Abstract
Three-dimensional (3D) organoids are a novel tool to model epithelial cell biology and human diseases of the esophagus. 3D organoid culture systems have been utilized to investigate the pathobiology of esophageal cancer, including both squamous cell carcinoma and adenocarcinoma. Additional organoid-based approaches for study of esophageal development and benign esophageal diseases have provided key insights into esophageal keratinocyte differentiation and mucosal regeneration. These investigations have implications for the identification of esophageal cancer stem cells, as well as the potential to halt malignant progression through induction of differentiation pathways. Patient-derived organoids (PDOs) from human tissue samples allow for unique and faithful in vitro modeling of esophageal cancers, and provide an exciting platform for investigation into personalized medicine and targeted treatment approaches, as well as new models for understanding therapy resistance and recurrent disease. Future directions include high-throughput genomic screening using PDOs, and study of tumor-microenvironmental interactions through co-culture with immune and stromal cells and novel extracellular matrix complexes.
Introduction
Over the past several years, the incidence of esophageal cancer has increased worldwide [1], underscoring the need for improved understanding of the pathogenesis of this disease. Animal models of esophageal cancer have been developed and provide insights into genetic and environmental contributions to malignant transformation, however the application of these findings to human disease is somewhat limited by interspecies differences in foregut anatomy and histologic characteristics [2]. 3D organoids derived from human cell lines or patient samples provide a novel and unique platform to model esophageal development, homeostatic regenerative differentiation, and benign and malignant esophageal diseases [3]. The use of 3D organoid technology in esophageal cancer has exciting implications for the development of new therapies for this disease. In this review, we highlight the use of esophageal 3D organoids to understand the biology of esophageal preneoplasia, cancers, and their cells of origin, and discuss the future translation of organoids for advanced personalized medicine approaches.
Structure and Function of the Esophagus in Humans and in Mice
The esophagus serves as the anatomic conduit for passage of solid and liquid food from the pharynx to the stomach after mechanical chewing and early enzymatic breakdown within the oral cavity. The luminal surface is exposed to ingested substances prior to initiation of most processes of enzymatic digestion or detoxification, which may place the esophagus at increased risk for development of benign or malignant diseases that can result from disruption of the protective mucosal barrier, toxin-induced stress, and immune-mediated acute and chronic inflammation [4–6]. The human esophagus is comprised of four distinct layers: the inner (luminal-facing) mucosa, the underlying supportive submucosa, the muscularis propria, which consists of an inner circular muscle layer surrounded by an outer longitudinal muscle layer, and the adventitia, which is the thin outer layer of the esophagus [7]. The circular and longitudinal muscle layers support swallowing function through peristalsis. The esophageal mucosa itself is comprised of three layers: the lumen-facing stratified squamous epithelium, the underlying subepithelial lamina propria, and an outer thin muscle layer, the muscularis mucosa [8]. Tight junctions between surface epithelial cells and thick mucus on the epithelial surface create a barrier between luminal contents and the deeper, proliferative cells of the stratified epithelium to protect against exposure and damage triggered by ingested toxins or infectious agents [6]. The lamina propria and the submucosa are both connective tissue layers that harbor fibroblasts, vascular and lymphatic channels, immune cells, nerves, mucin-secreting esophageal glands in the upper esophagus, and esophageal cardiac glands in the distal esophagus near the esophagogastric junction, all embedded within the collagen-rich and elastic extracellular matrix [8].
Of note, there are important differences in the esophageal anatomy between rodents and humans, which affect the development and use of murine models for study of human esophageal diseases. Rodents lack mucin-producing glands in the esophagus, and are instead protected by a thick acellular keratinized layer that overlies the mucosal surface and faces the esophageal lumen. While the glandular stomach in rodents and humans bear histologic similarities, rodents possess a forestomach that extends 3/5 of the way into the anatomic stomach, which is lined with keratinized squamous epithelium, and functions as a site for storage of ingested food [2]. As such, the transition between squamous and columnar epithelial cells that marks the esophagogastric junction in humans is located at the junction of the forestomach and glandular stomach in rodents.
Much of our knowledge regarding development of the mammalian esophagus has been performed using mouse embryonic models. While these models have provided key insights into the molecular pathways involved in esophageal development and cell fate specification, their direct translation to humans has been difficult to establish, in part due to the anatomic differences described above. Nevertheless, the findings from murine developmental models are summarized below.
In the mammalian embryo, the esophagus develops from the dorsal aspect of the anterior foregut, undergoing elongation and separation from the airway, which arises from the ventral anterior foregut to create the lungs and tracheobronchial tree [9, 10]. From mouse models, it has been found that the luminal surface of the esophageal mucosa is initially lined by ciliated columnar epithelium, which is later replaced by stratified squamous epithelium in the prenatal esophagus [11–13]. Proliferative basal cells of the epithelial layer, also called keratinocytes, permit epithelial renewal that continues throughout postnatal life to maintain normal homeostasis and initiate repair following mucosal injury [10, 14, 15]. In mice, basal cells express markers such as cytokeratin KRT5 and transcription factors SOX2 and TP63 [10–12, 16]. Basal cells undergo post-mitotic terminal differentiation within the suprabasal cellular layers and ultimately desquamate into the lumen. It remains controversial whether proliferating basal cells represent a molecularly heterogeneous population in mice or whether there is a single homogeneous subset of basal cells that retain proliferative capacity [17–21]. Meanwhile, early studies have suggested that the human basal cell population may be heterogeneous [22]. Nevertheless, the homeostatic proliferation-differentiation gradient of the esophageal epithelium is disrupted in many esophageal pathologies and most notably in cancer, where aberrant proliferation and differentiation can then lead to submucosal invasion, locoregional extension, and ultimately distant metastasis.
Epidemiology of Esophageal Cancer
Esophageal cancer is the eighth most common malignancy and sixth most common cause of cancer deaths worldwide, affecting roughly 5.9 per 100,000 people and resulting in 473,000 new cases in 2017 [1]. Despite advances in trimodality therapy, including chemotherapy, radiotherapy, and surgery, and expanded candidacy for both resection and neoadjuvant and adjuvant protocols, five-year survival remains extremely poor at 20%, with most patients developing distant metastatic disease [1, 23]. Esophageal cancer consists of two primary histologic subtypes, esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) [24]. ESCC cells display a variable degree of squamous-cell differentiation, while EAC cells resemble intestinal mucus-producing glandular cells. Both subtypes more commonly affect males over the age of 50 [1, 23].
ESCC Risk Factors and Cell of Origin
ESCC represents the majority of all esophageal cancer worldwide and is associated with several environmental risk factors, including use of tobacco, alcohol, and nutritional deficiencies [1]. Intake of mate, hot temperature liquids, areca nut, and nitrogenous compounds have all been linked to ESCC development [25]. Additionally, there is a geographic predisposition for ESCC, with high incidence in East Asia (China, Mongolia, and Japan), Russia and Central Asia, and Iran [1]. It is unknown whether this is primarily due to shared diet or lifestyle-related risk factors, or whether this may reflect common genetic risk factors through their shared history along the Silk Road [1]. The interplay of genetics and environmental factors likely plays a significant role, as ESCC in Japanese patients has been associated with single nucleotide polymorphisms in enzymes of alcohol metabolism, including alcohol dehydrogenase 1B and aldehyde dehydrogenase 2 family members [26, 27]. Genome-wide association studies in Chinese populations have also identified mutations in phospholipase C and in a specific region of chromosome 20 associated with ESCC development [28–30]. A second area of high ESCC incidence is termed the “African esophageal cancer corridor,” including eastern, central, and southern sub-Saharan Africa (Kenya, Uganda, Tanzania, Malawi, Zambia, and South Africa), again suspected to reflect a combination genetic and lifestyle-associated factors [1]. Though many molecular alterations are shared between ESCC and other squamous cell carcinomas (SCCs), including head and neck and anogenital SCC [31], the role for the human papilloma virus as a risk factor for ESCC remains controversial [32–35].
Genetic lesions commonly found in ESCC tumors include several oncogenes and tumor suppressors involved in the regulation of cell survival, pro-growth mitogenic signaling, and proliferation/cell cycle, including PI3KCA, EGFR, SOX2, cyclin D1/CCND1, p16/CDKN2A, and p53/TP53 [31]. Additionally, tumor suppressor roles for Notch and p120-catenin have been implicated in ESCC pathogenesis through their regulation of squamous-cell differentiation, cell adhesion, and epithelial barrier function [31, 36, 37]. ESCC is also associated with rare genetic disorders including Tylosis (RHBDF2 mutation), chronic mucocutaneous candidiasis (STAT1 mutation), and Fanconi anemia (FANCA, FANCC, and FANCG mutations), which are associated with geographically diverse global familial clusters, including several in Western Europe [38–40].
Progression from normal epithelium to squamous dysplasia, a histologic precursor for ESCC, and further progression to invasive ESCC correlates with accumulation of DNA damage [36]. Mutations in genes responsible for Fanconi anemia are essential for DNA repair following damage induced by acetaldehyde, a primary metabolite of alcohol and a constituent of tobacco smoke. Interestingly, Fanconi anemia-associated SCC occurs in young patients despite lower levels of exposure to alcohol or tobacco [41]. Genomic amplifications of SOX2 and TP63, markers of esophageal basal cells, are frequently present in ESCC [42, 43]. Murine cell-lineage tracing experiments have suggested that KRT5-expressing esophageal basal cells might be the cell of origin for ESCC [44]. While KRT5 is expressed in the human fetal esophagus and in esophageal progenitor cells derived from human pluripotent stem cells (hPSCs) in vitro [45], analogous lineage tracing experiments in human tumor models are lacking, and definitive identification of the cell of origin in human ESCC remains elusive.
EAC Risk Factors and Cell of Origin
In North America, Western Europe, and Australia, EAC represents the predominant form of esophageal cancer, primarily affecting Caucasian males [24]. Interestingly, among black Americans, the incidence of ESCC far exceeds that of EAC [46, 47], a disparity that warrants further investigation, but has been hypothesized to be related, in part, to genetic mutations in NRF2-mediated stress response pathways [48]. EAC often arises in the setting of Barrett’s esophagus (BE), an intestinal metaplasia that can progress to different degrees of dysplasia ranging from low grade to high grade/carcinoma in situ [49]. BE occurs more frequently in the distal esophagus in the setting of chronic gastroesophageal reflux disease (GERD) and is histopathologically defined as the replacement of the normal squamous epithelium with an incompletely intestinalized columnar epithelium with mucin-filled goblet cells [50].
Common risk factors for BE and EAC include age, male sex, and Caucasian ethnicity, as well as longstanding GERD, high fat diet, and central obesity [24]. EAC shares some common genetic lesions with ESCC, including EGFR and cyclin D1 overexpression, and p53 and p16 inactivation [51]. However, differential mutational signatures between EAC and ESCC include ERBB2, cyclin E1, and SMAD4, as well as increased chromosomal instability in EAC [52]. Whole genome analysis has also identified frequent amplifications of ERBB2, VEGFA, GATA4, and GATA6, and DNA hypermethylation in EAC patient samples [51, 52]. While suppression of Notch signaling may facilitate mucin-producing goblet cell differentiation in BE [53], Notch activation has been associated with the progression from BE to EAC [54].
Definitive identification of the cell of origin of BE and EAC remains elusive. Proposed theories regarding the BE cell of origin include stem/progenitor cells residing at the squamo-columnar junction, residual embryonic stem cells, and transdifferentiation of esophageal keratinocytes or esophageal cardiac glands [55]. While putative esophageal and BE stem/progenitor cells have been identified and characterized in mice [53, 56–60], their relevance to human pathology remains unclear, and identification of the human BE cell of origin is a key outstanding question in the field. In mice, basal progenitor cells marked by p63, KRT5 and KRT7 expression can repopulate the transitional epithelium at the squamocolumnar junction of the forestomach and glandular stomach, and when these cells are induced to express CDX2, they develop into an intestinal-like epithelium with goblet cells resembling Barrett’s metaplasia [57]. While analogous lineage tracing has not been performed in human samples, CDX2 overexpression has been found to induce intestinal metaplasia in human cells derived from the squamocolumnar junction in organoid models [57]. Meanwhile, integrated analyses of DNA methylation and transcriptome and genome profiling have identified four distinct subtypes of BE and EAC in patient samples [61], suggesting the potential for heterogeneity in the cell of origin across subtypes. Moreover, >50% of patients with EAC have no endoscopic or pathologic evidence of concurrent intestinal metaplasia, with some evidence that these patients demonstrate reduced overall survival compared to patients with concurrent BE [62]. These findings raise the possibility that a subset of EAC may arise directly from the squamous epithelium, a hypothesis supported by the unique subtype of adenosquamous carcinoma, a rare form of heterogeneous esophageal cancer containing both EAC and ESCC cells [63].
Animal and Cell Culture Models of Esophageal Cancer
Animal models of ESCC and EAC have been developed through the combinatorial approach of engineered genetic predisposition coupled with exposure to known risk factors, such as nitrosamines for ESCC and bile acids for EAC. Oncogenes and tumor suppressor genes such as cyclin D1, EGFR, p53, and p120-catenin as well as inflammatory cytokines such as interleukin-1β have been targeted in the esophageal epithelium in mice to model esophageal preneoplasia, invasive cancer, and the tumor microenvironment [37, 60, 64–66]. ESCC and preneoplastic lesions have been induced in mice exposed to 4-nitroquinoline 1-oxide (4NQO) in drinking water for 16 weeks [67]. 4NQO induces DNA lesions mimicking those induced by tobacco smoke through formation of DNA adducts and promoting mutations in p53, with reported 93.9% exome similarity between 4NQO mouse models of oral cancer and human head and neck SCC signatures [68]. 4NQO-induced esophageal carcinogenesis is accelerated in the setting of genetically engineered loss of p53 [44] or overexpression of cyclin D1 [69].
The initial rodent model of EAC was developed through modeling reflux-associated esophagitis in rats, where bile reflux was induced through surgical gastrectomy and esophagoduodenostomy [70]. EAC develops roughly 40 weeks following this procedure. Due to their difference in size, this surgical anatomic reconstruction is technically difficult to perform in mice, and therefore limits the utility of this approach. Additionally, while both gastric acid and bile reflux are known contributors to development of EAC in humans, this surgical model does not allow for direct study of gastric acid-associated changes, which are most common in EAC patients. Development of the L2-IL1B genetic mouse model of BE, in which the pro-inflammatory cytokine interleukin-1β is overexpressed in the esophageal epithelium, was a significant breakthrough for the study of BE and EAC [60]. These mice develop BE-like intestinal metaplasia by 12–15 months and severe dysplasia or EAC by 20 months, which can be accelerated by exposure to ingested bile acids and other toxins [60].
Larger animal models of EAC have been developed in animals that share more anatomic similarity with the foregut of humans. Induction of bile reflux through surgical gastrectomy and esophagojejunostomy and external acid perfusion studies have been performed in dogs, with BE developing in 1–3 years, and EAC after approximately 5 years [2, 71–74]. Pigs have esophageal submucosal glands that bear similarity to those of humans, and while they have spontaneous reflux events, they do not display histologic evidence of esophagitis [75]. Nevertheless, porcine models have provided unique insights into the role of submucosal glands in mucosal healing and tissue repair following esophageal injury [76], and have demonstrated the capacity for porcine esophageal submucosal glands to generate both squamous epithelium and glandular columnar epithelium that resembles BE in 3D culture systems [77]. Interestingly, baboons have naturally occurring chronic and continuous reflux, but have not been reported to develop EAC [78]. While challenging to study, more extensive investigation into both surgical and endogenously occurring large animal models may provide unique insights into the pathobiology of this disease as well as native protective mechanisms against adenocarcinoma development.
Many human esophageal cancer cell lines have been established since the 1980s and continue to serve as valuable tools to investigate ESCC and EAC tumor biology. Additionally, telomerase-immortalized human esophageal epithelial cell lines (e.g. EPC2-hTERT) became available in the early 2000s [79]. These cell lines have provided insights into the processes of esophageal proliferation, differentiation, and malignant transformation [80–82]. They have been extensively characterized in monolayer culture as well as organotypic 3D culture (OTC), a platform wherein esophageal epithelium is reconstituted at the air-liquid interface on top of a type 1 collagen-based matrix containing fibroblasts (see [83] for an extensive review). OTC has been used to study the homeostatic proliferation-differentiation gradient of the stratified squamous epithelium of the esophagus, as well as progression of normal and preneoplastic conditions utilizing esophageal cell lines engineered to express esophageal cancer-relevant oncogenes or loss of tumor suppressor genes. Co-culturing cancer cell lines and cancer-associated fibroblasts, OTC can serve as a powerful tool to model the invasive tumor front to study esophageal cancer cell invasion [84–86]. Unlike esophageal 3D organoids, OTC requires propagation of epithelial cells and fibroblasts grown in monolayer culture prior to 3D reconstitution.
Additional models for study of ESCC and EAC include subcutaneous xenograft models using tumor cell lines or patient biopsies for patient-derived xenograft (PDX) in immunodeficient host animals [87, 88]. Use of PDX models for esophageal cancer has provided some insights into tumor growth and metastasis in vivo, and has served as a platform to test tumor sensitivity to chemotherapeutics in preclinical models [89, 90].
Esophageal 3D Organoid Culture System
Organoids are 3D culture systems that develop from seeding single epithelial cells into extracellular basement membrane matrix, of which the most commonly used is Matrigel (Corning, USA). Matrigel is a combination of laminin, type IV collagen, heparin sulfate proteoglycans, entactin, and other growth factors that is derived from the Engelbreth-Holm-Swarm mouse sarcoma. 3D culture systems possess several advantages for studying cancer biology relative to 2D cell culture systems, including recapitulation of the layered stratified structure and differentiation gradient of the normal esophageal epithelium, and the cellular heterogeneity of tumors. From the single cell state, growth and differentiation occur inwardly such that the most differentiated cells are present at the center of the organoid structures, and the more basal “stem-like” cells are at the surface. 3D organoids can be generated from established esophageal carcinoma cell lines, including ESCC cell lines (TE4, TE5, TE8, TE9, TE10, and TE11) [91] and EAC lines (OE19 and OE33) [3, 92], as well as primary murine esophageal tissues and human patient-derived samples (PDOs) [3]. Organoids can therefore be generated from mouse models of dysplasia, ESCC, and EAC, as well as normal human esophagus, patient-derived esophageal tumors, and metastatic lesions for study in vitro (Figure 1). Furthermore, primary cells maintained in 2D culture can be seeded in 3D culture to recapitulate differentiation and heterogeneity, and organoid-derived cells can be dissociated for use in xenograft models of tumor initiation and metastasis [3, 93, 94].
Figure 1. Pathway of Organoid Generation for Scientific or Preclinical Studies.
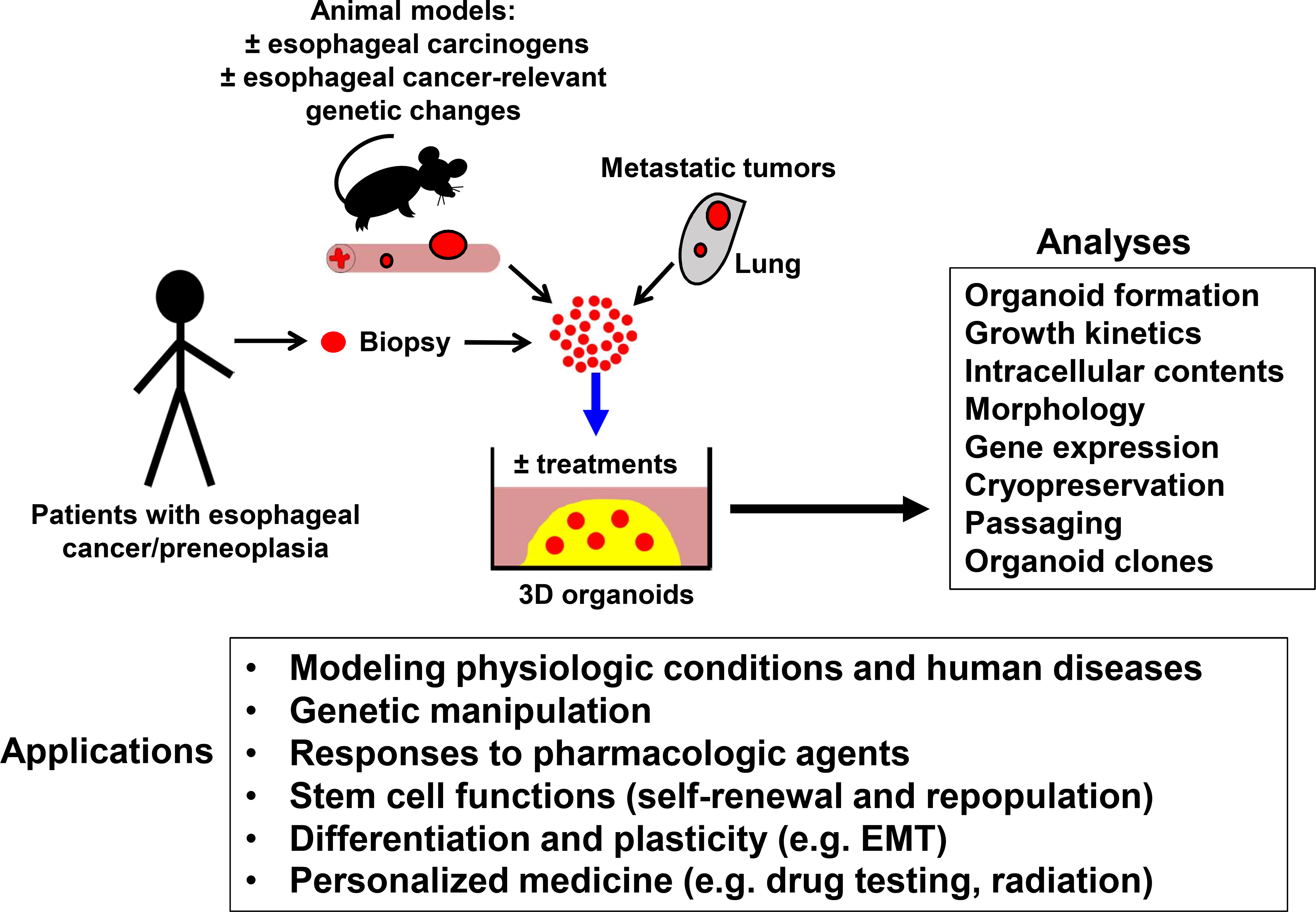
Organoids can be generated from patient biopsies or animal models of esophageal diseases, including normal or treated esophagus, primary esophageal cancer tissue, or metastases to lung or other organs. Esophageal epithelium or tumor tissues are dissociated into single cells and seeded in Matrigel. Organoids can be cultured in the presence of specific chemical treatments or subject to genetic manipulation in vitro, followed by cell-based analyses and in vitro or in vivo modeling of disease through culture-based or xenograft models.
Over the past five years, significant progress has been made in the use of 3D esophageal organoids for study of both ESCC and EAC, as well as esophageal development and epithelial differentiation using induced pluripotent stem cells in 3D organoid models. Therapy resistance and recurrence are attributed to intratumoral cancer cell heterogeneity, which in turn is influenced by genetic and environmental risk factors [95]. 3D organoids therefore represent a more faithful tumor model for modeling both disease progression and therapy resistance than traditional 2D culture systems by recapitulating epithelial architecture, interactions with the extracellular matrix (ECM), and tumor cell heterogeneity [93]. These novel approaches utilizing patient-derived tissues pave the way for personalized medicine protocols and targeted therapeutics. 3D organoids can be used to provide unique insights into tumor initiation, through quantification of organoid formation rate in vitro or PDX models in vivo, tumor proliferation and growth kinetics, cell morphology, and gene expression over a heterogeneous but clonal organoid population (Table 1) [96]. Applications of 3D organoids, which will be discussed more extensively below, include modeling of benign and malignant esophageal diseases, stem cell functions for tissue renewal, basal cell differentiation and epithelial plasticity, and personalized medicine approaches.
Table 1.
Use of 3D Organoids to Model Esophageal Cancer Development and Progression
| Advantages |
|---|
|
|
| Recapitulation of histology of primary tissue of origin (normal, dysplastic, cancer) |
| Recapitulation of genetic and epigenetic signature of original tissue/tumor |
| Recapitulation of tumor clonal heterogeneity |
| Evaluation of individual clones and tumor initiating cells |
| Long-term culture and passaging Biobanking of patient samples across institutions |
| Conversion to 2D culture High-throughput drug screening |
| High-throughput gene editing Personalized medicine using PDOs |
|
|
| Limitations |
|
|
| Lack diverse cell types and microenvironment of original tumors |
| Unknown genetic and epigenetic stability over multiple passages |
| Clonal drift over multiple passages |
3D Organoids for Study of Esophageal Development
Enhanced understanding of esophageal development and the pathways underlying keratinocyte differentiation may provide insights into the identity of a proliferative esophageal “cancer stem cell,” and may be exploited to induce differentiation of dysplastic squamous or BE lesions to suppress their progression to malignancy. 3D organoids provide a novel platform to study a wide spectrum of disorders of epithelial differentiation, including developmental defects such as esophageal atresia, basal cell hyperplasia underlying diseases such as eosinophilic esophagitis, and epithelial injury from toxic exposures such as alcohol.
Modeling Epithelial Differentiation and Basal Cell Hyperplasia Using 3D Organoids
3D organoid models have been used to study esophageal development, epithelial differentiation, and reactive inflammatory responses that are responsible both for benign esophageal pathologies, such as eosinophilic esophagitis, and for the predisposing injury and inflammation that enhance susceptibility for ESCC and EAC. Examination of the immortalized human esophageal epithelial cell line EPC2-hTERT grown in Keratinocyte Serum Free Medium revealed that these cells remain in a proliferative basaloid state when grown in 2D monolayer culture conditions, but recapitulate the differentiation gradient of the native esophageal mucosal epithelium when grown in 3D organoid structures [96, 97]. Activation of the Notch signaling pathway is required to promote squamous cell differentiation [82]. We have shown that inhibition of Notch signaling both pharmacologically and genetically results in a thick outer basaloid cell layer and hyperkeratosis of the organoid core [97]. Furthermore, Notch inhibition did not affect cell proliferation in organoid models, indicating that apparent basal cell hyperplasia may actually be the result of impaired differentiation. This finding is consistent with the histology of PDOs derived from patients with eosinophilic esophagitis [96], a condition characterized by basal cell hyperplasia and eosinophils within the epithelial layer of the esophagus. Using EPC2-hTERT organoids, we were able to reproduce basaloid cell expansion in response to inhibition of Notch signaling or treatment with pro-inflammatory cytokines known to play a role in development of eosinophilic esophagitis, including TNFα and interleukins 4, 5, and 13 [96]. These effects were also seen in organoids from patient-derived normal tissue biopsies, with decreased differentiation and expression of the differentiation marker Involucrin in response to cytokine treatment.
In addition to defining the role for Notch signaling in normal stratified squamous epithelial differentiation and dysregulated Notch signaling in conditions of basal cell hyperplasia, 3D organoids have been instrumental in elucidating mechanisms of epithelial stress responses in the setting of toxic and carcinogenic exposures, such as alcohol. Ethanol exposure decreased proliferation, organoid growth, and cell viability in EPC2-hTERT organoids, and resulted in mitochondrial dysfunction [98]. Using both 3D and 2D culture systems, these findings correlated with decreased intracellular ATP levels, decreased oxidative metabolism, and inhibition of MTORC1 signaling in conjunction with an upregulation of autophagy, consistent with previous findings linking autophagy to cell survival in esophageal epithelial cells [91]. Ongoing work to assess the effects of ethanol on ESCC development and progression is currently underway.
Esophageal organoid units (EOUs) have been derived by seeding dissociated whole murine esophagus in Matrigel, resulting in epithelial organoids co-cultured with neuromuscular cells and providing a complementary model to mimic whole-organ and functional esophageal development [99–101]. EOUs demonstrate an epithelial differentiation gradient from basaloid cells to mature squamous cells and exhibit spontaneous peristalsis, allowing for further insights into coordinated esophageal development and homeostasis [99]. The canonical bone morphogenetic protein (BMP) signaling pathway is essential for regulating dorsal-ventral patterning of the foregut into the dorsal esophagus and the ventral trachea, and is modulated by extracellular antagonists, including noggin, chordin, and follistatin, that bind BMP receptors to inhibit signaling [102]. Comparison of EOUs and tracheal organoid units from murine donors have identified a critical role for noggin in specifying esophageal rather than respiratory cell fates [103]. This finding correlates with enhanced DNA methylation at the noggin promoter and upstream regulatory sites in patients with esophageal atresia with or without tracheoesophageal fistula relative to control patient samples [103].
Human PSC-Derived Esophageal Organoids to Model Lineage Specification and Differentiation
Understanding the developmental pathways underlying esophageal epithelial specification and squamous cell differentiation have been greatly enhanced by the development of protocols for induction of human pluripotent stem cells (hPSCs) into esophageal cells in 3D cultures. The induction of hPSCs into esophageal progenitor cells (EPCs), then subsequently into fetal-like esophageal tissue, was described using organoid models in 2018 [45, 104]. This discovery overcame a critical barrier to developmental studies of the esophagus due to interspecies anatomic and histologic differences between the murine and human esophagus. The hPSC line RUES2 was initially differentiated into endoderm through exposure to several growth factors, including Activin A, BMP4, FGF2, and the ROCK inhibitor Y-27632, and subsequently differentiated into anterior foregut, followed by EPCs, through inhibition of canonical developmental signaling pathways, including BMP, TGFβ, and WNT [45]. EPCs were marked by expression of fetal esophageal markers, including Keratins 5 and 7. The hPSC-derived EPCs were then selected by sorting for cells expressing the epithelial-specific marker EPCAM and integrin β4, and were able to reform 3D organoids when seeded as single cells in culture. Basal esophageal cell differentiation was then induced from purified EPCs. Human PSC-derived organoids and EPCs are further induced towards esophageal differentiation in response to BMP and Notch activation, leading to increased expression of differentiation markers including Keratin 13 and Involucrin [45], demonstrating the utility of this hPSC-derived organoid protocol to model native human esophageal differentiation. Study of hPSC-derived esophageal organoids has led to new molecular insights, including identification of a role for the YAP protein in proliferation and differentiation of esophageal progenitor cells [105]. Inhibition of YAP with verteporfin reduced the number of p63-positive proliferative EPCs and reduced squamous stratification with reduced Keratin 13 expression [105]. Knockdown of YAP in EPCs resulted in decreased number and size of resulting organoids [105].
Using a similar but independently-developed differentiation protocol through sequential temporal manipulation of the BMP, WNT, and retinoic acid signaling pathways, hPSCs were induced to esophageal progenitor cells in 3D cultures, and were subsequently used to determine the role of SOX2 in esophageal development [104]. Early loss of SOX2 results in esophageal atresia in both humans and mouse models [11, 106]. Using CRISPR-mediated gene editing in the iPSC-derived organoid system, SOX2 knockdown in human dorsal anterior foregut cultures resulted in ectopic expression of NKX2–1, a marker of early respiratory rather than esophageal differentiation [104]. Transcriptome analysis of CRISPRi-SOX2 treated or control iPSC-derived organoids was able to identify differential roles for SOX2 and BMP signaling in dorsal versus anterior foregut specification [104]. These approaches can be applied to both human and murine iPSCs, and comparative analysis in the setting of targeted genetic manipulation has broad application for identification of novel developmental pathways, which may be exploited in the setting of dysplasia to induce differentiation and prevent carcinogenesis. Additionally, enhanced study of PSC differentiation may provide insights into basal cell hyperplasia and a putative “cancer stem cell” for ESCC and EAC.
Insights into the Cell of Origin for Esophageal Cancer
The utilization of hPSC-derived esophageal differentiation models may provide novel insights into the cell of origin for esophageal cancer. As previously discussed, hypotheses regarding the origin of BE cells include proximal migration of columnar epithelial cells from the esophagogastric junction or adjacent gastric mucosa, transdifferentiation of mature squamous epithelial cells, transcommitment of squamous progenitor cells from the basal cell or submucosal layers, or habitation by a circulating initiating cell from the blood [107]. Inhibition of Notch signaling combined with induction of MYC and CDX1 expression can convert EPC2-hTERT cells from a squamous histology to a more glandular histology, suggesting a process of transdifferentiation [53], and analysis of cells at the transition zone between regions of BE and normal esophageal mucosa have identified both ultrastructural and genetic elements of both squamous cell and intestinal cell identity [108]. Additionally, differentiated esophageal squamous cells have been shown to downregulate squamous markers such as p63 and upregulate intestinal markers including CDX1, CDX2, FOXA2, and SOX9 after exposure to acid or bile salts in vitro [109]. However, genomic lineage tracing based on single cell mutational analysis has identified shared mutations in p53 and p16 in Barrett’s regions and submucosal glands [110], supporting the hypothesis of a proximal or submucosal migratory cell of origin. This hypothesis was previously supported by animal studies of surgically-induced reflux in large animal models [73] and studies of porcine submucosal glands [77]. Transcommitment of hPSC-derived EPCs would imply the presence of an esophageal “stem cell” that can give rise to differentiated cells of multiple phenotypes. The regenerative basal cell population of the esophageal epithelium maintains cellular heterogeneity, with SOX2, WNT, and BMP signals promoting self-renewal [19]. Long-lived Keratin 15-expressing cells (Krt15+) that retain the ability to self-renew, proliferate, and differentiate, and are transcriptionally distinct from Keratin 15-negative basal cells, have been identified in the adult and fetal murine esophagus [111]. These Krt15+ cells in the adult murine esophagus demonstrate radioresistance and the ability to regenerate esophageal epithelium after mucosal injury [111]. The induction of hPSCs to esophageal cell fates, coupled with the ease of genetic manipulation of hPSC-derived organoid cultures, may provide key insights into the potential for transdifferentiation of squamous epithelial cells and transcommitment of basal cells and EPCs, as well as definitive identification of the human cell of origin for BE and EAC.
Development of Patient-Derived Esophageal Organoids for Study of Esophageal Cancer
PDOs from ESCC and Squamous Cell Dysplasia
Protocols for successful generation of PDOs from ESCC and normal esophageal mucosa (Figure 2) have been developed by our lab and others. We have demonstrated an ESCC organoid success rate of 60 percent, with 10% of ESCC PDOs able to be serially passaged over 5 times [3]. This protocol uses an advanced DMEM/F-12 basal medium, with supplemental nutrients and growth factors, including Noggin-R-Spondin conditioned medium, and 50 ng/ml EGF. This medium, while supporting ESCC PDO growth, has had less success supporting PDO growth from normal non-transformed esophageal epithelium. It has been an ongoing challenge of the field to identify a single medium that can support normal, ESCC, and EAC PDO growth, and current protocols use variable medium components based on the histology of the patient donor sample (Table 2). We have recently developed successful protocols for primary organoid generation from dysplastic head and neck and esophageal squamous cell samples to allow the study of squamous preneoplastic lesions in organoid culture.
Figure 2. Patient-Derived Organoids to Model Esophageal Cancer.
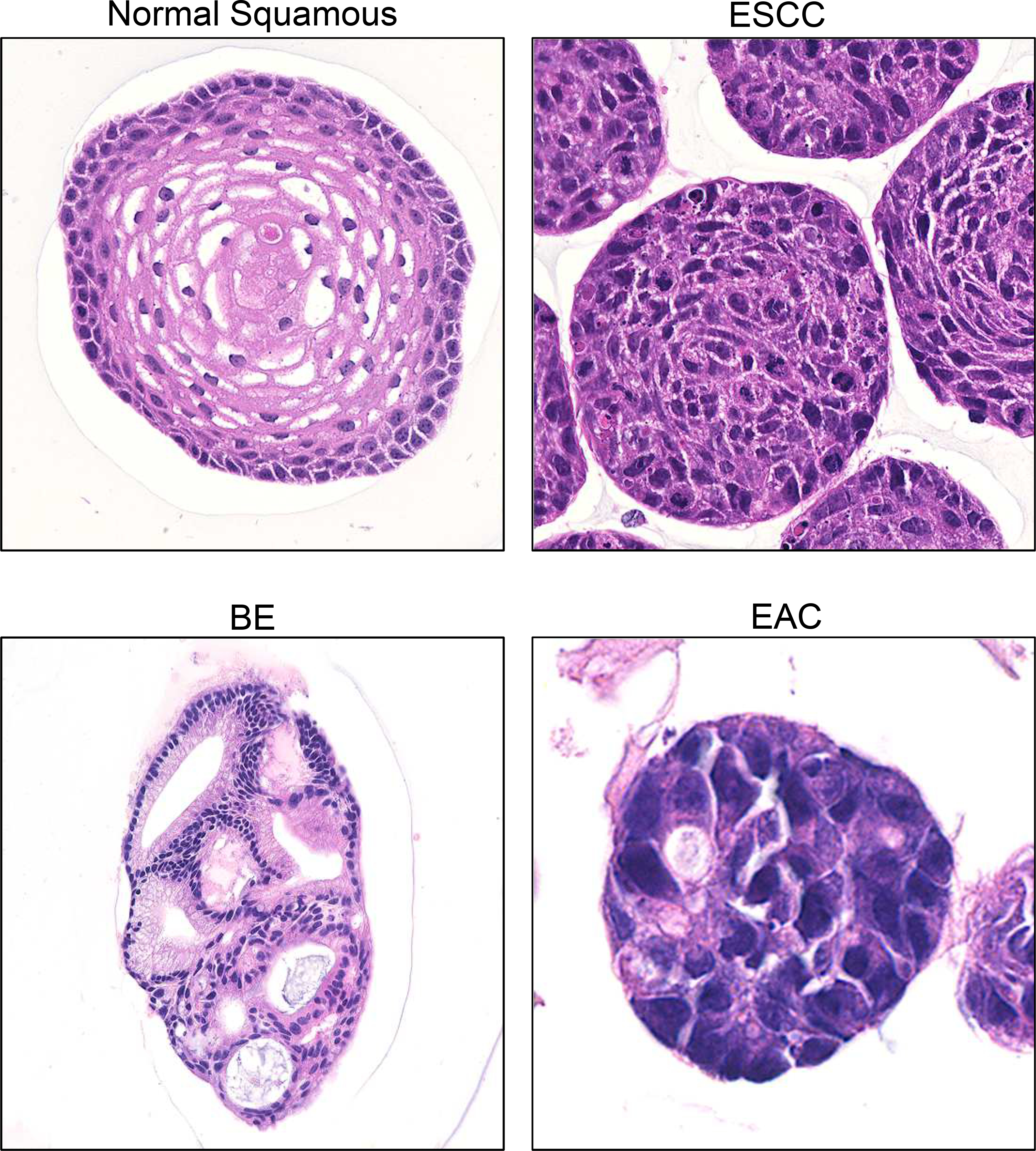
Hematoxylin and Eosin stained sections of fixed 3D organoids derived from the non-transformed human esophageal epithelial EPC2-hTERT cell line, and from ESCC, BE, and EAC PDO lines derived from human patient biopsies.
Table 2.
Comparison of Media Components for Normal Squamous Esophageal, ESCC, and EAC PDO Generation as Published.
| Media Component | Kasagi et al. | Kijima et al. | Karakasheva et al. | Karakasheva et al. |
|---|---|---|---|---|
| N | N/ESCC | ESCC | EAC | |
|
| ||||
| Basal media | KSFM1 | aDMEM2 | aDMEM2 | aDMEM2 |
| Glutamax | U | (+) | (+) | (+) |
| HEPES | U | (+) | (+) | (+) |
| B27 | U | (+) | (+) | (+) |
| N2 | U | (+) | (+) | (+) |
| NAC | U | (+) | (+) | (+) |
| CM 3 | U | RN4 | RN4 | WRN5 |
| Wnt3a | U | (+) | (−) | (−) |
| EGF | 1 ng/ml | 50 ng/ml | 50 ng/ml | 250 ng/ml |
| Nicotinamide | U | (+) | (−) | (+) |
| A83–01 | U | (+) | (−) | (+) |
| SB202190 | U | (+) | (−) | (+) |
| Gastrin | U | (+) | (−) | (+) |
| FGF10 | U | (−) | (−) | (+) |
| CHIR99021 | U | (−) | (−) | (+) |
| CaCl 2 | 0.6 mM | 1 mM | 1 mM | 1 mM |
| Y27632 | (+) | (+) | (+) | (+) |
|
| ||||
| Success rate | ||||
|
| ||||
| Normal | 100% | 66.7% | nd | nd |
| Cancer | nd | 71.4% | 60% | 80% |
Keratinocyte Serum-Free Medium,
advanced Dulbecco’s Modified Eagle Medium/F-12,
Conditioned Medium,
R-Spondin/Noggin,
Wnt/R-Spondin/Noggin. (N) normal; (U) unknown; (nd) not determined. All media are supplemented with antibiotics.
Comparing paired organoids generated from ESCC tumors and adjacent normal mucosa obtained from endoscopic biopsies, we have shown that resistance to 5-fluorouracil in ESCC PDOs is accompanied by enhanced CD44 expression and increased autophagy, identifying these as markers of survival following chemotherapy treatment [91]. Furthermore, CD44-high cells sorted from both PDO cultures and PDO-derived PDX tumor models demonstrated that these cells exhibit higher levels of autophagy induction and represent a resistant population in response to both chemotherapeutic agents [91] and alcohol-induced stress (ongoing work). PDOs have also been generated from oral cavity tumors, as well as pharynx, larynx, salivary gland, nasal cavity, and cervical regions with 60 percent success, along with successful serial passage, cryopreservation, and recovery [112]. These head and neck squamous cell carcinoma (HNSCC) organoids also retained the genetic and molecular characteristics of the original in situ tumors, recapitulating the stratified epithelial structure as evidenced by expression patterns of the basal cell marker TP63 and the differentiation marker KRT13, as well as mutations in TP53, PIK3CA, KRAS, BRAF, CDKN2A, FAT1, and PDGFRA [112]. Additionally, normal oral organoids were susceptible to infection by HPV and herpes simplex virus [112], demonstrating their potential use for study of viral-associated oncogenicity.
PDOs from EAC and BE
Development of 3D organoids from Barrett’s epithelium was first reported in 2011, where fragments of BE derived from endoscopic patient biopsies were partially dissociated to maintain Barrett’s crypts and seeded in Matrigel, with successful passage for over 3 months [113]. In the absence of fibroblast growth factor 10 (FGF10), BE organoids formed cystic structures reminiscent of senescent intestinal-derived organoids and stopped proliferating, while differentiation could be induced by inhibition of Notch signaling [113]. Further work to establish organoid cultures from EAC patient samples has shown that patient-derived organoids recapitulate the histology of the patient tumors from which they were derived, including apical/basal polarity and p53 status [93]. Furthermore, PDOs displayed concordance of driver somatic mutational events and whole genome mutational signatures, including mutations in p53, PIK3CA, and CDKN2A [93, 114]. Additionally, EAC PDO cultures retained the clonal heterogeneity that is a hallmark of EAC and contributes to chemotherapy resistance in the clinical setting and poor patient survival [93, 115]. Culture of EAC PDOs over multiple passages demonstrated clonal drift of the dominant clonal populations within the organoid culture [93, 114], illustrating a key advantage of using organoid culture over 2D culture systems for modeling esophageal cancer development, progression, and responses to treatment, including antineoplastic resistance mechanisms. We have established effective protocols to faithfully create organoids from EAC PDOs with 80 percent efficiency, as well as for passaging, cryopreservation, image-based monitoring of organoid size and proliferation, as well as harvesting for histology, immunoblotting, immunofluorescence and immunohistochemistry, bulk RNA seq, and flow cytometry [3]. EAC and BE PDOs (Figure 2) can be successfully grown with the same media components (Table 2), facilitating comparative analyses between dysplastic, preneoplastic, and malignant tissue samples. EAC PDOs were maintained for over 15 successive passages and successfully generated patient-derived xenograft mouse models. Ongoing work of our lab and others uses BE and EAC organoids for mutational profiling to better understand disease progression from dysplasia to cancer to clonal selection, as well as responses to novel therapeutics.
PDOs for Development of Personalized Medicine Approaches
Due to the faithful recapitulation of histologic and genomic characteristics of primary tumors, ESCC and EAC PDOs exhibit significant promise as avatars for testing tumor responses to treatment [116], including standard-of-care chemotherapies, radiation, molecularly targeted agents, and novel therapeutics. Similarly, the clonal heterogeneity of organoid cultures allows identification of uniquely resistant or susceptible clonal populations, which may provide key insights into residual disease and mechanisms of recurrence following induction chemoradiation and complete surgical resection.
PDOs for Personalized Therapy in ESCC
ESCC organoids have provided early insights into mechanisms of cancer cell survival and resistance to chemotherapy. High levels of CD44 expression and cellular autophagy are associated with resistance to 5-fluoruracil treatment in ESCC PDOs [91]. Exposure of HNSCC PDOs to Nutlin-3 demonstrated differential sensitivities based on p53 mutation status [117], providing evidence to support the use of PDOs for targeted drug sensitivity screening based on genomic mutational analysis. Differential responses of patient-derived HNSCC organoids to cisplatin, carboplatin, and the anti-EGFR antibody cetuximab, as well as ionizing radiation were observed, which correlated with clinical response to administered radiation therapy in seven of 31 cases [117]. HNSCC PDOs were also used to screen for sensitivity to novel therapeutics, including the radiosensitizing effect of Second Mitochondria-derived Activator of Caspase (SMAC), and evaluation of the mechanism of radiation-induced death by treatment with inhibitors of apoptosis or necrosis [112]. In vitro multi-drug screening and combinatorial regimens with both radiation and chemotherapeutic agents using ESCC and HNSCC PDOs have identified novel synergistic effects in a preclinical setting. Furthermore, novel therapies based on mutational signatures have been tested, such as the PIK3CA inhibitor alpelisib, the BRAF inhibitor vemurafenib, the FGFR inhibitor AZD4547, the PARP inhibitor niraparib, and the mTOR inhibitor everolimus, to determine whether these therapies may play a role in targeted treatment strategies in preclinical models [117]. Interestingly, PDOs with PIK3CA mutations did not uniformly display increased sensitivity to alpelisib, indicating the need for further investigations into the predictive potential for PDOs for personalized precision therapeutics based on targeted mutation analysis. Protocols for HNSCC and ESCC organoid generation and use for semi-automated drug screening and radiation treatment within a three-month timeframe have been published by several groups [3, 117].
PDOs for Personalized Medicine in EAC
Similar use of PDOs to test drug sensitivity in the preclinical setting has been demonstrated for EAC. EAC PDOs have been shown to retain stable drug sensitivity profiles for up to one year in serial cultures, and analogous to ESCC drug screens, similar sensitivities to drug treatments have been found for single cells seeded in Matrigel or pre-formed multicellular organoids. In the study by Li et al., nine EAC PDOs were tested with 24 compounds, including the MDM2 inhibitor Nutlin-3a, the MEK1/2 inhibitor trametinib, the EGFR inhibitor afatinib, and standard-of-care chemotherapeutic agents including 5-fluorouracil, epirubicin, and cisplatin [93]. EAC PDOs displayed sensitivities that could be grouped by their mutational subtype [93, 114]. Furthermore, most EAC PDOs tested displayed resistance to standard-of-care therapies in concordance with the donor patients’ poor clinical responses to therapy. Further studies have shown that PDOs derived from EAC patient biopsies treated with individual components based on CROSS (cisplatin and paclitaxel) or FLOT (5-fluorouracil, cisplatin and paclitaxel or epirubicin) regimens displayed sensitivities that correlated with patients’ clinical responses to neoadjuvant treatments [118]. To evaluate the use of PDOs for development of targeted personalized treatment strategies, one PDO line derived from a tumor displaying ERBB2 amplification was treated with the HER2-specific inhibitor mubritinib, resulting in moderate decrease in cell viability and organoid growth [118]. Interestingly, however, while three tested PDOs lacking HER2 amplification did not show sensitivity to mubritinib, one PDO lacking HER2 amplification also displayed sensitivity to mubritinib treatment, a finding that was attributed to HER2-independent effects of mubritinib on electron transport chain activity [118]. Thus, while PDOs represent a promising new tool for predictive and tailored treatment protocols for EAC, further investigations into the clinical utility and time required for completion of screening protocols are needed, as significant delays in standard-of-care therapy initiation in anticipation of tailored personalized strategies bear the significant potential to adversely affect progression of disease and patient survival.
Future perspectives
PDOs for High-Throughput Genomic Screens and Identification of Novel Targets
Given the ease of patient-specific in vitro drug testing using PDOs, as well as the cumulative data available through development of PDO biobanks, PDOs represent an ideal platform in which to evaluate sensitivities of EAC and ESCC to new therapies, including chemotherapeutic agents currently approved for use in other GI malignancies, molecularly targeted therapies based on PDO mutational analyses, optimal radiation dosage and delivery, and proton beam therapy. Additionally, esophageal cancer PDOs provide a promising platform for identification of novel genetic alterations that affect tumor survival, invasion, and treatment resistance using high-throughput gene editing strategies. CRISPR-Cas9 technology has allowed efficient knock-in or knock-down of targeted genes, and has been used successfully in GI epithelial organoids by several groups using liposomal transfection, electroporation, or viral vector-mediated transduction, followed by selection and expansion of single organoid clones [119]. This system has been exploited in human colorectal cancer organoids to study the specific role of oncogenes and driver mutations, and human intestinal organoids to determine the effects of sequential accumulation of directed mutations on carcinogenesis [120, 121]. Similar approaches can be applied to non-transformed esophageal epithelial organoids, BE and dysplastic squamous organoids, ESCC and EAC organoids, and hPSC-derived EPCs to gain novel insights into the progression from dysplasia to malignancy that underlies both major types of esophageal cancer [104, 122].
High-throughput genomic screens have identified novel drivers of tumor progression, suppression, and drug resistance in traditional 2D cultures [123]. CRISPR screens using single guide RNA (sgRNA) libraries applied to PDOs have the potential to identify novel pathways involved in drug resistance and to enhance personalized medicine approaches, particularly in the setting of highly resistant or metastatic disease [123]. However, CRISPR screens in 3D cultures pose unique challenges relative to 2D monolayer culture. 3D PDO cultures may not generate sufficient cell numbers to efficiently support traditional pooled genome-wide screening strategies [123, 124]. There are additional difficulties in predicting sgRNA efficacy in 3D culture systems, and false positives due to the integration of multiple sgRNAs into single organoids [124, 125]. Recently, however, significant progress has been made in screening strategies for gastrointestinal organoids to increase the efficiency of genome editing in PDOs. These strategies include: identifying the most effective sgRNAs by prescreening the CRISPR library in cells grown in 2D monolayer culture; decreasing the number of PDOs required by reducing sgRNA library size; and minimizing the number of false positives by capturing and removing passenger sgRNAs that co-integrate with functional sgRNAs in single organoids [124, 125]. Advances in single cell and nuclear sequencing protocols applied to 3D organoids [126] derived from dysplastic and preneoplastic patient samples will also allow further investigation into the cell of origin of ESCC and EAC by enabling lineage tracing experiments in vitro and in PDX models of tumor initiation.
Organoid and Immune Cell Co-Culture for Study of Tumor-Microenvironment Interactions
With the advent of immunotherapy and recent early data regarding the efficacy of immune checkpoint inhibitors in prolonging survival in patients with metastatic gastric and gastroesophageal cancers, the critical role of the immune landscape in the progression of both EAC and ESCC is increasingly apparent. Recent results of the CheckMate 649 Phase III clinical trial of programmed cell death 1 (PD-1) inhibitor nivolumab in combination with standard-of-care chemotherapy regimens in patients with unresectable gastric or esophageal adenocarcinoma [127] were recently presented at the European Society for Medical Oncology (ESMO) 2020 meeting. These results demonstrated significant improvement in survival in patients treated with the addition of nivolumab to chemotherapy versus chemotherapy alone, a finding that maintained significance across both randomly assigned patients and subgroup analyses of patients with elevated PD-L1 combined positive score. Early results of the Phase III KEYNOTE-590 study [128], also presented at ESMO 2020, assessed the effects of addition of pembrolizumab to a standard-of-care 5-fluorouracil and cisplatin regimen in patients with unresectable or metastatic EAC or ESCC. Overall survival was 12.4 months in the pembrolizumab group vs 9.8 months in the group treated with chemotherapy alone (ESMO 2020). The success of these and several other recent and ongoing trials has generated enthusiasm for the use of immune checkpoint inhibitors in the neoadjuvant setting for locally advanced esophageal cancers, with clinical trials currently underway at several institutions.
Immune profiling of resected ESCC surgical specimens has identified exhausted CD4 and CD8 T cells and natural killer (NK) cells as major proliferative components of the tumor microenvironment and suppressive crosstalk between macrophages and regulatory T cells, identifying mechanisms for tumor immune evasion through enrichment of immune-suppressive populations [129]. Additionally, loss of p53, which occurs early in the development of ESCC and EAC, has been shown to promote immune evasion through decreased expression of cytokines, decreased expression of major histocompatibility class I proteins, and recruitment of suppressive regulatory T cells [130], providing an additional mechanism by which esophageal cancers may bypass host immune surveillance. These studies underscore the need for tumor models with intact immunity to study mechanisms of host immune surveillance and tumor immune evasion, which continue to be a limitation of xenograft models that necessitate immunocompromised hosts. Co-culture of immune cell populations with esophageal epithelial cells in 3D culture systems using organotypic air-liquid interface culture has been used to model esophagitis, inducing a reactive proliferative response in the setting of oxidative stress and enhanced DNA damage [15, 131, 132]. Using analogous protocols in 3D organoid cultures derived from EAC and ESCC cell lines as well as PDOs from tissue biopsies in the presence of cultured T cells, macrophages, or patient-derived tumor-infiltrating lymphocytes will provide insights into intrinsic immune responses and their contribution to tumor progression, and has the potential to inform personalized treatment approaches.
Cancer associated fibroblasts (CAFs) within the tumor microenvironment have also been shown to play an important role in tumor development and progression in multiple GI malignancies through secretion of tumor-promoting cytokines. We have shown that interleukin 6 and chemokine C-C motif ligand 5 (CCL5) secreted by CAFs promote proliferation of both ESCC and EAC in 2D and 3D cultures through activation of the MEK/ERK and STAT3 signaling pathways [92]. CAFs also secrete plasminogen activator inhibitor-1 (PAI-1) that promotes migration and invasion of ESCC cells and macrophages through phosphorylation and activation of the Akt and Erk1/2 pathways [133]. Toll-like receptor-4 (TLR-4) expression by CAFs correlates with progression from reflux-associated esophagitis to BE to EAC through increased expression of pro-inflammatory and anti-apoptotic cyclo-oxygenase 2 (COX-2) [134–136]. Furthermore, increased intratumoral CAFs in ESCC surgical resection specimens, as identified by antifibroblast activation protein, was associated with decreased disease-free survival and overall survival, and increased lymph node metastases [137]. The further development of 3D co-culture models will better elucidate the mechanisms of crosstalk between CAFs and epithelial tumor cells that promote tumor growth and invasion in both EAC and ESCC.
Alternative Basement Membrane Composition and Use of Synthetic Hydrogels
While most ESCC and EAC organoid studies, as well as hPSC-derived esophageal cultures, to date have been performed by generating organoids in Matrigel basement membrane matrix, alternative extracellular matrix (ECM)-mimicking platforms, including synthetic hydrogels, can potentially allow tailored and modifiable ECM properties to study tumor growth and invasion, and esophageal development and differentiation. Indeed, synthetic hydrogels have already been used for the culture of tumor organoids, and to study the individual contributions of matrix properties to tumor progression and metastasis [138, 139]. Because Matrigel is derived from mouse sarcoma cells, its fractional composition of specific ECM proteins and concentration of embedded growth factors can vary significantly between lots, and its inherently limited by the inability to decouple biochemical and mechanical properties [140, 141]. Fully-defined synthetic hydrogels with modifiable mechanical properties and adhesive ligand types can provide a more controlled microenvironment for organoid culture models [142]. Synthetic hydrogels have also been shown to support growth of PSC-derived intestinal organoids, and can be safely injected in animal models, where they can facilitate organoid engraftment and repopulation of colonic epithelium in injury models [143]. Furthermore, because Matrigel is produced by mouse sarcoma cells, it has limited potential to be used in experimental therapeutics for treatment of human disease; thus, synthetic hydrogels provide an attractive alternative ECM for clinical applications [144]. Interestingly, oral treatment with porcine-derived hydrogel in combination with omeprazole has been reported to reduce esophagitis and decrease Barrett’s metaplasia in a canine model of BE [145]. Taken together, these studies suggest that combination of organoids with hydrogel ECMs may enable novel therapeutic strategies for cellular repopulation and tissue regeneration in the setting of esophageal inflammation and injury, as well as novel mechanisms for topical mucosal or injected submucosal drug delivery for treatment of dysplasia and early-stage cancers.
Conclusion
The generation of 3D organoids for study of ESCC and EAC tumor development and progression, normal esophageal epithelial homeostasis, inflammation and reactive changes, and esophageal development using hPSCs paves the way for major advances in our understanding of both benign and malignant esophageal diseases. The relative ease of genetic manipulation and the combination of CRISPR/Cas9 and 3D organoid technologies will allow for unique understanding of the progression from esophagitis, both reflux and toxin-induced, to BE-associated and squamous dysplasia, to the development of EAC and ESCC. Co-culture models of epithelial cells along with immune cells and stromal cells will provide novel insights into tumor-promoting interactions derived from the microenvironment, and elucidate crosstalk between tumor cells and stromal cells that may underlie immune evasion and cancer resistance. The development of synthetic hydrogels will allow for further precision experimentation through controlled modification of ECM composition and physicochemical properties, as well as introducing a novel mechanism for both cell delivery-based therapies and localized application of chemotherapeutics or radiosensitizing agents. By combining these approaches, PDOs can be used to tailor personalized therapeutic strategies for patients with ESCC or EAC for use in the neoadjuvant, adjuvant, or metastatic settings. Collaboration between clinicians, cancer biologists, computational geneticists, and bioengineers will be critically important to maximally leverage these powerful tools to advance the treatment and improve survival for patients with this aggressive disease.
Highlights.
3D organoid culture systems can model the esophageal stratified squamous epithelium
Esophageal organoid creation is possible from normal, dysplastic and cancer tissues
Induced pluripotent stem cells model esophageal development and differentiation
Patient-derived organoids provide a platform for screens and personalized medicine
Acknowledgments
We thank Dr. Andres Klein-Szanto (Fox Chase Cancer Center, Histopathology Facility) for histologic assessment of 3D organoids. We thank the Shared Resources (Flow Cytometry, Molecular Pathology, and Confocal & Specialized Microscopy) at the Herbert Irving Comprehensive Cancer Center at Columbia University for technical support. We thank Drs. Anil K. Rustgi, J. Alan Diehl, Adam J. Bass, and Kwok-Kin Wong (NCI P01 Mechanisms of Esophageal Carcinogenesis), Drs. Julian A Abrams and Timothy C. Wang (Columbia University Division of Digestive & Liver Diseases and the NCI U54 Barrett’s Esophagus Translational Research Network), Dr. Jianwen Que (Columbia University Division of Digestive & Liver Diseases and Center for Human Development), and members of the Rustgi laboratory for discussion.
Grant Support
This study was supported by the following NIH Grants: P01CA098101 (HN and RCA), U54CA163004 (HN and RCA), R01DK114436 (HN), R01AA026297 (MS, SF, and HN), and P30CA013696, and the following fellowships: Carolyn E. Reed Traveling Fellowship from the Thoracic Surgery Foundation/Women in Thoracic Surgery (UMS), and the Eleanor and Miles Shore Fellowship from Harvard Medical School/MGH (UMS).
Abbreviations
- BE
Barrett’s Esophagus
- BMP
bone morphogenetic protein
- EAC
esophageal adenocarcinoma
- EOU
esophageal organoid units
- EPC
esophageal progenitor cell
- ESCC
esophageal squamous cell carcinoma
- 4NQO
4-nitroquinoline 1-oxide
- GERD
gastroesophageal reflux disease
- HNSCC
head and neck squamous cell carcinoma
- hPSC
human pluripotent stem cell
- HPV
human papilloma virus
- OTC
organotypic culture
- PDO
patient-derived organoid
- PDX
patient-derived xenograft
- SCC
squamous cell carcinoma
- 3D
three-dimensional
- sgRNA
single guide RNA
- CAF
cancer associated fibroblast
Footnotes
Conflicts of interest
The authors have no conflicts of interest to disclose.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].G.B.D.O.C. Collaborators, The global, regional, and national burden of oesophageal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017, Lancet Gastroenterol Hepatol, 5 (2020) 582–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kapoor H, Lohani KR, Lee TH, Agrawal DK, Mittal SK, Animal Models of Barrett’s Esophagus and Esophageal Adenocarcinoma-Past, Present, and Future, Clin Transl Sci, 8 (2015) 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Karakasheva TA, Kijima T, Shimonosono M, Maekawa H, Sahu V, Gabre JT, Cruz-Acuna R, Giroux V, Sangwan V, Whelan KA, Natsugoe S, Yoon AJ, Philipone E, Klein-Szanto AJ, Ginsberg GG, Falk GW, Abrams JA, Que J, Basu D, Ferri L, Diehl JA, Bass AJ, Wang TC, Rustgi AK, Nakagawa H, Generation and Characterization of Patient-Derived Head and Neck, Oral, and Esophageal Cancer Organoids, Curr Protoc Stem Cell Biol, 53 (2020) e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Gunther C, Neumann H, Vieth M, Esophageal epithelial resistance, Dig Dis, 32 (2014) 6–10. [DOI] [PubMed] [Google Scholar]
- [5].Orlando RC, The integrity of the esophageal mucosa. Balance between offensive and defensive mechanisms, Best Pract Res Clin Gastroenterol, 24 (2010) 873–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Blevins CH, Iyer PG, Vela MF, Katzka DA, The Esophageal Epithelial Barrier in Health and Disease, Clin Gastroenterol Hepatol, 16 (2018) 608–617. [DOI] [PubMed] [Google Scholar]
- [7].Oezcelik A, DeMeester SR, General anatomy of the esophagus, Thorac Surg Clin, 21 (2011) 289–297, x. [DOI] [PubMed] [Google Scholar]
- [8].Zhang X, Patil D, Odze RD, Zhao L, Lisovsky M, Guindi M, Riddell R, Bellizzi A, Yantiss RK, Nalbantoglu I, Appelman HD, The microscopic anatomy of the esophagus including the individual layers, specialized tissues, and unique components and their responses to injury, Ann N Y Acad Sci, 1434 (2018) 304–318. [DOI] [PubMed] [Google Scholar]
- [9].Rosekrans SL, Baan B, Muncan V, van den Brink GR, Esophageal development and epithelial homeostasis, Am J Physiol Gastrointest Liver Physiol, 309 (2015) G216–228. [DOI] [PubMed] [Google Scholar]
- [10].Zhang Y, Jiang M, Kim E, Lin S, Liu K, Lan X, Que J, Development and stem cells of the esophagus, Semin Cell Dev Biol, 66 (2017) 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL, Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm, Development, 134 (2007) 2521–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yu WY, Slack JM, Tosh D, Conversion of columnar to stratified squamous epithelium in the developing mouse oesophagus, Dev Biol, 284 (2005) 157–170. [DOI] [PubMed] [Google Scholar]
- [13].Daniely Y, Liao G, Dixon D, Linnoila RI, Lori A, Randell SH, Oren M, Jetten AM, Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium, Am J Physiol Cell Physiol, 287 (2004) C171–181. [DOI] [PubMed] [Google Scholar]
- [14].Jovov B, Que J, Tobey NA, Djukic Z, Hogan BL, Orlando RC, Role of E-cadherin in the pathogenesis of gastroesophageal reflux disease, Am J Gastroenterol, 106 (2011) 1039–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kalabis J, Wong GS, Vega ME, Natsuizaka M, Robertson ES, Herlyn M, Nakagawa H, Rustgi AK, Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture, Nat Protoc, 7 (2012) 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Rodriguez P, Da Silva S, Oxburgh L, Wang F, Hogan BL, Que J, BMP signaling in the development of the mouse esophagus and forestomach, Development, 137 (2010) 4171–4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kalabis J, Oyama K, Okawa T, Nakagawa H, Michaylira CZ, Stairs DB, Figueiredo JL, Mahmood U, Diehl JA, Herlyn M, Rustgi AK, A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification, J Clin Invest, 118 (2008) 3860–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Frede J, Greulich P, Nagy T, Simons BD, Jones PH, A single dividing cell population with imbalanced fate drives oesophageal tumour growth, Nat Cell Biol, 18 (2016) 967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].DeWard AD, Cramer J, Lagasse E, Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population, Cell Rep, 9 (2014) 701–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Doupe DP, Alcolea MP, Roshan A, Zhang G, Klein AM, Simons BD, Jones PH, A single progenitor population switches behavior to maintain and repair esophageal epithelium, Science, 337 (2012) 1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu K, Jiang M, Lu Y, Chen H, Sun J, Wu S, Ku WY, Nakagawa H, Kita Y, Natsugoe S, Peters JH, Rustgi A, Onaitis MW, Kiernan A, Chen X, Que J, Sox2 cooperates with inflammation-mediated Stat3 activation in the malignant transformation of foregut basal progenitor cells, Cell Stem Cell, 12 (2013) 304–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Seery JP, Watt FM, Asymmetric stem-cell divisions define the architecture of human oesophageal epithelium, Curr Biol, 10 (2000) 1447–1450. [DOI] [PubMed] [Google Scholar]
- [23].Then EO, Lopez M, Saleem S, Gayam V, Sunkara T, Culliford A, Gaduputi V, Esophageal Cancer: An Updated Surveillance Epidemiology and End Results Database Analysis, World J Oncol, 11 (2020) 55–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rustgi AK, El-Serag HB, Esophageal carcinoma, N Engl J Med, 371 (2014) 2499–2509. [DOI] [PubMed] [Google Scholar]
- [25].Wheeler JB, Reed CE, Epidemiology of esophageal cancer, Surg Clin North Am, 92 (2012) 1077–1087. [DOI] [PubMed] [Google Scholar]
- [26].Sawada G, Niida A, Uchi R, Hirata H, Shimamura T, Suzuki Y, Shiraishi Y, Chiba K, Imoto S, Takahashi Y, Iwaya T, Sudo T, Hayashi T, Takai H, Kawasaki Y, Matsukawa T, Eguchi H, Sugimachi K, Tanaka F, Suzuki H, Yamamoto K, Ishii H, Shimizu M, Yamazaki H, Yamazaki M, Tachimori Y, Kajiyama Y, Natsugoe S, Fujita H, Mafune K, Tanaka Y, Kelsell DP, Scott CA, Tsuji S, Yachida S, Shibata T, Sugano S, Doki Y, Akiyama T, Aburatani H, Ogawa S, Miyano S, Mori M, Mimori K, Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population, Gastroenterology, 150 (2016) 1171–1182. [DOI] [PubMed] [Google Scholar]
- [27].Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T, Tsunoda T, Kamatani N, Kubo M, Nakamura Y, Matsuda K, Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk, Gastroenterology, 137 (2009) 1768–1775. [DOI] [PubMed] [Google Scholar]
- [28].Wu C, Hu Z, He Z, Jia W, Wang F, Zhou Y, Liu Z, Zhan Q, Liu Y, Yu D, Zhai K, Chang J, Qiao Y, Jin G, Liu Z, Shen Y, Guo C, Fu J, Miao X, Tan W, Shen H, Ke Y, Zeng Y, Wu T, Lin D, Genome-wide association study identifies three new susceptibility loci for esophageal squamous-cell carcinoma in Chinese populations, Nat Genet, 43 (2011) 679–684. [DOI] [PubMed] [Google Scholar]
- [29].Wang LD, Zhou FY, Li XM, Sun LD, Song X, Jin Y, Li JM, Kong GQ, Qi H, Cui J, Zhang LQ, Yang JZ, Li JL, Li XC, Ren JL, Liu ZC, Gao WJ, Yuan L, Wei W, Zhang YR, Wang WP, Sheyhidin I, Li F, Chen BP, Ren SW, Liu B, Li D, Ku JW, Fan ZM, Zhou SL, Guo ZG, Zhao XK, Liu N, Ai YH, Shen FF, Cui WY, Song S, Guo T, Huang J, Yuan C, Huang J, Wu Y, Yue WB, Feng CW, Li HL, Wang Y, Tian JY, Lu Y, Yuan Y, Zhu WL, Liu M, Fu WJ, Yang X, Wang HJ, Han SL, Chen J, Han M, Wang HY, Zhang P, Li XM, Dong JC, Xing GL, Wang R, Guo M, Chang ZW, Liu HL, Guo L, Yuan ZQ, Liu H, Lu Q, Yang LQ, Zhu FG, Yang XF, Feng XS, Wang Z, Li Y, Gao SG, Qige Q, Bai LT, Yang WJ, Lei GY, Shen ZY, Chen LQ, Li EM, Xu LY, Wu ZY, Cao WK, Wang JP, Bao ZQ, Chen JL, Ding GC, Zhuang X, Zhou YF, Zheng HF, Zhang Z, Zuo XB, Dong ZM, Fan DM, He X, Wang J, Zhou Q, Zhang QX, Jiao XY, Lian SY, Ji AF, Lu XM, Wang JS, Chang FB, Lu CD, Chen ZG, Miao JJ, Fan ZL, Lin RB, Liu TJ, Wei JC, Kong QP, Lan Y, Fan YJ, Gao FS, Wang TY, Xie D, Chen SQ, Yang WC, Hong JY, Wang L, Qiu SL, Cai ZM, Zhang XJ, Genome-wide association study of esophageal squamous cell carcinoma in Chinese subjects identifies susceptibility loci at PLCE1 and C20orf54, Nat Genet, 42 (2010) 759–763. [DOI] [PubMed] [Google Scholar]
- [30].Abnet CC, Freedman ND, Hu N, Wang Z, Yu K, Shu XO, Yuan JM, Zheng W, Dawsey SM, Dong LM, Lee MP, Ding T, Qiao YL, Gao YT, Koh WP, Xiang YB, Tang ZZ, Fan JH, Wang C, Wheeler W, Gail MH, Yeager M, Yuenger J, Hutchinson A, Jacobs KB, Giffen CA, Burdett L, Fraumeni JF Jr., Tucker MA, Chow WH, Goldstein AM, Chanock SJ, Taylor PR, A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma, Nat Genet, 42 (2010) 764–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dotto GP, Rustgi AK, Squamous Cell Cancers: A Unified Perspective on Biology and Genetics, Cancer Cell, 29 (2016) 622–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liyanage SS, Rahman B, Ridda I, Newall AT, Tabrizi SN, Garland SM, Segelov E, Seale H, Crowe PJ, Moa A, MacIntyre CR, The Aetiological Role of Human Papillomavirus in Oesophageal Squamous Cell Carcinoma: A Meta-Analysis, PLOS ONE, 8 (2013) e69238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bognár L, Hegedűs I, Bellyei S, Pozsgai É, Zoltán L, Gombos K, Horváth ÖP, Vereczkei A, Papp A, Prognostic role of HPV infection in esophageal squamous cell carcinoma, Infectious Agents and Cancer, 13 (2018) 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Michaelsen SH, Larsen CG, von Buchwald C, Human Papillomavirus Shows Highly Variable Prevalence in Esophageal Squamous Cell Carcinoma and No Significant Correlation to p16INK4a Overexpression: A Systematic Review, Journal of Thoracic Oncology, 9 (2014) 865–871. [DOI] [PubMed] [Google Scholar]
- [35].Ludmir EB, Stephens SJ, Palta M, Willett CG, Czito BG, Human papillomavirus tumor infection in esophageal squamous cell carcinoma, J Gastrointest Oncol, 6 (2015) 287–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Liu X, Zhang M, Ying S, Zhang C, Lin R, Zheng J, Zhang G, Tian D, Guo Y, Du C, Chen Y, Chen S, Su X, Ji J, Deng W, Li X, Qiu S, Yan R, Xu Z, Wang Y, Guo Y, Cui J, Zhuang S, Yu H, Zheng Q, Marom M, Sheng S, Zhang G, Hu S, Li R, Su M, Genetic Alterations in Esophageal Tissues From Squamous Dysplasia to Carcinoma, Gastroenterology, 153 (2017) 166–177. [DOI] [PubMed] [Google Scholar]
- [37].Stairs DB, Bayne LJ, Rhoades B, Vega ME, Waldron TJ, Kalabis J, Klein-Szanto A, Lee JS, Katz JP, Diehl JA, Reynolds AB, Vonderheide RH, Rustgi AK, Deletion of p120-catenin results in a tumor microenvironment with inflammation and cancer that establishes it as a tumor suppressor gene, Cancer Cell, 19 (2011) 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ellis A, Risk JM, Maruthappu T, Kelsell DP, Tylosis with oesophageal cancer: Diagnosis, management and molecular mechanisms, Orphanet J Rare Dis, 10 (2015) 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Humbert L, Cornu M, Proust-Lemoine E, Bayry J, Wemeau JL, Vantyghem MC, Sendid B, Chronic Mucocutaneous Candidiasis in Autoimmune Polyendocrine Syndrome Type 1, Front Immunol, 9 (2018) 2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Risitano AM, Marotta S, Calzone R, Grimaldi F, Zatterale A, Contributors R, Twenty years of the Italian Fanconi Anemia Registry: where we stand and what remains to be learned, Haematologica, 101 (2016) 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kutler DI, Auerbach AD, Satagopan J, Giampietro PF, Batish SD, Huvos AG, Goberdhan A, Shah JP, Singh B, High Incidence of Head and Neck Squamous Cell Carcinoma in Patients With Fanconi Anemia, Archives of Otolaryngology–Head & Neck Surgery, 129 (2003) 106–112. [DOI] [PubMed] [Google Scholar]
- [42].N. Cancer Genome Atlas Research, U. Analysis Working Group: Asan, B.C.C. Agency, Brigham, H. Women’s, Broad I, Brown U, U. Case Western Reserve, I. Dana-Farber Cancer, Duke U, C. Greater Poland Cancer, S. Harvard Medical, B. Institute for Systems, Leuven KU, Mayo C, C. Memorial Sloan Kettering Cancer, I. National Cancer, H. Nationwide Children’s, U. Stanford, A. University of, M. University of, C. University of North, P. University of, R. University of, C. University of Southern, M.D.A.C.C. University of Texas, W. University of, I. Van Andel Research, U. Vanderbilt, U. Washington, I. Genome Sequencing Center: Broad, L. Washington University in St, B.C.C.A. Genome Characterization Centers, Broad I, S. Harvard Medical, U. Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, C. University of North, C. University of Southern California Epigenome, M.D.A.C.C. University of Texas, I. Van Andel Research, I. Genome Data Analysis Centers: Broad, Brown U, S. Harvard Medical, B. Institute for Systems, C. Memorial Sloan Kettering Cancer, C. University of California Santa, M.D.A.C.C. University of Texas, C. Biospecimen Core Resource: International Genomics, H. Research Institute at Nationwide Children’s, S. Tissue Source Sites: Analytic Biologic, C. Asan Medical, Asterand B, H. Barretos Cancer, BioreclamationIvt, C. Botkin Municipal, S. Chonnam National University Medical, S. Christiana Care Health, Cureline, Duke U, Emory U, Erasmus U, M. Indiana University School of, M. Institute of Oncology of, C. International Genomics, Invidumed, H. Israelitisches Krankenhaus, M. Keimyung University School of, C. Memorial Sloan Kettering Cancer, G. National Cancer Center, B. Ontario Tumour, C. Peter MacCallum Cancer, S. Pusan National University Medical, S. Ribeirao Preto Medical, H. St. Joseph’s, C. Medical, U. St. Petersburg Academic, B. Tayside Tissue, D. University of, C. University of Kansas Medical, M. University of, H. University of North Carolina at Chapel, M. University of Pittsburgh School of, M.D.A.C.C. University of Texas, U. Disease Working Group: Duke, C. Memorial Sloan Kettering Cancer, I. National Cancer, M.D.A.C.C. University of Texas, M. Yonsei University College of, C.I. Data Coordination Center, H. Project Team: National Institutes of, Integrated genomic characterization of oesophageal carcinoma, Nature, 541 (2017) 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, Kim SY, Wardwell L, Tamayo P, Gat-Viks I, Ramos AH, Woo MS, Weir BA, Getz G, Beroukhim R, O’Kelly M, Dutt A, Rozenblatt-Rosen O, Dziunycz P, Komisarof J, Chirieac LR, Lafargue CJ, Scheble V, Wilbertz T, Ma C, Rao S, Nakagawa H, Stairs DB, Lin L, Giordano TJ, Wagner P, Minna JD, Gazdar AF, Zhu CQ, Brose MS, Cecconello I, Ribeiro U Jr., Marie SK, Dahl O, Shivdasani RA, Tsao MS, Rubin MA, Wong KK, Regev A, Hahn WC, Beer DG, Rustgi AK, Meyerson M, SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas, Nat Genet, 41 (2009) 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Natsuizaka M, Whelan KA, Kagawa S, Tanaka K, Giroux V, Chandramouleeswaran PM, Long A, Sahu V, Darling DS, Que J, Yang Y, Katz JP, Wileyto EP, Basu D, Kita Y, Natsugoe S, Naganuma S, Klein-Szanto AJ, Diehl JA, Bass AJ, Wong KK, Rustgi AK, Nakagawa H, Interplay between Notch1 and Notch3 promotes EMT and tumor initiation in squamous cell carcinoma, Nat Commun, 8 (2017) 1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang Y, Yang Y, Jiang M, Huang SX, Zhang W, Al Alam D, Danopoulos S, Mori M, Chen YW, Balasubramanian R, Chuva de Sousa Lopes SM, Serra C, Bialecka M, Kim E, Lin S, Toste de Carvalho ALR, Riccio PN, Cardoso WV, Zhang X, Snoeck HW, Que J, 3D Modeling of Esophageal Development using Human PSC-Derived Basal Progenitors Reveals a Critical Role for Notch Signaling, Cell Stem Cell, 23 (2018) 516–529 e515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ashktorab H, Nouri Z, Nouraie M, Razjouyan H, Lee EE, Dowlati E, El-Seyed E-W, Laiyemo A, Brim H, Smoot DT, Esophageal carcinoma in African Americans: a five-decade experience, Dig Dis Sci, 56 (2011) 3577–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pickens A, Orringer MB, Geographical distribution and racial disparity in esophageal cancer, Ann Thorac Surg, 76 (2003) S1367–1369. [DOI] [PubMed] [Google Scholar]
- [48].Erkizan HV, Johnson K, Ghimbovschi S, Karkera D, Trachiotis G, Adib H, Hoffman EP, Wadleigh RG, African-American esophageal squamous cell carcinoma expression profile reveals dysregulation of stress response and detox networks, BMC Cancer, 17 (2017) 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Shaheen NJ, What is behind the remarkable increase in esophageal adenocarcinoma?, Am J Gastroenterol, 109 (2014) 345–347. [DOI] [PubMed] [Google Scholar]
- [50].Falk GW, Jacobson BC, Riddell RH, Rubenstein JH, El-Zimaity H, Drewes AM, Roark KS, Sontag SJ, Schnell TG, Leya J, Chejfec G, Richter JE, Jenkins G, Goldman A, Dvorak K, Nardone G, Barrett’s esophagus: prevalence-incidence and etiology-origins, Annals of the New York Academy of Sciences, 1232 (2011) 1–17. [DOI] [PubMed] [Google Scholar]
- [51].Kim J, Bowlby R, Mungall AJ, Robertson AG, Odze RD, Cherniack AD, Shih J, Pedamallu CS, Cibulskis C, Dunford A, Meier SR, Kim J, Raphael BJ, Wu H-T, Wong AM, Willis JE, Bass AJ, Derks S, Garman K, McCall SJ, Wiznerowicz M, Pantazi A, Parfenov M, Thorsson V, Shmulevich I, Dhankani V, Miller M, Sakai R, Wang K, Schultz N, Shen R, Arora A, Weinhold N, Sanchez-Vega F, Kelsen DP, Zhang J, Felau I, Demchok J, Rabkin CS, Camargo MC, Zenklusen JC, Bowen J, Leraas K, Lichtenberg TM, Curtis C, Seoane JA, Ojesina AI, Beer DG, Gulley ML, Pennathur A, Luketich JD, Zhou Z, Weisenberger DJ, Akbani R, Lee J-S, Liu W, Mills GB, Zhang W, Reid BJ, Hinoue T, Laird PW, Shen H, Piazuelo MB, Schneider BG, McLellan M, Taylor-Weiner A, Cibulskis C, Lawrence M, Cibulskis K, Stewart C, Getz G, Lander E, Gabriel SB, Ding L, McLellan MD, Miller CA, Appelbaum EL, Cordes MG, Fronick CC, Fulton LA, Mardis ER, Wilson RK, Schmidt HK, Fulton RS, Ally A, Balasundaram M, Bowlby R, Carlsen R, Chuah E, Dhalla N, Holt RA, Jones SJM, Kasaian K, Brooks D, Li HI, Ma Y, Marra MA, Mayo M, Moore RA, Mungall AJ, Mungall KL, Robertson AG, Schein JE, Sipahimalani P, Tam A, Thiessen N, Wong T, Cherniack AD, Shih J, Pedamallu CS, Beroukhim R, Bullman S, Cibulskis C, Murray BA, Saksena G, Schumacher SE, Gabriel S, Meyerson M, Hadjipanayis A, Kucherlapati R, Pantazi A, Parfenov M, Ren X, Park PJ, Lee S, Kucherlapati M, Yang L, Baylin SB, Hoadley KA, Weisenberger DJ, Bootwalla MS, Lai PH, Van Den Berg DJ, Berrios M, Holbrook A, Akbani R, Hwang J-E, Jang H-J, Liu W, Weinstein JN, Lee J-S, Lu Y, Sohn BH, Mills G, Seth S, Protopopov A, Bristow CA, Mahadeshwar HS, Tang J, Song X, Zhang J, Laird PW, Hinoue T, Shen H, Cho J, Defrietas T, Frazer S, Gehlenborg N, Heiman DI, Lawrence MS, Lin P, Meier SR, Noble MS, Voet D, Zhang H, Kim J, Polak P, Saksena G, Chin L, Getz G, Wong AM, Raphael BJ, Wu H-T, Lee S, Park PJ, Yang L, Thorsson V, Bernard B, lype L, Miller M, Reynolds SM, Shmulevich I, Dhankani V, Abeshouse A, Arora A, Armenia J, Kundra R, Ladanyi M, Lehmann K-V, Gao J, Sander C, Schultz N, Sánchez-Vega F, Shen R, Weinhold N, Chakravarty D, Zhang H, Radenbaugh A, Hegde A, Akbani R, Liu W, Weinstein JN, Chin L, Bristow CA, Lu Y, Penny R, Crain D, Gardner J, Curley E, Mallery D, Morris S, Paulauskis J, Shelton T, Shelton C, Bowen J, Frick J, Gastier-Foster JM, Gerken M, Leraas KM, Lichtenberg TM, Ramirez NC, Wise L, Zmuda E, Tarvin K, Saller C, Park YS, Button M, Carvalho AL, Reis RM, Matsushita MM, Lucchesi F, de Oliveira AT, Le X, Paklina O, Setdikova G, Lee J-H, Bennett J, lacocca M, Huelsenbeck-Dill L, Potapova O, Voronina O, Liu O, Fulidou V, N. The Cancer Genome Atlas Research, U. Analysis Working Group: Asan, B.C.C. Agency, Brigham, H. Women’s, Broad I, Brown U, U. Case Western Reserve, I. Dana-Farber Cancer, Duke U, C. Greater Poland Cancer, S. Harvard Medical, B. Institute for Systems, Leuven KU, Mayo C, C. Memorial Sloan Kettering Cancer, I. National Cancer, H. Nationwide Children’s, U. Stanford, A. University of, M. University of, C. University of North, P. University of, R. University of, C. University of Southern, M.D.A.C.C. University of Texas, W. University of, I. Van Andel Research, U. Vanderbilt, U. Washington, I. Genome Sequencing Center: Broad, L. Washington University in St, B.C.C.A. Genome Characterization Centers, U. The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins, C. University of Southern California Epigenome, I. Genome Data Analysis Centers: Broad, Brown U, C. University of California Santa, C. Biospecimen Core Resource: International Genomics, H. The Research Institute at Nationwide Children’s, S. Tissue Source Sites: Analytic Biologic, C. Asan Medical, B. Asterand, H. Barretos Cancer, BioreclamationIvt, C. Botkin Municipal, S. Chonnam National University Medical, S. Christiana Care Health, Cureline, Integrated genomic characterization of oesophageal carcinoma, Nature, 541 (2017) 169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dulak AM, Stojanov P, Peng S, Lawrence MS, Fox C, Stewart C, Bandla S, Imamura Y, Schumacher SE, Shefler E, McKenna A, Carter SL, Cibulskis K, Sivachenko A, Saksena G, Voet D, Ramos AH, Auclair D, Thompson K, Sougnez C, Onofrio RC, Guiducci C, Beroukhim R, Zhou Z, Lin L, Lin J, Reddy R, Chang A, Landrenau R, Pennathur A, Ogino S, Luketich JD, Golub TR, Gabriel SB, Lander ES, Beer DG, Godfrey TE, Getz G, Bass AJ, Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity, Nat Genet, 45 (2013) 478–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vega ME, Giroux V, Natsuizaka M, Liu M, Klein-Szanto AJ, Stairs DB, Nakagawa H, Wang KK, Wang TC, Lynch JP, Rustgi AK, Inhibition of Notch signaling enhances transdifferentiation of the esophageal squamous epithelium towards a Barrett’s-like metaplasia via KLF4, Cell Cycle, 13 (2014) 3857–3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kunze B, Wein F, Fang HY, Anand A, Baumeister T, Strangmann J, Gerland S, Ingermann J, Munch NS, Wiethaler M, Sahm V, Hidalgo-Sastre A, Lange S, Lightdale CJ, Bokhari A, Falk GW, Friedman RA, Ginsberg GG, Iyer PG, Jin Z, Nakagawa H, Shawber CJ, Nguyen T, Raab WJ, Dalerba P, Rustgi AK, Sepulveda AR, Wang KK, Schmid RM, Wang TC, Abrams JA, Quante M, Notch Signaling Mediates Differentiation in Barrett’s Esophagus and Promotes Progression to Adenocarcinoma, Gastroenterology, 159 (2020) 575–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nakagawa H, Whelan K, Lynch JP, Mechanisms of Barrett’s oesophagus: intestinal differentiation, stem cells, and tissue models, Best practice & research. Clinical gastroenterology, 29 (2015) 3–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Giroux V, Lento AA, Islam M, Pitarresi JR, Kharbanda A, Hamilton KE, Whelan KA, Long A, Rhoades B, Tang Q, Nakagawa H, Lengner CJ, Bass AJ, Wileyto EP, Klein-Szanto AJ, Wang TC, Rustgi AK, Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration, J Clin Invest, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jiang M, Li H, Zhang Y, Yang Y, Lu R, Liu K, Lin S, Lan X, Wang H, Wu H, Zhu J, Zhou Z, Xu J, Lee DK, Zhang L, Lee YC, Yuan J, Abrams JA, Wang TC, Sepulveda AR, Wu Q, Chen H, Sun X, She J, Chen X, Que J, Transitional basal cells at the squamous-columnar junction generate Barrett’s oesophagus, Nature, 550 (2017) 529–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Lee Y, Urbanska AM, Hayakawa Y, Wang H, Au AS, Luna AM, Chang W, Jin G, Bhagat G, Abrams JA, Friedman RA, Varro A, Wang KK, Boyce M, Rustgi AK, Sepulveda AR, Quante M, Wang TC, Gastrin stimulates a cholecystokinin-2-receptor-expressing cardia progenitor cell and promotes progression of Barrett’s-like esophagus, Oncotarget, 8 (2017) 203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Quante M, Abrams JA, Lee Y, Wang TC, Barrett esophagus: what a mouse model can teach us about human disease, Cell Cycle, 11 (2012) 4328–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Quante M, Bhagat G, Abrams JA, Marache F, Good P, Lee MD, Lee Y, Friedman R, Asfaha S, Dubeykovskaya Z, Mahmood U, Figueiredo JL, Kitajewski J, Shawber C, Lightdale CJ, Rustgi AK, Wang TC, Bile acid and inflammation activate gastric cardia stem cells in a mouse model of Barrett-like metaplasia, Cancer Cell, 21 (2012) 36–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Jammula S, Katz-Summercorn AC, Li X, Linossi C, Smyth E, Killcoyne S, Biasci D, Subash VV, Abbas S, Blasko A, Devonshire G, Grantham A, Wronowski F, O’Donovan M, Grehan N, Eldridge MD, Tavare S, Oesophageal Cancer C, c. Molecular Stratification, R.C. Fitzgerald, Identification of Subtypes of Barrett’s Esophagus and Esophageal Adenocarcinoma Based on DNA Methylation Profiles and Integration of Transcriptome and Genome Data, Gastroenterology, 158 (2020) 1682–1697 e1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sawas T, Killcoyne S, Iyer PG, Wang KK, Smyrk TC, Kisiel JB, Qin Y, Ahlquist DA, Rustgi AK, Costa RJ, Gerstung M, Fitzgerald RC, Katzka DA, Consortium O, Identification of Prognostic Phenotypes of Esophageal Adenocarcinoma in 2 Independent Cohorts, Gastroenterology, 155 (2018) 1720–1728 e1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yendamuri S, Malhotra U, Hennon M, Miller A, Groman A, Halloon A, Reid ME, Clinical characteristics of adenosquamous esophageal carcinoma, J Gastrointest Oncol, 8 (2017) 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Nakagawa H, Wang TC, Zukerberg L, Odze R, Togawa K, May GH, Wilson J, Rustgi AK, The targeting of the cyclin D1 oncogene by an Epstein-Barr virus promoter in transgenic mice causes dysplasia in the tongue, esophagus and forestomach, Oncogene, 14 (1997) 1185–1190. [DOI] [PubMed] [Google Scholar]
- [65].Andl CD, Mizushima T, Nakagawa H, Oyama K, Harada H, Chruma K, Herlyn M, Rustgi AK, Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo, J Biol Chem, 278 (2003) 1824–1830. [DOI] [PubMed] [Google Scholar]
- [66].Opitz OG, Harada H, Suliman Y, Rhoades B, Sharpless NE, Kent R, Kopelovich L, Nakagawa H, Rustgi AK, A mouse model of human oral-esophageal cancer, J Clin Invest, 110 (2002) 761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Tang XH, Knudsen B, Bemis D, Tickoo S, Gudas LJ, Oral cavity and esophageal carcinogenesis modeled in carcinogen-treated mice, Clin Cancer Res, 10 (2004) 301–313. [DOI] [PubMed] [Google Scholar]
- [68].Wang Z, Wu VH, Allevato MM, Gilardi M, He Y, Luis Callejas-Valera J, Vitale-Cross L, Martin D, Amornphimoltham P, McDermott J, Yung BS, Goto Y, Molinolo AA, Sharabi AB, Cohen EEW, Chen Q, Lyons JG, Alexandrov LB, Gutkind JS, Syngeneic animal models of tobacco-associated oral cancer reveal the activity of in situ anti-CTLA-4, Nat Commun, 10 (2019) 5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wilkey JF, Buchberger G, Saucier K, Patel SM, Eisenberg E, Nakagawa H, Michaylira CZ, Rustgi AK, Mallya SM, Cyclin D1 overexpression increases susceptibility to 4-nitroquinoline-1-oxide-induced dysplasia and neoplasia in murine squamous oral epithelium, Mol Carcinog, 48 (2009) 853–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Hashimoto N, Expression of COX2 and p53 in Rat Esophageal Cancer Induced by Reflux of Duodenal Contents, ISRN Gastroenterol, 2012 (2012) 914824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kawaura Y, Tatsuzawa Y, Wakabayashi T, Ikeda N, Matsuda M, Nishihara S, Immunohistochemical study of p53, c-erbB-2, and PCNA in barrett’s esophagus with dysplasia and adenocarcinoma arising from experimental acid or alkaline reflux model, J Gastroenterol, 36 (2001) 595–600. [DOI] [PubMed] [Google Scholar]
- [72].Redo SF, Barnes WA, De La Sierra AO, Perfusion of the canine esophagus with secretions of the upper gastro-intestinal tract, Ann Surg, 149 (1959) 556–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Gillen P, Keeling P, Byrne PJ, West AB, Hennessy TP, Experimental columnar metaplasia in the canine oesophagus, Br J Surg, 75 (1988) 113–115. [DOI] [PubMed] [Google Scholar]
- [74].Bremner CG, Lynch VP, Ellis FH Jr., Barrett’s esophagus: congenital or acquired? An experimental study of esophageal mucosal regeneration in the dog, Surgery, 68 (1970) 209–216. [PubMed] [Google Scholar]
- [75].Kadirkamanathan SS, Yazaki E, Evans DF, Hepworth CC, Gong F, Swain CP, An ambulant porcine model of acid reflux used to evaluate endoscopic gastroplasty, Gut, 44 (1999) 782–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kruger L, Gonzalez LM, Pridgen TA, McCall SJ, von Furstenberg RJ, Harnden I, Carnighan GE, Cox AM, Blikslager AT, Garman KS, Ductular and proliferative response of esophageal submucosal glands in a porcine model of esophageal injury and repair, Am J Physiol Gastrointest Liver Physiol, 313 (2017) G180–G191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].von Furstenberg RJ, Li J, Stolarchuk C, Feder R, Campbell A, Kruger L, Gonzalez LM, Blikslager AT, Cardona DM, McCall SJ, Henning SJ, Garman KS, Porcine Esophageal Submucosal Gland Culture Model Shows Capacity for Proliferation and Differentiation, Cell Mol Gastroenterol Hepatol, 4 (2017) 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Glover EJ, Leland MM, Dick EJ Jr., Hubbard GB, Gastroesophageal reflux disease in baboons (Papio sp.): a new animal model, J Med Primatol, 37 (2008) 18–25. [DOI] [PubMed] [Google Scholar]
- [79].Harada H, Nakagawa H, Oyama K, Takaoka M, Andl CD, Jacobmeier B, von Werder A, Enders GH, Opitz OG, Rustgi AK, Telomerase induces immortalization of human esophageal keratinocytes without p16INK4a inactivation, Mol Cancer Res, 1 (2003) 729–738. [PubMed] [Google Scholar]
- [80].Harada H, Nakagawa H, Takaoka M, Lee J, Herlyn M, Diehl JA, Rustgi AK, Cleavage of MCM2 licensing protein fosters senescence in human keratinocytes, Cell Cycle, 7 (2008) 3534–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ohashi S, Natsuizaka M, Wong GS, Michaylira CZ, Grugan KD, Stairs DB, Kalabis J, Vega ME, Kalman RA, Nakagawa M, Klein-Szanto AJ, Herlyn M, Diehl JA, Rustgi AK, Nakagawa H, Epidermal growth factor receptor and mutant p53 expand an esophageal cellular subpopulation capable of epithelial-to-mesenchymal transition through ZEB transcription factors, Cancer Res, 70 (2010) 4174–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ohashi S, Natsuizaka M, Yashiro-Ohtani Y, Kalman RA, Nakagawa M, Wu L, Klein-Szanto AJ, Herlyn M, Diehl JA, Katz JP, Pear WS, Seykora JT, Nakagawa H, NOTCH1 and NOTCH3 coordinate esophageal squamous differentiation through a CSL-dependent transcriptional network, Gastroenterology, 139 (2010) 2113–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Whelan KA, Muir AB, Nakagawa H, Esophageal 3D Culture Systems as Modeling Tools in Esophageal Epithelial Pathobiology and Personalized Medicine, Cell Mol Gastroenterol Hepatol, 5 (2018) 461–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Okawa T, Michaylira CZ, Kalabis J, Stairs DB, Nakagawa H, Andl CD, Johnstone CN, Klein-Szanto AJ, El-Deiry WS, Cukierman E, Herlyn M, Rustgi AK, The functional interplay between EGFR overexpression, hTERT activation, and p53 mutation in esophageal epithelial cells with activation of stromal fibroblasts induces tumor development, invasion, and differentiation, Genes Dev, 21 (2007) 2788–2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Grugan KD, Miller CG, Yao Y, Michaylira CZ, Ohashi S, Klein-Szanto AJ, Diehl JA, Herlyn M, Han M, Nakagawa H, Rustgi AK, Fibroblast-secreted hepatocyte growth factor plays a functional role in esophageal squamous cell carcinoma invasion, Proc Natl Acad Sci U S A, 107 (2010) 11026–11031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Naganuma S, Whelan KA, Natsuizaka M, Kagawa S, Kinugasa H, Chang S, Subramanian H, Rhoades B, Ohashi S, Itoh H, Herlyn M, Diehl JA, Gimotty PA, Klein-Szanto AJ, Nakagawa H, Notch receptor inhibition reveals the importance of cyclin D1 and Wnt signaling in invasive esophageal squamous cell carcinoma, Am J Cancer Res, 2 (2012) 459–475. [PMC free article] [PubMed] [Google Scholar]
- [87].Kitamura M, Suda M, Nishihira T, Watanabe T, Kasai M, Heterotransplantation of human esophageal carcinoma to nude mice, Tohoku J Exp Med, 135 (1981) 259–264. [DOI] [PubMed] [Google Scholar]
- [88].Robinson KM, Bux S, Ultrastructural features of carcinomas induced in nude mice by the inoculation of human oesophageal carcinoma cell lines, J Pathol, 140 (1983) 193–207. [DOI] [PubMed] [Google Scholar]
- [89].Dodbiba L, Teichman J, Fleet A, Thai H, Starmans MH, Navab R, Chen Z, Girgis H, Eng L, Espin-Garcia O, Shen X, Bandarchi B, Schwock J, Tsao MS, El-Zimaity H, Der SD, Xu W, Bristow RG, Darling GE, Boutros PC, Ailles LE, Liu G, Appropriateness of using patient-derived xenograft models for pharmacologic evaluation of novel therapies for esophageal/gastro-esophageal junction cancers, PLoS One, 10 (2015) e0121872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Dodbiba L, Teichman J, Fleet A, Thai H, Sun B, Panchal D, Patel D, Tse A, Chen Z, Faluyi OO, Renouf DJ, Girgis H, Bandarchi B, Schwock J, Xu W, Bristow RG, Tsao MS, Darling GE, Ailles LE, El-Zimaity H, Liu G, Primary esophageal and gastro-esophageal junction cancer xenograft models: clinicopathological features and engraftment, Lab Invest, 93 (2013) 397–407. [DOI] [PubMed] [Google Scholar]
- [91].Kijima T, Nakagawa H, Shimonosono M, Chandramouleeswaran PM, Hara T, Sahu V, Kasagi Y, Kikuchi O, Tanaka K, Giroux V, Muir AB, Whelan KA, Ohashi S, Naganuma S, Klein-Szanto AJ, Shinden Y, Sasaki K, Omoto I, Kita Y, Muto M, Bass AJ, Diehl JA, Ginsberg GG, Doki Y, Mori M, Uchikado Y, Arigami T, Avadhani NG, Basu D, Rustgi AK, Natsugoe S, Three-Dimensional Organoids Reveal Therapy Resistance of Esophageal and Oropharyngeal Squamous Cell Carcinoma Cells, Cell Mol Gastroenterol Hepatol, 7 (2019) 73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Karakasheva TA, Lin EW, Tang Q, Qiao E, Waldron TJ, Soni M, Klein-Szanto AJ, Sahu V, Basu D, Ohashi S, Baba K, Giaccone ZT, Walker SR, Frank DA, Wileyto EP, Long Q, Dunagin MC, Raj A, Diehl JA, Wong KK, Bass AJ, Rustgi AK, IL-6 Mediates Cross-Talk between Tumor Cells and Activated Fibroblasts in the Tumor Microenvironment, Cancer Res, 78 (2018) 4957–4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Li X, Francies HE, Secrier M, Perner J, Miremadi A, Galeano-Dalmau N, Barendt WJ, Letchford L, Leyden GM, Goffin EK, Barthorpe A, Lightfoot H, Chen E, Gilbert J, Noorani A, Devonshire G, Bower L, Grantham A, MacRae S, Grehan N, Wedge DC, Fitzgerald RC, Garnett MJ, Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics, Nature Communications, 9 (2018) 2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Jin RU, Mills JC, Tumor organoids to study gastroesophageal cancer: a primer, J Mol Cell Biol, 12 (2020) 593–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Yan T, Cui H, Zhou Y, Yang B, Kong P, Zhang Y, Liu Y, Wang B, Cheng Y, Li J, Guo S, Xu E, Liu H, Cheng C, Zhang L, Chen L, Zhuang X, Qian Y, Yang J, Ma Y, Li H, Wang F, Liu J, Liu X, Su D, Wang Y, Sun R, Guo S, Li Y, Cheng X, Liu Z, Zhan Q, Cui Y, Multi-region sequencing unveils novel actionable targets and spatial heterogeneity in esophageal squamous cell carcinoma, Nat Commun, 10 (2019) 1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Nakagawa H, Kasagi Y, Karakasheva TA, Hara T, Aaron B, Shimonosono M, Kijima T, Giroux V, Bailey D, Wilkins B, Abrams JA, Falk GW, Aceves SS, Spergel JM, Hamilton KE, Whelan KA, Muir AB, Modeling Epithelial Homeostasis and Reactive Epithelial Changes in Human and Murine Three-Dimensional Esophageal Organoids, Curr Protoc Stem Cell Biol, 52 (2020) e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kasagi Y, Chandramouleeswaran PM, Whelan KA, Tanaka K, Giroux V, Sharma M, Wang J, Benitez AJ, DeMarshall M, Tobias JW, Hamilton KE, Falk GW, Spergel JM, Klein-Szanto AJ, Rustgi AK, Muir AB, Nakagawa H, The Esophageal Organoid System Reveals Functional Interplay Between Notch and Cytokines in Reactive Epithelial Changes, Cellular and Molecular Gastroenterology and Hepatology, 5 (2018) 333–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Chandramouleeswaran PM, Guha M, Shimonosono M, Whelan KA, Maekawa H, Sachdeva UM, Ruthel G, Mukherjee S, Engel N, Gonzalez MV, Garifallou J, Ohashi S, Klein-Szanto AJ, Mesaros CA, Blair IA, Pellegrino da Silva R, Hakonarson H, Noguchi E, Baur JA, Nakagawa H, Autophagy mitigates ethanol-induced mitochondrial dysfunction and oxidative stress in esophageal keratinocytes, PLoS One, 15 (2020) e0239625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Spurrier RG, Speer AL, Hou X, El-Nachef WN, Grikscheit TC, Murine and human tissue-engineered esophagus form from sufficient stem/progenitor cells and do not require microdesigned biomaterials, Tissue Eng Part A, 21 (2015) 906–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Trecartin A, Danopoulos S, Spurrier R, Knaneh-Monem H, Hiatt M, Driscoll B, Hochstim C, Al-Alam D, Grikscheit TC, Establishing Proximal and Distal Regional Identities in Murine and Human Tissue-Engineered Lung and Trachea, Tissue Eng Part C Methods, 22 (2016) 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Finkbeiner SR, Freeman JJ, Wieck MM, El-Nachef W, Altheim CH, Tsai YH, Huang S, Dyal R, White ES, Grikscheit TC, Teitelbaum DH, Spence JR, Generation of tissue-engineered small intestine using embryonic stem cell-derived human intestinal organoids, Biol Open, 4 (2015) 1462–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Que J, Choi M, Ziel JW, Klingensmith J, Hogan BL, Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps, Differentiation, 74 (2006) 422–437. [DOI] [PubMed] [Google Scholar]
- [103].Pinzon-Guzman C, Sangadala S, Riera KM, Popova EY, Manning E, Huh WJ, Alexander MS, Shelton JS, Boden SD, Goldenring JR, Noggin regulates foregut progenitor cell programming, and misexpression leads to esophageal atresia, The Journal of Clinical Investigation, 130 (2020) 4396–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Trisno SL, Philo KED, McCracken KW, Cata EM, Ruiz-Torres S, Rankin SA, Han L, Nasr T, Chaturvedi P, Rothenberg ME, Mandegar MA, Wells SI, Zorn AM, Wells JM, Esophageal Organoids from Human Pluripotent Stem Cells Delineate Sox2 Functions during Esophageal Specification, Cell Stem Cell, 23 (2018) 501–515 e507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Bailey DD, Zhang Y, van Soldt BJ, Jiang M, Suresh S, Nakagawa H, Rustgi AK, Aceves SS, Cardoso WV, Que J, Use of hPSC-derived 3D organoids and mouse genetics to define the roles of YAP in the development of the esophagus, Development, 146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Williamson KA, Hever AM, Rainger J, Rogers RC, Magee A, Fiedler Z, Keng WT, Sharkey FH, McGill N, Hill CJ, Schneider A, Messina M, Turnpenny PD, Fantes JA, van Heyningen V, FitzPatrick DR, Mutations in SOX2 cause anophthalmia-esophageal-genital (AEG) syndrome, Hum Mol Genet, 15 (2006) 1413–1422. [DOI] [PubMed] [Google Scholar]
- [107].Wang DH, The Esophageal Squamous Epithelial Cell-Still a Reasonable Candidate for the Barrett’s Esophagus Cell of Origin?, Cell Mol Gastroenterol Hepatol, 4 (2017) 157–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Shields HM, Zwas F, Antonioli DA, Doos WG, Kim S, Spechler SJ, Detection by scanning electron microscopy of a distinctive esophageal surface cell at the junction of squamous and Barrett’s epithelium, Dig Dis Sci, 38 (1993) 97–108. [DOI] [PubMed] [Google Scholar]
- [109].Wang DH, Souza RF, Transcommitment: Paving the Way to Barrett’s Metaplasia, Adv Exp Med Biol, 908 (2016) 183–212. [DOI] [PubMed] [Google Scholar]
- [110].Leedham SJ, Preston SL, McDonald SA, Elia G, Bhandari P, Poller D, Harrison R, Novelli MR, Jankowski JA, Wright NA, Individual crypt genetic heterogeneity and the origin of metaplastic glandular epithelium in human Barrett’s oesophagus, Gut, 57 (2008) 1041–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Giroux V, Lento AA, Islam M, Pitarresi JR, Kharbanda A, Hamilton KE, Whelan KA, Long A, Rhoades B, Tang Q, Nakagawa H, Lengner CJ, Bass AJ, Wileyto EP, Klein-Szanto AJ, Wang TC, Rustgi AK, Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration, J Clin Invest, 127 (2017) 2378–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Driehuis E, Kolders S, Spelier S, Lohmussaar K, Willems SM, Devriese LA, de Bree R, de Ruiter EJ, Korving J, Begthel H, van Es JH, Geurts V, He GW, van Jaarsveld RH, Oka R, Muraro MJ, Vivie J, Zandvliet M, Hendrickx APA, lakobachvili N, Sridevi P, Kranenburg O, van Boxtel R, Kops G, Tuveson DA, Peters PJ, van Oudenaarden A, Clevers H, Oral Mucosal Organoids as a Potential Platform for Personalized Cancer Therapy, Cancer Discov, 9 (2019) 852–871. [DOI] [PubMed] [Google Scholar]
- [113].Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, Clevers H, Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium, Gastroenterology, 141 (2011) 1762–1772. [DOI] [PubMed] [Google Scholar]
- [114].Secrier M, Li X, de Silva N, Eldridge MD, Contino G, Bornschein J, MacRae S, Grehan N, O’Donovan M, Miremadi A, Yang TP, Bower L, Chettouh H, Crawte J, Galeano-Dalmau N, Grabowska A, Saunders J, Underwood T, Waddell N, Barbour AP, Nutzinger B, Achilleos A, Edwards PA, Lynch AG, Tavare S, Fitzgerald RC, C. Oesophageal Cancer, C. Molecular Stratification, Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance, Nat Genet, 48 (2016) 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Dagogo-Jack I, Shaw AT, Tumour heterogeneity and resistance to cancer therapies, Nat Rev Clin Oncol, 15 (2018) 81–94. [DOI] [PubMed] [Google Scholar]
- [116].Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, Rata M, Koh DM, Tunariu N, Collins D, Hulkki-Wilson S, Ragulan C, Spiteri I, Moorcraft SY, Chau I, Rao S, Watkins D, Fotiadis N, Bali M, Darvish-Damavandi M, Lote H, Eltahir Z, Smyth EC, Begum R, Clarke PA, Hahne JC, Dowsett M, de Bono J, Workman P, Sadanandam A, Fassan M, Sansom OJ, Eccles S, Starling N, Braconi C, Sottoriva A, Robinson SP, Cunningham D, Valeri N, Patient-derived organoids model treatment response of metastatic gastrointestinal cancers, Science, 359 (2018) 920–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Driehuis E, Kretzschmar K, Clevers H, Establishment of patient-derived cancer organoids for drug-screening applications, Nature Protocols, 15 (2020) 3380–3409. [DOI] [PubMed] [Google Scholar]
- [118].Derouet MF, Allen J, Wilson GW, Ng C, Radulovich N, Kalimuthu S, Tsao M-S, Darling GE, Yeung JC, Towards personalized induction therapy for esophageal adenocarcinoma: organoids derived from endoscopic biopsy recapitulate the pre-treatment tumor, Scientific Reports, 10 (2020) 14514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Fujii M, Clevers H, Sato T, Modeling Human Digestive Diseases With CRISPR-Cas9-Modified Organoids, Gastroenterology, 156 (2019) 562–576. [DOI] [PubMed] [Google Scholar]
- [120].Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, Korving J, van de Wetering M, Schwank G, Logtenberg M, Cuppen E, Snippert HJ, Medema JP, Kops GJ, Clevers H, Sequential cancer mutations in cultured human intestinal stem cells, Nature, 521 (2015) 43–47. [DOI] [PubMed] [Google Scholar]
- [121].Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T, Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids, Nat Med, 21 (2015) 256–262. [DOI] [PubMed] [Google Scholar]
- [122].Liu X, Cheng Y, Abraham JM, Wang Z, Wang Z, Ke X, Yan R, Shin EJ, Ngamruengphong S, Khashab MA, Zhang G, McNamara G, Ewald AJ, Lin D, Liu Z, Meltzer SJ, Modeling Wnt signaling by CRISPR-Cas9 genome editing recapitulates neoplasia in human Barrett epithelial organoids, Cancer Lett, 436 (2018) 109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Driehuis E, Clevers H, CRISPR/Cas 9 genome editing and its applications in organoids, Am J Physiol Gastrointest Liver Physiol, 312 (2017) G257–G265. [DOI] [PubMed] [Google Scholar]
- [124].Ringel T, Frey N, Ringnalda F, Janjuha S, Cherkaoui S, Butz S, Srivatsa S, Pirkl M, Russo G, Villiger L, Rogler G, Clevers H, Beerenwinkel N, Zamboni N, Baubec T, Schwank G, Genome-Scale CRISPR Screening in Human Intestinal Organoids Identifies Drivers of TGF-beta Resistance, Cell Stem Cell, 26 (2020) 431–440 e438. [DOI] [PubMed] [Google Scholar]
- [125].Michels BE, Mosa MH, Streibl BI, Zhan T, Menche C, Abou-El-Ardat K, Darvishi T, Czlonka E, Wagner S, Winter J, Medyouf H, Boutros M, Farin HF, Pooled In Vitro and In Vivo CRISPR-Cas9 Screening Identifies Tumor Suppressors in Human Colon Organoids, Cell Stem Cell, 26 (2020) 782–792 e787. [DOI] [PubMed] [Google Scholar]
- [126].Brazovskaja A, Treutlein B, Camp JG, High-throughput single-cell transcriptomics on organoids, Curr Opin Biotechnol, 55 (2019) 167–171. [DOI] [PubMed] [Google Scholar]
- [127].Moehler MH, Janjigian YY, Adenis A, Aucoin J-S, Boku N, Chau I, Cleary JM, Feeney K, Franke FA, Mendez GA, Schenker M, Li M, Hitre E, Couture F, Karamouzis M, Etienne P-L, Ajani JA, CheckMate 649: A randomized, multicenter, open-label, phase III study of nivolumab (NIVO) + ipilimumab (IPI) or nivo + chemotherapy (CTX) versus CTX alone in patients with previously untreated advanced (Adv) gastric (G) or gastroesophageal junction (GEJ) cancer, Journal of Clinical Oncology, 36 (2018) TPS192–TPS192. [Google Scholar]
- [128].Kato K, Shah MA, Enzinger P, Bennouna J, Shen L, Adenis A, Sun JM, Cho BC, Özgüroglu M, Kojima T, Kostorov V, Hierro C, Zhu Y, McLean LA, Shah S, Doi T, KEYNOTE-590: Phase III study of first-line chemotherapy with or without pembrolizumab for advanced esophageal cancer, Future Oncol, 15 (2019) 1057–1066. [DOI] [PubMed] [Google Scholar]
- [129].Zheng Y, Chen Z, Han Y, Han L, Zou X, Zhou B, Hu R, Hao J, Bai S, Xiao H, Li WV, Bueker A, Ma Y, Xie G, Yang J, Chen S, Li H, Cao J, Shen L, Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment, Nat Commun, 11 (2020) 6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Blagih J, Buck MD, Vousden KH, p53, cancer and the immune response, J Cell Sci, 133 (2020). [DOI] [PubMed] [Google Scholar]
- [131].Laczko D, Wang F, Johnson FB, Jhala N, Rosztoczy A, Ginsberg GG, Falk GW, Rustgi AK, Lynch JP, Modeling Esophagitis Using Human Three-Dimensional Organotypic Culture System, Am J Pathol, 187 (2017) 1787–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Whelan KA, Muir AB, Nakagawa H, Esophageal 3D Culture Systems as Modeling Tools in Esophageal Epithelial Pathobiology and Personalized Medicine, Cellular and Molecular Gastroenterology and Hepatology, 5 (2018) 461–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Sakamoto H, Koma YI, Higashino N, Kodama T, Tanigawa K, Shimizu M, Fujikawa M, Nishio M, Shigeoka M, Kakeji Y, Yokozaki H, PAI-1 derived from cancer-associated fibroblasts in esophageal squamous cell carcinoma promotes the invasion of cancer cells and the migration of macrophages, Lab Invest, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Buttar NS, Wang KK, Leontovich O, Westcott JY, Pacifico RJ, Anderson MA, Krishnadath KK, Lutzke LS, Burgart LJ, Chemoprevention of esophageal adenocarcinoma by COX-2 inhibitors in an animal model of Barrett’s esophagus, Gastroenterology, 122 (2002) 1101–1112. [DOI] [PubMed] [Google Scholar]
- [135].Taddei A, Fabbroni V, Pini A, Lucarini L, Ringressi MN, Fantappie O, Bani D, Messerini L, Masini E, Bechi P, Cyclooxygenase-2 and inflammation mediators have a crucial role in reflux-related esophageal histological changes and Barrett’s esophagus, Dig Dis Sci, 59 (2014) 949–957. [DOI] [PubMed] [Google Scholar]
- [136].Verbeek RE, Siersema PD, Ten Kate FJ, Fluiter K, Souza RF, Vleggaar FP, Bus P, van Baal JW, Toll-like receptor 4 activation in Barrett’s esophagus results in a strong increase in COX-2 expression, J Gastroenterol, 49 (2014) 1121–1134. [DOI] [PubMed] [Google Scholar]
- [137].Kashima H, Noma K, Ohara T, Kato T, Katsura Y, Komoto S, Sato H, Katsube R, Ninomiya T, Tazawa H, Shirakawa Y, Fujiwara T, Cancer-associated fibroblasts (CAFs) promote the lymph node metastasis of esophageal squamous cell carcinoma, Int J Cancer, 144 (2019) 828–840. [DOI] [PubMed] [Google Scholar]
- [138].Beck JN, Singh A, Rothenberg AR, Elisseeff JH, Ewald AJ, The independent roles of mechanical, structural and adhesion characteristics of 3D hydrogels on the regulation of cancer invasion and dissemination, Biomaterials, 34 (2013) 9486–9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Fong ELS, Toh TB, Lin QXX, Liu Z, Hooi L, Mohd Abdul Rashid MB, Benoukraf T, Chow EK, Huynh TH, Yu H, Generation of matched patient-derived xenograft in vitro-in vivo models using 3D macroporous hydrogels for the study of liver cancer, Biomaterials, 159 (2018) 229–240. [DOI] [PubMed] [Google Scholar]
- [140].Hughes CS, Postovit LM, Lajoie GA, Matrigel: a complex protein mixture required for optimal growth of cell culture, Proteomics, 10 (2010) 1886–1890. [DOI] [PubMed] [Google Scholar]
- [141].Cruz-Acuna R, Garcia AJ, Engineered materials to model human intestinal development and cancer using organoids, Exp Cell Res, 377 (2019) 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Magno V, Meinhardt A, Werner C, Polymer Hydrogels to Guide Organotypic and Organoid Cultures, Advanced Functional Materials, 30 (2020) 2000097. [Google Scholar]
- [143].Cruz-Acuna R, Quiros M, Farkas AE, Dedhia PH, Huang S, Siuda D, Garcia-Hernandez V, Miller AJ, Spence JR, Nusrat A, Garcia AJ, Synthetic hydrogels for human intestinal organoid generation and colonic wound repair, Nat Cell Biol, 19 (2017) 1326–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Cruz-Acuna R, Quiros M, Huang S, Siuda D, Spence JR, Nusrat A, Garcia AJ, PEG-4MAL hydrogels for human organoid generation, culture, and in vivo delivery, Nat Protoc, 13 (2018) 2102–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Naranjo JD, Saldin LT, Sobieski E, Quijano LM, Hill RC, Chan PG, Torres C, Dziki JL, Cramer MC, Lee YC, Das R, Bajwa AK, Nossair R, Klimak M, Marchal L, Patel S, Velankar SS, Hansen KC, McGrath K, Badylak SF, Esophageal extracellular matrix hydrogel mitigates metaplastic change in a dog model of Barrett’s esophagus, Sci Adv, 6 (2020) eaba4526. [DOI] [PMC free article] [PubMed] [Google Scholar]


