Abstract
The majority of cells with latent human immunodeficiency virus 1 infection are located in lymphoid tissues that are difficult to access. In the current study, we used single-genome near-full-length proviral sequencing to evaluate intact and defective proviruses in blood and lymph node CD4 T cells enriched for specific functional polarizations. We observed minor variations between the frequencies of proviral sequences within individual CD4 T-cell subsets and across tissue compartments. However, we noted multiple clonal clusters of identical intact or defective proviral sequences from distinct compartments and CD4 T-cell subpopulations, suggesting frequent interchanges between viral reservoir cells in blood and tissues.
Keywords: HIV-1, reservoir, Lymph node, proviral sequencing, CD4 T cells
Using single-genome, near-full-length viral sequencing, we showed that clonal clusters of intact and defective human immunodeficiency virus 1 DNA sequences from patients receiving antiretroviral treatment patients consist of mixed proviral species from blood and lymph nodes.
Latently infected CD4 T cells harbor a transcriptionally silent, replication-competent, but antiretroviral treatment–unresponsive form of human immunodeficiency virus (HIV) 1 that can persist indefinitely owing to proviral integration into the host genome. Despite multiple attempts, recent efforts to specifically target or to reduce viral reservoir cells through clinical strategies in humans have not been successful. These disappointing clinical results may primarily reflect an incomplete insight in the physiology and evolutionary dynamics of such latently infected cells. Indeed, analyzing, quantitating, and isolating viral reservoir cells in antiretroviral-treated people living with HIV-1 has been extremely difficult, for several reasons. First, latently infected cells are very rare and specific surface markers or other biophysical characteristics that distinguish them from uninfected cells are yet to be discovered. In addition, the majority of virally infected cells harbor defective proviruses that result from errors during viral reverse-transcription and make it difficult to identify and target the even smaller subset of cells harboring genome-intact and replication-competent HIV-1 [1, 2]. Perhaps most prominently, the majority of viral reservoir cells are located in lymphoid tissues that are difficult to access for analytic purposes [3].
Prior studies have focused on identifying specific CD4 T-cell subsets that may represent preferential compartments for viral persistence during antiretroviral therapy. In peripheral blood, viral reservoir cells seem skewed toward a central memory or effector memory phenotype [4], and enriched within cells expressing specific activation markers [5] or displaying a T-helper (Th) 1 polarization profile [6, 7], although not all prior studies were able to distinguish between replication-competent and defective proviral sequences. In lymph nodes, viral outgrowth assays showed enrichment of cells containing inducible replication-competent virus within programmed cell death 1 (PD-1)/CXCR5+ CD4 T follicular helper (Tfh) cells [8, 9]. To what extent latently infected CD4 T cells in blood and lymphoid tissues intermingle, share signs of common clonal origin, or display features of distinct compartmentalization is less clear but may be critical for understanding the dynamics of viral reservoir cell homeostasis during suppressive antiretroviral therapy. In the current study, we sorted CD4 T cells enriched for distinct functional polarizations from peripheral blood and lymph nodes and analyzed near-full-length proviral genomes to investigate the dissemination and localization of genome-intact HIV-1 DNA sequences in individual CD4 T-cell populations.
METHODS
Study Participants
Study participants were recruited from the University of Lausanne, Switzerland, based on protocols approved by the local institutional review boards. Written informed consent was obtained from all study participants.
Cell Isolation
Inguinal lymph node biopsy specimens and blood samples were collected on the same day. Blood and lymph node mononuclear cells were isolated as described elsewhere [7].
Cell Sorting
CD4 T cells were enriched from blood mononuclear cells using EasySep Human CD4 T-cell enrichment kit (StemCell Technologies). Isolated blood CD4 T cells and lymph node mononuclear cells were then stained with Aqua LIVE/DEAD stain kit (at 4°C for 15 minutes), followed by incubation with fluorophore-labeled antibodies (CD3-Allophycocyanin-H7 [APCH7] [clone SK7], CD4–fluorescein isothiocyanate [clone RPA-T4], CD45RA-phycoerythrin-Texas Red [ECD] [clone 2H4], CXCR3-Pacific Blue [PB] [clone 1C6], CXCR5-allophycocyanin [clone J252D4], CCR4–phycoerythrin [PE]–cyanine 7 [clone 1G1], CCR6-PE [clone 11A9], PD-1–PE–cyanine 7 [clone EH12.1], CXCR5-PE [clone J252D4], at 4°C for 25 minutes). The grade of purity of the sorted cell populations was >97% in all sorting experiments.
Analysis of HIV-1 DNA
Proviral DNA was analyzed as described elsewhere [6, 10]; details are summarized in the Supplementary Methods.
Statistical Analysis
Data are summarized as individual data plots or pie charts. Differences were tested for statistical significance using 2-tailed Mann-Whitney or Kruskal-Wallis tests, followed by Dunn multiple-comparisons tests.
Data Availability
Viral sequencing data were deposited in GenBank (accession no. MT154920–MT155385).
RESULTS
For a detailed analysis of proviral sequences in individual CD4 T-cell subsets, we focused on 3 study participants who received suppressive antiretroviral therapy, and from whom simultaneously collected blood and lymph node samples were available for investigation. Clinical and demographic characteristics of these study participants are shown in Supplementary Table 1. In previous studies, Lee et al [6] analyzed CD4 T cells with a given polarization based on secretion of signature cytokines, but this approach is technically challenging when relatively small numbers of cells available from lymph node biopsies are analyzed. Therefore, we here used expression of chemokine receptors for distinguishing CD4 T cells enriched for a specific functional polarization. This approach is more inclusive, but only a small subset of cells with a given chemokine receptor expression profile are bona fide subpopulations of functionally polarized cells.
The following CD4 T-cell subpopulations were sorted from peripheral blood samples within the memory CD4 T-cell (CD45RA−) gate: CXCR3+CXCR5− (enriched for Th1 cells), CXCR3−CXCR5−CCR4+CCR6− (enriched for Th2 cells), CXCR3−CXCR5−CCR4+CCR6+ cells (enriched for Th17 cells), CXCR3−CXCR5+ (enriched for circulating Tfh-like cells), and CXCR3+CXCR5+ (enriched for Th1/Tfh-like cells). Lymph node memory CD4 T cells were sorted based on their CXCR5 and PD-1 expression patterns into the following 3 subsets: CXCR5+, PD-1+, and double-negative cells. For both blood and lymph node samples, CD45RA− cells were sorted as total memory cells; within lymph node samples, we also sorted naive cells (CD45RA+). Gating strategies are demonstrated in Supplementary Figure 1. DNA from sorted cells was extracted, followed by quantification of HIV-1 long terminal repeat–Gag copies by droplet digital polymerase chain reaction. DNA samples were then diluted to single viral genomes and subjected to amplification with primers spanning near-full-length HIV-1 DNA. Resulting amplification products were processed by next-general sequencing on the Illumina MiSeq platform; resulting proviral species were subjected to a previously described analysis algorithm to distinguish genome-intact and defective HIV-1 DNA sequences.
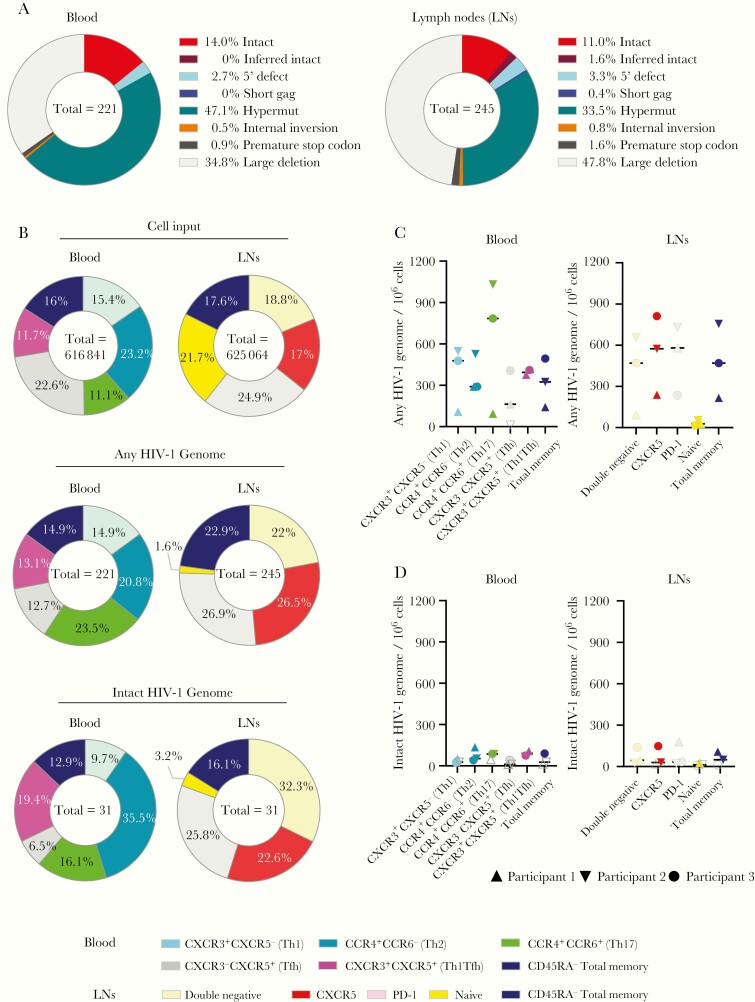
Analysis of a large number of individual proviral DNA sequences from blood (n = 231) and lymph nodes (n = 273) showed that the proviral landscape in both compartments were dominated by viral DNA sequences with large deletions, while genome-intact proviral species accounted for only a small fraction of all analyzed sequences, consistent with prior reports (Figure 1A and Supplementary Table 2). The frequency of intact and defective proviral sequences did not differ significantly between blood- and tissue-derived cells (P = .70 for intact HIV-1 genomes and P = .49 for any HIV-1 genome; Mann-Whitney test) (Figure 1B and 1C). Moreover, the relative distributions of total and intact HIV-1 sequences among the different CD4 T-cell subsets were remarkably similar, in both blood- and tissue-derived cells. (Figure 1B).
Figure 1.
Frequency of intact and defective human immunodeficiency virus (HIV) 1 sequences in CD4 T-cell subsets from peripheral blood and lymph node samples from antiretroviral-treated persons with HIV-1. A, Pie charts reflecting the relative proportion of intact and defective HIV-1 sequences isolated from lymph node and blood of the 3 study participants with HIV-1. B, Pie charts indicating the relative contribution of phenotypically distinct CD4 T-cell populations to the total number of cells analyzed (upper panels) and to the total number of any or intact HIV-1 sequences (lower panels) in blood and tissues. C, D, Frequencies of any HIV-1 sequences (C) or intact HIV-1 sequences (D) derived from indicated subsets of CD4 T cells from blood or lymph node compartments. Open symbols represent data below the limit of detection, calculated by assuming the presence of 0.2 viral copies in the number of cells in which no target was identified, and bars represent median values. Abbreviations: PD-1, programmed cell death 1; Tfh, T follicular helper; Th1, Th2, and Th17, T-helper 1, 2, and 17.
In lymph nodes, the majority of intact proviral sequences were harbored by CXCR5+, CXCR5−PD-1+, and CXCR5−PD-1− CD4 T cells, with roughly equal contributions made by each of these subsets; naive CD4 T cells from lymph nodes harbored the fewest intact proviral sequences (Figure 1B). In peripheral blood, no major differences were observed in absolute frequencies and relative proportions of total HIV-1 DNA sequences among the analyzed CD4 T-cell subsets; intact proviral sequences were slightly more frequent in CCR4+CCR6− CD4 T cells and slightly less frequent in cells enriched for Tfh-like cells, although none of these differences reached statistical significance (based on Kruskal-Wallis test followed by Dunn multiple-comparisons test).
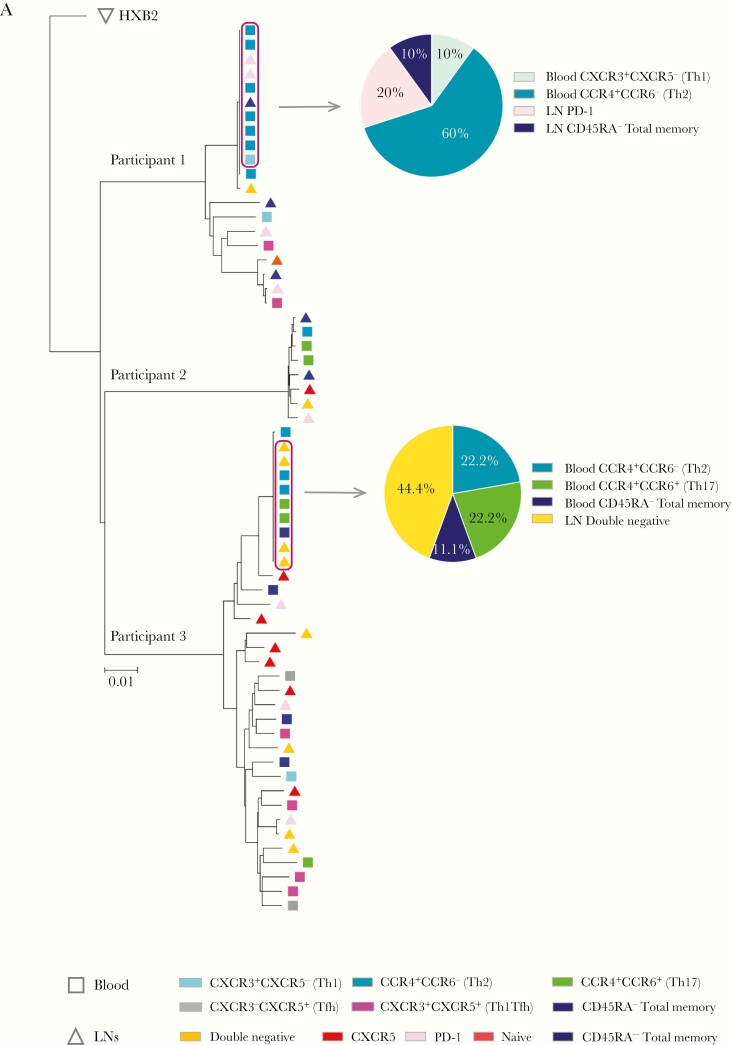
In our subsequent analysis, we focused on investigating phylogenetic relationships among all intact proviral sequences isolated from blood and lymph nodes from the 3 persons living with HIV-1 in our study. We noted that intact sequences from lymph nodes and blood were frequently phylogenetically intermingled, and there was no evidence for segregation or compartmentalization of intact proviruses from either anatomic compartment (Figure 2); in addition, intact proviral sequences from blood and lymph did not differ with regard to the frequency of sequence mutations associated with antiretroviral agents (Supplementary Table 3). In addition, intact proviral sequences retrieved form individual CD4 T-cell populations from blood or lymph nodes showed no evidence of enhanced phylogenetic association.
Figure 2.
Phylogenetic associations between intact proviruses isolated from individual subsets from blood and tissues (lymph nodes [LNs]). Figure shows neighborhood-joining phylogenetic tree of all intact proviral sequences (n = 62) isolated from indicated CD4 T-cell subsets from blood and tissue from 3 persons living with human immunodeficiency virus (HIV) 1. Clusters of completely identical HIV-1 sequences are highlighted with boxes; within each cluster, proportions of sequences derived from given CD4 T-cell subsets are shown in pie charts. If a near-full-length sequence showed a mapped 5’ long terminal repeat deletion with an absent or incomplete primer binding site, but otherwise displayed no lethal sequence defects, the missing 5’ long terminal repeat region was inferred to be present, and the sequence was termed “inferred intact.” Abbreviations: PD-1, programmed cell death 1; Tfh, T follicular helper; Th1, Th2, and Th17, T-helper 1, 2, and 17.
Remarkably, in 2 of our study participants, a large cluster of sequence-identical intact proviral species was noted. In both of these individuals, these clusters entailed a mixed combination of proviral sequences from blood and lymph nodes, and included sequences that were isolated from a diverse range of analyzed CD4 T-cell subpopulations from the 2 distinct tissue compartments. Multiple CD4 T-cell clones harboring sequence-identical defective proviral sequences were also noted; member sequences of these clones with defective proviruses also derived from a range of individual CD4 T-cell populations from blood and lymph nodes (Supplementary Figure 2 and Supplementary Table 4).
DISCUSSION
Characterizing HIV-1–infected CD4 T cells that resist currently available antiretroviral therapy and represent a long-lasting viral reservoir able to fuel rebound viremia is a critical step in developing strategies for HIV eradication and cure. Compared with peripheral blood reservoir cells, very little is known about viral reservoir cells in lymphoid tissues, which harbor the vast majority of an individual’s CD4 T-cell pool. Owing to the different immune environments and altered immunological selection forces in blood and tissues, it is possible that viral reservoir cells follow alternative evolutionary pathways in distinct anatomic compartments, such that virally infected clones adjusted to long-term survival in blood may be ill equipped to persist in lymphoid tissues, and vice versa. In particular, viral reservoir cells residing in immunologically privileged lymph node germinal centers may have a survival and selection advantage relative to circulating HIV-1–infected CD4 T cells [11, 12], which arguably are exposed to a more diversified spectrum of antiviral immune cells.
In the current study, we conducted a detailed analysis of intact proviral sequences at single-genome resolution, using next-generation sequencing approaches in autologous blood and tissue samples from individuals undergoing suppressive antiretroviral treatment. Consistent with prior studies, our work demonstrated clusters of intact proviral sequences that were completely identical, indicating that they are likely to originate from CD4 T cells that pass on proviral sequences during meiotic cell division [6, 13, 14]. Notably, these clones included a mixed combination of individual intact sequences isolated from blood and tissues, suggesting close interconnections and frequent interchanges between CD4 T cells from these 2 tissue compartments. Furthermore, we also frequently noted defective proviral sequence clusters consisting of individual member sequences derived both from blood and lymph nodes, and we generally failed to observe compartmentalized phylogenetic associations for clonal or nonclonal intact and defective proviral sequences from the 2 tissues analyzed.
In agreement with prior studies [15], these data strongly argue against segregated patterns of viral reservoir cell evolution in individual tissue compartments and do not support the hypothesis that lymphoid tissues represent a “sanctuary site” for persistence of distinct viral reservoir cell populations uniquely adjusted to survive long-term in the specific immune microenvironment of lymph nodes. Future studies involving alternative lymphoid tissue compartments and more HIV-1–infected persons will be informative for further investigating this question; in particular, samples from autopsies of antiretroviral-treated people with HIV-1 may allow for a more comprehensive evaluation of the evolutionary interchanges between viral reservoirs in blood and alternative anatomic compartments.
In addition to evaluating viral reservoirs in different tissues, our study also investigated viral reservoir sequences in CD4 T-cell populations with distinct chemokine receptor expression patterns known to enrich for defined functional polarizations. Within the relatively small numbers of cells and numbers of study participants available for our analysis, our study failed to discern notable differences in the frequency of intact or defective viral sequences within these phenotypically distinct populations. In contrast, functional outgrowth assays revealed that viral reservoir cells encoding for replication-competent HIV-1 in blood and lymph nodes were most dominantly encountered in cells enriched for Th1 [7] or Tfh [9] polarization, respectively.
Discrepancies between these studies and our work may primarily relate to the fact that only a fraction of genome-intact proviruses (arguably those with highest viral fitness) can be retrieved in viral outgrowth assays [1]. Moreover, it is important to recognize that isolation of different CD4 T-cell subsets in our work was based on chemokine receptor expression, which enriches for cells with a given polarization but also includes a large proportion of contaminating CD4 T cells lacking a defined functional lineage commitment. In contrast, previous studies used iterative sorting of cells with distinct secretion of signature cytokines to obtain highly purified populations of bona fide CD4 T cells sharing a specific functional polarization [6]. It is hoped that continuous progress in identifying the biophysical, functional, and molecular characteristics of viral reservoir cells will allow for targeting these cells more effectively in clinical settings.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Presented in part: Conference on Retroviruses and Opportunistic Infections, Boston, March 4–7, 2018.
Acknowledgments. We thank the Massachusetts General Hospital Center for Computational & Integrative Biology DNA Core, specifically Nicole Stange-Thomann, Amy Avery, Kristina Belanger, and Huajun Wang, for assistance with Illumina MiSeq deep sequencing.
Financial support . This work is supported by National Institutes of Health (grants AI098487, AI106468, AI114235, AI117841, AI120008, and AI124776 to M. L.), Massachusetts General Hospital (Tosteson Award for Medical Discovery to H. H. K.), the Swiss National Science Foundation (grant 320030_173071 to M. P.), Fondation Machaon (educational grant to R. B.), and the Center for AIDS Research, funded by the National Institutes of Health (developmental award P30-AI060354 to G. Q. L.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Ho YC, Shan L, Hosmane NN, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 2013; 155:540–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pinzone MR, VanBelzen DJ, Weissman S, et al. Longitudinal HIV sequencing reveals reservoir expression leading to decay which is obscured by clonal expansion. Nat Commun 2019; 10:728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Estes JD, Kityo C, Ssali F, et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med 2017; 23:1271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chomont N, El-Far M, Ancuta P, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 2009; 15:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee E, Bacchetti P, Milush J, et al. Memory CD4 + T-cells expressing HLA-DR contribute to HIV persistence during prolonged antiretroviral therapy. Front Microbiol 2019; 10:2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee GQ, Orlova-Fink N, Einkauf K, et al. Clonal expansion of genome-intact HIV-1 in functionally polarized Th1 CD4+ T cells. J Clin Invest 2017; 127:2689–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banga R, Procopio FA, Ruggiero A, et al. Blood CXCR3+ CD4 T cells are enriched in inducible replication competent HIV in aviremic antiretroviral therapy-treated individuals. Front Immunol 2018; 9:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Perreau M, Savoye AL, De Crignis E, et al. Follicular helper T cells serve as the major CD4 T cell compartment for HIV-1 infection, replication, and production. J Exp Med 2013; 210:143–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Banga R, Procopio FA, Noto A, et al. PD-1+ and follicular helper T cells are responsible for persistent HIV-1 transcription in treated aviremic individuals. Nat Med 2016; 22:754–61. [DOI] [PubMed] [Google Scholar]
- 10. Lee GQ, Reddy K, Einkauf KB, et al. HIV-1 DNA sequence diversity and evolution during acute subtype C infection. Nat Commun 2019; 10:2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fukazawa Y, Lum R, Okoye AA, et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med 2015; 21:132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Banga R, Rebecchini C, Procopio FA, et al. Lymph node migratory dendritic cells modulate HIV-1 transcription through PD-1 engagement. PLoS Pathog 2019; 15:e1007918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. McManus WR, Bale MJ, Spindler J, et al. HIV-1 in lymph nodes is maintained by cellular proliferation during antiretroviral therapy. J Clin Invest 2019; 130:4629–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mullins JI, Frenkel LM. Clonal expansion of human immunodeficiency virus-infected cells and human immunodeficiency virus persistence during antiretroviral therapy. J Infect Dis 2017; 215:119–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bozzi G, Simonetti FR, Watters SA, et al. No evidence of ongoing HIV replication or compartmentalization in tissues during combination antiretroviral therapy: Implications for HIV eradication. Sci Adv 2019; 5:eaav2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Viral sequencing data were deposited in GenBank (accession no. MT154920–MT155385).