Abstract
Macrocyclic peptides represent promising scaffolds for targeting biomolecules with high affinity and selectivity, making methods for the diversification and functional selection of these macrocycles highly valuable for drug discovery purposes. We recently reported a novel phage display platform (called MOrPH-PhD) for the creation and functional exploration of combinatorial libraries of genetically encoded cyclic peptides. In this system, spontaneous, post-translational peptide cyclization by means of a cysteine-reactive non-canonical amino acid is integrated with M13 bacteriophage display, enabling the creation of genetically encoded macrocyclic peptide libraries displayed on phage particles. Using this system, it is possible to rapidly generate and screen large libraries of phage displayed macrocyclic peptides (up to 108-1010 members) in order to identify high affinity binders of a target protein of interest. Herein, we describe step-by-step protocols for the production of MOrPH-PhD libraries, the screening of these libraries against an immobilized protein target, and the isolation and characterization of functional macrocyclic peptides from these genetically encoded libraries.
Keywords: Macrocyclic peptides, phage display, non-canonical amino acids, drug discovery, protein-protein interactions
1. Introduction
Macrocyclic peptides have emerged as a valuable class of molecules for modulating and disrupting protein-protein interactions (PPI), which have been notoriously difficult to target using small molecule agents [1–3]. Cyclization of peptide molecules has proven beneficial for enabling tight and specific interaction with extended protein-protein interfaces while conferring additional beneficial features such as improved proteolytic stability [4,5] and/or membrane permeability [6–8]. In light of the promise of cyclic peptides as chemical probes and potential therapeutic agents, there has been significant interest toward the development of combinatorial methods for the generation and screening of macrocyclic peptide libraries. Synthetic approaches have involved the use of DNA-encoded libraries [9]. For example, Liu and coworkers reported the isolation of a potent inhibitor of an insulin-degrading enzyme from a library of 13,824 peptide-based macrocycles constructed via DNA-templated synthesis [10], whereas one-bead–one-compound libraries [11,12] have been successfully applied for the identification of inhibitors of PPIs such as the NEMO-IκB kinase interaction and Ras-effector interactions [13,14]. Biological methods for the combinatorial synthesis and exploration of cyclic peptide libraries include the use of phage display [15–19], mRNA display [20–24], or the RaPID system [25–27]. Phage display, in particular, is an affinity-based selection system for the screening of polypeptide ligands from combinatorial libraries of up to 109-1010 members displayed on the surface of a M13 bacteriophage [28]. The application of this technique in combination with randomized peptide sequences flanked by two cysteine residues has enabled the identification of disulfide-bridged peptides capable of disrupting protein-protein interactions [29–34]. Unfortunately, the chemical instability of disulfide bonds, in particular under reducing conditions, limits the utility of these compounds beyond in vitro applications or imposes the need for replacing these linkages to overcome these limitations [35,36]. Chemical crosslinking of linear peptide sequences displayed on phage particles has provided an alternative means to obtain (bi)cyclic peptide ligands against a protein/enzyme of interest [37–42], but this process requires chemical manipulation of the phages and results in reduced infectivity of the phages during the selection process.
In the interest of making available new strategies for the combinatorial generation and exploration of genetically encoded peptide macrocycles, our group has introduced methodologies to generate Macrocyclic Organo-Peptide Hybrids (MOrPHs), in which ribosomally produced peptide sequences are cyclized by means of genetically encoded non-canonical amino acids (ncAA) incorporated into the precursor polypeptide via amber stop codon suppression [43–47]. Key advantages of these strategies include the possibility to obtain structurally diverse cyclic peptide scaffolds upon variation of the constituent components of these MOrPH scaffolds, such as the length and sequence of the peptide, the nature of the ncAA module, the cyclization chemistry and, when available, the nature of the non-peptidic moiety utilized to induce peptide cyclization [48]. According to one of these MOrPH synthesis methods, an electrophilic unnatural amino acid, i.e., O-2-bromoethyl-tyrosine (O2beY), is exploited to generate cyclic peptides constrained by a non-reducible, inter-side-chain thioether bond upon reaction with a nearby cysteine residue in the precursor peptide sequence [46,47]. This peptide cyclization strategy was exploited for the design and evolution of potent inhibitors of the Sonic Hedgehog/Patched interaction [49]. More recently, we have integrated O2beY-mediated peptide cyclization with M13 phage display in order to establish a high-throughput platform for generating and screening large combinatorial libraries of genetically encoded macrocycles against a target protein of interest (Figure 1) [50]. This system was applied to the discovery of a high-affinity binder for streptavidin and potent macrocyclic peptide inhibitors of Sonic Hedgehog and Keap1, demonstrating its value toward the discovery and evolution of functional macrocyclic peptides capable of targeting proteins and protein-mediated interactions [50]. This chapter describes methods and protocols for the design, preparation, and panning of MOrPH-PhD libraries generated using O2beY-mediated cyclization and procedures for deconvolution, production, and in vitro testing of the affinity-selected macrocyclic peptide binders, using streptavidin as a model target protein. This system can be readily adapted to target a variety of other target protein and/or protein-protein interactions.
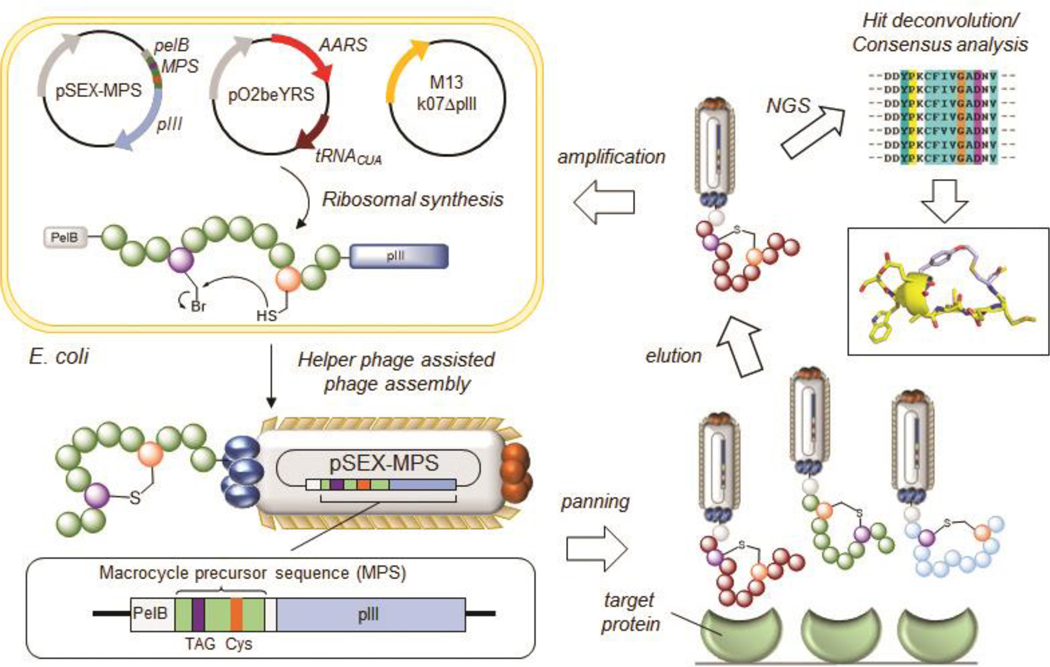
Figure 1. Overview MOrPH Phage Display (MOrPH-PhD) system.
A macrocycle precursor sequence (MPS) is fused to the N-terminal end of the M13 pIII protein encoded by a pSEX-based phagemid vector. A cysteine-reactive eUAA, such as O2beY, is introduced within the MPS via amber stop codon suppression utilizing an orthogonal AARS/tRNA pair. The eUAA mediates the spontaneous, post-translational cyclization of the MPS via a proximity-induced crosslinking reaction with a nearby cysteine residue. Phage production in the presence of a helper phage results in M13 phage particles displaying the thioether-bridged macrocycles on its surface. The phage-displayed peptide macrocycle library is panned and enriched against an immobilized target, followed by hit deconvolution via DNA sequencing of the MPS encoding gene contained in the bacteriophage. Reproduced from ref. 50 with permission from ACS Publications.
2. Materials
2.1. Reagents
Electrocompetent TOP10F’ cells
Chemically competent DH5a cells
Chemically competent BL21(DE3) cells
Ampicillin
Chloramphenicol
Tetracycline
Kanamycin
KOD Hot Start polymerase
Dpn I
Nco I/Nhe I restriction enzymes
10X Cutsmart buffer
Calf intestinal alkaline phosphatase
Polymerase Chain Reaction (PCR) Purification kit
Gel extraction kit
Plasmid Miniprep kit
Plasmid Midiprep kit
DNA clean & concentrator kit
Electroporation cuvettes
Agarose
Ethidium bromide
1X Tris-Acetate-EDTA (TAE) buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA)
5X DNA loading dye
1 kb DNA ladder
T4 DNA ligase (2,000,000 units/mL)
Luria Bertani (LB) media (per liter: 10 g tryptone, 5 g yeast extract, 10 g NaCl)
2xYT media (per liter: 16 g tryptone, 10 g yeast extract, 5 g NaCl)
Super optimal broth with catabolites (SOC) media (2% w/v tryptone, 0.5% w/v yeast extract, 10 mM NaCl, 2.5 mM KCl, 20 mM glucose)
M9 minimal media: 370 mL ddH2O, 100 mL of a 5X concentrated solution of M9 salts, 25 mL of a 10% (w/v) solution of yeast extract, 500 μL of a 2 M solution of MgSO4, 50 μL of a 1 M solution of CaCl2, and 5 mL glycerol.
Agar
Glycerol
M13 k07 ∆pIII Helper Phage
Arabinose
Isopropyl β-D-1-thiogalactopyranoside (IPTG)
O-2-bromoethyl-tyrosine (O2beY)
Spin filters, 30 kDa cutoff
Polyethylene glycol (PEG)
Sodium hydrogen phosphate (Na2HPO4)
Potassium dihydrogen phosphate (KH2PO4)
Sodium chloride (NaCl)
Potassium Chloride (KCl)
Immobilized tris(2-carboxyethyl)phosphine (TCEP) beads
Bovine serum albumin (BSA)
Biotin
Tween-20
Streptavidin immobilized magnetic beads
5x M9 salt solution (per liter: 64 g Na2HPO4, 15 g KH2PO4, 2.5 g NaCl, 5 g NH4Cl)
Monoclonal anti-FLAG M2 antibody
o-Phenylenediamine dihydrochloride (OPD) tablets
Ammonium chloride (NH4Cl)
Magnesium sulfate (MgSO4)
Calcium chloride (CaCl2)
Tris-hydrochloride (Tris-HCl)
Imidazole
Nickel nitrilotriacetic acid (NTA) resin
Mini Precast Gels
Prestained protein marker
Coomassie blue stain
Dithiothreitol (DTT)
1X SDS running buffer (3.0 g of Tris base, 14.0 g of glycine, and 1.0 g of SDS in 1000 ml of H2O)
Factor Xa protease
pSEX81 vector
Ni-NTA resin
A20 buffer: 50 mM Tris-HCl, 300 mM NaCl, 20 mM imidazole, pH 7.5.
A300 buffer: 50 mM Tris-HCl, 150 mM NaCl, 300 mM imidazole, pH 7.5.
Protein loading dye (4X): 0.2 M Tris-HCl, 0.4 M DTT, 8% w/v SDS, 6 mM bromophenol blue, 4.3 M glycerol.
SDS running buffer: 3.0 g of Tris base, 14.0 g of glycine, and 1.0 g of SDS in 1 L of H2O.
Coomassie blue staining solution: 0.1% (w/v) Coomassie Brilliant Blue, 20% (v/v) methanol, 10% (v/v) glacial acetic acid.
PBS buffer: 10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, pH 7.5.
Factor Xa buffer: 20 mM Tris, 100 mM NaCl, 2 mM CaCl2, pH=8.0.
Precipitation buffer: 20% polyethylene glycol 8000, 2.5 M NaCl.
pET22 plasmid
pEVOL_O2beY plasmid
Thermal cycler
UV-vis spectrophotometer
Refrigerated bench-top centrifuge
Electroporator
Sonicator
Incubator
DNA gel electrophoresis apparatus
Magnet
Graphing analysis software
pIII Reverse primer: 5’-CCTCAAGCTAGCTGATCATTAGCACAGG-3’
HPQ-NNK-Lib-1 forward primer 5’- TCGCCATGGCGGGCAGCTAG-NNKNNKCATCCGCAGNNKNNKTGCGGCAGCGCGGCCGCTGGATCCAAAG-3’
HPQ-NNK-Lib-2 forward primer: 5’-TCGCCATGGCGGGCAGCTGC-NNKNNKCATCCGCAGNNKNNKTAGGGCAGCGCGGCCGCTGGATCCAAAG-3’
2.2. Solvents
Double distilled water (ddH2O)
Ethanol
Trifluoroacetic acid (TFA)
Acetonitrile
Glacial acetic acid
3. Methods
3.1. Cloning of MOrPH-PhD Libraries
MOrPH-PhD libraries are created via insertion of a randomized macrocycle precursor sequence (MPS) between a PelB leader sequence and the pIII coat protein of an M13 bacteriophage in a pSEX81 phagemid vector (Figure 1) [50]. The PelB signal sequence is required for directing the cargo polypeptide to the periplasmic space of E. coli, where is it proteolytically cleaved by a signal peptidase [51]. N-terminal fusion of the MPS to the pIII coat protein encoded by the phagemid vector, along with the other viral proteins provided by the helper phage, enables incorporation of the MPS-pIII fusion into mature phage particles, where the macrocyclic peptide is displayed on the tip of the pIII coat proteins (5 copies/phage). The MPS is designed to consist of a fully or partially randomized peptide sequence, flanked by a Cys residue and an amber stop codon (TAG) for incorporation of O2beY [46] or other cysteine-reactive electrophilic unnatural amino acid (eUAA). The eUAA is genetically incorporated via amber stop codon (TAG) suppression [52] using an engineered aminoacyl-tRNA synthetase (AARS)/tRNACUA pair. For O2beY, the orthogonal AARS/tRNACUA pair is derived from Methanococcus jannaschii tyrosyl-tRNA synthetase and its cognate tRNA [46], whereas other variants of Mj TyrRS or other AARS/tRNACUA pairs can be used for alternative cysteine-reactive electrophilic eUAAs [53]. At the post-translational level, the eUAA reacts with the preinstalled Cys residue via a nucleophilic substitution reaction, forming a cyclic peptide constrained by a side-chain-to-side-chain thioether linkage (Figure 1). The peptide sequence library is generated via mutagenesis using randomized primers containing degenerate codons such as NNK, where N = A, C, G, T and K = G, T. O2beY-mediated peptide cyclization proceeds efficiently across peptide sequences encompassing up to 7–8 intervening residues between the fixed eUAA and cysteine residue [45]. Even longer peptide sequences (up to 15–20mer) can be efficiently cyclized using an expanded set of eUAAs recently introduced by our group [53]. In both cases, both orientations of the eUAA/Cys pair can be used to produce libraries of the format (eUAA)(Xaa)n(Cys) or (Cys)(Xaa)n(eUAA), in order to increase the structural diversity of the resulting MOrPH libraries [53]. In addition, this peptide cyclization method also allows the generation of libraries of ‘lariat’ peptides through randomization of positions within the N- and/or C-terminus of the peptide macrocycle (e.g., libraries of the format: (Xaa)n(eUAA)(Xaa)m(Cys) or (eUAA)(Xaa)n(Cys)(Xaa)m) [46].
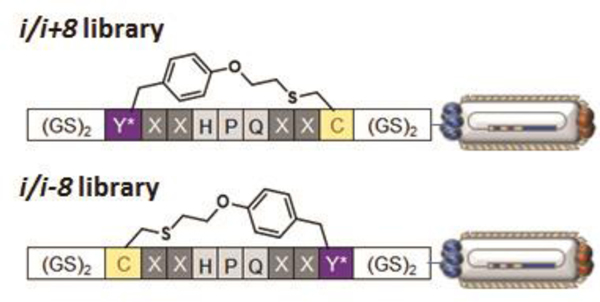
The following section describes a protocol for the production of a MOrPH-PhD library targeted against streptavidin. In this case, the library consists of a partially randomized MPS sequence in which a low-affinity (KD > 100 μM) streptavidin-binding motif, ‘HPQ’, is flanked by two fully randomized positions (using the NNK degenerate codon) and by the O2beY/Cys pair for peptide cyclization (Figure 2). Two libraries are prepared which explore two different orientations of the O2beY/Cys pair. The MOrPH-PhD library is generated via PCR and cloned into the Nco I/Nhe I cassette of the phagemid vector.
Figure 2. Libraries of semi-randomized O2beY-linked peptide macrocycles (X = NNK codon) displayed on phages.
The two macrocyclic peptide libraries differ in the relative arrangement of the O2beY/Cys pair (i/i+8 and i/i-8, where i is O2beY), providing a means to assess the effect of the thioether linkage orientation on the outcome of the affinity selection experiments.
Set up four parallel polymerase chain reactions (PCR) reactions for amplification of the insert corresponding to each of the two libraries (see Note 1). Set up the PCR reaction in a PCR tube by adding of the forward primer (HPQ-NNK-lib-1 or HPQ-NNK-lib-2) and the pIII reverse primer at a final concentration of 500 nM each in 28 μL ddH2O, 2 μL DMSO, 3 μL of a 25 mM solution of MgCl2, 5 μL of a 10X concentrated solution of KOD reaction buffer, 5 μL dNTP mix (2 mM each), 1 μL pSEX81 template (50 ng/μL), and 1 μL of KOD polymerase (1 unit). The reaction is mixed by pipetting and immediately incubated in a thermocycler (initial denaturation of 3 minutes at 95 °C; 31 cycles of 20 seconds at 95 °C, 10 seconds at 62 °C, 30 seconds at 70 °C).
Analyze the PCR product via gel electrophoresis on a 1.2% agarose gel in TAE buffer (100 V for ca. 30 min) for the correct insert size (~1,300 bp) (see Notes 2 and 3).
Pool the successful PCR reactions and add 2 μL of Dpn I (20 units) for digestion of the template plasmid. Incubate at 37°C for 1 hour. Purify the PCR product using a PCR Purification kit following the manufacturer’s instructions. In the final step of the purification, elute the PCR product with 40 μL of ddH2O.
Set up double digestion reactions for both the PCR insert and vector plasmid (pSEX81 template) using Nco I and Nhe I as the restriction enzymes:
4.1 PCR Insert: to 40 μL of eluted PCR insert, add 5 μL of 10X CutSmart buffer and bring the solution to a final volume of 47 μL with dd ddH2O. Add 1.5 μL of Nco I (15 units) and 1.5 μL Nhe I (15 units) restriction enzymes.
4.2 Vector: To a solution of pSEX81 vector (10 μg) (see Note 1), add 10 μL of 10X CutSmart buffer, and bring the reaction solution to a final volume of 94 μL with ddH2O. Add 3 μL of Nco I (30 units) and 3 μL of Nhe I (30 units) restriction enzymes.
5. Incubate the double digestion solutions at 37 °C for 4 hours. At the 3-hour mark, add 1 μL of calf intestinal phosphatase (CIP) (5 units) to the vector digestion (to minimize vector re-ligation) and allow the reaction to incubate at 37 °C for the last hour.
6. Purify both the digested insert and vector via electrophoresis using a 0.9% agarose gel in TAE buffer. Excise DNA bands corresponding to the proper size of the digested insert (~1300 bp) and vector (~2900 bp) using a 1 Kbp DNA ladder as reference. Extract the insert and vector using a QIAquick Gel Extraction kit following the manufacturer’s instructions. Elute both the digested insert and the vector in 30 μL of ddH2O.
7. Measure the concentration of both the insert and vector using a UV-vis spectrophotometer. Set up two separate ligation reactions using a total of 1.8 μg of DNA per reaction (500 ng vector; 1.3 μg of insert). To the insert and vector mixture, add 10 μL of 10X concentrated solution of T4 DNA ligase buffer and bring the final volume of the ligation to 96 μL using ddH2O. Add 4 μL of T4 DNA ligase (8000 units), mix the solution by pipetting and incubate at 16°C for 18 hours.
8. Set up a 5 mL overnight culture of TOP10F’ E. coli cells in 2xYT media supplemented with tetracycline (5 mg/L) for library transformation (see Note 5).
9. The next day, inoculate 500 mL of 2xYT media supplemented with tetracycline (5 mg/L) by adding the 5-mL cell culture from step 8. Grow the culture to an optical density at 600 nm (OD600) of 0.4 – 0.6 (determined with a UV-vis spectrometer) and pellet the cells in a centrifuge at 3,400 x g for 20 minutes at 4 °C and discard the supernatant. Note that cells should be kept at 4°C for the remainder of the procedure. Resuspend cells in 500 mL of a sterile, ice-cold solution of 10% (v/v) glycerol in ddH2O, then pellet the cells in a centrifuge at 3,400 x g for 20 minutes and discard the supernatant. Repeat this process using first 200 mL and then 50 mL of the ice-cold 10% glycerol solution. Resuspend the cells in 1 mL of ice-cold 10% glycerol solution. OD600 of the cell suspension should be 100~200 as calculated from an appropriate dilute sample with an OD600 between 0.1 and 1.0. Split the cells into 100 μL aliquots into 1.5-mL snap-top centrifuge tubes.
10. Heat inactivate the ligase from step 7 by heating the sample at 65°C for 5 minutes in a water bath to terminate the ligation reaction. Concentrate the reactions using a DNA concentrator kit following the manufacturer’s protocol. Elute the ligation product from the column using 10–15 μL of ddH2O. Add 2–3 μL of eluted DNA to freshly prepared, electrocompetent TOP10F’ cells from step 9 (4 transformations total) (see Note 1). Allow the cells to incubate on ice for 10 minutes and then transfer each aliquot to a separate ice-cold 100 μL electroporation cuvette. Electroporate the cells at 2.5 V following the manufacturer’s protocol and immediately resuspend the cells in 1 mL of 2xYT media prewarmed at 37 °C in a 1.5-mL snap-top conical tube. Allow the cells to recover with shaking at 200 rpm at 37 °C for 1 hour. Save a 10 μL aliquot.
11. Spread the transformation solutions from step 10 on two large (20 × 20 cm) 2xYT agar plates (see Note 3) supplemented with tetracycline (5 mg/L) and ampicillin (100 mg/L) (typically, 2 transformations per plate). Incubate the plates for 18 hours at 37 °C.
12. In parallel to step 11, dilute a 10 μL aliquot of the transformed cells from step 10 using 10-fold serial dilutions in PBS buffer (from 103 to 108) for measurement of the colony forming units (c.f.u.) and library diversity. Plate the serial dilution solutions on 2xYT agar plates supplemented with tetracycline (5 mg/L) and ampicillin (100 mg/L). Incubate the plates for 18 hours at 37 °C. The following day, estimate the c.f.u. by manually counting the colonies from plates containing ~100–200 colonies and extrapolating based on the dilution factor. For optimal coverage of the library, the observed c.f.u. should exceed the theoretical complexity of the DNA library by at least 3–5 folds (e.g., 3–5 × 106 c.f.u. are required for optimal coverage of a DNA library of 1 × 106 members).
13. Collect the cells from the plates in step 11 by adding 20 mL of 2xYT media to the plate. Collect the cell suspension in a 50-mL conical centrifuge tube and pellet the cells by centrifugation at 3,400 x g for 20 minutes. Extract the DNA library using a Plasmid Midi Kit following the manufacturer’s instructions.
14. It is recommended that the quality of library is assessed via DNA sequencing (see Note 4).
3.2. Production of MOrPH-PhD Library
The following section outlines protocols for the production of the MOrPH-PhD libraries generated in section 3.1. Phage particles are expressed and isolated from TOP10F’ E. coli cells utilizing a helper phage system [54]. The helper phage contains the genetic material for the production and assembly of the M13 bacteriophage, except for the pIII coat protein, which is encoded by the phagemid vector. This system allows for the incorporation of the MPS-pIII fusion into the phage particles, resulting in pentavalent display of the cyclic peptide library on the phage surface.
Add 1.5 μL of the DNA library at a concentration of ~500 ng/μL to a 100 μL aliquot of electrocompetent TOP10F’ cells containing the pEVOL_O2beY plasmid in a sterile 1.5-mL centrifuge tube [46]. Incubate the cells on ice for 10 minutes and transfer the cells to an ice-cold 100 μL electroporation cuvette. Electroporate the cells at 2.5 V and immediately resuspend the cells in 1 mL of 2xYT media prewarmed to 37 °C. Recover the cell solution with shaking at 200 rpm at 37 °C for 1 hour.
Prepare a sterile 125 mL Erlenmeyer flask containing 20 mL of 2xYT media supplemented with tetracycline (5 mg/L), ampicillin (100 mg/L), and chloramphenicol (34 mg/L). Inoculate the media by adding the cell solution from step 1 and incubate the cell culture with shaking at 200 rpm at 37 °C for 18 hours.
The next day, inoculate 5 mL of 2xYT media by adding 500 μL of the overnight culture to the 10-mL culture tube. Incubate the cell culture with shaking at 200 rpm at 37 °C until OD600 reaches 0.6.
Mix 2.5 mL of the cell culture from step 3 with 2.5 mL of fresh 2xYT media in a new culture tube. Infect the cell culture with M13 k07 ∆pIII helper phage at a multiplicity of infection (MOI) of 20. Allow the infection to take place at 37 °C for 1 hour with shaking. Pellet the culture at 3,400 x g for 10 minutes.
To a sterile 125 mL Erlenmeyer flask, add 20 mL of 2xYT media supplemented with tetracycline (5 mg/L), ampicillin (100 mg/L), chloramphenicol (34 mg/L), kanamycin (30 mg/L), arabinose (0.06%), and eUAA (2 mM). Transfer the cell pellet from step 4 to the cell culture and incubate with shaking at 200 rpm at 30 °C for 18 hours.
The next day, transfer the overnight culture from step 5 to a 50-mL conical centrifuge tube and pellet the culture at 3,400 x g for 30 minutes. Collect the supernatant in a separate 50-mL conical centrifuge tube and discard the cell pellet. Concentrate the supernatant at 3,400 x g for 45 minutes using a 30 kDa spin filter to a convenient volume (typically, 250–300 μL). Mix the concentrated supernatant with 1:4 (v/v) of precipitation buffer and incubate the phage solution overnight at 4 °C to precipitate the phage particles (see Note 6).
Pellet the precipitated phage by centrifugation at 18,000 x g for 30 minutes at 4 °C. Discard the supernatant and centrifuge again at 18,000 x g for 1 minute to discard any residual supernatant and resuspend the phage pellet in 200 μL of PBS buffer. Centrifuge the resuspended phage solution at 18,000 x g for an additional 5 minutes to remove any insoluble cellular debris and transfer the clarified phage solution supernatant to a clean 1.5 mL snap-top centrifuge tube.
Wash 50 μL of immobilized tris(2-carboxyethyl)phosphine (TCEP) bead suspension with 200 μL of PBS buffer. Centrifuge the suspension at 18,000 x g for 1 minute and remove the supernatant. Repeat the wash step two additional times, followed by the addition of the phage solution from step 7. Incubate for 2 hours at room temperature with gentle shaking (see Note 7).
Centrifuge the solution at 18,000 x g for 1 minute and transfer the supernatant to a separate 1.5-mL snap-top centrifuge tube. Wash the beads three times with 100 μL of PBS and pool the supernatant into the same centrifuge tube. Optionally, adjust the pH to 8.5–9.0 and incubate overnight at room temperature. This process is meant to maximize the amount of cyclized peptide sequences within the MOrPH-PhD library.
The next day, adjust the pH back to 7.0, if necessary, and precipitate the phage by adding 1:4 (v/v) of precipitation buffer (20% polyethylene glycol 8000, 2.5 M NaCl), followed by incubation at 4 °C for 2 hours.
Pellet the phage by centrifugation at 18,000 x g for 30 minutes at 4 °C and discard the supernatant.
Centrifuge again at 18,000 x g for 1 minute and discard any residual supernatant.
Resuspend the phage pellet in 100 μL PBS buffer. Centrifuge the resuspended phage solution at 18,000 x g for an additional 5 minutes to remove any insoluble cellular debris.
Transfer the clarified phage solution supernatant to a clean 1.5-mL snap-top centrifuge tube and store at 4 °C until the panning process.
3.3. Panning of MOrPH-PhD Library
This section outlines the procedure for panning the MOrPH-PhD library against an immobilized target protein of interest (biopanning). In the biopanning process, the library of phage-displayed MOrPHs is incubated with the immobilized target and the unbound phages are washed away. The protein-bound phages are eluted by lowering the pH and then amplified in E. coli. The resulting library is subjected to additional rounds of affinity selection and amplification (typically, 3–4 rounds in total) to enrich the pool with cyclic peptide sequences with high affinity toward the target protein. After the selection process, the individual clones are identified via DNA sequence and further characterized. In the protocol below, the MOrPH-PhD library from section 3.2 is panned against streptavidin immobilized on magnetic beads. For other target proteins, the protein of interest can be immobilized onto streptavidin- or neutravidin-coated (magnetic) beads via a biotin tag. Alternatively, the target protein can be immobilized on microtiter plate [50,53] or on other resins through non-specific adsorption, covalent capture, or through other affinity tags. After immobilization of the target protein, the capturing matrix (i.e. beads or plate) is blocked with bovine serum albumin (BSA) to minimize nonspecific binding of the phages to the matrix.
Transfer 10 μL of streptavidin immobilized magnetic beads to a clean 1.5-mL snap-top centrifuge tube and wash three times with 200 μL of PBS buffer. Between washes, separate the beads from the solution via magnetic separation to remove the supernatant.
Block the beads by adding 150 μL of PBS containing 0.5% (m/v) BSA and incubating for 2 hours at room temperature.
After blocking, wash the beads three times with 200 μL of PBS containing 0.05% (v/v) Tween-20. Between washes, separate the beads from the solution via magnetic separation to remove the supernatant.
Incubate the streptavidin beads with 90 μL of the phage solution in PBS buffer (1010 – 1011 plaque forming units or pfu) for 1 hour at room temperature with light shaking. Prior to incubation, remove 10 μL of the phage solution for determination of pfu as described in step 7.
After incubation with the phage solution, wash the beads three times with 200 μL of wash buffer. Separate the beads between wash steps via magnetic separation to remove the supernatant.
Add 50 μL of elution buffer (0.1 mM biotin in PBS) and incubate at room temperature for 30 minutes with light shaking. Separate the beads via magnetic separation and add the supernatant to a clean 1.5-mL snap-top centrifuge tube. Store the supernatant containing the eluted phage at 4 °C.
Start a 5 mL culture of TOP10F’ cells in 2xYT media supplemented with tetracycline (5 mg/L) for propagation of the enriched phage library and determination of pfu. Grow the cells to an OD600 of 0.6.
To determine pfu, take 10 μL of the phage solutions from both step 4 (pre-panning) and step 6 (eluted phages after panning) and perform 10-fold serial dilutions in 2xYT media (pre-panning: 107- to 1012- fold dilutions; post-panning: 103- to 108- fold dilutions). Add 90 μL of exponentially growing cells from step 4 to the phage dilutions and allow the phage to infect the TOP10F’ cells for 1 hour at 37 °C with shaking at 200 rpm. Plate the phage dilutions on 2xYT agar plates supplemented with tetracycline (5 mg/L) and ampicillin (100 mg/L) and incubate the plates at 37 °C for 18 hours. Count the colonies from plates containing ~100–500 colonies to determine pfu for the pre- and post-panning phage samples and the fraction of recovered phage after panning. (see Note 8)
Recover the enriched phage library from step 6 by the addition of 1 mL of exponentially growing TOP10F’ cells (OD600 ~ 0.6; from step 7) to the eluted phage solution. Allow the phage to infect the TOP10F’ cells for 1 hour at 37 °C with shaking.
Pellet the infected cells in a centrifuge at 3,400 x g for 5 minutes at 4 °C and discard the supernatant. Resuspend the pellet in 5 mL of 2xYT media supplemented with tetracycline (5 mg/L) and ampicillin (100 mg/L) and incubate at 37 °C for 18 hours.
Extract the enriched DNA library using a Plasmid Miniprep Kit following the manufacturer’s protocol and elute the DNA in 30 μL of ddH2O. Use the recovered DNA library to propagate new phage for additional rounds of affinity selection (Step 1 through 10).
3.4. Deconvolution of MOrPH-PhD Library via DNA Sequencing
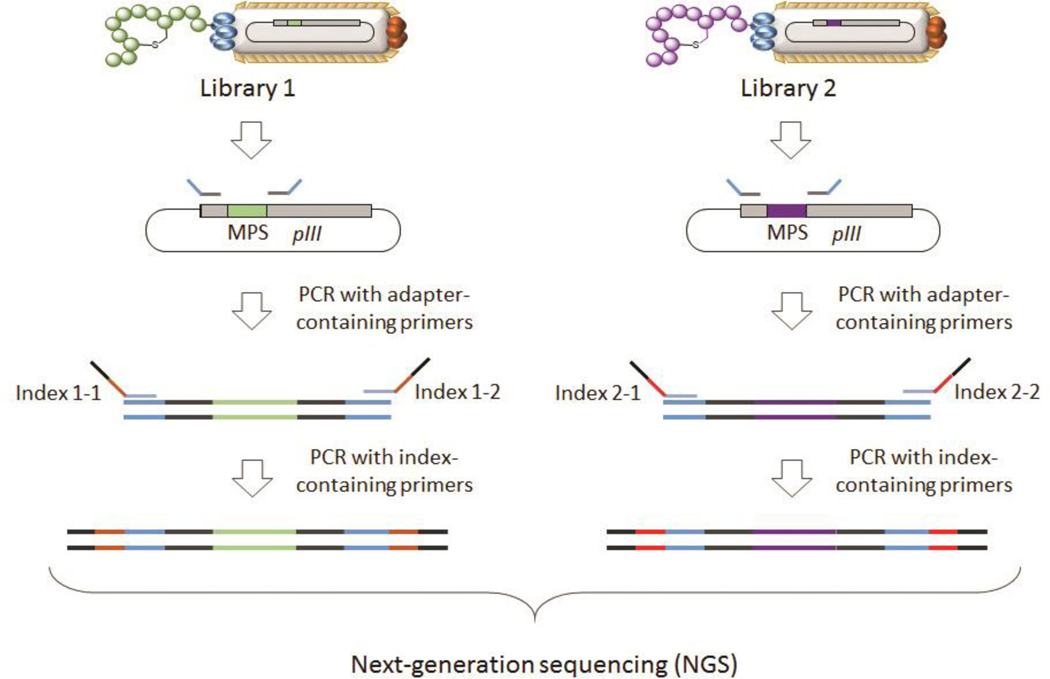
After multiple rounds of affinity selection and phage amplification (typically 3–4), the MOrPH-PhD library becomes enriched with high affinity binders for the target protein (streptavidin). For library deconvolution, the enriched sequences are identified via DNA sequencing (see Note 4). The latter is carried out via Sanger sequencing of 30–50 randomly chosen phages from the selected pool or, preferably, via next-generation sequencing (NGS), of the entire phage pool isolated after the biopanning process. NGS routinely yields ~ 1 million sequence reads in a single run55 and samples from multiple selection rounds and/or selection experiments can be sequenced simultaneously via multiplexing. In this section, we describe the use of the Illumina Miseq NGS platform for library sequencing. Amplicons are generated from enriched phage libraries via two PCR steps using Nextera-style PCR primers. The Illumina Nextera primers for amplicon generation are designed using Illumina’s 16s amplicon protocol. A first PCR reaction uses Nextera-style tag sequences containing a locus-specific sequence for annealing to the pSEX81 phagemid containing the enriched library. The second PCR reaction utilizes Nextera-style indexing sequences that allow for multiplexed analysis of multiple samples in a single run, as well as hybridization onto the sequencing flow cell (Figure 3). The sequencing data are then analyzed to identify the most highly enriched clones, as well as consensus sequences across the enriched sequences (Figure 4).
Figure 3. Workflow for generation of amplicons for next-generation sequencing.
A first PCR is used to amplify the MPS-encoding sequence from the phage library and add overhang adapters to the 5’ and 3’ ends of that region. A second PCR is applied to attach Illumina sequencing adapters using Nextera-style indexing primers.
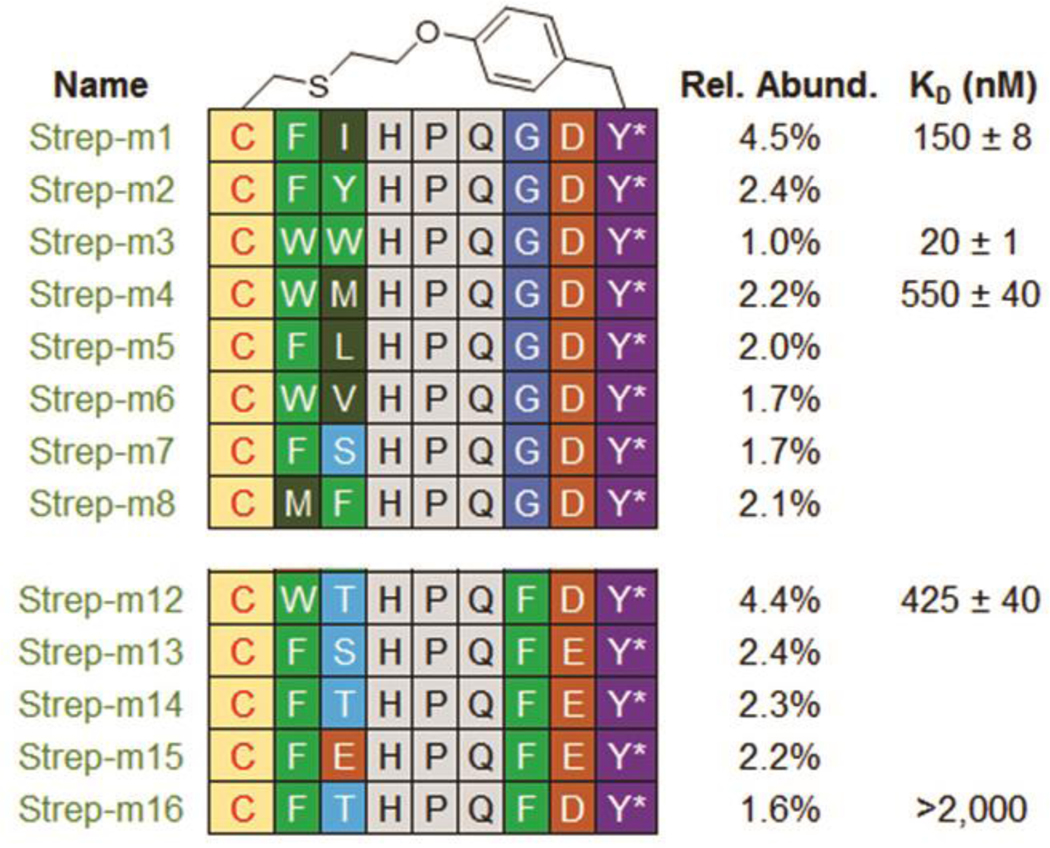
Figure 4. Hit sequences identified via next generation sequencing.
(relative abundance = n/54,000 sequences) Consensus is indicated by the color gradient. Aromatic amino acids (Y, F, W) are displayed in green, hydrophobic amino acids (V, L, I, M, P) are displayed in dark green, nucleophilic amino acids are displayed in blue (S, T), acidic amino acids (D, E) are displayed in brown, small amino acids (G, A) are displayed in navy blue. Adapted from ref. 50 with permission from ACS Publications.
In a first PCR reaction, the enriched phage libraries are amplified via PCR using primers containing (i) a Nextera-style tag sequence and (ii) a locus-specific sequence for annealing to the pSEX81 phagemid (see Note 9). Set up the PCR by the addition of both forward and reverse Nextera-style tag primers (500 nM each, final conc.), 28 μL ddH2O, 2 μL DMSO, 3 μL of a 25 mM solution of MgCl2, 5 μL of a 10X concentrated solution of KOD rection buffer, 5 μL dNTP mix (2 mM each), 1 μL of template plasmid library (50 ng/μL), and 1 μL of KOD polymerase (1 unit) in a PCR tube. The reaction is mixed by pipetting and immediately incubated in a thermocycler (initial denaturation of 3 minutes at 95 °C; 31 cycles of 20 seconds at 95 °C, 10 seconds at 62 °C, 30 seconds at 70 °C).
Purify the PCR reaction via electrophoresis on a 1.2% agarose gel in TAE buffer. Excise the DNA band corresponding to a size of 200 bp and extract the amplicon using a gel extraction kit following the manufacturer’s instructions and elute the purified amplicon in 30 μL of ddH2O.
In a second PCR reaction, Nextera-style index sequences are introduced into the amplicon (see Note 9) for multiplexed analysis of multiple libraries at once. Set up a PCR reaction in a similar manner as in step 1, this time using the Nextera-style indexing primers and the amplicon generated from step 1 as the template. Mix the reaction by pipetting and immediately incubate in a thermocycler (initial denaturation of 3 minutes at 95 °C; 31 cycles of 20 seconds at 95 °C, 10 seconds at 45 °C, 30 seconds at 70 °C).
Purify the PCR reaction via electrophoresis on a 1.2% agarose gel in TAE buffer. Excise the DNA band corresponding to a size of 250 bp and extract the amplicon using a QIAquick Gel Extraction kit following the manufacturer’s instructions. At the last step, elute the purified amplicon with 30 μL of ddH2O.
Submit the amplicon for Illumina MiSeq paired-end sequencing according to the service provider’s instructions.
3.5. Recombinant expression and isolation of the macrocyclic peptide hits
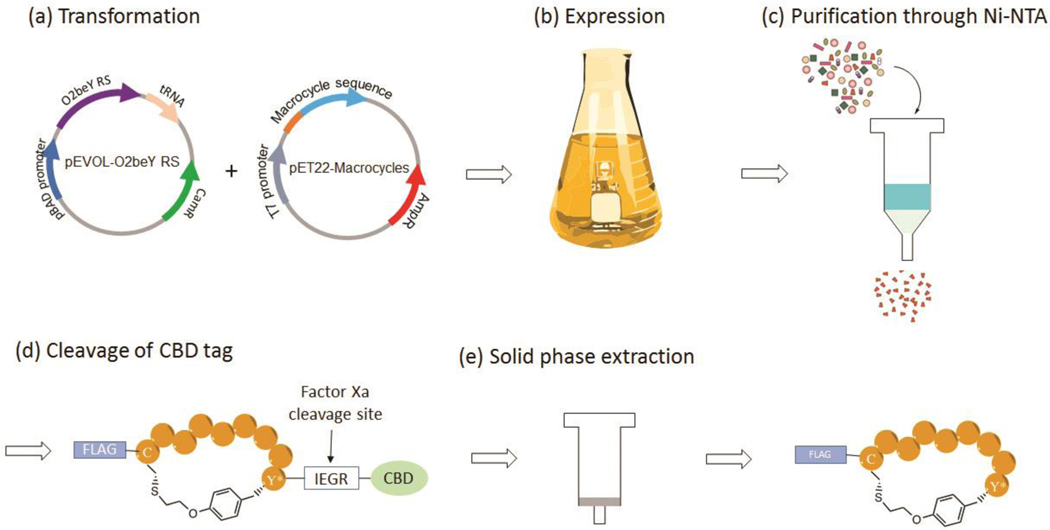
Whereas the cyclic peptides identified through this process can be synthesized via solid-phase peptide synthesis [49], a key advantage of the present method is that the O2beY-linked peptides can be efficiently prepared and isolated via recombinant expression in E. coli [50]. This is particularly useful for rapid characterization and validation of the cyclic peptide ‘hits’ identified from the biopanning process. Described below is a protocol for producing O2beY-linked macrocyclic peptides identified from the biopanning experiment against streptavidin. Briefly, the macrocyclic precursor sequence of the selected hit(s) is subcloned into a pET22 vector for expression of the corresponding macrocyclic peptide in E. coli [50]. The peptide is generated as a fusion protein containing an N-terminal FLAG tag and a C-terminal chitin binding domain (CBD) followed by a poly-histidine tag to facilitate purification and quantification. A Factor Xa cleavage site is introduced between the macrocycle precursor sequence and the CBD to facilitate proteolytic cleavage of the CBD tag with a Factor Xa protease. The recombinant expression of the CBD-fused MOrPH is carried out using E. coli BL21(DE3) cells co-transformed with (i) a pET22-based plasmid encoding the peptide sequence under the control of an ITPG-inducible T7 promoter, and (ii) a pEVOL plasmid encoding for two copies of the O2beY-specific Mj aminoacyl-tRNA synthetase under a constitutive GlnS promoter and an arabinose-inducible AraC promoter, respectively, along with a copy of the cognate Mj tRNATyrCUA (“pEVOL_O2beY plasmid”) [49,50]. After expression, purification of CBD-fused MOrPH is carried out via Ni-affinity chromatography using the polyhistidine tag. Protein purity and the occurrence of cyclization is confirmed by SDS-PAGE and MALDI-TOF-MS analysis, respectively. If desired, CBD-free MOrPHs can be isolated via proteolytic digestion with Factor Xa to remove the CBD tag, followed by solid-phase extraction (Figure 5) [50]. In this case, CBD is not cleaved from the streptavidin binding MOrPH since the CBD tag was determined to show no detectable binding to streptavidin [54].
Figure 5. Recombinant expression and isolation of the macrocycle peptides.
The plasmids encoding the CBD-fused macrocyclic peptide and the AARS/tRNA amber suppressor system are co-transformed into BL21(DE3) (a). The CBD-fused macrocyclic peptide is produced via expression in the presence of O2beY (b) and purified via Ni-affinity chromatography (c). The CBD tag is cleaved by digestion with Factor Xa protease (d) and the macrocyclic peptide is isolated via solid-phase extraction (e).
Add 1.0 μL of pET22 plasmid (50 ng/μL) encoding the hit peptide sequence and 1.0 μL of pEVOL_O2beY plasmid (50 ng/μL) for eUAA incorporation to a 100 μL aliquot of chemically competent BL21(DE3) cells. Incubate the cells on ice for 30 minutes.
Incubate the cells at 42 °C for 45 seconds in a water bath and transfer the tube back on ice for 5 minutes.
Add 200 μL of Super Optimal broth with Catabolites repression to the cells. Incubate at 37 °C for 60 minutes with shaking. Plate the cell solution on LB agar plates supplemented with ampicillin (100 mg/L) and chloramphenicol (34 mg/L) and incubate at 37°C overnight.
The next day, prepare 5 mL of LB media supplemented with ampicillin (100 mg/L) and chloramphenicol (34 mg/L) in a 10-mL culture tube. Inoculate the LB media with a single colony from step 3 and incubate at 37 °C for 16 hours with shaking at 200 rpm.
The next day, prepare 500 mL of M9 minimal media supplemented with ampicillin (100 mg/L) and chloramphenicol (34 mg/L) (see Note 10).
Inoculate the M9 culture by adding the 5 mL overnight culture from step 4 and incubate at 37 °C with shaking at 200 rpm until OD600 reaches 0.5–0.6.
Pellet the cell culture at 3,400 x g for 30 minutes and decant the supernatant. Resuspend the pellet in 100 mL of fresh M9 minimal media supplemented with ampicillin (100 mg/L), chloramphenicol (34 mg/L), arabinose (0.06% m/v) and eUAA (2 mM). Incubate the culture at 27 °C with shaking at 200 rpm for 1 hour.
Induce expression of the recombinant polypeptide by addition of IPTG to a final concentration of 1 mM from a sterile 0.5 M IPTG stock solution in water. Incubate the cell culture at 27 °C with shaking at 200 rpm for 16 hours.
Harvest the cells via centrifugation at 3,400 x g for 30 minutes. Resuspend the cell pellet with 25 mL A20 buffer in a 50-mL conical centrifuge tube. For long-term storage, store the cell suspension at −80 °C.
Lyse the cells via sonication (amplitude 50, 90 pulses, 1 second per pulse, and 10 seconds between pulses) with a temperature limit set at 18 °C to avoid overheating. Note that the cell lysate should be kept ice-cold from this point on to minimize protein degradation.
Clarify the cell lysate by centrifugation at 21,000 x g for 30 minutes at 4 °C. Separate the supernatant from the insoluble cellular debris and transfer it to a clean 50-mL conical tube.
Load the clarified cell lysate through a column containing 3 mL of Ni-NTA resin pre-equilibrated in A20 buffer. Wash the column with A20 buffer and then elute the bound protein using ice-cold A300 buffer. Pool the fractions containing the protein by monitoring the absorbance at 280 nm (see Note 11).
Buffer-exchange the protein solution with PBS buffer via dialysis or using a centrifugal filter. For the latter procedure, dilute the protein solution to 20 mL with PBS buffer. Concentrate the protein by ~100-fold via centrifugation at 3,400 x g using a 10-kDa cutoff centrifugal filter (final volume of 200–300 μL). Repeat the dilution/concentration procedure two more times to remove any residual imidazole. Transfer the concentrated peptide (200–300 μL) to a clean 1.5-mL snap-cap centrifuge tube and determine the protein concentration via UV-vis spectrometry. For long-term storage, flash-freeze and store the protein at −80 °C
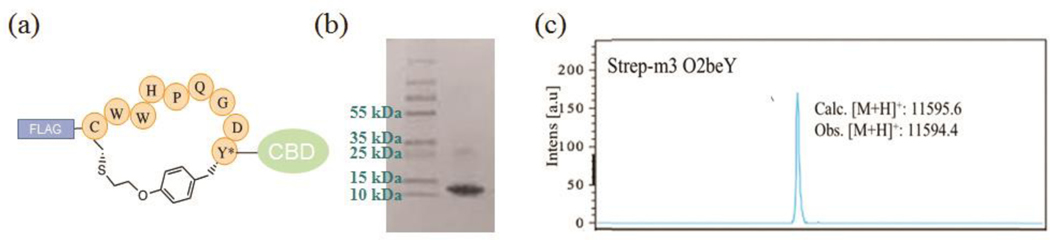
Determine protein purity by SDS-PAGE. Mix ca. 25 μg of the protein with 5 μL of the protein loading dye (4X) and dilute the solution to a final volume of 30 μL with ddH2O. Denature the protein by incubating the sample at 95 °C for 5 min. Load and run the sample on an acrylamide gel. Stain the gel with Coomassie blue staining solution and gentle shaking at room temperature for 2 hours. Destain the gel with a solution of 10% acetic acid overnight at room temperature with gentle shaking to visualize the protein bands (Figure 6b).
Use MALDI-TOF mass spectrometry (MS) or LC-MS analysis to confirm the identity of the protein and the occurrence of cyclization via the O2beY/Cys cross-linking reaction. The latter is evidenced by a mass difference of −82 Da (-HBr) compared to the mass of the linear CBD-fused peptide (Figure 6c). (see Note 12).
(Optional) For cleavage of the CBD tag, dilute purified CBD-fused macrocycle to a concentration of 250 μM with Factor Xa buffer. Add 5 μg/mL of Factor Xa protease (NEB) to the solution, incubate the digestion reaction overnight at room temperature.
Next day, acidify the reaction mixture with TFA (0.1%) and load it on the solid-phase column. Wash the column with water with 0.1% TFA, followed by eluting with a step gradient of acetonitrile (25%, 50 %, 75%, 100%) in water containing 0.1% TFA.
Fractions containing the peptide are identified by MALDI-TOF mass spectrometry (or LC-MS) and combined. The solution is lyophilized to obtain the macrocyclic peptide as in powder form.
(Optional) The procedure above typically yields the peptide in >90% purity. If necessary, the macrocyclic peptide can be further purified via high-pressure liquid chromatography (HPLC) using a reverse-phase (RP) C18 column and a gradient of acetonitrile (+0.1% TFA) in water (+0.1% TFA).
Figure 6. Purified macrocyclic peptide.
(a) Structure, (b) SDS-PAGE gel, and (c) MALDI-TOF spectrum of a representative CBD-fused macrocyclic peptide (CBD-Strep-m3(O2beY)). MS analysis confirms successful incorporation of O2beY and quantitative cyclization.
3.6. Streptavidin Binding Assay
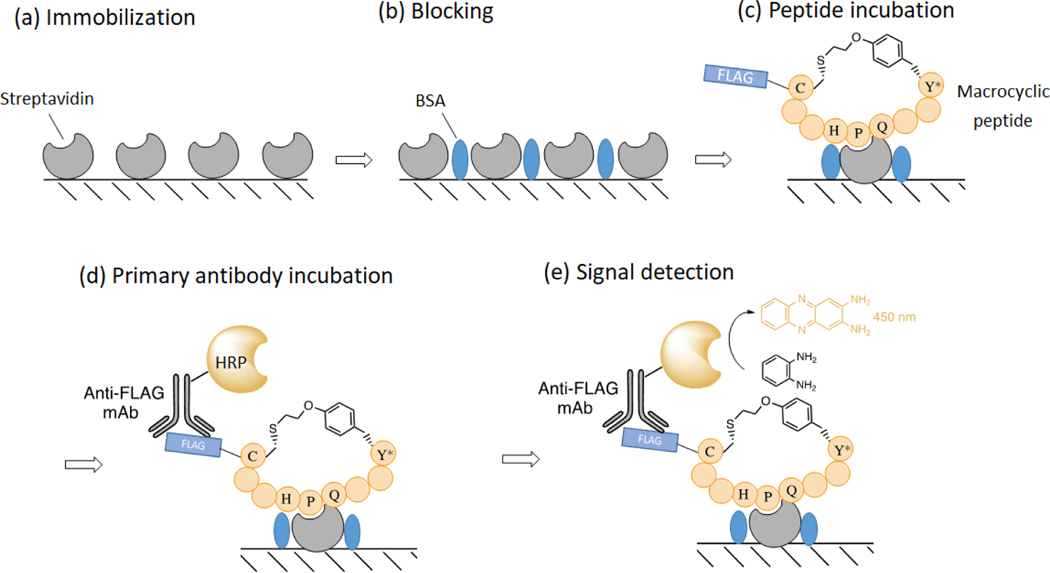
The binding affinity (KD) of the isolated MOrPHs for the target protein streptavidin is determined using an in vitro immunoassay as described in Figure 7 [49, 50]. In this assay, the FLAG-tagged macrocyclic peptide (with or without the CBD fusion tag) is incubated on a streptavidin coated plate at varying concentrations. The streptavidin-bound peptide is then quantified colorimetrically (λ450) using a horseradish peroxidase (HRP)-conjugated anti-FLAG antibody.
Figure 7. Streptavidin binding assay.
The target protein is immobilized on a 96-well polystyrene plate at 4 °C overnight (a), followed by blocking with BSA (b). Next, the macrocyclic peptide is incubated with the immobilized protein in varying concentration (c), followed by detection of the bound peptide with anti-FLAG-HRP conjugated antibody (d). Lastly, substrate o-phenylenediamine is added to detect peroxidase activity (e). The HRP catalyzed reaction of oxidizing o-phenylenediamine to 2,3-diaminophenazine exhibits a strong absorbance at 450 nm.
Prepare a 2- or 3-fold serial dilution of the peptide solution starting from 50–100 μM by diluting the peptide in PBS in 1.5-mL snap-cap centrifuge tubes (see Note 13).
Transfer 100 μL of the FLAG-tagged peptide solutions at different concentrations into 96-well streptavidin-coated polystyrene plates and incubate for 1 hour at room temperature. (see Note 14).
Wash the plate three times by adding 150 μL of wash buffer (PBS supplemented with 0.05% v/v Tween-20) and discarding the solution (see Note 15).
Prepare a solution of HRP-conjugated mouse anti-FLAG polyclonal antibody in a 1:2500 dilution with ice-cold PBS buffer. Add 100 μL of the antibody solution to each well and incubate at room temperature for 1 hour.
Wash the plate three times with 150 μL of wash buffer as in step 3.
Dissolve one o-Phenylenediamine dihydrochloride tablet and one urea hydrogen peroxide/buffer tablet in 20 mL of ddH2O (see Note 16). Add 100 μL of the solution to each well and incubate the plate at room temperature for 5–10 minutes until color develops.
Monitor the absorbance at 450 nm using a microtiter plate reader over the course of 30 min, taking a measurement every 5 minutes.
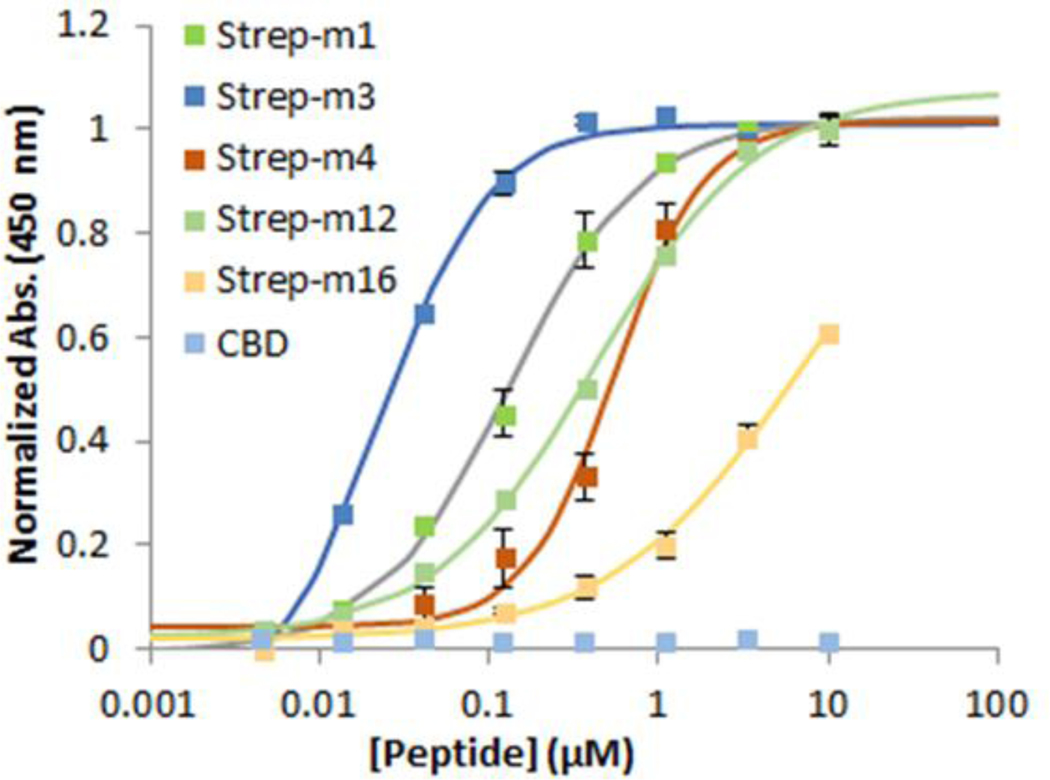
Determine the equilibrium dissociation constants (KD) for peptide binding to streptavidin by fitting the dose–response curves to a 1:1 binding isotherm equation via nonlinear regression using graphing analysis software. Mean values and standard deviations are calculated from experiments performed in triplicate (Figure 8).
Figure 8. Peptide binding curves.
Binding curves of the CDB-fused streptavidin-binding peptides to immobilized streptavidin as determined using the assay described in Figure 5. The CBD-only control shows no binding to streptavidin. Adapted from ref. 50 with permission from ACS Publications.
Acknowledgements
This research was supported by the U.S. National Institute of Health (NIH) grant R01 GM134076.
4. Notes
The scale of these reactions needs be adjusted based on the complexity of the gene/peptide libraries. For example, to prepare DNA libraries with a complexity of 108-109, it is recommended to obtain 8–10 μg of the insert DNA from multiple parallel PCR reactions (e.g. 6–8 reactions). The amount of recipient vector and transformations needs to be adjusted accordingly. For optimal coverage of a library, it is recommended that the number of c.f.u. obtained after transformation exceeds by at least 3–5 fold the overall complexity of the library (e.g., 3–5 × 109 c.f.u. for a DNA library of 1 × 109 members).
When casting an agarose gel, add ethidium bromide (EtBr) to a final concentration of approximately 0.2–0.5 μg/mL. EtBr is a known mutagen and should be handled with care. Wear a lab coat, eye protection and vinyl gloves when working with this chemical. All EtBr-contaminated material should be collected in dedicated containers for disposal.
For DNA gel electrophoresis techniques and agar plate preparation, refer to a basic molecular biology manual, e.g., H. B. Miller, D. S. Witherow, S. Carson, Molecular Biology Techniques: A Classroom Laboratory Manual, 3rd Edition, Academic Press, 2011.
Sanger sequencing and next-generation DNA sequencing services are available from commercial sources.
Transformation of the ligation reaction into freshly prepared electrocompetent cells greatly improves colony forming units (CFU).
After mixing with precipitation solution, the phage solution should turn cloudy. If not, add more precipitation solution until precipitation occurs.
This step reduces any disulfide bond potentially formed within the phage displayed randomized peptides during the phage production process.
This procedure derives the number of plaque forming units (pfu) from the number of colony forming units (cfu) as given by the infection of E. coli cells with the phages, which contain an ampicillin resistance marker. We determined that this approximation is valid as long as the MOI is kept low (>10−1-10−2), i.e., a large (>10- to 100-fold) excess of cells over phages is applied, which ensures that all infective phages are detected through the plate selection procedure. Alternatively, the conventional protocol for pfu determination using top agarose plates can be applied.
Nextera-style tag sequences and index sequences can be added using commercial kits. Refer to the manufacturer’s instructions for samples of tag and index sequences.
All of the ingredients for the M9 minimal media must be autoclaved or filter sterilized separately before combining. If reagents are mixed together before autoclaving, salts will precipitate from the solution.
It is recommended that the protein is not frozen before buffer exchange as the high concentration of imidazole can cause protein precipitation during the freezing/thawing process.
If incomplete cyclization is observed, peptide cyclization can be favored via incubation at slightly alkaline conditions. Adjust the pH of the peptide solution to 9 using a 0.2 M NaOH solution and incubate the solution overnight at room temperature. When cyclization is complete as determined by MALDI-TOF MS, adjust the pH of to 7.0 using a 0.5 M HCl solution.
Prepare 200 µL of protein solution at each fixed concentration for performing the experiment in duplicate. Add 100 μL of PBS buffer to a designated well as a blank.
Commercial streptavidin-coated plates are typically already pre-blocked with bovine serum albumin (BSA). If not, add a BSA blocking step prior to biopanning by incubating each well with 100 μL 5% (w/v) BSA in PBS for 30 min at room temperature, followed by washing.
The buffer solutions can be removed by tipping the plate over a sink. Residual buffer can be removed by gently patting the plate on a paper towel or by aspiration. Do not allow the wells to dry out at any time.
Dissolve the tablets in ddH2O 10 minutes prior to use and vortex vigorously.
5. References
- 1.Driggers EM, Hale SP, Lee J et al. (2008) The exploration of macrocycles for drug discovery--an underexploited structural class. Nat Rev Drug Discov 7: 608–624. [DOI] [PubMed] [Google Scholar]
- 2.Robinson JA, Demarco S, Gombert F et al. (2008) The design, structures and therapeutic potential of protein epitope mimetics. Drug Discov Today 13: 944–951. [DOI] [PubMed] [Google Scholar]
- 3.Marsault E, Peterson ML (2011) Macrocycles are great cycles: applications, opportunities, and challenges of synthetic macrocycles in drug discovery. J Med Chem 54: 1961–2004. [DOI] [PubMed] [Google Scholar]
- 4.Fairlie DP, Tyndall JD, Reid RC et al. (2000) Conformational selection of inhibitors and substrates by proteolytic enzymes: implications for drug design and polypeptide processing. J Med Chem 43: 1271–1281. [DOI] [PubMed] [Google Scholar]
- 5.Satoh T, Li S, Friedman TM et al. (1996) Synthetic peptides derived from the fourth domain of CD4 antagonize off function and inhibit T cell activation. Biochem Biophys Res Commun 224: 438–443. [DOI] [PubMed] [Google Scholar]
- 6.Walensky LD, Kung AL, Escher I et al. (2004) Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 305: 1466–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rezai T, Yu B, Millhauser GL et al. (2006) Testing the conformational hypothesis of passive membrane permeability using synthetic cyclic peptide diastereomers. J Am Chem Soc 128: 2510–2511. [DOI] [PubMed] [Google Scholar]
- 8.Gudmundsson OS, Vander Velde DG, Jois SD et al. (1999) The effect of conformation of the acyloxyalkoxy-based cyclic prodrugs of opioid peptides on their membrane permeability. J Pept Res 53: 403–413. [DOI] [PubMed] [Google Scholar]
- 9.Neri D, Lerner RA (2008) DNA-encoded chemical libraries: a selection system based on endowing organic compounds with amplifiable in formation. Annu Rev Biochem 87: 479–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maianti JP, McFedries A, Foda ZH et al. (2014) Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature 511: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam KS, Lebl M, Krchňák V (1997) The “one-bead-one-compound” combinatorial library method. Chem Rev 97: 411–448. [DOI] [PubMed] [Google Scholar]
- 12.Qian Z, Upadhyaya P, Pei D (2015) Synthesis and screening of one-bead-one-compound cyclic peptide libraries. Methods Mol Biol 1248: 39–53. [DOI] [PubMed] [Google Scholar]
- 13.Rhodes CA et al. (2018) Cell-permeable bicyclic peptidyl inhibitors against NEMO- IκB kinase interaction directly from a combinatorial library. J Am Chem Soc 140: 12102–12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trinh TB, Upadhyaya P, Qian Z et al. (2016) Discovery of a direct Ras inhibitor by screening a combinatorial library of cell-permeable bicyclic peptides. ACS Comb Sci 18: 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sidhu SS, Lowman HB, Cunningham BC et al. (2000) Phage display for selection of novel binding peptides. Methods Enzymol 328: 333–363. [DOI] [PubMed] [Google Scholar]
- 16.Clackson T, Wells JA (1995) A hot spot of binding energy in a hormone-receptor interface. Science 267: 383–386. [DOI] [PubMed] [Google Scholar]
- 17.Meyer SC, Shomin CD, Gaj T et al. (2007) Tethering small molecules to a phage display library: discovery of a selective bivalent inhibitor of protein kinase A. J Am Chem Soc 129: 13812–13813. [DOI] [PubMed] [Google Scholar]
- 18.Heinis C, Winter G (2015) Encoded libraries of chemically modified peptides. Curr Opin Chem Biol 26: 89–98. [DOI] [PubMed] [Google Scholar]
- 19.Rebollo IR, Heinis C (2013) Phage selection of bicyclic peptides. Methods 60: 46–54. [DOI] [PubMed] [Google Scholar]
- 20.Josephson K, Ricardo A, Szostak JW et al. (2014) mRNA display: from basic principles to macrocycle drug discovery. Drug Discov Today 19: 388–399. [DOI] [PubMed] [Google Scholar]
- 21.Howell SM, Fiacco SV, Takahashi TT et al. (2014) Serum stable natural peptides designed by mRNA display. Sci Rep 4: 6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nichols AL, Noridomi K, Hughes CR et al. (2018) alpha 1-FANGs: Protein ligands selective for the alpha-bungarotoxin site of the alpha 1-nicotinic acetylcholine receptor. ACS Chem Biol 13: 2568–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White ER, Sun LX, Ma Z et al. (2015) Peptide library approach to uncover phosphomimetic inhibitors of the BRCA1 C-terminal domain. ACS Chem Biol 10: 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nawatha M, Rogers JM, Bonn SM et al. (2019) De novo macrocyclic peptides that specifically modulate Lys48-linked ubiquitin chains. Nature Chem 11: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Passioura T, Katoh T, Goto Y et al. (2014) Selection-based discovery of druglike macrocyclic peptides. Ann Rev Biochem 83: 727–752. [DOI] [PubMed] [Google Scholar]
- 26.Hipolito CJ, Suga H (2012) Ribosomal production and in vitro selection of natural product-like peptidomimetics: The FIT and RaPID systems. Curr Opin Chem Biol 16: 196–203. [DOI] [PubMed] [Google Scholar]
- 27.Kawamura A, Munzel M, Kojima T et al. (2017) Highly selective inhibition of histone demethylases by de novo macrocyclic peptides. Nature Commun 8: 14773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parmley SF, Smith GP (1998) Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene 73:305–318. [DOI] [PubMed] [Google Scholar]
- 29.Wrighton NC, Farrell FX, Chang R et al. (1996) Small peptides as potent mimics of the protein hormone erythropoietin. Science 273(5274): 458–463. [DOI] [PubMed] [Google Scholar]
- 30.Lowman HB, Chen YM, Skelton NJ et al. (1998) Molecular mimics of insulin-like growth factor 1 (IGF-1) for inhibiting IGF-1: IGF-binding protein interactions. Biochemistry 37: 8870–8878. [DOI] [PubMed] [Google Scholar]
- 31.Fairbrother WJ, Christinger HW, Cochran AG et al. (1998) Novel peptides selected to bind vascular endothelial growth factor target the receptor-binding site. Biochemistry 37: 17754–17764. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura GR, Reynolds ME, Chen YM et al. (2002) Stable “zeta” peptides that act as potent antagonists of the high-affinity IgE receptor. Proc Natl Acad Sci USA 99: 1303–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckert DM, Malashkevich VN, Hong LH et al. (1998) Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99: 103–115. [DOI] [PubMed] [Google Scholar]
- 34.DeLano WL, Ultsch MH, de Vos AM et al. (2000) Convergent solutions to binding at a protein-protein interface. Science 287: 1279–1283. [DOI] [PubMed] [Google Scholar]
- 35.Dias RL, Fasan R, Moehle K et al. (2006) Protein ligand design: from phage display to synthetic protein epitope mimetics in human antibody Fc-binding peptidomimetics. J Am Chem Soc 128: 2726–2732. [DOI] [PubMed] [Google Scholar]
- 36.Quartararo JS, Wu P, Kritzer JA (2012) Peptide bicycles that inhibit the Grb2 SH2 domain, Chembiochem 13: 1490–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Angelini A, Heinis C (2011) Post-translational modification of genetically encoded polypeptide libraries. Curr Opin Chem Biol 15: 355–361. [DOI] [PubMed] [Google Scholar]
- 38.Heinis C, Rutherford T, Freund S et al. (2009) Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat Chem Biol 5: 502–507. [DOI] [PubMed] [Google Scholar]
- 39.Angelini A, Cendron L, Chen SY et al. (2012) Bicyclic peptide inhibitor reveals large contact interface with a protease target. ACS Chem Biol 7: 817–821. [DOI] [PubMed] [Google Scholar]
- 40.Baeriswyl V, Calzavarini S, Chen SY et al. (2015) synthetic factor XIIa inhibitor blocks selectively intrinsic coagulation initiation. ACS Chem Biol 10: 1861–1870. [DOI] [PubMed] [Google Scholar]
- 41.Baeriswyl V, Calzavarini S, Gerschheimer C et al. (2013) Development of a selective peptide macrocycle inhibitor of coagulation factor XII toward the generation of a safe antithrombotic therapy. J Med Chem 56: 3742–3746. [DOI] [PubMed] [Google Scholar]
- 42.Ng S, Derda R (2016) Phage-displayed macrocyclic glycopeptide libraries. Org Biomol Chem 14: 5539–5545. [DOI] [PubMed] [Google Scholar]
- 43.Smith JM, Vitali F, Archer SA et al. (2011) Modular assembly of Macrocyclic Organo-Peptide Hybrids using synthetic and genetically encoded precursors. Angew Chem Int Ed Engl 50: 5075–5080. [DOI] [PubMed] [Google Scholar]
- 44.Satyanarayana M, Vitali F, Frost JR et al. (2012) Diverse organo-peptide macrocycles via a fast and catalyst-free oxime/intein-mediated dual ligation. Chemical Commun 48: 1461–1463. [DOI] [PubMed] [Google Scholar]
- 45.Frost JR, Jacob NT, Papa LJ et al. (2015) Ribosomal synthesis of macrocyclic peptides in vitro and in vivo mediated by genetically encoded aminothiol unnatural amino acids. ACS Chem Biol 10: 1805–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bionda N, Cryan AL, Fasan R (2014) Bioinspired strategy for the ribosomal synthesis of thioether-bridged macrocyclic peptides in bacteria. ACS Chem Biol 9: 2008–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bionda N, Fasan R (2015) Ribosomal synthesis of natural product-like bicyclic peptides in Escherichia coli. Chembiochem 16: 2011–2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith JM, Frost JR, Fasan R (2013) Emerging Strategies to Access Peptide Macrocycles from Genetically Encoded Polypeptides. J Org Chem 78(8): 3525–3531. [DOI] [PubMed] [Google Scholar]
- 49.Owens AE, de Paola I, Hansen WA et al. (2017) Design and evolution of a macrocyclic peptide inhibitor of the Sonic Hedgehog/Patched interaction. J Am Chem Soc 139: 12559–12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Owen A, Iannuzzelli J, Gu Y et al. (2020) MOrPH-PhD: An Integrated Phage Display Platform for the Discovery of Functional Genetically-Encoded Peptide Macrocycles. ACS Cent Sci 6: 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pratap J, Dikshit KL (1997) Effect of signal peptide changes on the extracellular processing of streptokinase from Escherichia coli: requirement for secondary structure at the cleavage junction. Mol Gen Genet 258(4): 326–33. [DOI] [PubMed] [Google Scholar]
- 52.Young TS, Ahmad I, Yin JA et al. (2010) An enhanced system for unnatural amino acid mutagenesis in E. coli . J Mol Bio 395: 361–74. [DOI] [PubMed] [Google Scholar]
- 53.Iannuzzelli JA, Fasan R (2020) Expanded toolbox for directing the biosynthesis of macrocyclic peptides in bacterial cells. Chem Sci 11: 6202–6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rondot S, Koch S, Breitling F et al. (2010) A helper phage to improve single-chainantibody presentation in phage display. Nat Biotech 19: 75–78. [DOI] [PubMed] [Google Scholar]
- 55.MacLean D, Jones JDG, Studholme DJ (2009) Application of ‘next-generation’ sequencing technologies to microbial genetics. Nat Rev Microbiol 7: 287–296. [DOI] [PubMed] [Google Scholar]