Abstract
Background
Auto-aggregation is a desired property for probiotic strains because it is suggested to promote colonization of the human intestine, to prevent pathogen infections and to modulate the colonic mucosa. We recently reported the generation of adapted mutants of Lactiplantibacillus plantarum NZ3400, a derivative of the model strain WCFS1, for colonization under adult colonic conditions of PolyFermS continuous intestinal fermentation models. Here we describe and characterize the emerge of an auto-aggregating phenotype in L. plantarum NZ3400 derivatives recovered from the modelled gut microbiota.
Results
L. plantarum isolates were recovered from reactor effluent of four different adult microbiota and from spontaneously formed reactor biofilms. Auto-aggregation was observed in L. plantarum recovered from all microbiota and at higher percentage when recovered from biofilm than from effluent. Further, auto-aggregation percentage increased over time of cultivation in the microbiota. Starvation of the gut microbiota by interrupting the inflow of nutritive medium enhanced auto-aggregation, suggesting a link to nutrient availability. Auto-aggregation was lost under standard cultivation conditions for lactobacilli in MRS medium. However, it was reestablished during growth on sucrose and maltose and in a medium that simulates the abiotic gut environment. Remarkably, none of these conditions resulted in an auto-aggregation phenotype in the wild type strain NZ3400 nor other non-aggregating L. plantarum, indicating that auto-aggregation depends on the strain history. Whole genome sequencing analysis did not reveal any mutation responsible for the auto-aggregation phenotype. Transcriptome analysis showed highly significant upregulation of LP_RS05225 (msa) at 4.1–4.4 log2-fold-change and LP_RS05230 (marR) at 4.5–5.4 log2-fold-change in all auto-aggregating strains compared to non-aggregating. These co-expressed genes encode a mannose-specific adhesin protein and transcriptional regulator, respectively. Mapping of the RNA-sequence reads to the promoter region of the msa-marR operon reveled a DNA inversion in this region that is predominant in auto-aggregating but not in non-aggregating strains. This strongly suggests a role of this inversion in the auto-aggregation phenotype.
Conclusions
L. plantarum NZ3400 adapts to the in vitro colonic environment by developing an auto-aggregation phenotype. Similar aggregation phenotypes may promote gut colonization and efficacy of other probiotics and should be further investigated by using validated continuous models of gut fermentation such as PolyFermS.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12866-021-02331-x.
Keywords: Auto-aggregation, Lactiplantibacillus plantarum, In vitro gut microbiota, Adaptation, Transcriptome, DNA inversion
Background
Consumption of probiotics increased steadily over the past years and their application was recommended against a range of gastrointestinal tract related diseases [1]. Probiotics are defined as “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host “[2]. To exert a health benefit, a probiotic should exhibit certain properties such as survival during gastrointestinal passage, delivery of high viable-cell numbers to the colon and a certain degree of colonization in the gut environment [3, 4]. Successful colonization in the intestine is mediated via adherence to the intestinal epithelium or solid particles.
Auto-aggregation occurs between genetically identical cells and co-aggregation between genetically different cells [5]. Auto-aggregation is a well-studied phenomenon that leads to the formation of a community structure that facilitates interaction and communication between cells, genetic exchange, adherence and colonization in different environments [6–11]. Auto-aggregation ability correlates positively with adherence to human epithelial cell lines and the ability to co-aggregate with Listeria monocytogenes, Staphylococcus aureus and pathogenic Escherichia coli [8, 12–16]. Auto-aggregation is frequently assessed as feature in studies evaluating putative probiotic strains [17–19].
Although auto-aggregation is a common phenomenon, there is still no complete understanding of underlying mechanisms and triggers, possibly impeded by species-specific differences. Triggering factors of auto-aggregation so far identified include intestinal, nutritive, chemical, and oxidative stresses, changes in temperature and nutrient availability [20–25]. Several auto-aggregation mechanisms have been reported, including cell-surface properties, −structures and -enzymes. Cell surface hydrophobicity for example is positively linked to auto-aggregation [14, 26, 27]. Molecules involved in auto-aggregation, so called autoagglutinins, include cell-surface proteins, exopolysaccharides, carbohydrates, glycoproteins, teichoic and lipoteichoic acid secreted proteins that act as aggregation promoting factors [7, 23, 28, 29].
Auto-aggregation was reported for Lactobacillus spp. strains isolated from distinct environments like the piglet, chicken and murine gastrointestinal tract, vaginal tract, dairy and fermented foods [12, 30–37]. However, not much is known about bacterial auto-aggregation and its role in the human intestinal tract. Lactiplantibacillus plantarum WCFS1 is a model strain for probiotic lactobacilli. It harbors possible genetic predisposition for auto-aggregation in form of a serine/threonine rich domain in LP_RS01260 with affinity to mucin which was identified to be involved in auto-aggregation of L. plantarum NCIMB 8826, the mother strain of WCFS1 [38, 39]. However, there is no description of a WCFS1 auto-aggregation phenotype yet. Recently, we supplemented L. plantarum NZ3400, a derivative strain of WCFS1, to different in vitro human colonic microbiota, continuously cultivated in the PolyFermS model in the context of an evolutionary engineering experiment [40]. L. plantarum NZ3400 derivatives were recovered from the microbiota and phenotypically and genotypically characterized. Here, we describe the emerge of an auto-aggregation phenotype in a non-aggregating strain after exposure to human colon conditions. The novel phenotype was characterized, and transcriptome analysis was performed to investigate the underlying mechanisms.
Results
Development of a high-throughput screening for auto-aggregation
A high-throughput screening method based on a yeast-agglutination assay was set-up to detect auto-aggregating L. plantarum strains [41]. Microscopic investigation of 96 L. plantarum cultures grown in a 96-well tissue culture test plate revealed that auto-aggregating L. plantarum cultures produced a sediment of white clumps, while non-aggregating cultures formed a homogenous white layer at the bottom of the wells (Fig. 1A). Therefore, detection of clumps can be used to distinguish rapidly between auto-aggregating and non-aggregating strains.
Fig. 1.
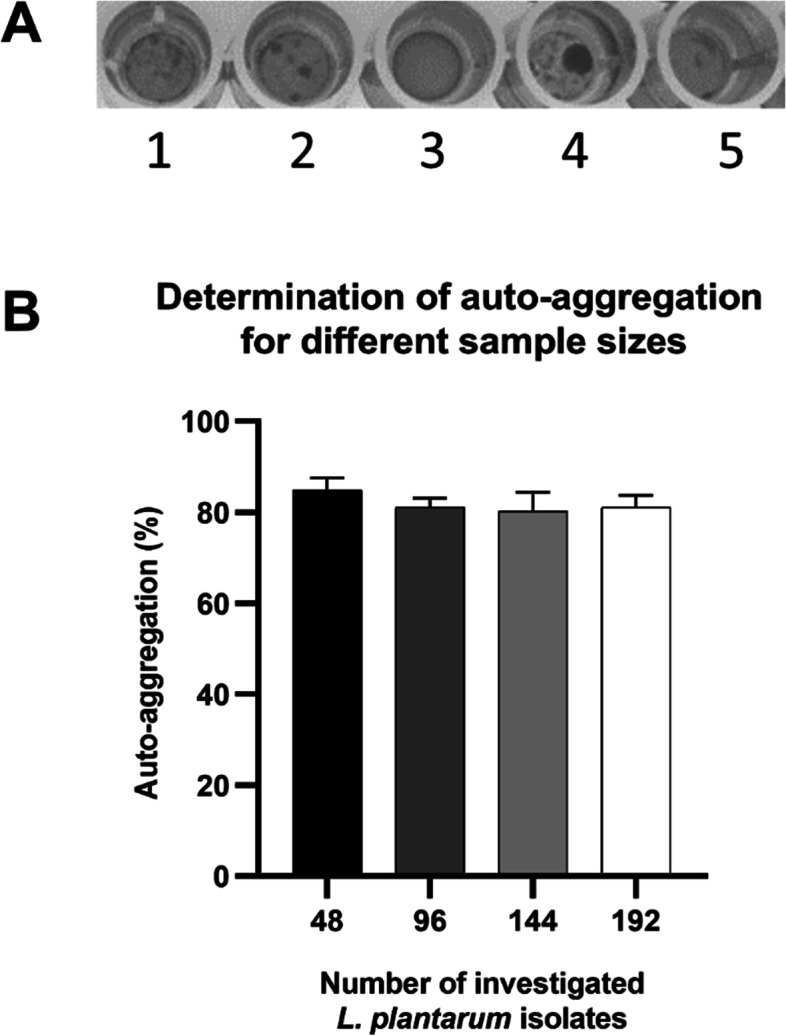
Visual auto-aggregation detection and determination of the number of colonies to investigate auto-aggregation. A 96-well plate well bottom appearance of non-aggregating (well 3) and auto-aggregating (well 1–2,4–5) L. plantarum isolates. B Percentage of auto-aggregation after testing different numbers of L. plantarum colonies
To determine how many L. plantarum isolates have to be analyzed to obtain a reliable percentage of the auto-aggregation subpopulation, different sets of colonies isolated from a single reactor and time point were visually assigned as aggregating or not. Testing 48, 96, 144 and 192 colonies did not significantly change aggregation percentage (Fig. 1B). Therefore, 48 colonies were investigated in all further experiments to determine auto-aggregation percentages.
L. plantarum auto-aggregation increases in biofilm, during starvation and during cultivation in in vitro colonic microbiota
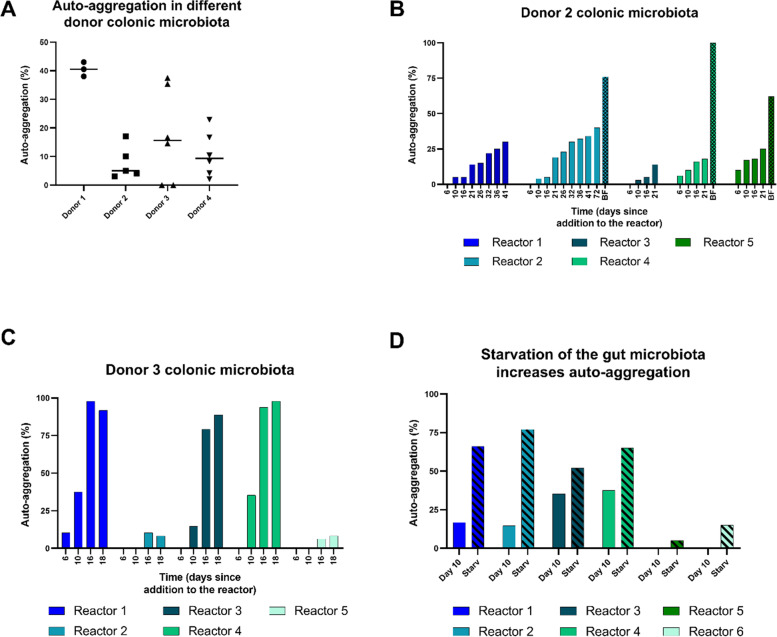
To investigate if auto-aggregation is microbiota-dependent, L. plantarum NZ3400 was supplemented to four PolyFermS models inoculated with different colonic microbiota and recovered after 10 days. Auto-aggregation was detected in L. plantarum isolates recovered from all four microbiota at levels between 8 and 41% (Fig. 2A). Auto-aggregation percentage steadily increased over time in five different reactors containing microbiota of donor 2, reaching approximately 15 and 30% after 21 and 42 days post supplementation, respectively (Fig. 2B). An increase in auto-aggregation over time was also observed for donor 3 microbiota. It was more pronounced compared to donor 2 microbiota in the reactors 1, 3 and 4 with 88–89% aggregation 18 days post L. plantarum supplementation, but lower in the reactors 2 and 5 with only 8% aggregation after 18 days (Fig. 2C).
Fig. 2.
Influence of different microbiota, biofilm and starvation on L. plantarum auto-aggregation percentage. L. plantarum strains were recovered from four different in vitro human adult colonic microbiota. A Auto-aggregation percentage of L. plantarum strains recovered after 10 days of cultivation in reactors inoculated with colonic microbiota from four different donors. Each data point represents auto-aggregation percentage in one reactor. B Auto-aggregation percentage of L. plantarum strains recovered from five reactors inoculated with microbiota of donor 2 over time and from reactor biofilms (BF). C Auto-aggregation percentage of L. plantarum recovered from five reactors containing colonic microbiota of donor 3. D Auto-aggregation percentage at the day before and one day after starvation of the microbiota of donor 4 (Starv)
Further, biofilms spontaneously formed on the reactor walls were collected at the end of fermentations. The auto-aggregation percentage of L. plantarum strains from biofilms was at least two times higher than from reactor effluent at the final day of fermentation (Fig. 2B), suggesting a role of auto-aggregation in biofilm establishment. However, no correlation was found between auto-aggregation percentage and L. plantarum colonization level in the reactor (data not shown). Since aggregation might facilitate the formation of biofilms, biofilm formation ability of the wild type and the aggregating, isogenic strain PA4_02 on polystyrene was assessed using crystal violet and optical density measurement. Strains were grown compared in MRS and minimal medium containing different carbon sources. The aggregating strain PA4_02 showed clearly increased biofilm formation compared to the wild type when grown in minimal medium supplemented with maltose but not in any other conditions (Additional file 1: Figure S1).
Auto-aggregation is suggested to be linked to cell-proximity during nutrient exchange or to nutritive stress [23]. Therefore, starvation of colonic microbiota was simulated by interrupting the medium inflow for 24 h in six TRs containing microbiota of donor 4. Starvation led to accumulation of isobutyrate, isovalerate and valerate, which indicates enhanced protein fermentation (Additional file 2: Figure S2). Auto-aggregation percentage increased during starvation in all reactors (Fig. 2D). Moreover, after starvation, auto-aggregation was even detected in reactors where no auto-aggregation was observed before the starvation period (Fig. 2D, reactor 5 and 6). Simultaneously, L. plantarum viable cell counts decreased by 1–2 logs during starvation (data not shown).
Bistability of auto-aggregation phenotype depends on nutrient availability
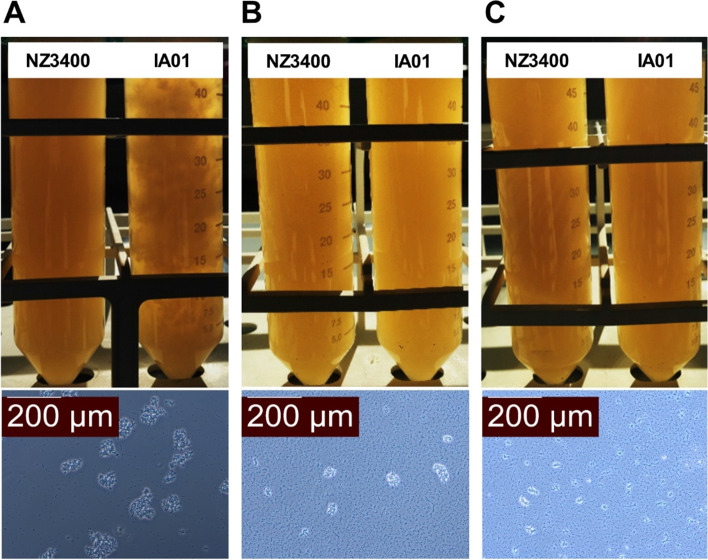
To investigate the stability of the auto-aggregation phenotype, fifteen L. plantarum NZ3400 derivatives with auto-aggregation phenotype, recovered from all four microbiota, were serially cultured in MRS medium daily for 3 days. All strains still exhibited a strong auto-aggregation phenotype that was clearly visible after the first overnight culture (Fig. 3A). Auto-aggregation was not visually detectable anymore after the second overnight culture, yet small aggregates were still observed microscopically (Fig. 3B). Finally, aggregates were only barely detectable microscopically in the third culture (Fig. 3C). Conclusively, auto-aggregation is instable under nutrient-rich cultivation conditions, hinting again towards a role of nutrient availability in auto-aggregation.
Fig. 3.
Auto-aggregation phenotype diminishes during consecutive cultivation in MRS broth. Visual comparison between L. plantarum NZ3400 (left tube) and IA01 (right tube) and microscopic images of IA01 after the first (A), second (B) and third (C) consecutive culture in MRS broth. Results are representative for 15 tested L. plantarum cultures with auto-aggregation phenotype
Thereafter, L. plantarum strains that lost their aggregation capacity upon consecutive cultivation in MRS medium were grown in a minimal medium supplemented with different carbon sources and in EMS, a medium that simulates the abiotic environment of the reactor gut microbiota [40]. L. plantarum strains IA01, PA4_02 and PA1.2_01 lost their auto-aggregation capacity in repeated MRS cultures but recovered this capacity when regrown in EMS medium and in minimal medium containing sucrose or maltose, but not in the same medium with glucose as sole carbon source. Remarkably, L. plantarum NZ3400 and four isolates (Table 1) recovered from the modelled microbiota of donor 1 and 2 that never showed auto-aggregation, did not auto-aggregate in any of the tested conditions. These results show that different nutrient conditions as such do not result in an auto-aggregation phenotype in L. plantarum NZ3400 and its derivatives, but that auto-aggregation is only observed in strains that have been exposed to the gut microbiota.
Table 1.
L. plantarum NZ3400 derivative strains recovered from human in vitro microbiota
| Strain | Description | Auto-aggregation | Reference |
|---|---|---|---|
| IA01 | NZ3400 derivative, CmR, C979T in LP_RS14990, isolated from in vitro fermentation | + | Isenring et al., 2021 [40] |
| IA01t0 | Strain IA01 grown after one overnight culture in MRS | + | This study |
| IA01t4 | Strain IA01 grown after four consecutive cultures in MRS | – | This study |
| PA4_02 | NZ3400 derivative, CmR, isolated from in vitro fermentation | + | Isenring et al., 2021 [40] |
| PA1.2_01 | NZ3400 derivative, CmR, C979T in LP_RS14990 and G382A in LP_RS01530, isolated from in vitro fermentation | – | Isenring et al., 2021 [40] |
| IA10 | NZ3400 derivative, CmR, C569A in LP_RS14255, isolated from in vitro fermentation | – | Isenring et al., 2021 [40] |
| PA2_04 | NZ3400 derivative, CmR, C837A in LP_RS15205, isolated from in vitro fermentation | – | Isenring et al., 2021 [40] |
| PA2_06 | NZ3400 derivative, CmR, C837A in LP_RS15205, isolated from in vitro fermentation | – | Isenring et al., 2021 [40] |
Genome and transcriptome analyses of auto-aggregating L. plantarum isolates
The possibility to reestablish auto-aggregation only in isolates that were exposed to modelled colonic microbiota suggests a lasting genomic modification responsible for the phenotype. However, whole genome sequencing analysis did not reveal any shared single nucleotide polymorphisms nor genetic reorganization in seven auto-aggregating isolates compared to NZ3400. Moreover, three thereof did not harbor any mutation and were thus isogenic to L. plantarum NZ3400 (data not shown). Because genome sequences did not provide an explanation for the auto-aggregation phenotype, transcriptome analysis using RNA sequencing was performed to identify genes possibly involved in the auto-aggregation phenotype.
The transcriptome of the wild type strain NZ3400 was compared to that of the strain IA01t0 during growth in MRS, conditions under which AI01 aggregates and NZ3400 does not. The comparison revealed 192 significantly up- and 289 downregulated genes in IA01 compared to NZ3400 (FDR < 0.05; Additional file 3: Table S1). In addition, IA01 was grown in four consecutive overnight cultures in MRS, which led to loss of the auto-aggregation phenotype, resulting in the strain IA01t4. Comparison of aggregating IA01t0 and non-aggregating IA01t4 resulted in 583 significantly up- and 632 downregulated genes in IA01t0 (Additional file 4: Table S2). Further, 94 up- and 123 downregulated genes were shared in both comparisons, being primary candidate genes for involvement in auto-aggregation. To narrow down these candidate genes, RNA sequencing data of the auto-aggregating strain PA4_02 and the wild type strain NZ3400 were compared. A total of 138 genes were significantly upregulated and 143 were significantly downregulated in PA4_02 (Additional file 5: Table S3). Combining all three comparisons revealed 50 significantly up- and 62 downregulated genes in auto-aggregating strains versus non-aggregating strains (Additional file 6: Table S4). Thereof, only genes with a │log2 ratio│ > 1 amongst all three comparisons were considered as possible candidate genes involved in auto-aggregation (Table 2). Up-regulated genes in this data set encoded for two hypothetical proteins, three proteins with domains of unknown function (DUF), a haloacid dehalogenase-like hydrolase (HAD), the Multiple Antibiotic Resistance Regulator (MarR), and a mucin-binding protein (MucBP) domain (Table 2). The latter is known to be a mannose-specific adhesion protein encoded by msa [42]. The MarR family transcriptional regulator (LP_RS05230, marR) and the MucB domain-containing protein (LP_RS05225, msa) were strongest regulated by far with a log-fold change of 4.5–5.4 and 4.1–4.4, respectively (Table 2). The marR gene is located 11 bp downstream of the msa gene and in the same direction, suggesting that they belong to one operon.
Table 2.
Genes significantly up- and down-regulated (│Ratio(log2)│ > 1) in auto-aggregating compared to non-aggregation strains*
| ORF | Product | Ratio (log2) | ||
|---|---|---|---|---|
| PA4_02a) | IA01t0b) | IA01t0c) | ||
| Upregulated | ||||
| LP_RS05230 | MarR family transcriptional regulator | −5.17 | −4.54 | − 5.37 |
| LP_RS05225 | MucBP domain-containing protein | −4.23 | −4.10 | − 4.42 |
| LP_RS02885 | Hypothetical protein | −2.17 | −1.54 | −1.25 |
| LP_RS10335 | DUF4355 domain-containing protein | −2.04 | −1.53 | −1.29 |
| LP_RS02870 | DUF4355 domain-containing protein | −1.90 | −1.18 | − 1.22 |
| LP_RS10300 | Hypothetical protein | −1.55 | −1.03 | − 1.07 |
| LP_RS14830 | HAD family hydrolase | −1.52 | −1.11 | − 1.28 |
| LP_RS10310 | DUF3168 domain-containing protein | −1.49 | −1.58 | − 1.20 |
| Downregulated | ||||
| LP_RS01470 | Hypothetical protein | 1.05 | 1.27 | 1.22 |
| LP_RS00795 | Cna B-type domain-containing protein | 1.08 | 1.02 | 1.04 |
aAuto-aggregating PA4_02 compared to the wild type NZ3400
bAuto-aggregating IA01t0 compared to IA01t4
cAuto-aggregating IA01t0 compared to the wild type NZ3400
*phage-related genes were omitted. ORF: open reading frame
Further, 2 genes encoding a hypothetical protein and Can B-type domain-containing protein, were significantly down-regulated in all three analyses (Table 2). However, the magnitude of change (maximum of 1.27 log-fold change) was small compared to the msa-marR operon (Table 2).
DNA inversion predominates in auto-aggregating L. plantarum strains
The very strong regulation of the msa-marR operon makes this operon the primary candidate responsible for the auto-aggregation phenotype. The msa promoter region contains two repeats that can invert, thereby changing the promoter activity of the operon [41]. To test whether such inversion occurred in auto-aggregating strains, the RNAseq reads were plotted on the genome of WCFS1 and checked carefully for any genetic reorganization in the msa region. Indeed, an alternative junction was identified in all samples of the aggregating strains IA01t0 and PA4_02, exactly at the repeat in the promoter of msa. This new junction was not identified in the transcriptome samples from the wild type, IA01t4, nor in the samples of a previously constructed non-aggregating lamC knockout strain L. plantarum ΔLP_RS14990 [40] (Isenring, unpublished results).
Next, we quantified the occurrence of the inverse promoter using the RNAseq data reads. The ratio between the native and the inverted promoter was 8.3 ± 1.2 in the wild type and 9.5 ± 1.4 in the ΔLP_RS14990 strain, showing high prevalence of the native promoter in both non-aggregating strains. In contrast, the auto-aggregating strains A01t0 and PA4_02 had a significantly lower ratio of 0.56 ± 0.04 and 0.54 ± 0.07 than the wild type strain and ΔLP_RS14990 (p = 0.001). Strain IA01t4 lost its auto-aggregating phenotype and had a significantly lower a ratio of 5.3 ± 0.9 than the wild type (p = 0.03) suggesting that the inverted promoter is still present in strains that have lost the phenotype recently.
Last, we investigated whether the inverted promoter region is already present in a subpopulation of the wild type strain and in the non-aggregating strain IA10 (Table 1). A PCR specific for the native promoter resulted in amplification for the wild type strain NZ3400, strain IA10 and for both aggregating strains PA4_02 and AI01 (Additional file 7: Figure S3), supporting that the native promoter is present in all L. plantarum cultures. However, the PCR specific for the inverted promoter did not result in amplification for the wild type and IA10 strains but produced a clear amplicon in the cultures of PA4_02 and IA01.
Discussion
Auto-aggregation is a desired property for probiotics which is associated with enhanced colonization, inhibition of pathogenic infections and immunomodulation of the intestinal mucosa [12, 13, 28, 43, 44]. In this study, we describe for the first time the emerge of an auto-aggregating phenotype in the non-aggregating L. plantarum NZ3400 after exposure to modelled human colonic microbiota. Percentage of auto-aggregation was highest for L. plantarum isolated from reactor biofilms. Biofilm formation can be facilitated by auto-aggregation [5, 45], however, quantification of biofilm formation on a polystyrene surface via crystal violet showed that biofilm formation of the aggregating strain PA4–02 was only enhanced compared to the wild type in minimal medium containing maltose. Aggregates were reported to form a multi-species cell-network that facilitates nutrient exchange, provides protection and creates a novel niche that aids microbiota colonization [23, 46]. L. plantarum cells in aggregates might therefore be able to survive better during starvation. It should be noted that starvation did not induce changes in propionate and butyrate concentrations and glucose, galactose and lactate were not detectable in both, control, and starved conditions. Therefore, these factors and the pH, which was kept constant during fermentation, are not explaining the effects observed on aggregation.
Transcriptome analysis of the wild type strain and auto-aggregating L. plantarum strains showed that the genes encoded by LP_RS05225 (msa) and LP_RS05230 (marR) were both highly overexpressed in the auto-aggregation phenotype. LP_RS05230 (previously known as lp_1230) is a transcriptional regulator that is located 11 base pairs downstream of LP_RS05225. In silico analysis of the genes showed that they form a bicistronic operon [41, 42], which is also supported by our data. The msa gene was identified as mannose-specific adhesin gene and is responsible for the agglutination of L. plantarum with Saccharamyces cerevisiae [41, 42]. Mannose-specific adherence mediates adherence of L. plantarum to human colonic cell line HT-29, epithelial Caco-2 cells and intestinal mucus in rats and thus assists in intestinal colonization [41, 47, 48]. In addition, pathogens such as Vibrio cholerae, Salmonella or Pseudomonas aeruginosa bind to mannose-containing glycoconjugates on the host intestinal surface [49–51]. Adherence of L. plantarum by mannose-specific adhesins to such glycoconjugates might therefore prevent binding of the pathogens. Furthermore, mannose-specific adhesins reduce pathogen colonization due to co-aggregation with mannose-containing surface structures of the pathogen [52–54]. Hence, activation of msa may be a desirable property for probiotics.
Identification of marR and msa did not reveal the mechanism sustaining the auto-aggregation phenotype, since genome sequencing did not reveal any mutation or other variation in the auto-aggregating strains. However, plotting RNAseq reads on the wild type genome revealed two novel DNA-junctions in the promoter region of LP_RS05225, that apparently could only be detected with the high read numbers produced in RNAseq experiments and the dedicated breseq software with a low detection limit for identification of variations in the genome [55]. The junction is identical to a previously reported inversion that is responsible for a strong yeast-agglutination phenotype in L. plantarum WCFS1, the parental strain of NZ3400 [41]. The inversion causes prevention of a stem-loop structure increasing both, transcription and translation of the msa gene. The inversion was predominant in aggregation cultures and still more present in IA01t4 than in non-aggregating stains. The occurrence of the aggregating phenotype over several generations can be explained by the presence of this relatively stable inversion. Moreover, the detected remains of the inversion in cultures that lost their aggregation phenotype may explain the rapid reverse of these cultures to an aggregating phenotype under selected conditions.
Aggregation was activated in absence of detectable glucose in fermentation reactors and in minimal medium without glucose. In addition, aggregation disappeared in glucose containing MRS medium. This suggests a role of glucose mediated catabolite repression via the canonical regulatory carbon catabolite protein A (CcpA) [56]. CcpA binds to catabolite response elements (cre) in the proximity to the regulated promoter, thereby regulating gene expression [57]. However, involvement of CcpA seems unlikely because reestablishment of auto-aggregation also occurred in the glucose-containing EMS medium. Further, no cre-elements were identified in the promoter region of msa and transcriptome analysis of a L. plantarum WCFS1 ccpa-knockout strain did not reveal differential expression in LP_RS05225 and LP_RS05230 [58]. Other regulators might be involved in the msa promoter inversion, yet the exact regulation remains unclear.
The observed loss and reestablishment of the auto-aggregation phenotype could be explained by phase variation, which leads to an heterogenous culture via an ON/OFF switch [59]. DNA inversions cause phase-variable surface protein expression [60–66] partially via genetic modifications by the activity of recombinases [60, 61, 67–70]. Indeed, recombinases lead to DNA inversion in Bacteroides fragilis resulting in phase-variable expression of surface proteins responsible for auto-aggregation [71]. The higher expression of the recombinase RecT (Additional file 6: Table S4) in auto-aggregating strains might induce DNA inversion in the upstream region of msa. However, classical phase variation in the sense of an ON-OFF switch does not explain the slow disappearance of auto-aggregation during three consecutive MRS cultures.
Alternatively, growth in MRS medium might select for the native promoter, which is shown by the presence of the native orientation in the aggregating strains IA01 and PA4_02 (Additional file 7: Figure S3). Overexpression of the mannose-specific adhesin is toxic in L. plantarum 299v, but not WCFS1, yet might lead to slower growth [41], explaining the vanishing of the auto-aggregating phenotype. Growth in MRS medium can then select for the fast-growing, non-aggregating fraction resulting in outcompeting the aggregating fraction during consecutive culturing. Slower growth of L. plantarum on sucrose, maltose [58] and EMS (data not shown) would allow outgrowth of the auto-aggregating fraction. Strong supporting evidence for this explanation is the difference in inverse and native promoter prevalence between the wild type and auto-aggregating variants.
The presence of both promoter orientations in aggregating strain cultures shows that an aggregating subculture proliferates in the fermenter. The inverse promoter orientation could not be detected in non-aggregating strains, suggesting strongly that the inversion is not present in non-aggregating cultures but is rather a relatively rare event and must be selected for to become visible. However, whether the inversion occurs spontaneously or is induced by the fermenter environment remains unclear.
Conclusion
We showed that cultivation of L. plantarum in a modelled in vitro human colonic microbiota leads to an auto-aggregating subpopulation. This novel auto-aggregating phenotype is caused by a high expression of the mannose-specific adhesion gene msa. Both, auto-aggregation and mannose-mediated adhesion are desired properties of probiotics. Our data demonstrate that the PolyFermS fermentation model is suitable to detect and select for novel phenotypes related to the intestinal environment. This could further be applied to elucidate phenotypic adaptations and their underlying mechanisms of both probiotics and gut pathogens.
Methods
Strains and growth conditions
L. plantarum NZ3400, a WCFS1 derivative harboring a chloramphenicol resistance cassette in a neutral locus on the chromosome, was used as wild type strain [72]. NZ3400 derivative strains were recovered from in vitro human colonic microbiota after cultivation for up to 100 generations as described previously [40] (Table 1).
L. plantarum strains were cultivated in De Man, Rogosa and Sharpe (MRS, Labo-Life Sàrl, Pully, Switzerland) broth at 37 °C, overnight, unless stated otherwise. To investigate the influence of different carbon sources on auto-aggregation, a minimal medium was designed based on the MRS composition [73] by omitting beef extract, peptone, glucose, Tween 80 and reducing yeast extract from 0.4 to 0.2% (w/v). This minimal medium did not support L. plantarum growth and metabolic activity. Glucose, sucrose, maltose and fructose were added as individual carbon source at 1% (w/v). In addition, the previously designed Effluent-MacFarlane-Sugar (EMS) medium [40], mimicking the abiotic environment of in vitro colonic microbiota, was used to test the effect on auto-aggregation in absence of microbiota. EMS medium consisted of sterile filtered effluent from the in vitro colonic microbiota, MacFarlane medium [74], which mimics the chyme entering the colon (ratio 9:1), and glucose (0.75%, w/v).
Establishment of a high-throughput screening method for auto-aggregation
A high-throughput screening method to detect auto-aggregating L. plantarum strains from colonic in vitro human gut microbiota was developed by modifying an existing agglutination assay [41]. MRS broth in 96-well tissue culture test plates (Bioswisstec AG, Schaffhausen, Switzerland) was inoculated with L. plantarum strains and grown overnight. Cultures were analyzed microscopically in biological triplicates (Leica DM1000, Leica Microsystems, Wetzlar, Germany) for aggregate formation.
Characterization of L. plantarum auto-aggregation in the in vitro colonic microbiota
The design and implementation of the continuous intestinal fermentation model PolyFermS was presented in detail previously [40]. In short, the inoculum reactors (IR) of four models were inoculated (30%, v/v) with distinct fecal microbiota from healthy adults immobilized in polysaccharide gel beads. A detailed description of the PolyFermS set-up and conditions, microbiota compositions and fermentation profiles are reported in Isenring et al. [40]. The four IRs used for auto-aggregation experiments were operated for time periods ranging from 10 to 72 days. Second-stage treatment reactors (TRs) were continuously inoculated by IR effluent (5%) and fed with fresh MacFarlane medium (95%), formulated to mimic the chime entering the colon [74]. Metabolite concentrations of the continuous fermentation were determined by high-performance liquid chromatography as described previously [40]. To investigate the influence of colonic microbiota on auto-aggregation, L. plantarum NZ3400 was supplemented to TRs connected to the IR and operated with identical conditions. Derivatives were isolated from the effluent after 10 days from three TRs containing microbiota of donor 1, five TRs of donor 2, five TRs of donor 3 and six of donor 4.
L. plantarum strains were recovered from the biofilm in three reactors collected at the end of the fermentation containing microbiota of donor 2 as described previously [40] and compared to L. plantarum isolated from reactor effluent harvested on the same day. Nutritive stress was simulated by starvation of the modelled gut microbiota by medium inflow interruption. L. plantarum strains were recovered before and 24 h after starvation.
Biofilm assay
Biofilm formation capability of the non-aggregating strain NZ3400 and its aggregating derivative strain PA4_02 was assessed in MRS, minimal medium containing glucose, sucrose, fructose or maltose as sole carbon source and EMS medium as described previously with minor modifications [75]. Wells of 96-well tissue culture test plates (Bioswisstec AG, Schaffhausen, Switzerland) containing 200 μl of corresponding growth medium were inoculated with 106 CFU/ml L. plantarum. Wells only containing the medium served as control. The plates were incubated at 37 °C for 24 h. The liquid was removed, and the wells were washed three times with phosphate-buffered saline (PBS), pH 6.2. After drying, the wells were supplemented with crystal violet (0.1%, w/v) and incubated for 30 min. The stained biofilm was washed three times with PBS, the dye was resolved in ethanol (99%) and absorbance was measured at 595 nm (PowerWaveTMXS; Bio-Tek Instrument Inc., Winooski, VT, USA).
Stability of auto-aggregation phenotype
The stability of auto-aggregation in 15 L. plantarum isolates recovered from all four microbiota was tested in three consecutive overnight cultures in MRS broth inoculated at 1% (v/v), corresponding to approximately 25 generations. Auto-aggregation was assessed visually and microscopically.
RNA isolation and sequencing
RNA sequencing was performed on the wild type strain L. plantarum NZ3400, the auto-aggregating strains IA01t0 and PA4_02 and the non-aggregating IA01t4. Pre-cultures were done in MRS broth at 30 °C overnight. Experiments were performed in triplicates for NZ3400, IA01t0 and IA01t4 and in duplicates for PA4_02. Cultures were inoculated from the pre-culture and grown until OD600nm = 2.6–2.7, with final measurement of the pH, glucose utilization and lactate production. Total RNA was extracted based on chloroform/phenol extraction followed by purification using the High Pure RNA isolation kit (Roche Diagnostics, Rotkreuz, Switzerland) as described previously (58). RNA quantity, purity and integrity were verified using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). Samples with an RNA integrity number (RIN) > 9 and a 16S/23S-rRNA ratio > 1.5 were selected for rRNA depletion. Thereafter, EDTA was added to 1 mM and depletion was performed using the MICROBExpress™ Bacterial mRNA Enrichment Kit (Life Technologies Europe BV, Zug, Switzerland) following the manufacturer’s instructions. Concentrations of depleted samples were determined in a TapeStation and normalized to 100 ng/μl using in Tris-HCl (10 mM, pH = 8.5).
Sequencing of 100 bp single reads was done on Illumina Novaseq 6000 (Illumina Inc., California, USA) at the Functional Genomics Center Zurich (FGCZ). The library was prepared according to the Illumina Truseq Total RNA protocol.
Analyses of DNA inversion upstream of LP_RS05225
The pipeline for the analysis of short-read re-sequencing breseq [76] was used to identify possible new junctions in the upstream region of msa (LP_RS05225). A 9439-bp fragment of the WCFS1 genome 7000 bp upstream and 2000 bp downstream of the marR gene (LP_RS05230) was used as reference and the RNAseq reads as re-sequencing reads. Standards settings were applied, and each set of reads was analyzed separately.
To quantify the amount of standard and reversed promoter regions of msa in the strains, a fragment containing the 234-bp upstream regions of the msa start with both promotor orientations was constructed and stored as two sequences in a single fasta file. RNAseq reads were plotted to the sequences using Bowtie2 [77] with standard settings. The Bowtie2 output was further processed to a reads-per-gene spreadsheet as described above. The ratio between both promoter regions was calculated and averaged. A two-tailed t-test was used to calculate the p value.
To identify a possible subpopulation harboring the msa-MarR-inversion, primers were designed (Additional file 8: Figure S4) to amplify the native sequence (Native_fw: 5′-GGGAGTAAAGCGTGCAATGT-3′; Native_rev: 5′-GCATTACCTATTTGATAACGCAGA-3′) and the inverted sequence (Inversion_fw: 5′-TCATGCGAAAGGATAGGTGTAA-3′; Inversion_rev:5′- TTGAGATGCTGAATCGTTCG-3′) in the promoter region. DNA of the non-aggregating wild type NZ3400 and IA10, and the aggregating PA4_02 and IA01 was extracted as described previously using lysozyme-cell-lysis and purification using the Wizard Genomic DNA purification kit (Promega, Dübendorf, Switzerland) [40]. PCR reactions were performed in a volume of 25 μl containing 20 ng DNA, 12.5 μl 2 x PCR Master Mix (Fermentas, Le Mont-sur-Lausanne, Switzerland), 1 μM of each primer (Microsynth, Balgach, Switzerland) and sterile, DNase-free water (Fermentas). PCR was performed with an initial denaturation (95 °C, 2 min), followed by 30 or 40 cycles of denaturation (95 °C, 30 s), annealing (51 °C, 30 s) and replication (72 °C, 25 s) and subsequent final replication (72 °C, 7 min) in a Biometra® T3000 Thermocycler (Labgene, Châtel-Saint-Denis, Switzerland). Amplified products and DNA marker (100 bp, BioConcept, Allschwil, Switzerland) were analyzed via agarose (2%, w/v) gel electrophoresis and visualized with gel red.
Data analysis
Graphs were created using GraphPad Prism® version 8 (GraphPad Software Inc., San Diego, CA, USA). Analysis of RNAseq reads was performed as described previously [78]. Shortly, reads were mapped on the chromosome and plasmid of L. plantarum WCFS1 (accession numbers NC_006375.1, NC_006376.1, NC_006377.1, and NC_004567.2) using Bowtie2. Data filtering, normalization and analysis was done in R (version 3.6.2) using the packages DESeq and EdgeR. Only genes with a false discovery rate (FDR) < 0.05 and a differential expression of minimum 2 fold (│ratio(log2) > 1│) were considered as significant differently expressed.
Supplementary Information
Additional file 1: Figure S1. Biofilm formation ability of the non-aggregating wild type strain L. plantarum NZ3400 (blue) and the isogenic aggregating strain PA4_02 (red). Biofilm formation was assessed in MRS and EMS medium and minimal medium containing sucrose (mm-sucrose), fructose (mm-fructose), glucose (mm-glucose) or maltose (mm-maltose) as carbon source. Data represent mean value ± standard deviation of biological triplicates.
Additional file 2: Figure S2. Fermentation metabolite profile before (blue) and after (red) microbiota starvation. Bars represent mean ± standard deviation of metabolite abundance relative to the total produced metabolites of six parallel operated TRs the day before and 24 h after starvation of the gut microbiota. Significance was calculated by paired-sample t test: ** < 0.01, **** p < 0.0001.
Additional file 3: Table S1. Significantly up- and downregulated genes in the auto-aggregating L. plantarum IA01 compared to NZ3400.
Additional file 4: Table S2. Significantly up- and downregulated genes in the auto-aggregating L. plantarum IA01t0 compared to the non-aggregating IA01t4.
Additional file 5: Table S3. Significantly up- and downregulated genes in the auto-aggregating L. plantarum PA4_02 compared to NZ3400.
Additional file 6: Table S4. Significantly up- and downregulated genes in all auto-aggregating L. plantarum.
Additional file 7: Figure S3. Visualization of DNA fragments (agarose gel, 2%) obtained by PCR amplification using 30 (A) and 40 (B) cycles of denaturation, annealing and replication. NZ3400: L. plantarum wild type strain (non-aggregating); IA10: L. plantarum recovered from the gut microbiota (non-aggregating); PA4_02 and IA01: L. plantarum recovered from the gut microbiota (auto-aggregating).
Additional file 8: Figure S4. Scheme of the PCR-set-up to detect the native and inversed msa-marR promoter region. A) Native orientation: DNA will be amplified using the primer combination Native_fw/Native_rev, resulting in a 225 bp amplicon. B) Inverted DNA will be amplified using the primer combination Inverse_fw/Inverse_rev, resulting in a 313 bp amplicon. P: promoter.
Acknowledgements
We thank Alfonso Die for assistance during HPLC measurements and Luc Jaquenod and Ilsabe Wiebecke for experimental assistance.
Authors’ contributions
J.I., A.G., C.L. and M.J.A.S. designed the study. Experiments were performed by J.I. Data analyses were done by J.I. and M.J.A.S. J.I. and M.J.A.S. drafted the manuscript and all authors critically reviewed the manuscript. The authors read and approved the final manuscript.
Funding
This study was funded by the ETH Zurich research grant program (ETH-42 16–1).
Availability of data and materials
Gene expression data are directly accessible through GEO (series accession number GSE172351, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE172351).
Declarations
Ethics approval and consent to participate
This study was exempted from review by the Ethics Committee of ETH Zurich because sample collection procedure has not been performed under intervention conditions. Informed written consent was obtained from fecal donors.
Consent for publication
Not applicable.
Competing interests
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Puebla-Barragan S, Reid G. Forty-five-year evolution of probiotic therapy. Microb Cell. 2019;6:184–196. doi: 10.15698/mic2019.04.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506-14. [DOI] [PubMed]
- 3.Tuomola E, Crittenden R, Playne M, Isolauri E, Salminen S. Quality assurance criteria for probiotic bacteria. Am J Clin Nutr. 2001;73:393–398. doi: 10.1093/ajcn/73.2.393s. [DOI] [PubMed] [Google Scholar]
- 4.Charteris WP, Kelly PM, Morelli L, Collins JK. Development and application of an in vitro methodology to determine the transit tolerance of potentially probiotic Lactobacillus and Bifidobacterium species in the upper human gastrointestinal tract. J Appl Microbiol. 1998;84:759–768. doi: 10.1046/j.1365-2672.1998.00407.x. [DOI] [PubMed] [Google Scholar]
- 5.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/S0966-842X(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 6.Caldwell DE, Atuku E, Wilkie DC, Wivcharuk KP, Karthikeyan S, Korber DR, et al. Germ theory vs. community theory in understanding and controlling the proliferation of biofilms. Adv Dent Res. 1997;11:4–13. doi: 10.1177/08959374970110011501. [DOI] [PubMed] [Google Scholar]
- 7.Eboigbodin KE, Newton JRA, Routh AF, Biggs CA. Role of nonadsorbing polymers in bacterial aggregation. Langmuir. 2005;21:12315–12319. doi: 10.1021/la051740u. [DOI] [PubMed] [Google Scholar]
- 8.Cesena C, Morelli L, Alander M, Siljander T, Tuomola E, Salminen S, et al. Lactobacillus crispatus and its nonaggregating mutant in human colonization trials. Int J Dairy Sci. 2001;84:1001–1010. doi: 10.3168/jds.S0022-0302(01)74559-6. [DOI] [PubMed] [Google Scholar]
- 9.Reniero R, Cocconcelli P, Bottazzi V, Morelli L. High-frequency of conjugation in Lactobacillus mediated by an aggregation-promoting factor. J Gen Microbiol. 1992;138:763–768. doi: 10.1099/00221287-138-4-763. [DOI] [Google Scholar]
- 10.Janković T, Frece J, Abram M, Gobin I. Aggregation ability of potential probiotic Lactobacillus plantarum strains. Int J Sanit Eng Res. 2012;6:19–24. [Google Scholar]
- 11.Gomaa EZ. Antimicrobial and anti-adhesive properties of biosurfactant produced by lactobacilli isolates, biofilm formation and aggregation ability. J Gen Appl Microbiol. 2013;59:425–436. doi: 10.2323/jgam.59.425. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Cayuela T, Korany AM, Bustos I, de Cadinanos LPG, Requena T, Pelaez C, et al. Adhesion abilities of dairy Lactobacillus plantarum strains showing an aggregation phenotype. Food Res Int. 2014;57:44–50. doi: 10.1016/j.foodres.2014.01.010. [DOI] [Google Scholar]
- 13.Collado MC, Meriluoto J, Salminen S. Adhesion and aggregation properties of probiotic and pathogen strains. Eur Food Res Technol. 2008;226:1065–1073. doi: 10.1007/s00217-007-0632-x. [DOI] [Google Scholar]
- 14.Del Re B, Sgorbati B, Miglioli M, Palenzona D. Adhesion, autoaggregation and hydrophobicity of 13 strains of Bifidobacterium longum. Lett Appl Microbiol. 2000;31:438–442. doi: 10.1046/j.1365-2672.2000.00845.x. [DOI] [PubMed] [Google Scholar]
- 15.Tulumoglu S, Yuksekdag ZN, Beyatli Y, Simsek O, Cinar B, Yasar E. Probiotic properties of lactobacilli species isolated from children's feces. Anaerobe. 2013;24:36–42. doi: 10.1016/j.anaerobe.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Vlkova E, Rada V, Smehilova M, Killer J. Auto-aggregation and co-aggregation ability in bifidobacteria and clostridia. Folia Microbiol. 2008;53:263–269. doi: 10.1007/s12223-008-0040-z. [DOI] [PubMed] [Google Scholar]
- 17.Topcu KC, Kaya M, Kaban G. Probiotic properties of lactic acid bacteria strains isolated from pastirma. LWT. 2020;134:110216. doi: 10.1016/j.lwt.2020.110216. [DOI] [Google Scholar]
- 18.Fonseca HC, Melo DD, Ramos CL, Dias DR, Schwan RF. Probiotic properties of lactobacilli and their ability to inhibit the sdhesion of enteropathogenic bacteria to Caco-2 and HT-29 cells. Probiotics Antimicro. 2021;13:102–112. doi: 10.1007/s12602-020-09659-2. [DOI] [PubMed] [Google Scholar]
- 19.Wang J, Wang J, Yang K, Liu MM, Zhang J, Wei XY, et al. Screening for potential probiotic from spontaneously fermented non-dairy foods based on in vitro probiotic and safety properties. Ann Microbiol. 2018;68:803–813. doi: 10.1007/s13213-018-1386-3. [DOI] [Google Scholar]
- 20.Binti Wan Mohd Noor WSA . An investigation of the phenotypic and genotypic changes in Pseudomonas putida CP1 following substrate-dependent autoaggregation. Dublin: Dublin City University; 2013. [Google Scholar]
- 21.Zuo FL, Yu R, Xiao M, Khaskheli GB, Sun XF, Ma HQ, et al. Transcriptomic analysis of Bifidobacterium longum subsp longum BBMN68 in response to oxidative shock. Sci Rep. 2018;8:1–12. doi: 10.1038/s41598-018-35286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hevia A, Martinez N, Ladero V, Alvarez MA, Margolles A, Sanchez B. An extracellular serine/threonine-rich protein from Lactobacillus plantarum NCIMB 8826 is a novel aggregation-promoting factor with affinity to mucin. Appl Environ Microbiol. 2013;79:6059–6066. doi: 10.1128/AEM.01657-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trunk T, Khalil HS, Leo JC. Bacterial autoaggregation. AIMS Microbiol. 2018;4:140–164. doi: 10.3934/microbiol.2018.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bossier P, Verstraete W. Triggers for microbial aggregation in activated sludge? Appl Microbiol Biot. 1996;45:1–6. doi: 10.1007/s002530050640. [DOI] [Google Scholar]
- 25.Farrell A, Quilty B. Substrate-dependent autoaggregation of pseudomonas putida CP1 during the degradation of mono-chlorophenols and phenol. J Ind Microbiol Biot. 2002;28:316–324. doi: 10.1038/sj.jim.7000249. [DOI] [PubMed] [Google Scholar]
- 26.Perez PF, Minnaard Y, Disalvo EA, De Antoni GL. Surface properties of bifidobacterial strains of human origin. Appl Environ Microb. 1998;64:21–26. doi: 10.1128/AEM.64.1.21-26.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wadstrom T, Andersson K, Sydow M, Axelsson L, Lindgren S, Gullmar B. Surface properties of lactobacilli isolated from the small intestine of pigs. J Appl Bacteriol. 1987;62:513–520. doi: 10.1111/j.1365-2672.1987.tb02683.x. [DOI] [PubMed] [Google Scholar]
- 28.Goh YJ, Klaenhammer TR. Functional roles of aggregation-promoting-like factor in stress tolerance and adherence of Lactobacillus acidophilus NCFM. Appl Environ Microb. 2010;76:5005–5012. doi: 10.1128/AEM.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boris S, Suarez JE, Barbes C. Characterization of the aggregation promoting factor from Lactobacillus gasseri, a vaginal isolate. J Appl Microbiol. 1997;83:413–420. doi: 10.1046/j.1365-2672.1997.00250.x. [DOI] [PubMed] [Google Scholar]
- 30.Malik S, Petrova MI, Claes IJJ, Verhoeven TLA, Busschaert P, Vaneechoutte M, et al. The highly autoaggregative and adhesive phenotype of the vaginal Lactobacillus plantarum strain CMPG5300 is sortase dependent. Appl Environ Microb. 2013;79:4576–4585. doi: 10.1128/AEM.00926-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krausova G, Hyrslova I, Hynstova I. In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation. 2019;5:100. doi: 10.3390/fermentation5040100. [DOI] [Google Scholar]
- 32.Spencer RJ, Chesson A. The effect of Lactobacillus spp. on the attachment of enterotoxigenic Escherichia coli to isolated porcine enterocytes. J Appl Bacteriol. 1994;77:215–220. doi: 10.1111/j.1365-2672.1994.tb03066.x. [DOI] [PubMed] [Google Scholar]
- 33.Nath S, Sikidar J, Roy M, Deb B. In vitro screening of probiotic properties of Lactobacillus plantarum isolated from fermented milk product. Food Qual Saf. 2020;4:213–223. doi: 10.1093/fqsafe/fyaa026. [DOI] [Google Scholar]
- 34.Abushelaibi A, Al-Mahadin S, El-Tarabily K, Shah NP, Ayyash M. Characterization of potential probiotic lactic acid bacteria isolated from camel milk. Lwt-Food Sci Technol. 2017;79:316–325. doi: 10.1016/j.lwt.2017.01.041. [DOI] [Google Scholar]
- 35.Aziz G, Fakhar H, Rahman SU, Tariq M, Zaidi A. An assessment of the aggregation and probiotic characteristics of Lactobacillus species isolated from native (desi) chicken gut. J Appl Poult Res. 2019;28:846–857. doi: 10.3382/japr/pfz042. [DOI] [Google Scholar]
- 36.Sui Y, Liu J, Liu Y, Wang Y, Xiao Y, Gao B, et al. In vitro probiotic characterization of Lactobacillus strains from fermented tangerine vinegar and their cholesterol degradation activity. Food Biosci. 2021;39:100843. doi: 10.1016/j.fbio.2020.100843. [DOI] [Google Scholar]
- 37.Sheth RU, Li MQ, Jiang WQ, Sims PA, Leong KW, Wang HH. Spatial metagenomic characterization of microbial biogeography in the gut. Nat Biotechnol. 2019;37:877. doi: 10.1038/s41587-019-0183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Nieuwboer M, van Hemert S, Claassen E, de Vos WM. Lactobacillus plantarum WCFS1 and its host interaction: a dozen years after the genome. Microb Biotechnol. 2016;9:452–465. doi: 10.1111/1751-7915.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R, et al. Complete genome sequence of Lactobacillus plantarum WCFS1. PNAS. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Isenring J, Geirnaert A, Hall AR, Jans C, Lacroix C, Stevens MJA. In vitro gut modeling as a tool for adaptive evolutionary engineering of Lactiplantibacillus plantarum. mSystems. 2021;6:e01085–e01020. doi: 10.1128/mSystems.01085-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holst B, Glenting J, Holmstrom K, Israelsen H, Vrang A, Antonsson M, et al. Molecular switch controlling expression of the mannose-specific adhesin, Msa, in Lactobacillus plantarum. AEM. 2019;85:e02954–e02918. doi: 10.1128/AEM.02954-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pretzer G, Snel J, Molenaar D, Wiersma A, Bron PA, Lambert J, et al. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J Bacteriol. 2005;187:6128–6136. doi: 10.1128/JB.187.17.6128-6136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Galdiero F, Romano Carratelli C, Nuzzo I, Bentivoglio C, Galdiero M. Phagocytosis of bacterial aggregates by granulocytes. Eur J Epidemiol. 1988;4:456–460. doi: 10.1007/BF00146398. [DOI] [PubMed] [Google Scholar]
- 44.Voltan S, Castagliuolo I, Elli M, Longo S, Brun P, D'Inca R, et al. Aggregating phenotype in Lactobacillus crispatus determines intestinal colonization and TLR2 and TLR4 modulation in murine colonic mucosa. Clin Vaccine Immunol. 2007;14:1138–1148. doi: 10.1128/CVI.00079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorroche FG, Spesia MB, Zorreguieta A, Giordano W. A positive correlation between bacterial autoaggregation and biofilm formation in native Sinorhizobium meliloti isolates from Argentina. Appl Environ Microb. 2012;78:4092–4101. doi: 10.1128/AEM.07826-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tytgat HLP, Nobrega FL, van der Oost J, de Vos WM. Bowel biofilms: tipping points between a healthy and compromised gut? Trends Microbiol. 2019;27:17–25. doi: 10.1016/j.tim.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 47.Adlerberth I, Ahrne S, Johansson ML, Molin G, Hanson LA, Wold AE. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl Environ Microbiol. 1996;62:2244–2251. doi: 10.1128/aem.62.7.2244-2251.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun J, Le GW, Shi YH, Su GW. Factors involved in binding of Lactobacillus plantarum Lp6 to rat small intestinal mucus. Lett Appl Microbiol. 2007;44:79–85. doi: 10.1111/j.1472-765X.2006.02031.x. [DOI] [PubMed] [Google Scholar]
- 49.Aslanzadeh J, Paulissen LJ. Role of type 1 and type 3 fimbriae on the adherence and pathogenesis of Salmonella enteritidis in mice. Microbiol Immunol. 1992;36:351–359. doi: 10.1111/j.1348-0421.1992.tb02034.x. [DOI] [PubMed] [Google Scholar]
- 50.Imberty A, Wimmerova M, Mitchell EP, Gilboa-Garber N. Structures of the lectins from Pseudomonas aeruginosa: insights into the molecular basis for host glycan recognition. Microbes Infect. 2004;6:221–228. doi: 10.1016/j.micinf.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 51.Bhattacharjee JW, Srivastava BS. Mannose-sensitive haemagglutinins in adherence of vibrio cholerae eltor to intestine. J Gen Microbiol. 1978;107:407–410. doi: 10.1099/00221287-107-2-407. [DOI] [PubMed] [Google Scholar]
- 52.Johansson ML, Nobaek S, Berggren A, Nyman M, Bjorck I, Ahrne S, et al. Survival of Lactobacillus plantarum DSM 9843 (299v), and effect on the short-chain fatty acid content of faeces after ingestion of a rose-hip drink with fermented oats. Int J Food Microbiol. 1998;42:29–38. doi: 10.1016/S0168-1605(98)00055-5. [DOI] [PubMed] [Google Scholar]
- 53.Wold AE, Thorssen M, Hull S, Eden CS. Attachment of Escherichia coli via mannose- or gal alpha1->4Gal beta-containing receptors to human colonic epithelial cells. Infect Immun. 1988;56:2531–2537. doi: 10.1128/iai.56.10.2531-2537.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leon-Romero A, Dominguez-Manzano J, Garrido-Fernandez A, Arroyo-Lopez FN, Jimenez-Diaz R. Formation of in vitro mixed-species biofilms by Lactobacillus pentosus and yeasts isolated from spanish-style green table olive fermentations. Appl Environ Microbiol. 2016;82:689–695. doi: 10.1128/AEM.02727-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deatherage DE, Traverse CC, Wolf LN, Barrick JE. Detecting rare structural variation in evolving microbial populations from new sequence junctions using breseq. Front Genet. 2015;5:468. doi: 10.3389/fgene.2014.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Henkin TM, Grundy FJ, Nicholson WL, Chambliss GH. Catabolite repression of alpha amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacl and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 57.Weickert MJ, Chambliss GH. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. P Natl Acad Sci USA. 1990;87:6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stevens MJ. Transcriptome response of Lactobacillus plantarum to global regulator deficiency, stress and other environmental conditions. Netherlands: Wageningen University; 2008. [Google Scholar]
- 59.van der Woude MW. Epigenetic phase variation in bacterial pathogens. In: Doerfler W, Casadesús J, editors. Epigenetics of infectious diseases. Cham: Springer; 2017. pp. 159–173. [Google Scholar]
- 60.Dybvig K. DNA rearrangements and phenotypic switching in prokaryotes. Mol Microbiol. 1993;10:465–471. doi: 10.1111/j.1365-2958.1993.tb00919.x. [DOI] [PubMed] [Google Scholar]
- 61.Vanputte P, Goosen N. DNA inversions in phages and bacteria. Trends Genet. 1992;8:457–462. doi: 10.1016/0168-9525(92)90331-W. [DOI] [PubMed] [Google Scholar]
- 62.Henderson IR, Owen P, Nataro JP. Molecular switches - the ON and OFF of bacterial phase variation. Mol Microbiol. 1999;33:919–932. doi: 10.1046/j.1365-2958.1999.01555.x. [DOI] [PubMed] [Google Scholar]
- 63.Dworkin J, Blaser MJ. Molecular mechanisms of campylobacter fetus surface layer protein expression. Mol Microbiol. 1997;26:433–440. doi: 10.1046/j.1365-2958.1997.6151958.x. [DOI] [PubMed] [Google Scholar]
- 64.Lenich AG, Glasgow AC. Amino-acid-sequence homology between Piv, an essential protein in site-specific DNA inversion in Moraxella lacunata, and transposases of an unusual family of insertion elements. J Bacteriol. 1994;176:4160–4164. doi: 10.1128/jb.176.13.4160-4164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moses EK, Good RT, Sinistaj M, Billington SJ, Langford CJ, Rood JI. A multiple site-specific DNA inversion model for the control of Omp1 phase and antigenic variation in Dichelobacter nodosus. Mol Microbiol. 1995;17:183–196. doi: 10.1111/j.1365-2958.1995.mmi_17010183.x. [DOI] [PubMed] [Google Scholar]
- 66.Bhugra B, Voelker LL, Zou NX, Yu HL, Dybvig K. Mechanism of antigenic variation in mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol Microbiol. 1995;18:703–714. doi: 10.1111/j.1365-2958.1995.mmi_18040703.x. [DOI] [PubMed] [Google Scholar]
- 67.Rojo F, Alonso JC. A novel site-specific recombinase encoded by the streptococcus pyogenes plasmid pSM19035. J Mol Biol. 1994;238:159–172. doi: 10.1006/jmbi.1994.1278. [DOI] [PubMed] [Google Scholar]
- 68.Alonso JC, Weise F, Rojo F. The Bacillus subtilis histone-like protein Hbsu is required for DNA resolution and DNA inversion mediated by the beta recombinase of plasmid pSM19035. J Biol Chem. 1995;270:2938–2945. doi: 10.1074/jbc.270.7.2938. [DOI] [PubMed] [Google Scholar]
- 69.Canosa I, Lurz R, Rojo F, Alonso JC. Beta recombinase catalyzes inversion and resolution between two inversely oriented six sites on a supercoiled DNA substrate and only inversion on relaxed or linear substrates. J Biol Chem. 1998;273:13886–13891. doi: 10.1074/jbc.273.22.13886. [DOI] [PubMed] [Google Scholar]
- 70.Hallet B, Sherratt DJ. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol Rev. 1997;21:157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 71.Weinacht KG, Roche H, Krinos CM, Coyne MJ, Parkhill J, Comstock LE. Tyrosine site-specific recombinases mediate DNA inversions affecting the expression of outer surface proteins of Bacteroides fragilis. Mol Microbiol. 2004;53:1319–1330. doi: 10.1111/j.1365-2958.2004.04219.x. [DOI] [PubMed] [Google Scholar]
- 72.Remus DM, van Kranenburg R, van Swam II, Taverne N, Bongers RS, Wels M, et al. Impact of 4 Lactobacillus plantarum capsular polysaccharide clusters on surface glycan composition and host cell signaling. Microb Cell Factories. 2012;11:1–10. doi: 10.1186/1475-2859-11-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Man J, Rogosa D, Sharpe ME. A medium for the cultivation of lactobacilli. J Appl Microbiol. 1960;23:130–135. [Google Scholar]
- 74.Macfarlane GT, Macfarlane S, Gibson GR. Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol. 1998;35:180–187. doi: 10.1007/s002489900072. [DOI] [PubMed] [Google Scholar]
- 75.O'Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, Kolter R. Genetic approaches to study of biofilms. Biofilms. 1999;310:91–109. doi: 10.1016/S0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 76.Barrick JE, Colburn G, Deatherage DE, Traverse CC, Strand MD, Borges JJ, et al. Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics. 2014;15:1039. doi: 10.1186/1471-2164-15-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Horlbog JA, Stevens MJA, Stephan R, Guldimann C. Global transcriptional response of three highly acid-tolerant field strains of listeria monocytogenes to HCl stress. Microorganisms. 2019;7:455. doi: 10.3390/microorganisms7100455. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Biofilm formation ability of the non-aggregating wild type strain L. plantarum NZ3400 (blue) and the isogenic aggregating strain PA4_02 (red). Biofilm formation was assessed in MRS and EMS medium and minimal medium containing sucrose (mm-sucrose), fructose (mm-fructose), glucose (mm-glucose) or maltose (mm-maltose) as carbon source. Data represent mean value ± standard deviation of biological triplicates.
Additional file 2: Figure S2. Fermentation metabolite profile before (blue) and after (red) microbiota starvation. Bars represent mean ± standard deviation of metabolite abundance relative to the total produced metabolites of six parallel operated TRs the day before and 24 h after starvation of the gut microbiota. Significance was calculated by paired-sample t test: ** < 0.01, **** p < 0.0001.
Additional file 3: Table S1. Significantly up- and downregulated genes in the auto-aggregating L. plantarum IA01 compared to NZ3400.
Additional file 4: Table S2. Significantly up- and downregulated genes in the auto-aggregating L. plantarum IA01t0 compared to the non-aggregating IA01t4.
Additional file 5: Table S3. Significantly up- and downregulated genes in the auto-aggregating L. plantarum PA4_02 compared to NZ3400.
Additional file 6: Table S4. Significantly up- and downregulated genes in all auto-aggregating L. plantarum.
Additional file 7: Figure S3. Visualization of DNA fragments (agarose gel, 2%) obtained by PCR amplification using 30 (A) and 40 (B) cycles of denaturation, annealing and replication. NZ3400: L. plantarum wild type strain (non-aggregating); IA10: L. plantarum recovered from the gut microbiota (non-aggregating); PA4_02 and IA01: L. plantarum recovered from the gut microbiota (auto-aggregating).
Additional file 8: Figure S4. Scheme of the PCR-set-up to detect the native and inversed msa-marR promoter region. A) Native orientation: DNA will be amplified using the primer combination Native_fw/Native_rev, resulting in a 225 bp amplicon. B) Inverted DNA will be amplified using the primer combination Inverse_fw/Inverse_rev, resulting in a 313 bp amplicon. P: promoter.
Data Availability Statement
Gene expression data are directly accessible through GEO (series accession number GSE172351, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE172351).