Abstract
Penicillin resistance among Streptococcus pneumoniae isolates has rapidly emerged in Poland during the last decade and has reached prevalence levels of up to 14.4% in 1997. In order to investigate the nature of this increase, a molecular epidemiological analysis of non-penicillin-susceptible multidrug-resistant pneumococci isolated in 1995 and 1996 was conducted. Thirty-seven patients who suffered mainly from upper respiratory tract infections and pneumococcal pneumonia were enrolled in this study. The medical centers to which the patients were admitted were located in 16 Polish towns across the country. Eight distinct BOX PCR types were observed, representing 14 subtypes. Restriction fragment end labeling (RFEL) analysis divided the pneumococcal strains into 16 distinct types. By combining the BOX PCR and RFEL data, four genetically distinct clusters of strains were identified. Two clusters represented the genetic clones 23F and 9V, which have recently emerged all over the world. The two other genetic clusters, which represented serotypes 23F and 6B, clearly predominated in the analyzed collection of Polish non-penicillin-susceptible pneumococcal strains. Since the latter clusters did not match any of the 133 RFEL types of non-penicillin-susceptible pneumococci collected in 15 other countries, their Polish clonal origin is most likely.
Streptococcus pneumoniae (pneumococcus) continues to be a common cause of serious and life-threatening infections, such as pneumonia, bacteremia, and meningitis, and of noninvasive infections, such as otitis media and sinusitis (3). In addition, pneumococci are often part of the normal nasopharyngeal flora. Especially in young children, pneumococcal colonization often occurs. Colonization is an important risk factor: children colonized with S. pneumoniae more often develop acute otitis media than children who are not colonized (9, 18, 33, 43).
Until the 1960s, pneumococci were considered uniformly susceptible to penicillin, and sensitivity tests were hardly ever performed. In 1967, the first isolation of penicillin-resistant pneumococci was reported in Australia (12). Subsequently, penicillin-resistant pneumococci have been isolated in Papua New Guinea and South Africa (20, 22, 31). In the late 1970s and 1980s, rates of resistance, including multiple resistance, have increased in Western countries, particularly in Spain, with resistance levels of up to 50% (11, 20, 22, 31). A recent epidemiological study performed in the United States has demonstrated that 25% of invasive pneumococci are non-penicillin susceptible (17). The emergence of high-level resistance to penicillin, particularly in combination with other resistance determinants, is a serious threat for current treatment strategies.
Several investigators have reported the international spread of multiply resistant pneumococcal clones. Soares and coworkers have reported the spread of a drug-resistant clone of serotype 6B from Spain to Iceland in the late 1980s (37). This has resulted in an epidemic of this clone, which was isolated with frequencies of up to 12% in 1992 (21). In 1991, Munoz and colleagues reported evidence for the intercontinental spread of a multiply resistant clone of S. pneumoniae serotype 23F from Spain to the United States (24). This clone has subsequently disseminated through the United States (23). Finally, in 1995 Gasc and colleagues reported the spread of a multiply resistant pneumococcal clone of serogroup 9 from Spain to France (10). The international clones 23F and 9V have been identified in many countries in different parts of the world (14, 28). Besides the international spread of the clones 6B, 23F, and 9V, novel penicillin-resistant and multiply resistant clones that tend to spread in an epidemic manner have been observed in the former Czechoslovakia, Spain, Japan, and South Africa (4, 30, 34, 42).
Penicillin resistance among pneumococcal isolates has rapidly emerged in Poland during the last decade. The percentage of non-penicillin-susceptible strains has increased from 0 to 3% in 1990 to 1993 (1, 19) to 14.3% in 1996 (39) and 14.4% in 1997 (38). In order to identify the nature of the increase in the prevalence of non-penicillin-susceptible pneumococci in Poland, a molecular epidemiological study was conducted. To this end, strains were isolated in 1995 and 1996 from 37 patients who suffered mainly from upper respiratory tract infections and pneumococcal pneumonia and who were diagnosed in 19 different clinical centers located in 16 towns across the country. The pneumococcal isolates were characterized by BOX PCR typing, restriction fragment end labeling (RFEL) analysis, penicillin-binding protein (PBP) genotyping, serotyping, and drug susceptibility testing.
MATERIALS AND METHODS
Bacterial isolates.
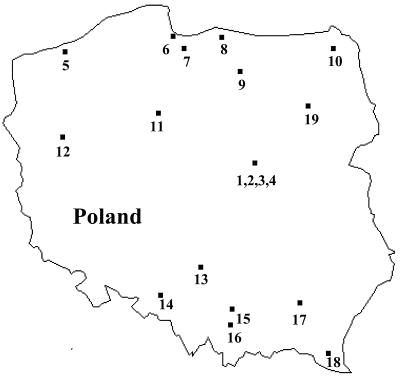
Non-penicillin-susceptible S. pneumoniae strains were isolated from 37 Polish patients in 1995 and 1996. A single pneumococcal isolate per patient per infection was analyzed. Bacteriological diagnosis was carried out in 19 distinct medical centers. These centers were located in 16 Polish towns across the country (Fig. 1). The clinical, bacteriological, and demographic parameters are listed in Table 1.
FIG. 1.
Map of Poland. The medical centers collaborating in this study are as follows: 1, Mother and Child Institute, Warsaw; 2, Health Care Center Praga-Polnoc, Warsaw; 3, Nieklanska Street Hospital, Warsaw; 4, Sienna Street Hospital, Warsaw; 5, Health Care Center, Kolobrzeg; 6, Regional Hospital, Gdansk; 7, Health Care Center, Tczew; 8, Health Care Center, Braniewo; 9, Regional Children’s Hospital Olsztyn; 10, Regional Hospital, Suwalki; 11, Medical University, Bydgoszcz; 12, Health Care Center, Gorzow Wielkopolski; 13, Health Care Center, Czestochowa; 14, Health Care Center, Kedzierzyn Kozle; 15, Jan Bober Center for Microbiology and Autovaccines, Krakow; 16, Health Care Center, Myslenice; 17, Health Care Center, Mielec; 18, Health Care Center, Sanok; and 19, Regional Hospital, Lomza.
TABLE 1.
Microbiological parameters of 37 penicillin-resistant pneumococcal isolates and clinical and demographic data for the patients
| Patient
|
Pneumococcal strain
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (yr) | Sex | Clinical diagnosisa | Centerb | Strain no. | Clinical originc | Penicillin MIC (μg/ml) | Resistance profiled | Serotype | BOX PCR type | RFEL typee (cluster)f | PBP genotype |
| <1 | Female | URTI | 7 | 39 | Nose | 4.0 | PAXCDELT | 23F | 1 | 106 (I) | 6-2-53 |
| <1 | Male | URTI | 7 | 40 | Nose | 4.0 | PAXCDELT | 23F | 1 | 106 (I) | 6-1-53 |
| Child | —g | URTI | 6 | 32 | Nose | 4.0 | PAXCDELT | 23F | 1 | 106 (I) | 6-1-53 |
| 1 | Male | URTI | 7 | 26 | Nose | 4.0 | PAXCDELT | 23F | 1 | 105 (I) | 6-18-53 |
| Child | Male | URTI | 17 | 29 | Nose | 4.0 | PAXCDELT | 23F | 1 | 105 (I) | 6-18-56 |
| 1 | Male | LRTI | 1 | 6 | Throat | 8.0 | PAXCDET | 23F | 1 | 105 (I) | 16-3-1 |
| <1 | Male | LRTI | 7 | 9 | Blood | 4.0 | PAXCDELT | 23F | 1 | 105 (I) | 6-18-53 |
| 1 | Male | URTI | 14 | 22 | Nose | 2.0 | PAXCDELT | 23F | 1 | 145 (I) | 6-14-43 |
| 6 | Male | Otitis media | 4 | 151 | Ear | 4.0 | PA*CDE** | 23F | 1 | 44 (I) | 6-14-43 |
| 4 | Female | URTI | 3 | 178 | Nose | 8.0 | PA*CDE** | 23F | 1 | 44 (I) | 6-14-1 |
| 4 | Male | URTI | 4 | 233 | Throat | 8.0 | PA*CDE** | 23F | 1 | 45 (I) | 6-14-1 |
| 5 | Female | URTI | 4 | 234 | Throat | 8.0 | PA*CDE** | 23F | 1 | 45 (I) | 6-14-1 |
| Adult | Female | LRTI | 2 | 4 | Sputum | 2.0 | PAXCDET | 23F | 2 | 15 (II) | 1-1-1 |
| 13 | Female | LRTI | 18 | 16 | BAL | 2.0 | PAXDET | 23F | 2.1 | 15 (II) | 1-1-1 |
| 3 | Male | URTI | 16 | 33 | Throat | 2.0 | PAXCDET | 23F | 2 | 15 (II) | 1-18-1 |
| 57 | Male | Brain abscess | 12 | 8 | CSF | 0.5 | PCDELT | 19A | 3 | 104 | 7-17-52 |
| 55 | Male | LRTI | 10 | 2 | BAL | 1.0 | PCT | 6A | 4 | 97 | 2-5-51 |
| — | — | — | 8 | 31 | — | 1.0 | PD | Rough | 5 | 148 | 0h-2-54 |
| 3 | Female | URTI | 7 | 36 | Nose | 1.0 | PCDELT | 6B | 6 | 147 | 0-1-54 |
| 14 | Male | LRTI | 9 | 12 | Sputum | 0.12 | PCDELT | 6B | 7 | 98 (III) | 2-2-9 |
| 7 | Male | LRTI | 9 | 13 | Sputum | 0.12 | PCDELT | 6B | 7 | 98 (III) | 2-2-9 |
| 3 | Female | LRTI | 9 | 14 | Blood | 0.12 | PCDELT | 6B | 7.1 | 98 (III) | 2-2-9 |
| 1 | Male | URTI | 13 | 35 | Nose | 0.12 | PCDELT | 6B | 7.2 | 98 (III) | 2-18-9 |
| 6 | Male | URTI | 16 | 34 | Nose | 0.12 | PCDELT | 6B | 7.1 | 98 (III) | 2-18-9 |
| Adult | — | LRTI | 11 | 17 | Sputum | 0.12 | PCDELT | 6B | 7.3 | 102 (III) | 2-2-9 |
| 2 | Male | URTI | 14 | 23 | Nose | 0.12 | PDE | 6B | 7.3 | 102 (III) | 2-2-9 |
| Child | Male | Conjunctivitis | 2 | 30 | Eye | 0.12 | PDEL | 6B | 7.1 | 102 (III) | 2-2-9 |
| 12 | Male | LRTI | 9 | 15 | Blood | 0.12 | PDEL | 6B | 7.2 | 101 (III) | 2-2-9 |
| <1 | — | LRTI | 2 | 5 | Throat | 0.12 | PDEL | 6B | 7.3 | 99 (III) | 2-2-9 |
| 5 | Male | URTI | 17 | 27 | Nose | 0.12 | PCDET | 6B | 7.4 | 99 (III) | 2-2-9 |
| Child | Female | URTI | 7 | 24 | Nose | 1.0 | PACDT | 9V | 8 | 23 (IV) | 1-1-1 |
| Adult | Male | URTI | 17 | 38 | Nose | 1.0 | PAXCDET | 9V | 8.1 | 23 (IV) | 1-2-1 |
| Adult | Female | LRTI | 6 | 1 | Pleural fluid | 1.0 | PAXCDET | 9V | 8.1 | 23 (IV) | 1-1-1 |
| Child | Male | URTI | 5 | 10 | Nose | 1.0 | PAXCDET | 9V | 8 | 23 (IV) | 1-5-1 |
| Adult | — | LRTI | 15 | 11 | Sputum | 2.0 | PAXCDELT | 14 | 8 | 23 (IV) | 1-5-1 |
| Child | Female | URTI | 3 | 37 | Throat | 0.5 | PXCDT | 9V | 8.1 | 23 (IV) | 1-2-1 |
| 10 | Female | URTI | 19 | 181 | Throat | 1.0 | PA*C** | 9V | 8.1 | 140 (IV) | 1-1-1 |
URTI, upper respiratory tract infection; LRTI, lower respiratory tract infection.
See Fig. 1.
BAL, broncheoalveolar lavage; CSF, cerebrospinal fluid.
P, penicillin G; A, amoxicillin; X, cefotaxime, C, chloramphenicol; D, doxycycline; E, erythromycin; L, lincomycin; T, trimethoprim-sulfamethoxazole (co-trimoxazole); ∗, susceptibility to cefotaxime, lincomycin, and co-trimoxazole was not tested.
The arbitrary cutoff value for an identical RFEL type is 95%; RFEL codes refer exclusively to the genetic distances calculated by using the entire collection of 147 pneumococcal RFEL types, in which the Polish genotypes were included (see Materials and Methods) (13, 14).
See Fig. 2.
—, data not available.
Unknown PBP genotype.
Biochemical characterization, serotyping, and susceptibility testing.
The isolates were determined to be S. pneumoniae by investigating their optochin susceptibility and bile solubility (25). All pneumococci were serotyped in the Sera and Vaccines Central Research Laboratory in Warsaw on the basis of capsular swelling (Quellung reaction) observed microscopically after suspension in antisera prepared at Statens Seruminstitut, Copenhagen, Denmark (8).
The susceptibilities of pneumococcal strains were determined in the Sera and Vaccines Central Research Laboratory in Warsaw according to the guidelines of the National Committee for Clinical Laboratory Standards (26) by the broth microdilution method in cation-adjusted Mueller-Hinton II broth (Beckton Dickinson, Oxford, United Kingdom) supplemented with 5% lysed horse blood. The following panel of antimicrobial agents was tested: penicillin (Polfa, Poznan, Poland), amoxicillin (SmithKline Beecham, Brentford, United Kingdom), cefotaxime (Roussel, Paris, France), doxycycline (Polfa), chloramphenicol (Polfa), erythromycin (Polfa), lincomycin (Upjohn, Kalamazoo, Mich.), vancomycin (Eli Lilly, Indianapolis, Ind.), rifampin (Polfa), ofloxacin (Hoechst-Roussel, Kansas City, Kans.), sparfloxacin (Rhone-Poulenc, Courbevoie cedex, France), and co-trimoxazole (trimethoprim with sulfamethoxazole) (Polfa). For doxycycline, lincomycin, and sparfloxacin susceptibility testing, French interpretative criteria were applied (6). The microdilution panels were prepared by a single senior technician. The quality control strains S. pneumoniae ATCC 49619 and Staphylococcus aureus ATCC 29213 were included in each run.
RFEL analysis and BOX PCR typing.
Typing of pneumococcal strains by RFEL analysis was performed as described by Van Steenbergen et al. (41) and adapted by Hermans et al. (16). Briefly, purified pneumococcal DNA was digested with the restriction enzyme EcoRI. The DNA restriction fragments were end labeled at 72°C with [α-32P]dATP by using DNA polymerase (Goldstar; Eurogentec, Seraing, Belgium). The radiolabeled fragments were denatured and separated electrophoretically on a 6% polyacrylamide sequencing gel containing 8 M urea. Subsequently, the gel was transferred onto filter paper, vacuum dried (HBI, Saddlebrook, N.Y.), and exposed for variable lengths of time at room temperature to ECL Hyperfilms (Amersham, Bucks, United Kingdom).
BOX PCR typing was carried out in a single laboratory as described before (40). Briefly, 50 ng of pneumococcal DNA was amplified by PCR (4 min at 94°C [predenaturation]; 40 cycles of 1 min at 94°C, 1 min at 60°C, and 2 min at 74°C; and 2 min at 74°C [extension]), using primer BOX-A (5′ATACTCTTCGAAAATCTCTTCAAAC), which was designed from the primary structure of the BOX repeat motif. The amplified products were separated on a 1.5% agarose gel. Gels were stained with ethidium bromide, after which the banding patterns were evaluated visually. BOX PCR patterns showing a single band difference were defined as nonidentical types (e.g., 1, 2, and 3). Identical banding patterns varying in the intensity of one or more bands were defined as subtypes (e.g., 7, 7.1, and 7.2).
PBP genotyping.
Genetic polymorphism of the penicillin resistance genes pbp1a, pbp2b, and pbp2x was investigated by restriction fragment length polymorphism (RFLP) analysis. To this end, we amplified the genes by PCR and analyzed the digested DNA products by agarose gel electrophoresis. PCR amplification of the PBP-encoding genes was performed in a 50-μl PCR buffer system containing 75 mM Tris-HCl (pH 9.0), 20 mM (NH4)2SO4, 0.01% (wt/vol) Tween 20, 1.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, 10 pmol of the individual primers, 0.5 U of DNA polymerase (Eurogentec), and 10 ng of purified chromosomal DNA. Cycling was performed in a PTC-100 Programmable Thermal Controller (MJ Research, Watertown, Mass.) and consisted of the following steps: predenaturation at 94°C for 1 min; 30 cycles of 1 min at 94°C, 1 min at 52°C, and 2 min at 72°C; and a final extension at 72°C for 3 min. The primers used to amplify the genes pbp1a, pbp2b, and pbp2x were described previously (5, 7, 24).
The amplification products (5 μl) were digested with the restriction endonuclease HinfI and separated by electrophoresis in 2.5% agarose gels containing 0.5× Tris-borate-EDTA and 0.1 μg of ethidium bromide per ml (5 mm thick) (Agarose MP; Boehringer Mannheim, Almere, The Netherlands). Gels were run in 0.5× Tris-borate-EDTA containing 0.1 μg of ethidium bromide per ml at 20 mA for 4 h. Prior to electrophoresis, samples were mixed with a 5×-concentrated layer mix consisting of 50% glycerol in water and 0.8 mg of bromophenol blue per ml. Gels were photographed with a Polaroid MP4 Landcamera and Polaroid 667 films. The different PBP genotypes are represented by a three-number code (e.g., 6-14-43, referring to the RFLP patterns of the genes pbp1a [pattern 6], pbp2b [pattern 14], and pbp2x [pattern 43], respectively.
Computer-assisted analysis of the DNA banding patterns.
The RFEL types were analyzed by using the Windows version of the Gelcompar software version 4 (Applied Maths, Kortrijk, Belgium) after imaging the RFEL autoradiograms with the Image Master DTS (Pharmacia Biotech, Uppsala, Sweden). To this end, the DNA fragments in the molecular size range of 160 to 400 bp were explored. The DNA banding patterns were normalized by using pneumococcus-specific bands present in the RFEL banding patterns of all strains. Comparison of the banding patterns was performed by the unweighted pair group method with arithmetic averages (29) and with the Jaccard similarity coefficient applied to peaks (36). Computer-assisted analysis and the methods and algorithms used in this study were according to the instructions of the manufacturer of Gelcompar. A tolerance of 1.5% in band positions was applied during comparison of the DNA patterns. Identical DNA types were arbitrarily defined as those with RFEL homologies higher than 95%. Cluster codes I to IV refer to clusters of pneumococcal strains displaying RFEL types with homologies higher than 80% that were confirmed to be genetically related by using BOX PCR typing (identical BOX PCR types or subtypes).
RESULTS
The molecular epidemiology of non-penicillin-susceptible multidrug-resistant pneumococci in Poland was investigated. Thirty-seven patients who suffered mainly from upper and lower respiratory tract infections (n = 20 and 13, respectively) were enrolled in this study. In addition, three patients suffered from otitis media, a brain abscess, and conjunctivitis, respectively. The clinical diagnosis of one patient was unknown. The medical centers to which the patients were referred were located in 16 Polish towns across the country (Fig. 1). The clinical and demographic parameters are listed in Table 1.
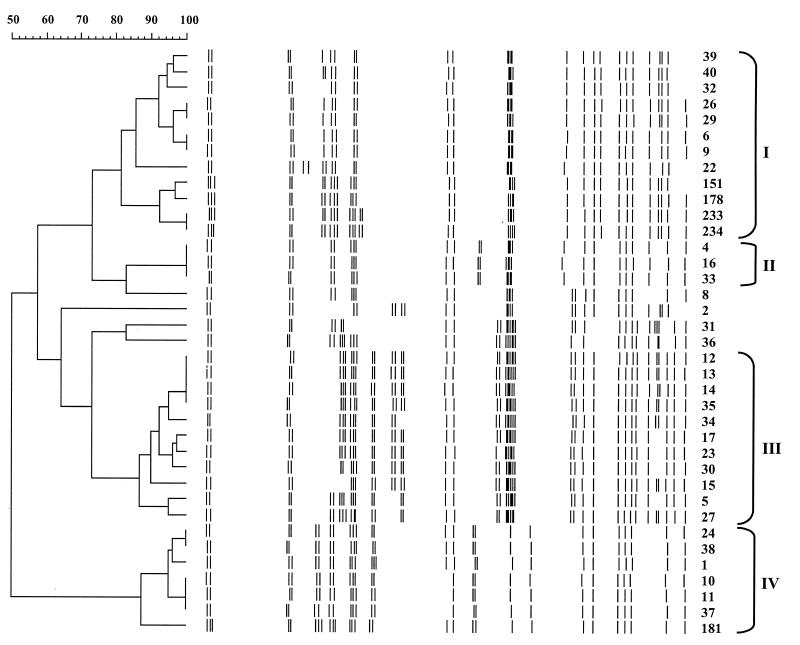
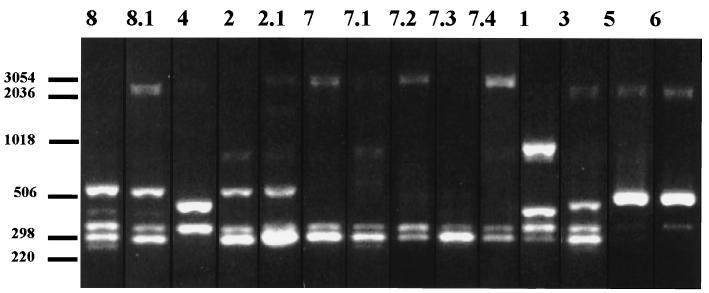
The 37 non-penicillin-susceptible pneumococcal strains were characterized by RFEL analysis and BOX PCR typing. RFEL analysis divided the 37 pneumococcal strains into 16 distinct types (Fig. 2; Table 1). Eight distinct BOX PCR types were observed, representing 14 subtypes (Fig. 3; Table 1). When RFEL and BOX PCR analyses were combined, four genetically distinct clusters of strains (designated I, II, III, and IV) were identified. Within these clusters, genetic relatedness of the strains was demonstrated by both RFEL analysis and BOX PCR (sub)typing. Clusters I, II, III, and IV consisted of 12, 3, 11, and 7 strains, respectively. Each cluster was represented by strains that originated from different centers (Table 1). The remaining four strains did not fulfill the cluster criteria.
FIG. 2.
Genetic relatedness of 37 Polish non-penicillin-susceptible pneumococcal strains based on the RFEL banding patterns of the isolates. The RFEL fingerprints, their relatedness (dendrogram), strain numbers, and genetic cluster codes (Roman numerals) are depicted. Cluster codes I to IV refer to clusters of pneumococcal strains displaying RFEL types with homologies higher than 80% that are confirmed to be genetically related by BOX PCR typing (identical BOX PCR types or subtypes).
FIG. 3.
DNA fingerprint patterns of the 14 distinct BOX PCR (sub)types represented by the 37 Polish non-penicillin-susceptible pneumococcal strains. Lane numbers indicate BOX PCR type codes. Numbers at the left indicate the sizes of standard DNA fragments in base pairs.
Comparison of the Polish RFEL types with 133 genotypes present in the international RFEL data library representing 15 other countries (13, 14) revealed that the genetic clusters II and IV matched the Spanish pandemic clone 23F (RFEL type 15) and the Spanish-French international clone 9V (RFEL type 23), respectively (data not shown). The remaining 14 Polish RFEL types did not match any of the 133 non-Polish types present in the international library.
The 37 non-penicillin-susceptible pneumococci from Poland represented seven distinct serotypes (Table 1). The most predominant serotypes were 23F (n = 15), 6B (n = 12), and 9V (n = 6). Serotypes 6A, 14, and 19A were all observed once in the Polish collection. Strain 31 displayed a rough phenotype and could not be serotyped. Serotypes 23F, 6B, and 9V included 6, 5, and 2 RFEL types, respectively, whereas the remaining three serotypes (6A, 14, and 19A) were restricted to single RFEL types. The clusters I, II (pandemic clone 23F), and III displayed the serotypes 23F, 23F, and 6B, respectively. Genetic cluster IV (pandemic clone 9V) harbored both serotypes 9V (n = 6) and 14 (n = 1), indicating horizontal transfer of capsular genes within this cluster.
The penicillin MICs of the pneumococcal strains varied from 0.12 to 8.0 μg/ml. Within the genetic clusters, various resistance profiles were observed, ranging from lack of susceptibility to penicillin and doxycycline only to multidrug resistance against penicillins, cefotaxime, co-trimoxazole, doxycycline, chloramphenicol, erythromycin, and lincomycin. Genetic cluster I had the highest penicillin MICs (up to 8 μg/ml), as well as the greatest multidrug resistance (Table 1). The lowest penicillin MICs were observed for isolates of genetic cluster III. The resistance patterns of these strains demonstrated their relative susceptibilities to the antimicrobial agents tested. Nevertheless, all members of cluster III were multidrug resistant, i.e., resistant to three or more antibiotics. None of the isolates were resistant to vancomycin, rifampin, and fluoroquinolones.
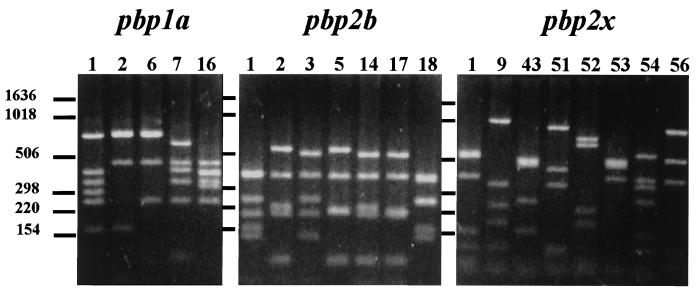
The penicillin resistance profiles of the pneumococcal strains were investigated in further detail by PBP genotyping. To this end, the penicillin resistance genes pbp1a, pbp2b, and pbp2x were subjected to RFLP analysis (Fig. 4). The genetic clusters II (pandemic clone 23F) and IV (pandemic clone 9V) included two and three distinct PBP genotypes, respectively (Table 1). The polymorphism was restricted to pbp2b. Similar results were observed for cluster III: there were two distinct PBP genotypes that differed only in the pbp2b gene. For cluster I, seven distinct PBP genotypes were observed; in contrast to the case for the other three genetic clusters, genetic polymorphism was demonstrated in pbp1a (two types), pbp2b (five types), and pbp2x (four types). Except for clusters II and IV, representing pandemic clones 23F and 9V, respectively, no overlap was observed between the PBP genotypes of the four clusters. Similarly, the four genotypes that were observed only once in the Polish collection also displayed unique PBP genotypes. The polymorphism of the PBP-encoding genes within clusters II and IV was remarkable, as the majority of the pandemic 23F and 9V isolates that are now present in the international data library (51 23F strains from 10 countries [96%] and 31 9V strains from 5 countries [97%]) display PBP genotype 1-1-1 (13, 14).
FIG. 4.
DNA fingerprint patterns of the pbp1a (n = 5), pbp2b (n = 7), and pbp2x (n = 8) genotypes represented by the 37 Polish non-penicillin-susceptible pneumococcal strains. Lane numbers indicate PBP genotype codes. Numbers at the left indicate the sizes of standard DNA fragments in base pairs.
Comparison of the PBP genotypes of the individual genes pbp1a, pbp2b, and pbp2x between the Polish non-penicillin-susceptible strains and 185 Dutch penicillin-susceptible pneumococcal meningitis isolates (15, 32) revealed the exclusive presence of penicillin-susceptible pbp1a and pbp2b genotypes among the Polish clusters I, III, and IV. Two strains of cluster IV (pandemic clone 9V; strains 37 and 38 with penicillin MICs of 0.5 and 1.0 μg/ml, respectively) displayed the predominant penicillin-susceptible pbp2b genotype 2 (Fig. 4; Table 1). In addition, the majority of the strains belonging to cluster III displayed the predominant penicillin-susceptible pbp1a-pbp2b genotype 2-2 (penicillin MIC of 0.12 μg/ml). Finally, the majority of cluster I displayed the penicillin-susceptible pbp1a genotype 6 (penicillin MICs ranging from 2.0 to 8.0 μg/ml); this genotype was observed only once in the collection of 185 Dutch penicillin-susceptible pneumococci.
Among the pbp1a, pbp2b, and pbp2x genotypes that are restricted to non-penicillin-susceptible pneumococci (13, 14), the presence of pbp1a genotype 1 was observed among clusters II (clone 23F) and IV (clone 9V) (Table 1). In addition, the pbp2b genotypes 1 and 18 and the pbp2x genotype 1 were observed in three genetic clusters. These data indicate horizontal exchange of penicillin resistance genes between pneumococcal strains of the different Polish clusters.
DISCUSSION
Penicillin resistance among pneumococcal isolates has rapidly emerged in Poland during the last decade. The percentage of non-penicillin-susceptible strains has increased from 0 to 3% in 1990 to 1993 (1, 19) to 14.3% in 1996 (39) and 14.4% in 1997 (38). In order to identify the nature of the increase in the prevalence of non-penicillin-susceptible pneumococci in Poland, a molecular epidemiological study was conducted. The emergence of four genetically distinct non-penicillin-susceptible multidrug-resistant pneumococcal clusters was clearly demonstrated. Clusters II and IV represented the genetic clones 23F (24) and 9V (11), respectively, which have recently emerged all over the world (13, 14). In addition, the two other clusters, I and III, predominated in the collection of non-penicillin-susceptible pneumococci from Poland. Since these clusters did not match any of the 133 RFEL types of non-penicillin-susceptible pneumococci collected in 15 other countries and present in the international data library of RFEL types (13, 14), their Polish origin is most likely.
Various researchers have reported horizontal transfer of capsular genes (2, 13–15). The high frequency of capsular transfer may have consequences for the outcome of current vaccine strategies, which focus on the use of a restricted number of distinct capsular polysaccharides. The use of multivalent conjugate vaccines may shift the capsular distribution towards capsular types that are not present in these vaccines. Such a shift might be enhanced by the frequent horizontal exchange of capsular genes. Genetic cluster IV (pandemic clone 9V) harbored both serotypes 9V (n = 6) and 14 (n = 1). These serotypes are known to occur in this international clone (13, 14). The Polish strains that were genetically related to the pandemic clone 23F (cluster II), which harbors serotypes 23F, 19F, 14, 9N, 19A, and 3 (13, 14, 27), displayed only the ancestral serotype 23F.
Within the four genetic clusters, RFLP of the PBP-encoding genes was often observed, in particular in pbp2b. This finding indicates frequent horizontal transfer of the penicillin resistance genes. PBP genotype 1-1-1 was observed in clusters II (pandemic clone 23F) and IV (pandemic clone 9V). In a recent study by Hermans and coworkers (13), the vast majority of isolates of the pandemic clones 23F and 9V also displayed the PBP genotype 1-1-1. In that study, PBP genotype 1-1-1 was displayed by 41% of the strains collected in 15 other countries and represented 20 of the 133 distinct RFEL types. In addition, genetic polymorphism in the penicillin resistance genes of the pandemic clones 9V and 23F was rarely observed. However, in 5 of the 10 Polish strains that were related to the pandemic clones 9V and 23F, differences in the PBP genotypes were observed, suggesting that in Poland horizontal transfer of penicillin resistance genes, in particular pbp2b, occurs more frequently in these clones than in those present in the other countries. The pbp2b genotypes 18 (observed only in Poland so far) and 1 and the pbp2x genotype 1 were observed in three of the four genetic clusters. These data suggest that these PBP genotypes have spread in the Polish population of non-penicillin-susceptible pneumococci by horizontal gene transfer.
The strains belonging to cluster III invariably displayed low penicillin MICs of 0.12 μg/ml. In addition, the majority of these strains displayed the penicillin-susceptible pbp1a-pbp2b genotype 2-2. Although the predictive value of PBP genotyping for pneumococcal penicillin resistance can be debated for methodological reasons, this observation is in concordance with data from Smith and coworkers (35), who have demonstrated that in pneumococci with low penicillin MICs, pbp2b is usually not affected by mutations.
In summary, penicillin resistance has rapidly emerged in Poland over the last decade. Although the pandemic clones 9V and 23F are present in this country, the nationwide emergence of two novel multidrug-resistant pneumococcal clones that are presumably of Polish origin has clearly contributed to the increase in the prevalence of non-penicillin-susceptible pneumococci in Poland.
ACKNOWLEDGMENTS
We thank Melanie Srodzinski and Daniëlle van Nispen for technical assistance.
This study was sponsored by grants from the Sophia Foundation for Medical Research, Rotterdam, The Netherlands (to K.O.) and from the Polish Committee of Scientific Studies, Warsaw, Poland (Microbank Project) (to K.T. and W.H.).
P.W.M.H. and K.T. contributed equally to this study.
REFERENCES
- 1.Appelbaum P C, Gladkova C, Hryniewicz W, Kojourahov B, Kotulova D, Mihalcu F, Shindler J, Setchanova L, Semina N, Trupl J, Tyski S, Urbaskova P, Jacobs M R. Carriage of antibiotic-resistant Streptococcus pneumoniae by children in Eastern and Central Europe—a multicenter study using standardized methods. Clin Infect Dis. 1996;23:712–717. doi: 10.1093/clinids/23.4.712. [DOI] [PubMed] [Google Scholar]
- 2.Barnes D M, Whittier S, Gilligan P H, Soares S, Tomasz A, Henderson F W. Transmission of multidrug-resistant serotype 23F Streptococcus pneumoniae in group day care: evidence suggesting capsular transformation of the resistant strain in vivo. J Infect Dis. 1995;171:890–896. doi: 10.1093/infdis/171.4.890. [DOI] [PubMed] [Google Scholar]
- 3.Caputo G M, Appelbaum P C, Liu H H. Infections due to penicillin-resistant pneumococci. Arch Intern Med. 1993;153:1301–1310. [PubMed] [Google Scholar]
- 4.Coffey T J, Berron S, Daniels M, Garcia-Leoni E, Cercenado E, Bouza E, Fenoll A, Spratt B G. Multiply antibiotic-resistant Streptococcus pneumoniae recovered from Spanish hospitals (1988–1994): novel major clones of serotypes 14, 19F and 15F. Microbiology. 1996;142:2747–2757. doi: 10.1099/13500872-142-10-2747. [DOI] [PubMed] [Google Scholar]
- 5.Coffey T J, Dowson C G, Daniels M, Zhou J, Martin C, Spratt B G, Musser J M. Horizontal transfer of multiple penicillin-binding protein genes, and capsular biosynthetic genes, in natural populations of Streptococcus pneumoniae. Mol Microbiol. 1991;5:2255–2260. doi: 10.1111/j.1365-2958.1991.tb02155.x. [DOI] [PubMed] [Google Scholar]
- 6.Courvalin P, Soussy C-J. Report of the Comité de l’Antibiogramme de la Societé Francaise de Microbiologie. Clin Microbiol Infect. 1996;2(Suppl.):S46–S49. doi: 10.1007/BF01974543. [DOI] [PubMed] [Google Scholar]
- 7.Dowson C G, Hutchison A, Spratt B G. Extensive re-modelling of the transpeptidase domain of penicillin-binding protein 2B of a penicillin-resistant South African isolate of Streptococcus pneumoniae. Mol Microbiol. 1989;3:95–102. doi: 10.1111/j.1365-2958.1989.tb00108.x. [DOI] [PubMed] [Google Scholar]
- 8.Facklam R R, Washington J A. Streptococcus and related catalase-negative gram-positive cocci. In: Balows A, Hausler W J, Hermann R L, Isenberg H D, Shadomy H J, editors. Manual of clinical microbiology. 5th ed. Washington, D.C: American Society for Microbiology; 1991. pp. 238–257. [Google Scholar]
- 9.Faden H, Duffy L, Wasielewski R, Wolf J, Krystofik D, Tung Y. Relationship between nasopharyngeal colonization and the development of otitis media in children. J Infect Dis. 1997;175:1440–1445. doi: 10.1086/516477. [DOI] [PubMed] [Google Scholar]
- 10.Gasc A M, Geslin P, Sicard A M. Relatedness of penicillin-resistant Streptococcus pneumoniae serogroup 9 strains from France and Spain. Microbiology. 1995;141:623–627. doi: 10.1099/13500872-141-3-623. [DOI] [PubMed] [Google Scholar]
- 11.Gómez J, Ruiz-Gómez J, Hernández-Cardona N, Canteras M, Valdés M. Antibiotic resistance patterns of Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis: a prospective study in Murcia, Spain, 1983–1992. Chemotherapy. 1994;40:299–303. doi: 10.1159/000239210. [DOI] [PubMed] [Google Scholar]
- 12.Hansman D, Bullen M M. A resistant pneumococcus. Lancet. 1967;ii:264–265. doi: 10.1016/s0140-6736(75)91547-0. [DOI] [PubMed] [Google Scholar]
- 13.Hermans, P. W. M., K. Overweg, M. Sluijter, and R. de Groot. Penicillin-resistant Streptococcus pneumoniae: an international molecular epidemiological study. In A. Tomasz (ed.), Streptococcus pneumoniae: molecular biology and mechanisms of disease—update for the 1990s, in press. Mary Ann Liebert Inc., New York, N.Y.
- 14.Hermans P W M, Sluijter M, Dejsirilert S, Lemmens N, Elzenaar K, van Veen A, Goessens W H F, de Groot R. Molecular epidemiology of drug-resistant pneumococci: toward an international approach. Microb Drug Resist. 1997;3:243–251. doi: 10.1089/mdr.1997.3.243. [DOI] [PubMed] [Google Scholar]
- 15.Hermans P W M, Sluijter M, Elzenaar K, van Veen A, Schonkeren J J M, Nooren F M, van Leeuwen W J, de Neeling A J, van Klingeren B, Verbrugh H A, de Groot R. Penicillin-resistant Streptococcus pneumoniae in The Netherlands: results of a 1-year molecular epidemiologic survey. J Infect Dis. 1997;175:1413–1422. doi: 10.1086/516474. [DOI] [PubMed] [Google Scholar]
- 16.Hermans P W M, Sluijter M, Hoogenboezem T, Heersma H, van Belkum A, de Groot R. Comparative study of five different DNA fingerprint techniques for molecular typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1995;33:1606–1612. doi: 10.1128/jcm.33.6.1606-1612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann J, Cetron M S, Farley M M, Baughman W S, Facklam R R, Elliott J A, Deaver K A, Breiman R F. The prevalence of drug resistant Streptococcus pneumoniae in Atlanta. N Engl J Med. 1995;333:481–486. doi: 10.1056/NEJM199508243330803. [DOI] [PubMed] [Google Scholar]
- 18.Homoe P, Prag J, Farholt S, Henrichsen J, Hornsleth A, Kilian M, Jensen J S. High rate of nasopharyngeal carriage of potential pathogens among children in Greenland: results of a clinical survey of middle-ear disease. Clin Infect Dis. 1996;23:1081–1090. doi: 10.1093/clinids/23.5.1081. [DOI] [PubMed] [Google Scholar]
- 19.Hryniewicz W. Bacterial resistance in Eastern Europe—selected problems. Scand J Infect Dis. 1994;93(Suppl.):33–39. [PubMed] [Google Scholar]
- 20.Klugman K P. Pneumococcal resistance to antibiotics. Clin Microbiol Rev. 1990;3:171–196. doi: 10.1128/cmr.3.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kristinsson K G, Hjalmarsdottir M A, Steingrimsson O. Increasing penicillin resistance in pneumococci in Iceland. Lancet. 1992;339:1606–1607. doi: 10.1016/0140-6736(92)91868-9. [DOI] [PubMed] [Google Scholar]
- 22.Lister P D. Multiply-resistant pneumococcus: therapeutic problems in the management of serious infection. Eur J Clin Microbiol Infect Dis. 1995;4(Suppl.):18–25. [PubMed] [Google Scholar]
- 23.McDougal L K, Facklam R R, Reeves M, Hunter S, Swenson J M, Hill B C, Tenover F C. Analysis of multiply antimicrobial-resistant isolates of Streptococcus pneumoniae from the United States. Antimicrob Agents Chemother. 1992;36:2176–2184. doi: 10.1128/aac.36.10.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Munoz R, Coffey T R, Daniels M, Dowson C G, Laible G, Casal J, Hakenbeck R, Jacobs M, Musser J M, Spratt B G, Tomasz A. Intercontinental spread of a multiresistant clone of serotype 23F Streptococcus pneumoniae. J Infect Dis. 1991;164:302–306. doi: 10.1093/infdis/164.2.302. [DOI] [PubMed] [Google Scholar]
- 25.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 6th ed. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 26.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. Approved standard M7-A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 27.Nesin M, Ramirez M, Tomasz A. Capsular transformation of a multidrug-resistant Streptococcus pneumoniae in vivo. J Infect Dis. 1998;177:707–713. doi: 10.1086/514242. [DOI] [PubMed] [Google Scholar]
- 28.Reichmann P, Varon E, Günther E, Reinert R R, Lüttiken R, Marton A, Geslin P, Wagner J, Hakenbeck R. Penicillin-resistant Streptococcus pneumoniae in Germany: genetic relationship to clones from other European countries. J Med Microbiol. 1995;43:377–385. doi: 10.1099/00222615-43-5-377. [DOI] [PubMed] [Google Scholar]
- 29.Romesburg H C. Cluster analysis for researchers. Malabar, Fla: Krieger; 1990. pp. 9–28. [Google Scholar]
- 30.Sa Figueiredo A M, Austrian R, Urbaskova P, Teixeira L A, Tomasz A. Novel penicillin-resistant clones of Streptococcus pneumoniae in the Czech Republic and in Slovakia. Microb Drug Resist. 1995;1:71–78. doi: 10.1089/mdr.1995.1.71. [DOI] [PubMed] [Google Scholar]
- 31.Schreiber J, Jacobs M. Antibiotic-resistant pneumococci. Pediatr Clin N Am. 1995;42:519–537. doi: 10.1016/s0031-3955(16)38977-5. [DOI] [PubMed] [Google Scholar]
- 32.Sluijter, M. Unpublished observations.
- 33.Sluijter M, Faden H, de Groot R, Lemmens N, Goessens W H F, van Belkum A, Hermans P W M. Molecular characterization of pneumococcal nasopharynx isolates in children during the first two years of life. J Clin Microbiol. 1998;36:2248–2253. doi: 10.1128/jcm.36.8.2248-2253.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith A M, Klugman K P. Three predominant clones identified within penicillin-resistant South-African isolates of Streptococcus pneumoniae. Microb Drug Resist. 1997;3:385–389. doi: 10.1089/mdr.1997.3.385. [DOI] [PubMed] [Google Scholar]
- 35.Smith A M, Klugman K P, Coffey T J, Spratt B G. Genetic diversity of penicillin-binding protein 2B and 2X genes from Streptococcus pneumoniae in South Africa. Antimicrob Agents Chemother. 1993;37:1938–1944. doi: 10.1128/aac.37.9.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sneath, P. H. A., and R. R. Sokal. Numerical taxonomy, p. 131–132. Freeman, San Francisco, Calif.
- 37.Soares S, Kristinsson K G, Musser J M, Tomasz A. Evidence for the introduction of a multiresistant clone of serotype 6B Streptococcus pneumoniae from Spain to Iceland in the late 1980s. J Infect Dis. 1993;168:158–163. doi: 10.1093/infdis/168.1.158. [DOI] [PubMed] [Google Scholar]
- 38.Trzcinski, K. Unpublished observations.
- 39.Trzcinski K, Hryniewicz W. Antimicrobial susceptibility of common bacterial pathogens isolated from lower respiratory tract infections in Poland in 1996—The Alexander Project. Med Sci Monitor. 1997;3:714–722. [Google Scholar]
- 40.Van Belkum A, Sluijter M, de Groot R, Verbrugh H A, Hermans P W M. A novel BOX-repeat PCR assay for high-resolution typing of Streptococcus pneumoniae strains. J Clin Microbiol. 1996;34:1176–1179. doi: 10.1128/jcm.34.5.1176-1179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Steenbergen T J M, Colloms S D, Hermans P W M, de Graaff J, Plasterk R H A. Genomic DNA fingerprinting by restriction fragment end labeling (RFEL) Proc Natl Acad Sci USA. 1995;92:5572–5576. doi: 10.1073/pnas.92.12.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshida R, Hirakata Y, Kaku M, Takemura H, Tanaka H, Tomono K, Koga H, Kohno S, Kamahira S. Genetic relationship of penicillin resistant Streptococcus pneumoniae serotype 19B strains in Japan. Epidemiol Infect. 1997;118:105–110. doi: 10.1017/s0950268896007273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zenni M K, Cheatham S H, Thompson J M, Reed G W, Batson A B, Palmer P S, Holland K L, Edwards K M. Streptococcus pneumoniae colonization in the young child: association with otitis media and resistance to penicillin. J Pediatr. 1995;127:533–537. doi: 10.1016/s0022-3476(95)70108-7. [DOI] [PubMed] [Google Scholar]