Two structures of human 3-hydroxyanthranilate 3,4-dioxygenase, one with iron bound at the active site and one with zinc bound at the active site, are reported.
Keywords: 3-hydroxyanthranilate 3,4-dioxygenase; oxidoreductase; kynurenine pathway; quinolinic acid; neurodegeneration
Abstract
3-Hydroxyanthranilate 3,4-dioxygenase (3HAO) is an enzyme in the microglial branch of the kynurenine pathway of tryptophan degradation. 3HAO is a non-heme iron-containing, ring-cleaving extradiol dioxygenase that catalyzes the addition of both atoms of O2 to the kynurenine pathway metabolite 3-hydroxyanthranilic acid (3-HANA) to form quinolinic acid (QUIN). QUIN is a highly potent excitotoxin that has been implicated in a number of neurodegenerative conditions, making 3HAO a target for pharmacological downregulation. Here, the first crystal structure of human 3HAO with the native iron bound in its active site is presented, together with an additional structure with zinc (a known inhibitor of human 3HAO) bound in the active site. The metal-binding environment is examined both structurally and via inductively coupled plasma mass spectrometry (ICP-MS), X-ray fluorescence spectroscopy (XRF) and electron paramagnetic resonance spectroscopy (EPR). The studies identified Met35 as the source of potential new interactions with substrates and inhibitors, which may prove useful in future therapeutic efforts.
1. Introduction
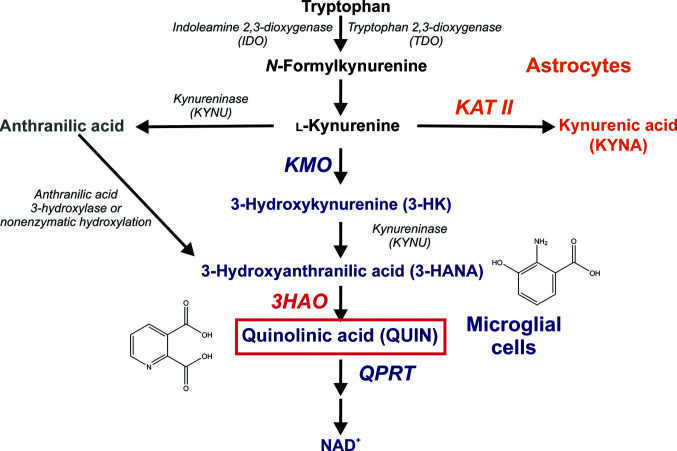
The kynurenine pathway (KP) accounts for most of the tryptophan metabolism in both peripheral organs and the brain, shuttling tryptophan to de novo NAD+ synthesis for use in fundamental metabolic processes (Leklem, 1971 ▸). In the brain, the pathway is compartmentalized (Fig. 1 ▸), with the kynurenine aminotransferase (KAT)-containing branch residing primarily in astrocytes and the 3-hydroxyanthranilate 3,4-dioxygenase (3HAO)-containing branch residing primarily in the microglia. The respective roles of these two metabolic arms are not well understood. What is known is that the KP plays a key role in a network of immune and inflammatory processes that mediate both normal and pathological neurological states (Schwarcz et al., 2012 ▸). Such regulatory control of the metabolic landscape of the brain stems in part from the production by the KP of several neuroactive metabolites, all of which need to be maintained in balance to protect neurons and preserve cognitive functions. In particular, the KAT branch produces kynurenic acid (KYNA), which is neuroprotective at normal levels (Hardeland et al., 1999 ▸; Lugo-Huitrón et al., 2011 ▸) but when elevated antagonizes receptors that are required for learning and memory, resulting in cognitive deficits (Chess et al., 2007 ▸; Perkins & Stone, 1982 ▸). The 3HAO branch produces the N-methyl-d-aspartate (NMDA) receptor agonist quinolinic acid (QUIN) and its bioprecursor, the free-radical generator 3-hydroxykynurenine (3-HK), which are toxic to neurons if present at abnormally high levels (Goldstein et al., 2000 ▸; Rios & Santamaria, 1991 ▸). These metabolites are potential causative agents in acute and chronic brain diseases, making understanding how the balance of metabolites is maintained in the KP critical for both understanding and treating a number of neuropathological conditions.
Figure 1.
The major reactions that occur via the kynurenine pathway of tryptophan degradation. The enzyme 3-hydroxyanthranilic acid dioxygenase (3HAO, red) generates the potent excitotoxin quinolinic acid (QUIN, red box) as part of the microglial branch of the kynurenine pathway.
Of the KP metabolites, QUIN has been implicated in a number of neurodegenerative conditions (Giorgini et al., 2005 ▸; Guidetti et al., 2004 ▸; Gulaj et al., 2010 ▸; Pearson & Reynolds, 1992 ▸; Tai et al., 2007 ▸; Toyn et al., 2010 ▸), making it a leading KP target for pharmacological downregulation. When elevated, QUIN exerts neurotoxic effects by a number of mechanisms. One is via the generation of free radicals which destroy neurons (Rios & Santamaria, 1991 ▸; Santamaría et al., 2001 ▸). In addition, QUIN is a selective NMDA receptor agonist that causes excitotoxic lesions when administered intracerebrally (Schwarcz et al., 1983 ▸). Thirdly, too much QUIN triggers excessive glutamate release as another route to neurotoxicity (Tavares et al., 2000 ▸, 2002 ▸; Ting et al., 2009 ▸). These and other neurotoxic effects are exacerbated by the lack of an effective QUIN removal system, which also contributes to its potent excitotoxic properties in vivo (Foster et al., 1984 ▸). Moreover, since QUIN is produced by microglia, which constitute the main active immune response in the central nervous system, its levels can be expected to increase in areas of the brain that have been subjected to an insult via infection, trauma or underlying pathology. Thus, such events, either alone or in conjunction with dysregulated QUIN metabolism, can significantly elevate local QUIN concentrations (Kita et al., 2002 ▸), thereby potentiating a number of neurodegenerative pathologies such as Huntington’s disease (Schwarcz et al., 2010 ▸), Alzheimer’s disease (Toyn et al., 2010 ▸), HIV-induced dementia (Drewes et al., 2015 ▸) and secondary brain damage following traumatic brain injury (Darlington et al., 2007 ▸; Saito, Nowak, Markey et al., 1993 ▸; Saito, Nowak, Suyama et al., 1993 ▸). Quinolinic acid is formed by 3-hydroxyanthranilic acid oxygenase, which is a non-heme iron-containing, ring-cleaving extradiol dioxygenase that catalyzes the addition of both atoms of O2 to the KP metabolite 3-hydroxyanthranilic acid (3-HANA). Structural and biochemical data for Ralstonia metallidurans 3HAO (Colabroy et al., 2005 ▸; Zhang et al., 2005 ▸) indicate that the 3HAO reaction is initiated by substrate binding, which induces closure of the active site. Oxygen then binds to the active-site iron. Transfer of an electron from the active-site iron to oxygen creates an oxygen radical, thereby facilitating the addition of both O atoms to 3-HANA and forming 2-amino-3-carboxymuconic 6-semialdehyde (ACMS), an active intermediate (Foster et al., 1986 ▸; Long et al., 1954 ▸). ACMS then spontaneously cyclizes to form QUIN, which can in turn be catabolized by quinolinic acid phosphoribosyltransferase (QPRT) as part of the de novo NAD+-biosynthetic pathway.
Because of its biological impact, it is important to understand the structure and function of 3HAO, particularly the human enzyme. Some bacterial species, such as R. metallidurans and Cupriavidus metallidurans, have 3HAO homologues that have been studied extensively (Colabroy et al., 2005 ▸; Liu et al., 2015 ▸; Zhang et al., 2005 ▸). However, unlike mammalian 3HAO, these enzymes are dimeric and contain an iron–sulfur cluster that appears to act as a metal reservoir for the enzyme. A structure has also been reported for yeast 3HAO (Li et al., 2006 ▸) which, like its bacterial counterparts, is dimeric and contains an iron–sulfur cluster. One structure of a mammalian 3HAO, that of the bovine enzyme (Đilović et al., 2009 ▸), has been published, and a human 3HAO crystal structure with a non-native nickel ion coordinated has been deposited in the PDB (PDB entry 2qnk; E. Bitto, C. A. Bingman, G. E. Wesenberg & G. N. Phillips Jr, unpublished work) without an associated publication. Here, we present two high-resolution structures of human 3HAO (h3HAO). In one structure the native iron is present in the active site, while the other contains zinc, which is known to inhibit 3HAO activity (Calderone et al., 2002 ▸). Zinc is a physiologically relevant metal ion, particularly in the brain, where 3HAO plays a crucial role in the kynurenine pathway. In particular, zinc has been implicated in several conditions that promote neurodegeneration, such as cerebral ischemia (Thompson, 2012 ▸) and Alzheimer’s disease (Li & Wang, 2016 ▸). Moreover, zinc has been shown to directly stimulate microglial activation (Higashi et al., 2011 ▸; Kauppinen et al., 2008 ▸), a process that when dysregulated can induce neurodegeneration, in part via increased production of quinolinic acid by 3HAO (Kita et al., 2002 ▸). The role of zinc in pathophysiology of the brain is multi-factorial and any role that its inhibitory effect on h3HAO activity might play in such conditions remains to be elucidated.
2. Materials and methods
2.1. Protein expression and purification
The expression plasmid for h3HAO was purchased from the Arizona State University protein structure initiative. It contains a 6×His tag/maltose-binding protein (6×His-MBP) fusion followed by a Tobacco etch virus (TEV) protease cleavage site preceding the insert. The expression plasmid was transformed into the BL21 (DE3) strain of Escherichia coli. The cells were grown at 37°C to an OD600 of 0.6 and induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at 25°C overnight. The cells were harvested at 4000 rev min−1 for 20 min and resuspended in lysis buffer consisting of 20 mM NaH2PO4, 500 mM NaCl. The cells were lysed by sonication and soluble proteins were separated by centrifugation at 15 000 rev min−1 for 45 min. The clarified lysate was first purified using nickel-affinity chromatography and then buffer-exchanged into TEV protease cleavage buffer (20 mM NaH2PO4, 150 mM NaCl). The 6×His-MBP tag was removed from the purified h3HAO by TEV protease cleavage according to the manufacturer’s instructions. Cleaved h3HAO was isolated from the 6×His-MBP tag by successive rounds of nickel-affinity and amylose-affinity chromatography. The partially purified 3HAO was further subjected to size-exclusion chromatography as a final purification step. The purified protein was concentrated to 15 mg ml−1 using a centrifugal concentrator.
2.2. Metal loading, crystallization, data collection and processing
Purified 3HAO was incubated with a 1.25 molar excess of either ZnSO4 or (NH4)2Fe(SO4)2 overnight prior to crystallization. We note that, based on our biophysical characterization below, any iron-loaded 3HAO not prepared in an anaerobic environment is likely to contain iron(III) in the active site. Thus, all iron-loaded 3HAO not prepared in an anaerobic environment will be referred to as Fe(III)-3HAO throughout this manuscript. The best crystals from our screens were obtained in a condition consisting of 0.1 M HEPES pH 7.5, 2% PEG 400, 2.0 M ammonium sulfate. Crystals appeared after 2–3 d and grew to maximum size in a week. These crystals belonged to space group P6522 and have unit-cell dimensions a = b = 57.3, c = 417.1 Å, α = β = 90, γ = 120°. Crystals were transferred to 15% glycerol in the crystallization condition and were flash-cooled in liquid nitrogen prior to data collection. Native data to 1.88 Å (Fe) and 1.75 Å (Zn) resolution were collected on beamline 12-2 at Stanford Synchrotron Radiation Laboratory (SSRL). The data were reduced using autoXDS (Kabsch, 2010 ▸) and AIMLESS (Evans & Murshudov, 2013 ▸) from the CCP4 program suite (Winn et al., 2011 ▸). A summary of the data-collection statistics for each crystal is provided in Table 1 ▸.
Table 1. Data-collection and refinement statistics.
| Fe(III)-h3HAO | Zn(II)-h3HAO | |
|---|---|---|
| PDB code | 5tk5 | 5tkq |
| Data collection | ||
| Wavelength (Å) | 1.5498 | 1.12709 |
| Space group | P6522 | P6522 |
| Unit-cell parameters (Å) | a = 57.08, b = 57.08, c = 415.87 | a = 57.34, b = 57.34, c = 417.10 |
| Resolution (Å) | 38.0–1.88 | 38.15–1.75 |
| Unique reflections | 34383 | 42900 |
| Completeness (%) | 99.2 (87.2) | 99.7 (100) |
| Multiplicity | 32.9 (15.6) | 10.1 (10.0) |
| 〈I/σ(I)〉 | 8.8 (1.8) | 11.7 (0.4) |
| R p.i.m. | 0.084 (1.220) | 0.099 (2.061) |
| CC1/2 | 0.982 (0.063) | 0.995 (0.126) |
| Wilson B factor (Å2) | 19.9 | 23.8 |
| Refinement | ||
| R work (%) | 20.3 | 20.2 |
| R free (%) | 21.8 | 22.4 |
| No. of non-H atoms | ||
| Protein | 2225 | 2234 |
| Waters | 173 | 170 |
| Ligand | 1 | 1 |
| Sulfate | 15 | 25 |
| Average B factor (Å2) | 20.7 | 28.2 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.01 | 0.01 |
| Angles (°) | 1.1 | 1.0 |
| Ramachandran favored (%) | 97.5 | 97.9 |
| Ramachandran outliers (%) | 0 | 0 |
2.3. Structure determination, model building and refinement
Initial phases for both Fe(III)-h3HAO and zinc-bound h3HAO [Zn(II)-h3HAO] were determined by molecular replacement using Phaser (Storoni et al., 2004 ▸; McCoy et al., 2005 ▸, 2007 ▸; McCoy, 2007 ▸) using the coordinates of nickel-bound h3HAO [Ni(II)-h3HAO; PDB entry 2qnk]. Prior to molecular replacement, all alternate conformations, solvent molecules and the active-site nickel were removed from the search model. The structure was initially refined using REFMAC5 (Murshudov et al., 1997 ▸, 1999 ▸, 2011 ▸; Winn et al., 2001 ▸, 2003 ▸) from the CCP4 program suite. Iterative cycles of model building using Coot (Brown et al., 2015 ▸; Debreczeni & Emsley, 2012 ▸; Emsley et al., 2010 ▸; Emsley & Cowtan, 2004 ▸) and TLS-restrained refinement using BUSTER (Bricogne et al., 2011 ▸) in the final stages of refinement yielded structures with an R work and R free of 0.20 and 0.22, respectively, for Fe(III)-h3HAO and 0.20 and 0.22, respectively, for Zn(II)-h3HAO. A summary of the refinement statistics for both structures is provided in Table 1 ▸. The refined structures were deposited in the PDB [Fe(III)-h3HAO, PDB entry 5tk5; Zn(II)-h3HAO, PDB entry 5tkq].
2.4. Inductively coupled plasma mass spectrometry (ICP-MS) and electron paramagnetic resonance (EPR)
2.4.1. Sample preparation for ICP-MS analysis
Purified protein samples were received frozen and were thawed on ice in a Coy anaerobic chamber (3% H2, 97% N2 atmosphere). Freshly prepared ascorbic acid (0.01%) and ammonium iron(II) sulfate (1.2 equivalents) or zinc sulfate (1.2 equivalents) were added to the thawed protein solutions and left to incubate for 4 h. Once incubation was complete, dialysis was performed to remove excess (noncoordinating) metal ions. The samples were then diluted to a concentration of 1 µM protein with 2% HNO3 (trace metal grade). The metal content of the samples was then analyzed by ICP-MS.
2.4.2. Iron and zinc detection by ICP-MS
The concentrations of metal ions in the protein samples were determined by injecting samples into an Agilent 7700x ICP-MS system (Agilent Technologies, Santa Clara, California, USA). The metal-ion levels were detected via an Octopole Reaction System (ORS) cell in helium mode to remove ArO interference. The ICP-MS parameters used for the analysis were an RF power of 1550 W, an argon carrier gas flow of 1.01 l min−1, an argon make-up gas flow of 0.1 l min−1, a helium gas flow of 4.3 ml min−1, an octopole RF of 200 V and an OctP bias of −18 V. Samples were directly infused using the 7700x peristaltic pump with a speed of 0.1 rotations per second and a micromist nebulizer. To determine the iron and zinc concentrations present in the protein samples, a calibration curve of corresponding iron and zinc atomic absorption standards (Fluka Analytical) prepared in the same matrix as the samples was utilized. Data analysis was performed using the Agilent MassHunter software.
2.4.3. Sample preparation for EPR
The Fe(II)-h3HAO protein solutions (400 µl) that were analyzed by ICP-MS were mixed with 50% glycerol solution (200 µl) to give a final protein concentration of 157 µM. The solution was then transferred to quartz EPR tubes, flash-frozen in liquid nitrogen and stored at −80°C until EPR analysis. EPR spectra were collected on a Bruker EMX EPR spectrometer controlled with a Bruker ER 041 microwave bridge at 12 K. The temperature was maintained with a continuous-flow liquid He cryostat and an ITC503 temperature controller (Oxford Instruments).
2.5. X-ray fluorescence
An EDAX Eagle 3 microbeam X-ray fluorescence spectrometer was used for the detection of metals bound to the prepared Fe(III)-h3HAO crystals. Crystals of Fe(III)-h3HAO were added to a quartz capillary and were placed into the sample chamber of the Eagle 3. The quartz capillary was positioned horizontally and a 3.8 × 2.9 mm map was taken of the crystals present inside the capillary. Maps for Co, Fe, Mn, Ni, Si, W and Zn were collected with a pixel spacing of approximately 30 µm and a detector live time of 8 s per pixel. During the analysis the X-ray tube voltage and current were set to 30 kV and 1000 mA, respectively, and the sample chamber was kept at atmospheric pressure to prevent damage to or loss of the crystals. After the maps had been collected, they were visually searched for any localized enhancements of signal indicating the presence of metals. Upon observation of signal enhancement consistent with the presence of metals, the sample stage was moved to the spot of interest and a spectrum was collected with the same settings, except that the detector live time was extended to 1 h to enhance the sensitivity. The sample stage was also moved to a location where crystals were present but no signal enhancement was observed for any metals in order to collect a blank spectrum, also for 1 h. All data collection was performed using EDAX Vision32 software v.4.992-977R on a PC running Windows XP.
3. Results and discussion
3.1. Overall structures of human 3HAO with iron and zinc bound
We determined the crystal structures of human 3HAO with iron and zinc present in the active site to resolutions of 1.88 and 1.75 Å, respectively. Human 3HAO, like its bovine counterpart (Đilović et al., 2009 ▸), is a monomer containing two domains. The first domain is a cupin fold which contains residues 1–150 (Fig. 2 ▸). An extended random coil containing residues 151–181 follows the cupin domain and connects it to a small domain of unknown function which contains residues 182–286. Both domains of h3HAO contain core jelly-roll motifs. The jelly roll for the cupin fold encompasses residues 32–85 and the jelly roll for the C-terminal domain contains residues 209–276 and makes up the majority of this domain. The models for both iron-bound h3HAO [Fe(III)-h3HAO] and zinc-bound h3HAO [Zn(II)-h3HAO] contain residues 3–285. The model for Fe(III)-h3HAO also contains 173 waters and three sulfate ions, while the model for Zn(II)-h3HAO contains 170 waters and five sulfate ions.
Figure 2.
The overall structure of human 3HAO (a) compared with R. metallodurans 3HAO (b). Fe atoms are shown as orange spheres. The cupin domain of human 3HAO (red) has the same fold as R. metallodurans 3HAO (dark red). Human 3HAO contains an additional domain (blue) which contains a core jelly-roll motif. The function of this domain is unknown.
3.2. Comparison of the active sites of Fe(III)-h3HAO and Zn(II)-h3HAO
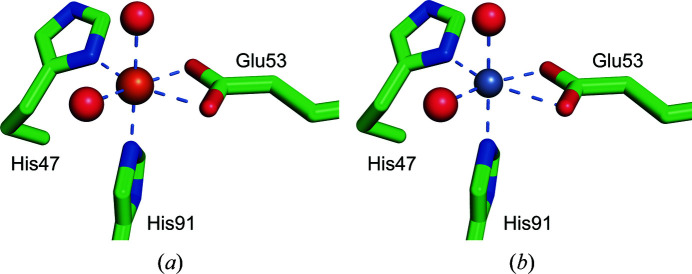
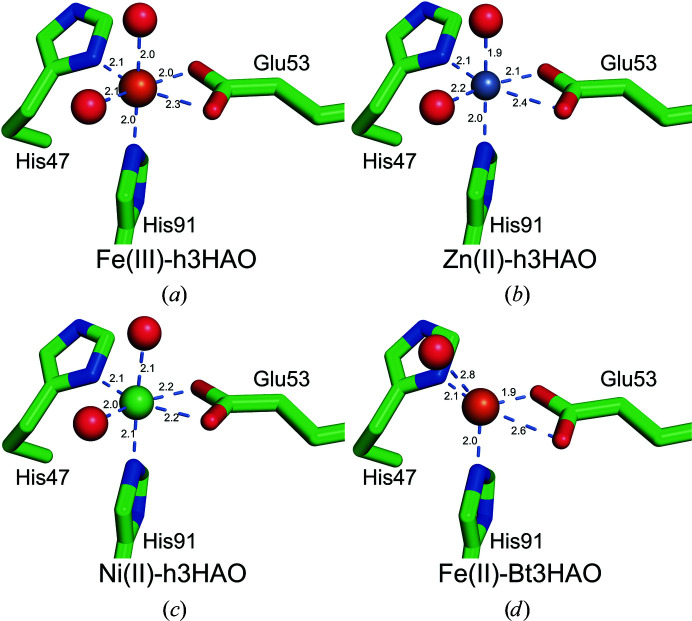
The h3HAO active site contains a single non-heme iron coordinated by a 2-His-1-carboxylate facial triad motif. Two water molecules complete the distorted octahedral coordination geometry (Fig. 3 ▸). In this arrangement, the NE2 atom of His91 and the ND1 atom of His47 bind at distances of 2.0 and 2.1 Å, respectively. Glu53 provides a bidentate ligand with asymmetric bond distances (2.0 and 2.3 Å), and the two water molecules reside at distances of 2.0 and 2.1 Å. In addition to the water molecules coordinated to the active-site iron, there are seven water molecules in the active site.
Figure 3.
Coordination of iron (left, orange sphere) and zinc (right, gray sphere) in the h3HAO active site. In each, His46 and His91 contribute one nitrogen ligand, Glu53 forms a bidentate interaction and two tightly bound water molecules complete the coordination sphere. Metal–ligand interactions are indicated by light blue dashed lines.
Similarly, the Zn(II)-h3HAO active site contains a single zinc ion in a distorted octahedral coordination. The bidentate Glu53 ligand binds asymmetrically, with bonding distances of 2.1 and 2.4 Å. The His47 ND1 atom resides 2.1 Å away from the bound zinc, while the NE2 atom of His91 is closer (2.0 Å). Two water molecules complete the coordination sphere, binding at distances of 1.9 and 2.1 Å. The active site of Zn(II)-3HAO contains an additional nine water molecules and one bound sulfate anion. These studies show that zinc binds directly to the h3HAO active site, as proposed previously, and that it binds with a coordination geometry that closely matches that of the native iron. Moreover, based on what is known about the catalytic mechanism of 3HAO (Colabroy et al., 2005 ▸; Zhang et al., 2005 ▸), the observed inhibitory effects are most likely owing to the inability of zinc to adopt the +3 oxidation state. The relative affinities of iron and zinc for h3HAO are not known, so it is unclear at present whether zinc binds more avidly to the h3HAO active site or has an off-rate that facilitates inhibition. However, our qualitative data from dialysis/ICP-MS experiments (see below) suggest that zinc does not bind as tightly to h3HAO as iron does. It should be noted that the availability of iron and zinc in microglia will significantly impact the potential inhibitory effects of zinc. Evidence exists for free zinc in cell models of ischemia (McCranor et al., 2012 ▸) and hypoosmotic stress (Segawa et al., 2014 ▸), but it is not clear whether free iron is present in the brain under conditions relevant to microglial activation. Further study of the inhibition of 3HAO by zinc is required to determine the origins of this effect.
3.3. Biophysical characterization of the active-site metals
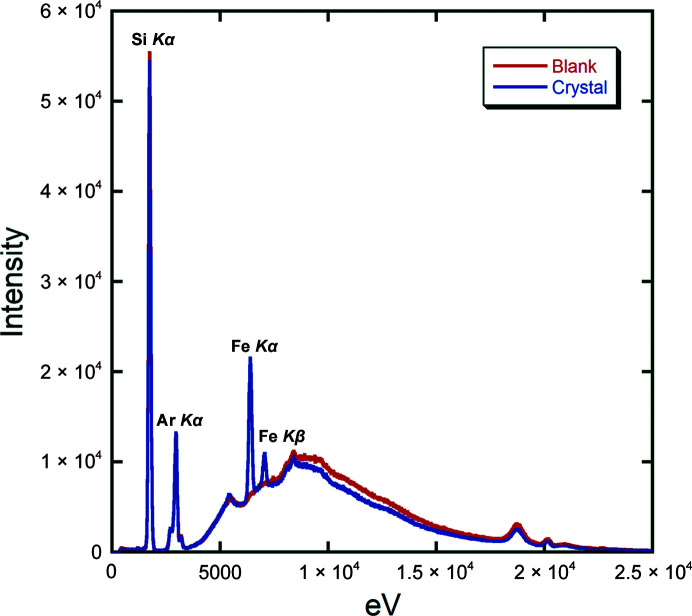
To determine the effectiveness of our metal-loading protocols, we first examined the samples using inductively coupled plasma mass spectrometry (ICP-MS). As a control, we determined that the protein samples prior to loading with either iron or zinc contained 0.7 equivalents of nickel, which is consistent with observations from both the yeast 3HAO structure (Li et al., 2006 ▸) and the unpublished human 3HAO structure (PDB entry 2qnk) that 3HAO acquires nickel during purification. Upon incubation with 1.25 and 2.5 molar equivalents of iron, 0.23–1.3 equivalents of iron per protein were measured following buffer exchange. After incubation with zinc but prior to buffer exchange, only zinc was detected, indicating that the nickel is displaced. Interestingly, after buffer exchange the zinc content was measured at between 0.18 and 0.45 molar equivalents per protein. While qualitative, these data suggest that zinc is more easily displaced from the h3HAO active site than its native iron. In addition to examining Fe(III)-h3HAO in solution, we used X-ray fluorescence spectroscopy to examine crystals of the enzyme which were mounted in quartz capillaries. Based upon a comparison of the spectra of the crystal versus a region in the capillary containing only buffer (Fig. 4 ▸), we can only detect the presence of iron, as indicated by the presence of the Kα and Kβ peaks at 6404 and 7058 eV, respectively. These data indicate that our Fe(III)-h3HAO crystals are exclusively iron-bound and lack contaminating ions from the purification procedure or metal-incubation protocol.
Figure 4.
X-ray fluorescence spectrum of an Fe(III)-h3HAO crystal. Overlay of the spectrum of the crystal (blue) versus the blank (red) clearly shows the presence of Fe Kα and Kβ peaks. No other metals were detected. Peaks for Si and Ar are present owing to the glass capillary and the air around the sample, respectively.
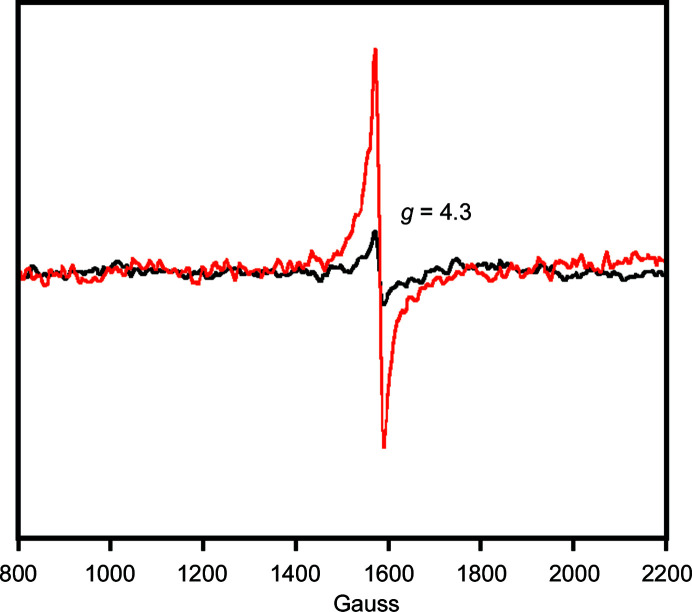
Thus far, no biophysical examination of the active-site metal of a monomeric 3HAO that typifies the enzyme from higher eukaryotes has been performed. Therefore, we characterized the active-site iron of h3HAO by electron paramagnetic resonance (EPR). The EPR of Fe(II)-3HAO was measured (at 12 K) and no signal was observed, as expected [iron(II) is d 6, diamagnetic]. The sample was then warmed, exposed to air, refrozen to 12 K and the EPR spectrum was recorded. A peak centered at g = 4.3 [indicative of iron(III), d 5] was observed (Fig. 5 ▸). The signal was not intense, as expected, and matches that reported for R. metallidurans 3HAO (Colabroy et al., 2005 ▸). In particular, the level of autooxidation [i.e. from iron(II) to iron(III)] observed for R. metallidurans 3HAO in the absence of substrate or inhibitor after a brief exposure to air was approximately 10%. By contrast, in the presence of the inhibitor 4-chloro-3-hydroxyanthranilate (4-Cl-3-HANA), approximately 75% of the active-site iron population was in the iron(III) state after exposure to air (Colabroy et al., 2005 ▸). Similarly, when using nitric oxide as a spin probe and oxygen analog, the signal indicative of an {Fe-NO}7 complex increased markedly upon incubation with the substrate 3-HANA (Colabroy et al., 2005 ▸). The behavior exhibited by R. metallidurans 3HAO is consistent with previous observations that substrate binding increases oxygen affinity in extradiol dioxygenases (Arciero et al., 1985 ▸). The low level of autooxidation observed in h3HAO is consistent with a mechanism in which substrate binding is required for oxygen activation by the active-site metal (Arciero & Lipscomb, 1986 ▸; Arciero et al., 1985 ▸), a property that is conserved not just in 3HAO, but in extradiol dioxygenases from bacteria to humans.
Figure 5.
EPR spectra of air-free (black) and air-exposed (red) Fe(II)-h3HAO. Spectra were collected at a temperature of 12 K with a microwave frequency of 9.43 GHz, a microwave power of 20 mW, a modulation amplitude of 8 G and a sweep time of 41.9 s. The lack of signal in the anaerobic sample and the peak centered at g = 4.3 are indicative of the expected d 6 and d 5 electronic configurations.
3.4. Comparison of Fe(III)-h3HAO and Zn(II)-h3HAO with Ni(II)-h3HAO and bovine 3HAO
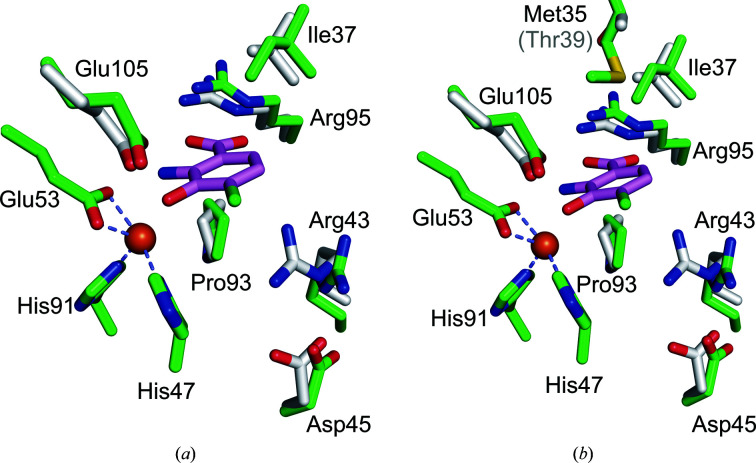
In general, the backbones of our Fe(III)-h3HAO and Zn(II)-h3HAO structures agree quite well with the deposited Ni(II)-h3HAO crystal structure (PDB entry 2qnk). In particular, the r.m.s.d.s (Cα to Cα) for our two structures versus 2qnk are 0.6 and 0.5 Å, respectively. This is not surprising because we are using the same expression construct and 2qnk was the molecular-replacement search model. However, there are 12 residues for which the r.m.s.d. (Cα to Cα) is greater than 1 Å. Of those 12, the positional shifts for all but residues Glu152, Arg155 and Arg244 can be attributed to differences in crystal packing. These residues are distal from the active site and the observed shifts are likely to be owing to inherent conformational flexibility in these regions. Compared with bovine 3HAO, the overall r.m.s.d. (Cα to Cα) is 1.1 Å. Similar to the Ni(II)-h3HAO structure, there are discrete groups of residues which exhibit large positional shifts relative to that expected given the r.m.s.d. These residues mostly reside in flexible loop regions with different crystal-packing environments between the human and bovine crystal forms. The only exceptions are residues 137 and 138, which are modeled as part of the first turn of a helix in h3HAO and as part of the loop that precedes this helix in the bovine enzyme. In contrast, the residues that coordinate the bound metal in the active sites of all of the human and bovine structures superpose almost exactly, with less than 0.1 Å r.m.s.d. for all atoms for Zn(II)-h3HAO versus Ni(II)-h3HAO and 0.1 Å r.m.s.d. for all atoms for Zn(II)-h3HAO versus bovine 3HAO. In addition, the metal coordination distances exhibit minor differences between the different crystal structures. In general, Ni(II)-h3HAO shows the most uniform distribution of bond distances (Fig. 6 ▸). Bovine 3HAO is a notable exception, however, in that the metal coordination sphere appears to be incomplete, with four ligands in a tetrahedral geometry with an additional water ligand residing at an unusually long distance (2.8 Å) from the iron for a ligand (Harding, 2006 ▸). One common feature among the structures is an asymmetric bidentate coordination by the conserved glutamate (Glu53 in h3HAO) in which one bond is longer than the other. This feature is also present in bacterial and yeast 3HAOs. Once again, Ni(II)-h3HAO is the outlier, as the bond distances between the OE1 and OE2 atoms and the metal are in closer agreement with each other than for the other structures. A summary of the comparison of the metal coordination for these structures is provided in Fig. 6 ▸. In addition to the residues in direct contact with iron, residues shown to interact with ligands in the R. metallidurans 3HAO structures are for the most part structurally conserved. In particular, Ile37 (Val in bovine 3HAO), Pro93 and Glu105 align well in the three h3HAO and bovine 3HAO structures. Likewise, Asn24 and Leu141 structurally align, with bovine 3HAO showing the largest deviation but still less than a 1.5 Å distance between Cα atoms. Arg43 aligns less well in the case of bovine 3HAO, with a distance of 2.2 Å between Cα atoms. However, the interaction between Asp45 and Arg43 is preserved in all four structures. Arg43 (Arg47 in R. metallodurans 3HAO) is important for catalysis and is implicated in binding oxygen in the active site (Zhang et al., 2005 ▸). Thus, regulation of its orientation via hydrogen bonding to Asp45 (Asp49 in R. metallodurans 3HAO) appears to be an important feature that is conserved throughout evolution. In contrast, Leu137 shows a nearly 3 Å distance between Cα atoms in the human and bovine 3HAO structures and Val22, because it is in a flexible loop near crystal contacts, deviates significantly among the four mammalian structures.
Figure 6.
Comparison of the metal–ligand distances for all of the available mammalian 3HAO crystal structures: Fe(III)-h3HAO (this work; PDB entry 5tk5), Zn(II)-h3HAO (this work; PDB entry 5tkq), Ni(II)-h3HAO (PDB entry 2qnk) and bovine 3HAO [Fe(III)-Bt3HAO; Đilović et al., 2009 ▸]. Fe atoms are shown as orange spheres, the Ni atom is shown as a green sphere and the Zn atom is shown as a gray sphere. Metal–ligand interactions are indicated by light blue dashed lines.
3.5. Comparison of Fe(III)-h3HAO and Zn(II)-h3HAO with bacterial and yeast 3HAO structures
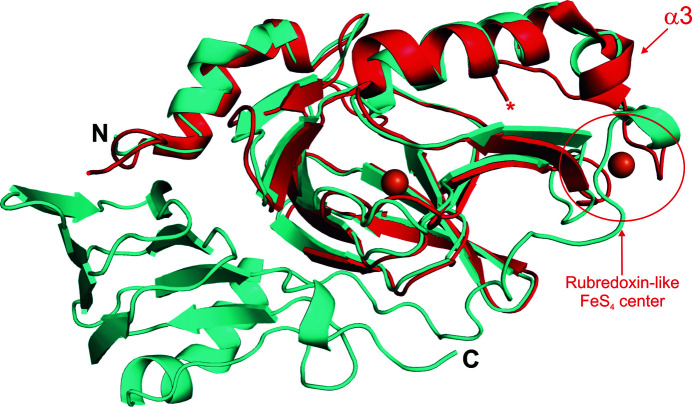
The primary differences between the human 3HAO structures and those of the bacterial and yeast homologues are the oligomeric state (i.e. human 3HAO is monomeric whereas the yeast and bacterial enzymes are dimeric), the presence of a rubredoxin-like iron reservoir in the yeast and bacterial homologues, and the presence of an additional jelly-roll-containing domain in the human enzyme which is absent in the yeast and bacterial homologs (Fig. 7 ▸). Despite these differences, the cupin folds of the respective enzymes superpose with low r.m.s.d. values, indicating a high degree of structural similarity. In particular, the r.m.s.d. between 149 equivalent Cα atoms of human and yeast 3HAO is 1.3 Å and the r.m.s.d.s between the equivalent Cα atoms of the human and R. metallidurans (146 Cα atoms) and C. metallidurans (145 Cα atoms) 3HAOs are 1.0 Å. The greatest structural divergences occur, as one would expect, near the termini, in loops and in helix α3, which in the yeast and bacterial structures is adjacent to the rubredoxin motif that is absent in the human enzyme (Fig. 7 ▸). Similar to the mammalian enzymes, the metal-coordinating residues of the bacterial 3HAOs superpose almost exactly with the equivalent residues from Fe(III)-h3HAO. In particular, the all-atom r.m.s.d.s are 0.2 Å for yeast 3HAO and 0.3 Å for both R. metallidurans and C. metallidurans 3HAO. All of the residues identified as interacting with 4-Cl-3-HANA in the bacterial complex structure are either absolutely conserved or highly conserved [e.g. Ile37 (human) versus Val41 (bacterial) and Leu137 (human) versus Ile142 (bacterial)]. Similarly, the residues surrounding the active-site ligand-binding pocket are generally well conserved. One notable exception residing near the ligand-binding pocket is Met35, which is Thr39 in the bacterial enzymes. Met35 projects its SD and CE atoms into the ligand-binding pocket, providing extra hydrophobic character to the region of the pocket that would interact with the hydrophobic face of either the 3-HANA substrate or the 4-Cl-3-HANA inhibitor (Fig. 8 ▸). In the superposition, the distance between Met35 and 4-Cl-3-HANA (4.6 Å) is just outside the distance at which a van der Waals interaction could occur. However, a minor alteration to the orientation of Met35 could result in a new interaction between mammalian 3HAO and substrates or inhibitors. Further studies are required to determine whether this nonconserved residue impacts substrate and/or inhibitor binding.
Figure 7.
Superposition of Fe(III)-h3HAO (cyan) and R. metallodurans 3HAO (red). Fe atoms are shown as orange spheres. The N- and C-termini of Fe(III)-h3HAO are labeled. The C-terminus of R. metallodurans 3HAO is labeled with a red asterisk. The locations of both the rubredoxin-like FeS4 center and helix α3 in R. metallodurans 3HAO are indicated.
Figure 8.
Superposition of key active-site residues of Fe(III)-h3HAO (green C atoms) and R. metallodurans 3HAO (gray C atoms) in complex with the inhibitor 4-Cl-3-HANA (magenta C atoms). Fe atoms are shown as orange spheres. (a) The active sites superimposed showing that all of the residues implicated in inhibitor binding are structurally well conserved, with the exception that Arg47 in R. metallodurans 3HAO, which interacts with bound oxygen in the complex (not shown), shifts relative to its Fe(III)-h3HAO counterpart Arg43. (b) The same view but with the nonconserved Met35 shown in comparison to its R. metallodurans 3HAO counterpart Thr39. Met35 projects into the active site near, but not close enough to interact with, a bound ligand. However, a minor reorientation of Met35 could result in a van der Waals interaction with a substrate or inhibitor.
Supplementary Material
PDB reference: human 3HAO with zinc bound in the active site, 5tkq
PDB reference: with iron bound in the active site, 5tk5
Acknowledgments
The authors would like to acknowledge the staff at SSRL beamline 12-2 for their assistance in data collection. We would also like to acknowledge the DNASU plasmid repository for providing the human 3HAO expression plasmid used in these studies. The plasmid was deposited by the University of Wisconsin-Madison. Certain commercial equipment, instruments or materials are identified in this paper to foster understanding. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
Funding Statement
This work was funded by National Science Foundation, Directorate for Mathematical and Physical Sciences grant CHE1306208.
References
- Arciero, D. M. & Lipscomb, J. D. (1986). J. Biol. Chem. 261, 2170–2178. [PubMed]
- Arciero, D. M., Orville, A. M. & Lipscomb, J. D. (1985). J. Biol. Chem. 260, 14035–14044. [PubMed]
- Bricogne, G., Blanc, E., Brandl, M., Flensburg, C., Keller, P., Paciorek, W., Roversi, P., Sharff, A., Smart, O. S., Vonrhein, C. & Womack, T. O. (2011). BUSTER v.2.10.2. Cambridge: Global Phasing Ltd.
- Brown, A., Long, F., Nicholls, R. A., Toots, J., Emsley, P. & Murshudov, G. (2015). Acta Cryst. D71, 136–153. [DOI] [PMC free article] [PubMed]
- Calderone, V., Trabucco, M., Menin, V., Negro, A. & Zanotti, G. (2002). Biochim. Biophys. Acta, 1596, 283–292. [DOI] [PubMed]
- Chess, A. C., Simoni, M. K., Alling, T. E. & Bucci, D. J. (2007). Schizophr. Bull. 33, 797–804. [DOI] [PMC free article] [PubMed]
- Colabroy, K. L., Zhai, H., Li, T., Ge, Y., Zhang, Y., Liu, A., Ealick, S. E., McLafferty, F. W. & Begley, T. P. (2005). Biochemistry, 44, 7623–7631. [DOI] [PubMed]
- Darlington, L. G., Mackay, G. M., Forrest, C. M., Stoy, N., George, C. & Stone, T. W. (2007). Eur. J. Neurosci. 26, 2211–2221. [DOI] [PubMed]
- Debreczeni, J. É. & Emsley, P. (2012). Acta Cryst. D68, 425–430. [DOI] [PMC free article] [PubMed]
- Đilović, I., Gliubich, F., Malpeli, G., Zanotti, G. & Matković-Čalogović, D. (2009). Biopolymers, 91, 1189–1195. [DOI] [PubMed]
- Drewes, J. L., Meulendyke, K. A., Liao, Z., Witwer, K. W., Gama, L., Ubaida-Mohien, C., Li, M., Notarangelo, F. M., Tarwater, P. M., Schwarcz, R., Graham, D. R. & Zink, M. C. (2015). J. Neurovirol. 21, 449–463. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. R. & Murshudov, G. N. (2013). Acta Cryst. D69, 1204–1214. [DOI] [PMC free article] [PubMed]
- Foster, A. C., Miller, L. P., Oldendorf, W. H. & Schwarcz, R. (1984). Exp. Neurol. 84, 428–440. [DOI] [PubMed]
- Foster, A. C., White, R. J. & Schwarcz, R. (1986). J. Neurochem. 47, 23–30. [DOI] [PubMed]
- Giorgini, F., Guidetti, P., Nguyen, Q., Bennett, S. C. & Muchowski, P. J. (2005). Nature Genet. 37, 526–531. [DOI] [PMC free article] [PubMed]
- Goldstein, L. E., Leopold, M. C., Huang, X., Atwood, C. S., Saunders, A. J., Hartshorn, M., Lim, J. T., Faget, K. Y., Muffat, J. A., Scarpa, R. C., Chylack, L. T. Jr, Bowden, E. F., Tanzi, R. E. & Bush, A. I. (2000). Biochemistry, 39, 7266–7275. [DOI] [PubMed]
- Guidetti, P., Luthi-Carter, R. E., Augood, S. J. & Schwarcz, R. (2004). Neurobiol. Dis. 17, 455–461. [DOI] [PubMed]
- Gulaj, E., Pawlak, K., Bien, B. & Pawlak, D. (2010). Adv. Med. Sci. 55, 204–211. [DOI] [PubMed]
- Hardeland, R., Zsizsik, B. K., Poeggeler, B., Fuhrberg, B., Holst, S. & Coto-Montes, A. (1999). Adv. Exp. Med. Biol. 467, 389–395. [DOI] [PubMed]
- Harding, M. M. (2006). Acta Cryst. D62, 678–682. [DOI] [PubMed]
- Higashi, Y., Segawa, S., Matsuo, T., Nakamura, S., Kikkawa, Y., Nishida, K. & Nagasawa, K. (2011). Glia, 59, 1933–1945. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kauppinen, T. M., Higashi, Y., Suh, S. W., Escartin, C., Nagasawa, K. & Swanson, R. A. (2008). J. Neurosci. 28, 5827–5835. [DOI] [PMC free article] [PubMed]
- Kita, T., Morrison, P. F., Heyes, M. P. & Markey, S. P. (2002). J. Neurochem. 82, 258–268. [DOI] [PubMed]
- Leklem, J. E. (1971). Am. J. Clin. Nutr. 24, 659–672. [DOI] [PubMed]
- Li, L.-B. & Wang, Z.-Y. (2016). Histol. Histopathol. 31, 623–627. [DOI] [PubMed]
- Li, X., Guo, M., Fan, J., Tang, W., Wang, D., Ge, H., Rong, H., Teng, M., Niu, L., Liu, Q. & Hao, Q. (2006). Protein Sci. 15, 761–773. [DOI] [PMC free article] [PubMed]
- Liu, F., Geng, J., Gumpper, R. H., Barman, A., Davis, I., Ozarowski, A., Hamelberg, D. & Liu, A. (2015). J. Biol. Chem. 290, 15621–15634. [DOI] [PMC free article] [PubMed]
- Long, C. L., Hill, H. N., Weinstock, I. M. & Henderson, L. M. (1954). J. Biol. Chem. 211, 405–417. [PubMed]
- Lugo-Huitrón, R. et al. (2011). Neurotoxicol. Teratol. 33, 538–547. [DOI] [PubMed]
- McCoy, A. J. (2007). Acta Cryst. D63, 32–41. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- McCranor, B. J., Bozym, R. A., Vitolo, M. I., Fierke, C. A., Bambrick, L., Polster, B. M., Fiskum, G. & Thompson, R. B. (2012). J. Bioenerg. Biomembr. 44, 253–263. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. (1999). Acta Cryst. D55, 247–255. [DOI] [PubMed]
- Pearson, S. J. & Reynolds, G. P. (1992). Neurosci. Lett. 144, 199–201. [DOI] [PubMed]
- Perkins, M. N. & Stone, T. W. (1982). Brain Res. 247, 184–187. [DOI] [PubMed]
- Rios, C. & Santamaria, A. (1991). Neurochem. Res. 16, 1139–1143. [DOI] [PubMed]
- Saito, K., Nowak, T. S. Jr, Markey, S. P. & Heyes, M. P. (1993). J. Neurochem. 60, 180–192. [DOI] [PubMed]
- Saito, K., Nowak, T. S. Jr, Suyama, K., Quearry, B. J., Saito, M., Crowley, J. S., Markey, S. P. & Heyes, M. P. (1993). J. Neurochem. 61, 2061–2070. [DOI] [PubMed]
- Santamaría, A., Galván-Arzate, S., Lisý, V., Ali, S. F., Duhart, H. M., Osorio-Rico, L., Ríos, C. & Sut’astný, F. (2001). Neuroreport, 12, 871–874. [DOI] [PubMed]
- Schwarcz, R., Bruno, J. P., Muchowski, P. J. & Wu, H.-Q. (2012). Nature Rev. Neurosci. 13, 465–477. [DOI] [PMC free article] [PubMed]
- Schwarcz, R., Guidetti, P., Sathyasaikumar, K. V. & Muchowski, P. J. (2010). Prog. Neurobiol. 90, 230–245. [DOI] [PMC free article] [PubMed]
- Schwarcz, R., Whetsell, W. O. Jr & Mangano, R. M. (1983). Science, 219, 316–318. [DOI] [PubMed]
- Segawa, S., Nishiura, T., Furuta, T., Ohsato, Y., Tani, M., Nishida, K. & Nagasawa, K. (2014). Life Sci. 94, 137–144. [DOI] [PubMed]
- Storoni, L. C., McCoy, A. J. & Read, R. J. (2004). Acta Cryst. D60, 432–438. [DOI] [PubMed]
- Tai, Y. F., Pavese, N., Gerhard, A., Tabrizi, S. J., Barker, R. A., Brooks, D. J. & Piccini, P. (2007). Brain, 130, 1759–1766. [DOI] [PubMed]
- Tavares, R. G., Tasca, C. I., Santos, C. E., Alves, L. B., Porciúncula, L. O., Emanuelli, T. & Souza, D. O. (2002). Neurochem. Int. 40, 621–627. [DOI] [PubMed]
- Tavares, R. G., Tasca, C. I., Santos, C. E., Wajner, M., Souza, D. O. & Dutra-Filho, C. S. (2000). Neuroreport, 11, 249–254. [DOI] [PubMed]
- Thompson, R. B. (2012). Metal Ions in Stroke, 1st ed., edited by Y. V. Li & J. H. Zhang, pp. 209–226. New York: Springer-Verlag.
- Ting, K. K., Brew, B. J. & Guillemin, G. J. (2009). J. Neuroinflammation, 6, 36. [DOI] [PMC free article] [PubMed]
- Toyn, J. H., Lin, X.-A., Thompson, M. W., Guss, V., Meredith, J. E. Jr, Sankaranarayanan, S., Barrezueta, N., Corradi, J., Majumdar, A., Small, D. L., Hansard, M., Lanthorn, T., Westphal, R. S. & Albright, C. F. (2010). BMC Neurosci. 11, 143. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Winn, M. D., Isupov, M. N. & Murshudov, G. N. (2001). Acta Cryst. D57, 122–133. [DOI] [PubMed]
- Winn, M. D., Murshudov, G. N. & Papiz, M. Z. (2003). Methods Enzymol. 374, 300–321. [DOI] [PubMed]
- Zhang, Y., Colabroy, K. L., Begley, T. P. & Ealick, S. E. (2005). Biochemistry, 44, 7632–7643. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: human 3HAO with zinc bound in the active site, 5tkq
PDB reference: with iron bound in the active site, 5tk5