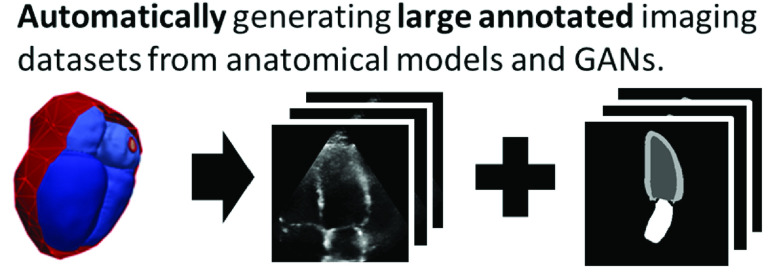
Abstract
Deep learning can bring time savings and increased reproducibility to medical image analysis. However, acquiring training data is challenging due to the time-intensive nature of labeling and high inter-observer variability in annotations. Rather than labeling images, in this work we propose an alternative pipeline where images are generated from existing high-quality annotations using generative adversarial networks (GANs). Annotations are derived automatically from previously built anatomical models and are transformed into realistic synthetic ultrasound images with paired labels using a CycleGAN. We demonstrate the pipeline by generating synthetic 2D echocardiography images to compare with existing deep learning ultrasound segmentation datasets. A convolutional neural network is trained to segment the left ventricle and left atrium using only synthetic images. Networks trained with synthetic images were extensively tested on four different unseen datasets of real images with median Dice scores of 91, 90, 88, and 87 for left ventricle segmentation. These results match or are better than inter-observer results measured on real ultrasound datasets and are comparable to a network trained on a separate set of real images. Results demonstrate the images produced can effectively be used in place of real data for training. The proposed pipeline opens the door for automatic generation of training data for many tasks in medical imaging as the same process can be applied to other segmentation or landmark detection tasks in any modality. The source code and anatomical models are available to other researchers.1
Keywords: Data generation, echocardiography, generative adversarial networks, segmentation, synthesis
I. Introduction
Medical imaging provides a window to capture the structure and function of internal anatomies. Imaging modalities such as ultrasound, computed tomography (CT) or magnetic resonance imaging (MRI) can be used to measure physical and physiological parameters. Accurate automation of these measurements would provide significant time-savings for clinical practitioners.
Convolutional neural networks (CNNs), have become the candidates of choice for measurement automation because they can accurately learn complex relevant features. However, CNNs require large sets of labeled data to learn and they are limited in accuracy by the quality of labels used in training. Inter-observer errors can be high in medical imaging tasks, especially when there is noise or other artifacts in the image. For example, in cardiovascular ultrasound (echocardiography or ‘echo’), inter-observer errors for labeling common measurements can range from 4-22% even for experienced cardiologists [1], [2]. The variability in measurements is due to (a) the difficulty of accurately interpreting signals delineating structures amid image noise, and (b) differences in implementation between different acquisition machines and between practitioners in different institutions. A second problem when building datasets to automate tasks in medical imaging is labeling is time-consuming and expensive since quality annotations require experienced medical professionals. Finally, manual labels are inflexible and adapting them based on new insights requires a significant amount of time.
While CNNs have been at the forefront of automating diagnostic measurements, anatomical models are progressing the personalization of treatments. Simulations from “digital twins” (models with patient-specific parameters) are increasingly being used to guide therapies and develop new treatments [3]. As with the revolution in statistical inferencing led by deep learning, larger computational resources have allowed the growth in complexity and realism of these anatomical models [4], [5]. While originally developed for personalized simulation of mechanics and biophysics, anatomical models are also a valuable source of high-quality shape information. We propose a method to solve the labeling challenges for medical deep learning by harnessing the information contained in anatomical models. Instead of labeling images, we let these models represent ground-truth anatomical shapes and generate task-specific paired realistic images as summarized in Fig. 1.
Fig. 1.
Using anatomical models as high quality ground truth annotations, we propose a pipeline to generate large synthetic datasets for training convolutional neural networks.
In particular, we demonstrate the usefulness of this pipeline for the task of segmenting parts of the left heart in echo images and thus make use of a set of cardiac models developed for electromechanical simulations of the heart. Similar anatomical models have been developed for a wide range of anatomies and most are free for academic use [4], [6]. The pipeline described here could readily be applied to those models as well with some application specific modifications. Section V-E provides more details on extensions to new anatomies.
A. Contributions
The proposed pipeline shifts the focus from annotating images to ensuring a CNN trained on synthetic images will generalize to real images. We test our pipeline by generating synthetic data for echo segmentation. Our main contributions are three-fold:
-
1)
We present a pipeline to generate realistic synthetic images with paired labels using anatomical models and a CycleGAN [7]. The pipeline can generate datasets of arbitrary size and include labels from any region included in the original anatomical models.
-
2)
We demonstrate the utility of the pipeline by building annotated synthetic 2D echo images from cardiac models. We show these synthetic images can be used for training deep learning algorithms, specifically by demonstrating accurate segmentation without any real labeled images. We present extensive validation of the proposed pipeline by testing on multiple datasets of real images from different clinical sites and annotators that were completely unseen during development.
-
3)
We present an analysis of the sources of error in the segmentation including differences in image texture, tissue shape, and annotator style. We show that differences in the segmentations primarily come from differences in annotator bias, highlighting the need for standardized annotations.
B. Related Work
Because there are often only a few accurate anatomical models available, we first experiment with using shape analysis techniques to expand the available set of ground truth models. Shape analysis has previously been used in medical imaging for improving segmentations as well as for pathology detection and registration [8]–[11].
The proposed work translates labels from a source domain (slices from anatomical models) to a target domain (echo in the example application). Domain adaptation is a similar task, but uses labels from a different imaging modality instead of models. Recently, CycleGANs have facilitated domain adaptation with unpaired images by using two sets of generative and discriminative networks, one for each transformation direction [7]. Kazeminia et al. [12] and Taghanaki et al. [13] provide overviews of CycleGANs in medical imaging. So far CycleGANs have primarily been used for realistic cross-modality translation to CT or MRI images whereas this work focuses on echo. Generating echo images is challenging because of the complex noise patterns. These patterns change dramatically between images and even within a single image following the acquisition settings of the user and the stretching/squeezing of the scan-conversion process. Compared to echo, the well-defined boundaries in MRI or CT represent a more similar domain to the anatomical model images. The cone in echo images is also a consistent defining feature in the image which degrades the translational invariance of convolutional networks. CycleGANs have been applied in echo for segmentation with image quality improvement [14] and view conversion [15], but these works used two real datasets of echo images and thus did not have to address the above challenges of translating from a different modality to echo.
Others have developed alternative strategies for surmounting limited datasets in medical imaging and Tajbakhsh et al. provide an overview of different strategies for segmentation with unlabeled or limited data [16]. Specifically relevant to this work, several groups have proposed strategies using GANs to generate synthetic data. Eschweiler et al. proposed a CycleGAN strategy for synthesizing a microscopy cell image and location dataset [17]. However, their labels are randomly generated, which loses the key advantage of ground truth anatomical models and is not applicable to most other applications in medical imaging where anatomies cannot be randomly generated from scratch. Huo et al. proposed SynSeg-Net, a similar pipeline using unpaired labels from MRI to train networks on CT images using a CycleGAN [18]. While some of the methodologies are similar, the central difference is that our ground truth annotations come from 3D anatomical models rather than unpaired images from another modality. Because detailed 3D annotations are an intrinsic part of each anatomical model, our pipeline is applicable to any segmentation or landmark detection task in any modality with no additional labeling required. Our approach is focused on image synthesis rather than domain adaptation.
Previous works generating echocardiography images have primarily used physics simulators to exactly replicate speckle creation from a set of reflectors. In general, these approaches have focused on generating a few specific images rather than large datasets. For example, Alessandrini et al. demonstrated a full pipeline for generation of 3D echo video loops that were realistic enough to trick some human observers [19]. While useful for providing a ground truth of myocardial motion for strain estimation, this pipeline and similar approaches [20]–[22], are ill-suited for generating training data for deep learning algorithms because it does not scale well to larger datasets. Each new generated image requires manual initialization and computationally heavy simulation. Other groups have used generative adversarial networks (GANs) for echo image synthesis. For example, Abdi et al. sampled new echo images from labels after conditioning a GAN on a paired dataset [23] an approach also demonstrated for skin lesions [24]. The key drawback is this approach can only be used to augment existing, already annotated datasets.
II. Methods
The proposed pipeline consists of two primary steps as shown in Fig. 2. First, pseudo images and paired labels are generated from 3D anatomical models as described in Sec. II-A. Second, the pseudo images are transformed into realistic synthetic ultrasound images using a CycleGAN and a set of real images as described in Sec. II-B. Afterwards, in Sec. II-C, we test the utility of the generated datasets by comparing segmentation networks trained with synthetic images to those trained with real images when testing on real images. The proposed pipeline is general to any medical imaging application, although models for the relevant anatomy are needed and application specific parameters are required in the extraction. Sec. III-A describes the models used for this application and Sec. III-B provides details on the example application (echo segmentation) and parameters. We demonstrate the method using several datasets as described in Sec. III-D.
Fig. 2.
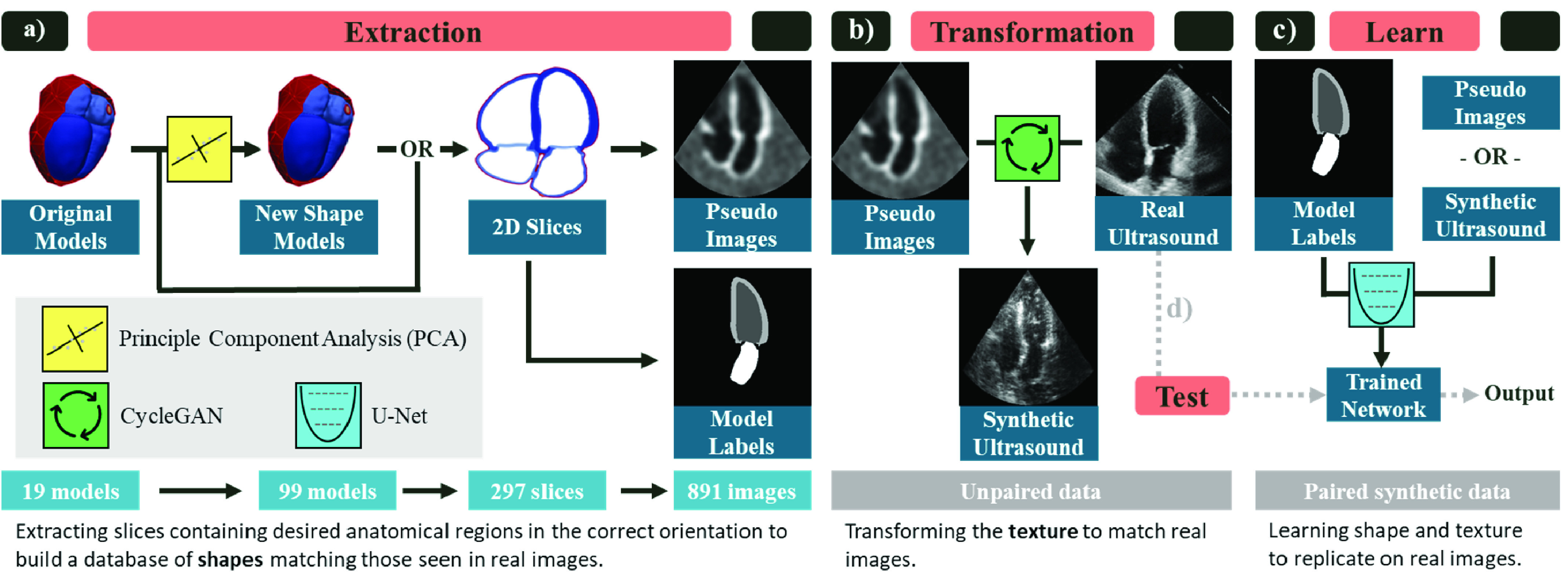
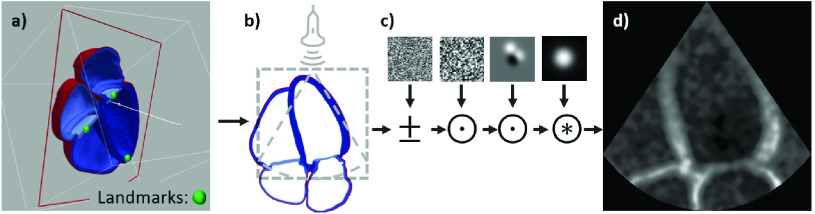
Overview of proposed pipeline implemented for echocardiography segmentation. a) Extraction: pseudo images and ground truth labels are built from the 3D anatomical models. First, a larger cohort of shapes is generated by building a statistical shape model from the original anatomies and sampling new 3D instances using principle component analysis (PCA). Next, 2D slices of the desired view (apical four chamber shown) are sampled. Finally, pseudo-ultrasound images and the corresponding labels are built. Each step expands the size of the dataset. b) Transformation: The pseudo images and a dataset of unlabeled real echo images are used to train a CycleGAN to transform the pseudo images into synthetic ultrasound images. c) Learn: The paired synthetic ultrasound images and model labels are used to train a U-Net segmentation network. d) Test: The network trained on synthetic images is tested on real images to evaluate the utility of the pipeline. The creation of new shape models as well as the transformation module are optional extensions. The slicing can be performed on the original models and the segmentation network can be trained using pseudo images instead. We evaluate the effectiveness of these components in Sec. IV.
A. Extraction
The input to the pipeline is a small set of anatomical models (“Original Models”) which contain labels for the attributes that will be segmented or detected.
1). New Shape Models:
While anatomical models provide excellent ground truth, there are often few available which may not provide sufficient anatomical variability to build a heterogeneous dataset. We experiment with building additional anatomically realistic models using statistical shape analysis. The primary modes of variation are deconstructed from the original models using principle component analysis. New models are generated by randomly sampling from the first 9 modes of variation (capturing 90% of the total variation) within two standard deviations of the mean model. We repeat this procedure to generate 99 new models in total. Each of the synthetic models is still an anatomically plausible shape, but adds a heterogeneous example to our dataset. Full details of the construction are given in Appendix A.
2). Pseudo Images and Model Labels:
A CycleGAN learns to transform the appearance from one imaging set to another. To generate the input for the CycleGAN, a “pseudo” database is generated where the anatomical shapes present in the pseudo database generally match the shape distribution found in an equivalent database of real images. Therefore the necessary functions here are application specific and are discussed in detail in Sec. III-B. The output of this step is both a pseudo image and a label image which contains ground truth for the learning step. Synthetic labels are generated from the original model to match the chosen task and selecting the task simply involves choosing the relevant regions in the model.
B. Transform
A CycleGAN [7] is trained to transform the pseudo images into synthetic ultrasound images using an unlabeled set of real ultrasound images. The default CycleGAN architecture and hyper-parameters are used except the generator network is replaced with a U-Net with 8 down-sampling levels [25] because it trains faster and gives equivalent results. The CycleGAN is trained for 200 epochs. Network weights are saved every 5 epochs. We select the best epoch by manually reviewing a sample result from each epoch (typically around epoch 180) but the exact epoch chosen did not have a significant impact on results in preliminary experiments. The selected network is used to save a synthetic image and paired label for each pseudo image.
C. Learn
A segmentation network is trained from the set of generated synthetic images and labels. The same U-Net architecture from the transform step is used. The network is trained for 30 epochs using cross-entropy loss. While the segmentation network can be included within the CycleGAN for end-to-end training [18], [26], we found the segmentation network was able to consistently achieve very good results on the synthetic images in preliminary results and did not find value in including this as a loss term within the transformation process. Additionally, splitting these two steps allowed us to develop an equal comparison between the synthetic and real data.
III. Example Application: Echo Segmentation
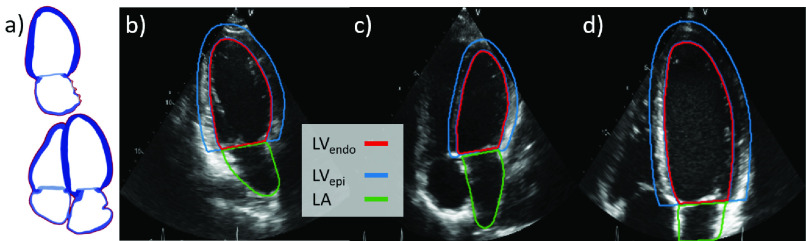
The feasibility of the pipeline is proven by building synthetic datasets for 2D echo segmentation. This application was chosen to enable comparison against existing real datasets. Two task variants are tested. First, matching all overlapping constraints of the synthetic and real datasets presented in Sec. III-D, a network was trained to segment the left ventricle endocardial border (LVendo) from apical four chamber images taken from the end diastole phase of the cardiac cycle. Second, the task was extended to include the left ventricle epicardial border (LVepi) and left atrium (LA) border from both four chamber and apical two chamber views and both end diastole and end systole phases. Fig. 3 shows examples of apical four/two chamber images extracted from the anatomical model as well as examples of performing the relevant annotations in real ultrasound images.
Fig. 3.
Example application: echo segmentation. a) Apical two chamber (top) and apical four chamber (bottom) views as shown in an anatomical model. The right images show example real apical two chamber (b) and apical four chamber (c, d) echo images with task labels. (b, c) show the full heart while d) is zoomed to focus on the left ventricle.
A. Original Models
The original models for this application were a set of 19 3D heart models derived semi-automatically from CT images. CT images have high contrast and spatial resolution which enables accurate delineations of structure boundaries. These models were built for electromechanical simulations and contain a complete set of tissue labels. Each model contains labels for both ventricles, both atria, aorta, pulmonary artery and veins, and both venae cavae. Additional details on the model creation process are given in Appendix A and by Rodero et al. [27] (currently under review, the model construction matches that described in [28]).
B. Task-Specific Data Generation
To generate a dataset for this task, 2D slices were sampled from the anatomical models and masked to mimic ultrasound images. A perfect 2D apical four chamber image is defined as the plane intersecting the apex, mitral valve center, and aortic valve center [29]. These landmarks were extracted from each model to define the optimal plane. Apical two chamber images were extracted by performing a 70 degree rotation counter-clockwise around the apical long axis from the four chamber landmarks (see Fig. 3). Although clinical guidelines suggest rotating the probe 60 degrees [29], using 70 degrees gave a better cut plane for the models from qualitative evaluation. To mimic natural variation in acquisition, random rotations of the cut planes around the long and short axes of the LV were sampled so that some slices are foreshortened or off-plane.
The 2D slices were transformed into pseudo images which mimic the appearance of ultrasound images. One of the most distinguishing features of an ultrasound image is the ‘cone’ marking the boundaries of imaging data. This is a consistent strong feature in all images and we found that the translational invariance of CNNs is degraded because the network could learn relationships between the cone boundary and structures. In other words, the CycleGAN discriminators could find difference between real and synthetic images from differences in structure location. In response, the generators would hallucinate structures in random locations. For the CycleGAN to properly transform structures as well as appearance, it is important that the distributions of locations of different anatomical structures are equally represented in both datasets.
To match this constraint, a series of affine transformations were applied to mimic the different LV orientations found in real images. This primarily consisted of masking the image with a cone and randomly cropping to either the entire heart (‘whole heart’ image) or the LV (‘LV focused’ image). The different crops are shown in Fig. 3 and match the image types suggested in clinical guidelines [29]. After cropping, other affine transforms such as rotations and squeezing were applied to ensure the region of interest remains inside the cone and add variance to the dataset (see Appendix A). Hard edges also decreased the realism of the generated images (see Appendix F) so random uniform noise and shadowing was added and the images were blurred by convolving with a Gaussian kernel. This process is shown in Fig. 4. To introduce additional variety, the slicing and pseudo extraction processes were repeated 3 times each for a total of 891 images (99  3
3  3). The entire process is fully automated.
3). The entire process is fully automated.
Fig. 4.
Extraction details for echocardiography. To extract pseudo images a) a 2D plane is defined from a set of landmarks, b) the plane is rotated and cropped to match standard acquisition parameters and positioned to match standard positioning in real images, c) random noise and shadows are added and the slice is blurred yielding d) the final pseudo image. Additional details are given in Appendix A.
C. Segmentation Evaluation and Network Selection
Several metrics were used to evaluate the accuracy of the trained segmentation networks. First, the Dice score was measured where 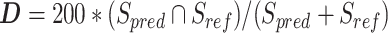 and measures the overlap between a predicted segmentation
and measures the overlap between a predicted segmentation 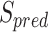 and a reference segmentation
and a reference segmentation 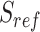 . Second, following [30], we analyzed the convexity and simplicity of the output as criteria which identify successful annotations. Because we found these two metrics vary together, only simplicity is reported. Simplicity is defined as
. Second, following [30], we analyzed the convexity and simplicity of the output as criteria which identify successful annotations. Because we found these two metrics vary together, only simplicity is reported. Simplicity is defined as 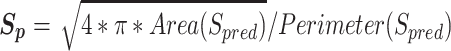 [31]. Note that simplicity relies only on the segmentation mask output from the network
[31]. Note that simplicity relies only on the segmentation mask output from the network 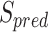 and not the label mask
and not the label mask 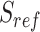 .
.
For the task of LVendo segmentation, differences between annotators are often because of differing placements of the endocardial border within the myocardial tissue rather than differing ventricular shapes. According to guidelines [29], the LVendo border falls at the interface between the non-compacted and compacted myocardium. If this border cannot be determined then the border falls at the blood-tissue interface. In noisy ultrasound images it can be difficult to accurately label this border, and there may be disagreement about which criteria should be used. There are no clear guidelines established for labeling LVepi and LA borders for segmentation [30] which can lead to differences between annotators for those tasks as well.
To capture these potential disagreements, we calculated several additional metrics: mean distance between the contours and Bias. Mean distance (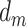 ) is the distance between two contours
) is the distance between two contours 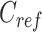 and
and 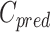 averaged across their length.
averaged across their length. 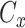 indicates the border of
indicates the border of 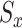 . Bias is the percentage error between the segmentation areas and is defined as:
. Bias is the percentage error between the segmentation areas and is defined as:
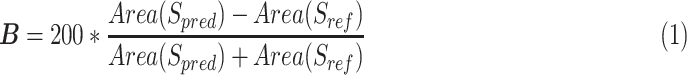 |
A high average Bias (positive or negative) across a dataset indicates a systematic difference in the labeling since the predicted results are consistently larger/smaller than the reference. Mean distance is calculated in pixels since we do not have access to image sizes in mm for all datasets. All other metrics are unit-less.
All networks were able to achieve high Dice scores on the synthetic data in preliminary experiments so selecting the network based on best Dice on a synthetic validation set lead to over-fitting to the synthetic data. Simplicity is a marker of the annotation quality that relies only on the network output and does not require a label. Therefore simplicity was tracked on an unlabeled set of real images (separate from the test set) through the course of training and the network with the highest simplicity was selected for final testing. This choice encouraged networks that generalized well to real data without requiring labels.
Median metrics were calculated in all cases since the distribution of scores was not normal. Therefore median absolute deviation was used as a measure of variance where 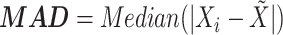 . The Wilcoxon signed-rank test was used to calculate statistical significance between different results [32].
. The Wilcoxon signed-rank test was used to calculate statistical significance between different results [32].
D. Real Datasets
Validating a dataset on a single source can lead to implicit bias in the developed methods [33]. For example, Degel et al. showed a decrease from 0.75 to 0.10 in Dice score for a CNN trained on one machine and tested on another for 3D left atrial segmentation. To account for this we validated the pipeline using a selection of real datasets. The characteristics of each dataset are described below and full details are listed in Appendix A.
1). Camus:
The Camus dataset was introduced by Leclerc et al. [30]. It consists of apical four and two chamber images with segmentation labels for LVendo, LVepi, and LA at end diastole and end systole time points in the cardiac cycle. The images also include quality labels, and following the authors we limit our analysis to images of good or medium quality, leaving 1,600 images. The images are divided into training, validation and test splits of 80%, 10%, and 10% respectively, keeping images from the same patient in the same split.
2). EchoNet:
The EchoNet dataset was introduced by Ouyang et al. [34]. It consists of 10,024 apical four chamber video loops with LVendo segmentation labels for end diastole and end systole The images were divided into training, validation and test splits of 80%, 10%, and 10% respectively, keeping images from the same patient in the same split.
3). Additional Real Datasets:
Since EchoNet contains only LVendo annotations in apical four chamber images, additional real images were labeled with a full set of annotations, views and cardiac phases. Mixed apical four and two chamber videos from two different clinical sites were annotated by two experienced cardiologists ( and
and  ). Both cardiologists use echo as a part of their daily practice. To annotate the images they used the whole loop to check myocardial movement to find the correct structures and annotated LVendo, LVepi, and LA labels at end diastole and end systole ensuring that the labels between phases matched. The datasets were split by institution, Site A contains 336 images and was further divided into training and validation splits of 80% and 20% respectively. Site B contains 229 images and was left exclusively as a test set. SiteA was labeled by
). Both cardiologists use echo as a part of their daily practice. To annotate the images they used the whole loop to check myocardial movement to find the correct structures and annotated LVendo, LVepi, and LA labels at end diastole and end systole ensuring that the labels between phases matched. The datasets were split by institution, Site A contains 336 images and was further divided into training and validation splits of 80% and 20% respectively. Site B contains 229 images and was left exclusively as a test set. SiteA was labeled by  and SiteB was labeled by
and SiteB was labeled by  .
.
4). Pathological Dataset:
The anatomical models were derived from asymptomatic patients and the aforementioned datasets contain no information on patient diagnosis. Therefore a set of pathological images was also gathered to test how well the networks trained on real and synthetic images would be able to adapt to pathological cases. 61 exams were gathered from patients diagnosed with severe functional mitral regurgitation, which is correlated with significant changes in LV shape [35], [36]. A severe diagnosis corresponds to a rating of 4 on a 4 point scale of severity. A random apical four chamber image was selected for each patient and  labeled LVendo, LVepi, and LA areas at end diastole and end systole (yielding 122 images total) using the same criteria as above. All images were used exclusively for testing.
labeled LVendo, LVepi, and LA areas at end diastole and end systole (yielding 122 images total) using the same criteria as above. All images were used exclusively for testing.
E. Synthetic Datasets
Synthetic versions of the Camus, EchoNet, and SiteA datasets were generated using the pipeline in Fig. 2. No synthetic dataset was generated for the SiteB or Pathological datasets since both were used for testing. Extraction and transformation were performed individually for each dataset and separately for each view. We predicted that using separate CycleGANs for each dataset and view would enhance the quality of the generated views and would allow the learned image features to be specific to the relevant dataset/view. Although customization of datasets and views could likely be combined into a single transformation process (using for instance an additional conditional input to the network), the focus of this work was on evaluating the feasibility of the pipeline rather than optimizing the generation process for multiple views and datasets. In most use cases all available datasets could be combined, however they were left separate here for evaluation purposes. Since SiteA contained fewer images, the CycleGAN for that dataset was initialized from the final trained CycleGAN from the Camus dataset and it was trained for only 100 epochs. The models were built for only a single time step so the synthetic datasets contain only end diastole images.
To test the impact of the new shape models, a synthetic EchoNet dataset was created without using the additional models generated in the shape extension described in Sec. II-A.1. To maintain dataset size, the extraction part was modified to extract 5 2D slices per anatomical model and 9 pseudo images per slice (for a total of 855 images). This set is denoted with an * in the experiments in Sec. IV.
F. Inter-Observer Study
To analyze label variability, an inter-observer study was conducted for a subset of each dataset. 20 random images were selected from the test set (or validation if no test set was created) for the Camus, EchoNet, SiteA, Synthetic Camus, Synthetic EchoNet and Synthetic SiteA datasets. To minimize the possible sources of variability, and match the overlapping constraints of the datasets, only apical four chamber end diastole images were selected.  and
and  annotated all images (except only
annotated all images (except only  labeled the EchoNet sets) with LVendo, LVepi and LA labels. The second round of labeling was conducted at least 2 months after the first round for SiteA.
labeled the EchoNet sets) with LVendo, LVepi and LA labels. The second round of labeling was conducted at least 2 months after the first round for SiteA.
G. Implementation Details
Hyperparameters for the segmentation such as the learning rate and loss function were tuned on the synthetic validation sets. All approaches were evaluated on the Camus validation set to ensure proper convergence and several different validation runs were run in the course of building the extraction and transformation steps. In general, the goal of this work was to evaluate the synthetic dataset construction using standard segmentation approaches rather than tuning an optimal segmentation network for the given application. The unlabeled EchoNet and SiteA validation datasets were used only for network selection (see II-C) so the labels and metrics for these sets were never seen (and thus cannot influence design choices). This allows us to detect implicit bias in the design choices or training datasets. The test sets (Camus, EchoNet, and SiteB) were used only once during final testing for the results presented below. Additional details on implementation and hyperparameters can be found in the supplementary material.
IV. Results
The pipeline is evaluated first in Sec. IV-A by comparing expert cardiologist’s annotations to those produced by the proposed pipeline. Next, since the aim of this pipeline is primarily to generate image/label pairs that are suitable for deep learning training, we check if a CNN can effectively learn from synthetic images in Sec. IV-B and compare to networks trained on real data. Finally, various versions of the synthetic dataset are analyzed in Sec. IV-C to determine which factors contributed to accurate segmentations.
A. Generated Images and Annotations
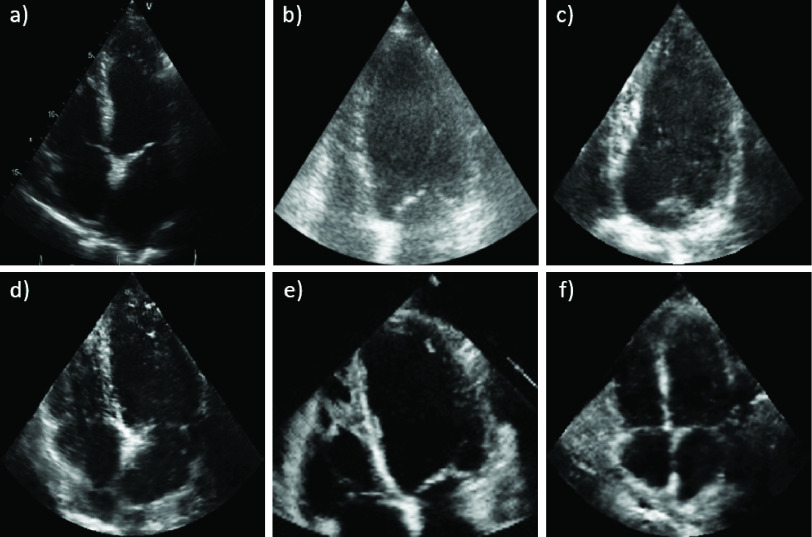
Images from the randomly selected inter-observer set are shown in Fig. 5 to demonstrate the realistic output of the generation pipeline. The synthetic images closely match their real counterparts in appearance. The GAN generates this appearance while maintaining the ground truth cardiac structures from the anatomical models. Generating a single ultrasound image from the prepared slice takes 81 ms.
Fig. 5.
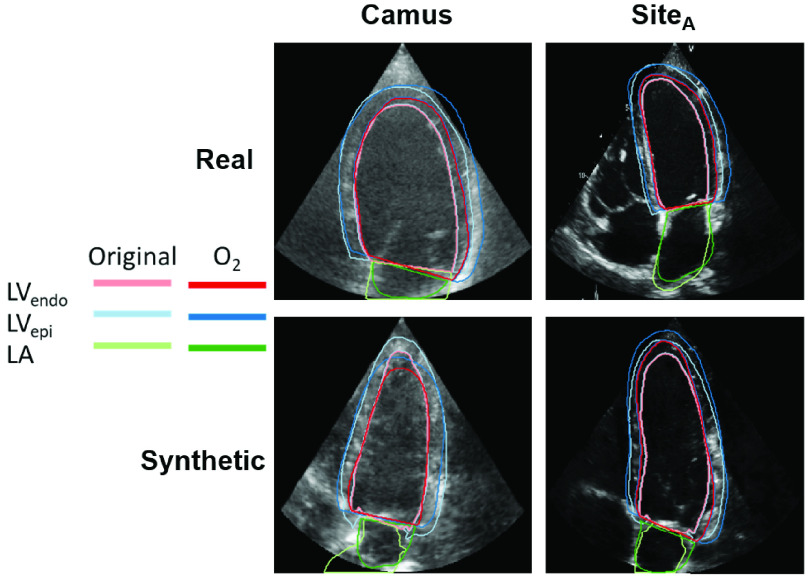
Synthetic images closely match real images. Can you guess which images are synthetic? Answers below.3
Next, we checked if experts agreed with the pipeline-generated annotations. Metrics from the inter-observer study are shown at the top of Table I. 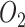 had higher Dice scores on synthetic images than real images on LVendo segmentations, was comparable for LVepi and had higher scores on real images on LA segmentations. The median image in LVendo Dice score between
had higher Dice scores on synthetic images than real images on LVendo segmentations, was comparable for LVepi and had higher scores on real images on LA segmentations. The median image in LVendo Dice score between 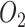 and the original annotator is shown in Fig. 6. Overall,
and the original annotator is shown in Fig. 6. Overall, 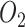 closely matched the pipeline-generated labels although there was some disagreement in the apical region. Fig. 6 also shows that while structure consistency between pseudo and synthetic images was not explicitly forced in the CycleGAN, the synthetic structures remain true to the original annotation mask. Only the results from
closely matched the pipeline-generated labels although there was some disagreement in the apical region. Fig. 6 also shows that while structure consistency between pseudo and synthetic images was not explicitly forced in the CycleGAN, the synthetic structures remain true to the original annotation mask. Only the results from 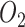 are used for comparison here for simplicity and because there was a large intra-observer bias in the results for
are used for comparison here for simplicity and because there was a large intra-observer bias in the results for 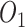 . The results from
. The results from 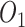 are presented in Appendix C and showed the same patterns as
are presented in Appendix C and showed the same patterns as 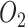 between synthetic and real. Finally, Fig. 5 and Fig. 6 shows the difference in appearance between the different datasets for both synthetic and real images. The Camus images are typically cloudier in appearance while the EchoNet/SiteA images usually have a higher gain setting and are thus clearer.
between synthetic and real. Finally, Fig. 5 and Fig. 6 shows the difference in appearance between the different datasets for both synthetic and real images. The Camus images are typically cloudier in appearance while the EchoNet/SiteA images usually have a higher gain setting and are thus clearer.
TABLE I. Synthetic Data Can Effectively be Used in Place of Real Data: Median Metrics Comparing Training With Real Datasets to Training With Synthetic Datasets. The First Section Compares Inter-Observer Results for 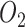 on Real and Synthetic Data. The Next Section Shows Networks Trained on Real and Synthetic Data for LVendo Segmentation in A4C ED Images and Tested on EchoNet. The Final Section Compares Networks Trained on Real and Synthetic Data for All Annotations/Views/Phases. All Dice Results Are Statistically Different With a p-Value < 0.05 (Computed With a Wilcoxon Signed-Rank Test) Except for the Marked Comparison (†). Results are Ordered by Dice Score and Bold Shows the Best Result in Each Section.
on Real and Synthetic Data. The Next Section Shows Networks Trained on Real and Synthetic Data for LVendo Segmentation in A4C ED Images and Tested on EchoNet. The Final Section Compares Networks Trained on Real and Synthetic Data for All Annotations/Views/Phases. All Dice Results Are Statistically Different With a p-Value < 0.05 (Computed With a Wilcoxon Signed-Rank Test) Except for the Marked Comparison (†). Results are Ordered by Dice Score and Bold Shows the Best Result in Each Section.
| Task | Training Data | Testing Data |
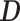 (%) [ (%) [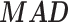 ] ] |
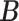 (%) (%) |
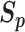 |
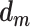 |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| OR inter-observer comparison | LVendo | LVepi | LA | LVendo | LVepi | LA | LVendo | |||
| Inter- |
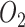 vs. Camus/SiteA vs. Camus/SiteA
|
87.9 [4.4] | 93.4 [2.2] | 87.5 [6.1] | 24 | 8 | −3 | 0.83 | 5.0 | |
| Observer |
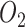 vs. proposed pipeline vs. proposed pipeline |
90.8 [2.3] | 92.0 [2.1] | 83.0 [8.0] | 9 | 4 | 14 | 0.80 | 3.9 | |
| LVendo in A4C ED | EchoNet (base) | EchoNet | 94.0 [1.6] | n/a | n/a | 0 | n/a | n/a | 0.85 | 2.7 |
| Camus | 89.6 [2.6] | −2 | 0.87 | 4.8 | ||||||
| SiteA | 87.7 [3.8] | 14 | 0.86 | 5.6 | ||||||
| Synth EchoNet | 87.1 [3.7] | 15 | 0.85 | 5.9 | ||||||
| All | Camus (base) | Camus | 93.3 [2.3] | 95.7 [1.0] | 92.0 [3.2] | 1 | 0 | 1 | 0.84 | 2.6 |
| SiteA | 88.9 [3.4] | 92.3 [2.1] | 85.4 [5.3] | 10 | −2 | 14 | 0.85 | 4.8 | ||
| Synth Camus | 81.2 [5.4] | 90.3 [2.8] | 79.6 [8.3] | 33 | 7 | −5 | 0.83 | 7.5 | ||
| Synth SiteA | SiteB | 88.4 [3.8] | 90.7 [3.4] | 83.1 [9.4] | −9 | −1 | −11 | 0.83 | 5.4 | |
| SiteA (base) | 85.1 [4.0] | 91.3 [2.0]† | 84.4 [4.7] | −27 | −8 | 8 | 0.84 | 7.5 | ||
| Camus | 81.8 [4.9] | 91.9 [1.8]† | 86.6 [4.4] | −35 | −10 | −10 | 0.83 | 8.7 | ||
| Camus | Pathological | 89.8 [2.6] | 92.5 [1.8] | 92.2 [2.0] | −9 | −2 | 8 | 0.86 | 4.1 | |
| SiteA (base) | 89.0 [3.3] | 92.0 [2.9] | 87.5 [3.1] | −4 | 5 | 19 | 0.85 | 4.2 | ||
| Synth SiteA | 88.3 [4.9] | 87.6 [4.1] | 84.3 [6.1] | 9 | 5 | −2 | 0.84 | 4.6 | ||
LVendo = left ventricle endocardium, LVepi = left ventricle epicardium, LA = left atrium, A4C = apical four chamber, A2C = apical two chamber, ED = end diastole, ES = end systole. All refers to all annotations (LVendo, LVepi, and LA), views (A4C and A2C), and phases (ED and ES). 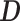 = Dice score,
= Dice score, 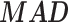 = median absolute deviation,
= median absolute deviation, 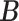 = Bias percentage,
= Bias percentage, 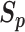 = simplicity, and
= simplicity, and 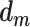 = mean average distance. For inter-observer
= mean average distance. For inter-observer 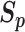 is listed for the second round annotations and
is listed for the second round annotations and 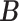 is calculated as
is calculated as 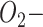 Original.
Original.
Not statistically different with a P-value < 0.05.
Fig. 6.
Expert annotations on synthetic images match the anatomical model annotations at a level equal to inter-observer error on real images: A sample image with included labels from the Camus and SiteA images used for the inter-observer study, chosen by taking the median Dice score between 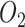 and the original labeler. For the real datasets the original labeler was [30] and
and the original labeler. For the real datasets the original labeler was [30] and 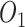 for Camus and SiteA respectively. For the synthetic datasets the original label comes from the anatomical models.
for Camus and SiteA respectively. For the synthetic datasets the original label comes from the anatomical models.
B. Learning From Synthetic Data
Networks were trained on Camus, EchoNet, SiteA, and each of the synthetic datasets for the task of LVendo segmentation in apical four chamber end diastole images. Networks were then tested on the EchoNet, Camus, SiteB, and Pathological test sets. Results for EchoNet are shown in Table I and for the other three sets in Appendix D. On the EchoNet test set the networks trained on real EchoNet data unsurprisingly achieved the best results, but the network trained on synthetic data was comparable to both the networks trained on separate real datasets (Camus and SiteA).
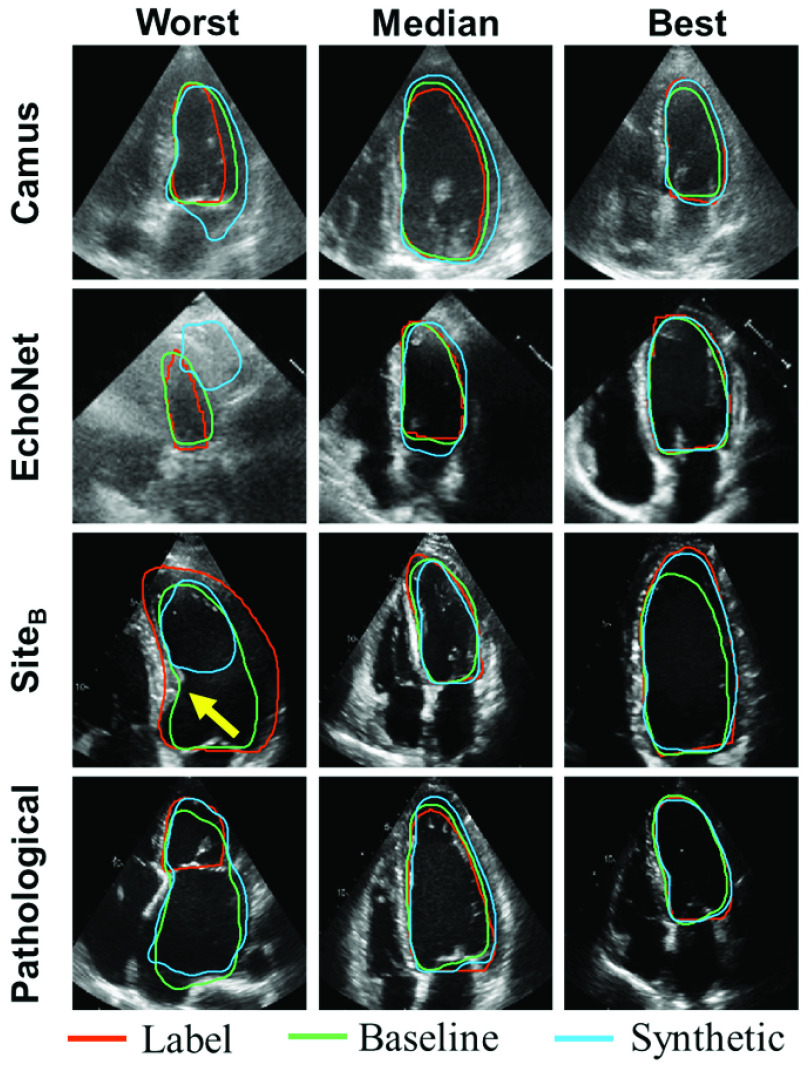
Qualitative results are shown in Fig. 7. In some cases the networks trained on synthetic data performed poorly. For example, in the worst case for Camus the network did not find the correct mitral valve cut-off plane. In the worst case for EchoNet the network found the wrong chamber, likely fooled by the strong reflective signal just beneath that resembles a valve. This image is also poor quality. In the worst case for SiteB the network misread the bulging septum (yellow arrow) as the mitral valve and cut off the segmentation there. The anatomical models were originally built from CT scans of asymptomatic patients and thus the segmentation network from synthetic images was not exposed to pathological cases (such as those with a bulging septum) during training. This was shown explicitly on the Pathological test set where the network failed to identify the LV given an enlarged LA (although the baseline network also failed in this case). However, these results were outliers. In most cases the network trained on synthetic data performed well with annotations that are similar to the manual labels and baseline.
Fig. 7.
Networks trained on synthetic data produce accurate segmentations in most cases: Worst, median, and best LVendo segmentation results on the Camus, EchoNet, SiteB, and Pathological test sets for the network trained on the baseline real data and the synthetic data. The task for all networks was LVendo segmentation in apical four chamber end diastole images. Images were ranked by Dice score for the network trained on synthetic data. The baseline and synthetic networks are always specific to the dataset (so for Camus the baseline network was trained on Camus and the synthetic network was trained on synthetic Camus). The yellow arrow points to a bulging septum in that image (see text). The baseline for SiteB and Pathological was SiteA.
Next, the robustness of the synthetic data was tested by extending the task to all annotations, phases, and views. We evaluated end diastole and end systole although the synthetic datasets do not contain end systole images. Results testing on the Camus, SiteB, and Pathological test sets are shown in Table I. The network trained on the synthetic data performed worse in both cases on LA segmentation and for LVendo segmentation in the Camus dataset. However, on SiteB the synthetic network outperformed all real datasets in LVendo Dice and distance scores. There was a high positive Bias for the synthetically trained networks on Camus and a strong negative Bias for SiteA and Camus on SiteB. The network trained on synthetic data was able to achieve similar performance to the networks trained on real datasets on the Pathological dataset for LVendo segmentation, although LVepi and LA Dice scores were slightly lower.
C. Variability Analysis
To test the impact of parameters in the pipeline, synthetic datasets with tweaked parameters were generated and a segmentation network was trained for each. To test the effect of the transformation process, the pseudo dataset (before transformation with the CycleGAN) was compared to the synthetic dataset (after transformation with the CycleGAN). The Camus pseudo and synthetic datasets were compared to the EchoNet pseudo and synthetic dataset to analyze the effect of different parameters in the extraction process and different real datasets in the transformation process respectively. To test whether including additional variability helped, datasets extracted from just the 19 original anatomical models (Pseudo EchoNet* and Synth EchoNet*) were compared to datasets extracted from the set of 99 new shape models (Pseudo EchoNet and Synth EchoNet). To simplify results, all networks were trained for LVendo segmentation only and were tested on the EchoNet test set since it was the largest.
Results are shown in Table II. Using the pseudo images provided a good baseline result even without the transformation process. Extending the anatomical model set as well as using dataset specific extraction processes slightly helped, but did not make a large difference. The transformation process did increase performance in the case that the correct dataset or a similar dataset was used (EchoNet/SiteA). However, using the Camus dataset actually significantly degraded the results.
TABLE II. Evaluating Data Generation Variability: Median Results on the EchoNet Test Set for LVendo Segmentation While Changing Various Parts of the Generation Pipeline. All Dice Results Except Pseudo Vs. Pseudo* are Statistically Different With a p-Value < 0.05 (Computed With a Wilcoxon Signed-Rank Test). Results are Ordered by Dice Score and Bold Shows the Best Result.
| Train | Test |
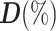 [ [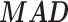 ] ] |
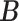 (%) (%) |
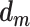 |
|---|---|---|---|---|
| Synth EchoNet | EchoNet | 87.1 [3.7] | 15 | 5.9 |
| Synth SiteA | 86.8 [4.0] | 14 | 6.2 | |
| Synth EchoNet* | 86.5 [4.0] | 17 | 6.1 | |
| Pseudo EchoNet* | 84.4 [5.3]† | 14 | 6.7 | |
| Pseudo EchoNet | 84.1 [4.5]† | 18 | 6.7 | |
| Pseudo Camus | 83.3 [4.7] | 9 | 7.0 | |
| Synth Camus | 81.9 [6.9] | 13 | 7.3 |
these datasets were extracted from only the original 19 anatomical models rather than the extended set including the new shape models from PCA.
Not statistically different with a P-value < 0.05.
V. Discussion
We developed a fully automated4 pipeline for generating large annotated datasets for training CNNs from anatomical models. The generated synthetic images look realistic and expert annotations on the synthetic images closely matched those from the pipeline. Moreover, segmentation networks trained from the synthetic datasets produced accurate segmentations on real images in most cases. Dice scores from the synthetically trained networks were comparable to inter-observer errors and networks trained on a separate set of real data.
A. Generated Images and Annotations
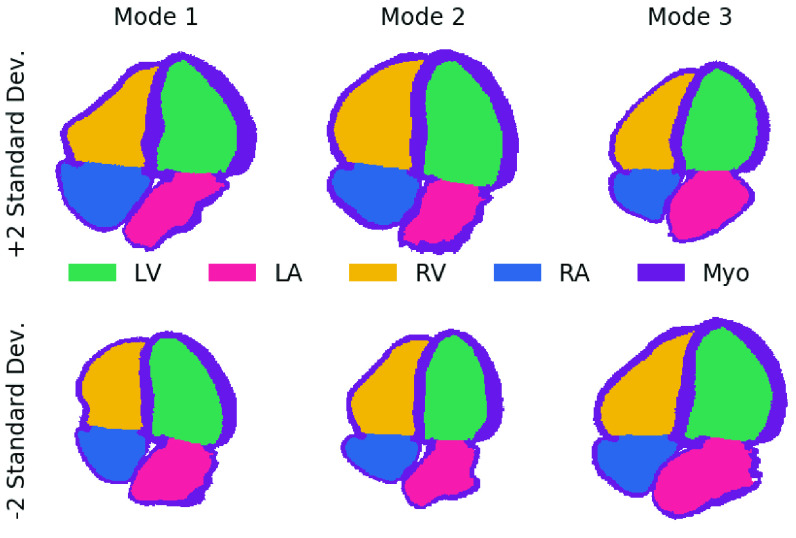
We found that the expert annotations on synthetic images closely matched the ones generated by the pipeline. This indicates the paired synthetic images and labels are accurately delineating the LV in a manner consistent with expert expectations. Dice scores between experts and the anatomical model were lower (although still comparable) for the LA. To explain this, Fig. 8 shows samples from the first several modes of the shape analysis described in Sec. II-A.1. The anatomical models show complex LA shapes as well high variability in shapes between different models. However, the LA is typically still annotated as a half-ellipsoid shape by the annotators (similar to the LV - see Fig. 3) in images and we hypothesize the lower scores were due to this difference in annotation complexity. Apical images are typically optimized for image quality in the LV rather than the LA, which may hinder accurate labeling of detailed LA shapes.
Fig. 8.
Shape variations are mainly seen in the LA, not the LV: Four chamber slices showing ± 2 standard deviations from the mean model for the first three modes calculated using principal component analysis (see Sec. II-A.1). LV = left ventricle, LA = left atrium, RV = right ventricle, RA = right atrium, and Myo = myocardium.
The inter-observer LVendo Dice scores for real images presented here are lower than those presented by Leclerc et al. on the same tasks [30]. There are two likely contributors to this. First, as discussed in Sec. III-C, a lack of explicit guidelines can cause differences in standard practice at different clinics and our results measure experts practicing in different sites. Second, in our inter-observer study the annotators were only given access to a single frame during the second round. This was necessary since the current pipeline only generates a single frame, but the lack of myocardial movement inhibits accurate detection of the compacted myocardium and other features. While more difficult, it also matches the task of the segmentation network, which is given a single frame only, and thus represents a better comparison for the pipeline.
B. Learning From Synthetic Data
We evaluated segmentation networks trained from synthetic data. First, we tested LVendo segmentation in apical four chamber end diastole images and then extended the task to LVepi and LA segmentation in apical four chamber and two chamber views and end diastole and end systole phases. Since there are numerous examples of deep learning methods failing once deployed due to implicit bias in the training dataset, we extensively validated our approach using five different datasets from various institutions and annotators. All hyperparameter tuning and initial tests were conducted using only a single dataset (Camus) and we then tested the same pipeline on additional unseen datasets. In some cases implicit bias towards the Camus dataset in the pseudo generation step were observed (see Appendix G), but the pipeline is still able to adapt and produce good results across datasets. This robustness is a strength of our work.
The network was able to achieve comparable results to a network trained on a separate real dataset. In a review of the results, failure cases primarily occurred when the network struggled to properly identify the mitral valve plane in real images (such as the worst case in SiteB of Fig. 7). Since the valve is included in the anatomical models as a flat disk, the synthetic images do not contain the same variation of valve appearances of real datasets. Including a variety of valve structures in the synthetic images is one way the proposed pipeline could be improved. The network trained on synthetic data was generally able to segment images from the Pathological dataset well, but could not properly identify the LV in cases with an enlarged LA (shown in Fig. 7 and in supplementary material). However, networks trained on real data also struggled on these images indicating that these cases would likely require expert review and adjustments regardless of the dataset used. If a known pathology should be handled, the models could also be adjusted to include this by including a single anatomical model exhibiting this pathology and using the PCA shape analysis to generate variations compared to a healthy normal model.
C. Clinical Applicability
LVendo segmentation is used clinically for an estimation of volumes and ejection fraction which are important measures of the efficiency of heart function. Clinical measures are not presented here because metric pixel sizes are not given for the datasets. However, previous studies have shown a strong correlation between the accuracy of Dice scores and the accuracy of predictions of clinical parameters across multiple algorithms and inter-/intra-observer studies (correlation coefficient of −0.92 between Dice scores at end-diastole/end-systole and ejection fraction mean average error across 12 experiments) [30]. Thus, the small decrease in accuracy of Dice scores presented here would likely result in a small decrease in accuracy of clinical metrics. The Dice scores obtained with the synthetically trained networks are still within the range of inter-observer error, indicating the same would likely be true for clinical metrics. Annotators rely on visually tracking the same point across the cycle to ensure consistency between predictions at end-diastole and end-systole and ejection fraction prediction could also be improved by including this temporal coherence between the predictions at different phases in the segmentation networks (using recursive neural networks for example).
D. Variability Analysis
We also analyzed potential sources of error for the networks. When testing images trained on one dataset on a different dataset there are two primary elements that cause decreased performance:
-
1)
Texture differences: In echo these are linked to acquisition changes such as varying ultrasound machines, gain, focus, resolution, and other imaging parameters. In the proposed pipeline, texture primarily comes from the transformation step.
-
2)
Shape differences: Due to a) differences in the width/depth of the acquisition which change tissue shape in the produced image, b) changes in the underlying tissue shape, or c) differences in annotation style. In the proposed pipeline, shape changes comes from the extraction step. Annotation style is linked to the original anatomical models.
Texture and shape differences were previously explored in object recognition where Geirhos et al., who showed that CNNs trained on ImageNet for classification were more biased towards changes in texture than shape [37]. We tested these differences in echo segmentation in Table II, using the generation pipeline to isolate the impact of each component.
Changes in shape due to imaging parameters were isolated by varying the width/depth/percentage of LV focused images in the two pseudo datasets and had a very small effect. Changes in underlying tissue shape were isolated by comparing the datasets built from the original models (Pseudo EchoNet*/Synth EchoNet*) to the set of models containing additional variability from the shape extension in Sec. II-A.1 (Pseudo EchoNet/Synth EchoNet). Changes in results were small and reversed between the pseudo and synthetic sets. This is likely because there were minimal variations in LV shape. As shown in Fig. 8, the largest changes in the LV are variations in size and width. Modifications to these parameters are already included in the pseudo image generation process, thus the shape extension did not add significant new variations of LV shape to the dataset. Pathological changes in the underlying shape (such as the bulging septum or enlarged left atrium in Fig. 7) do seem to reduce segmentation accuracy. To include these elements in the pipeline, new models could be built from pathological cases as discussed above.
Texture changes were isolated by comparing different synthetic datasets using CycleGANs tuned to different real datasets since the same underlying shape was used in all cases. Results showed that image appearance could make a significant difference as the Synth Camus network performed significantly worse than Synth EchoNet/Synth SiteA. This matches the qualitative appearance difference between EchoNet/SiteA and Camus in Fig. 7. Results here also showed that solid performance could be obtained with only the pseudo network. This is an encouraging result indicating that applications without high accuracy needs could further simplify the pipeline by removing the transformation step.
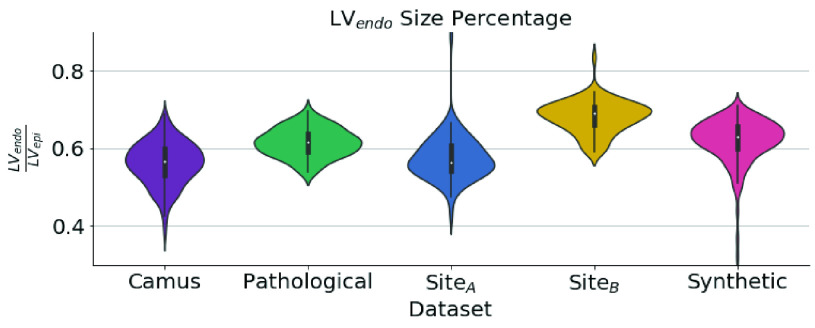
Assuming that human observers are adept at adapting to differences in texture and shape, differences in annotator style can be isolated from the inter-observer study presented in Table I. Differences between observers were substantial both in terms of Dice score and Bias, indicating a systematic difference between annotators. Although there were various constraints in this study (as discussed above), this difference was also clearly present in the original datasets without those constraints. Fig. 9 shows the ratio of LVendo area to LVepi area for the labels of each dataset which generally corresponds to the thickness of the labeled myocardium. This percentage is much higher (indicating a thinner myocardium) for the synthetic datasets than all the other datasets excluding SiteB. While this difference could instead indicate the prevalence of pathologies (e.g. hypertension) in the dataset, we present additional validation in Appendix E that the differences in Fig. 9 are primarily due to changes in annotation style. Results also match previous studies showing echo measurements typically overestimate the thickness of the myocardium [38]–[40].
Fig. 9.
Annotation bias can yield large differences: Violin plot showing the amount of LVepi area comprised of the LVendo area for the labels in each dataset. A lower value indicates a thicker myocardium. EchoNet does not have LVepi labels.
Our segmentation results also point towards annotation style as the critical factor in determining accuracy. Bias was high for LVendo results for networks trained on synthetic data on all other datasets than SiteB. On the other hand, networks trained on real datasets had a high negative Bias when tested on SiteB. The increase in Bias was correlated with lower Dice sores and higher mean distances, but not with simplicity, showing that the segmentations were still well-formed. This Bias was not observed for LVepi in Table I indicating that the variation comes purely from the differences in LVendo annotation style. The high performance of the synthetic network on SiteB matches both Fig. 9 and the low bias with 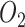 in the inter-observer study since
in the inter-observer study since 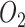 labeled SiteB.
labeled SiteB.
Therefore, the primary reason for decreased performance in our experiments (for networks trained on both synthetic and real data) was differences in annotation style, with texture differences playing a secondary role. Other than several outlier cases, the networks trained on synthetic data performed well and produced well-formed segmentations. One of the advantages of the pipeline proposed in this work is that the same annotation style can be applied to images from any dataset which will bring consistent performance for a network implemented in clinical practice. Given that the synthetic images are built from anatomical models derived from CT images, the synthetic images generated can be used to standardize annotation style.
E. Extensions and Future Applications
An abundance of augmentation techniques exist specifically for improving segmentation performance on limited datasets. For example, several authors introduced method based on statistical models to modify images following the deconstructed natural shape variation [8], [9]. Methods such as Jafari et al. [41] or Shin et al. [42] use GANs to expand the dataset with new natural images. This work focuses on the performance of the standard pipeline rather than one with augmentations tuned for a specific application, but these techniques, as well as any other task-specific augmentation techniques (or loss functions), could readily be applied here to improve results.
While we implemented the pipeline for 2D LV/LA echo segmentation to enable comparison against existing techniques, one of the strengths of our method is that the anatomical models are 3D and contain annotations for a variety of tissue types. Moreover, our method is not limited to ultrasound and a paired database of CT or MRI images could also be generated using this method. The pipeline is theoretically extensible to any segmentation or landmark detection task. Extension requires a) a small set of anatomical shape models similar to those described in Section III-A, b) a real dataset of unlabeled images from the relevant modality and view, and c) code to extract a slice from the anatomical models matching real images. Part c) can be accomplished through an analysis of important landmarks present in the relevant images that are also defined in the model. Additional unforeseen challenges likely exist for adapting to new anatomies and modalities, but we anticipate the ability to overcome these.
In addition to testing the pipeline on novel applications, future work will focus on adapting the pipeline to 3D, which is increasingly being used in clinical practice, but where manual labeling is even more difficult. The difficulty of manual labeling has thus far limited the development of benchmark datasets which is why the focus of this validation work is limited to 2D images. While challenging, other groups have previously shown the ability to adapt generative networks for 3D medical image synthesis (for example [42] and [43]). Due to GPU memory constraints these works required use of lower resolution volumes, a challenge for adapting the existing pipeline as well. The anatomical models could also be used as context for generation and/or segmentation as was proposed in [44]. Additionally, one of the strengths of echo is the high temporal resolution. Future work will also focus on extending image generation techniques to include labels and images across the cardiac cycle.
VI. Conclusion
Building large annotated datasets can be difficult and time-consuming. For cases where a small percentage of outliers are acceptable, or a confidence metric can be designed to catch outliers, we present a method to train a cardiac segmentation network with zero manual labeling required. The generated labels represent an accurate ground truth, can be rapidly built, and grant additional flexibility since the anatomical models providing the ground truth can be automatically adjusted as required. By eliminating or reducing labeling requirements, the proposed pipeline enables greatly accelerated deep learning algorithm development in cardiac imaging.
Acknowledgment
The authors thank S. A. Aase and J. Schnabel for guidance and feedback throughout this work and also D. Kulikova and A. Novikova for assistance with annotating images.
Funding Statement
This work was supported by the European Union’s Horizon 2020 research and Innovation program under the Marie Sklodowska-Curie under Grant 764738. The work of Pablo Lamata was supported by the Wellcome Trust Senior Research Fellowship under Grant 209450/Z/17/Z.
Footnotes
a) Real SiteA b) Real Camus c) Synthetic Camus d) Synthetic SiteA e) Real EchoNet f) Synthetic EchoNet.
Other than the manual step of selecting the CycleGAN epoch, which does not significantly impact results.
Contributor Information
Andrew Gilbert, Email: andrew.gilbert@ge.com.
Maciej Marciniak, Email: maciej.marciniak@kcl.ac.uk.
Cristobal Rodero, Email: rodero_gomez@kcl.ac.uk.
Pablo Lamata, Email: pablo.lamata@kcl.ac.uk.
Eigil Samset, Email: eigil.samset@ge.com.
Kristin Mcleod, Email: kristin.mcleod@ge.com.
References
- [1].Armstrong A. C.et al. , “Quality control and reproducibility in M-mode, two-dimensional, and speckle tracking echocardiography acquisition and analysis: The CARDIA study, year 25 examination experience,” Echocardiography, vol. 32, no. 8, pp. 1233–1240, Aug. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Thorstensen A., Dalen H., Amundsen B. H., Aase S. A., and Stoylen A., “Reproducibility in echocardiographic assessment of the left ventricular global and regional function, the HUNT study,” Eur. J. Echocardiography, vol. 11, no. 2, pp. 149–156, Mar. 2010. [DOI] [PubMed] [Google Scholar]
- [3].Corral-Acero J.et al. , “The ‘Digital Twin’ to enable the vision of precision cardiology,” Eur. Heart J., vol. 31, no. 48, pp. 4556–4564, Dec. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kainz W.et al. , “Advances in computational human phantoms and their applications in biomedical Engineering—A topical review,” IEEE Trans. Radiat. Plasma Med. Sci., vol. 3, no. 1, pp. 1–23, Jan. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xu X. G., “An exponential growth of computational phantom research in radiation protection, imaging, and radiotherapy: A review of the fifty-year history,” Phys. Med. Biol., vol. 59, no. 18, pp. R233–R302, Sep. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Caon M., “Voxel-based computational models of real human anatomy: A review,” Radiat. Environ. Biophys., vol. 42, no. 4, pp. 229–235, Feb. 2004. [DOI] [PubMed] [Google Scholar]
- [7].Jay F., Renou J.-P., Voinnet O., and Navarro L., “Unpaired image-to-image translation using cycle-consistent adversarial networks Jun-Yan,” in Proc. IEEE Int. Conf. Comput. Vis., Oct. 2017, pp. 183–202. [Google Scholar]
- [8].Corral Acero J.et al. , “SMOD—Data augmentation based on statistical models of deformation to enhance segmentation in 2D cine cardiac MRI,” in Functional Imaging and Modeling of the Heart (Lecture Notes in Computer Science: Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 11504. Cham, Switzerland: Springer, 2019, pp. 361–369. [Google Scholar]
- [9].Bhalodia R., Elhabian S. Y., Kavan L., and Whitaker R. T., “DeepSSM: A deep learning framework for statistical shape modeling from raw images,” in Proc. Int. Conf. Med. Image Comput. Comput. Interv., vol. 11167. Cham, Switzerland: Springer, 2018, pp. 244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Tavakoli V. and Amini A. A., “A survey of shaped-based registration and segmentation techniques for cardiac images,” Comput. Vis. Image Understand., vol. 117, no. 9, pp. 966–989, Sep. 2013. [Google Scholar]
- [11].Allan G.et al. , “Simultaneous analysis of 2D echo views for left atrial segmentation and disease detection,” IEEE Trans. Med. Imag., vol. 36, no. 1, pp. 40–50, Jan. 2017. [DOI] [PubMed] [Google Scholar]
- [12].Kazeminia S.et al. , “GANs for medical image analysis,” Artif. Intell. Med., vol. 109, 2020, Art. no. 101938. [DOI] [PubMed] [Google Scholar]
- [13].Taghanaki S. A., Abhishek K., Cohen J. P., Cohen-Adad J., and Hamarneh G., “Deep semantic segmentation of natural and medical images: A review,” Artif. Intell. Rev., pp. 1–42, 2020.
- [14].Jafari M. H.et al. , “Cardiac point-of-care to cart-based ultrasound translation using constrained CycleGAN,” Int. J. Comput. Assist. Radiol. Surgery, vol. 15, no. 5, pp. 877–886, May 2020. [DOI] [PubMed] [Google Scholar]
- [15].Abdi A. H., Jafari M. H., Fels S., Tsang T., and Abolmaesumi P., “A study into echocardiography view conversion,” 2019, arXiv:1912.03120. [Online]. Available: http://arxiv.org/abs/1912.03120
- [16].Tajbakhsh N., Jeyaseelan L., Li Q., Chiang J. N., Wu Z., and Ding X., “Embracing imperfect datasets: A review of deep learning solutions for medical image segmentation,” Med. Image Anal., vol. 63, Jul. 2020, Art. no. 101693. [DOI] [PubMed] [Google Scholar]
- [17].Eschweiler D., Klose T., Muller-Fouarge F. N., Kopaczka M., and Stegmaier J., “Towards annotation-free segmentation of fluorescently labeled cell membranes in confocal microscopy images,” in Proc. Int. Workshop Simulation Synth. Med. Imag., 2019, pp. 81–89. [Google Scholar]
- [18].Huo Y.et al. , “SynSeg-Net: Synthetic segmentation without target modality ground truth,” IEEE Trans. Med. Imag., vol. 38, no. 4, pp. 1016–1025, Apr. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Alessandrini M.et al. , “A pipeline for the generation of realistic 3D synthetic echocardiographic sequences: Methodology and open-access database,” IEEE Trans. Med. Imag., vol. 34, no. 7, pp. 1436–1451, Jul. 2015. [DOI] [PubMed] [Google Scholar]
- [20].De Craene M.et al. , “3D strain assessment in ultrasound (Straus): A synthetic comparison of five tracking methodologies,” IEEE Trans. Med. Imag., vol. 32, no. 9, pp. 1632–1646, Sep. 2013. [DOI] [PubMed] [Google Scholar]
- [21].Zhou Y.et al. , “A framework for the generation of realistic synthetic cardiac ultrasound and magnetic resonance imaging sequences from the same virtual patients,” IEEE Trans. Med. Imag., vol. 37, no. 3, pp. 741–754, Mar. 2018. [DOI] [PubMed] [Google Scholar]
- [22].Duan Q., Moireau P., Angelini E. D., Chapelle D., and Laine A. F., “Simulation of 3D ultrasound with a realistic electro-mechanical model of the heart,” in Functional Imaging and Modeling of the Heart (Lecture Notes in Computer Science: Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 4466. Berlin, Germany: Springer, May 2007, pp. 463–473. [Google Scholar]
- [23].Abdi A. H., Tsang T., and Abolmaesumi P., “GAN-enhanced conditional echocardiogram generation,” 2019, arXiv:1911.02121. [Online]. Available: https://arxiv.org/abs/1911.02121
- [24].Abhishek K. and Hamarneh G., “Mask2Lesion: Mask-constrained adversarial skin lesion image synthesis,” in Proc. Int. Workshop Simulation Synth. Med. Imag. Cham, Switzerland: Springer, 2019, pp. 71–80. [Google Scholar]
- [25].Ronneberger O., Fischer P., and Brox T., “U-Net: Convolutional networks for biomedical image segmentation,” in Proc. Int. Conf. Med. Image Comput. Comput. Interv., 2015, pp. 234–241. [Google Scholar]
- [26].Jafari M. H.et al. , “Echocardiography segmentation by quality translation using anatomically constrained cyclegan,” in Proc. Int. Conf. Med. Image Comput. Comput. Interv. Cham, Switzerland: Springer, 2019, pp. 655–663. [Google Scholar]
- [27].Rodero C.et al. , “Anatomical changes influences mechanics, electrophysiologyand haemodynamics in a complementary and localised wayin the healthy adult human heart,” PLOS Comput. Biol., submitted for publication.
- [28].Strocchi M.et al. , “A publicly available virtual cohort of four-chamber heart meshes for cardiac electro-mechanics simulations,” PLoS ONE, vol. 15, no. 6, pp. 1–26, Jun. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mitchell C.et al. , “Guidelines for performing a comprehensive transthoracic echocardiographic examination in adults: Recommendations from the American society of echocardiography,” J. Amer. Soc. Echocardiography, vol. 32, no. 1, pp. 1–64, Jan. 2019. [DOI] [PubMed] [Google Scholar]
- [30].Leclerc S.et al. , “Deep learning segmentation in 2D echocardiography using the camus dataset: Automatic assessment of the anatomical shape validity,” in Proc. Int. Conf. Med. Imag. Deep Learn. Ext. Abstr. Track, 2019, pp. 1–5. [Google Scholar]
- [31].Zhu Y., Tian Y., Metaxas D., and Dollár P., “Semantic amodal segmentation,” in Proc. IEEE Conf. Comput. Vis. Pattern Recognit. (CVPR), Jul. 2017, pp. 3001–3009. [Google Scholar]
- [32].Wilcoxon F., “Individual comparisons by ranking methods,” Biometrics Bull., vol. 1, no. 6, pp. 80–83, 1945. [Google Scholar]
- [33].Torralba A. and Efros A. A., “Unbiased look at dataset bias,” in Proc. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recognit., Jun. 2011, pp. 1521–1528. [Google Scholar]
- [34].Ouyang D.et al. , “Video-based AI for beat-to-beat assessment of cardiac function,” Nature, vol. 580, no. 7802, pp. 252–256, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sabbah H. N., Kono T., Rosman H., Jafri S., Stein P. D., and Goldstein S., “Left ventricular shape: A factor in the etiology of functional mitral regurgitation in heart failure,” Amer. Heart J., vol. 123, no. 4, pp. 961–966, Apr. 1992. [DOI] [PubMed] [Google Scholar]
- [36].Kono T., Sabbah H. N., Stein P. D., Brymer J. F., and Khaja F., “Left ventricular shape as a determinant of functional mitral regurgitation in patients with severe heart failure secondary to either coronary artery disease or idiopathic dilated cardiomyopathy,” Amer. J. Cardiol., vol. 68, no. 4, pp. 355–359, Aug. 1991. [DOI] [PubMed] [Google Scholar]
- [37].Geirhos R., Michaelis C., Wichmann F. A., Rubisch P., Bethge M., and Brendel W., “ImageNet-trained CNNs are biased towards texture; increasing shape bias improves accuracy and robustness,” in Proc. 7th Int. Conf. Learn. Represent. (ICLR), 2019, pp. 1–22. [Google Scholar]
- [38].Devereux R. B.et al. , “Echocardiographic assessment of left ventricular hypertrophy: Comparison to necropsy findings,” Amer. J. Cardiol., vol. 57, no. 6, pp. 450–458, Feb. 1986. [DOI] [PubMed] [Google Scholar]
- [39].Malm S., Frigstad S., Sagberg E., Larsson H., and Skjaerpe T., “Accurate and reproducible measurement of left ventricular volume and ejection fraction by contrast echocardiography: A comparison with magnetic resonance imaging,” J. Amer. Coll. Cardiol., vol. 44, no. 5, pp. 1030–1035, 2004. [DOI] [PubMed] [Google Scholar]
- [40].Mor-Avi V.et al. , “Real-time 3-dimensional echocardiographic quantification of left ventricular volumes,” JACC, Cardiovascular Imag., vol. 1, no. 4, pp. 413–423, Jul. 2008. [DOI] [PubMed] [Google Scholar]
- [41].Jafari M. H.et al. , “Semi-supervised learning for cardiac left ventricle segmentation using conditional deep generative models as prior,” in Proc. IEEE 16th Int. Symp. Biomed. Imag. (ISBI), Apr. 2019, pp. 649–652. [Google Scholar]
- [42].Shin H. C.et al. , “Medical image synthesis for data augmentation and anonymization using generative adversarial networks,” in Simulation and Synthesis in Medical Imaging (Lecture Notes in Computer Science: Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 11037. Cham, Switzerland: Springer, 2018, pp. 1–11. [Google Scholar]
- [43].Zhang Z., Yang L., and Zheng Y., “Translating and segmenting multimodal medical volumes with cycle-and shape-consistency generative adversarial network,” in Proc. IEEE/CVF Conf. Comput. Vis. Pattern Recognit., Jun. 2018, pp. 9242–9251. [Google Scholar]
- [44].Wang C. and Smedby O., “Automatic whole heart segmentation using deep learning and shape context,” in Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges (Lecture Notes in Computer Science: Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol. 10663. Cham, Switzerland: Springer, Mar. 2018, pp. 242–249. [Google Scholar]