Abstract
Previously, we have used a chromatin cross-linking and immunoprecipitation protocol for the analysis of Myc and USF binding to the cad promoter. The adaptation of this technique for the study of mammalian transcription factors was a big step forward in the analysis of transcription factor family member specificity, allowing for the first time a definitive knowledge of which factor binds to a promoter region under normal physiological conditions. However, due to limitations of the assay, our previous studies could not definitively prove that both Myc and USF bound to the exact same site on the cad promoter, nor could we directly correlate loss of in vivo binding of a particular factor with loss of transcriptional activity. Therefore, we have further modified the chromatin immunoprecipitation protocol to alleviate these problems. We have now shown that it is possible to coexamine growth-regulated transcriptional activity and promoter occupancy by using stably integrated promoter constructs. We show that both Myc and USF bind to the exact same E box on the cad promoter, suggesting that competition between these two factors for a single site occurs in living cells. We also find that cad promoter constructs that retain USF binding but lose Myc binding in vivo no longer display an increase in transcriptional activity in mid- to late G1 phase of the cell cycle. Finally, we propose that cell cycle-regulated transcriptional activation of the cad promoter may be a stochastic, rather than a predetermined, process.
In mammalian cells, gene expression is directly regulated by DNA-binding transcription factors and their associated cofactors. Most DNA-binding transcription factors can be grouped into large families of related proteins which have similar DNA-binding domains. Each member of a family displays conserved sets of amino acids within the DNA-binding domain which, in cases where the crystal structure of the protein-DNA complex has been solved, are known to contact the DNA. Conservation of the amino acids involved in DNA recognition between family members suggests that the binding sites of each member will also be conserved. In vitro binding studies have shown that this sequence commonality between members of the same family does indeed result in similar DNA-binding specificities. For example, members of the bHLHzip family, which include the proto-oncoprotein c-Myc and its heterodimeric partner Max as well as the USF proteins, recognize a common core sequence (5, 13, 23). In vitro gel shift experiments have defined the consensus binding site for these factors as CACGTG (known as an E box) and have shown that subtle changes in the consensus sequence or in the nucleotides which flank the E box can greatly influence protein binding. For example, inversion of the internal 2 nucleotides results in a loss of binding by factors from the Myc family but confers binding by members of the distantly related MyoD family (26). Similarly, positioning of the E box between a 5′ T and a 3′ A (instead of C and G nucleotides) abolishes binding of c-Myc as assayed in vitro but enhances binding of the distantly related Microphthalmia protein (1). Based on these examples, one straightforward mechanism by which certain transcription factors may be excluded from regulating specific target genes is through subtle variations in binding site sequences. However, in vivo, chromosomal binding sites are not isolated but are located adjacent to core promoter elements and binding sites for other factors. Little is known about how subtle variations in E-box sequences will influence bHLHzip factor binding in this complex environment.
Although the sequence CACGTG was initially identified as the highest-affinity binding site for both c-Myc and USF1, more-recent studies indicate that these factors can bind additional sequences. For example, several studies indicate that c-Myc can bind to a variety of noncanonical E-box elements such as the sequence CATGTG (4, 11). USF has also been shown to bind variant E boxes (3). Another alternative binding site is the positioning element involved in selection of the transcription start site termed the initiator (Inr). Both c-Myc- and USF1-associated complexes have been shown to bind to the initiator elements from the TdT (terminal deoxynucleotidyl transferase) and Ad-ML (adenovirus major late) promoters in vitro (17, 20, 21). Binding to initiator elements is believed to involve interaction with the basal transcription factor TFII-I and result in transcriptional activation by USF1 and transcriptional repression by c-Myc. It has been proposed that c-Myc may bind the 5′ end of an Inr which resembles an optimal half-site site for c-Myc binding such as CAC or CAT (20). Finally, Myc1, an alternatively translated and longer form of the c-Myc protein, has been shown to bind the C/EBP consensus element (TTATGCAAT), which is completely unrelated to an E box (12). Although the in vitro binding data strongly suggests that c-Myc may regulate gene expression by binding to sites other than consensus E boxes, binding of c-Myc or USF1 to these sites in vivo has not been directly confirmed. However, binding of c-Myc to chromosomally located nonconsensus E boxes has been inferred through binding site cloning experiments (11) and formaldehyde cross-linking studies with intact cells (7).
Previously, we have used the cad promoter as a model system for studying Myc target genes. CAD is a trifunctional protein (carbamoyl-phosphate synthetase–aspartate carbamoyltransferase–dihydroorotase) the gene for which encodes the first three rate-limiting steps of de novo pyrimidine biosynthesis. We have shown that the mouse cad promoter contains a consensus E-box element which is required for activating cad transcription in response to growth signals (6, 7). Using a chromatin cross-linking and immunoprecipitation assay, we have previously shown that, within the context of living cells, both c-Myc and USF1 are bound to DNA fragments of approximately 500 bp in length containing the cad promoter during times of elevated cad transcription (7). However, in these earlier binding studies, we were not able to prove that c-Myc or USF1 bound specifically to the E box responsible for cad growth regulation in living cells. Due to the plethora of sites to which these factors have been shown to bind in vitro, it was necessary to determine if the E box located at +65 was indeed the Myc and/or USF1 binding site in the cad promoter. Here, we have now analyzed site-specific binding by c-Myc (referred to as Myc) and USF1 in living cells by formaldehyde cross-linking and immunoprecipitation. Through the generation of stable cell lines which contain different cad promoter constructs, we have shown that both factors absolutely require the consensus E box in order to bind to the cad promoter and that neither factor associates with the cad initiator element. We also demonstrate that an E-box element bound specifically by USF1 in intact cells cannot support activated cad transcription. Thus, we have shown a correlation between in vivo occupancy of the cad promoter by Myc and cell cycle-regulated transcription of the cad gene.
MATERIALS AND METHODS
Cell culture.
Subconfluent cultures of NIH 3T3 cells were maintained in Dulbecco’s modified Eagle’s medium with high glucose (DMEM/HG) (GIBCO) supplemented with 5% (vol/vol) defined-supplemented bovine calf serum (HyClone), 100 U of penicillin per ml, and 100 μg of streptomycin (GIBCO) per ml at 37°C and 5% CO2. NIH 3T3 cells were passaged at 70% confluence with 0.05% trypsin-EDTA (GIBCO) in phosphate-buffered saline (PBS). Serum-synchronized cultures of NIH 3T3 cells used for formaldehyde cross-linking were prepared by plating cells directly into starvation medium (0.5% bovine calf serum in DMEM/HG) and incubating them for 48 to 60 h until the cultures were quiescent. Cells were then stimulated to reenter the cell cycle by the addition of 10% bovine calf serum into the culture medium. Growth cycle progression was monitored by flow cytometric analysis of propidium iodide-stained cells as previously described (24).
Stable transfections.
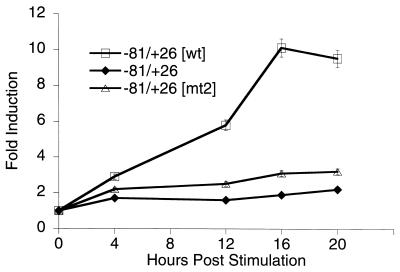
Construction of the cad reporter plasmids cad−81/+26, cad−81/+26[wt], and cad−81/+26[mt2], which contain hamster cad promoter fragments cloned upstream of the luciferase cDNA in the pGL2Basic vector (Promega), have been described previously (15). Stably transfected NIH 3T3 cells were prepared by transfecting 60-mm-diameter dishes of 1.5 × 105 NIH 3T3 cells with 9 μg of cad reporter plasmid (cad−81/+26, cad−81/+26[wt], or cad−81/+26[mt2]) and 1 μg of pcDNA3 (Invitrogen) as a neomycin resistance marker. Following transfection and glycerol shock, cells were incubated in maintenance medium (5% bovine calf serum in DMEM/HG). One day following transfection, cells were reseeded into 150-mm-diameter dishes in maintenance medium supplemented with 1 mg of G418 sulfate (GIBCO) per ml. Following approximately 2 weeks of selection, individual clones were generated. For each construct, 10 to 15 clones were picked and expanded. All clones were carried as subconfluent monolayers in maintenance medium with 1 mg of G418 sulfate per ml. To screen for luciferase activity, clones were plated at 1.5 × 105 cells/60-mm-diameter dish in maintenance medium lacking G418 and harvested the following day. Clones expressing luciferase were also analyzed for growth regulation by plating them at 1.5 × 105 cells/60-mm-diameter dish into low-serum medium (0.5% bovine calf serum in DMEM/HG) lacking G418 for 48 h and then harvesting them at intervals following the addition of high-serum (10%)-containing medium. Luciferase activity of total cell lysates was measured by using a luminometer (Analytical Luminescence Laboratory). The clones used for Fig. 4 produce very similar luciferase units; e.g., in serum-starved cells, the amount of luciferase activity of the −81/+26, −81/+26[wt], and −81/+26[mt2] clones varied about twofold. Thus, the site of integration did not greatly alter promoter strength in these particular clones.
FIG. 4.
Analysis of growth-regulated expression from integrated cad promoter constructs. Graphical representation of relative fold induction of stably integrated cad promoter-reporter activity throughout the growth cycle. NIH 3T3 cells were transfected with the indicated constructs, and stably transfected clones were selected with G418. Clones were serum starved for 48 h and then stimulated to grow by the addition of 10% serum to the culture medium. Cells were harvested for luciferase activity at the points indicated. Relative fold activation was calculated by normalizing the luciferase activity from serum-stimulated cells to the activity of the same clone prior to serum treatment. Data represents the average activity of one clone for each construct obtained from three independent time course experiments. Error bars represent the standard errors of the means.
Electromobility shift assays.
Electromobility shift assays were performed as previously described (18) with the following modifications. Where specified, binding reaction mixtures were incubated for 20 min with either a 50-fold molar excess of unlabeled probe oligonucleotide as a competitor or 2 μg of polyclonal antibody prior to the addition of double-stranded oligonucleotide probes, which were end labeled with [γ-32P]ATP by T4 polynucleotide kinase (22). Upon addition of the probe, binding reaction mixtures were incubated for an additional 20 min at room temperature and then resolved by electrophoresis on a 5% nondenaturing polyacrylamide gel (29:1 acrylamide/bisacrylamide ratio) for 2 h. The gel was preelectrophoresed for 60 min. Gels were dried, and protein-DNA interactions were visualized by autoradiography. The gel and running buffer was 22.5 mM Tris-morpholinepropanesulfonic acid (MOPS, pH 7.0) and 0.5 mM EDTA.
Cross-linking and immunoprecipitation of chromatin.
The formaldehyde cross-linking and immunoprecipitation protocol was adapted from references 2 and 8 with modifications. Formaldehyde (37% solution; Fisher Scientific) was added directly to cell culture medium at a final concentration of 1% at 0, 4, 8, or 12 h following serum addition to serum-starved NIH 3T3 cells. Fixation proceeded at 22°C for 10 min and was stopped by the addition of glycine to a final concentration of 0.125 M. To harvest cross-linked NIH 3T3 cells, plates were rinsed with cold PBS, incubated with 5 ml of trypsin-EDTA (GIBCO) in PBS, and then scraped. Cells were collected by centrifugation and washed in cold PBS plus 0.5 mM phenylmethylsulfonyl fluoride (PMSF). Pellets from approximately 108 cells were resuspended in 3 ml of swelling buffer (5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid); pH 8.0], 85 mM KCl, 0.5% NP-40, 0.5 mM PMSF, and 100 ng of leupeptin and aprotinin per ml) and incubated on ice for 20 min. NP-40 was added to 0.5%, and the cells were Dounce homogenized in a B Dounce homogenizer. Nuclei were collected by microcentrifugation at 5,000 rpm, resuspended in 1 to 2 ml of sonication buffer (1% sodium dodecyl sulfate [SDS], 10 mM EDTA, 50 mM Tris-HCl [pH 8.1], 0.5 mM PMSF, and 100 ng of leupeptin and aprotinin per ml), and incubated on ice for 10 min. Samples were sonicated with an Ultrasonics sonicator at full power for three 30-s pulses on ice to an average length of 200 to 500 bp and then microcentrifuged at 14,000 rpm. The chromatin solution was precleared with the addition of Staph A cells (prepared as described previously [6, 7]) for 15 min at 4°C. Prior to use, Staph A cells were blocked with 1 μg of sheared herring sperm DNA per ml and 1 μg of bovine serum albumin per ml for at least 4 h at 4°C. Precleared chromatin from 2.5 × 107 cells was incubated with 1 μg of affinity-purified rabbit polyclonal antibody (Santa Cruz; anti-Myc sc-764-X) or 1 μl of anti-human USF1 rabbit antiserum (gift of E. H. Bresnick) or no antibody and rotated at 4°C for 12 h. Immunoprecipitation, washing, and elution of immune complexes were carried out as described previously (6, 7). Prior to the first wash, one-half of the supernatant from the no-primary-antibody reaction for each time point was saved as 50% total input chromatin and was processed with the eluted immunoprecipitates beginning at the cross-link reversal step.
After addition of NaCl to 200 mM and 10 μg of RNase A, samples were incubated at 65°C for 5 h to reverse the cross-links. Samples were then precipitated at −20°C overnight by the addition of 2 volumes of ethyl alcohol (EtOH) and then pelleted by microcentrifugation at 15,000 rpm. Samples were resuspended in 100 μl of Tris-EDTA (pH 7.5)–25 μl of 5× proteinase K buffer (1.25% SDS, 50 mM Tris [pH 7.5], and 25 mM EDTA)–1.5 μl of proteinase K solution (Boehringer Mannheim) and incubated at 42°C for 2 h. Samples were extracted with phenol-chloroform-isoamyl alcohol (25:24:1) and then precipitated with a 1/10 volume of 3 M NaOAc (pH 5.3), 5 μg of tRNA, and 2 volumes of EtOH at −20°C overnight. Pellets were collected by microcentrifugation at 15,000 rpm, resuspended in 30 μl of H2O, and analyzed by PCR. Total input samples were resuspended in 100 μl of H2O and then diluted 1:100 prior to PCR. PCR mixtures contained 2 μl of immunoprecipitate or diluted total input; 50 ng of each primer; 0.88 mM MgCl2; 2 mM (each) dATP, dCTP, dGTP, and dTTP; 1× thermophilic buffer (Promega); and 1.25 U of Taq DNA polymerase (Promega) in a total volume of 20 μl. Integrated copies of cad were analyzed by PCR with the primers −81cad (5′CATGGTCCCGCCCCTTACGT) and goodluc (5′GGCGTCTTCCATTTTACCAACAGTACCGG), and endogenous cad was analyzed with the primers mcadA (5′TGACTAGCGGTACCGGGGTTGCTGCTGTGGAACC) and 3′cad (5′CGGGCTTGCTTACCCACCTTCCCCAGCAGTCGACAC). Following 32 to 35 or 13 to 15 cycles of amplification, PCR products were run on a 1.5% agarose gel and analyzed by ethidium bromide (EtBr) staining or Southern blot analysis, respectively. Primers were obtained from the University of Wisconsin Biotechnology Center.
Southern blot analysis.
Ten microliters of each PCR mixture was electrophoresed on a 1.5% agarose gel (no EtBr), stained with 1.5 μg of EtBr per ml in 1× Tris-EDTA-acetate for 10 min, destained in 1× Tris-EDTA-acetate for 5 min, and photographed alongside a fluorescent ruler. The gel was denatured for 30 min in 1.5 M NaCl–0.5 M NaOH and neutralized for 30 min in 1.5 M NaCl–0.5 M Tris (pH 7.2)–1 mM EDTA (pH 8.0). Samples were transferred onto a Hybond-N membrane (Amersham) with 20× SSPE (3.6 M NaCl, 0.2 M NaPO4, 20 mM EDTA) overnight by standard capillary transfer. The blot was baked at 80°C for 30 min, UV cross-linked at 120,000 μJ, and incubated at 42°C overnight in prehybridization solution (50% formamide, 3.4× SSPE, 100 μg of sonicated salmon sperm DNA per ml, 50 μg of boiled sonicated salmon sperm DNA per ml, 5× Denhardt’s solution, 10% dextran sulfate, 5% SDS, and 1% Sarkosyl). Labeled probe was added to the prehybridization solution (8 × 105 cpm/ml) and incubated at 42°C for 48 h. Blots were washed in 2× SSPE–0.1% SDS at room temperature for 30 min, 1× SSPE–0.1% SDS at 65°C for 15 min, and 0.5× SSPE–0.2% SDS at 65°C for 1 h. Blots were exposed to BioMax film (Kodak) for 24 to 48 h. The gel was stained, denatured, and transferred as described above. A 659-bp EcoRI/BamHI fragment of the mouse cad promoter from the plasmid mcad−440/+219 was labeled by nick translation. Two hundred nanograms of DNA was incubated in 1× labeling buffer (50 mM Tris [pH 7.5], 10 mM MgSO4, 100 mM dithiothreitol) with 50 nM (each) dATP, dGTP, and dTPP; 2.5 μl of [α-32P]dCTP (3,000 Ci/mmol; 10 mCi/ml); 3 × 10−4 U of DNase I; and 5 U of DNA polymerase I at 16°C for 1 h. The reaction was stopped by the addition of 25 mM EDTA and precipitated by the addition of a 1/2 volume of 7.5 M NH4OAc and 3 volumes of EtOH. Approximately 5 × 106 cpm of labeled probe was boiled and added to the prehybridization fluid.
RESULTS
The E-box element is required for Myc and USF1 to bind the proximal cad promoter in living cells.
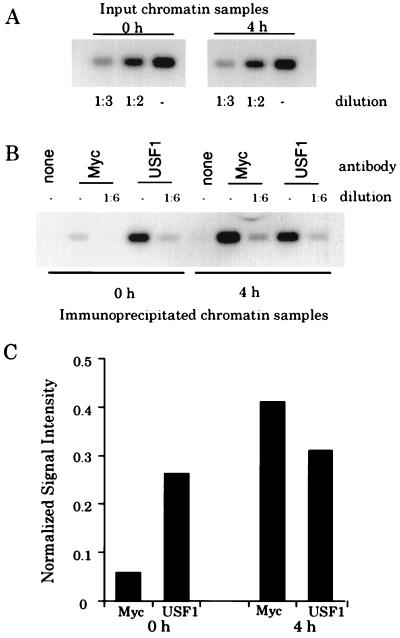
Within the context of living cells, we have previously observed that the cad promoter is bound by USF1 in the G0 phase of the growth cycle and by both Myc and USF1 during mid- to late G1 phase and early S phase (7). Although one might expect that recruitment of Myc to the cad promoter would result in displacement of the bound USF, it appeared as if the amount of USF bound to the cad promoter was not reduced when Myc was recruited. However, these previous studies were performed with a large number of PCR cycles and therefore we could not be sure that the USF signal was in the linear range. To obtain a more quantitative answer to the question as to whether USF binding was reduced when Myc binding increased, our approach was to analyze the immunoprecipitated samples by using a limited number of PCR cycles, Southern blotting the products, and quantitating the signals with a phosphorimager.
NIH 3T3 cells were serum starved to induce a quiescent state, and then serum was added to the medium and cells were harvested 4 h later. The 4-h time point was chosen because we have previously shown that Myc protein displays a dramatic increase in abundance at this time (18) and that both Myc and USF can be detected on the cad promoter at this time (7). Before analyzing the immunoprecipitated samples, we first performed a serial dilution of the input chromatin, beginning with 0.06% of the total input chromatin and then diluting this 1:2 or 1:3 (Fig. 1A). Dilution of the sample resulted in the respective reduction in the amount of PCR product generated, indicating that the signal obtained is proportional to the amount of input DNA. For example, quantitation of the signals from the two- and threefold-diluted input chromatin for the 0- and 4-h time points showed a 1.9- and 2.7-fold and a 2.1- and 3.2-fold decrease, respectively, relative to the signal from the undiluted total input. Next, we analyzed two dilutions of the immunoprecipitated samples. A sixfold dilution of the Myc and USF1 immunoprecipitates reduced the signal intensity an average of fivefold relative to the undiluted samples; thus, signal is also proportional to input in the immunoprecipitates (Fig. 1B). As shown in Fig. 1C, the cad signal immunoprecipitated by the Myc antibody increases eightfold by 4 h following serum stimulation (7). The cad signal immunoprecipitated by the USF1 antibody varies very little between the 0- and 4-h chromatin. In summary, analysis of the amounts of USF1 and Myc bound in the different stages of the cell cycle indicates that recruitment of Myc does not result in the displacement of USF1 from the cad promoter.
FIG. 1.
Myc does not displace USF from the cad promoter. (A) Fourteen cycles of PCR amplification were performed on chromatin from quiescent cells (0 h) or from cells which had been stimulated with serum for 4 h. Prior to PCR amplification, samples were diluted as indicated. PCR products were electrophoresed on an agarose gel, Southern blotted, hybridized with a radiolabeled cad probe, and analyzed with the phosphorimager with ImageQuant software. (B) PCRs were performed and analyzed as described above for immunoprecipitates from reactions containing no primary antibody (none), c-Myc antibody (Myc), or USF antibody (USF1). (C) Graphical analysis of cad PCR signals from the Southern blot shown in panel B. Normalized signal intensity is the quantitated numerical value of the signals from the diluted (1:6) anti-Myc and -USF1 lanes at 0 and 4 h normalized to the signal intensity of the input chromatin (1:2 diluted) for the 0- and 4-h samples, respectively. The signal intensity of the input chromatin (1:2 diluted) was arbitrarily set to a value of 1, and the normalized signals are presented as a fraction of this value.
Our results showing that Myc does not displace USF from the cad promoter raise the possibility that Myc and USF1 can bind simultaneously at different sites within the promoter. The cad promoter contains both an E box and a consensus initiator element. Since Myc and USF1 have been shown to bind initiator elements present in other promoters (17, 21), it was possible that one of these factors also binds the cad initiator. However, previous evidence suggests that the cad initiator is not a good candidate site for Myc binding. First, growth-regulated cad expression, which requires c-Myc protein (9, 18), is not affected by mutation of the cad initiator element (6). Second, Myc has been reported to repress transcription through initiator elements (16, 17), and yet our studies (7) and those by Bush et al. (9) suggest that Myc activates cad expression. Therefore, it seemed more likely that USF1, rather than Myc, might bind to the cad initiator.
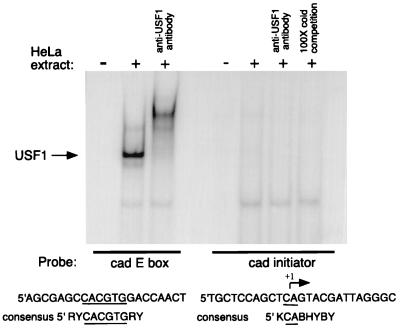
To evaluate whether USF1 can bind the cad initiator, we first performed in vitro gel shift assays (Fig. 2). A double-stranded oligonucleotide probe corresponding to the cad initiator sequence was radiolabeled and incubated with HeLa cell nuclear extract. As a positive control, the double-stranded cad E-box probe was also used in binding reactions. As expected, USF1 complexes bound efficiently to the E-box probe and were supershifted by the addition of anti-USF1 antibody. Under the same conditions, the probe containing the cad initiator sequences was not bound by USF1. Our observation that USF1 does not bind to the cad initiator is in agreement with studies of Roy et al. (21) which suggest that specific nucleotides within the initiator element are required for USF1 binding. Alignment of the cad initiator sequence with the consensus initiator binding site reveals that the cad sequence is not a perfect match to the consensus. These results suggest that the cad promoter-USF1 interaction detected in our cross-linking studies is likely to occur through DNA-binding sites other than the initiator element.
FIG. 2.
USF1 does not bind the cad initiator element in vitro. Gel shift analysis of USF1 binding at the cad E box and cad initiator elements. Ten micrograms of HeLa cell nuclear extract was incubated with radiolabeled cad E-box and cad initiator probes. Binding complexes were supershifted by the addition of anti-USF1 antibody or competed by the addition of excess unlabeled probe. Samples were resolved on a 6% native polyacrylamide gel. The sequence of each gel shift probe is shown below the gel, along with the USF1 consensus binding sequence. The arrow indicates the nucleotide where transcription initiates. Y represents pyrimidine nucleotides, and R represents purine nucleotides. The E-box element is underlined.
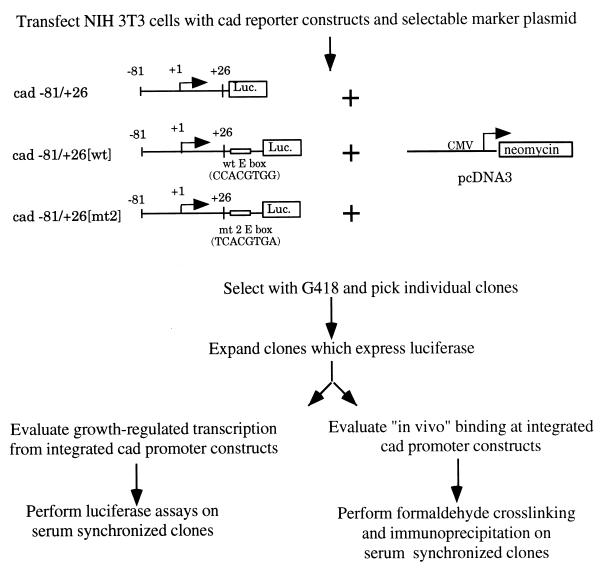
The results of our previous in vitro binding studies (6) indicate that both Myc and USF1 can bind the cad E-box element. However, in vitro experiments do not reveal whether these factors directly bind the cad E box within the context of living cells. To test the hypothesis that the cad E box is the specific binding site for Myc and/or USF1, we used the approach outlined in Fig. 3. The ultimate goal of these experiments was to examine transcription factor binding to cad promoters which either contain or lack the E-box element, by the formaldehyde cross-linking and immunoprecipitation technique. However, cell lines containing a natural mutation of the +65 E box within the cad loci have not been identified. Therefore, we created cell lines which contain stably integrated copies of the cad promoter. The −81/+26[wt] plasmid contains the cad E-box element which is bound by both Myc and USF1 in vitro, whereas no E box is present in the −81/+26 construct. The −81/+26[mt2] construct contains an E box which we have previously shown to be bound preferentially by USF1 in vitro (6). All three constructs drive expression of the luciferase reporter, enabling characterization of growth-regulated transcription from the integrated promoters. Following selection in G418, several individual clones of cells harboring each construct were expanded and analyzed for luciferase expression under logarithmic growth conditions. Since the site of plasmid integration within a chromosome may influence transcriptional regulation, we chose clones expressing both high and low levels of luciferase activity to further examine in serum synchronization assays. The variation in expression levels ensures that different clones were indeed unique integration events and allowed us to assess whether the results obtained in the regulation assays were consistent within a wide range of gene expression. Results from three representative clones are shown in Fig. 4. In general, we observed that clones which harbored the −81/+26[wt] construct displayed increasing luciferase activity as serum-stimulated cells progressed through the growth cycle, while those containing either the −81/+26 or −81/+26[mt2] construct showed little change in luciferase activity. Therefore, transcriptional regulation of the integrated cad promoter constructs recapitulates what had been observed by transient-transfection assays (6) and reinforces the conclusion that the wild-type E-box sequence is required for growth-regulated cad expression.
FIG. 3.
Experimental approach to studying site-specific binding of Myc and USF1 to the E-box element in the cad promoter. See the text for details. Luc., luciferase; wt, wild type; CMV, cytomegalovirus.
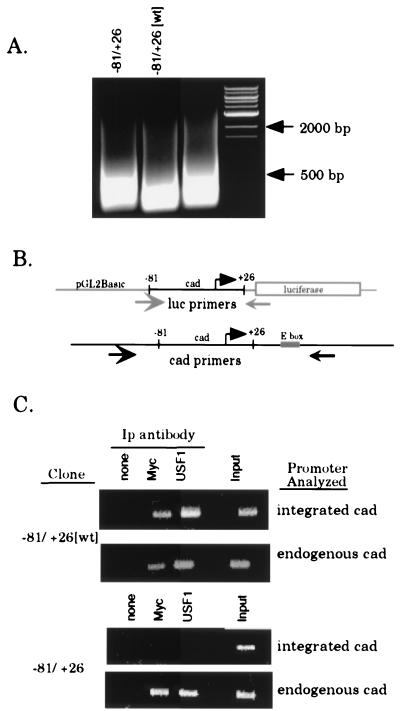
Next, we used the stable cell lines to evaluate whether Myc and/or USF1 specifically binds the cad promoter in vivo through the E-box motif. The first experiment was to determine if the 130-bp segment of the cad promoter contained within the −81/+26[wt] construct is bound by both Myc and USF1. If the binding sites for Myc and/or USF1 are not within the fragment of the cad promoter which is sufficient to confer growth regulation on a reporter construct, then it would be necessary to reinterpret the in vivo binding observed on the endogenous cad gene. Clones containing the −81/+26[wt] construct were treated with formaldehyde at 8 h following serum stimulation of quiescent cultures, since this corresponds to a time when the endogenous cad promoter is bound by both Myc and USF1 (7). Prior to immunoprecipitation with antibodies against Myc and USF1, cross-linked chromatin was sonicated to an average length of 200 to 500 bp (Fig. 5A). This step is critical to the success of these experiments, since the integrated copies of the cad promoter may reside near other endogenous E-box elements. However, by sonicating the chromatin to a small size, we can be sure that we monitor binding of Myc and USF only to the cad promoter and not to surrounding chromatin. Immunoprecipitates were assayed by using PCR primers (Fig. 5B) specific for either the integrated cad plasmid or the endogenous cad promoter which serves as an internal control. As shown in the top panel of Fig. 5C, we observed that both the endogenous cad promoter and the integrated −81/+26[wt] cad promoter were bound by Myc and USF1. This confirms that binding of both factors is localized to a DNA fragment containing the 130-bp integrated cad promoter.
FIG. 5.
Myc and USF1 bind specifically to the E box within the cad promoter. Clones of stably transfected cells (−81/+26[wt] clone 1 and −81/+26 clone 5) were serum starved for 48 h, then serum stimulated for 8 h, and cross-linked with formaldehyde. Cross-linked chromatin from each line was prepared and immunoprecipitated with antibodies against Myc and USF1. (A) EtBr-stained agarose gel showing the size of the DNA fragments following sonication. Lanes were loaded with 2.5% input chromatin from the specified clone after reversal of the cross-links and proteinase K treatment. DNA size markers are as indicated. (B) Schematic of the annealing position of PCR primers used to specifically amplify either integrated or endogenous cad promoters. The luciferase and cad primers amplify products of 200 and 350 bp, respectively. (C) PCR analysis of immunoprecipitation reactions with luciferase or cad primers to amplify the integrated or endogenous cad promoter, respectively. For all stable clones, immunoprecipitates were resuspended in 30 μl of H2O and input chromatin samples were diluted to 1% in 100 μl of H2O. The copy number of the integrated plasmid was normalized by utilizing appropriate sample volumes such that the input chromatin signal for all clones analyzed was approximately equivalent (last lane of each upper panel, integrated cad). Volumes used for both input chromatin and the immunoprecipitates (Ip) were as follows: 3 μl of the −81/+26-5 samples and 2 μl of −81/+26[wt]-1 samples. PCR mixtures with the cad primers (endogenous cad) contained equivalent sample volumes (2 μl) for all clones examined. PCR products were electrophoresed on a 1.5% agarose gel and stained with EtBr.
The next step was to determine if the E box within the cad promoter was the direct binding site for either Myc or USF1. Therefore, stable cell clones containing the −81/+26 construct, which lacks the E box, were also analyzed by the formaldehyde cross-linking procedure. We found that in the −81/+26 stable cell lines, binding by Myc and USF1 was no longer readily detected at levels over background (Fig. 5C) on the integrated cad promoter. However, within these same chromatin samples, the endogenous cad promoter shows clear binding by Myc and USF1, which verifies that the cross-linking and immunoprecipitation were successful. Therefore, loss of binding is specific to the −81/+26 cad promoter. The results presented in Fig. 5C, which are representative of several different clones examined, demonstrate that the E box is required for both Myc and USF1 to bind the cad promoter in living cells. In addition, the finding that Myc and USF1 no longer bind the −81/+26 promoter which contains the cad initiator element supports the conclusion that neither USF1 nor Myc associates with the cad initiator in intact cells. Finally, the finding that stably integrated promoter-reporter constructs can be used successfully to coexamine transcription factor binding and activity within the same cells should prove generally useful for future studies of gene expression. Accordingly, in the following section, we have used this approach to examine transcriptional activity and binding of Myc and USF1 at E-box elements with various flanking sequences.
Variation of the sequences flanking the E box can influence relative levels of Myc versus USF1 binding in intact cells.
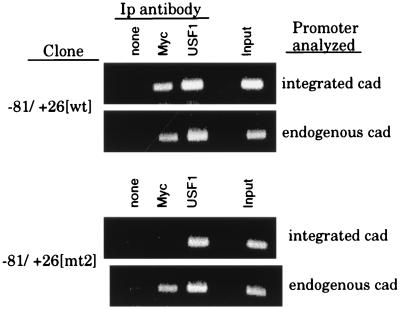
Previously, we presented evidence that the nucleotides flanking the E box could influence binding of Myc in vitro (6). In particular, arrangement of the E-box sequence CACGTG between a 5′ T and a 3′ A abolishes Myc binding in vitro but has no effect on USF1, which readily binds this sequence. As shown in Fig. 4, an E-box element which has these flanking nucleotides does not confer growth-regulated expression on the cad promoter. Although these results are consistent with the conclusion that USF1 could not activate cad transcription, we had no formal proof that the E box, 5′ TCACGTGA, was bound by USF1, but not Myc, in vivo. Since we had shown that stably integrated promoter-reporter constructs can be used successfully to coexamine factor binding and activity, it was now possible to evaluate more directly the consequence of changing the E-box-flanking nucleotides on Myc and USF1 binding in intact cells by formaldehyde cross-linking. For this experiment, binding was compared between cell lines harboring the −81/+26[wt] plasmid (which contains the endogenous CCACGTGG cad E box and displays growth regulation) and the −81/+26[mt2] plasmid (which contains a TCACGTGA E box and lacks growth-regulated activity). As shown in Fig. 5 and 6, the E box in the −81/+26[wt] cad promoter can be bound by both Myc and USF1 in living cells. Notably, similar signals in the anti-Myc lanes, relative to the input signals, were observed for both the integrated and endogenous cad promoters. In contrast, very little Myc binding was detected on the integrated −81/+26[mt2] cad promoter compared to that bound to the endogenous cad E box within the same cells, indicating that the TCACGTGC E box is a poor binding site for Myc in vivo (Fig. 6). However, the ratio of anti-USF1 signal to the input signal was similar on each of the [wt], [mt2], and endogenous cad E-box elements. These results indicate that, in intact cells, USF1 binding is relatively insensitive to changes in the nucleotides flanking the E box. Importantly, we now have direct evidence that although the cell lines harboring the integrated −81/+26[mt2] cad promoter do not display growth regulation, USF1 is bound to the [mt2] E box following serum stimulation. Together, these results indicate that USF1 binding does not correlate with transcriptional activation of the cad promoter.
FIG. 6.
USF1, but not Myc, binds the 5′ TCACGTGA E box in intact cells. The stable cell lines −81/+26[wt] clone 1 (containing the CCACGTGG E box) and −81/+26[mt2] clone 2 (containing the TCACGTGA E box) were serum synchronized, cross-linked, and immunoprecipitated as described in the legend to Fig. 5. PCR analysis was performed with luciferase primers (integrated cad) and cad primers (endogenous cad). The copy number of the integrated plasmids was normalized by utilizing appropriate sample volumes so that the input signals for each clone were equivalent, as described in the legend to Fig. 5. Volumes used were as follows: 2 μl of the −81/+26[wt] samples and 1 μl of the −81/+26[mt2] samples. PCRs with the cad primers were performed with equivalent sample volumes for all clones examined. PCR products were electrophoresed on a 1.5% agarose gel and stained with EtBr. Ip, immunoprecipitation.
DISCUSSION
Using the formaldehyde cross-linking and immunoprecipitation technique on cells which contain stably integrated promoter constructs, we have shown that site-specific transcription factor binding can be directly correlated with promoter activity in parallel cultures of cells. By comparison of various cell lines harboring cad promoter constructs differing only in the E-box sequence, we have demonstrated that both Myc and USF1 bind specifically to a single site in the cad promoter. Although previous studies have suggested that genes containing E boxes may be regulated by both Myc and USF, our studies are the first to show that both proteins have the potential to be recruited to a specific basal promoter complex under normal physiological conditions. However, our results with a change-of-specificity mutation in the E box indicate that binding of USF1 cannot confer growth regulation on the cad promoter. Thus, the main determinant of Myc versus USF1 activity on the cad promoter appears to be a post-DNA-binding mechanism. It is likely that differences between transactivation by Myc and that by USF1 are due to differences in protein-protein interactions. For example, we have recently found that Myc, but not USF1, binds the coactivator CREB-binding protein (9b).
The observations that both Myc and USF1 bind to the same site in the cad promoter and that recruitment of Myc in S phase does not displace USF suggest that Myc likely binds to a different subset of cad alleles than does USF. Using antibodies to Max, the heterodimeric partner of Myc, in the chromatin immunoprecipitation assays, we have observed that Max is bound to the cad promoter in quiescent cells, even though Myc protein is not expressed (data not shown). The Max-containing complexes may represent either homodimers or Max complexed to a member of the Mad family. Regardless, we suggest that Myc does not displace USF because Myc-Max heterodimers exchange places with the existing Max complexes. Recent studies (25) have shown that Max-containing complexes (Max-Max, Max-Myc, and Max-Mad) all have a high off-rate from DNA (half-life of 10 to 20 min), supporting our hypothesis that exchange of Max-containing complexes occurs as Myc-Max complexes become more abundant in mid- to late G1 phase. Preliminary analysis of the dissociation rate of USF from the cad E box by gel mobility shift assays indicates that USF binds quite stably to the site in the cad promoter (half-life of ∼60 min) (9a). Thus, the differences between the stability of USF bound to an E box and that of Myc-Max bound to an E box, as measured by in vitro DNA-binding assays, are consistent with our finding that USF is not displaced by Max-Myc in cells.
Because the chromatin immunoprecipitation procedure measures protein-DNA interactions in a population of cells, we cannot determine if, in a single cell, one cad allele is bound by Myc-Max and the other is bound by USF or if both cad alleles within a given cell are bound by the same factor, the identity of which differs between neighboring cells. Regardless, our results suggest that, under physiological conditions in a clonally derived synchronized cell population, all alleles of a given gene do not have the same profile of bound transcription factors and, therefore, may not exhibit identical patterns of gene expression. The concept that sister cad alleles or cad alleles in neighboring cells may not display the same transcriptional profile in response to environmental signals such as serum growth factors suggests that cell cycle-regulated transcriptional activation may be a stochastic, rather than a predetermined, process. Previous studies have also suggested that a cell-to-cell variation in a transcriptional response may occur. For example, Newlands et al. (19) have shown that not all nuclei in a muscle fiber transactivate a particular gene at the same time, even though the nuclei have a common cytoplasm. Other studies showing that individual promoter templates having almost identical genetic and physiological conditions can be induced to different levels in different cells also support the hypothesis that alternative forms of transcription complexes can form on a given promoter (14). Previous studies relied on correlations between in vitro binding and in vivo gene activation (10); our results now provide evidence suggesting that two different transcription complexes can be formed on the cad promoter in living cells. Our future work will be focused on further analysis of the consequences of Myc versus USF1 binding to a specific E box in the context of neoplastic transformation of cells.
ACKNOWLEDGMENTS
We thank David Allis and Richard Treisman for sharing their formaldehyde cross-linking protocols, Stephanie Bartley and Julie Wells for technical assistance, and Emery Bresnick for anti-USF1 rabbit antisera.
This work was supported in part by Public Health Service grants CA45240 and CA07175; K.E.B. was supported by Public Health Service Training Grant CA09135.
REFERENCES
- 1.Aksan I, Goding C R. Targeting the microphthalmia basic helix-loop-helix–leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol Cell Biol. 1998;18:6930–6938. doi: 10.1128/mcb.18.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts A S, Geneste O, Treisman R. Activation of SRF-regulated chromosomal templates by Rho-family GTPases requires a signal that also induces H4 hyperacetylation. Cell. 1998;92:475–487. doi: 10.1016/s0092-8674(00)80941-1. [DOI] [PubMed] [Google Scholar]
- 3.Bendall A J, Molloy P L. Base preferences for DNA binding by the bHLH-Zip protein USF: effects of MgCl2 on specificity and comparison with binding of Myc family members. Nucleic Acids Res. 1994;22:2801–2810. doi: 10.1093/nar/22.14.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackwell T K, Huang J, Ma A, Kretzner L, Alt F, Eisenman R, Weintraub H. Binding of Myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blackwell T K, Kretzner L, Blackwood E M, Eisenman R N, Weintraub H. Sequence-specific DNA-binding by the c-Myc protein. Science. 1990;250:1149–1151. doi: 10.1126/science.2251503. [DOI] [PubMed] [Google Scholar]
- 6.Boyd K E, Farnham P J. Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol Cell Biol. 1997;17:2529–2537. doi: 10.1128/mcb.17.5.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd K E, Wells J, Gutman J, Bartley S M, Farnham P J. c-Myc target gene specificity is determined by a post-DNA-binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braunstein M, Rose A B, Holems S G, Allis C D, Broach J R. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- 9.Bush A, Mateyka M, Dugan K, Obaya A, Adachi S, Sedivy J, Cole M. c-myc null cells misregulate cad and gadd45 but not other proposed c-Myc targets. Genes Dev. 1998;12:3797–3802. doi: 10.1101/gad.12.24.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.D’Cunha, C., and P. J. Farnham. Unpublished data.
- 9b.Eberhardy, S., and P. J. Farnham. Unpublished data.
- 10.Fiering S, Northrop J P, Nolan G P, Mattila P S, Crabtree G R, Herzenberg L A. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- 11.Grandori C, Mac J, Siëbelt F, Ayer D E, Eisenman R. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- 12.Hann S R, Dixit M, Sears R C, Sealy L. The alternatively initiated c-Myc proteins differentially regulate transcription through a non-canonical DNA binding site. Genes Dev. 1994;8:2441–2452. doi: 10.1101/gad.8.20.2441. [DOI] [PubMed] [Google Scholar]
- 13.Hodgkinson C A, Moore K J, Nakayama A, Steingrimsson E, Copeland N G, Jenkins N A, Arnheiter H. Mutations of the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- 14.Ko M S H, Nakauchi H, Takahashi N. The dose dependence of glucocorticoid-inducible gene expression results from changes in the number of transcriptionally active templates. EMBO J. 1990;9:2835–2842. doi: 10.1002/j.1460-2075.1990.tb07472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kollmar R, Sukow K A, Sponagle S K, Farnham P J. Start site selection at the TATA-less carbamoyl-phosphate synthase (glutamine-hydrolyzing)/aspartate carbamoyltransferase/dihydroorotase promoter. J Biol Chem. 1994;269:2252–2257. [PubMed] [Google Scholar]
- 16.Li L-H, Nerlov C, Prendergast G, MacGregor D, Ziff E B. c-Myc represses transcription in vivo by a novel mechanism dependent on the initiator element and Myc box II. EMBO J. 1994;13:4070–4079. doi: 10.1002/j.1460-2075.1994.tb06724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mai S, Martensson I L. The c-myc protein represses the lambda5 and TdT initiators. Nucleic Acids Res. 1995;23:1–9. doi: 10.1093/nar/23.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miltenberger R J, Sukow K, Farnham P J. An E-box-mediated increase in cad transcription at the G1/S-phase boundary is suppressed by inhibitory c-Myc mutants. Mol Cell Biol. 1995;15:2527–2535. doi: 10.1128/mcb.15.5.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newlands S, Levitt L K, Robinson C S, Carmen Karpf A B, Hodgson V R M, Wade R P, Hardeman E C. Transcription occurs in pulses in muscle fibers. Genes Dev. 1998;12:2748–2758. doi: 10.1101/gad.12.17.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy A L, Carruthers C, Gutjahr T, Roeder R G. Direct role for Myc in transcription initiation mediated by interaction with TFII-I. Nature. 1993;365:359–361. doi: 10.1038/365359a0. [DOI] [PubMed] [Google Scholar]
- 21.Roy A L, Meisterernst M, Pognonec P, Roeder R G. Cooperative interaction of an initiator-binding transcription factor and the helix-loop-helix activator USF. Nature. 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Sirito M, Lin Q, Maity T, Sawadogo M. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 1994;22:427–433. doi: 10.1093/nar/22.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slansky J E, Li Y, Kaelin W G, Farnham P J. A protein synthesis-dependent increase in E2F1 mRNA correlates with growth regulation of the dihydrofolate reductase promoter. Mol Cell Biol. 1993;13:1610–1618. doi: 10.1128/mcb.13.3.1610. (Author’s correction, 13:7201.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sommer A, Boussett K, Kremmer E, Austen M, Lusher B. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J Biol Chem. 1998;273:6632–6642. doi: 10.1074/jbc.273.12.6632. [DOI] [PubMed] [Google Scholar]
- 26.Van Antwerp M E, Chen D G, Chang C, Prochownik E V. A point mutation in the MyoD basic domain imparts c-Myc-like properties. Proc Natl Acad Sci USA. 1992;89:9010–9014. doi: 10.1073/pnas.89.19.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]