Abstract
The full range of sequences that constitute nuclear localization signals (NLSs) remains to be established. Even though the sequence of the classical NLS contains polybasic residues that are recognized by importin-α, this import receptor can also bind cargo that contains no recognizable signal, such as STAT1. The situation is further complicated by the existence of six mammalian importin-α family members. We report the identification of an unusual type of NLS in human Ran binding protein 3 (RanBP3) that binds preferentially to importin-α3. RanBP3 contains a variant Ran binding domain most similar to that found in the yeast protein Yrb2p. Anti-RanBP3 immunofluorescence is predominantly nuclear. Microinjection of glutathione S-transferase–green fluorescent protein–RanBP3 fusions demonstrated that a region at the N terminus is essential and sufficient for nuclear localization. Deletion analysis further mapped the signal sequence to residues 40 to 57. This signal resembles the NLSs of c-Myc and Pho4p. However, several residues essential for import via the c-Myc NLS are unnecessary in the RanBP3 NLS. RanBP3 NLS-mediated import was blocked by competitive inhibitors of importin-α or importin-β or by the absence of importin-α. Binding assays using recombinant importin-α1, -α3, -α4, -α5, and -α7 revealed a preferential interaction of the RanBP3 NLS with importin-α3 and -α4, in contrast to the simian virus 40 T-antigen NLS, which interacted to similar extents with all of the isoforms. Nuclear import of the RanBP3 NLS was most efficient in the presence of importin-α3. These results demonstrate that members of the importin-α family possess distinct preferences for certain NLS sequences and that the NLS consensus sequence is broader than was hitherto suspected.
Transport of proteins and nucleic acids into and out of the nucleus occurs through nuclear pore complexes (NPCs), which are plugged through the double membrane of the nuclear envelope. Small molecules and ions can diffuse passively through the NPC, but macromolecules larger than about 50 kDa require a facilitated mechanism. Soluble receptors mediate macromolecular transport through the NPC (reviewed in reference 16). These receptors recognize and bind to signal sequences that mark proteins destined for nuclear import or export (reviewed in reference 45).
The classical nuclear localization signal (NLS) was the first such sequence to be identified and is characterized by a string of basic amino acid residues. An example is the simian virus 40 (SV40) T-antigen NLS (PKKKRKV, in the single-letter amino acid code) (35). Bipartite NLSs exist that comprise two strings of basic amino acid residues separated by a short intervening sequence (reviewed in reference 19). The receptor protein importin-α (also called p54/p56 and karyopherin-α, among other names) (1, 13, 15, 26, 40, 51, 79) recognizes both types of NLS. A second component of the receptor, importin-β (also called p97 and karyopherin-β) (11, 61), binds importin-α (20, 22, 80) and permits transit through the NPC (27, 51). Accumulation within the nucleus requires that the NLS cargo be released from its receptor so that the receptor can recycle back to the cytosol. This release is triggered by Ran-GTP (25, 50). Ran is localized predominantly to the nucleoplasm in intact cells (7). The distribution of regulatory factors for Ran is asymmetric (31), in that the Ran exchange factor, RCC1 (6), is nuclear while the GTPase-activating protein, RanGAP (5), is cytosolic or is associated with the cytoplasmic face of the NPC (42, 47, 67). The intrinsic nucleotide dissociation and hydrolysis rates for Ran are low but are stimulated many thousandfold by these factors. Consequently, much of the nuclear Ran is probably GTP bound while Ran present in the cytoplasm is bound to GDP. This steep Ran-GTP gradient across the NPC may determine the vectorality of nuclear transport and allow the accumulation of cargo proteins against their concentration gradients. NTF2 facilitates uptake of Ran into the nucleus to maintain the Ran-GTP gradient (63, 72). NLS cargo docked at the NPC in a complex with importin-α and -β is dissociated by Ran-GTP, releasing the cargo into the nucleoplasm (25, 50).
Importin-β is the archetype of a family of receptors that are involved in many nucleocytoplasmic transport pathways (21, 81). Several members of the family, including RanBP7, transportin, and importin-β, recognize a highly basic region (the basic importin-β binding domain [BIB]) that is present in many ribosomal proteins and permits the transport of these proteins into the nucleus (32). The N terminus of importin-α contains an importin-β binding domain (IBB) that is distinct from the C-terminal NLS cargo binding site (22, 80).
Several mammalian paralogs of importin-α have been recently discovered, leading to great confusion in the nomenclature. For consistency, this report uses numerical designations described by Köhler et al. (36, 37). To provide unambiguous identification of the paralogs, the importin-αs are listed in Table 1 together with the sequences of the first 10 residues and their alternative names. Each importin-α consists of a core of eight armadillo repeats that are highly related to one another. Further, both the order and sequences of these armadillo repeats have been highly conserved throughout evolution, indicating that the repeats are not functionally interchangeable (44). The major source of diversity between the importin-αs lies within the amino- and carboxy-terminal regions. Based on the similarities of their primary structures, the importin-α proteins have been separated into three subfamilies: importin-α1 (15) is the lone member of one family, importin-α3 (36, 49, 70) and importin-α4 (36, 53, 73) form a second family, and importin-α5 (13, 57), importin-α6, and importin-α7 (36, 37) form the third family. The crystal structure of yeast karyopherin-α (Srp1p), soaked with peptide corresponding to the SV40 NLS, has been determined (12; reviewed in reference 18). Armadillo repeats 5 to 7 form a groove within which the polybasic NLS binds.
TABLE 1.
Descriptions of importin-αs used in this study
| Importin | N-terminal sequencea | Other name(s) | Reference(s) | Mouse homolog |
|---|---|---|---|---|
| α1 | MSTNENANTP | hSrp1α, karyopherin-α2, Rch1 | 15, 79 | αP1 |
| α3 | MADNEKLDNQ | Qip1, | 36, 70 | αQ1 |
| α4 | MAENPSLENH | hSrp1γ, karyopherin-α3 | 36, 53, 73 | αQ2 |
| α5 | MTTPGKENFR | hSrp1, NPI-1, karyopherin-α1 | 13, 51, 57 | αS1 |
| α6 | MASPGKDNYR | Karyopherin-α5 | 36 | |
| α7 | METMASPGKD | 37 | αS2 |
Amino acids 1 to 10.
Recently, a number of NLS sequences have been identified that do not conform to the classical NLS consensus motif. For example, the M9 sequence which is recognized by transportin (karyopherin-β2) is glycine rich rather than basic in character (8, 9, 71), whereas the ribosomal protein BIB domain, recognized by RanBP5, RanBP7, transportin, and importin-β, is highly basic (32). A unique signal within the hnRNP K protein (called KNS) has been described that permits nuclear transport via a mechanism independent of soluble factors (48). These findings highlight the complexity of the nuclear import pathway.
A highly conserved Ran binding domain (RanBD) of about 135 amino acid residues is present in RanBP1 (14) and its Saccharomyces cerevisiae homolog, Yrb1p (69), plus RanBP2/NUP358 (82, 84) and the Caenorhabditis elegans protein Ranup96 (4). This domain is necessary and sufficient to bind Ran-GTP and coactivate RanGAP (4). A second S. cerevisiae protein, Yrb2p, possesses a variant RanBD and binds Gsp1p—the yeast Ran homolog—with much lower affinity than does Yrb1p (55). The YRB2 gene is not essential, but its deletion results in a cold-sensitive phenotype (55). Recent studies suggest that Yrb2p plays a role in nuclear protein export via the Crm1p pathway (56, 74). A human gene product (RanBP3) has been identified that more closely resembles Yrb2p than Yrb1p or RanBP1 (52). Two isoforms, RanBP3a and RanBP3b, were isolated by using the two-hybrid system with RCC1 as the bait (52). The RanBP3 proteins preferentially bind to Ran-GTP and can form an in vitro trimeric complex with Ran-GTP and RCC1 (52). We independently cloned RanBP3b from human expressed sequence tags (ESTs) that were identified in a database search for RanBDs. We also found an alternate splice variant, RanBP3c, that lacks an internal sequence encoding 30 amino acid residues. RanBP3 is a nuclear protein of about 50 kDa that contains a double FXFG motif upstream of the variant RanBD. FXFG motifs have been identified as frequent repeats within a subset of nucleoporins (66).
We show here that a discrete region of RanBP3b, N terminal to the FXFG repeat, is required for correct nuclear localization of the protein. Microinjection assays showed further that residues 40 to 57 are necessary and sufficient for nuclear import. This motif, although related to the NLSs of c-Myc and Pho4p, was shown to possess significant differences. Transport of the RanBP3 NLS into nuclei of permeabilized cells was mediated more efficiently by importin-α3 than by the other importin-αs tested.
MATERIALS AND METHODS
DNA manipulations.
The human cDNA clones 53258, IB1953, and 376469 (EST clones with GenBank accession numbers for the 5′ sequences of R16269, T15830, and AA041393, respectively) were obtained from the American Type Culture Collection and sequenced, and the cDNA inserts were subcloned into pBluescript II KS−. A RanBP3 construct (encoding residues 57 to 499) was created by subcloning the HindIII-NcoI fragment of IB1953 into 53258. A full-length RanBP3 plasmid was made by sequential subcloning of PCR products encoding residues 1 to 150 and 150 to 499 into pGEX-2T (Pharmacia). The other plasmids used in this study were constructed by subcloning of PCR products into pGEX-2T, pGEX-GFP (64), or pKH3 (46) using the BamHI site within the 5′ oligonucleotide and the EcoRI site in the 3′ oligonucleotide. The mutations in full-length glutathione S-transferase (GST)–green fluorescent protein (GFP)–RanBP3 (GGR fusion protein) were made by using a QuikChange site-directed mutagenesis kit (Stratagene). Plasmid DNA was sequenced by using a United States Biochemical Sequenase 2.0 kit or on an ABI automated sequencer.
Recombinant protein expression and purification.
GST fusion proteins were expressed in Escherichia coli DH5α and purified by attachment to glutathione-Sepharose beads (Pharmacia). After elution with glutathione, the buffer was exchanged and the proteins were concentrated by using a Centricon 30 (Amicon). Recombinant expression and purification of the importin-α3, -α4, -α5, and -α7 proteins were done as described previously (37). Briefly, C-terminally His-tagged proteins were expressed in E. coli XL1/pSB161 and purified by using nickel agarose. Following elution using an imidazole gradient, the importin-α proteins (except importin-α5, which was dialyzed against sonication buffer and stored at −80°C after addition of 250 mM sucrose) were loaded onto a Mono Q column and eluted in 50 mM Tris-HCl (pH 7.5)–5% glycerol using an NaCl gradient. Peak fractions were pooled again and stored at −80°C after addition of 250 mM sucrose. Preparation of importin-α1 was done as previously described (23).
Sequence analysis and homology searches.
Database searches were performed by using BLAST (3). DNA sequences obtained by sequencing of EST clones were compiled into a contiguous fragment using Sequencher software.
Preparation of anti-RanBP3 antibodies.
A GST fusion protein containing residues 410 to 499 of RanBP3 was expressed in E. coli DH5α and purified by attachment to glutathione-Sepharose beads (Pharmacia). These beads were washed extensively and then boiled in Laemmli sample buffer, and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The band corresponding to the GST-RanBP3 fusion was excised and used as antigens for the production of rabbit polyclonal antisera (Cocalico Biologicals Inc.). The resulting antisera were affinity purified against recombinant RanBP3. GST-RanBP3 was cleaved with thrombin to remove the GST, and the purified RanBP3 fragment was covalently attached to CNBr-activated Sepharose. Antiserum was applied to the RanBP3 column, and after extensive washing, the antibodies were eluted as described by Harlow and Lane (28) and then concentrated by using a Centricon 30 (Amicon).
Mammalian cell culture and transfection.
Cells were maintained in Dulbecco’s minimal essential medium supplemented with 10% calf serum plus penicillin and streptomycin at 37°C in 5% CO2 and grown on 100-mm-diameter plates or two-well glass slides (LabTek). Transfections were performed by using a calcium phosphate precipitation method (68), with 15 μg of DNA per 100-mm plate or 3 μg per well. Cells were rinsed in phosphate-buffered saline (PBS) at 16 h posttransfection and incubated with fresh medium for a further 24 to 30 h before analysis.
Immunoblotting.
Protein samples from mammalian cells were prepared by incubating cells on ice in lysis buffer (50 mM HEPES [pH 7.4], 5 mM MgCl2, 1 mM dithiothreitol, 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 1% Triton X-100) for 5 min. The cells were scraped off the plate and incubated for a further 20 min on ice before centrifugation at 600 × g for 2 min to remove large bits of cell debris. Proteins separated by SDS-PAGE were transferred onto nitrocellulose and immunoblotted by using a standard procedure (28). The antibodies were diluted as follows: 12CA5, 1:7,000; anti-RanBP3 1:500; antiserum or purified antibodies, 1:50,000; anti-GFP (Clontech), 1:2,000; anti-His6 (Babco), 1:1,000. Horseradish peroxidase-coupled secondary antibodies (Jackson Laboratories or Sigma) and chemiluminescence reagents (KPL) were used for detection.
Immunofluorescence assay.
Cells were fixed with 4% paraformaldehyde in PBS for 15 min. Fixed cells were then permeabilized with −20°C methanol for 2 min and blocked in 3% bovine serum albumin (BSA) in PBS plus 0.1% Tween 20 (PBS-T) for at least 1 h. Cells were stained for endogenous RanBP3 by using either RanBP3 antiserum (1:5) or purified antibodies (1:500), and hemagglutinin (HA)-tagged proteins were detected by using 12CA5 (1:400). After washing with PBS-T, detection of the antibody was accomplished by incubation of the cells with a secondary antibody (1:1500) coupled to Cy3, rhodamine, or fluorescein isothiocyanate (FITC) and counterstaining for DNA with 4′,6′-diamidino-2-phenylindole (DAPI) at 75 μg/ml. Cells were washed with PBS-T and mounted with Gel/Mount (Biomedia Corp.). Fluorescence was detected by using a Nikon microscope with a 60× objective water immersion lens. Images were captured by using a Hamamatsu charge-coupled device camera and processed by using Openlab and Adobe Photoshop software.
Microinjection of GGR fusion proteins.
Baby hamster kidney (BHK21) cells were grown on CELLocate gridded coverslips (Eppendorf). Prior to microinjection, the medium was changed to Ringer’s solution (25 mM HEPES [pH 7.2], 110 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 10 mM glucose, 1 mM KH2PO4, 1 mg of BSA/ml). Bacterially expressed GGR fusion proteins were purified on glutathione-Sepharose beads (Pharmacia), concentrated by use of a Centricon 30, and then exchanged into microinjection buffer (10 mM NaPO4 [pH 7.2], 70 mM KCl, 1 mM MgCl2) (10). By use of an Eppendorf 5242 system, cells were injected with GGR fusion proteins plus tetramethyl rhodamine isothiocyanate (TRITC)-dextran as an injection site marker. Injections were performed at room temperature over a period of 15 to 30 min. The cells were fixed in 4% paraformaldehyde–PBS after a total elapsed time of 30 min and then analyzed by fluorescence microscopy. GGR fusion protein was injected at 20 to 40 μM in the RanBP3 fragment experiment and at 50 to 70 μM in the mutant experiment.
In vitro import assay.
Nuclear import was assayed by the method of Adam et al. (2) using BHK21 cells. Briefly, cells were permeabilized by using 0.008% digitonin in import buffer (20 mM HEPES-KOH [pH 7.5], 100 mM potassium acetate, 2 mM magnesium acetate, 1 mM EGTA, 250 mM sucrose, 2 mM dithiothreitol) and incubated on ice for 5 min. Permeabilization was stopped by addition of import buffer containing 10 mg of BSA/ml and incubation for 20 min at 20°C. Either reticulocyte lysate (Promega) diluted 1:1 in 2× import buffer containing 20 mg of BSA/ml) or a recombinant protein mixture (1.5 μM Ran, 150 nM RanBP1, 150 nM RanGAP, 150 nM NTF2, 3 μM importin-α1, 1 μM importin-β, and 1 mg of nucleoplasmin core/ml [final concentrations]) was used (32). The import reaction mixture also contained import substrate (1 μM GGR fusion protein) and an energy-regenerating system (1 mM ATP, 0.2 mM GTP, 10 mM creatine phosphate, 100 μg of creatine kinase/ml). The energy depletion system (50 U of hexokinase/ml, 12.5 mM glucose) and other factors to be assayed were added, and the mixture was incubated at 20°C for 1 h. Concentrations of added transport inhibitors were as follows: wheat germ agglutinin (WGA), 0.25 mg/ml; importin-β (amino acids 45 to 462), 1 μM; Ran (wild type or G19V mutant), 4 μM; CBP80 peptide, 100 μM; rabphilin peptide, 100 μM; TC10 peptide, 100 μM; BIB domain, 100 μM; IBB domain, 100 μM. The cells were then washed in import buffer, fixed in 4% paraformaldehyde–import buffer, and visualized by fluorescence microscopy.
For import competition assays, HeLa cells were grown on 12-mm-diameter coverslips to 40 to 80% confluence, washed once in ice-cold PBS, and permeabilized for 8 min as described previously (37). Coverslips were incubated with 20 μl of import mixture containing Texas red-labeled nucleoplasmin (TR-NPL) and GGR(1-150) for 8 min at room temperature. Reactions were stopped by fixation with 4% paraformaldehyde in PBS. After washing in PBS and water, the coverslips were mounted by using Vectorshield mounting medium (Vector) and analyzed by confocal microscopy (Leica TCS NT). The import reaction mixtures consisted of an energy-regenerating system (0.5 mM ATP, 0.5 mM GTP, 10 mM creatine phosphate, creatine kinase at 50 μg/ml), core buffer (nucleoplasmin core at 2 mg/ml, 20 mM HEPES-KOH [pH 7.5], 140 mM potassium acetate, 6 mM magnesium acetate, 250 mM sucrose), 0.5 mM EGTA, 3 μM RanGDP, 0.2 μM Rna1p, 0.3 μM RanBP1, 0.4 μM NTF2, 1 μM importin-β, importin-α protein at 2 μM, 10% reticulocyte lysate (37).
Solution binding assay.
GGR fusion protein (12 μg) and His-tagged importin-α (3.5 μg) were incubated with glutathione-Sepharose beads in the presence of bacterial lysate (to minimize nonspecific binding) in binding buffer (20 mM morpholinepropanesulfonic acid [MOPS; pH 7.1], 100 mM K acetate, 5 mM Mg acetate, 5 mM DTT, 0.05% Tween 20, 50 μM phenylmethylsulfonyl fluoride at 4°C for 3 h. A fraction of the supernatant and all of the beads (after three washes in binding buffer) were then separately boiled in Laemmli sample buffer. The proteins were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted as described above.
RESULTS
A novel isoform of human RanBP3.
A search of the human EST database revealed several clones that encode a protein motif more similar to the RanBD of the budding yeast gene product Yrb2p than to that of human RanBP1. One such EST, 52358, was obtained and sequenced. Further database searches identified overlapping clones IB1953 and 376469, from which a full-length cDNA could be constructed. The 5′ end of the resulting open reading frame contains an initiation codon that obeys Kozak’s rule (38), upstream of which are several in-frame stop codons. The full-length cDNA encodes a protein of 499 amino acid residues with a predicted molecular mass of 54 kDa. This 499-residue protein corresponds to human RanBP3 isoform b (RanBP3b), which was recently identified through its ability to interact with RCC1 in the yeast two-hybrid system (52). Several additional ESTs were identified that all lacked a 90-bp sequence corresponding to 30 amino acid residues (V321 to W350) within the variant RanBD and which probably represents a splice variant. Consistent with the terminology of Mueller et al., we named the new splice variant RanBP3c.
Northern blot analysis revealed that transcripts of about 4.0 and 2.2 kb were present in all of the tissues analyzed (data not shown). The larger transcript corresponds in length to the complete cDNA sequenced from the overlapping ESTs and supports the conclusion that we have identified the full-length open reading frame. We have been unable to find any ESTs in the database that correspond to the insert region of the isoform RanBP3a.
The overall structure of the RanBP3 protein is similar to that of Yrb2p. Both proteins contain a variant RanBD within the C-terminal half of the protein sequence and two copies of the FXFG nucleoporin peptide motif N terminal to the RanBD. The two variant RanBDs are more similar to one another than to the consensus classical RanBDs found in vertebrate RanBP2 and in all identified eukaryotic RanBP1 sequences. Gene products with greater similarity to the RanBP3/Yrb2p variant RanBDs than to the classical RanBD consensus were also identified in C. elegans (open reading frame RC12C12.2), Schizosaccharomyces pombe (hba1), and Arabidopsis thaliana (GenBank accession no. U23510, U38783, and U62742, respectively), indicating that this domain has been conserved throughout eukaryotic evolution. The RanBP3b sequence also contains a highly acidic C-terminal sequence (EEDDSDDDDV) reminiscent of the acidic C-terminal tail of the Ran GTPase (DEDDDL).
RanBP3 is a predominantly nuclear protein.
In vitro solution assays using recombinant proteins indicate that RanBP3 binds RanGTP with much lower affinity than proteins containing the classical RanBD (52, 80a). These other proteins are predominantly cytosolic (RanBP1) or are associated with the cytoplasmic face of the nuclear pores (RanBP2/NUP358) (reviewed in reference 45). To determine the subcellular distribution of RanBP3b, rabbit antiserum was raised against a GST-RanBP3b(410-499) fusion protein. This C-terminal region of RanBP3b is not similar to any other open reading frames found in the GenBank database but is conserved between RanBP3 isoforms. Anti-RanBP3 antibodies were affinity purified using recombinant RanBP3b coupled to CNBr-Sepharose. The affinity-purified antibodies detected recombinant RanBP3b but not GST by immunoblot assay (not shown).
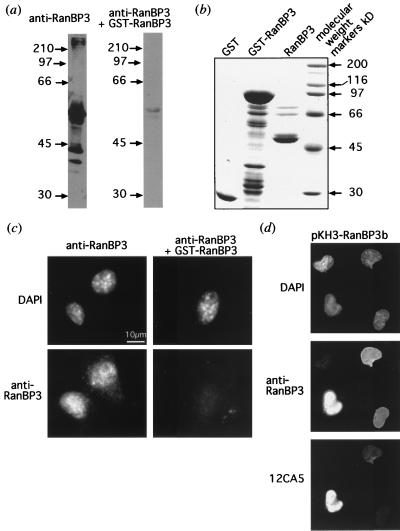
When whole-cell lysate from NIH 3T3 fibroblasts was immunoblotted using anti-RanBP3, a major band of approximately 55 kDa and minor bands of about 40 and 42 kDa were detected in the soluble fraction (Fig. 1a). These bands were almost completely eliminated by competing for the antibody with a protein with GST fused to the carboxy-terminal portion of RanBP3. The major band corresponds in size to that predicted from the sequenced open reading frame of RanBP3, confirming that the cDNA encodes the full-length protein. The minor band at 42 kDa may correspond to the putative splice variant (Fig. 1a).
FIG. 1.
RanBP3b is localized to the cell nucleus. An antibody prepared to the carboxy-terminal 90 residues of RanBP3 (anti-RanBP3) was utilized in immunofluorescence and immunoblot analyses. (a) As described in Materials and Methods, an NIH 3T3 whole-cell lysate was subjected to immunoblot analysis. The anti-RanBP3 was used either alone or in the presence of GST-RanBP3 (amino acids 182 to 499) protein to act as a competitor for the antibodies (the same exposure time was used for both blots). The values beside the lanes are molecular size markers in kilodaltons. (b) Bacterially expressed recombinant proteins, GST, GST-RanBP3, and RanBP3 from which the GST had been cleaved by thrombin, were separated by SDS-PAGE and stained with Coomassie blue. (c) NIH 3T3 cells were fixed, permeabilized, and then incubated with anti-RanBP3 or with anti-RanBP3 plus GST-RanBP3 (amino acids 182 to 499) protein, followed by a secondary fluorescent antibody (Cy3) and DAPI (as described in Materials and Methods). (d) COS cells transfected with pKH3-RanBP3b were treated as described for panel b, except that affinity-purified anti-RanBP3 antibody was used. A 12CA5 (anti-HA) antibody with a secondary fluorescent antibody (FITC conjugated) to visualize the HA-tagged expressed protein was included. The same fields are presented in each of the three panels to show DAPI staining, endogenous RanBP3 (anti-RanBP3), and expressed HA-RanBP3b (12CA5).
Mueller et al. (52) had determined apparent sizes for recombinant RanBP3a and RanBP3b of about 100 and 80 kDa, respectively, by SDS-PAGE, and their antibodies detected bands of 100 and 80 kDa in cell lysates. In our study, RanBP3b does not migrate anomalously on SDS-PAGE and we were unable to detect any bands of 80 to 100 kDa cross-reacting with our anti-RanBP3b antibody. Further, bacterially expressed GST-RanBP3b migrated as expected, at 80 kDa, and after thrombin cleavage to remove the GST, the recombinant protein migrated at about 55 kDa (Fig. 1b). The reason for this discrepancy with the data of Mueller et al. is not known. As noted above, however, although multiple RanBP3b and RanBP3c ESTs are present in the database, we have been unable to find any ESTs that correspond to the insert in RanBP3a. These observations suggest that RanBP3a expression is at a very low level or is confined to minor tissues.
Indirect immunofluorescence of NIH 3T3 cells using anti-RanBP3 serum produced a rather weak nuclear signal, showing diffuse staining with a faint punctate pattern (Fig. 1c). COS and BHK21 cells showed identical staining patterns (data not shown). Anti-RanBP3 staining was largely abolished in the presence of GST-RanBP3b, indicating that this staining pattern was specific. The immunofluorescence indicates that RanBP3 is present within the nucleus rather than being associated with the nuclear pore complexes, despite the presence in RanBP3 of two FXFG nucleoporin repeat motifs. To confirm antibody specificity, COS cells were transfected with pKH3-RanBP3b. pKH3 provides a triple, N-terminal HA1 epitope tag to expressed proteins, which is detectable by the 12CA5 monoclonal antibody. The transfected cell cultures were stained with both 12CA5 and the anti-RanBP3 antibody. Those cells that expressed HA-tagged RanBP3b exhibited a much more intense nuclear fluorescence than did neighboring cells lacking detectable HA tag staining (Fig. 1d). These data confirm that the anti-RanBP3 antibody can detect RanBP3b and that the protein is nuclear.
The nuclear targeting signal of RanBP3 resides in the N-terminal region.
The RanBP3 protein sequence does not possess an obvious polybasic NLS sequence or sequences related to the BIB (32), IBB (22, 80), M9 (71, 78), and KNS (48) domains. RanBP3 is too large to diffuse rapidly through the nuclear pores. We reasoned, therefore, that RanBP3 either binds to another protein with an NLS and enters the nucleus as a passenger or contains an unusual nuclear import signal.
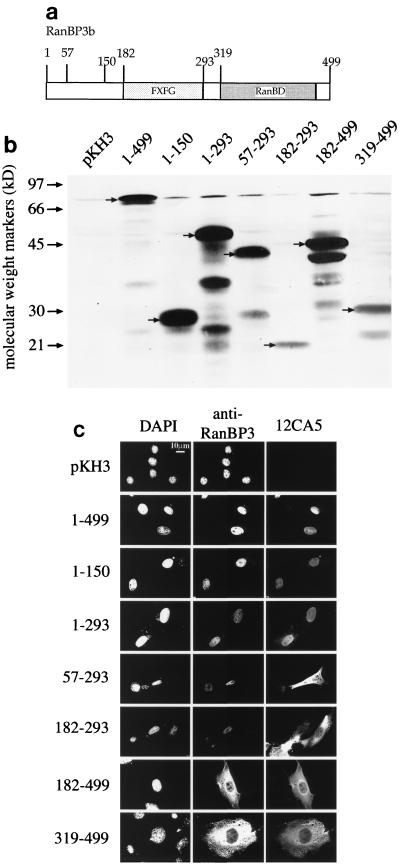
To identify the region of RanBP3 necessary for nuclear localization, fragments of the open reading frame were subcloned into pKH3. The fragments contained one or more of the domains depicted in Fig. 2a. Following transfection, BHK21 cells were fixed and visualized by indirect immunofluorescence using 12CA5 for detection of the triple-HA tag and anti-RanBP3 for detection of endogenous RanBP3 (and expressed fragments of RanBP3b that contained the C-terminal epitope). Note that the endogenous level of RanBP3 is considerably lower than that of the expressed proteins, which made it difficult to image simultaneously both the transfected and untransfected cells in those cases in which the overexpressed protein contained the anti-RanBP3 epitope. Proteins from an extract of transfected cells were separated by SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-HA antibody to confirm that the expressed HA-RanBP3b protein fragments were of the expected size (Fig. 2b; note that the triple-HA sequence reduced the mobility of tagged proteins by about 10 kDa).
FIG. 2.
The amino-terminal domain of RanBP3b is required for nuclear localization. (a) The RanBP3b structural domains are diagrammed with boxes representing the nucleoporin motif domain (FXFG) and the RanBD. The amino acid residue number is shown at each end of the domain. (b) BHK21 cells transfected with fragments of RanBP3b expressed from the pKH3 vector were used in immunofluorescence and immunoblot analyses. Whole-cell lysates were subjected to immunoblot analysis as described in Materials and Methods, using the 12CA5 antibody in conjunction with a secondary antibody labeled with horseradish peroxidase for detection of the expressed HA-tagged proteins. (c) Immunofluorescence of the transfected cells was performed as described in Materials and Methods. Cells were incubated with DAPI, anti-RanBP3 antibodies, and the 12CA5 (anti-HA) antibody, followed by secondary antibodies conjugated to rhodamine and FITC.
Only those fragments that contained the N-terminal region of RanBP3b were localized to the nucleus (Fig. 2c), whereas fragments missing the first 57 or more residues were cytoplasmic. This result suggests that the initial 57 residues of RanBP3b contain either a nuclear targeting signal or a nuclear retention signal. We could not distinguish between these two possibilities because many of the fragments are small enough—in principle—to diffuse through the nuclear pores. They might then be retained within the nucleoplasm by binding to other nuclear proteins. The ectopic expression of HA-RanBP3 did not compromise the nuclear localization of endogenous RanBP3.
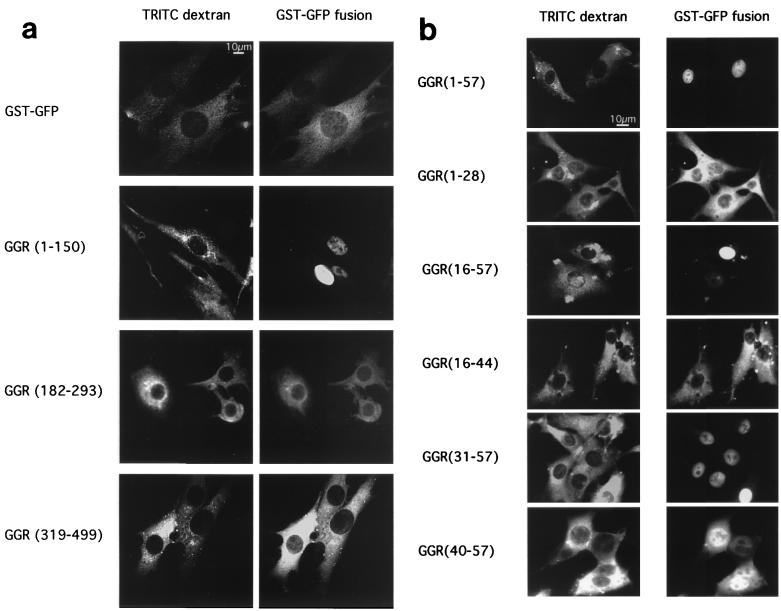
We therefore created recombinant GGR fusion proteins and tested the hypothesis that the N-terminal region of RanBP3b contains an import signal by using a microinjection assay. As described previously (64), GST-GFP contains no intrinsic signal sequences and is incapable of diffusing through the nuclear pore. It also has the advantage of being detectable directly by fluorescence microscopy. BHK21 cells were coinjected with GGR fusion proteins and TRITC-dextran (a marker for the site of injection) in the cytoplasm and maintained at room temperature for 30 min. The fragment containing the N-terminal region [GGR(1-150)] translocated into the nucleus, whereas both the FXFG domain [GGR(182-293)] and the RanBD [GGR(319-499)] remained in the cytoplasm (Fig. 3a). These data confirm that the N-terminal residues of RanBP3b contain a nuclear targeting signal (RanBP3 NLS).
FIG. 3.
Delimitation of the nuclear targeting signal of RanBP3b. Microinjections into the cytoplasm of BHK21 cells were performed as described in Materials and Methods. (a) TRITC-dextran (site-of-injection marker) was coinjected with GGR. Cells were incubated at room temperature for 30 min and then observed by fluorescence microscopy for TRITC or GFP. Representative cells are shown for GST-GFP, GGR(1-150), GGR(182-293), and GGR(319-499). (b) Similarly, GGR with a RanBP3b fragment containing residues 1 to 57, 1 to 28, 16 to 57, 16 to 44, 31 to 57, or 40 to 57 was coinjected with TRITC-dextran as described for panel a. Each fusion protein was injected into at least 100 cells in two separate experiments.
Residues 40 to 57 of RanBP3 contain the nuclear targeting signal.
To further characterize the RanBP3 NLS, GST-GFP fused to smaller fragments of this region was used in the microinjection assay. We found that GGR(1-57) was capable of nuclear import (Fig. 3b), but GGR(58-150) remained cytoplasmic (data not shown). Further constructs containing fragments within residues 1 to 57 narrowed the region required for nuclear import to between residues 40 and 57. Fragments consisting of amino acids 1 to 28, 16 to 44, or 1 to 51 remained in the cytoplasm after 30 min at room temperature (Fig. 3b), a result which suggests that residues between 44 and 57 play a critical role in defining the import signal. Interestingly, both GGR(31-57) and GGR(40-57) entered the nucleus, albeit more slowly than GGR(1-57), which contains the complete N-terminal region. By monitoring of GFP fluorescence at intervals after microinjection, GGR(31-57) and GGR(40-57) were found to require about 25 min (at room temperature) for complete redistribution into the nucleus. However, GGR(16-57) was nuclear within 10 min and the GGR(1-150) construct took only 5 min for the majority of the protein to enter the nucleus (data not shown). Import of proteins containing the RanBP3 NLS is therefore highly efficient.
Unique features of the RanBP3 NLS.
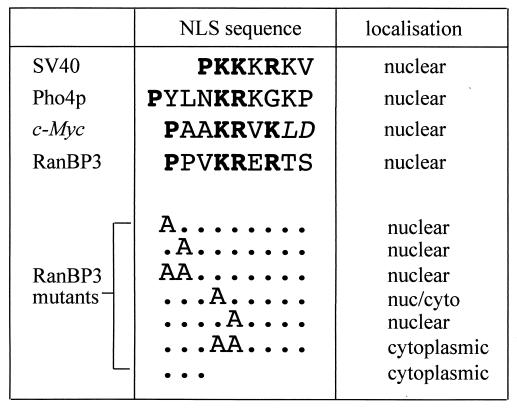
The region of the RanBP3 NLS which has been shown to be essential for nuclear localization, SDREDGNYCPPVKRERTS (residues 40 to 57), contains several features common to classical NLSs (in boldface) (19, 33). It has three basic residues, preceded by two prolines, and another basic residue 10 residues amino terminal to this basic domain, which is the typical spacing for a bipartite NLS, although normally at least two basic residues are present. The crystal structure of karyopherin-α (Srp1p) from yeast revealed two NLS peptide binding pockets. The larger binding site is optimized for the recognition of five lysine or arginine residues, while the smaller binding site can hold two basic residues. The binding sites are separated such that bipartite NLS peptides span both sites. A helix-breaking residue such as proline, prior to the basic stretch of the NLS, helps the basic helix fit into the importin-α groove (12, 18).
To test the possibility that the RanBP3 NLS is a variant of a bipartite NLS, the amino-terminal basic residue, arginine, was mutated to an alanine (R42A). When GGR(40-57) and GGR(40-57, R42A) were microinjected separately into the cytoplasm of BHK21 cells, the results were indistinguishable. In each case, the GGR protein was predominantly nuclear after 20 to 30 min (data not shown). Therefore, R42 is not essential for nuclear import of RanBP3 and the RanBP3 NLS is not bipartite.
The basic domain of the RanBP3 NLS shows the closest similarity to the c-Myc NLS (Fig. 4). Mutational analysis of the c-Myc NLS (43) revealed that the presence of a proline residue amino terminal to the basic residues is essential for efficient import. The RanBP3 NLS has two proline residues adjacent to the basic domain. These were mutated, singly and together, to alanine by PCR mutagenesis to construct further mutant GGR proteins [GGR(1-57, P49A), GGR(1-57, P50A), and GGR(1-57, P49A, P50A)]. Surprisingly, all three recombinant proteins were imported into the nucleus when microinjected into the cytoplasm of BHK21 cells. However, GGR(1-57) showed import within 5 min but the mutant proteins were slower, requiring 20 min for nuclear accumulation to be visible.
FIG. 4.
Mutational analysis of sequence requirements for import function by the RanBP3 NLS. RanBP3 NLS residues 49 to 57 are shown aligned with the NLSs from the SV40 T antigen, Pho4p, and c-Myc. PCR was used to create GGR fusion proteins with the first 57 amino acids of RanBP3 containing the mutations shown. The letters A represent changes to alanine residues, and dots represent the wild-type residues. The last mutation (three dots) represents a deletion of the last six residues. The GGR fusion proteins were microinjected into the cytoplasm of BHK21 cells with TRITC-dextran as a site-of-microinjection marker. After a 30-min incubation at room temperature, the subcellular localization was visualized by direct fluorescence microscopy. Each fusion protein was injected into at least 100 cells in two separate experiments.
The consensus monopartite NLS sequence is KKXK (where X is any amino acid). To determine the importance of the basic residues in the RanBP3 NLS, further mutants were constructed [GGR(1-57, K52A), GGR(1-57, R53A), and GGR(1-57, K52A, R53A)]. In the microinjection assay, GGR(1-57, K52A) showed only very slight nuclear accumulation after 30 min. GGR(1-57, R53A) was imported into the nucleus at the reduced rate obtained with the proline mutant proteins. This result was particularly surprising because in classical NLSs, such as those of SV40 T antigen and c-Myc, the second basic residue is strictly required for nuclear import. This basic residue provides the most energetically favorable interaction in the importin-α binding site (12). The double mutant GGR(1-57, K52A, R53A) completely inhibited import. These mutation results are summarized in Fig. 4.
To test the effects of the point mutations in the context of the full-length protein, GGR(1-499) was produced. GGR(1-499) entered the nucleus more slowly than the N-terminal fragment GGR(1-57), requiring 20 min for complete nuclear accumulation. Either the K52A or the R53A mutation was sufficient to prevent nuclear import of the full-length protein, even 2 h postmicroinjection (data not shown). These data suggest that full-length RanBP3 interacts less efficiently with the import machinery than does the isolated N-terminal domain.
The RanBP3 nuclear import pathway mediated by an importin-α-dependent pathway.
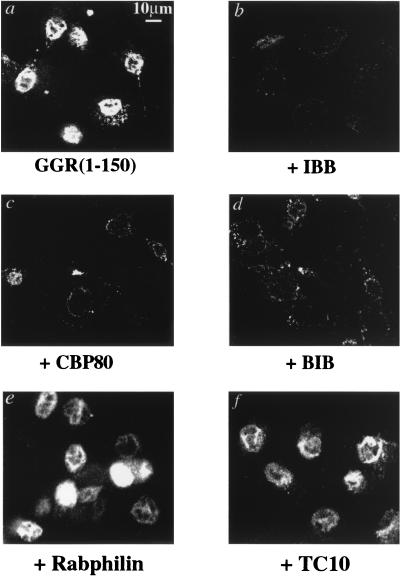
Both the RanBP3 NLS and the NLS of c-Myc are related to an import signal sequence recently identified in the yeast protein Pho4p (Fig. 4). This protein is translocated into the nucleus by Pse1p/Kap121p, a member of the importin-β family of import receptors (34). A mammalian homolog of Pse1p, named RanBP5 or karyopherin-β3, has been identified (17, 83). Neither Pse1p/Kap121p nor RanBP5/karyopherin-β3 interacts with importin-α. Thus, NLSs containing similar sequence motifs can be imported by different mechanisms. It was therefore of interest to identify the receptor pathway that mediates nuclear import of the RanBP3 NLS. To this end, we utilized digitonin-permeabilized BHK21 cells for an in vitro nuclear transport assay, with GGR(1-150) as the import substrate, which was directly visualized by fluorescence microscopy. After 1 h at room temperature in the presence of reticulocyte lysate and an ATP energy-regenerating system, GGR(1-150) was clearly visible within the cell nuclei (Fig. 5a). Another construct, GGR(58-150), which does not contain the nuclear targeting signal, was used as a negative control and produced diffuse background staining (Fig. 5b). When the reaction mixture with GGR(1-150) was incubated on ice, import of the substrate was almost completely abolished (Fig. 5e). When the permeabilized cells were incubated with hexokinase and glucose to deplete residual ATP and the import assay was performed in the absence of an ATP-regenerating system, nuclear accumulation was blocked but the GGR(1-150) substrate remained detectable as a sharply defined ring around the nucleus (Fig. 5c).
FIG. 5.
In vitro assay of RanBP3 NLS nuclear import. In vitro import assays were performed on digitonin-permeabilized BHK21 cells as described in Materials and Methods. Import substrates GGR(1-150) (a) and GGR(58-150) (b) were incubated with reticulocyte lysate and an energy-regenerating system at room temperature for 1 h prior to fixing with paraformaldehyde (4%). The GGR fusion proteins were visualized by fluorescence microscopy. By using GGR(1-150) as the substrate, a number of conditions were tested. Energy was depleted by using hexokinase and glucose (c), the reaction mixture contained no reticulocyte lysate (d), or the reaction mixture was incubated on ice for 1 h (e). Wild-type (wt) Ran-GTP (f), Ran(G19V)-GTP (g), importin-β (amino acids 45 to 462) (h), or WGA (i) was added as described in Materials and Methods. Each experiment was performed twice.
To test the requirement for soluble factors, GGR(1-150) was incubated with permeabilized cells in the absence of reticulocyte lysate. These conditions did not permit nuclear accumulation of the substrate (Fig. 5d). To test whether import was mediated by a Ran-dependent mechanism, the effect of a dominant interfering Ran mutant, Ran G19V, was assayed. Addition of this mutant protein to the in vitro import assay completely blocked accumulation of the GGR(1-150) substrate (Fig. 5g). As a control, we also tested the effect of a similar concentration of wild-type Ran-GTP, which had no discernible effect on nuclear import (Fig. 5f).
Importin-β(45-462) has previously been shown to interfere dominantly with nuclear import of both NLS and M9 target proteins (41). Inhibition likely occurs because the importin-β(45-462) fragment binds to a docking site on the nuclear pore but cannot be released by Ran-GTP. When we included recombinant importin-β(45-462) in the in vitro assay, the nuclear accumulation of GGR(1-150) was almost completely inhibited (Fig. 5h). WGA, a factor that associates with glycosylated nucleoporins and inhibits nuclear pore complex function, had a similar inhibitory effect, although substrate docking remained visible in some cells (Fig. 5i). Therefore, RanBP3 NLS nuclear import is most likely via a Ran-dependent pathway and is receptor mediated.
The most well-defined soluble transport factors are importin-α, importin-β, and transportin. To further investigate the import mechanism utilized by the RanBP3 NLS, a number of factors known to block the importin-α–importin-β pathway were used in permeabilized-cell import assays. The amino terminus of importin-α contains the IBB domain, which acts as a competitive inhibitor of importin-α-mediated import (22). Similarly, the BIB domain from ribosomal protein L23 (32) can act as an inhibitor of importin-β-mediated transport. A peptide containing the importin-α binding region of CBP80 (24) is also an inhibitor of importin-α-mediated nuclear import. Addition of the IBB domain (Fig. 6b), the CBP80 peptide (Fig. 6c), or the BIB domain (Fig. 6d) to the reticulocyte lysate in the permeabilized-cell import assay blocked import of GGR(1-150). As negative controls, peptides corresponding to rabphilin amino acids 611 to 630 and TC10 amino acids 193 to 205 were utilized under the same conditions. Although each contains several basic residues, neither peptide was able to block nuclear import of the RanBP3 NLS (Fig. 6e and f). The use of these unrelated peptides confirms the specificity of the importin binding domains. The same result was obtained when a classical NLS from the SV40 T antigen fused to GST-GFP (GG-NLS) was used as the import substrate (data not shown). These data suggest that RanBP3 NLS nuclear import occurs via an importin-α-dependent pathway.
FIG. 6.
Inhibitors of importin-α and importin-β block RanBP3 NLS nuclear import. In vitro import assays were performed on digitonin-permeabilized BHK21 cells as described in the legend to Fig. 5. The IBB domain of importin-α (b), a synthetic CBP80 peptide (c), the BIB domain of ribosomal protein L23 (d), a synthetic rabphilin peptide (e), or a synthetic TC10 peptide (f) was added in excess to the import substrate, GGR(1-150), as described in Materials and Methods to test for the ability to act as a competitive inhibitor of RanBP3 NLS nuclear import. Each experiment was performed twice.
The RanBP3 NLS is imported more efficiently by importin-α3 than by other members of the importin-α family.
The previous data suggest that import of the RanBP3 NLS is mediated by an importin-α-dependent pathway, and in initial experiments, neither GGR(1-150) nor GG-NLS was imported into the nucleus in the absence of importin-α (data not shown). The unusual sequence of the RanBP3 NLS raised the possibility that it is recognized preferentially by one or another of the different importin-α isoforms present in mammalian cells.
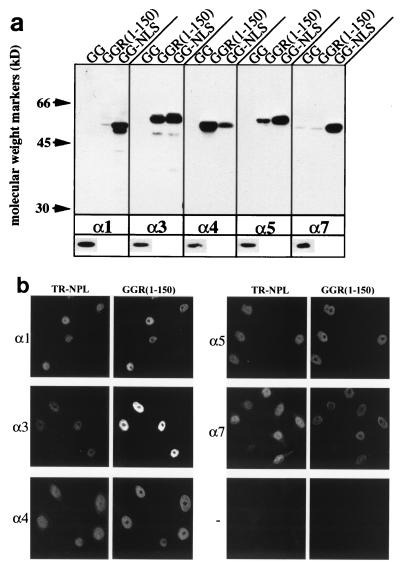
Six human importin-α paralogs have been identified. The nomenclature has become confusing because several of the importins have two or more names. Therefore, we used the numerical nomenclature which best fits the phylogeny of the importin-αs. To clarify the relationship among the various naming systems, Table 1 provides the first 10 amino acid residues for each importin-α, together with the alternative names and the names of the mouse homologs. The importin-αs segregate into three subfamilies, namely, importin-α1, importin-α3 and -α4, and importin-α5, -α6, and -α7 (36, 37). To determine whether the RanBP3 NLS interacts differentially with these proteins, importin-α1, -α3, -α4, -α5, and -α7 (recombinantly expressed importin-α6 is insoluble) were expressed as C-terminal His-tagged fusion proteins in E. coli and used in binding assays. The proteins were incubated with either GGR(1-150) or a GST-GFP (GG) (as a negative control) or GST-GFP fusion to the SV40 NLS (GG-NLS) (as a positive control) in the presence of glutathione-Sepharose beads. After washing, the proteins bound to the beads were separated by SDS-PAGE and then immunoblotted with an anti-His6 antibody to detect the importin-α proteins (Fig. 7a). All importin α proteins bound to GG-NLS, although importin-α4 bound at a reduced level. Both importin-α3 and -α4 bound significantly to GGR(1-150), whereas importin-α5 had reduced binding and neither importin-α1 nor -α7 bound above background levels. There was no detectable background binding to GG, indicating that the interactions are specific.
FIG. 7.
The RanBP3 NLS is preferentially imported by importin-α3. (a) Solution binding assays were performed as described in Materials and Methods. (a) His-tagged importin-α1, -α3, -α4, -α5, or -α7 was incubated with GGR fusion proteins attached to glutathione-Sepharose beads. After extensive washing, the bead-associated proteins were subjected to SDS-PAGE, transferred to nitrocellulose, and immunoblotted with an anti-His6 antibody. The lower panel shows a loading control, 1/20 of the original sample incubated with beads. Each experiment was performed twice with similar results. (b) In vitro import competition assays were performed on digitonin-permeabilized HeLa cells as described in Materials and Methods. GGR(1-150) and TR-NPL were used as competitive import substrates for each importin-α or in the absence of importin-α (−).
To test for functional differences in the interaction of the RanBP3 NLS with importin-α paralogs, a nuclear import assay was performed with GGR(1-150) as the cargo. In this assay, importin-α3 was found to be much more effective than importin-α1 (data not shown). To investigate this differential effect further, a competition assay was performed with permeabilized HeLa cells. In this assay, TR-NPL is provided as an internal control cargo. Nucleoplasmin contains a classical bipartite NLS and is imported with similar efficiency by all importin-αs (37). Differential import of the test cargo can be ascertained by comparison of the fluorescence intensity of TR-NPL with that of the test cargo. As can be seen in Fig. 7b, the RanBP3 NLS (GGR(1-150) was preferentially imported by importin-α3. These data demonstrate the differential binding and import capabilities of the NLS and the RanBP3 NLS for importin-α paralogs.
DISCUSSION
We have shown that nuclear RanBP3 contains an unusual NLS sequence near its N terminus. Residues 40 to 57 of RanBP3 are sufficient to target a carrier protein into the cell nucleus upon injection into the cytoplasm. Although this stretch contains four basic residues, mutational analysis indicated that it does not conform to a classical, polybasic NLS. Interestingly, although imported via the same importin-α/β pathway as the polybasic NLSs, the RanBP3 NLS binds preferentially to specific importin-α paralogs, importin-α3, and -α4 (36, 70). This finding is supported by in vivo import assays in which importin-α3 more efficiently transported the RanBP3 NLS into the nucleus. Thus, the six known mammalian paralogs of importin-α may function in part to discriminate between subsets of NLSs, as has been recently demonstrated in vitro for different substrates (37), and this signaling region may be rather more diverse than has been previously assumed.
RanBP3b was identified as a human open reading frame that resembles Yrb2p, a yeast protein with a variant RanBD and two FXFG nucleoporin repeat motifs. Yrb2p is a nuclear protein that binds Ran-GTP, but only with very low affinity (55). It can also associate with the Ran nucleotide exchange factor RCC1 (called Prp20p in S. cerevisiae) (75). Deletion of the YRB2 gene results in a cold-sensitive phenotype in yeast. The structural similarities between Yrb2p and RanBP3 suggested that they are functional homologs. However, RanBP3b was unable to reverse the cold-sensitive phenotype of the Δyrb2 cells even when expressed from a potent yeast promoter (80a).
As described by Mueller et al. (52), RanBP3 binds to Ran-GTP with an affinity about 3 orders of magnitude lower than that of RanBP1 or the RanBDs of RanBP2 (39, 76). However, the estimated concentration of Ran within the nucleus is 15 to 20 μM and if about 30% of this is in the GTP-bound state (62), then a substantial fraction of RanBP3 is likely to be in a complex with Ran-GTP. The low affinity is most likely a consequence of RanBP3 lacking at least five residues that in the RanBD1 structure interact with Ran-GppNHp (76). Surprisingly, the RanBP3c isoform, which appears to be a splice variant of RanBP3b, lacks much of the variant RanBD and may therefore be unable to interact with Ran at all. The biochemistry and function of this isoform remain to be investigated. One other isoform, RanBP3a, has been described by Mueller et al. and contains a proline-rich sequence near the N terminus. We have not detected a protein of the size reported for RanBP3a in any immunoblots using our anti-RanBP3 antibody (against a constant C-terminal epitope). In addition, we could not locate any human ESTs corresponding to RanBP3a in the database, despite the occurrence of many RanBP3b ESTs and four RanBP3c ESTs. We therefore concluded that RanBP3b is the primary transcript. If RanBP3a is a true isoform rather than a cloning artifact, it may only be present in a limited range of tissues or be very low in abundance.
Yrb2p, like RanBP3, is nuclear and possesses two potential basic NLSs which may direct it to the nucleus (55). The functional significance of the nuclear localization of RanBP3 remains to be determined. However, nuclear export is disrupted in Δyrb2 mutant cells and the overexpression of Xpo1p/Crm1p, a receptor for nuclear export signals, can rescue the Δyrb2-encoded phenotype (56, 74). Crm1p has been shown to bind directly to Yrb2p (56). This failure in nuclear protein export may be the cause of a prolonged delay in the short-spindle phase of mitosis that occurs in Δyrb2 mutant cells (56). Despite the inability of RanBP3 to complement a YRB2 deletion, the overall structural similarities and subcellular distribution argue for a homology of function, a hypothesis that we are currently investigating.
In classical nuclear import, cargo containing a polybasic NLS forms a complex with importin-α and importin-β. Importin-β binds to the NPC, initiating translocation into the nucleus. The high concentration of Ran-GTP in the nucleus causes dissociation of the complex, whereupon the importins are exported back into the cytoplasm for further rounds of import (reviewed in reference 45). Two forms of basic NLS have been well characterized: the first, like that in the SV40 T antigen, is a monopartite sequence with a minimal consensus of KKXK; the second, like that in nucleoplasmin, is bipartite, consisting of two or three basic residues followed by a spacer, followed by a cluster of three to five basic residues (reviewed in references 33 and 45).
The minimum defined sequence of the RanBP3 NLS identified in this study is residues 40 to 57 (SDREDGNYCPPVKRERTS). Deletion of the last six amino acids abolishes nuclear import. Therefore, residues critical for function are present in the C-terminal half of this sequence. Although this region contains a basic-basic-X-basic motif, mutational studies showed that the resemblance to a mono- or bipartite NLS is minimal. In contrast to classical NLSs, in which the mutation of the second basic residue leads to cytoplasmic localization, the 53R mutation reduced the rate of RanBP3 NLS import but did not abolish nuclear localization. Loss of two basic residues, 52K and 53R, was required to completely abolish nuclear import in the RanBP3 NLS. In the context of the full-length protein [GGR(1-499)], either mutation was sufficient to block nuclear import of full-length RanBP3. However, in this construct, the NLS is situated near the center of the polypeptide sequence, rather than near one end [as in the GGR(1-57) construct or native RanBP3] and hence may be less accessible to the transport machinery. In support of this view, import of the unmutated GGR(1-499) construct was unusually slow.
Mutational analysis of the c-Myc NLS has revealed that the sequence flanking the basic region plays an important part in recognition by the import machinery and can override the omission of a basic residue. In particular, a proline residue just upstream of the basic sequence was critical for NLS function and the presence of an acidic amino acid residue C terminal to the NLS also strongly enhanced function (43). However, we found that the mutation of either or both the prolines (49P and 50P) in the RanBP3 NLS resulted in only a moderate reduction in the rate of nuclear import and the C-terminal residues are not related to those of c-Myc. Thus, despite the similarities between the c-Myc and RanBP3 NLS sequences, the level of functional relatedness between key residues in the two sequences is rather low.
Canine parvovirus capsid protein VP1 also contains a sequence which resembles that of the RanBP3 NLS, namely, PAKRARRGYKC. However, mutational analysis has shown that the loss of any one of the basic residues, except the last, resulted in cytoplasmic localization, again suggesting that the basic residues are more central to VP1 NLS function than is the case for the RanBP3 NLS (77). The receptor for VP1 transport is unknown. Another NLS with significant similarity to the RanBP3 NLS is that of the yeast protein Pho4p. Remarkably, Pho4p binds not to importin-α but directly to Pse1p/Kap121p, a member of the karyopherin-β family (34, 65). Thus, similar basic motifs can be distinguished from one another and recognized by distinct importin receptor proteins. Such discrimination may rely on the identities of specific flanking sequences. This idea is supported by observations that the phosphorylation of residues close to a classical NLS can enhance or abolish nuclear import function. The replacement of the phosphorylated residue with an acidic amino acid, so as to maintain the negative charge, mimics this behavior (reviewed in reference 33). Thus, the flanking residues may play a greater role than previously considered and may even dictate receptor specificity.
As discussed in Results, six distinct importin-α proteins have been identified in mammals (13, 15, 36, 53, 70, 73). Although quite divergent near both the N and C termini, all possess closely related armadillo repeats that in yeast karyopherin-α has been shown to form a groove within which the SV40 T-antigen NLS peptide can bind (12, 18). Whether there are distinct but overlapping binding sites on individual importin-αs is still not clear, although there is some evidence in favor of this hypothesis. For example, the SV40 T-antigen NLS binding site on importin-α1 was mapped to residues 81 to 362 whereas the NLS of lymphoid enhancer factor 1 bound to residues 250 to 470 (30). Interestingly, both binding sites incorporate part of the variable sequence outside the armadillo repeats.
Previous studies have found differential binding of nuclear cargoes to the isoforms of importin-α. For example, when NLS-conjugated affinity columns were used to precipitate importin-α5 and importin-α1 from cytosolic extracts, differential binding patterns were observed, dependent on the NLS used for the pull-down and the cell line used as a source of extract (54). Importin-α3 was cloned in a two-hybrid screen by its interaction with DNA helicase Q1. The same screen isolated importin-α1 but not importin-α5, and further testing showed that importin-α5 did not interact with DNA helicase Q1 (70). Further analysis showed that importin-α3 preferentially bound and imported the DNA helicase Q1 NLS (CYGSKNTGAKKRKIDDA) (49). The NLS of lymphoid enhancer factor 1, KKKKRKREK, but not the very similar NLS of T-cell factor 1, KKKRRSREK, bound to importin-α5 and importin-α1 (60). The receptor for T-cell factor 1 is unidentified. Furthermore, in vitro studies showed that some proteins are imported preferentially by one specific importin-α isoform whereas other substrates, like nucleoplasmin, can be imported by all soluble importin-α proteins with only marginal differences (37).
Importin-α3 has been shown to bind three very diverse proteins, RanBP3 (this study), DNA helicase Q1 (49, 70), and tissue transglutaminase (59). No obvious sequence similarity, which may constitute an importin-α3-specific NLS, exist among these three proteins, although they all contain a stretch of three or four basic residues. It may be that the other importin-α3 subfamily member, importin-α4, will also bind these proteins.
The definition of a minimal importin-α NLS consensus sequence has proven much more difficult than had been initially assumed. While many proteins contain recognizable polybasic sequences that function as nuclear targeting signals, some—such as those in several ribosomal proteins and histones—interact directly with certain members of the importin-β family rather than importin-α (9, 32). Arginine-rich sequences found in importin-α itself, in Rev, and in the Rex protein of human T-cell leukemia virus type 1 are transported into the nucleus by direct binding to importin-β (29, 58). The c-Myc NLS, which does bind to importin-α, requires an upstream proline in addition to basic residues (43). We have now found that the RanBP3 NLS does not require an upstream proline and that even the second basic residue—known to be critical in other NLSs—is not essential. Determination of a consensus NLS has been complicated by the discovery of a family of at least six mammalian importin-αs. It is notable that the yeast genome contains only 1 importin-α gene and 14 importin-β family genes. The human EST database contains sequences related to these 14 importin-βs, but there is no evidence for a divergence of these genes into large families during metazoan evolution. Rather, the importin-α family has diversified, perhaps to accommodate and discriminate among a broadening array of cargoes that need to be transported in a regulated fashion into the nucleus.
ACKNOWLEDGMENTS
We thank Dirk Gorlich, Steve Adam, Mark Rush, and Bryce Paschal for the kind gifts of expression clones and proteins necessary for performing in vitro import assays (nucleoplasmin, nucleoplasmin core, human Ran, Rna1p, RanBP1, NTF2, importin-α1, importin-β, and the IBB and BIB domains). We also thank Amy Brownawell, Mike Nemergut, Clark Peterson, Kendra Plafker, and Alicia Smith from the Macara laboratory for reagents and helpful suggestions.
This work was supported by a grant from the National Institutes of Health, DHHS (GM-50526).
REFERENCES
- 1.Adam S A, Gerace L. Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell. 1991;66:837–847. doi: 10.1016/0092-8674(91)90431-w. [DOI] [PubMed] [Google Scholar]
- 2.Adam S A, Sterne-Marr R, Gerace L. Nuclear protein import using digitonin-permeabilized cells. Methods Enzymol. 1992;219:97–110. doi: 10.1016/0076-6879(92)19013-v. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Boguski M S, Gish W, Wootton J C. Issues in searching molecular sequence databases. Nat Genet. 1994;6:119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- 4.Beddow A L, Richards S A, Orem N R, Macara I G. The Ran/TC4 GTPase-binding domain: identification by expression cloning and characterization of a conserved sequence motif. Proc Natl Acad Sci USA. 1995;92:3328–3332. doi: 10.1073/pnas.92.8.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischoff F R, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bischoff F R, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff F R, Ponstingl H. Mitotic regulator protein RCC1 is complexed with a nuclear Ras-related polypeptide. Proc Natl Acad Sci USA. 1991;88:10830–10834. doi: 10.1073/pnas.88.23.10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogerd H P, Benson R E, Truant R, Herold A, Phingbodhipakkiya M, Cullen B R. Definition of a consensus transportin-specific nucleocytoplasmic transport signal. J Biol Chem. 1999;274:9771–9777. doi: 10.1074/jbc.274.14.9771. [DOI] [PubMed] [Google Scholar]
- 9.Bonifaci N, Moroianu J, Radu A, Blobel G. Karyopherin beta-2 mediates nuclear import of a mRNA binding protein. Proc Natl Acad Sci USA. 1997;94:5055–5060. doi: 10.1073/pnas.94.10.5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carey K L, Richards S A, Lounsbury K M, Macara I G. Evidence using a green fluorescent protein-glucocorticoid receptor chimera that the Ran/TC4 GTPase mediates an essential function independent of nuclear protein import. J Cell Biol. 1996;133:985–996. doi: 10.1083/jcb.133.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chi N C, Adam E J, Adam S A. Sequence and characterization of cytoplasmic nuclear protein import factor p97. J Cell Biol. 1995;130:265–274. doi: 10.1083/jcb.130.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- 13.Cortes P, Ye Z S, Baltimore D. RAG-1 interacts with the repeated amino acid motif of the human homologue of the yeast protein SRP1. Proc Natl Acad Sci USA. 1994;91:7633–7637. doi: 10.1073/pnas.91.16.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coutavas E, Ren M, Oppenheim J D, D’Eustachio P, Rush M G. Characterization of proteins that interact with the cell-cycle regulatory protein Ran/TC4. Nature. 1993;366:585–587. doi: 10.1038/366585a0. [DOI] [PubMed] [Google Scholar]
- 15.Cuomo C A, Kirch S A, Gyuris J, Brent R, Oettinger M A. Rch1, a protein that specifically interacts with the RAG-1 recombination-activating protein. Proc Natl Acad Sci USA. 1994;91:6156–6160. doi: 10.1073/pnas.91.13.6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis L I. The nuclear pore complex. Annu Rev Biochem. 1995;64:865–896. doi: 10.1146/annurev.bi.64.070195.004245. [DOI] [PubMed] [Google Scholar]
- 17.Deane R, Schafer W, Zimmermann H P, Mueller L, Gorlich D, Prehn S, Ponstingl H, Bischoff F R. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-beta but interacts differentially with RanBP1. Mol Cell Biol. 1997;17:5087–5096. doi: 10.1128/mcb.17.9.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dingwall C, Laskey R A. Nuclear import: a tale of two sites. Curr Biol. 1998;8:R922–R924. doi: 10.1016/s0960-9822(98)00010-4. [DOI] [PubMed] [Google Scholar]
- 19.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 20.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 21.Gorlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. A novel class of RanGTP binding proteins. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorlich D, Henklein P, Laskey R A, Hartmann E. A 41 amino acid motif in importin-alpha confers binding to importin-beta and hence transit into the nucleus. EMBO J. 1996;15:1810–1817. [PMC free article] [PubMed] [Google Scholar]
- 23.Gorlich D, Kostka S, Kraft R, Dingwall C, Laskey R A, Hartmann E, Prehn S. Two different subunits of importin cooperate to recognize nuclear localization signals and bind them to the nuclear envelope. Curr Biol. 1995;5:383–392. doi: 10.1016/s0960-9822(95)00079-0. [DOI] [PubMed] [Google Scholar]
- 24.Gorlich D, Kraft R, Kostka S, Vogel F, Hartmann E, Laskey R A, Mattaj I W, Izaurraide E. Importin provides a link between nuclear protein import and U snRNA export. Cell. 1996;87:21–32. doi: 10.1016/s0092-8674(00)81319-7. [DOI] [PubMed] [Google Scholar]
- 25.Gorlich D, Pante N, Kutay U, Aebi U, Bischoff F R. Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J. 1996;15:5584–5594. [PMC free article] [PubMed] [Google Scholar]
- 26.Gorlich D, Prehn S, Laskey R A, Hartmann E. Isolation of a protein that is essential for the first step of nuclear protein import. Cell. 1994;79:767–778. doi: 10.1016/0092-8674(94)90067-1. [DOI] [PubMed] [Google Scholar]
- 27.Gorlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Distinct functions for the two importin subunits in nuclear protein import. Nature. 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 28.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- 29.Henderson B R, Percipalle P. Interactions between HIV Rev and nuclear import and export factors—the Rev nuclear localisation signal mediates specific binding to human importin-beta. J Mol Biol. 1997;274:693–707. doi: 10.1006/jmbi.1997.1420. [DOI] [PubMed] [Google Scholar]
- 30.Herold A, Truant R, Wiegand H, Cullen B R. Determination of the functional domain organization of the importin alpha nuclear import factor. J Cell Biol. 1998;143:309–318. doi: 10.1083/jcb.143.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Izaurralde E, Kutay U, Vonkobbe C, Mattaj I W, Gorlich D. The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J. 1997;16:6535–6547. doi: 10.1093/emboj/16.21.6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakel S, Gorlich D. Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jans D A, Hubner S. Regulation of protein transport to the nucleus—central role of phosphorylation. Physiol Rev. 1996;76:651–685. doi: 10.1152/physrev.1996.76.3.651. [DOI] [PubMed] [Google Scholar]
- 34.Kaffman A, Rank N M, Oshea E K. Phosphorylation regulates association of the transcription factor Pho4 with its import receptor Pse1/Kapl21. Genes Dev. 1998;12:2673–2683. doi: 10.1101/gad.12.17.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalderon D, Roberts B L, Richardson W D, Smith A E. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- 36.Köhler M, Ansieau S, Prehn S, Leutz A, Haller H, Hartmann E. Cloning of two novel human importin-alpha subunits and analysis of the expression pattern of the importin-alpha protein family. FEBS Lett. 1997;417:104–108. doi: 10.1016/s0014-5793(97)01265-9. [DOI] [PubMed] [Google Scholar]
- 37.Köhler M, Speck C, Christiansen M, Bischoff F R, Prehn S, Haller H, Gorlich D, Hartmann E. Evidence for distinct substrate specificities of importin α-family members in nuclear protein import. Mol Cell Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozak M. Structural features in eukaryotic mRNAs that modulate the initiation of translation. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 39.Kuhlmann J, Macara I, Wittinghofer A. Dynamic and equilibrium studies on the interaction of Ran with its effector, RanBP1. Biochemistry. 1997;36:12027–12035. doi: 10.1021/bi970524k. [DOI] [PubMed] [Google Scholar]
- 40.Kussel P, Frasch M. Yeast Srp1, a nuclear protein related to Drosophila and mouse pendulin, is required for normal migration, division, and integrity of nuclei during mitosis. Mol Gen Genet. 1995;248:351–363. doi: 10.1007/BF02191602. [DOI] [PubMed] [Google Scholar]
- 41.Kutay U, Izaurralde E, Bischoff F R, Mattaj I W, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahajan R, Delphin C, Guan T L. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/s0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 43.Makkerh J P S, Dingwall C, Laskey R A. Comparative mutagenesis of nuclear localization signals reveals the importance of neutral and acidic amino acids. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- 44.Malik H S, Eickbush T H, Goldfarb D S. Evolutionary specialization of the nuclear targeting apparatus. Proc Natl Acad Sci USA. 1997;94:13738–13742. doi: 10.1073/pnas.94.25.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattaj I, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 46.Mattingly R R, Sorisky A, Brann M R, Macara I G. Muscarinic receptors transform NIH 3T3 cells through a Ras-dependent signalling pathway inhibited by the Ras-GTPase-activating protein SH3 domain. Mol Cell Biol. 1994;14:7943–7952. doi: 10.1128/mcb.14.12.7943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matunis M J, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michael W M, Eder P S, Dreyfuss G. The K nuclear shuttling domain—a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO J. 1997;16:3587–3598. doi: 10.1093/emboj/16.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyamoto Y, Imamoto N, Sekimoto T, Tachibana T, Seki T, Tada S, Enomoto T, Yoneda Y. Differential modes of nuclear localization signal (NLS) recognition by three distinct classes of NLS receptors. J Biol Chem. 1997;272:26375–26381. doi: 10.1074/jbc.272.42.26375. [DOI] [PubMed] [Google Scholar]
- 50.Moore M S, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 51.Moroianu J, Blobel G, Radu A. Previously identified protein of uncertain function is karyopherin alpha and together with karyopherin beta docks import substrate at nuclear pore complexes. Proc Natl Acad Sci USA. 1995;92:2008–2011. doi: 10.1073/pnas.92.6.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mueller L, Cordes V C, Bischoff F R, Ponstingl H. Human RanBP3, a group of nuclear RanGTP binding proteins. FEBS Lett. 1998;427:330–336. doi: 10.1016/s0014-5793(98)00459-1. [DOI] [PubMed] [Google Scholar]
- 53.Nachury M V, Ryder U W, Lamond A I, Weis K. Cloning and characterization of hSrp1-gamma, a tissue-specific nuclear transport factor. Proc Natl Acad Sci USA. 1998;95:582–587. doi: 10.1073/pnas.95.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nadler S G, Tritschler D, Haffar O K, Blake J, Bruce A G, Cleaveland J S. Differential expression and sequence-specific interaction of karyopherin alpha with nuclear localization sequences. J Biol Chem. 1997;272:4310–4315. doi: 10.1074/jbc.272.7.4310. [DOI] [PubMed] [Google Scholar]
- 55.Noguchi E, Hayashi N, Nakashima N, Nishimoto T. Yrb2p, a Nup2p-related yeast protein, has a functional overlap with Rna1p, a yeast Ran-Gtpase-activating protein. Mol Cell Biol. 1997;17:2235–2246. doi: 10.1128/mcb.17.4.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noguchi E, Saitoh Y H, Sazer S, Nishimoto T. Disruption of the YRB2 gene retards nuclear protein export, causing a profound mitotic delay, and can be rescued by overexpression of XPO1/CRM1. J Biochem. 1999;125:574–585. doi: 10.1093/oxfordjournals.jbchem.a022323. [DOI] [PubMed] [Google Scholar]
- 57.O’Neil R E, Palese P. NPI-1, the human homolog of SRP-1, interacts with influenza virus nucleoprotein. Virology. 1995;206:116–125. doi: 10.1016/s0042-6822(95)80026-3. [DOI] [PubMed] [Google Scholar]
- 58.Palmeri D, Halim M H. Importin beta can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Peng X J, Zhang Y H, Zhang H F, Graner S, Williams J F, Levitt M L, Lokshin A. Interaction of tissue transglutaminase with nuclear transport protein importin-alpha 3. FEBS Lett. 1999;446:35–39. doi: 10.1016/s0014-5793(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 60.Prieve M G, Guttridge K L, Munguia J, Waterman M L. Differential importin-α recognition and nuclear transport by nuclear localization signals within the high-mobility-group DNA binding domains of lymphoid enhancer factor 1 and T-cell factor 1. Mol Cell Biol. 1998;18:4819–4832. doi: 10.1128/mcb.18.8.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Radu A, Blobel G, Moore M S. Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA. 1995;92:1769–1773. doi: 10.1073/pnas.92.5.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A. The 1.7-angstrom crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature. 1998;392:97–101. doi: 10.1038/32204. [DOI] [PubMed] [Google Scholar]
- 63.Ribbeck K, Lipowsky G, Kent H M, Stewart M, Gorlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Richards S A, Carey K L, Macara I G. Requirement of guanosine triphosphate-bound Ran for signal-mediated nuclear protein export. Science. 1997;276:1842–1844. doi: 10.1126/science.276.5320.1842. [DOI] [PubMed] [Google Scholar]
- 65.Rout M P, Blobel G, Aitchison J D. A distinct nuclear import pathway used by ribosomal proteins. Cell. 1997;89:715–725. doi: 10.1016/s0092-8674(00)80254-8. [DOI] [PubMed] [Google Scholar]
- 66.Rout M P, Wente S R. Pores for thought: nuclear pore complex proteins. Trends Cell Biol. 1994;4:357–365. doi: 10.1016/0962-8924(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 67.Saitoh H, Sparrow D B, Shiomi T, Pu R T, Nishimoto T, Mohun T J, Dasso M. Ubc9p and the conjugation of Sumo-1 to RanGAP1 and RanBP2. Curr Biol. 1997;8:121–124. doi: 10.1016/s0960-9822(98)70044-2. [DOI] [PubMed] [Google Scholar]
- 68.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 69.Schlenstedt G, Wong D H, Koepp D M, Silver P A. Mutants in a yeast Ran binding protein are defective in nuclear transport. EMBO J. 1995;14:5367–5378. doi: 10.1002/j.1460-2075.1995.tb00221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seki T, Tada S, Katada T, Enomoto T. Cloning of a cDNA encoding a novel importin-alpha homologue, Qip1: discrimination of Qip1 and Rch1 from hSrp1 by their ability to interact with DNA helicase Q1/RecQL. Biochem Biophys Res Commun. 1997;234:48–53. doi: 10.1006/bbrc.1997.6535. [DOI] [PubMed] [Google Scholar]
- 71.Siomi H, Dreyfuss G. A nuclear localization domain in the hnRNP A1 protein. J Cell Biol. 1995;129:551–560. doi: 10.1083/jcb.129.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smith A, Brownawell A, Macara I G. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- 73.Takeda S, Fujiwara T, Shimizu F, Kawai A, Shinomiya K, Okuno S, Ozaki K, Katagiri T, Shimada Y, Nagata M, Watanabe T, Takaichi A, Kuga Y, Suzuki M, Hishigaki H, Takahashi E, Shin S, Nakamura Y, Hirai Y. Isolation and mapping of karyopherin alpha 3 (KPNA3), a human gene that is highly homologous to genes encoding Xenopus importin, yeast SRP1 and human RCH1. Cytogenet Cell Genet. 1997;76:87–93. doi: 10.1159/000134521. [DOI] [PubMed] [Google Scholar]
- 74.Taura T, Krebber H, Silver P A. A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc Natl Acad Sci USA. 1998;95:7427–7432. doi: 10.1073/pnas.95.13.7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taura T, Schlenstedt G, Silver P A. Yrb2p is a nuclear protein that interacts with Prp20p, a yeast Rcc1 homologue. J Biol Chem. 1997;272:31877–31884. doi: 10.1074/jbc.272.50.31877. [DOI] [PubMed] [Google Scholar]
- 76.Vetter I R, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 77.Vihinenranta M, Kakkola L, Kalela A, Vilja P, Vuento M. Characterization of a nuclear localization signal of canine parvovirus capsid proteins. Eur J Biochem. 1997;250:389–394. doi: 10.1111/j.1432-1033.1997.0389a.x. [DOI] [PubMed] [Google Scholar]
- 78.Weighardt F, Biamonti G, Riva S. Nucleo-cytoplasmic distribution of human hnRNP proteins: a search for the targeting domains in hnRNP A1. J Cell Sci. 1995;108:545–555. doi: 10.1242/jcs.108.2.545. [DOI] [PubMed] [Google Scholar]
- 79.Weis K, Mattaj I W, Lamond A I. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science. 1995;268:1049–1053. doi: 10.1126/science.7754385. [DOI] [PubMed] [Google Scholar]
- 80.Weis K, Ryder U, Lamond A I. The conserved amino-terminal domain of hSRP1 alpha is essential for nuclear protein import. EMBO J. 1996;15:1818–1825. [PMC free article] [PubMed] [Google Scholar]
- 80a.Welch, K., and I. G. Macara. Unpublished data.
- 81.Wozniak R W, Rout M P, Aitchison J D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 82.Wu J, Matunis M J, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–14213. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 83.Yaseen N R, Blobel G. Cloning and characterization of human karyopherin beta-3. Proc Natl Acad Sci USA. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–188. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]