The CAMP factor of group B streptococcus, a pore-forming toxin, was crystallized with an MBP tag. Structural and functional analysis suggests that the basic patch formed by helices 6 and 7 within the C-terminal domain is critical for its activity.
Keywords: group B streptococcus, CAMP factor, hemolytic mechanism, MBP tag, structure determination, Streptococcus agalactiae
Abstract
CAMP factor is a unique α-helical bacterial toxin that is known for its co-hemolytic activity in combination with staphylococcal sphingomyelinase. It was first discovered in the human pathogen Streptococcus agalactiae (also known as group B streptococcus), but homologous genes have been found in many other Gram-positive pathogens. In this study, the efforts that led to the determination of the first structure of a CAMP-family toxin are reported. Initially, it was possible to produce crystals of the native protein which diffracted to near 2.45 Å resolution. However, a series of technical obstacles were encountered on the way to structure determination. Over a period of more than five years, many methods, including selenomethionine labeling, mutations, crystallization chaperones and heavy-atom soaking, were attempted, but these attempts resulted in limited progress. The structure was finally solved using a combination of iodine soaking and molecular replacement using the crystallization chaperone maltose-binding protein (MBP) as a search model. Analysis of native and MBP-tagged CAMP-factor structures identified a conserved interaction interface in the C-terminal domain (CTD). The positively charged surface may be critical for binding to acidic ligands. Furthermore, mutations on the interaction interface at the CTD completely abolished its co-hemolytic activities. This study provides novel insights into the mechanism of the membrane-permeabilizing activity of CAMP factor.
1. Introduction
During infection, pathogenic bacteria secrete proteins into the environment to suppress, modulate or damage the immune system of the host. Pore-forming toxins are very common among these bacterial cytotoxic proteins and are required for virulence in a large number of pathogens, including Mycobacterium tuberculosis, Escherichia coli, Staphylococcus aureus and Streptococcus pneumoniae, as well as group A and group B streptococci (Los et al., 2013 ▸).
The CAMP reaction was first described in clinical isolates of Streptococcus agalactiae (group B streptococcus) in 1944 (Christie et al., 1944 ▸; Munch-Petersen et al., 1945 ▸): hemolytic zones were observed when S. agalactiae was grown on sheep-blood agar plates next to colonies of S. aureus. This co-hemolytic effect is brought about by S. aureus sphingomyelinase and S. agalactiae CAMP factor. The gene that encodes CAMP factor, cfb, was cloned in 1988 (Schneewind et al., 1988 ▸). The CAMP test is often used in clinical microbiology laboratories to presumptively identify S. agalactiae and other pathogens. In addition to S. agalactiae, genes homologous to cfb have been found in several other bacteria, including streptococci such as S. pyogenes (group A streptococcus; GAS; Gase et al., 1999 ▸) and those from other taxa such as Mobiluncus curtisii and Propionibacterium acnes (Valanne et al., 2005 ▸; Sörensen et al., 2010 ▸). On the other hand, some species that have been reported to be positive in the CAMP test, including Listeria monocytogenes (McKellar, 1994 ▸) and Bartonella henselae (Litwin & Johnson, 2005 ▸), do not have genes homologous to that for CAMP factor, suggesting the presence of other gene products with similar activity.
We previously found that the group B streptococcus (GBS) CAMP factor can form oligomeric membrane pores; however, the structural details of the membrane-inserted oligomer remained a mystery (Lang & Palmer, 2003 ▸). In our efforts to explore the structure–function relationship of the CAMP factor, we sought to determine its crystal structure. Unfortunately, we encountered a series of obstacles during the course of solving this structure. After multiple failed attempts, we were able to determine the novel structure of this important virulence factor. We report on these experiments in detail as we believe that the approaches used may be useful to other researchers. In addition, new additional functional insights into the role of the C-terminal domain (CTD) are discussed.
2. Materials and methods
2.1. Protein expression and purification
The wild-type (WT) CAMP-factor protein (Ala29–Lys255; NCBI accession code AEO12680) from S. agalactiae was expressed with a thrombin-cleavable GST tag using pGEX vector in bacteria and was purified as described previously (Lang & Palmer, 2003 ▸). In order to crystallize CAMP factor with a maltose-binding protein (MBP) tag, the CAMP-coding region was re-cloned into pET-30a vector with a non-cleavable N-terminal MBP tag. The MBP tag was designed with the following surface entropy-reducing mutations to enhance crystallization: D82A/K83A/E172A/N173A/K239A (Moon et al., 2010 ▸; Jin, Perry et al., 2013 ▸; Jin et al., 2017 ▸). A series of fusion constructs was created in which the N-terminal residue of the CAMP-factor reading frame was varied systematically in two-residue increments from Ala29 to Met50 in order to increase the chance of fusion-protein crystallization. The expression and purification of MBP-fusion proteins has been reported (Jin, Curry et al., 2013 ▸; Fang et al., 2018 ▸). Selenomethionine (Se-M)-labeled WT CAMP and MBP-CAMP were expressed as described previously (Jin et al., 2005 ▸). The purification procedure was identical to that used for the native protein.
A mutation of the CAMP gene (CAMPF-9XA) at nine critical residues at its CTD interface on helices 6 and 7 (R199A, R202A, L207A, F212A, Y215A, N219A, T223A, V226A and L230A) was generated using the joining PCR-based method with Phusion DNA polymerase (NEB, Ipswich, Massachusetts, USA). The CAMPF-9XA mutant and its CTD (9XA-CTD) were expressed and purified in the same way as the WT protein.
2.2. Hemolysis assay
Sheep red blood cells were sensitized to CAMP factor by pretreatment with staphylococcal sphingomyelinase (Sigma; 50 mU ml−1) in HEPES-buffered saline (HBS) with 10 mM MgCl2, washed in the same buffer by centrifugation and resuspended at 1%(v/v). An equal volume of the cell suspension was added to samples of CAMP-factor proteins (WT or mutant, alone or in combination) in the wells of a 96-well microtiter plate. The progress of hemolysis was monitored using the optical density at 650 nm, a wavelength that lies outside the absorption band of hemoglobin and thus reflects cell turbidity alone. These measurements were performed using a SpectraMax Plus 384 microplate spectrophotometer (Molecular Devices, Sunnyvale, California, USA).
To measure the inhibitory effect of WT CTD and 9XA-CTD on WT CAMP factor, 1 µg ml−1 of both WT CAMP factor and CTD or 9XA-CTD were used in the hemolysis assay.
2.3. Crystallization
WT native CAMP factor was crystallized in a well solution consisting of 0.2 M ammonium nitrate, 20% PEG 3350, 0.1 M MES pH 6.0. Thin plate-shaped crystals appeared after incubation for one week at room temperature. The well solution was supplemented with 20%(v/v) ethylene glycol and 1%(w/v) glucose as cryoprotectants. Se-M-labeled nontagged CAMP factor was crystallized in 1.8 M ammonium sulfate, 5% PEG 550, 0.1 M MES pH 6.5. An extra 20% of ethylene glycol was added to the crystallization solution before flash-cooling the crystals in liquid nitrogen for X-ray diffraction.
With MBP-CAMP, pyramid-shaped crystals [Fig. 1 ▸(a)] grew after one week at 18°C in a well solution consisting of 1.5 M ammonium sulfate, 0.1 M HEPES pH 7.0. 20%(w/v) maltose was added to the reservoir solution as a cryoprotectant. Se-M-labeled MBP-CAMP was crystallized in 2 M lithium sulfate, 0.1 M Tris–HCl pH 8.0. 3.6% maltose was added to the solution as a cryoprotectant.
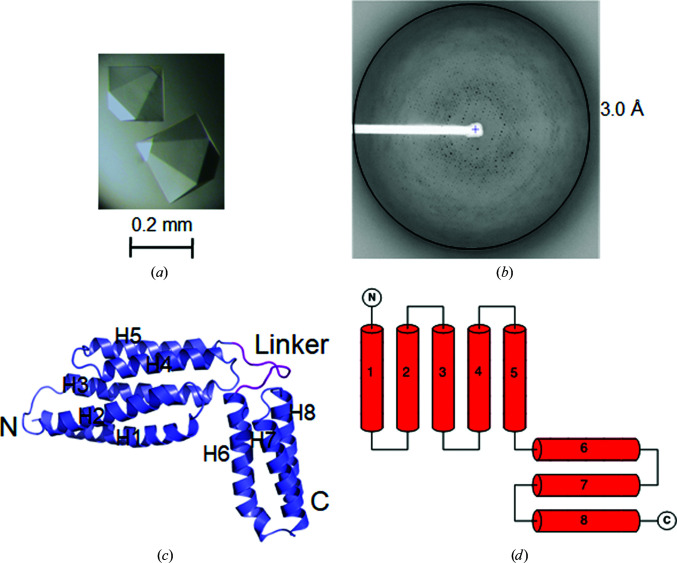
Figure 1.
Structure of CAMP factor within MBP-CAMP crystals. (a) Single hexagonal pyramid-shaped crystals of MBP-CAMP. The crystals reached a size of 0.2–0.4 mm in each dimension after two weeks at 18°C. (b) A typical diffraction image of MBP-CAMP. (c) Cartoon representation of CAMP factor. Its N- and C-termini and eight helices are labeled. (d) Topology of CAMP factor. It has two well separated domains composed of five and three helices.
2.4. X-ray diffraction, structure determination and refinement
X-ray diffraction data were collected at SER-CAT and GM/CA-CAT at the Advanced Photon Source, Argonne National Laboratory (ANL) and the BL17U, BL18U and BL19U1 beamlines at the Shanghai Synchrotron Radiation Facility (SSRF). Data were processed with the HKL-2000 program suite (Otwinowski & Minor, 1997 ▸) and XDS (Kabsch, 2010 ▸). The best native CAMP crystal diffracted to 2.45 Å resolution. The Se-M-labeled CAMP crystals diffracted poorly (data not shown). Over 100 MBP-CAMP crystals were screened. The best crystals diffracted to 3.0 Å resolution (Table 1 ▸).
Table 1. X-ray data-collection and refinement statistics for native MBP-CAMP.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | P6522 |
| a, b, c (Å) | 145.64, 145.64, 156.38 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 50–3.00 (3.18–3.00) |
| No. of reflections (total/unique) | 419717/20079 |
| Multiplicity | 20.9 (21.4) |
| Completeness (%) | 99.5 (99.7) |
| 〈I/σ(I)〉 | 32.0 (1.9) |
| R meas † (%) | 6.3 (195.9) |
| CC1/2 (%) | 100.0 (77.5) |
| Refinement | |
| Resolution (Å) | 50–3.0 |
| No. of protein atoms | 4502 |
| No. of solvent/heteroatoms | 38 |
| R.m.s.d., bond lengths (Å) | 0.0102 |
| R.m.s.d., bond angles (°) | 1.221 |
| R work ‡ (%) | 23.80 |
| R free § (%) | 31.53 |
| Ramachandran plot¶ | |
| Favored | 87.6 |
| Disallowed | 0 |
| PDB code | 5y2g |
Rmeas = \textstyle \sum_{hkl}\{N(hkl)/[N(hkl)-1]\}^{1/2}\sum_{i}|I_{i}(hkl)- \langle I(hkl)\rangle|/\textstyle \sum_{hkl}\sum_{i}I_{i}(hkl), where I i(hkl) and 〈I(hkl)〉 are the ith and the mean measurement of the intensity of reflection hkl.
Rwork = \textstyle \sum_{hkl}\big ||F_{\rm obs}|-|F_{\rm calc}|\big |/ \textstyle \sum_{hkl}|F_{\rm obs}|, where F obs and F calc are the observed and calculated structure factors, respectively. No I/σ(I) cutoff was applied.
Rfree is the R value obtained for a test set of reflections consisting of a randomly selected 5% subset of the data set excluded from refinement.
Values from the MolProbity server (http://molprobity.biochem.duke.edu/).
2.5. Phasing, model building and refinement
In order to solve the first structure of CAMP factor, several different types of crystals were grown and X-ray diffraction data were collected [Fig. 2 ▸(a)]. The MBP-CAMP structure was determined by molecular replacement with Phaser (McCoy et al., 2007 ▸) using the closed-form MBP structure (PDB entry 4ifp; Jin, Huang et al., 2018 ▸) as a search template. After molecular replacement, electron density corresponding to the first five helices of the CAMP molecule was clearly identifiable. Guided by the electron-density map from Se-M-labeled MBP-CAMP crystals and from iodine-soaked native MBP-CAMP crystals, the CAMP region including the C-terminal helices was manually built in Coot. This low-resolution CAMP-factor model was then used as a template in molecular-replacement calculations with native data, which led to its final structural determination [Fig. 2 ▸(b)]. The structural models were improved by several rounds of manual model fitting in Coot and structural optimization in phenix.refine in PHENIX (Adams et al., 2010 ▸). The final structural models were validated by the MolProbity server (Chen et al., 2010 ▸) and the ADIT validation server at the RCSB PDB deposition website (Berman et al., 2002 ▸). Molecular graphics were displayed with PyMOL (Schrödinger). Structure superpositions were performed and the r.m.s.d.s of the structures were calculated in PyMOL.
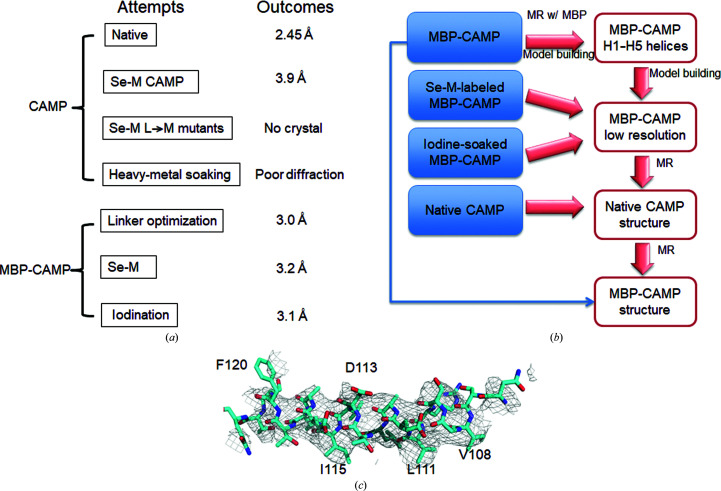
Figure 2.
A flowchart for the structural determination of CAMP factor. (a) Data sets collected to solve the CAMP structure. Seven types of protein crystals were tested for X-ray diffraction and data collection. (b) A flowchart describing CAMP structure determination. Native MBP-CAMP fusion crystals diffracted to 3.0 Å resolution and the structure was solved by molecular replacement using MBP as a search model. The heavy atoms introduced by Se-M labeling and iodine soaking also provided guidance for manual model building and side-chain assignment of the CAMP region. (c) Partial difference electron-density map from MBP-CAMP data at 3.0 Å resolution after molecular replacement with MBP. The F o − F c map of the H5 region is contoured at 1σ. The map is superimposed with the final model of H5 (cyan).
3. Results
3.1. Structure-determination attempts for CAMP factor
We tested a number of methods for the structure determination of CAMP factor, as summarized in Figs. 2 ▸(a) and 2 ▸(b). Of the several different types of crystals that we collected data from, the native CAMP-factor crystal diffracted to a best resolution of 2.45 Å, while MBP-tagged fusion-protein crystals diffracted to around 3.0 Å resolution. A BLAST search did not identify any high-homology structure for the CAMP toxin, which made structure determination by molecular replacement (MR) very challenging. Numerous attempts with low sequence-similarity models failed (data not shown). We also tried ab initio structure prediction with the QUARK server (Xu & Zhang, 2012 ▸). However, our attempts to use the predicted structure as a molecular-replacement search model were unsuccessful. Structure comparisons showed that these models were very different from the solved structure (data not shown).
As an alternate strategy, we sought to solve the CAMP structure by single-wavelength anomalous dispersion (SAD) with Se-M labeling. The best Se-M-labeled CAMP crystals diffracted to 3.9 Å resolution (Supplementary Table S1) and their anomalous signals were weak at resolutions beyond 8 Å (data not shown). Moreover, there are only two methionine residues in the total of 226 residues, and one of these residues was shown to be exposed to solvent in the solved structure; both of these factors made it challenging to solve this structure using SAD.
To overcome this problem, we introduced three extra methionine sites by point mutations (L61M, L250M and L254M). Unfortunately, we were not able to obtain crystals of this CAMP mutant. In addition, we tried heavy-metal soaking for MAD/SAD phasing. For the native crystals, we screened a series of common heavy-metal compounds containing lead, mercury, platinum or iridium, and the iodide-containing compounds I3C and BIC. The native CAMP crystals, which grew in thin plate shapes, did not survive in the soaking solutions and the soaked crystals diffracted poorly. Consequently, we were not able to collect a useful data set (data not shown).
3.2. MBP fusion tag-aided crystallization
Inspired by the successful use of MBP as a crystallization chaperone for crystallizing challenging targets by our group and several others (Potter et al., 2008 ▸; Moon et al., 2010 ▸; Jin et al., 2017 ▸), we decided to try the MBP-fusion tag to solve the phase problem of CAMP crystals. The use of a crystallization tag to aid target-protein crystallization usually requires a reasonable structural model. The lack of even a homology model makes construct design challenging. Firstly, we performed a secondary-structure prediction for CAMP factor. We realized that the linker sequence between the fusion tag (MBP) and the protein of interest (in this case, CAMP factor) is most critical for the success of such an application. A long flexible linker could be detrimental to crystallization. Accordingly, we designed a set of 14 MBP-fusion proteins. In the first fusion protein the CAMP-factor portion started at residue Ala22, while in the other variants 2–3 residues were successively removed from the N-terminus, with the shortest variant starting at Met50. It is worth noting that the N-terminal region of GBS CAMP is not conserved, and most homologous proteins to CAMP factor have shorter native N-termini. Two of these initial 14 fusion proteins crystallized in the first round. One of them starting at Asn42 produced hexagonal pyramid crystals that diffracted to ∼3.0 Å resolution [Figs. 1 ▸(a), 1 ▸(b) and 2 ▸(c), Table 1 ▸]. In addition, we were able to obtain data of similar quality from a subsequently produced fusion construct starting at Ser43.
Using this data set from the native MBP-tagged CAMP fusion-protein crystals, a solution for MBP was readily found in a molecular-replacement calculation using Phaser (McCoy et al., 2007 ▸) from the CCP4 program suite (Winn et al., 2011 ▸). After several cycles of rigid-body refinement, electron-density maps calculated using the MBP model clearly showed extra positive densities for the CAMP protein, which is mostly composed of α-helices. Fig. 2 ▸(c) shows the F o − F c electron-density map contoured at 1σ in the H5 region after MR calculation superimposed with the final structural model (cyan). A partial Cα model of α-helices (H1–H5) was built manually in Coot (Emsley et al., 2010 ▸) and was followed by cycles of manual model building in Coot and refinement in phenix.refine (Adams et al., 2010 ▸). At this stage, we were not able to build the loops and assign the side chains with confidence owing to the combination of low resolution, the lack of a reference structure and the unique structural features of this protein.
3.3. Model building and structure determination
We also obtained Se-M-labeled MBP-fusion protein crystals. The best crystals diffracted to a similar resolution to the native crystals. Even though the phasing power was not sufficient to solve the structure by itself, the two Se-M residues can be seen clearly in the CAMP density after molecular replacement with MBP, which helped in sequence assignment.
Furthermore, iodine derivatives of the fusion-protein crystals were prepared by vapor diffusion of elemental iodine by adding KI3 solution to the reservoir solution. The crystals turned a brown color overnight. The diffraction of the iodinated MBP-CAMP fusion crystals was still low, which made it less valuable to collect data at around the peak energy near 7 keV for SAD phasing at the synchrotron. The best data set, which yielded data to 3.1 Å resolution, was collected at an energy of 12 keV (Supplementary Table S1). Molecular replacement with MBP showed 12 copies of MBP in space group P1. At 3.1 Å resolution, automatic model building with SOLVE/RESOLVE failed. A partial model was built and some iodide ions were clearly visible in the density map (data not shown).
On the other hand, for the most ambiguous region of the model, helices H6–H8 of the CTD, we were able to assign the sequence based on the information from the Se-M map and the iodinated map. Using this partial model, we succeeded in molecular replacement of the native 2.45 Å resolution data; the model obtained was in turn used as an MR model for MBP-fusion proteins [Fig. 2 ▸(b)].
3.4. The MBP-fusion tag did not interfere with the folding of the CAMP factor
The final refined MBP-CAMP fusion-protein structure showed that the CAMP factor possesses eight helices and represents a novel structure [Figs. 1 ▸(c) and 1 ▸(d)]. The MBP tag is well separated from the C-terminal CAMP factor [Fig. 3 ▸(a)]. A preliminary functional assignment of its two well separated functional domains, i.e. the N-terminal domain (NTD) and the CTD, has recently been reported (Jin, Brefo-Mensah et al., 2018 ▸).
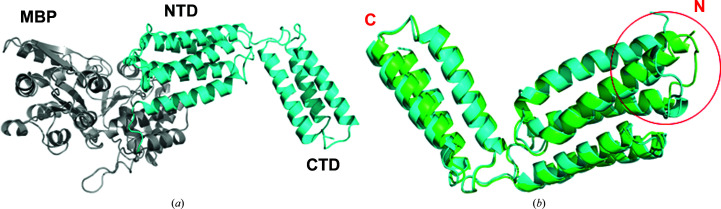
Figure 3.
Structural comparisons of native CAMP and MBP-CAMP. (a) Cartoon representation of MBP-CAMP. The N-terminal MBP tag is colored gray and the CAMP factor is in cyan. (b) Structural superposition of CAMP factor crystallized with and without an MBP tag. The overall structures of CAMP are highly similar, with some minor conformational variations in the loop region L23 in the NTD. The r.m.s.d. of 1424 aligned atoms is 1.12 Å. The structure of native CAMP factor is colored green and that of MBP-CAMP is in cyan.
To assess whether the addition of a fusion tag affected the structure of the target protein, we superimposed the native CAMP structure crystallized by itself (PDB entry 5h6i) with the MBP-CAMP fusion structure. The overall structures of CAMP factor are highly similar. The r.m.s.d. between the 1424 aligned atoms is 1.12 Å. The most variable region lies in the L23 region (the loop region between helices H2 and H3) [Fig. 3 ▸(b)]. Considering that its NTD functions as a membrane-insertion domain (Jin, Brefo-Mensah et al., 2018 ▸), the potentially important role that the L23 region plays in membrane insertion needs further investigation.
3.5. Native CAMP and MBP-CAMP proteins have similar crystal-packing interfaces
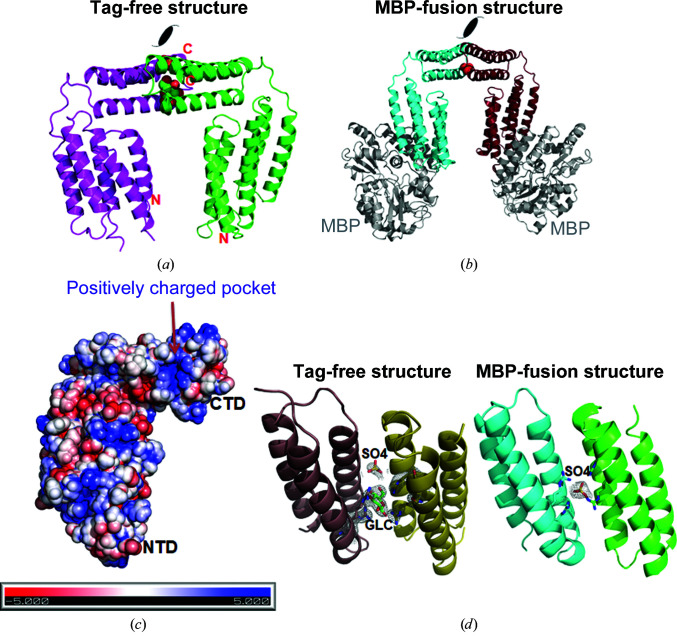
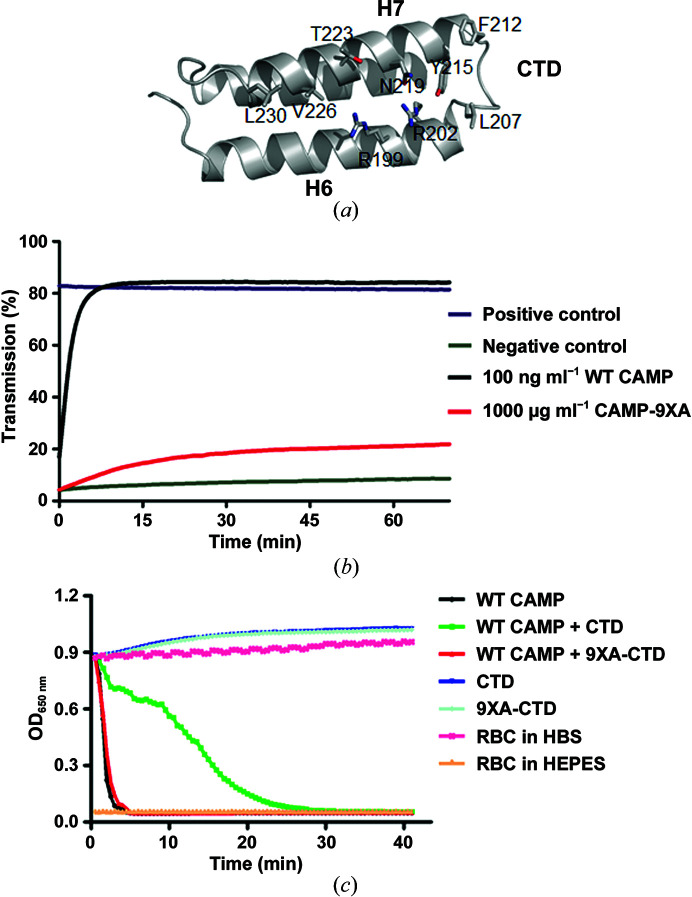
Interestingly, a conserved protein–protein interaction interface centered on the CTD was found in both tag-free and MBP-fusion CAMP crystals [Figs. 4 ▸(a) and 4 ▸(b)]. Two of the three molecules in one asymmetric unit of tag-free native CAMP crystals make intensive interactions [Fig. 4 ▸(a)]. The CTD interacts symmetrically with the identical surface, composed of helices H6–H7, of a second CAMP-factor molecule. On each CAMP-factor molecule, 16 residues (both charged and hydrophobic) are involved in this interaction. Judging from the free energy of this interaction (Δi G = −8.3 kcal mol−1), such a molecular interaction is not favorable in solution, which is in agreement with our data that both native and MBP-fusion CAMP factor exist as monomers in solution. This made us wonder why such an interface is observed in both crystal forms and what its potential functional relevance may be. A close inspection of the CTD packing interface identified positively charged residues composed of four arginine residues in the center and revealed it to be decorated with hydrophobic and hydrophilic residues [Fig. 4 ▸(c)].
Figure 4.
Identification of the conserved CTD interaction interface. (a) The packing interface in native CAMP crystals. The two molecules are colored magenta and green. One sulfate ion and one sugar molecule were modeled into the pocket between the symmetric CTD interfaces. (b) Packing interface in MBP-CAMP crystals. MBP is colored gray and the two CAMP molecules are colored cyan and ruby. The red sphere indicates a sulfate ion. (c) Electrostatic charges of the CTD interaction interface calculated by APBS. The orientation of CAMP is the same as for the magenta molecule in (a). The interface is composed of a positively charged pocket and is decorated by several hydrophobic residues. (d) The 2F o − F c difference electron-density map of the sulfate ions in both native and MBP-fused CAMP and of the glucose in the native structure. The map is contoured at 2σ.
Unexpectedly, we found the presence of some extra density in this CTD interface in both the tag-free and MBP-tagged crystals [Fig. 4 ▸(d)]. Since a high concentration of ammonium sulfate was present in both of the crystallization conditions, we built the density with sulfate ions. Indeed, sulfate salts were required for crystal formation for both proteins. We also included the sugar glucose in the cryoprotectant solution for the native CAMP crystals. As a result, some extra density was built as a glucose molecule in the native structure. Density for both sulfate ions and glucose fitted well to their assigned positions in the electron-density map. The stereochemistry is also reasonable [Fig. 4 ▸(d)]. The identification of acidic ligands in the CTD interfaces made us hypothesize that the CAMP factor may use the H6–H7 surface to bind both acidic ligands and cell-surface glycosylphosphatidylinositol (GPI) anchors, which are composed primarily of lipids, phosphate and sugar chains (Gao et al., 2017 ▸), with the lipid chains inserting into the cell membrane and the protruding phosphosugar group used to bind CAMP factor.
3.6. Mutation at the H6–H7 surface abolished hemolytic activity
In order to assess the importance of the CTD H6–H7 surface for the hemolytic activity of CAMP, we generated a mutant with the nine interacting residues mutated to alanines [Fig. 5 ▸(a), see Section 2], which we named CAMPF-9XA for the full-length protein and 9XA-CTD for the CTD. While alanine has a high propensity towards helix formation, this mutation is expected to retain the same secondary and tertiary structures as the WT protein. A hemolytic assay on RBCs showed that the CAMPF-9XA mutant (red) has a much lower activity compared with WT CAMP [Fig. 5 ▸(b)]. As shown in Fig. 5 ▸(c), both WT CTD and 9XA-CTD lack hemolytic activity, and WT CTD reduced the hemolytic activity of WT CAMP, while 9XA-CTD lacked the inhibitory effect of the WT CTD protein towards full-length CAMP factor. This result demonstrates that the conserved CTD H6–H7 packing interface is important for modulating the hemolytic activity of CAMP factor.
Figure 5.
The conserved CTD packing interface is important for the hemolytic activity of CAMP factor. (a) The nine residues at the H6–H7 surface involved in CTD packing are shown as sticks. (b) The CAMPF-9XA mutant (red) is virtually devoid of hemolytic activity. (c) 9XA-CTD, but not WT CTD, lacks the inhibitory effect on full-length CAMP factor. RBC, red blood cells; HEPES, 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid; HBS, HEPES-buffered saline.
4. Discussion
4.1. An MBP-fusion tag aided the crystallization and structure determination of CAMP factor
Our initial efforts to use MR and SAD/MAD to determine the structure of CAMP factor were unsuccessful. In contrast, an MBP-fusion protein with an optimized linker sequence formed sizable crystals that were suitable for further structural determination. The strategy of fusing a nonrelated protein, i.e. MBP, to a target protein is often used when the target protein fails to form crystals on its own. In this case, even though the native CAMP factor can be crystallized and diffracts well, the structure was not solved owing to the lack of a suitable model for MR and the poor diffraction of heavy-metal derivatives for SAD/MAD. The MBP crystallization tag was successfully used to aid in the structure determination of our target protein. After molecular replacement using the MBP tag and combining information from Se-M and iodine derivatives, the structure could finally be solved. Our study showed that such a crystallization-tag strategy could be used to solve novel structures, either by themselves by molecular replacement or in combination with other phasing methods such as SAD/MAD.
4.2. Functional role of the CTD interface
Our study also has important implications for understanding the exact mechanism of the pore-forming function of CAMP. Previously, we determined that its NTD is a membrane-permeabilization domain and that its CTD is likely to be responsible for receptor binding (Jin, Brefo-Mensah et al., 2018 ▸). In addition, the CTD inhibits the hemolytic activity of full-length CAMP factor. In this study, we identified a conserved CTD interaction interface via which it forms a positively charged ligand-binding pocket in both crystal forms. In our crystals, we could fit the extra density in the pocket with one molecule of glucose and one sulfate ion, both of which were used in the crystallization and cryoprotection solutions. This suggests that the interface may be involved in GPI binding. The CAMPF-9XA mutant has a much decreased hemolytic activity. With this observation, the previously observed inhibitory effect of the CTD on full-length CAMP factor (Jin, Brefo-Mensah et al., 2018 ▸) can be explained as excessive CTD competes with the full-length protein for GPI anchor binding.
4.3. Functional mechanism of CAMP factors
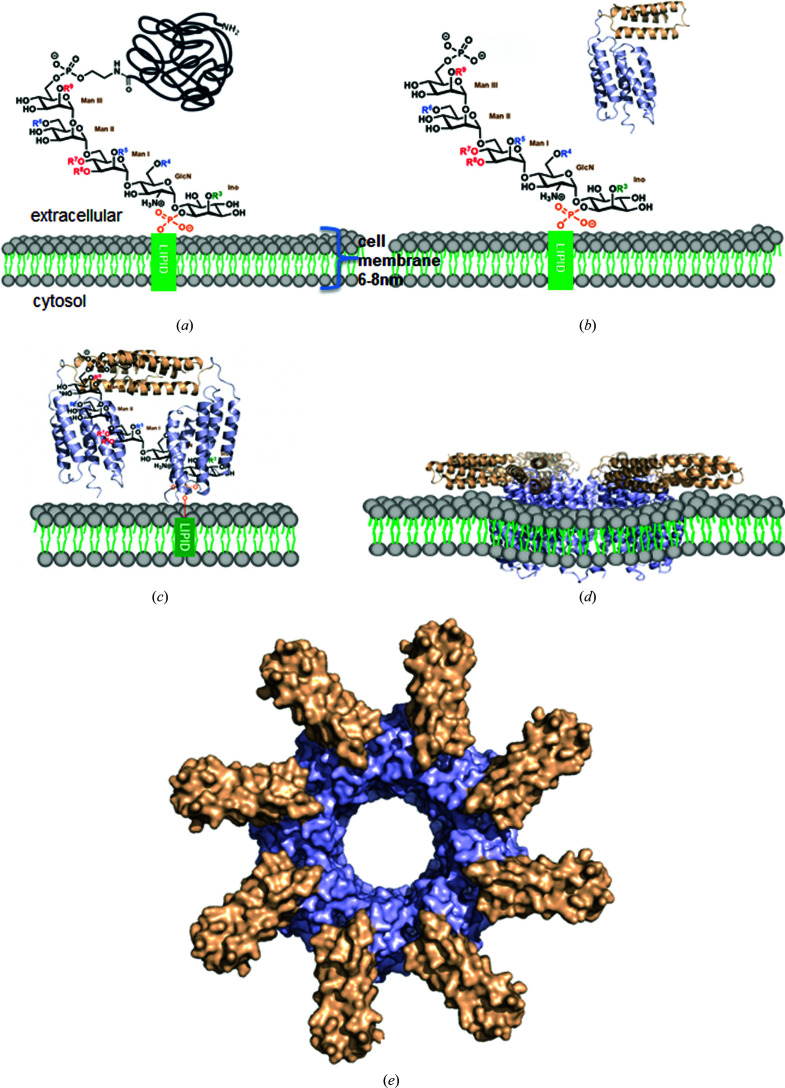
Finally, based on our results, we propose a working model for the CAMP toxin (Fig. 6 ▸). The mammalian cell membrane is inserted with proteins linked by glycosylphosphatidylinositol (GPI) anchors. The GPI anchors are composed of lipids, phosphate and sugar chains (Gao et al., 2017 ▸). During infection, CAMP factor is secreted by GBS into the extracellular space and behaves as a monomer in solution [Fig. 6 ▸(b)]. The membranes of some eukaryotic cells, i.e. red blood cells, are enriched with phosphatidylcholine (PC) and/or sphingomyelin (SM) (Montes et al., 2008 ▸), both of which have phosphocholine headgroups on the outer surface; the negative charge of the phosphate and the positive charge of the amine group together result in a net neutral charge at pH 7.0. The cleavage of the phosphatidylcholine headgroup by sphingomyelinase potentially alters the structure and surface-charge distribution of the cell membrane and exposes negatively charged headgroups in other phospholipids, which attract CAMP factor.
Figure 6.
Proposed working model for CAMP factor. (a) Chemical structure of a GPI anchor on the cell surface. (b) The action of sphingomyelinase cleaves neutrally charged phosphatidylcholine in the cell membrane, which exposes the negatively charged outer surface of the cell membrane and facilitates the binding of monomeric soluble CAMP factors to the negatively charged phosphate-sugar moieties of GPI. (c) Attachment to the cell surface through binding of the CTD to the GPI anchor. The positively charged patches on the H6–H7 surface of the CTD bind to phosphate-sugar moieties of the GPI anchor, which leads to the dimerization of CAMP-factor molecules through the CTD along with bound GPI anchors. The NTD is oriented towards the membrane, suitable for membrane insertion. (d) Membrane insertion of the NTD and structural rearrangement in a lipid bilayer with unknown mechanism. (e) A hypothetical model of the membrane-inserted pore. The model was generated by symmetric docking of eight CAMP-factor molecules through NTD interactions.
In fact, the avid association between a cationic protein and negatively charged acidic phospholipids, such as phosphatidylserine (PS), phosphatidylglycerol, phosphatidic acid (PA) and cardiolipin, is well known (Escribá et al., 2008 ▸). On the cell surface, the cationic H6–H7 surface of the CTD binds specifically to negatively charged phosphate-sugar chains of GPI anchors localized on the lipid rafts of the cell membrane to facilitate the membrane insertion of the NTD (Fig. 6 ▸ c). The enrichment of CAMP factor on the cell membrane induces its oligomerization and further conformational changes [Fig. 6 ▸(d)] by an unknown mechanism, which leads to the formation of membrane pores and the leakage of cellular contents. A hypothetical octamer pore is presented in Fig. 6 ▸(e). The structure of the membrane-inserted conformation of CAMP toxin is critical to understanding its mechanism and awaits future investigations.
Supplementary Material
PDB reference: MBP-CAMP, 5y2g
Supplementary Table S1. DOI: 10.1107/S205979831901057X/qh5057sup1.pdf
Acknowledgments
X-ray diffraction data were collected at the Southeast Regional Collaborative Access Team (SER-CAT) 22-ID and GM/CA-CAT 23-ID-D beamlines at the Advanced Photon Source (APS), Argonne National Laboratory and the BL17U, BL18U and BL19U1 beamlines at the Shanghai Synchrotron Radiation Facility (SSRF). We thank the staff at these beamlines for their support and assistance. We also wish to thank Professor Zhongwu Guo at the University of Florida and Professor Jian Gao at Shandong University for providing GPI ligands. The authors declare that they have no conflicts of interest with the contents of this article.
Funding Statement
This work was funded by National Natural Science Foundation of China, Young Scientists Fund grants 31600598 and 31870731 to Yajuan Li; National Natural Science Foundation of China, Major Research Plan grant U1732109 to Tengchuan Jin; Chinese Academy of Sciences grant XDB29030104 to Tengchuan Jin; Fundamental Research Funds for the Central Universities grants WK2070000108 and WK2070000097.
References
- Adams, P. D., Afonine, P. V., Bunkóczi, G., Chen, V. B., Davis, I. W., Echols, N., Headd, J. J., Hung, L.-W., Kapral, G. J., Grosse-Kunstleve, R. W., McCoy, A. J., Moriarty, N. W., Oeffner, R., Read, R. J., Richardson, D. C., Richardson, J. S., Terwilliger, T. C. & Zwart, P. H. (2010). Acta Cryst. D66, 213–221. [DOI] [PMC free article] [PubMed]
- Berman, H. M., Battistuz, T., Bhat, T. N., Bluhm, W. F., Bourne, P. E., Burkhardt, K., Feng, Z., Gilliland, G. L., Iype, L., Jain, S., Fagan, P., Marvin, J., Padilla, D., Ravichandran, V., Schneider, B., Thanki, N., Weissig, H., Westbrook, J. D. & Zardecki, C. (2002). Acta Cryst. D58, 899–907. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Christie, R., Atkins, N. E. & Munch-Petersen, E. (1944). Aust. J. Exp. Biol. Med. 22, 197–200.
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Escribá, P. V., González-Ros, J. M., Goñi, F. M., Kinnunen, P. K., Vigh, L., Sánchez-Magraner, L., Fernández, A. M., Busquets, X., Horváth, I. & Barceló-Coblijn, G. (2008). J. Cell. Mol. Med. 12, 829–875. [DOI] [PMC free article] [PubMed]
- Fang, K., Li, Y., Yin, X., Samad, A. & Jin, T. (2018). Clin. Lab. 64, 467–475. [DOI] [PubMed]
- Gao, J., Zhou, Z., Guo, J. & Guo, Z. (2017). Chem. Commun. 53, 6227–6230. [DOI] [PMC free article] [PubMed]
- Gase, K., Ferretti, J. J., Primeaux, C. & McShan, W. M. (1999). Infect. Immun. 67, 4725–4731. [DOI] [PMC free article] [PubMed]
- Jin, T., Brefo-Mensah, E., Fan, W., Zeng, W., Li, Y., Zhang, Y. & Palmer, M. (2018). J. Biol. Chem. 293, 11867–11877. [DOI] [PMC free article] [PubMed]
- Jin, T., Chuenchor, W., Jiang, J., Cheng, J., Li, Y., Fang, K., Huang, M., Smith, P. & Xiao, T. S. (2017). Sci. Rep. 7, 40991. [DOI] [PMC free article] [PubMed]
- Jin, T., Curry, J., Smith, P., Jiang, J. & Xiao, T. S. (2013). Proteins, 81, 1266–1270. [DOI] [PMC free article] [PubMed]
- Jin, T., Howard, A., Golemis, E. A., Wang, Y. & Zhang, Y.-Z. (2005). Acta Cryst. F61, 531–533. [DOI] [PMC free article] [PubMed]
- Jin, T., Huang, M., Jiang, J., Smith, P. & Xiao, T. S. (2018). PLoS One, 13, e0190547. [DOI] [PMC free article] [PubMed]
- Jin, T., Perry, A., Smith, P., Jiang, J. & Xiao, T. S. (2013). J. Biol. Chem. 288, 13225–13235. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Lang, S. & Palmer, M. (2003). J. Biol. Chem. 278, 38167–38173. [DOI] [PubMed]
- Litwin, C. M. & Johnson, J. M. (2005). Infect. Immun. 73, 4205–4213. [DOI] [PMC free article] [PubMed]
- Los, F. C., Randis, T. M., Aroian, R. V. & Ratner, A. J. (2013). Microbiol. Mol. Biol. Rev. 77, 173–207. [DOI] [PMC free article] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McKellar, R. C. (1994). Appl. Environ. Microbiol. 60, 4219–4225. [DOI] [PMC free article] [PubMed]
- Montes, L.-R., López, D. J., Sot, J., Bagatolli, L. A., Stonehouse, M. J., Vasil, M. L., Wu, B. X., Hannun, Y. A., Goñi, F. M. & Alonso, A. (2008). Biochemistry, 47, 11222–11230. [DOI] [PMC free article] [PubMed]
- Moon, A. F., Mueller, G. A., Zhong, X. & Pedersen, L. C. (2010). Protein Sci. 19, 901–913. [DOI] [PMC free article] [PubMed]
- Munch-Petersen, E., Christie, R., Simmons, R. & Beddome, H. (1945). Aust. J. Exp. Biol. Med. 23, 193–195. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Potter, J. A., Randall, R. E. & Taylor, G. L. (2008). BMC Struct. Biol. 8, 11. [DOI] [PMC free article] [PubMed]
- Schneewind, O., Friedrich, K. & Lütticken, R. (1988). Infect. Immun. 56, 2174–2179. [DOI] [PMC free article] [PubMed]
- Sörensen, M., Mak, T. N., Hurwitz, R., Ogilvie, L. A., Mollenkopf, H. J., Meyer, T. F. & Brüggemann, H. (2010). J. Microbiol. Methods, 83, 211–216. [DOI] [PubMed]
- Valanne, S., McDowell, A., Ramage, G., Tunney, M. M., Einarsson, G. G., O’Hagan, S., Wisdom, G. B., Fairley, D., Bhatia, A., Maisonneuve, J.-F., Lodes, M., Persing, D. H. & Patrick, S. (2005). Microbiology, 151, 1369–1379. [DOI] [PubMed]
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Xu, D. & Zhang, Y. (2012). Proteins, 80, 1715–1735. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: MBP-CAMP, 5y2g
Supplementary Table S1. DOI: 10.1107/S205979831901057X/qh5057sup1.pdf