Abstract
The processing and methylation of precursor rRNA is mediated by the box C/D small nucleolar RNAs (snoRNAs). These snoRNAs differ from most cellular RNAs in that they are not exported to the cytoplasm. Instead, these RNAs are actively retained in the nucleus where they assemble with proteins into mature small nucleolar ribonucleoprotein particles and are targeted to their intranuclear site of action, the nucleolus. In this study, we have identified the cis-acting sequences responsible for the nuclear retention of U3 box C/D snoRNA by analyzing the nucleocytoplasmic distributions of an extensive panel of U3 RNA variants after injection of the RNAs into Xenopus oocyte nuclei. Our data indicate the importance of two conserved sequence motifs in retaining U3 RNA in the nucleus. The first motif is comprised of the conserved box C′ and box D sequences that characterize the box C/D family. The second motif contains conserved box sequences B and C. Either motif is sufficient for nuclear retention, but disruption of both motifs leads to mislocalization of the RNAs to the cytoplasm. Variant RNAs that are not retained also lack 5′ cap hypermethylation and fail to associate with fibrillarin. Furthermore, our results indicate that nuclear retention of U3 RNA does not simply reflect its nucleolar localization. A fragment of U3 containing the box B/C motif is not localized to nucleoli but retained in coiled bodies. Thus, nuclear retention and nucleolar localization are distinct processes with differing sequence requirements.
In all eukaryotic cells, there exist a multitude of RNAs which perform diverse functions at distinct subcellular locations. Since RNAs function at sites in the cell which are remote from where they are synthesized, RNA transport and localization are obligatory steps in gene expression for all eukaryotic cells. Despite the fundamental importance of RNA transport and localization, we have a limited understanding of the cellular mechanisms that ensure that individual RNAs are sorted to the intracellular destinations where they function.
We are investigating the mechanisms controlling the intracellular trafficking of small nucleolar RNAs (snoRNAs). snoRNAs consist of a family of more than 150 molecules that are known to function in the processing and modification of rRNA within the nucleolus (2, 18, 47, 68, 74). The snoRNAs are categorized almost exclusively into two classes based on sequence homology, secondary structure, function, and binding of common proteins. The two major classes of snoRNAs are known as the box C/D snoRNAs and the box H/ACA snoRNAs (3, 16). In contrast to most other cellular RNAs, newly synthesized snoRNAs are not exported from the nucleus to the cytoplasm (52, 71, 73). Rather, these RNAs remain in the nucleus and are selectively targeted from their sites of synthesis within the nucleoplasm (17, 69) to their intranuclear site of function, the nucleolus. At least some snoRNAs associate with nucleoplasmic structures known as coiled bodies (5, 32, 53, 61, 65). Since the association of snoRNAs with coiled bodies is transient and precedes localization of these RNAs to nucleoli (53), coiled bodies likely play a key role in the biogenesis and/or intranuclear transport of snoRNAs.
U3 snoRNA, the focus of this investigation, is the best characterized snoRNA and is a member of the box C/D family of snoRNAs. U3 is required in the nucleolus for several pre-rRNA endonucleolytic cleavage events that selectively lead to the generation of 18S rRNA, a component of the small subunit of ribosomes (13, 25, 33, 50, 62). Phylogenetic analysis of sequences from a wide range of eukaryotic organisms has revealed that all U3 RNAs contain six short sequence elements known as boxes A, A′, B, C, C′, and D (see Fig. 1). Detailed structural studies and functional mapping of U3 RNAs in diverse organisms reveal that many features of U3 RNA structure are conserved (21, 23, 26, 31, 35, 42, 48, 49, 54, 55, 58, 60). A common two-domain secondary structure exists in which a short 5′ domain is linked to a larger 3′ domain via a single-stranded sequence called the hinge region (see Fig. 1). The 5′ domain and hinge region contain sequences with complementarity to pre-rRNA (within box A, box A′, and the hinge) and act as a pre-rRNA binding domain (6, 7, 24, 49, 64). The 3′ domain contains box B, C, C′, and D elements, which serve as conserved protein binding sites (4, 23, 31, 44, 49, 55). Significantly, these four box elements of the 3′ domain of U3 RNA, which are separated in the primary sequence of the RNA, are consistently assembled together in the folded RNA to form the box B/C motif and the box C′/D motif (60). In these motifs, box B and box C, or box C′ and box D, are situated on opposite strands of a loop formed as the result of base-pairing of flanking sequences. The motif comprised of box C′ and box D of U3 is functionally equivalent to the box C/D motif present in all other members of the box C/D family (49, 53, 60). In contrast, the box B/C motif appears to be a unique structural feature of U3 RNA.
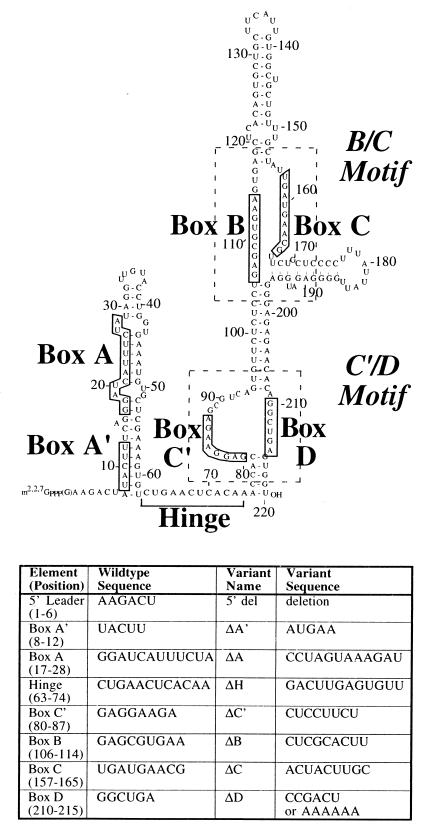
FIG. 1.
Predicted secondary structure of the U3 snoRNA. The 220-nucleotide sequence of X. laevis U3 snoRNA is shown. The boxed nucleotides are phylogenetically conserved sequences. The hinge region connects the 5′ and 3′ domains of U3 and is marked by a square bracket. The general regions of the C′/D and B/C motifs are shown with dashed lines. The table lists the block substitution changes (indicated as Δ) made in the U3 mutants used in this study.
The biogenesis and intracellular transport of U3 RNA have been investigated. The 7-methylguanosine (m7G) 5′ cap structure of newly synthesized U3 RNA is hypermethylated to a 2,2,7-trimethylguanosine (m2,2,7G) cap structure by a nuclear methyl transferase (71, 73). Cellular U3 RNA is complexed with several proteins and is found in 12S to 15S and ∼80S ribonucleoprotein particles (RNPs) which likely correspond to free U3 snoRNPs and U3 snoRNPs associated with pre-rRNA and other nucleolar components, respectively (14, 34, 76). Like all box C/D snoRNAs, U3 associates with three nucleolar proteins known as fibrillarin, Nop56, and Nop58 (4, 37, 41, 63, 78). Additional proteins including p55, p50, p15, Sof1, mpp10, nucleolin, Imp3p, Imp4p, and LCP5 appear to selectively associate with U3 RNA (12, 19, 29, 43, 44, 57, 77). cis-acting sequences required for the nucleolar localization of U3, and other box C/D snoRNAs, have been studied in Xenopus oocytes, mammalian cells, and yeast (38–40, 53, 61). We have found that the box C′/D motif of U3 RNA, including box C′, box D, and a nearby 3′-terminal stem structure, is both necessary and sufficient for the nucleolar localization of U3 RNA (53). Studies with other box C/D snoRNAs support the idea that nucleolar localization of all box C/D snoRNAs is dependent upon the box C/D motif (53, 61).
The mechanism which selectively retains U3 and other snoRNAs within the nucleus has not been extensively characterized. In vivo competition studies performed with Xenopus oocytes have shown that U3 and other box C/D snoRNAs are actively retained in the nucleus by a mechanism which is saturable (73). These studies indicate that snoRNAs are normally prevented from leaving the nucleus because they bind titratable specific RNA binding factors. In the study presented here, we have carried out a detailed mutational analysis of U3 RNA to determine the cis-acting sequences required for nuclear retention of U3 RNA. We have defined the sequences that are necessary and sufficient for the retention of U3 within the nucleus by assaying the nucleocytoplasmic distribution of a large panel of U3 RNA variants following injection into the nucleus. Our results show that the box B/C motif and the box C′/D motif each retain U3 in the nucleus. Furthermore, these two motifs appear to mediate the nuclear retention of U3 RNA through distinct mechanisms.
MATERIALS AND METHODS
Generation of U3 mutant constructs.
Most mutant constructs contained block substitution mutations in which all nucleotides in a conserved box element were replaced as described previously (see Fig. 1) (53). Single box variants of U3 as well as the U3 3′ domain fragment were generated by PCR approaches which are described elsewhere (53). The production of U3 double box variants was accomplished by similar PCR strategies utilizing single box mutant templates to produce the desired double mutation. The construction of the B/C subfragment (nucleotides 93 to 208) was also accomplished by PCR with a wild-type U3 template and primers 2 plus 3 (see below). The U3 subfragment C′/D (nucleotides 1 to 6, 75 to 104, a GCUU tetraloop, and 198 to 220) was constructed with primer 4 as a template and primers 5 plus 6. The U3 5′ deletion mutant was produced with a wild-type U3 template and primers 1 plus 7. All U3 mutant DNA fragments produced were subcloned into the SmaI site of pUC19 and sequenced. All PCRs were performed with Pfu DNA polymerase (Stratagene) as the enzyme and an annealing temperature of 52°C. The deoxyoligonucleotide primers used to prepare the mutated Xenopus U3 templates are as follows, with SP6 or T7 promoter sequences underlined: 1, ACCACTCAGCCTGTGTTCTCTCCCTCC; 2, GATTTAGGTGACACTATAGAGTGTTCTCTCCTGAGCG; 3, GTGTTCTCTCCCTCCATCTCC; 4, TAATACGACTCACTATAGGGAAGACTACCACGAGGAAGAGCGTCAGTGTTCTCTCCTTCGGGAGAGAACACAGGCTGAGTGGT; 5, CACGGATCCTAATACGACTCACTATAGGG; 6, ACCACTCAGCCTGTGTTC; 7, GATTTAGGTGACACTATAGATACTTTCAGGGATCA.
In vitro transcription.
Linearized plasmids or PCR products were utilized as transcription templates. The reaction conditions used to generate m7G-capped, 32P-labeled RNAs by SP6 or T7 RNA polymerase were carried out essentially as previously described (72). Fluorescein-labeled U3 variants were generated by in vitro transcription with an equal mixture (250 μM each) of UTP and fluorescein-12-UTP (Boehringer Mannheim) as described elsewhere (53). Control RNAs, as follows, were prepared by in vitro transcription as previously described: Xenopus U8 (56), Xenopus U1, U1sm−, and U6 (72).
Injection of RNAs into Xenopus oocytes.
A detailed description of the procedure by which we microinjected and micromanipulated oocytes has been reported previously (53, 70). In brief, the nuclei of stage V and VI Xenopus laevis oocytes were injected with 10 nl of a 20-mg/ml blue dextran solution which contained 1 fmol of each 32P-labeled RNA (fluorescein-labeled RNA experiments were done with 5 fmol of RNA/10 nl). After injection, oocytes were maintained in MBSH buffer at 18°C until being manually dissected in J buffer (8) into nuclear and cytoplasmic fractions. The RNAs were isolated from two to four dissected nuclear and cytoplasmic fractions by proteinase K digestion, phenol extraction, and ethanol precipitation. One nuclear or one cytoplasmic equivalent was then resolved on an 8% denaturing polyacrylamide gel and detected by autoradiography.
Cytoplasmic injections were accomplished with 10 nl of solution containing 1 fmol of each 32P-labeled RNA. The injections were done at the equator between the animal and vegetal poles of the oocyte. Isolation of the RNAs after injection was performed as described above.
Immunoprecipitations.
Polyclonal antibodies directed against either the m7G (51) or m2,2,7G (10) cap and a monoclonal antibody (72B9) directed against fibrillarin (59) were utilized as previously described (53), except that Net-2 buffer (150 mM NaCl, 50 mM Tris HCl [pH 7.5], and 0.05% NP-40) was used for fibrillarin immunoprecipitations.
Nuclear spreads, immunofluorescence, and microscopy.
Preparation of nuclear spreads and image collection were performed as previously described (53). Indirect immunofluorescence with an antibody against p80 coilin was performed as previously described (53).
RESULTS
Nuclear retention of U3 snoRNA is mediated by both the box B/C and box C′/D motifs.
U3 RNA injected into Xenopus oocyte nuclei was previously shown to be stable and retained within the nucleus (71, 73). To identify the cis-acting sequences within U3 RNA necessary for its nuclear retention, we injected an extensive panel of U3 RNA sequence variants into Xenopus oocyte nuclei. In these experiments, we hoped to identify nuclear retention elements as sequences within U3 RNA that when disrupted by mutation would lead to mislocalization of U3 RNA from the nucleus to the cytoplasm.
To test the idea that one of the phylogenetically conserved box elements of U3 was required for nuclear retention, we introduced block substitution mutations into the A, A′, B, C, C′, and D box elements. In each case, every nucleotide in the box element was replaced by its Watson-Crick complement (Fig. 1) (see Materials and Methods). Similar base substitutions were introduced into the hinge region of U3 RNA (Fig. 1). Although the hinge region is not conserved in primary sequence, it plays an important functional role in rRNA processing (6, 49). In addition, the first six nucleotides of U3 were deleted to determine if this single-stranded 5′ leader sequence was essential for nuclear retention.
32P-labeled, m7G-capped U3 RNA variants were synthesized by in vitro transcription and coinjected into oocyte nuclei with U1sm−, U8, and U6 control RNAs. The control RNAs were utilized to show that cellular transport was functioning (U1sm− is exported to the cytoplasm) and that the nuclear injections and dissections were precise (U6 and U8 RNAs are normally retained in the nucleus). The use of U8, which is also a member of the box C/D family of snoRNAs, allowed us to determine if changes observed with U3 variants were specific to those molecules and not due to a global affect on box C/D snoRNA family members. After injection, the oocytes were manually dissected into nuclear and cytoplasmic fractions at specific times after injection (1, 4, and 8 h). The RNAs in each fraction were purified and analyzed by electrophoresis on denaturing polyacrylamide gels, followed by autoradiography.
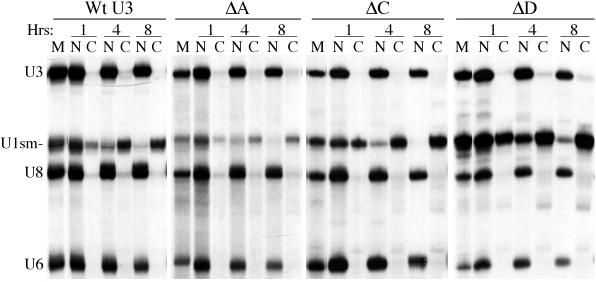
We found that all of the single box substitution mutants, the 5′ deletion mutant, and the hinge mutant were retained in the oocyte nucleus at all time points analyzed, like wild-type U3 RNA (Fig. 2 and Table 1). The control RNAs were exported (U1sm−) or retained (U6 and U8) as expected in every experiment, and all U3 RNA variants were sufficiently stable to allow analysis (Fig. 2 and data not shown). Thus, none of the tested sequence elements is individually essential for retention of U3 in the nucleus. These results suggest that the nuclear retention of U3 is not dependent upon any single, dominantly acting nuclear retention element.
FIG. 2.
Nuclear retention of wild-type U3 and single box mutants of U3. 32P-labeled U3, U3 variant, U1sm−, U8, and U6 RNAs were synthesized by in vitro transcription. A mixture of U3 or a U3 mutant and control RNAs (U1sm−, U8, and U6) was injected into the nuclei of X. laevis oocytes. After 1, 4, and 8 h, the radiolabeled RNAs present in the nuclear (N) and cytoplasmic (C) fractions of the oocytes were isolated and analyzed by electrophoresis in a denaturing polyacrylamide gel. Autoradiography shows the nucleocytoplasmic distribution of the RNAs after incubation. Marker lanes (M) show the RNAs prior to injection.
TABLE 1.
Summary of nucleocytoplasmic distributions of U3 variantsa
| U3 RNA | Retention in nucleus |
|---|---|
| Wild type | Yes |
| Single variants | |
| 5′ deletion | Yes |
| ΔA′ | Yes |
| ΔA | Yes |
| ΔH | Yes |
| ΔC′ | Yes |
| ΔB | Yes |
| ΔC | Yes |
| ΔD | Yes |
| Double variants | |
| ΔA′B | Yes |
| ΔA′C | Yes |
| ΔAH | Yes |
| ΔAC′ | Yes |
| ΔAC | Yes |
| ΔAD | Yes |
| ΔHC | Yes |
| ΔHD | Yes |
| ΔC′B | No |
| ΔC′C | No |
| ΔC′D | Yes |
| ΔBC | Yes |
| ΔBD | No |
| ΔCD | No |
The distribution of each RNA was analyzed over an 8-h time course (as described for Fig. 2).
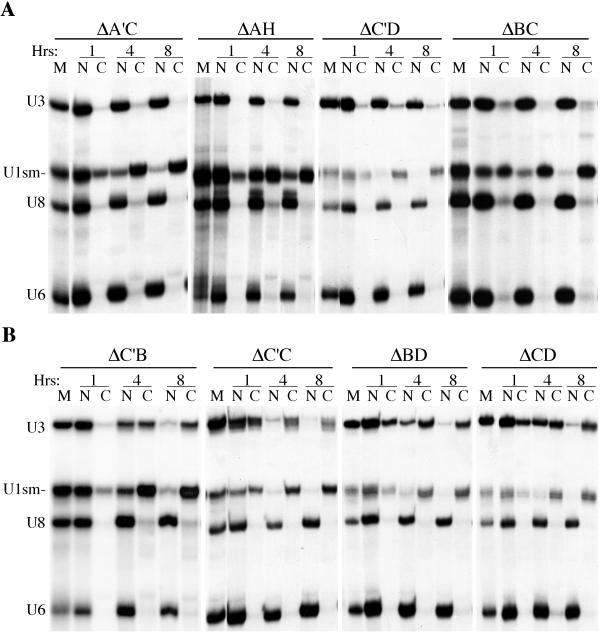
The finding that mutation of no single element affected nuclear retention of U3 prompted us to analyze double mutants in which combinations of two conserved elements were altered (Fig. 3). The majority of the double mutants were retained in the oocyte nucleus (Fig. 3A and Table 1). In contrast, a loss of nuclear retention was observed with a specific subset of U3 double mutants (ΔC′B, ΔC′C, ΔBD, and ΔCD [Fig. 3B and Table 1]). These double mutants accumulated in the cytoplasm in a time-dependent manner following nuclear injection and were exported out of the nucleus at a rate comparable to that of U1sm− RNA (Fig. 3B). Eight hours following injection, essentially all of the mutant U3 RNAs were present in the cytoplasm.
FIG. 3.
Nucleocytoplasmic distribution of U3 double box variants. RNAs were prepared, injected, and analyzed as described in the legend to Fig. 2. Nuclear (N) and cytoplasmic (C) distribution of RNAs at 1, 4, and 8 h after nuclear injection are shown. Marker lanes (M) show the RNAs prior to injection. (A) Double box variants of U3 that are retained in the nucleus. (B) The four double box variants that are not retained in the nucleus.
Loss of nuclear retention was observed only with those double mutants involving elements confined to the 3′ domain of U3 (boxes B, C, C′, and D [Fig. 1]). Furthermore, only particular combinations of the double mutations in the 3′ domain box elements disrupted nuclear retention. The results indicate that both the box B/C and box C′/D motifs are important for nuclear retention and that each motif is independently capable of supporting the nuclear retention of U3 RNA. Nuclear retention was not disrupted when both sequence elements of either the box B/C or the box C′/D motif were mutated (ΔBC and ΔC′D, respectively [Fig. 3A]). However, simultaneous mutation of one box element from each motif (i.e., ΔC′B, ΔC′C, ΔBD, and ΔCD) results in loss of nuclear retention of that molecule (Fig. 3B). The results indicate that box B and box C, as well as box C′ and box D, function as pairs and that the structurally conserved (see the introduction) box B/C and box C′/D motifs function separately to retain U3 RNA in the nucleus.
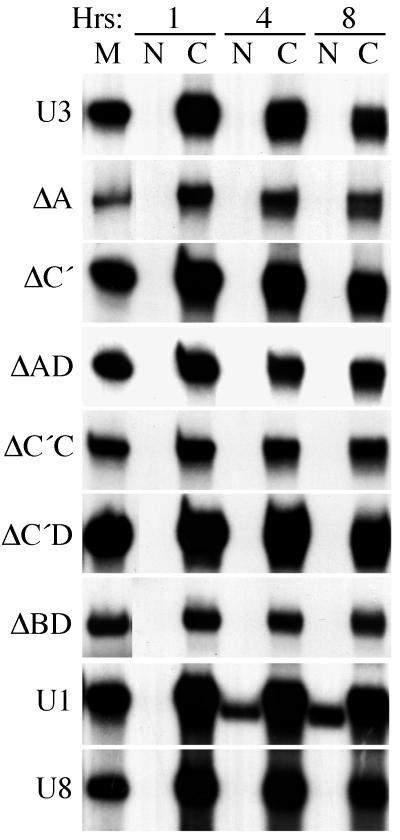
U3 RNA variants are stable and remain in the cytoplasm following injection into the cytoplasm.
While wild-type U3 RNA is normally retained within the nucleus (71, 73), control experiments were performed to test for nuclear import of the U3 variants upon direct injection of the RNAs into the cytoplasmic compartment (Fig. 4). RNA mixtures containing U3, or a U3 variant, and control RNAs (U1 and U8) were injected into the cytoplasm of Xenopus oocytes. The nucleocytoplasmic distributions of the RNAs were determined at 1, 4, and 8 h after injection. Neither the wild-type U3 or U8 RNAs nor any of the variant U3 RNAs exhibited any significant import to the nucleus after 8 and even 24 h (Fig. 4 and data not shown). Importantly, the coinjected U1 control RNA was imported into the nucleus in a time-dependent manner, as expected (Fig. 4). The same results were observed multiple times with oocytes from many different frogs. Furthermore, the variant U3 RNAs were stable in the cytoplasm (Fig. 4 and data not shown). Since the RNAs are stable and not imported from the cytoplasm, any U3 variant RNA exported from the nucleus should have been observed in the cytoplasm. These control experiments indicate that the variant U3 RNAs that appeared to be retained were retained in the nucleus, not exported and rapidly reimported. Finally, any decrease in stability of any of the variant U3 RNAs relative to the wild type (e.g., ΔD [Fig. 2] and ΔC′D [Fig. 3]) was due to degradation within the nucleus, not degradation in the cytoplasm following export.
FIG. 4.
Absence of detectable nuclear import of U3, U3 mutants, or U8 following injection into oocyte cytoplasms. Mixtures of U3 or a U3 variant and control RNAs (U1 and U8) were injected into the cytoplasm of Xenopus oocytes. After 1, 4, and 8 h, the nuclear and cytoplasmic distributions of RNAs were determined as described for the previous figures. The nucleocytoplasmic distributions of U3 and each of several U3 mutants are shown. One example each of the typical nucleocytoplasmic distributions of U1 and U8 from these experiments are also shown.
Export of U3 RNA variants is affected by temperature and the 5′ cap structure.
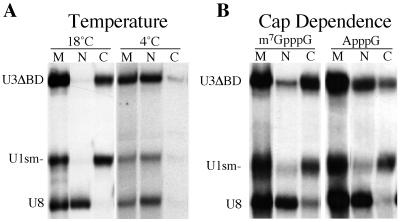
For those U3 RNA variants that failed to be retained in the nucleus, we sought to better understand the mechanism by which they were transported out of the nucleus (Fig. 5). Low temperatures generally inhibit active, energy-dependent transport processes in cells. To determine if the nuclear export of the U3 variants is a temperature-dependent process, we examined the nucleocytoplasmic distribution of the injected RNAs at reduced temperatures (Fig. 5A). We injected a variant U3 RNA that failed to be retained (ΔBD) and control RNAs into oocytes that were preincubated and maintained at 4°C or at the normal temperature of 18°C. Incubation at 4°C resulted in a dramatic reduction in the nuclear export of ΔBD and U1sm− RNAs relative to incubation at 18°C. Thus, nuclear export of the U3 mutants is a temperature-dependent process.
FIG. 5.
Temperature and cap dependence of nuclear export of U3 mutants. (A) 32P-labeled U3ΔBD and control RNAs (U1sm− and U8) were injected into the nuclei of Xenopus oocytes. The oocytes were incubated for 8 h at 4 or 18°C. After incubation, the radiolabeled RNAs present in the nuclear (N) and cytoplasmic (C) fractions of the oocytes were isolated and analyzed. (B) 32P-labeled U3ΔBD with either an m7G or ApppG cap structure and control RNAs (U1sm− and U8) were injected into the nucleus. After 8 h of incubation, the radiolabeled RNAs present in the nuclear and cytoplasmic fractions of the oocytes were isolated and analyzed. Marker lanes (M) show the RNAs prior to injection.
Another indication that the U3 RNA variants do not simply passively diffuse out of the nucleus was the finding that the export rate of the RNA is influenced by the 5′ cap structure. It is known that an m7G cap structure is an important determinant for RNA export (22, 27, 28, 72). We tested the effect of the m7G cap structure of U3 on the rate of export of mutant U3 snoRNAs that failed to be retained by examining the transport of ApppG-capped U3ΔBD. The U3ΔBD variant RNA with the ApppG cap structure was exported from the nucleus but at a reduced rate relative to the m7G-capped RNA (Fig. 5B and data not shown). Thus, as has been observed for other RNAs (22, 72), the m7G cap enhances the rate of export of the U3 RNA variant but it is not essential for the transport.
Variant U3 RNAs that fail to be retained in the nucleus also fail to associate with fibrillarin and to undergo 5′ cap hypermethylation.
Like all members of the box C/D family of snoRNAs, U3 RNA associates with the nucleolar protein fibrillarin (4). In addition, U3 snoRNA is synthesized with an m7G cap which is hypermethylated within the nucleus to an m2,2,7G cap structure (71, 73). We tested whether the variant U3 RNAs that failed to be retained in the nucleus were able to associate with fibrillarin and to undergo 5′ cap hypermethylation prior to exiting the nucleus (Fig. 6).
FIG. 6.
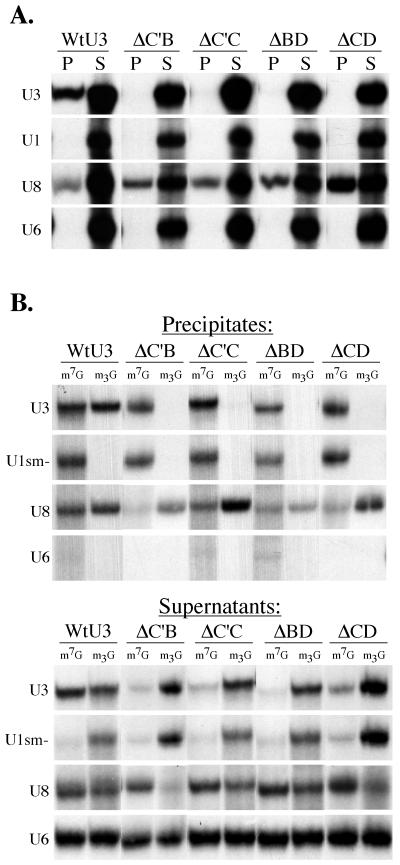
Exported U3 mutants fail to bind fibrillarin and fail to undergo 5′ cap hypermethylation. (A) Immunoprecipitations with antifibrillarin antibodies (72B9) were performed on nuclear extracts prepared 3 h after injection of RNAs into oocyte nuclei. RNAs present in the precipitate (P) and 20% of the supernatant (S) were analyzed by gel electrophoresis followed by autoradiography. (B) The RNAs present in the nucleus 3 h after injection were precipitated with anti-m7G or anti-m2,2,7G cap (m3G) antibodies, as indicated. RNAs present in the precipitates are shown in the top panel, and those in the supernatants are shown in the bottom panel.
To determine if the U3 RNA variants associated with fibrillarin, we assayed whether these RNAs could be coimmunoprecipitated by using a monoclonal antibody (72B9) against fibrillarin (59) (Fig. 6A). Radiolabeled RNAs were injected into oocyte nuclei, and nuclear extracts were prepared 3 h after injection (when approximately 50% of exported variant U3 RNA was still present in the nucleus). As expected, the box C/D snoRNAs, U3 and U8, were coimmunoprecipitated by the antifibrillarin antibodies while U1 and U6 were not (Fig. 6A). Each of the U3 variants that exhibited a loss of nuclear retention (ΔC′B, ΔC′C, ΔBD, and ΔCD) failed to be coimmunoprecipitated (Fig. 6A), indicating that these RNAs had lost the ability to associate with the common box C/D snoRNA binding protein fibrillarin.
To test for 5′ cap hypermethylation of the variant U3 RNAs, nuclear RNAs were isolated 3 h after injection and subsequently immunoprecipitated with antibodies that specifically recognize either the m7G cap (51) or the m2,2,7G cap (10) (Fig. 6B). Each experiment included the use of coinjected control RNAs U1sm−, U8, and U6. As expected, the hypermethylation-defective U1sm− RNA was recognized only by the m7G antibody (46, 72), and U6 snRNA was not recognized by either antibody since it has a methyl-pppG cap structure (67). The m7G cap of wild-type U3 snoRNA showed extensive hypermethylation following a 3-h nuclear incubation. However, the U3 RNA variants that are not retained were not appreciably hypermethylated. Importantly, coinjected m7G-capped U8 RNA was hypermethylated in every experiment. In summary, our results show that the U3 RNA variants that have lost the ability to be retained in the nucleus are not hypermethylated and do not associate with the snoRNA binding protein fibrillarin.
Both the box B/C motif and the box C′/D motif are sufficient for nuclear retention of U3 RNA fragments, but the RNAs localize to different intranuclear structures.
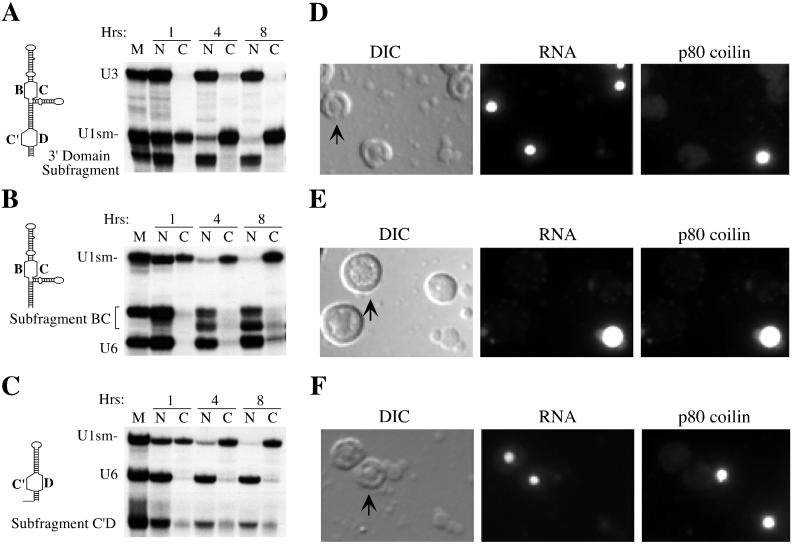
To further characterize the sequence elements responsible for nuclear retention, we tested fragments of U3 RNA for the ability to remain in the nucleus following nuclear injection (Fig. 7). Deletion of the entire 5′ domain and hinge region (nucleotides 1 to 75) did not affect the ability of the RNA to be retained in the nucleus (Fig. 7A), indicating that the 3′ domain of U3 RNA is sufficient for retention in the nucleus. Our mutational analysis indicated that both the box B/C and box C′/D motifs in the 3′ domain functioned in nuclear retention (Fig. 3 and Table 1). Subfragments of the 3′ domain were then tested to determine if the box B/C motif and/or the box C′/D motif were sufficient for nuclear retention. A fragment consisting of box B, box C, and the flanking stem regions was retained within the nucleus (Fig. 7B). This RNA underwent 3′ trimming in the oocyte to produce a smaller, stable RNA species that can be observed as a faster-migrating RNA (Fig. 7B) with an intact 5′ end, as assayed by immunoprecipitation with m7G cap antibodies (data not shown). A fragment containing box C′, box D, and flanking stems demonstrated reduced stability over time but was retained within the nucleus (Fig. 7C). As described for other variant U3 RNAs (Fig. 4), these subfragments were stable and remained in the cytoplasm when injected into the cytoplasm (data not shown). Thus, the box B/C motif and the box C′/D motif are each sufficient for nuclear retention.
FIG. 7.
Fragments of U3 that are sufficient for nuclear retention are not all localized to nucleoli. The diagrams on the left represent the RNA fragments injected (see Materials and Methods for sequences). The nucleocytoplasmic distributions of the 3′ domain fragment (A), the B/C subfragment (B), and the C′/D subfragment (C) are shown next to the corresponding diagrams. The adjacent panels on the right show nuclear spreads prepared 4 h after injection of fluorescein-labeled 3′ domain fragment (D), B/C subfragment (E), and C′/D subfragment (F) RNAs. Each set of three images includes differential interference contrast (DIC), the fluorescein-labeled RNAs (RNA), and indirect immunofluorescence with antibody H1 directed against the coiled-body marker protein p80 coilin (75a) from the same field. The arrow in each DIC image points to a representative nucleolus.
We also examined the localization of the U3 fragments within the nucleus. We assayed the localization of injected, fluorescently labeled RNAs in Xenopus oocyte nuclear spreads (Fig. 7). Briefly, this technique involves centrifuging the contents of manually isolated oocyte nuclei onto microscope slides and determining the localizations of the fluorescent RNAs relative to nuclear structures such as nucleoli, chromosomes, and coiled bodies by fluorescence and light microscopy. We have shown previously that labeling of U3 RNA with fluorescein does not affect properties of the RNA, including cap hypermethylation, fibrillarin binding, nuclear retention, or nucleolar localization (52, 53). We also found that U3 RNA (and other box C/D snoRNAs) transiently localizes to coiled bodies prior to nucleoli (53). Coiled bodies are conserved nuclear structures of uncertain function (15, 45) that can be readily identified with antibodies against the coiled-body marker protein, p80 coilin (1) (Fig. 7D to F).
The 3′ domain fragment of U3 and the subfragment containing the box C′/D motif were both targeted specifically to nucleoli (Fig. 7D and F, respectively). Their transport to nucleoli is preceded by localization to coiled bodies at early time points (15 min [data not shown]), as we observed previously for full length U3 (53). In contrast, the subfragment containing the box B/C motif fails to localize to nucleoli but is clearly retained in coiled bodies (Fig. 7E). Full-length U3 RNAs in which the box C′/D motif is disrupted but the box B/C motif remains intact (ΔC′ or ΔD) are also retained in the nucleus (Fig. 3A) and not targeted to nucleoli but retained in coiled bodies (53). These results demonstrate that nucleolar localization is not necessary for nuclear retention of U3 RNA.
DISCUSSION
We have found that U3 snoRNA contains two independent structural motifs that each contribute to nuclear retention of the molecule (see Fig. 8). Both the box B/C motif and the box C′/D motif are capable of retaining U3 RNA within the nucleus; disruption of one motif does not prevent nuclear retention by the other (Fig. 3A). Furthermore, U3 RNA fragments consisting of either motif are sufficient for nuclear retention (Fig. 7). However, disruption of both motifs results in loss of the RNA from the nucleus (Fig. 3B). The entire 5′ domain and hinge region of U3, although required for U3 interaction with pre-rRNA (6, 24, 49), is dispensable for nuclear retention of the molecule (Fig. 2 and 7 and Table 1).
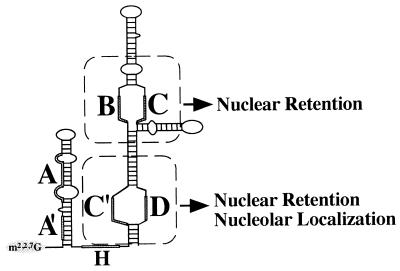
FIG. 8.
Model of U3 snoRNA depicting the positions and roles of the box B/C and box C′/D motifs in nuclear retention and nucleolar localization.
While either the box B/C motif or the box C′/D motif can function to retain U3 RNA in the nucleus, each retention motif may mediate nuclear retention via a distinct mechanism. The box C′/D motif of U3 RNA has been shown to be both necessary and sufficient for nucleolar localization of the RNA (53); the box B/C motif is neither necessary (53) nor sufficient (Fig. 7) for nucleolar targeting. Nuclear retention may be accomplished by sequestering RNA in the nucleolus in the case of the box C′/D motif, but the box B/C motif retains RNA within the nucleus without targeting it to the nucleolus (Fig. 8). Interestingly, the box B/C (and the box C′/D) subfragment associates with coiled bodies (Fig. 7E), suggesting that coiled bodies may be important for retention. On the other hand, the U3 variants that are not retained (e.g., ΔC′B, ΔC′C, ΔBD, and ΔCD) also transiently associate with coiled bodies prior to export (our unpublished data), indicating that association with coiled bodies is not sufficient for nuclear retention.
Our finding that U3 RNA contains two nuclear retention motifs, one specific to U3 (B/C) and the other shared among box C/D snoRNA family members (C′/D), sheds new light on results obtained in previous studies aimed at understanding the nuclear retention of U3 RNA. Previous studies with Xenopus oocytes revealed that box D was essential for nuclear retention of U8 snoRNA but not for U3 (73). It is clear from the present study that box D also functions in the retention of U3 RNA in the nucleus but that mutation of box D is insufficient to disrupt nuclear retention because the box B/C nuclear retention motif remains functional. Furthermore, previous in vivo competition experiments showed that high levels of U3 RNA effectively compete for nuclear retention of both the U3 and U8 snoRNAs. In contrast, U8 RNA was an effective competitor for its own retention but was not able to compete for the nuclear retention of U3 RNA (73). Taken together with the results of this study, these observations indicate that nuclear retention of U3 RNA depends upon binding of both trans-acting factors that are common to U3 and U8 snoRNAs (at the box C/D motif) and factors specific to U3 RNA (at the box B/C motif). Thus, the box C/D motif is very likely of general importance for the nuclear retention (and nucleolar localization) of all box C/D snoRNA family members. It will be interesting to determine whether the related box C′/D′ motif, found in addition to the box C/D motif in the majority of box C/D snoRNAs other than U3 and U8 (36), also functions as a nuclear retention signal. There is evidence that redundant nuclear retention signals may also exist in the spliceosomal snRNA U6 (9).
We observed a correlation between the loss of nuclear retention of four variants of U3 RNA and an inability to undergo normal 5′ cap hypermethylation and to bind fibrillarin (Fig. 6). However, several observations indicate that neither cap hypermethylation nor fibrillarin binding alone mediate nuclear retention of U3 RNA. For example, it was previously demonstrated that U3 RNA remains in the nucleus irrespective of the nature of its 5′ cap structure (73). Furthermore, mutation of the box D element of U3 prevents hypermethylation but does not affect nuclear retention (73) (Fig. 2). Also, mutant U3 RNAs that fail to interact with fibrillarin (4) are nevertheless retained within the nucleus (ΔC [Fig. 2]). Finally, genetic depletion of fibrillarin in yeast cells does not prevent nucleolar localization of snoRNAs, including U3 (75). It is possible that hypermethylation and fibrillarin binding together mediate the retention of U3 or that the failure of these U3 RNA variants to be hypermethylated and to bind fibrillarin is a secondary defect.
The box B/C and the box C′/D motif are likely highly conserved among U3 RNAs from diverse organisms despite functional redundancy in nuclear retention because the motifs provide other, nonredundant functions. The box C′/D motif but not the box B/C motif is required for the stability, hypermethylation, and nucleolar localization of U3 (49, 53, 60, 73). Furthermore, both motifs are important for the function of U3 in rRNA processing (49, 60). The box C′/D motif likely binds common box C/D family snoRNA binding proteins (see above) such as Nop56 and Nop58 (37, 78). Several proteins that appear to selectively associate with U3 snoRNA have been identified, and among these may be box B/C motif binding factors. We have found that the U3-specific protein p55 (44, 57) specifically interacts with sequence elements in the box B/C motif of U3 in vivo (44a), suggesting that p55 may play a role in nuclear retention of U3 snoRNA.
While U3 RNA is normally not exported to the cytoplasm in oocytes or somatic cells (11, 71, 73), it has been reported that a fraction of U3 RNA may leave the nucleus during serum starvation in certain cell types (20, 66). It was of interest to characterize the manner in which the exported mutant U3 RNAs left the nucleus. Several lines of evidence lead us to suggest that upon disruption of nuclear retention, the U3 variants access a cellular export pathway that normally exists to transport spliceosomal snRNAs. First, the export of both the mutant U3 RNAs and snRNAs is temperature dependent, indicating an active rather than passive transport process (Fig. 5A). Second, different classes of RNAs are transported from the oocyte nucleus with distinct kinetics (30, 72), and the rate at which the variant U3 RNAs were exported was nearly identical to that of the control U1 snRNA (Fig. 3B and 5A and data not shown). Finally, as has been observed with U1 snRNA (72), replacement of the 5′ m7G cap with the nonphysiological cap structure ApppG greatly reduces the rate of variant U3 RNA export (Fig. 5B). The presence of U3 in the cytoplasm under special circumstances raises the interesting possibility that nuclear retention of U3 RNA may be a regulated process. Altering the functional pool of U3 RNAs within the nucleus may provide a way to regulate ribosome production in response to extracellular signals.
ACKNOWLEDGMENTS
We kindly thank Reinhard Lührmann (m2,2,7G cap), Elsebet Lund (m7G cap), Joseph Gall (p80 coilin monoclonal antibody H1), and Michael Pollard and Eng Tan (fibrillarin monoclonal antibody 72B9) for providing antibodies used in this study. We are grateful to Claiborne V. C. Glover III and members of our laboratory for critical reading of the manuscript and to James Griffith, Thomas Eades, and Ellie Kalwerisky for assistance in generating many of the mutant templates used in this work.
This work was supported in part by a Basil O’Conner Starter Scholar Research Award from the March of Dimes Birth Defects Foundation and by a grant from the National Institutes of Health (GM54682) to M.P.T.
REFERENCES
- 1.Andrade L, Chan E, Raska I, Peebles C, Roos G, Tan E. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachellerie J, Michot B, Nicoloso M, Balakin A, Ni J, Fournier M. Antisense snoRNAs: a family of nucleolar RNAs with long complementarities to rRNA. Trends Biochem Sci. 1995;20:261–264. doi: 10.1016/s0968-0004(00)89039-8. [DOI] [PubMed] [Google Scholar]
- 3.Balakin A, Smith L, Fournier M. The RNA world of the nucleolus: two major families of small RNAs defined by different box elements with related functions. Cell. 1996;86:823–834. doi: 10.1016/s0092-8674(00)80156-7. [DOI] [PubMed] [Google Scholar]
- 4.Baserga S J, Yang X D, Steitz J A. An intact Box C sequence in the U3 snRNA is required for binding of fibrillarin, the protein common to the major family of nucleolar snRNPs. EMBO J. 1991;10:2645–2651. doi: 10.1002/j.1460-2075.1991.tb07807.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauer D, Murphy C, Wu Z, Wu C, Gall J. In vitro assembly of coiled bodies in Xenopus egg extract. Mol Biol Cell. 1994;5:633–644. doi: 10.1091/mbc.5.6.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beltrame M, Tollervey D. Mutational analysis of an essential binding site for the U3 snoRNA in the 5′ external transcribed spacer of yeast pre-rRNA. Nucleic Acids Res. 1994;22:5139–5147. doi: 10.1093/nar/22.23.5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkenmeier E H, Brown D D, Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978;15:1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- 9.Boelens W C, Palacios I, Mattaj I W. Nuclear retention of RNA as a mechanism for localization. RNA. 1995;1:273–283. [PMC free article] [PubMed] [Google Scholar]
- 10.Bringmann P, Rinke J, Appel B, Reuter R, Luhrmann R. Purification of snRNPs U1, U2, U4, U5, and U6 with 2,2,7-trimethylguanosine-specific antibody and definition of their constituent proteins reacting with anti-Sm and anti-(U1) RNP antisera. EMBO J. 1983;2:1129–1135. doi: 10.1002/j.1460-2075.1983.tb01557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng Y, Dahlberg J E, Lund E. Diverse effects of the guanine nucleotide exchange factor RCC1 on RNA transport. Science. 1995;267:1807–1810. doi: 10.1126/science.7534442. [DOI] [PubMed] [Google Scholar]
- 12.Dunbar D, Wormsley S, Agentis T, Baserga S. Mpp10p, a U3 small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. Mol Cell Biol. 1997;17:5803–5812. doi: 10.1128/mcb.17.10.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enright C, Maxwell E, Sollner-Webb B. 5′ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3. RNA. 1996;2:1094–1099. [PMC free article] [PubMed] [Google Scholar]
- 14.Epstein P, Reddy R, Busch H. Multiple states of U3 RNA in Novikoff hepatoma nucleoli. Biochemistry. 1984;23:5421–5425. doi: 10.1021/bi00318a007. [DOI] [PubMed] [Google Scholar]
- 15.Gall J, Tsvetkov A, Wu Z, Murphy C. Is the sphere organelle/coiled body a universal nuclear component? Dev Genet. 1995;16:25–35. doi: 10.1002/dvg.1020160107. [DOI] [PubMed] [Google Scholar]
- 16.Ganot P, Caizergues F M, Kiss T. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev. 1997;11:941–956. doi: 10.1101/gad.11.7.941. [DOI] [PubMed] [Google Scholar]
- 17.Gao L, Frey M, Matera A. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res. 1997;25:4740–4747. doi: 10.1093/nar/25.23.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerbi S. Small nucleolar RNA. Biochem Cell Biol. 1995;73:845–858. doi: 10.1139/o95-092. [DOI] [PubMed] [Google Scholar]
- 19.Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–1486. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glibetic M, Larson D E, Sienna N, Bachellerie J P, Sells B H. Regulation of U3 snRNA expression during myoblast differentiation. Exp Cell Res. 1992;202:183–189. doi: 10.1016/0014-4827(92)90418-8. [DOI] [PubMed] [Google Scholar]
- 21.Greenwood S J, Schnare M N, Gray M W. Molecular characterization of U3 small nucleolar RNA from the early diverging protist, Euglena gracilis. Curr Genet. 1996;30:338–346. doi: 10.1007/s002940050142. [DOI] [PubMed] [Google Scholar]
- 22.Hamm J, Mattaj I W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 23.Hartshorne T, Agabian N. A common core structure for U3 small nucleolar RNAs. Nucleic Acids Res. 1994;22:3354–3364. doi: 10.1093/nar/22.16.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes J. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 25.Hughes J, Ares M J. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes J M, Konings D A, Cesareni G. The yeast homologue of U3 snRNA. EMBO J. 1987;6:2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izaurralde E, McGuigan C, Mattaj I W. Nuclear localization of a cap-binding protein complex. Cold Spring Harbor Symp Quant Biol. 1995;60:669–675. doi: 10.1101/sqb.1995.060.01.072. [DOI] [PubMed] [Google Scholar]
- 28.Izaurralde E, Stepinski J, Darzynkiewicz E, Mattaj I. A cap binding protein that may mediate nuclear export of RNA polymerase II-transcribed RNAs. J Cell Biol. 1992;118:1287–1295. doi: 10.1083/jcb.118.6.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jansen R, Tollervey D, Hurt E C. A U3 snoRNP protein with homology to splicing factor PRP4 and G beta domains is required for ribosomal RNA processing. EMBO J. 1993;12:2549–2558. doi: 10.1002/j.1460-2075.1993.tb05910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jarmolowski A, Boelens W, Izaurralde E, Mattaj I. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeppesen C, Stebbins B B, Gerbi S. Nucleotide sequence determination and secondary structure of Xenopus U3 snRNA. Nucleic Acids Res. 1988;16:2127–2148. doi: 10.1093/nar/16.5.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jimenez-Garcia L, Segura-Valdez M, Ochs R, Rothblum L, Hannan R, Spector D. Nucleologenesis: U3 snRNA-containing prenucleolar bodies move to sites of active pre-rRNA transcription after mitosis. Mol Biol Cell. 1994;5:955–966. doi: 10.1091/mbc.5.9.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kass S, Tyc K, Steitz J A, Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- 34.Kiss T, Marshallsay C, Filipowicz W. Alteration of the RNA polymerase specificity of U3 snRNA genes during evolution and in vitro. Cell. 1991;65:517–526. doi: 10.1016/0092-8674(91)90469-f. [DOI] [PubMed] [Google Scholar]
- 35.Kiss T, Solymosy F. Molecular analysis of a U3 RNA gene locus in tomato: transcription signals, the coding region, expression in transgenic tobacco plants and tandemly repeated pseudogenes. Nucleic Acids Res. 1990;18:1941–1949. doi: 10.1093/nar/18.8.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiss-Laszlo Z, Henry Y, Kiss T. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 1998;17:797–807. doi: 10.1093/emboj/17.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafontaine D L J, Tollervey D. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA. 1999;5:455–467. doi: 10.1017/s135583829998192x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lange T, Borovjagin A, Gerbi S. Nucleolar localization elements in U8 snoRNA differ from sequences required for rRNA processing. RNA. 1998;4:789–800. doi: 10.1017/s1355838298980438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lange T, Borovjagin A, Maxwell E, Gerbi S. Conserved Boxes C and D are essential nucleolar localization elements of U14 and U8 snoRNAs. EMBO J. 1998;17:3176–3187. doi: 10.1093/emboj/17.11.3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lange T, Ezrokhi M, Borovjagin A, Rivera-Leon R, North M, Gerbi S. Nucleolar localization elements of xenopus laevis U3 small nucleolar RNA. Mol Biol Cell. 1998;9:2973–2985. doi: 10.1091/mbc.9.10.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lapeyre B, Mariottini P, Mathieu C, Ferrer P, Amaldi F, Amalric F, Caizergues-Ferrer M. Molecular cloning of Xenopus fibrillarin, a conserved U3 small nuclear ribonucleoprotein recognized by antisera from humans with autoimmune disease. Mol Cell Biol. 1990;10:430–434. doi: 10.1128/mcb.10.1.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leader D J, Connelly S, Filipowicz W, Brown J W. Characterisation and expression of a maize U3 snRNA gene. Biochim Biophys Acta. 1994;1219:145–147. doi: 10.1016/0167-4781(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 43.Lee S J, Baserga S J. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lubben B, Marshallsay C, Rottmann N, Luhrmann R. Isolation of U3 snoRNP from CHO cells: a novel 55 kDa protein binds to the central part of U3 snoRNA. Nucleic Acids Res. 1993;21:5377–5385. doi: 10.1093/nar/21.23.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44a.Lukowiak, A., S. Mattox, R. Terns, and M. P. Terns. Unpublished data.
- 45.Matera A. Of coiled bodies, gems, and salmon. J Cell Biochem. 1998;69:1–13. [PubMed] [Google Scholar]
- 46.Mattaj I W. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell. 1986;46:905–911. doi: 10.1016/0092-8674(86)90072-3. [DOI] [PubMed] [Google Scholar]
- 47.Maxwell E S, Fournier M J. The small nucleolar RNAs. Annu Rev Biochem. 1995;35:897–933. doi: 10.1146/annurev.bi.64.070195.004341. [DOI] [PubMed] [Google Scholar]
- 48.Mazan S, Gulli M P, Joseph N, Bachellerie J P. Structure of the differentially expressed mouse U3A gene. Eur J Biochem. 1992;205:1033–1041. doi: 10.1111/j.1432-1033.1992.tb16871.x. [DOI] [PubMed] [Google Scholar]
- 49.Mereau A, Fournier R, Gregoire A, Mougin A, Fabrizio P, Luhrmann R, Branlant C. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 50.Mougey E B, Pape L K, Sollner-Webb B. A U3 small nuclear ribonucleoprotein-requiring processing event in the 5′ external transcribed spacer of Xenopus precursor rRNA. Mol Cell Biol. 1993;13:5990–5998. doi: 10.1128/mcb.13.10.5990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munns T W, Liszewski M K, Tellam J T, Sims H F, Rhoads R E. Antibody-nucleic acid complexes. Immunospecific retention of globin messenger ribonucleic acid with antibodies specific for 7-methylguanosine. Biochemistry. 1982;21:2922–2928. doi: 10.1021/bi00541a018. [DOI] [PubMed] [Google Scholar]
- 52.Narayanan A, Lukowiak A, Jady B E, Dragon F, Kiss T, Terns R M, Terns M P. Nucleolar localization signals of box H/ACA small nucleolar RNAs. EMBO J. 1999;18:5120–5130. doi: 10.1093/emboj/18.18.5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Narayanan A, Speckmann W, Terns R, Terns M. Role of the box C/D motif in the localization of small nucleolar RNAs to nucleoli and coiled bodies. Mol Biol Cell. 1999;10:2131–2147. doi: 10.1091/mbc.10.7.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Orum H, Nielsen H, Engberg J. Sequence and proposed secondary structure of the Tetrahymena thermophila U3-snRNA. Nucleic Acids Res. 1993;21:2511. doi: 10.1093/nar/21.10.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parker K, Steitz J. Structural analysis of the human U3 ribonucleoprotein particle reveal a conserved sequence available for base pairing with pre-rRNA. Mol Cell Biol. 1987;7:2899–2913. doi: 10.1128/mcb.7.8.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peculis B A, Steitz J A. Sequence and structural elements critical for U8 snRNP function in Xenopus oocytes are evolutionarily conserved. Genes Dev. 1994;8:2241–2255. doi: 10.1101/gad.8.18.2241. [DOI] [PubMed] [Google Scholar]
- 57.Pluk H, Soffner J, Luhrmann R, Van V W. cDNA cloning and characterization of the human U3 small nucleolar ribonucleoprotein complex-associated 55-kilodalton protein. Mol Cell Biol. 1998;18:488–498. doi: 10.1128/mcb.18.1.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Porter G L, Brennwald P J, Holm K A, Wise J A. The sequence of U3 from Schizosaccharomyces pombe suggests structural divergence of this snRNA between metazoans and unicellular eukaryotes. Nucleic Acids Res. 1988;16:10131–10152. doi: 10.1093/nar/16.21.10131. . (Erratum, 19:3484, 1991.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reimer G, Pollard K M, Penning C A, Ochs R L, Lishwe M A, Busch H, Tan E M. Monoclonal autoantibody from a (New Zealand black X New Zealand white) F1 mouse and some human scleroderma sera target an Mr 34,000 nucleolar protein of the U3 RNP particle. Arthritis Rheum. 1987;30:793–800. doi: 10.1002/art.1780300709. [DOI] [PubMed] [Google Scholar]
- 60.Samarsky D, Fournier M. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:3431–3444. doi: 10.1128/mcb.18.6.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samarsky D, Fournier M, Singer R, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Savino R, Gerbi S A. In vivo disruption of Xenopus U3 snRNA affects ribosomal RNA processing. EMBO J. 1990;9:2299–2308. doi: 10.1002/j.1460-2075.1990.tb07401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schimmang T, Tollervey D, Kern H, Frank R, Hurt E. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989;8:4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shaw P, Beven A, Leader D, Brown J. Localization and processing from a polycistronic precursor of novel snoRNAs in maize. J Cell Sci. 1998;111:2121–2128. doi: 10.1242/jcs.111.15.2121. [DOI] [PubMed] [Google Scholar]
- 66.Sienna N, Larson D E, Sells B H. Altered subcellular distribution of U3 snRNA in response to serum in mouse fibroblasts. Exp Cell Res. 1996;227:98–105. doi: 10.1006/excr.1996.0254. [DOI] [PubMed] [Google Scholar]
- 67.Singh R, Reddy R. Gamma-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci USA. 1989;86:8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith C, Steitz J. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 69.Suh D, Busch H, Reddy R. Human U3 small nucleolar RNA genes are localized to the nucleoplasm. Biochem Biophys Res Commun. 1987;143:658–664. doi: 10.1016/0006-291x(87)91404-5. [DOI] [PubMed] [Google Scholar]
- 70.Terns M, Goldfarb D. Nuclear transport of RNAs in microinjected Xenopus oocytes. Methods Cell Biol. 1998;53:559–589. doi: 10.1016/s0091-679x(08)60895-x. [DOI] [PubMed] [Google Scholar]
- 71.Terns M P, Dahlberg J E. Retention and 5′ cap trimethylation of U3 snRNA in the nucleus. Science. 1994;264:959–961. doi: 10.1126/science.8178154. [DOI] [PubMed] [Google Scholar]
- 72.Terns M P, Dahlberg J E, Lund E. Multiple cis-acting signals for export of pre-U1 snRNA from the nucleus. Genes Dev. 1993;7:1898–1908. doi: 10.1101/gad.7.10.1898. [DOI] [PubMed] [Google Scholar]
- 73.Terns M P, Grimm C, Lund E, Dahlberg J E. A common maturation pathway for small nucleolar RNAs. EMBO J. 1995;14:4860–4871. doi: 10.1002/j.1460-2075.1995.tb00167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 75.Tollervey D, Lehtonen H, Carmo-Fonseca M, Hurt E C. The small nucleolar RNP protein NOP1 (fibrillarin) is required for pre-rRNA processing in yeast. EMBO J. 1991;10:573–583. doi: 10.1002/j.1460-2075.1991.tb07984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75a.Tuma R S, Stolk J A, Roth M B. Identification and characterization of a sphere organelle protein. J Cell Biol. 1993;122:767–773. doi: 10.1083/jcb.122.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tyc K, Steitz J A. U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J. 1989;8:3113–3119. doi: 10.1002/j.1460-2075.1989.tb08463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wiederkehr T, Prétôt R F, Minvielle-Sebastia L. Synthetic lethal interactions with conditional poly(A) polymerase alleles identify LCP5, a gene involved in 18S rRNA maturation. RNA. 1998;4:1357–1372. doi: 10.1017/s1355838298980955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu P, Brockenbrough J, Metcalfe A, Chen S, Aris J. Nop5p is a small nucleolar ribonucleoprotein component required for pre-18S rRNA processing in yeast. J Biol Chem. 1998;273:16453–16463. doi: 10.1074/jbc.273.26.16453. [DOI] [PMC free article] [PubMed] [Google Scholar]