Abstract
Purpose:
Intratumoral heterogeneity (ITH) challenges the molecular characterization of clear cell renal cell carcinoma (ccRCC) and is a confounding factor for therapy selection. Most approaches to evaluate ITH are limited by two-dimensional ex vivo tissue analyses. Dynamic Contrast Enhanced Magnetic Resonance Imaging (DCE-MRI) can noninvasively assess the spatial landscape of entire tumors in their natural milieu. To assess the potential of DCE-MRI, we developed a vertically-integrated radiogenomics co-localization approach for multi-region tissue acquisition and analyses. We investigated the potential of spatial imaging features to predict molecular subtypes using histopathologic and transcriptome correlatives.
Experimental Design:
We report the results of a prospective study of 49 patients with ccRCC who underwent DCE-MRI prior to nephrectomy. Surgical specimens were sectioned to match the MRI acquisition plane. RNA sequencing data from multi-region tumor sampling (80 samples) were correlated with percent enhancement on DCE-MRI in spatially co-localized regions of the tumor. Independently, we evaluated clinical applicability of our findings in 19 metastatic RCC patients (39 metastases) treated with first-line anti-angiogenic drugs or checkpoint inhibitors.
Results:
DCE-MRI identified tumor features associated with angiogenesis and inflammation, which differed within and across tumors, and likely contribute to the efficacy of anti-angiogenic and immuno-therapies. Our vertically-integrated analyses show that angiogenesis and inflammation frequently co-exist and spatially anti-correlate in the same tumor. Furthermore, MRI contrast enhancement identifies phenotypes with better response to anti-angiogenic therapy among metastatic RCC patients.
Conclusion:
These findings have important implications for decision models based on biopsy samples and highlight the potential of more comprehensive imaging-based approaches.
Keywords: Dynamic contrast enhanced MRI, Radiogenomics, Renal cell carcinoma, tumor heterogeneity, angiogenesis, inflammation
Introduction
Clear cell renal cell carcinoma (ccRCC), the most common subtype of kidney cancer (1–3), is considered both an angiogenic (4) and immunogenic tumor (5). Inactivation of the von Hippel-Lindau (VHL) gene in the majority of tumors (6) leads to upregulation of hypoxia inducible factors (HIFs), vascular endothelial growth factor (VEGF), and angiogenesis (7,8). This pathway paved the way for the development of molecularly targeted antiangiogenic therapies (AA; e.g. tyrosine kinase inhibitors [TKIs] targeting VEGF receptor 2 and neutralizing VEGF antibodies). More recently, immuno-oncology (IO) drugs (e.g., check-point inhibitors) have shown superior response rates and overall survival in metastatic ccRCC compared to TKIs (9–11). Yet, a percentage of patients, in particular those with low risk disease, may experience superior oncologic outcomes with TKI treatment compared to IO (11). Indeed, we recently showed that ccRCC that metastasize to the pancreas, responds better to TKIs than IO (12). Intriguingly, these tumors are characterized by intense arterial enhancement on contrast-enhanced imaging (12). These data suggest that imaging may be able to discriminate different subtypes of ccRCC. However, the role of imaging to predict response to therapy in metastatic ccRCC has been previously reported with conflicting and variable success (13–16). Nevertheless, these reports preceded the development of predictive molecular biomarkers of response in ccRCC. Here, we exploit a novel, vertically-integrated radiogenomics co-localization approach for multi-region tissue acquisition and analyses to assess the potential of spatial imaging features to predict molecular subtypes correlating with treatment response.
Growing evidence in ccRCC, indicates that angiogenesis and inflammation may be prognostic biomarkers (17,18). However, the lack of predictive biomarkers of response is arguably the most crucial knowledge gap in the management of ccRCC patients. Furthermore, a key confounding factor in developing tissue-based predictive biomarkers of therapy response is the intrinsic intra-tumoral heterogeneity (ITH) of ccRCC (19). Classification of ccRCC into two distinct subgroups, angiogenic and immunogenic, is likely an oversimplification (20,21). Angiogenic and immunogenic pathway activation likely coexist in many tumors and combination therapy with pembrolizumab (IO) and axitinib (AA) in patients with advanced kidney cancer demonstrated an impressive 59% objective response rate across all risk groups (22).
Dynamic contrast-enhanced (DCE) magnetic resonance imaging (MRI) can provide an evaluation of the entire tumor in vivo enabling non-invasive assessments of vascular tumor characteristics in the natural milieu (23). DCE MRI exploits the paramagnetic effect of extracellular gadolinium chelates, which shorten the T1 relaxation times of blood and tissues thereby increasing signal intensity on T1-weighted images (24). DCE MRI data can be analyzed using both quantitative and model-based (e.g., Tofts) approaches to extract anatomical variations of tumor vascular density and permeability (23,25).
We hypothesized that variations in MRI contrast enhancement in different regions within a given tumor could predict differences in molecular and histopathological features inherent to those regions. To test this hypothesis, we applied a radiomic strategy. We used first order statistics of the distribution of signal intensities within each tumor after administration of intravenous contrast as a surrogate of molecular ITH. Further, we implemented a radiogenomics platform using DCE MRI-based targeted tissue procurement combined with a comprehensive genomic and pathologic assessment to non-invasively decipher ITH in ccRCC. We demonstrate the ability of DCE MRI to predict activation of both angiogenic and inflammatory pathways in the same tumor, which may underlie therapeutic effectiveness of AA and IO combinations.
Patients and Methods
Patient selection criteria
This Health Insurance Portability and Accountability Act–compliant prospective study was approved by the Institutional Review Board (IRB # STU 052011–080) and carried out in accordance with the principles expressed in the Declaration of Helsinki. Written informed consent was obtained from all subjects prior to enrollment which included consent for molecular analysis of tissue samples, pathology, and DCE MRI, and their records were de-identified prior to the analysis. Inclusion criteria were: 1) ≥ 18 years of age, 2) known renal mass ≥ 2.5 cm scheduled for partial or radical nephrectomy, and 3) confirmed diagnosis of ccRCC at histopathology after surgery. Exclusion criteria included: 1) inability to undergo MRI, 2) renal insufficiency (estimated glomerular filtration rate < 30 mL/min/1.73 m2), 3) pregnancy, 4) limited image quality, 5) predominantly cystic (i.e., <25% enhancing) renal mass. 79 patients were prospectively enrolled and underwent MRI between August 2012 and March 2016. ccRCC was confirmed histologically after surgery in 58/79 patients. Tumor cellularity was adequate in 51/58 tumors. Tumor samples passed the quality control for RNA analyses in 49/58 patients (34 males) and constitute our study cohort. Pre-operative MRI included T1- and T2-weighted sequences, arterial spin labeled (ASL), diffusion weighed imaging (DWI), and dynamic contrast-enhanced (DCE) MRI. Mean age was 58 ± 9.8 (range 32–82). Mean time between MRI and surgery was 5 ± 4 days (range 1–19 days). This work presents analysis of DCE MRI data. MRI protocol is described in Supplementary Methods.
MR Image analysis
MR images were analyzed on the open-source Digital Imaging and Communications in Medicine (DICOM) viewer (OsiriX, version 5.6, 64 bit, Bernex, Switzerland). All tumors were manually segmented, and regions of interest (ROIs) were drawn by an MRI-fellowship trained radiologist (I.P.), in the center slice, along the boundary of the entire tumor and in areas subjectively exhibiting high and low enhancement levels. Non-enhancing areas were avoided. In homogeneous tumors, a single ROI was drawn. From the DCE MRI datasets, percentage tumor enhancement in ROIs was extracted at four phases (26): 1) before administration of gadolinium-based contrast agent (GBCA) (PRE); 2) during the first pass, where much of the contrast material resides within the renal capillary network in the cortex, approximately 30 seconds after initiation of its I.V. administration (corticomedullary, CM), 3) early nephrographic enhancement, where the contrast material filters through renal tubules, approximately 15 seconds after CM (eNG), and 4) frank nephrographic enhancement, where the contrast material reaches equilibrium in the renal parenchyma, approximately 55 seconds after CM (NG). Percentage (%) enhancement was defined as 100 × (SIpost – SIpre)/SIpre, where SIpost is the mean signal intensity within the ROI on each of the post-contrast phases (CM, eNG, and NG) and SIpre is the mean pre-contrast signal intensity within the ROI. Since Ktrans and Kep did not provide additional value, the rest of the analysis focused on % enhancement.
Retrospective analysis of metastatic RCC patients
Patient cohort:
Under IRB approval (IRB# STU 062012–017), we used Kidney Cancer Explorer, a i2b2-based database that integrates clinicopathologic data, to identify metastatic RCC patients who received first line TKI monotherapy or IO and underwent pre-treatment contrast-enhanced MRI. An ROI was drawn around metastases boundary by a 5th year radiology resident (C.S.), who was blinded to the therapy, in up to 5 metastases per patient in the pre-treatment NG-phase MRI. Metastases were classified as High (HE) or Low (LE) enhancement relative to the median % area enhancement across all tumors (Supplementary Methods). The largest dimension for each metastasis at pre-treatment MRI and available follow-up imaging (i.e., contrast-enhanced MRI or computed tomography) up to 3 months before any treatment changes were recorded. Progression per metastatic site was calculated based on RECIST 1.1 (27). A secondary analysis defining partial response as ≥10% decrease in largest diameter (28) was completed.
Histopathology analysis and immunostaining
Histopathology served as reference standard. Final diagnosis was provided by a uropathologist (P.K.). For accurate co-localization post radical or partial nephrectomy, the surgical specimen was positioned to match its anatomic orientation in vivo using fiducial markers placed during surgery by a urologist (23,29), and/or with a 3D-printed mold (30). Tumors were bivalved, and co-localized samples were collected from one half of the specimen and snap frozen. A section from the other half of the specimen was submitted for processing into formalin fixed paraffin embedded (FFPE) blocks. Depending on tumor size, several sections or a whole mount were generated. Snap frozen tumor samples were inked in different colors, and thin flanking sections were collected for FFPE blocks for tumor adequacy assessment by hematoxylin and eosin (H&E) staining. Tumor adjacent to flanking sections with >70% tumor cellularity was used for RNA extractions. To assess expression levels of various protein markers, tissue blocks were sliced in 4 μm thick sections and stained using H&E, and immunohistochemistry (IHC) was performed according to standard protocols (17,31) (Supplementary methods).
cDNA library construction, RNA sequencing, and analyses
Total RNA was isolated from tumor according to an established protocol (32). RNA samples with RIN >7 were used for further RNA sequencing. Sequencing data obtained from 180 tissue samples (49 patients) (Admera Health Inc. Austin, TX) were used. RNA sequencing libraries were prepared using the Illumina TruSeq Stranded mRNA high throughput sample preparation kit following the manufacturers’ protocol (Supplementary Methods). Sequencing data for patients specifically consenting to have their genomic data shared in a public database are deposited at the European Genome-phenome Archive (EGA), which is hosted by the EBI and the CRG, under accession number EGAS00001003846 with controlled access (https://ega-archive.org).
Statistical analysis
Patient and tumor characteristics were summarized with means ± standard deviations (SD), counts, and percentages. Semi-supervised category identification and assignment (SCINA) algorithm (https://github.com/jcao89757/SCINA) (33) was applied for tumor subtype prediction based on 7 transcriptional subclusters described by Motzer et al. (20) using gene expression of 10 different pathway specific gene sets. The 10 sets of signature genes were provided as input to SCINA. Six angiogenic genes (VEGFA, KDR, ESM1, CD34, PECAM1, and ANGPTL4) in clusters 1 and 2 were used to calculate pathway activity scores, which were utilized to separate angiogenic tumor samples from non-angiogenic samples. Similarly, 28 genes from T-effector, Cell Cycle, FAS/Pentose Phosphate, and FAO/AMPK signatures (clusters 4 and 5) were used to separate the T-effector/proliferative group from other groups. The same sets of angiogenic or T-effector/proliferative expression genes were used to calculate the Euclidean distances between samples and between tumors.
For the DCE heatmap, Spearman correlation analysis was applied between expression values of each gene in selected tumor samples (i.e., areas) and the corresponding % eNG enhancement. Genes were ranked based on the Spearman correlation coefficient (ρ) and overrepresentation enrichment analysis was performed on genes with highest positive/negative ρ values to identify the most correlated gene regulatory pathways with the web-tool (http://www.webgestalt.org/option.php). We selected genes within the top 10 correlated pathways from both sides and plotted scaled gene expression levels on the heatmap. For each gene, the expression levels were normalized by a min-max scaler. The gene expression mean value was calculated, and the expression values subtracted and then divided by this mean. For immune infiltration analyses, we applied eTME gene signatures coupled with the ssGSEA method, to predict the infiltration of immune cells (17,34). All reported p-values were obtained from one-tailed student’s t-tests at the 0.05 significance level, and false discovery rate (FDR) adjustments were applied to p-values (0.05 and 0.1 thresholds of significance, as indicated). ρ values were derived for all 27,051 genes. Statistical analyses were performed using SAS 9.4 (SAS Institute Inc., Cary, NC, Statistical Analysis System, RRID:SCR_008567) and R computing environment (version 3.3.2)(35).
Results
Patient Cohort and Implementation of Radiogenomics Platform for DCE MRI-based Tissue Assessment
Fig. 1 is a schematic representation of the workflow for image acquisition, tissue procurement, processing, and analyses. Tumors were classified based on the International Society of Urological Pathology (ISUP) grade. Patient and tumor characteristics are provided in Supplementary Table S1. Overall, 144 samples (49 tumors) were collected.
Fig. 1. Schematic of radiogenomics platform for DCE MRI-based tissue procurement and genomic analysis.
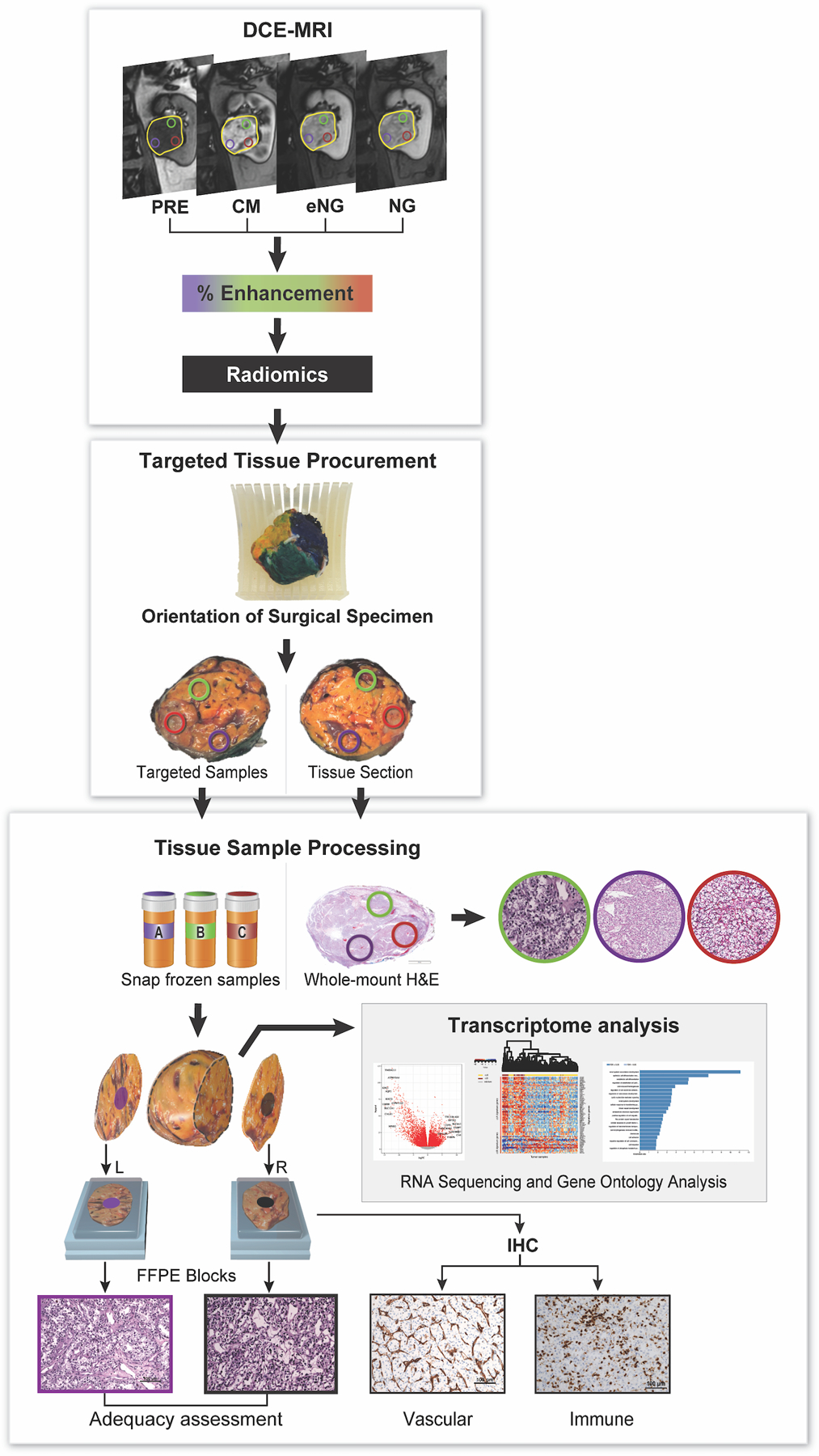
Patients underwent magnetic resonance imaging (MRI), including dynamic contrast-enhanced (DCE) MRI, for vascularity determination. Yellow circle delineates the entire tumor. Other colored circles represent the targeted regions of interest (tROIs) corresponding to areas of high and low enhancement on the DCE MRI. After surgical resection, each tumor was anatomically co-registered and sliced to match the imaging plane. Tissues samples from the same location in the tumor specimen and approximate size as the tROIs were collected as targeted samples. These samples were snap frozen and further processed for RNA extraction, and genomic analysis as indicated. The results of the gene expression analysis were then correlated with the MRI-derived measures. Immunohistochemistry (IHC) slides were generated from flanking sections of each targeted tissue sample for assessment of vascular and immune features. Tumors were also graded based on the International Society of Urological Pathology (ISUP) grading system. Overall, 45 out of 49 primary tumors had more than one tROI: 16 tumors had 2 tROIs, 17 tumors had 3 tROIs, 7 tumors had 4 tROIs, 3 tumors had 5 tROIs, 1 tumor had 6 tROIs and 1 tumor had 8 tROIs.
DCE MRI Identifies Angiogenic and Inflammatory Regions in ccRCC
We hypothesized that MRI-based tissue procurement of different tumor regions exhibiting high and low MRI enhancement in vivo would result in quantifiable differences in gene expression. First, we assessed the differences in gene expression patterns of primary tumors and renal parenchyma using all samples. Principal component analyses (PCA) confirmed distinct gene expression patterns in primary tumors (N= 49 tumors, 144 samples) compared to renal parenchyma (N = 36 samples) (Fig. 2A). We detected 3,296 genes differentially expressed in ccRCC (1,525 up-regulated and 1,771 down-regulated; FDR<0.05; fold change threshold, FC > 2) (Supplementary Table S2).
Fig. 2. Dissecting associations between DCE-MRI and angiogenic/inflammatory regions in ccRCC tumor samples.
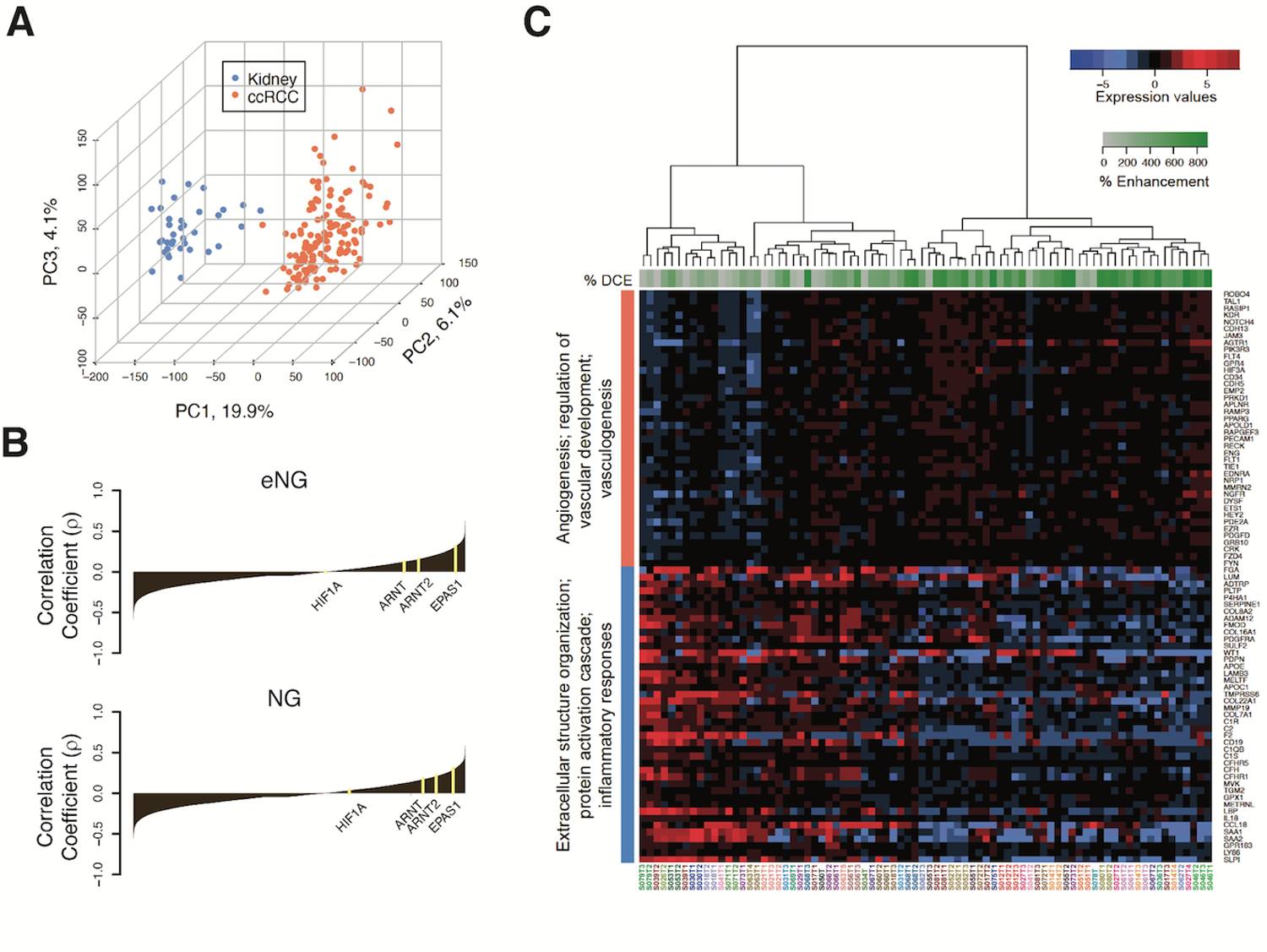
(A) Principal component analysis (PCA) plot generated by analyzing 27,051 genes using unsupervised multivariate method. Points represent 144 ccRCC samples (orange) and 36 kidney (uninvolved renal parenchyma) samples (blue), with axes showing the values of the first 3 PCs for every point, and percentage of variance indicated. (B) Waterfall plot of Spearman ranked correlation coefficients (ρ) between all genes analyzed and percent enhancement level for early nephrographic (eNG), and frank nephrographic (NG) post-contrast DCE phases. The ρ values were calculated from correlating percent enhancement on MRI with expression levels for 27,051 genes detected by RNA sequencing across all tumor samples. (C) Heatmap of semi-supervised hierarchical clustering of top 5 positive and top 5 negative enriched gene processes obtained through over representation gene ontology analysis. Normalized RNA expression values of non-redundant genes that correlated positively (coral vertical bar) and negatively (blue vertical bar) with % eNG enhancement are indicated. Genes are shown on the right side of the heatmap. Tumor samples (each column) are indicated below the heatmap and are color matched for samples from the same tumor. The % eNG enhancement for each sample is indicated with a green horizontal bar on the top. The green scale bar indicates the percent Enhancement values, and the blue-to-red scale indicates scaled gene expression values.
After careful evaluation of MRI, pictures taken during specimen anatomic orientation after surgery, gross appearance and shape of the tumor after sectioning, and location of tumor samples, we identified 37 tumors (80 samples) with at least 1 tumor sample with optimal co-localization between imaging and pathology. Using these 80 tumor samples, we correlated % enhancement in the tumor location corresponding to each biopsy with the entire gene expression profile. First order statistics were computed to obtain the % CM, eNG, and NG enhancement (Supplementary Table S3). Fig. 2B shows a waterfall plot of correlation coefficient values.
We then studied the ability MRI enhancement to identify areas with activation of angiogenic pathways (17,18). Analyses showed significant enrichment of RCC-specific processes such as angiogenesis and vascular development in both the eNG (Supplementary Table S4a), and NG phase images (Supplementary Table S4b). Analysis of the CM phase did not demonstrate enrichment of these processes (Supplementary Fig. S1; Table S4c), possibly due to technical factors (e.g., low temporal resolution of DCE-MRI). The signal intensity changes during the eNG and NG phases were more relevant surrogates for variations in gene expression.
A semi-supervised hierarchical clustering of top 5 positive and top 5 negative enriched gene processes obtained through over representation gene ontology analysis indicate that increased eNG enhancement reflects increased expression of genes associated with vascular development and angiogenic processes (FDR<0.1). Conversely, decreased eNG enhancement involves increased expression of genes associated with protein activation cascade, extracellular structure organization, and inflammatory response (FDR<0.1) (Fig. 2C, Supplementary Table S4a).
Top ten genes with highest positive correlation with eNG enhancement were further evaluated to assess their potential clinical significance. Analysis of the ccRCC cohort in The Cancer Genome Atlas (TCGA-KIRC) confirmed that high expression of SCN4A, PDZD2, RAVER2, and BTNL9 is associated with better outcomes. Similarly, we analyzed ten genes with the highest negative correlation with eNG enhancement using the TCGA-KIRC cohort. Eight of these 10 genes, KDELR3, MCTS1, UCHL1, UCN2, KIAA1211, CCNO, SAA1, and PP1R1A were negative prognostic biomarkers, where higher expression results in unfavorable outcomes (Supplementary Fig. S2).
DCE MRI Identifies Regions with Defined Gene Expression Signatures Predictive of Response to Anti-angiogenic Therapy
We hypothesized that differences in MRI enhancement would be predictive of distinct molecular alterations known to correlate with response to AA and IO therapies. We applied recently reported transcriptional signatures predicting differential clinical outcomes (20) to classify all of our 144 samples (49 tumors) into 7 subtypes with similar molecular features using SCINA (33) and calculated their pathway activity scores (Fig. 3A). Cluster proportions in our samples closely match those reported at the signature level (20): 1) Angio/Stromal (21 samples), 2) Angiogenic (55 samples), 3) Complement/Omega-ox (8 samples), 4) Proliferative (5 samples); 5) SnoRNA (4 samples), 6) Stromal/Proliferative (40 samples), and 7) T-eff/Proliferative (11 samples). Activity scores are overall consistent with reported pathway activation in each subtype (20), with observed differences likely attributed to small sample size and predominantly early stage tumors in our cohort.
Fig. 3. DCE MRI serves as a predictive marker for detecting tumor regions enriched for defined molecular signatures of therapy response.
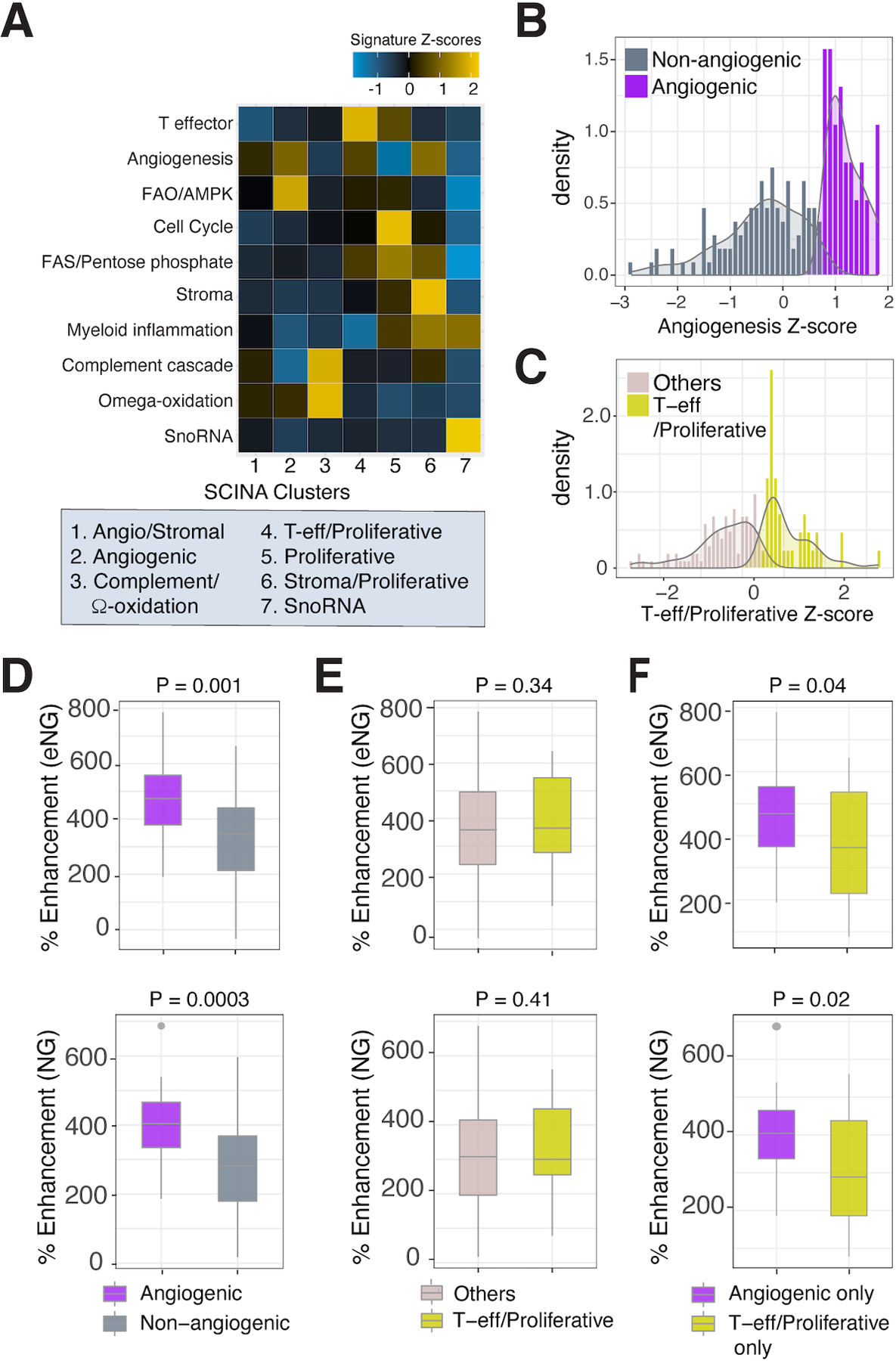
(A) Heatmap showing expression levels of transcriptome signature identified by Motzer et al (20) in SCINA clustering groups. Samples were clustered using the mean across samples for each gene, and the mean Z score for each signature was calculated, resulting in one Z score per signature per SCINA cluster. Histogram plots for the density of angiogenesis (B) and T-effector/Proliferative (C) Z-scores of all 144 tumor samples. (D) Box plot comparing % enhancement in the early (eNG) and frank(NG) nephrographic phases of DCE MRI between angiogenic and non angiogenic sample groups (80 tumor samples). (E) Box plot comparing % enhancement in the eNG and NG phases between T-effector/Proliferative and other genes group. (F) Box plots comparing % enhancement in the eNG (top) and NG (bottom) phases between exclusively angiogenic or T-effector/Proliferative signature enriched sample groups. All p-values are indicated in the figure.
Next, we evaluated whether MRI enhancement, a physiologic readout of microvessel density in ccRCC (23), can predict angiogenic and inflammatory features using prognostic transcription signatures (20). Given the relatively small number of samples in each of the 7 clusters, we selected 6 angiogenic signature genes (VEGFA, KDR, ESM1, CD34, PECAM1, ANGPTL4) shared between clusters 1 and 2 (i.e., enriched in responders to sunitinib (20)) to calculate the angiogenesis pathway activity scores at the sample level (144 samples). The histogram for density of angiogenesis Z-scores demonstrated a bimodal distribution (Fig. 3B), with two peaks and one valley. We selected the valley as a cutoff and classified samples with z-scores above/below the cutoff as ‘angiogenic’ (38/144) or ‘non-angiogenic’ (106/144) groups, respectively. We then used clusters 4 and 5 (i.e., enriched in responders to atezolizumab + bevacizumab (20)), to calculate the T-effector/Proliferative pathway activity scores. For this, the Cell Cycle and FAS/Pentose Phosphate signatures were found to be shared between the two clusters. Additionally, we included the highly enriched T-effector signature in cluster 4 and FAO/AMPK signature in cluster 5. Thus, these four signatures were combined to create a T-effector/Proliferative group. Our data again showed a bimodal distribution (Fig. 3C). Applying the valley as cut-off, tumor samples were divided into ‘T-effector/Proliferative’ (42/144) and ‘Other genes’ (102/144) groups.
We then focused on 80/144 samples (37 tumors) with precise radiology-pathology co-localization, and correlated the % CM, eNG and NG enhancement in the tumor location corresponding to each biopsy and molecular profiling of the same biopsy. As anticipated, we found higher % enhancement in samples classified as angiogenic (20/80) compared to non-angiogenic (60/80), especially in the eNG (p=0.001) and NG (p =0.0003) phases (Fig 3D, Supplementary Fig. S3). In contrast, we did not observe significant differences in % enhancement between T-effector/Proliferative and other genes (P>0.05) (Fig. 3E, Supplementary Fig S3). We also observed higher % enhancement in the eNG (p = 0.04) and NG (p=0.02) phases in samples assigned only to the angiogenic group (17/80) compared to those assigned only to T-effector/Proliferative (21/80) group (Fig 3F, Supplementary Table S5). Interestingly, 3/80 samples with DCE correlation (5/144 total) were co-enriched for both angiogenic and T-effector/proliferative signatures suggesting the possibility of heterogenous transcriptome even at the sample level. Overall, our data suggest that MRI enhancement can distinctly identify angiogenic molecular subtypes previously shown to correlate with improved response to AA therapy.
Lastly, we evaluated ITH in tumors with more than one sample (45/49). Using 7 molecular clusters (20), 17 tumors (38%) exhibited 1 cluster (i.e., homogeneous tumors), 22 (49%) showed 2 clusters, and 6 (13%) had 3 clusters. Thus, two thirds of tumors were heterogeneous when classified with the 7 molecular subcluster scheme (20). Using our angiogenic and non-angiogenic groups, 5 (11%) tumors were angiogenic, 25 (56%) were non-angiogenic, and 15 (33%) were mixed. In the T-effector/Proliferative vs. other signatures classification, 21 tumors (47%) were T-effector/proliferative, 6 tumors (13%) were classified as others, and 18 tumors (40%) were mixed. Thus, more than one third of tumors were heterogeneous using each of our clustering schemes. To assess the inter-tumor heterogeneity and ITH, we calculated the Euclidean distance between different samples within the same tumor and different tumors (Supplemental Fig. S4). Substantially smaller Euclidean distances within the same patients than across patients were observed for the angiogenic genes, while less so for non angiogenic genes. A similar trend was observed for the T-effector/proliferative compared with other genes group. These observations indicate that although overall transcriptomic features could be highly heterogeneous across different regions of the same tumors, angiogenesis and inflammation are relatively more stable and homogeneous within the same tumors.
DCE-MRI Enhancement Predicts Gene Expression of Vascular Markers and HIF Response Genes.
We next evaluated whether eNG enhancement can predict changes at the gene expression level of known vascular markers. We observed positive correlation between enhancement levels and established vascular markers (Supplementary Table S6). Since activation of the HIF-VEGF pathway is a key feature in ccRCC development, we specifically looked at the correlation between eNG enhancement and expression of reported HIF-2 (n=439), and HIF-1 (n=475) target genes (31,36). Six HIF-2 target genes correlated positively (FDR <0.05) with % eNG enhancement (Fig. 4A, B) and two of them (PDGFD, CD34) (37,38) have established roles in angiogenesis. Two HIF-2 target genes (MYBL2 and STEAP3) associated with cell proliferation and iron homeostasis (39,40) correlated negatively (FDR <0.05) with % eNG enhancement although have no established role in angiogenesis.
Fig. 4. Contrast enhancement on MRI predicts expression of HIF target genes.
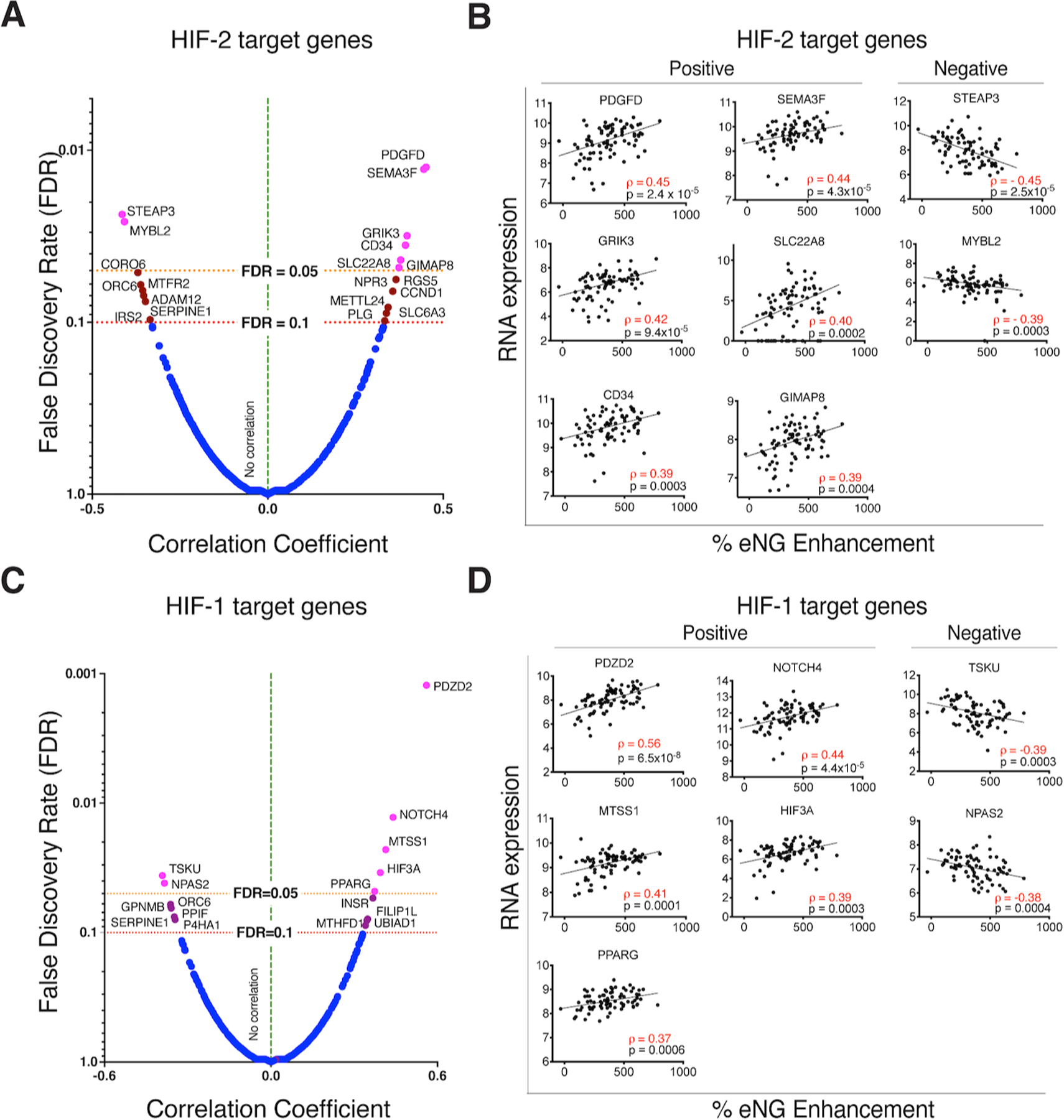
Volcano plot of ranked correlation coefficient values and corresponding false discovery rate (FDR) values between early nephrographic (eNG) enhancement and HIF-2 (A) and HIF-1 (C) target gene expression levels. Genes displayed to the right and left side of the vertical dotted line represent those exhibiting a positive and negative correlation, respectively. HIF-2 target genes that passed FDR<0.05 (magenta), and FDR <0.1 (brown) cutoffs are indicated. Correlation graphs between expression level and percent enhancement on MRI for top positively correlated, and top negatively correlated HIF-2 (B) and HIF-1 (D) target genes that passed FDR<0.1.
Similarly, we determined if % eNG enhancement can predict expression of HIF-1 target genes (36) (Fig. 4C, D). Five HIF-1 target genes correlated positively and two negatively (FDR<0.05) with eNG enhancement. The role of these in angiogenesis is controversial (41–44) or not established in ccRCC (45–47). We observed a significant positive correlation between eNG enhancement and RNA expression of HIF3A (rho=0.4, FDR=0.03). We did not find similar correlation for the other paralogs (HIF1A [rho=−0.003, FDR=0.99], HIF2A [rho=0.31, FDR=0.13], or the two HIF-β paralogs (ARNT [rho=0.09, FDR=0.85], ARNT2 [rho=0.15, FDR=0.65]. A trend toward positive correlation between HIF-2A and eNG enhancement (rho=0.31, p= 0.005, FDR=0.13) was not confirmed by IHC analysis (Supplementary Fig. S5). These results suggest that eNG enhancement can potentially serve as a readout for downstream physiological response to HIF targeting, rather than direct HIF expression itself. Moreover, HIF-2 may be a more relevant driver of tumor angiogenesis than HIF-1 in ccRCC. Alternatively, these genes may be uncoupled from HIF-2 and regulated via other processes.
DCE MRI Reports on ccRCC Tumor Microenvironment (TME)
We next sought to determine whether DCE MRI could reveal differences in immune cell infiltration within particular tumor areas. RNA-Seq data from the 80 co-localized samples (37 tumors) were analyzed using eTME gene signatures to predict immune-cell infiltration levels in tumor microenvironment. eTME signatures refer to a set of RCC specific immune cell/stroma markers derived empirically from comparison of expression data of human tumors and corresponding patient-derived xenografts (17). We classified our samples as ‘high’ or ‘low’ based on their % eNG enhancement being above or below the median % eNG enhancement (median cutoff 364%) (Fig. 5). Differences in eTME cell types based in mRNA expression (Fig. 5A) were confirmed at the protein expression level by IHC for samples in the top quartile (75 – 100 percentile; n = 19 samples) and bottom quartile (0–25 percentile; n=20 samples) % eNG enhancement. We confirmed higher enrichment of pericytes and endothelial cell type specific genes in high enhancing samples (Fig. 5A, P-values <0.02) compared to low enhancing samples; protein expression of CD31 and CD34 (vascular markers) were consistent with this finding (Fig. 5B). Similarly, we found increased expression of mast cell type-specific genes in high enhancing regions compared to low enhancing regions (P-value = 0.016). We found a subtle increase in cells expressing CD117, a mast cell marker, in high enhancing samples (1–2% staining positive) compared to low enhancing samples (<1% staining positive) (Fig. 5C). The role of infiltrating mast cells as promoter of angiogenesis in RCC TME was recently described (38). Notably, CD8+ T cell enrichment did not correlate with % eNG enhancement (Fig. 5A, C). These findings suggest that T-cell infiltration was more uniform in our cohort with higher representation of smaller ccRCCs, compared to heterogeneous T-cell infiltration commonly present in larger tumors. Further, low enhancing samples showed higher enrichment of B-cells, macrophages, and eosinophils (P-values <0.05) (Fig. 5A) supporting the hypothesis that decreased tumor vascularity is associated with inflammation. A higher percentage of cells staining positively for CD20 (B-cell marker) and CD163 (macrophage marker) in low enhancing samples compared to high enhancing samples was not statistically significant (Fig. 5C).
Fig. 5. DCE-MRI enhancement predicts enrichment of eTME gene signatures.
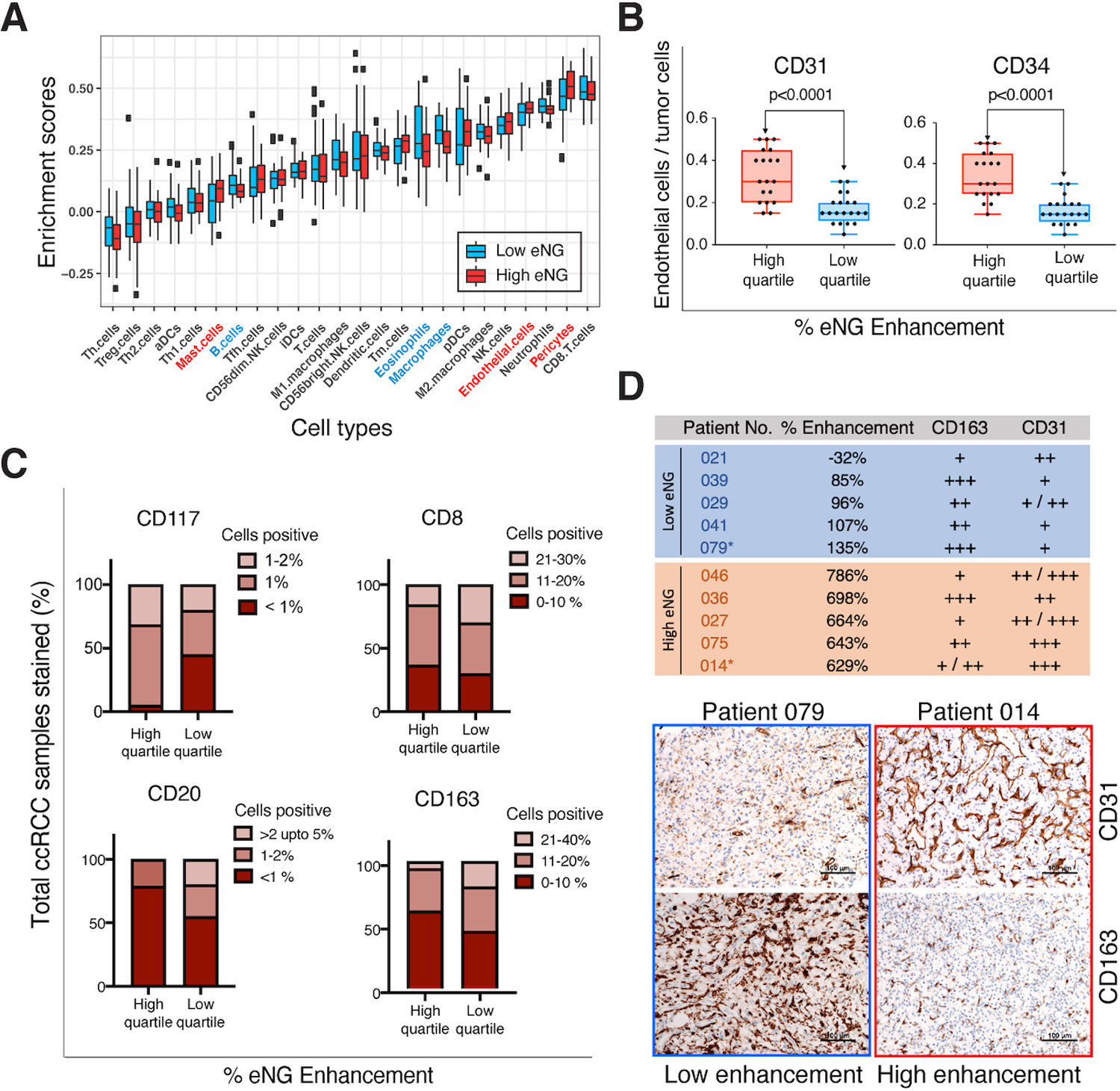
(A) Box plot of eTME gene clusters with corresponding ssGSEA enrichment score. The enrichment scores were calculated independently for each cell type based on specific gene signatures unique to those cell types. Blue and red boxes represent median low and high early nephrographic (eNG) enhancement level on DCE, respectively. The immune cell types that were statistically enriched (p<0.05) in the tissue samples corresponding to low enhancing tumor regions are represented in blue, and those enriched in tissue samples obtained in high enhancing areas on DCE are represented in red. The p-values for the enriched cell types that are significantly different between tumor samples corresponding to high and low enhancing areas on MRI are as follows: 1) Macrophages (p = 0.003), 2) Pericytes (p = 0.003), 3) Endothelial cells (p = 0.02), 4) Eosinophils (p = 0.02), 5) Mast cells (p = 0.02), and 6) B-cells (p = 0.03). Th.cells: T-helper cells, Tfh.cells: T follicular helper cells, Treg.cells: regulatory T-cells, aDCs: activated dentritic cells, iDCs: immature dentritic cells, pDCs: plasmacytoid dentritic cells, Tm cells: memory T cells. NK cells: natural killer cells. (B) Tumors were classified into high and low enhancing based on median percent enhancement (Median cutoff: 364% increased signal between pre-contrast [PRE] and early nephrographic [eNG phases]). Histological validation of differences in vascular marker expression (CD31, and CD34) in the study cohort grouped into high enhancing (High quartile) and low enhancing samples (Low quartile). (C) Similarly, protein expression level differences in eTME cell type markers were assessed by IHC for samples in the top quartile enhancement (n = 19 samples), and bottom quartile enhancement (n=20 samples) values as indicated in the figure. (D) Top panel indicates the pathologist’s assessment of CD31 and CD163 expression levels from IHC slides for 5 representative high and low enhancing tumor samples on the early nephrographic (eNG) phase. Patient number and corresponding eNG enhancement values derived from first order statistics are as indicated. Bottom panel shows the representative IHC images of CD31 and CD163 expression in one sample from each group as indicated. High enhancing tumor sample indicated in yellow, and low enhancing tumor sample indicated in blue.
Next, we explored the relationship of tumor angiogenesis and inflammation within areas of high and low-level % eNG enhancement at the histopathology level. Fig. 5D shows the qualitative assessment of CD31 and CD163 expression levels in representative tissue samples corresponding to those with the highest (n=5), and lowest (n=5) % eNG enhancement. Low enhancing samples exhibited less vascular staining (CD31) and more prominent inflammatory staining (CD163), whereas high enhancing samples exhibited prominent vascular and weak inflammatory IHC staining. This is consistent with a previously described association between a ‘hypoxic gene signature’, and a high innate and adaptive gene expression signature in ccRCC (48). Together, these results confirm that ITH in specific TME gene signatures can be captured by changes in DCE MRI. Furthermore, our data indicate that angiogenesis and inflammation tend to anti-correlate within the same tumor.
MRI Contrast Enhancement Identifies Phenotypes with Better Response to AA Therapy Among Metastatic clear cell renal cell carcinoma (ccRCC) Patients
We hypothesized that ccRCC exhibiting more robust enhancement would have heightened angiogenesis and be more responsive to TKI therapy. Contrarily, tumors exhibiting less enhancement would be less angiogenic, with increased inflammatory mediators and benefit from IO therapy. We identified 19 consecutive metastatic ccRCC patients who received first line AA monotherapy (Sunitinib, Sorafenib, Pazopanib, or Axitinib) or IO (Nivolumab as monotherapy or in combination with Ipilimumab) and underwent pre-treatment multiphasic contrast-enhanced MRI (Supplementary Table S7). Of these, 12 patients with 22 metastases received AA and 7 patients with 17 metastases received IO. Median follow-up after initiation of therapy was 92 days (range 21–182 days). We classified metastases as High (HE), or Low (LE) enhancement based on the median percent area of NG enhancement of all metastases (Supplementary Methods). Overall, 10/22 and 12/22 metastases receiving AA were classified as HE and LE, respectively. Out of 17 metastases treated with IO, 9 were HE and 8 were LE. Overall, 8 metastases showed progressive disease (PD), 24 stable disease (SD), 7 partial response (PR), and none complete response (CR). 4/22 metastases exhibited PR to AA whereas 3/17 metastases showed PR to IO (Supplementary Table S7).
There was significant association between different groups (HE AA, LE AA, HE IO, LE IO) and response to therapy (p<0.0001), which remained significant when adjusted for IMDC and MSKCC scores (p<0.0001). However, there was no correlation between the IMDC and MSKCC inputs and likelihood of response to AA and IO. No HE AA metastases (0 out of 10) progressed while 5 out of 9 HE IO metastases progressed. On a per-lesion basis, HE AA metastases had longer PFS than HE IO metastases (adjusted p <0.0001; Supplementary Table S7). HE AA metastases also exhibited a longer PFS than LE AA metastases (adjusted p <0.0001). Notably, only 1 of the 8 LE IO metastases progressed (Fig. 6A) although differences in PFS between LE IO and HE IO did not reach statistical significance (Adj P = 0.62; Supplementary Table S7). There was no significant difference in tumor response by RECIST 1.1 (p value = 0.12). The HE AA metastases exhibited the highest PR rate using 10% decrease in longest diameter (6 out of 10 metastatic lesions), a superior reported criteria for metastatic RCC receiving TKI (28). However, these differences were not statistically significant (P=0.17) (Fig. 6B). Representative images in three metastatic ccRCC patients are shown in Fig 6C.
Fig. 6. MRI Contrast Enhancement identifies phenotypes with better response to anti-angiogenic (AA) therapy among metastatic clear cell renal cell carcinoma (ccRCC) patients.
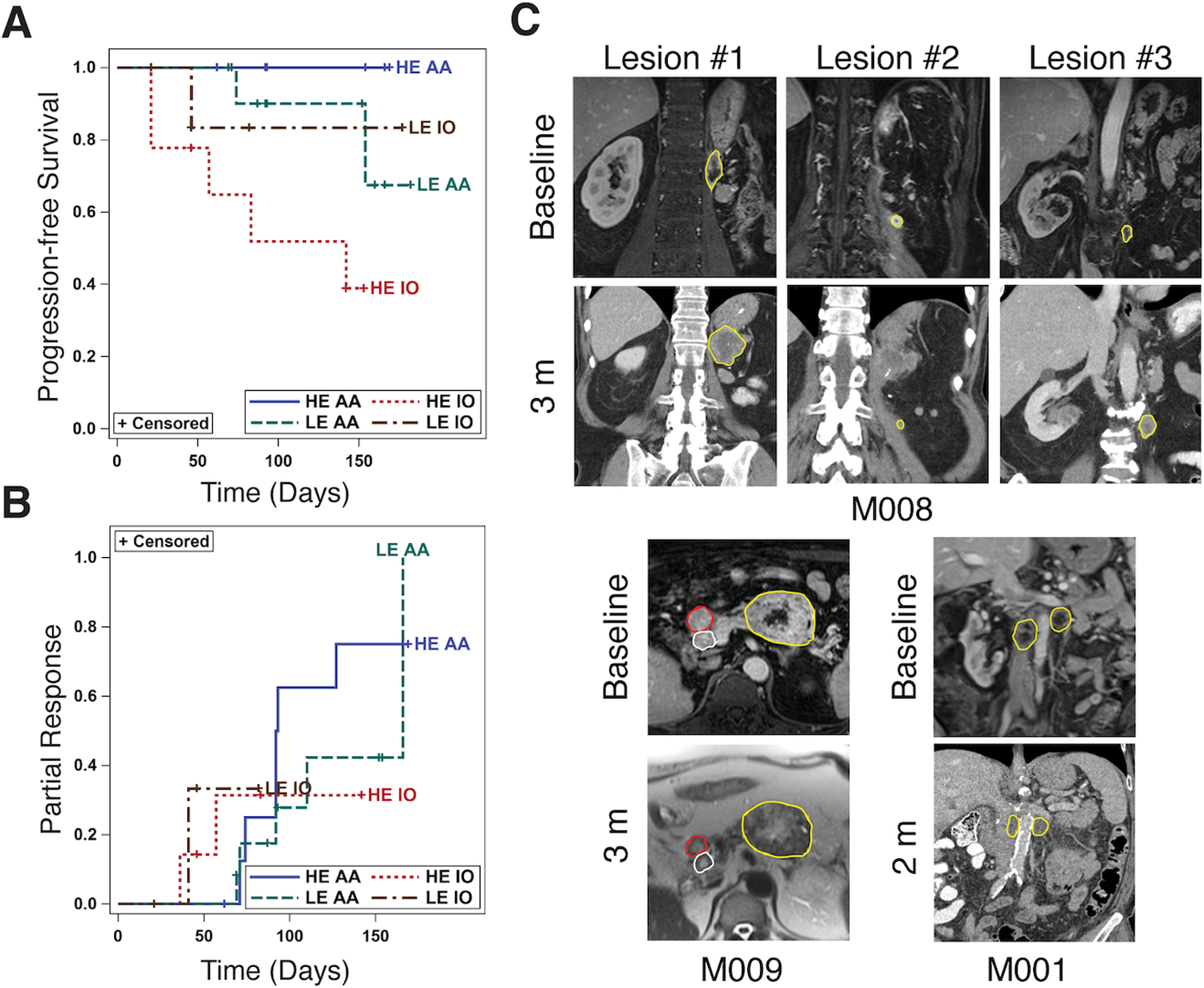
(A) Kaplan-Meier plot of the progression-free survival in different subgroups based on % area enhancement (High enhancement [HE] and Low enhancement [LE]) on nephrographic (NG) phase at DCE MRI; HE AA represents lesions with high % area enhancement treated with AA (brown); LE AA represents lesions with low % area enhancement treated with AA (red); HE IO represents lesions with high % area enhancement treated with IO (green); LE IO represents lesions with low % area enhancement treated with IO (blue). The number of lesions that progressed (defined as >20% increase in longest lesion diameter compared to baseline), out of total number of lesions for each subgroup are: 1) LE IO: 1/8 (13%), 2) HE IO: 5/9 (55%), 3) LE TKI: 2/12 (15%), and HE TKI: 0/10 (0%). (B) Kaplan-Meier plot of the secondary analyses of the probability of partial response. The number of lesions that partially responded (defined as at least 10% decrease in longest lesion diameter compared to baseline), out of total number of lesions in each subgroup are: 1) LE IO: 2/8 (25%), 2) HE IO: 2/9 (22%), 3) LE TKI: 5/12, and 4) HE TKI: 6/10 (60%). (C) Representative images in 3 patients with metastatic ccRCC. Metastatic lesions are delineated with colored regions of interest (ROI). Top panel: Patient M008 had 3 metastatic deposits in the retroperitoneum at baseline. Lesion 1 and 3 were classified as LE whereas lesion 2 was classified as HE at baseline. Patient received AA therapy with Pazopanib and 3 months after initiation of therapy, a contrast enhanced computed tomography (CT) shows marked increase in size of lesions 1 and 3 (progressive disease) but mild decrease in size of lesion 2 representing partial response. Left panel: Patient M009 had 3 metastases in the pancreas, two of them were used as target lesions (red and yellow ROI). At baseline, the largest mass (yellow ROI) was classified as LE whereas the smaller pancreatic metastasis (red ROI) was classified as HE. Patient received AA therapy with Pazopanib and 3 months after initiation of therapy, a non-contrast MRI revealed unchanged size of the largest mass (yellow ROI, stable disease) and decrease in size of the smaller lesion (red ROI, partial response). A third lesion exhibiting intense enhancement at baseline (white ROI) also exhibited partial response at 3 months. Right panel: Patient M001 had two retroperitoneal lymph nodes classified as LE at baseline. Patient received IO with Ipilimumab/Nivolumab and a two month follow up contrast-enhanced CT shows decrease in size of both lymph nodes (partial response), which remain hypoenhancing.
Discussion
Our analyses support the ability of DCE MRI to detect co-existence of angiogenic and inflammatory areas within the same tumor. We observed increased expression of several genes involved in vascular processes in tumors samples obtained in areas with intense early (eNG) and frank (NG) nephrographic enhancement. Furthermore, assessment of regions with low eNG enhancement revealed higher expression of complement genes and a rich inflammatory signature (49). Our data are particularly relevant in patients with advanced RCC, in whom a decision among available first line therapeutic regimens remain empirical. Whereas combination AA/IO regimens may be able to overcome the challenge of tumor heterogeneity, this approach offers no benefit for tumors that are resistant to one of the two drugs and results in unnecessary side effects. Based on our findings, it is plausible that patients with homogeneous hyper-enhancing tumors may respond more favorably to AA whereas IO would be preferable in those with extensive hypo-enhancement.
Previous DCE MRI reports have focused on differentiating renal masses with different histology (50,51). Studies in metastatic RCC preceding the era of checkpoint inhibitors demonstrated that DCE MRI-derived metrics are pharmacodynamic biomarkers for the TKI, sorafenib (15). However, conflicting results were found in their ability to predict PFS (13,15). There are important differences in our study. First, sorafenib is a first generation TKI with proven inferior oncologic benefit compared to newer TKIs. Second, prior studies included non-clear cell histology. Third, to our knowledge, we report the first correlation between DCE MRI enhancement and molecular signatures in ccRCC
Intratumor heterogeneity (ITH) is thought to be associated with treatment failure and therapy resistance (19,52,53). Swanton and colleagues (52) reported an exon-capture multi-region sequencing in ccRCC, where primary nephrectomy tumor samples showed only 34% of the detected mutations shared between sample sites. Similarly, the TRACERx Renal study (54) proposed grouping ccRCCs genomically into different evolutionary subtypes that exhibit different clinical outcomes. However, clinical implementation of such approach is not without challenges. Characterization of ITH with multiregional sequencing to guide clinical decision making would be cost-prohibitive and technically difficult. Indeed, TRACERx data suggest an average of 7 biopsies per tumor to detect >75% of all driver variants in the same tumor. Our data using modern predictive gene signatures (20) indicate that up to two thirds of renal masses would be incorrectly characterized using a single biopsy due to ITH, despite enrichment of small renal masses (i.e., presumably more homogeneous) in our cohort. Thus, the variable molecular landscape is not completely reflected with clinical biopsies offering limited tissue samples. Moreover, although histologic evaluation of tumor vascularity can delineate blood vessel density, it does not offer a true physiologic assessment of the vascular network and blood flow. Whole-tumor imaging and a more comprehensive histopathologic analysis of available tissue samples will undoubtedly help reveal and decipher ITH allowing for better predictive and prognostic models.
Recently, metastatic patients in angiogenic and angiogenic/stromal subsets (20) showed improved PFS with AA, while patients in T-effector/proliferative subsets showed improved PFS with IO. Our results are consistent with those findings and suggest that angiogenic clusters can be identified non-invasively. High-enhancing untreated metastases showed better PFS when treated with AA than hypoenhancing metastases receiving AA or any metastases receiving IO. Similarly, we recently demonstrated that high baseline perfusion measured by arterial spin labeled MRI, a technique that estimates tissue perfusion in ccRCC (23,55), correlates with response to AA therapy in metastatic RCC (56). Furthermore, these data are also consistent with our finding in ccRCC patients with pancreatic metastases, which exhibit intense enhancement on contrast-enhanced imaging, indolent oncologic behavior and histopathology, prominent vasculature, and a strikingly favorable response to frontline AA therapy but are refractory to IO therapy (12). In contrast, more aggressive forms of ccRCC have higher levels of immune cell infiltration, lower angiogenesis, and respond better to IO therapy (17,18). We noted longer PFS and higher responses in metastases with low enhancement undergoing IO therapy although the small sample size precluded reaching statistical significance. Other factors may have contributed to these findings and more sophisticated image analysis, including radiomics and machine learning, may offer better assessment of disease response. Combination therapies may be particularly helpful in patients exhibiting both imaging phenotypes although further validation is needed.
Our study has some limitations. First, the proposed image-based tissue procurement relies heavily on the co-localization efforts between pre-surgical imaging and post-surgical histologic analysis. Despite using fiducial markers placed during surgery to orient the specimen, some misalignment between imaging and the tissue specimen became apparent in some cases. We addressed this by including only the 37 tumors (80 samples) for which we had high confidence in the co-localization of imaging and biopsy sites. Second, our study cohort was enriched for small renal tumors, which tend to exhibit low grade histology and indolent behavior. Further studies will focus on larger tumors and assessment of imaging features as prognostic biomarkers. Lastly, clinical validation of our findings in patients with metastatic ccRCC receiving AA or IO therapy was completed in a small cohort and require further validation.
In conclusion, we have established a radiogenomics platform using DCE MRI, co-localized tissue procurement methods, correlation of transcriptome levels with imaging features, and immunohistochemical validation to detect, for the first time, molecular heterogeneity in ccRCC non-invasively. We have demonstrated that imaging can differentiate heterogeneity in both angiogenic and immunologic features within the same tumor. Our platform provides a first step to overcome limitations of percutaneous biopsies in informing physicians regarding prognosis and treatment of heterogeneous ccRCC tumors.
Supplementary Material
Translational Relevance.
We have implemented a novel vertically-integrated radiogenomics platform to non-invasively decipher tumor heterogeneity in clear cell renal cell carcinoma (ccRCC). This prospective study combined the analyses of in vivo phenotypes from dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) with histopathological and molecular assessment of multi-region tumor samples obtained from spatially co-localized tumor areas. We postulated that differences in enhancement at DCE-MRI reflect unique underlying histopathological and molecular features. Using an MRI-driven targeted tissue procurement and analysis, we demonstrate that imaging phenotypes correlate with heterogenous molecular signatures associated with angiogenesis and inflammation. Notably, MRI phenotypes identify a subgroup of metastatic lesions that are more likely to respond to antiangiogenic therapy. This study uncovers the potential of non-invasive, image-based whole-tumor predictive biomarkers to assist in the selection of optimal therapeutic regimens.
Acknowledgements
We thank the patients who provided their time to be scanned for MRI studies, and generously provided tissues for analyses. The authors would like to thank Trevor Wigal RT(R), MR, Kelli Key, Ph.D., and Christina Carrigan R.N., for patient recruitment and MRI scans, Admera Health/Genohub for RNA sequencing analyses, Oreoluwa Onabolu B.A., for his technical assistance with the histology, Stacy Wang who assisted with data uploading, and Erin Moore M.A., Senior graphic designer, UTSW Radiology creative services for assistance with Fig. 1 illustration.
Financial support:
This work was partially funded by grants NIH # 5R01CA154475 (D. Udayakumar, Y. Xi, D. Dwivedi, P. Kapur, M. Fulkerson, S. Haldeman, Q. Yuan, J. Guo, J. Cadeddu, I. Pedrosa), P50CA196516 (J. Cadeddu, P. Kapur, A. Christie, R. McKay, I. Pedrosa, J. Brugarolas), U01CA207091 (A. Madhuranthakam, I. Pedrosa), R01CA258584 (Z. Zhang, T. Wang), CPRIT RP190208 (T. Wang), and Cancer Center Support Grant 5P30CA142543.
Footnotes
Conflict of interest disclosure statement:
I. P. served on scientific advisory boards of Bayer Healthcare and Merck, holds stocks options at Health Tech International, and holds joint patents with Philips Healthcare without royalties. I.P. and A.J.M. report receiving institutional non-financial research support from Philips Healthcare. No potential competing interests were disclosed by the other authors.
References
- 1.Kondo K, Kim WY, Lechpammer M, Kaelin WG, Jr. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol 2003;1(3):E83 doi 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen C, Kaelin WG, Jr. The VHL/HIF axis in clear cell renal carcinoma. Semin Cancer Biol 2013;23(1):18–25 doi 10.1016/j.semcancer.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 2007;11(4):335–47 doi 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gnarra JR, Zhou S, Merrill MJ, Wagner JR, Krumm A, Papavassiliou E, et al. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci U S A 1996;93(20):10589–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chevrier S, Levine JH, Zanotelli VRT, Silina K, Schulz D, Bacac M, et al. An Immune Atlas of Clear Cell Renal Cell Carcinoma. Cell 2017;169(4):736–49e18 doi 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clinical cancer research : an official journal of the American Association for Cancer Research 2008;14(15):4726–34 doi 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giaccia AJ, Simon MC, Johnson R. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev 2004;18(18):2183–94 doi 10.1101/gad.1243304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruick RK, McKnight SL. Building better vasculature. Genes Dev 2001;15(19):2497–502 doi 10.1101/gad.931601. [DOI] [PubMed] [Google Scholar]
- 9.Alsharedi M, Katz H. Check point inhibitors a new era in renal cell carcinoma treatment. Med Oncol 2018;35(6):85 doi 10.1007/s12032-018-1147-y. [DOI] [PubMed] [Google Scholar]
- 10.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med 2015;373(19):1803–13 doi 10.1056/NEJMoa1510665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motzer RJ, Tannir NM, McDermott DF, Aren Frontera O, Melichar B, Choueiri TK, et al. Nivolumab plus Ipilimumab versus Sunitinib in Advanced Renal-Cell Carcinoma. N Engl J Med 2018;378(14):1277–90 doi 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Singla N, Xie Z, Zhang Z, Gao M, Yousuf Q, Onabolu O, et al. Pancreatic tropism of metastatic renal cell carcinoma. JCI Insight 2020;5(7) doi 10.1172/jci.insight.134564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaherty KT, Rosen MA, Heitjan DF, Gallagher ML, Schwartz B, Schnall MD, et al. Pilot study of DCE-MRI to predict progression-free survival with sorafenib therapy in renal cell carcinoma. Cancer Biol Ther 2008;7(4):496–501 doi 10.4161/cbt.7.4.5624. [DOI] [PubMed] [Google Scholar]
- 14.Fournier LS, Oudard S, Thiam R, Trinquart L, Banu E, Medioni J, et al. Metastatic renal carcinoma: evaluation of antiangiogenic therapy with dynamic contrast-enhanced CT. Radiology 2010;256(2):511–8 doi 10.1148/radiol.10091362. [DOI] [PubMed] [Google Scholar]
- 15.Hahn OM, Yang C, Medved M, Karczmar G, Kistner E, Karrison T, et al. Dynamic contrast-enhanced magnetic resonance imaging pharmacodynamic biomarker study of sorafenib in metastatic renal carcinoma. J Clin Oncol 2008;26(28):4572–8 doi 10.1200/JCO.2007.15.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith AD, Zhang X, Bryan J, Souza F, Roda M, Sirous R, et al. Vascular Tumor Burden as a New Quantitative CT Biomarker for Predicting Metastatic RCC Response to Antiangiogenic Therapy. Radiology 2016;281(2):484–98 doi 10.1148/radiol.2016160143. [DOI] [PubMed] [Google Scholar]
- 17.Wang T, Lu R, Kapur P, Jaiswal BS, Hannan R, Zhang Z, et al. An Empirical Approach Leveraging Tumorgrafts to Dissect the Tumor Microenvironment in Renal Cell Carcinoma Identifies Missing Link to Prognostic Inflammatory Factors. Cancer Discov 2018;8(9):1142–55 doi 10.1158/2159-8290.CD-17-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDermott DF, Huseni MA, Atkins MB, Motzer RJ, Rini BI, Escudier B, et al. Clinical activity and molecular correlates of response to atezolizumab alone or in combination with bevacizumab versus sunitinib in renal cell carcinoma. Nat Med 2018;24(6):749–57 doi 10.1038/s41591-018-0053-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlinger M, Horswell S, Larkin J, Rowan AJ, Salm MP, Varela I, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nature genetics 2014;46(3):225–33 doi 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motzer RJ, Banchereau R, Hamidi H, Powles T, McDermott D, Atkins MB, et al. Molecular Subsets in Renal Cancer Determine Outcome to Checkpoint and Angiogenesis Blockade. Cancer cell 2020;38(6):803–17e4 doi 10.1016/j.ccell.2020.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brugarolas J, Rajaram S, Christie A, Kapur P. The Evolution of Angiogenic and Inflamed Tumors: The Renal Cancer Paradigm. Cancer cell 2020;38(6):771–3 doi 10.1016/j.ccell.2020.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rini BI, Plimack ER, Stus V, Gafanov R, Hawkins R, Nosov D, et al. Pembrolizumab plus Axitinib versus Sunitinib for Advanced Renal-Cell Carcinoma. N Engl J Med 2019;380(12):1116–27 doi 10.1056/NEJMoa1816714. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Kapur P, Yuan Q, Xi Y, Carvo I, Signoretti S, et al. Tumor Vascularity in Renal Masses: Correlation of Arterial Spin-Labeled and Dynamic Contrast-Enhanced Magnetic Resonance Imaging Assessments. Clin Genitourin Cancer 2016;14(1):e25–36 doi 10.1016/j.clgc.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao D, Ai T, Goerner F, Hu X, Runge VM, Tweedle M. MRI contrast agents: basic chemistry and safety. J Magn Reson Imaging 2012;36(5):1060–71 doi 10.1002/jmri.23725. [DOI] [PubMed] [Google Scholar]
- 25.Tofts PS. Modeling tracer kinetics in dynamic Gd-DTPA MR imaging. J Magn Reson Imaging 1997;7(1):91–101 doi 10.1002/jmri.1880070113. [DOI] [PubMed] [Google Scholar]
- 26.Zhen Jane Wang MSD, Silverman Stuart G., Chandarana Hersh, Doshi Ankur, Israel Gary M., Leyendecker John R., Pedrosa Ivan, Raman Steve, Remer Erick M., Shinagare Atul B., Smith Andrew D., Vikram Raghu. MRI renal mass protocol v1.0. https://cdn.ymaws.com/www.abdominalradiology.org/resource/resmgr/education_dfp/RCC/RCC.MRIprotocolfinal-7-15-17.pdf.
- 27.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). European journal of cancer 2009;45(2):228–47 doi 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 28.Krajewski KM, Guo M, Van den Abbeele AD, Yap J, Ramaiya N, Jagannathan J, et al. Comparison of four early posttherapy imaging changes (EPTIC; RECIST 1.0, tumor shrinkage, computed tomography tumor density, Choi criteria) in assessing outcome to vascular endothelial growth factor-targeted therapy in patients with advanced renal cell carcinoma. European urology 2011;59(5):856–62 doi 10.1016/j.eururo.2011.01.038. [DOI] [PubMed] [Google Scholar]
- 29.Yuan Q, Kapur P, Zhang Y, Xi Y, Carvo I, Signoretti S, et al. Intratumor Heterogeneity of Perfusion and Diffusion in Clear-Cell Renal Cell Carcinoma: Correlation With Tumor Cellularity. Clin Genitourin Cancer 2016;14(6):e585–e94 doi 10.1016/j.clgc.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dwivedi DK, Chatzinoff Y, Zhang Y, Yuan Q, Fulkerson M, Chopra R, et al. Development of a Patient-specific Tumor Mold Using Magnetic Resonance Imaging and 3-Dimensional Printing Technology for Targeted Tissue Procurement and Radiomics Analysis of Renal Masses. Urology 2018;112:209–14 doi 10.1016/j.urology.2017.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen W, Hill H, Christie A, Kim MS, Holloman E, Pavia-Jimenez A, et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 2016;539(7627):112–7 doi 10.1038/nature19796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pena-Llopis S, Brugarolas J. Simultaneous isolation of high-quality DNA, RNA, miRNA and proteins from tissues for genomic applications. Nat Protoc 2013;8(11):2240–55 doi 10.1038/nprot.2013.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, Luo D, Zhong X, Choi JH, Ma Y, Wang S, et al. SCINA: A Semi-Supervised Subtyping Algorithm of Single Cells and Bulk Samples. Genes (Basel) 2019;10(7) doi 10.3390/genes10070531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462(7269):108–12 doi 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S ppcor: An R Package for a Fast Calculation to Semi-partial Correlation Coefficients. Commun Stat Appl Methods 2015;22(6):665–74 doi 10.5351/CSAM.2015.22.6.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, et al. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 2005;105(2):659–69 doi 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- 37.Lopez JI, Erramuzpe A, Guarch R, Cortes JM, Pulido R, Llarena R, et al. CD34 immunostaining enhances a distinct pattern of intratumor angiogenesis with prognostic implications in clear cell renal cell carcinoma. APMIS 2017;125(2):128–33 doi 10.1111/apm.12649. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Yuan W, Wu L, Tang Q, Xia Q, Ji J, et al. PDGF-D promotes cell growth, aggressiveness, angiogenesis and EMT transformation of colorectal cancer by activation of Notch1/Twist1 pathway. Oncotarget 2017;8(6):9961–73 doi 10.18632/oncotarget.14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han M, Xu R, Wang S, Yang N, Ni S, Zhang Q, et al. Six-Transmembrane Epithelial Antigen of Prostate 3 Predicts Poor Prognosis and Promotes Glioblastoma Growth and Invasion. Neoplasia 2018;20(6):543–54 doi 10.1016/j.neo.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Musa J, Aynaud MM, Mirabeau O, Delattre O, Grunewald TG. MYBL2 (B-Myb): a central regulator of cell proliferation, cell survival and differentiation involved in tumorigenesis. Cell Death Dis 2017;8(6):e2895 doi 10.1038/cddis.2017.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bonyadi Rad E, Hammerlindl H, Wels C, Popper U, Ravindran Menon D, Breiteneder H, et al. Notch4 Signaling Induces a Mesenchymal-Epithelial-like Transition in Melanoma Cells to Suppress Malignant Behaviors. Cancer Res 2016;76(7):1690–7 doi 10.1158/0008-5472.CAN-15-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hainaud P, Contreres JO, Villemain A, Liu LX, Plouet J, Tobelem G, et al. The role of the vascular endothelial growth factor-Delta-like 4 ligand/Notch4-ephrin B2 cascade in tumor vessel remodeling and endothelial cell functions. Cancer Res 2006;66(17):8501–10 doi 10.1158/0008-5472.CAN-05-4226. [DOI] [PubMed] [Google Scholar]
- 43.Wu G, Chen Z, Li J, Ye F, Chen G, Fan Q, et al. NOTCH4 Is a Novel Prognostic Marker that Correlates with Colorectal Cancer Progression and Prognosis. J Cancer 2018;9(13):2374–9 doi 10.7150/jca.26359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dufraine J, Funahashi Y, Kitajewski J. Notch signaling regulates tumor angiogenesis by diverse mechanisms. Oncogene 2008;27(38):5132–7 doi 10.1038/onc.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vattulainen-Collanus S, Akinrinade O, Li M, Koskenvuo M, Li CG, Rao SP, et al. Loss of PPARgamma in endothelial cells leads to impaired angiogenesis. J Cell Sci 2016;129(4):693–705 doi 10.1242/jcs.169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yuan P, Yang T, Mu J, Zhao J, Yang Y, Yan Z, et al. Circadian clock gene NPAS2 promotes reprogramming of glucose metabolism in hepatocellular carcinoma cells. Cancer Lett 2020;469:498–509 doi 10.1016/j.canlet.2019.11.024. [DOI] [PubMed] [Google Scholar]
- 47.Mouchiroud M, Camire E, Aldow M, Caron A, Jubinville E, Turcotte L, et al. The hepatokine Tsukushi is released in response to NAFLD and impacts cholesterol homeostasis. JCI Insight 2019;4(15) doi 10.1172/jci.insight.129492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Senbabaoglu Y, Gejman RS, Winer AG, Liu M, Van Allen EM, de Velasco G, et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol 2016;17(1):231 doi 10.1186/s13059-016-1092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turajlic S, Litchfield K, Xu H, Rosenthal R, McGranahan N, Reading JL, et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: a pan-cancer analysis. Lancet Oncol 2017;18(8):1009–21 doi 10.1016/S1470-2045(17)30516-8. [DOI] [PubMed] [Google Scholar]
- 50.Chandarana H, Amarosa A, Huang WC, Kang SK, Taneja S, Melamed J, et al. High temporal resolution 3D gadolinium-enhanced dynamic MR imaging of renal tumors with pharmacokinetic modeling: preliminary observations. J Magn Reson Imaging 2013;38(4):802–8 doi 10.1002/jmri.24035. [DOI] [PubMed] [Google Scholar]
- 51.Wang HY, Su ZH, Xu X, Huang N, Sun ZP, Wang YW, et al. Dynamic Contrast-enhanced MRI in Renal Tumors: Common Subtype Differentiation using Pharmacokinetics. Sci Rep 2017;7(1):3117 doi 10.1038/s41598-017-03376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366(10):883–92 doi 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei EY, Hsieh JJ. A river model to map convergent cancer evolution and guide therapy in RCC. Nat Rev Urol 2015;12(12):706–12 doi 10.1038/nrurol.2015.260. [DOI] [PubMed] [Google Scholar]
- 54.Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell 2018;173(3):595–610e11 doi 10.1016/j.cell.2018.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schor-Bardach R, Alsop DC, Pedrosa I, Solazzo SA, Wang X, Marquis RP, et al. Does arterial spin-labeling MR imaging-measured tumor perfusion correlate with renal cell cancer response to antiangiogenic therapy in a mouse model? Radiology 2009;251(3):731–42 doi 10.1148/radiol.2521081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsai LL, Bhatt RS, Strob MF, Jegede OA, Sun MRM, Alsop DC, et al. Arterial Spin Labeled Perfusion MRI for the Evaluation of Response to Tyrosine Kinase Inhibition Therapy in Metastatic Renal Cell Carcinoma. Radiology 2021;298(2):332–40 doi 10.1148/radiol.2020201763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


