L, Satyananda V, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am J Cancer Res. 2020;10:897–907. [PMC free article] [PubMed] [Google Scholar]
- 33.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, Endo I, Takabe K. G2M cell cycle pathway score as a prognostic biomarker of metastasis in estrogen receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21:2921. doi: 10.3390/ijms21082921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, Endo I, Takabe K. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9:1643. doi: 10.3390/cells9071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21:6708. doi: 10.3390/ijms21186708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K. High G2M pathway score pancreatic cancer is associated with worse survival, particularly after margin-positive (R1 or R2) resection. Cancers (Basel) 2020;12:2871. doi: 10.3390/cancers12102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulze A, Oshi M, Endo I, Takabe K. MYC targets scores are associated with cancer aggressiveness and poor survival in ER-positive primary and metastatic breast cancer. Int J Mol Sci. 2020;21:8127. doi: 10.3390/ijms21218127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, Takabe K. Degree of early estrogen response predict survival after endocrine therapy in primary and metastatic ER-positive breast cancer. Cancers (Basel) 2020;12:3557. doi: 10.3390/cancers12123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tokumaru Y, Katsuta E, Oshi M, Sporn JC, Yan L, Le L, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of miR-34a associated with less aggressive cancer biology but not with survival in breast cancer. Int J Mol Sci. 2020;21:3045. doi: 10.3390/ijms21093045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tokumaru Y, Asaoka M, Oshi M, Katsuta E, Yan L, Narayanan S, Sugito N, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of microRNA-143 is associated with favorable tumor immune microenvironment and better survival in estrogen receptor positive breast cancer. Int J Mol Sci. 2020;21:3213. doi: 10.3390/ijms21093213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katsuta E, Rashid OM, Takabe K. Fibroblasts as a biological marker for curative resection in pancreatic ductal adenocarcinoma. Int J Mol Sci. 2020;21:3890. doi: 10.3390/ijms21113890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tokumaru Y, Oshi M, Katsuta E, Yan L, Huang JL, Nagahashi M, Matsuhashi N, Futamura M, Yoshida K, Takabe K. Intratumoral adipocyte-high breast cancer enrich for metastatic and inflammation-related pathways but associated with less cancer cell proliferation. Int J Mol Sci. 2020;21:5744. doi: 10.3390/ijms21165744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, Takabe K. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21:6968. doi: 10.3390/ijms21186968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2019;51:411–412. doi: 10.1016/j.immuni.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Asaoka M, Patnaik SK, Zhang F, Ishikawa T, Takabe K. Lymphovascular invasion in breast cancer is associated with gene expression signatures of cell proliferation but not lymphangiogenesis or immune response. Breast Cancer Res Treat. 2020;181:309–322. doi: 10.1007/s10549-020-05630-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takahashi H, Kawaguchi T, Yan L, Peng X, Qi Q, Morris LGT, Chan TA, Tsung A, Otsuji E, Takabe K. Immune cytolytic activity for comprehensive understanding of immune landscape in hepatocellular carcinoma. Cancers (Basel) 2020;12:1221. doi: 10.3390/cancers12051221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chouliaras K, Tokumaru Y, Asaoka M, Oshi M, Attwood KM, Yoshida K, Ishikawa T, Takabe K. Prevalence and clinical relevance of tumor-associated tissue eosinophilia (TATE) in breast cancer. Surgery. 2021;169:1234–1239. doi: 10.1016/j.surg.2020.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oshi M, Newman S, Murthy V, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. ITPKC as a prognostic and predictive biomarker of neoadjuvant chemotherapy for triple negative breast cancer. Cancers (Basel) 2020;12:2758. doi: 10.3390/cancers12102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, Matsuhashi N, Futamura M, Ishikawa T, Yoshida K, Endo I, Takabe K. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. 2020;10:16554. doi: 10.1038/s41598-020-73624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K. Abundance of regulatory T Cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel) 2020;12:3038. doi: 10.3390/cancers12103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, Takabe K. Plasmacytoid dendritic cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in triple negative breast cancer (TNBC) more strongly than conventional dendritic cell (cDC) Cancers (Basel) 2020;12:3342. doi: 10.3390/cancers12113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. Inflammation is associated with worse outcome in the whole cohort but with better outcome in triple-negative subtype of breast cancer patients. J Immunol Res. 2020;2020:5618786. doi: 10.1155/2020/5618786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bao M, Chen Z, Xu Y, Zhao Y, Zha R, Huang S, Liu L, Chen T, Li J, Tu H, He X. Sphingosine kinase 1 promotes tumour cell migration and invasion via the S1P/EDG1 axis in hepatocellular carcinoma. Liver Int. 2012;32:331–338. doi: 10.1111/j.1478-3231.2011.02666.x. [DOI] [PubMed] [Google Scholar]
- 56.Liang J, Nagahashi M, Kim EY, Harikumar KB, Yamada A, Huang WC, Hait NC, Allegood JC, Price MM, Avni D, Takabe K, Kordula T, Milstien S, Spiegel S. Sphingosine-1-phosphate links persistent STAT3 activation, chronic intestinal inflammation, and development of colitis-associated cancer. Cancer Cell. 2013;23:107–120. doi: 10.1016/j.ccr.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park ES, Choi S, Shin B, Yu J, Yu J, Hwang JM, Yun H, Chung YH, Choi JS, Choi Y, Rho J. Tumor necrosis factor (TNF) receptor-associated factor (TRAF)-interacting protein (TRIP) negatively regulates the TRAF2 ubiquitin-dependent pathway by suppressing the TRAF2-sphingosine 1-phosphate (S1P) interaction. J Biol Chem. 2015;290:9660–9673. doi: 10.1074/jbc.M114.609685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nagahashi M, Tsuchida J, Moro K, Hasegawa M, Tatsuda K, Woelfel IA, Takabe K, Wakai T. High levels of sphingolipids in human breast cancer. J Surg Res. 2016;204:435–444. doi: 10.1016/j.jss.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Donoviel MS, Hait NC, Ramachandran S, Maceyka M, Takabe K, Milstien S, Oravecz T, Spiegel S. Spinster 2, a sphingosine-1-phosphate transporter, plays a critical role in inflammatory and autoimmune diseases. FASEB J. 2015;29:5018–5028. doi: 10.1096/fj.15-274936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagahashi M, Yamada A, Katsuta E, Aoyagi T, Huang WC, Terracina KP, Hait NC, Allegood JC, Tsuchida J, Yuza K, Nakajima M, Abe M, Sakimura K, Milstien S, Wakai T, Spiegel S, Takabe K. Targeting the SphK1/S1P/S1PR1 axis that links obesity, chronic inflammation, and breast cancer metastasis. Cancer Res. 2018;78:1713–1725. doi: 10.1158/0008-5472.CAN-17-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tsuchida J, Nagahashi M, Nakajima M, Moro K, Tatsuda K, Ramanathan R, Takabe K, Wakai T. Breast cancer sphingosine-1-phosphate is associated with phospho-sphingosine kinase 1 and lymphatic metastasis. J Surg Res. 2016;205:85–94. doi: 10.1016/j.jss.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aoki H, Aoki M, Katsuta E, Ramanathan R, Idowu MO, Spiegel S, Takabe K. Host sphingosine kinase 1 worsens pancreatic cancer peritoneal carcinomatosis. J Surg Res. 2016;205:510–517. doi: 10.1016/j.jss.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aoki M, Aoki H, Ramanathan R, Hait NC, Takabe K. Sphingosine-1-phosphate signaling in immune cells and inflammation: roles and therapeutic potential. Mediators Inflamm. 2016;2016:8606878. doi: 10.1155/2016/8606878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagahashi M, Ramachandran S, Kim EY, Allegood JC, Rashid OM, Yamada A, Zhao R, Milstien S, Zhou H, Spiegel S, Takabe K. Sphingosine-1-phosphate produced by sphingosine kinase 1 promotes breast cancer progression by stimulating angiogenesis and lymphangiogenesis. Cancer Res. 2012;72:726–735. doi: 10.1158/0008-5472.CAN-11-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jin J, Liao W, Yao W, Zhu R, Li Y, He S. Aldo-keto reductase family 1 member B 10 mediates liver cancer cell proliferation through sphingosine-1-phosphate. Sci Rep. 2016;6:22746. doi: 10.1038/srep22746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Refolo MG, Messa C, Guerra V, Carr BI, D’Alessandro R. Inflammatory mechanisms of HCC development. Cancers (Basel) 2020;12:641. doi: 10.3390/cancers12030641. [DOI] [PMC free article] [PubMed] [Google Scholar]