Abstract
Background
The new SARS-CoV-2 variant VOC (202012/01), identified recently in the United Kingdom (UK), exhibits a higher transmissibility rate compared to other variants, and a reproductive number 0.4 higher. In the UK, scientists were able to identify the increase of this new variant through the rise of false negative results for the spike (S) target using a three-target RT-PCR assay (TaqPath kit).
Methods
To control and study the current coronavirus pandemic, it is important to develop a rapid and low-cost molecular test to identify the aforementioned variant. In this work, we designed primer sets specific to the VOC (202012/01) to be used by SYBR Green-based RT-PCR. These primers were specifically designed to confirm the deletion mutations Δ69/Δ70 in the spike and the Δ106/Δ107/Δ108 in the NSP6 gene. We studied 20 samples from positive patients, detected by using the Applied Biosystems TaqPath RT-PCR COVID-19 kit (Thermo Fisher Scientific, Waltham, USA) that included the ORF1ab, S, and N gene targets. 16 samples displayed an S-negative profile (negative for S target and positive for N and ORF1ab targets) and four samples with S, N and ORF1ab positive profile.
Results
Our results emphasized that all S-negative samples harbored the mutations Δ69/Δ70 and Δ106/Δ107/Δ108. This protocol could be used as a second test to confirm the diagnosis in patients who were already positive to COVID-19 but showed false negative results for S-gene.
Conclusions
This technique may allow to identify patients carrying the VOC (202012/01) or a closely related variant, in case of shortage in sequencing.
Keywords: B.1.1.7 lineage, VOC (202012/01), COVID-19 pandemic, SARS CoV-2, SYBR Green-based RT-PCR
Introduction
As the pandemic of SARS-CoV-2 continues to affect the planet, researchers around the world are monitoring the virus and detecting acquired mutations that may lead to higher threats of spreading COVID-19 [1]. A new variant was recently reported in the UK as a VOC 202012/01 or B.1.1.7 lineage [2]. Its rate of transmission is estimated to be >70%, and its reproductive number (Ro) seems to be up to 0.4 higher [3]. This variant harbors 14 non-synonymous mutations, 6 synonymous mutations and 3 deletions [2, 3]. A wide variety of diagnostic tests have been used by high-throughput national testing systems around the world, to monitor the SARS-CoV-2 infection [4]. The arising prevalence of new SARS-CoV-2 variants such as B.1.1.7 has become of great concern, as most of the existing RT-PCR tests will not be able to specifically distinguish these new variants because they were not designed for such a purpose. Therefore, public health officials most rely on their current testing systems and their sequencing results to draw conclusions on the prevalence of new variants in their territories [2, 5]. An example of such cases has been seen in the UK. In fact, scientists were able to identify the augmentation of the B.1.1.7 SARS-CoV-2 variant infection in the population through an increase in the S-gene target failure in their three target gene assay (N+, ORF1ab+, S−) when using the Applied Biosystems TaqPath RT-PCR COVID-19 kit (Thermo Fisher Scientific, Waltham, USA) that included the ORF1ab, S, and N gene targets [1, 6, 7].
In 24 December 2020, Thermo Fisher Scientific confirmed that the S deletion Δ69/Δ70 was in the area targeted by the TaqPath Kit. Whilst other variants with Δ69/70 are also circulating worldwide, the absence of detection of the S gene target increasingly appears to be a highly specific marker for the new variant [2, 7]. In addition, the European CDC recommended that multi-target RT-PCR assays that included an S gene target affected by the deletions could be used as a signal for the presence of the Δ69/Δ70 mutation for further investigations and could be helpful to keep tracking these mutant strains [8].
Genome sequencing is the gold method to confirm the new variant, but observational studies provide also stronger evidence if similar models are observed in multiple countries, especially when randomized studies are not possible. In Lebanon, surveillance data from Beirut Medical Center and Bahman Hospital showed a rapid and dramatic increase in S-negative profile in PCR testing for SARS-CoV-2 in the first twelve days in January, reaching approximately 60% of positive cases and 95% in February [9, 10].
In 15 January 2021, in GISAID we found 374,613 SARS-CoV-2 sequences, among them 17,473 sequences belonged to the new variant (VOC 202012/01). It is important to notice that the co-occurrence of mutations Δ69/Δ70 in the spike and Δ106/Δ107/Δ108 in the NSP6, was present in this new variant. We then decided to use these two deletions as targets for our rapid and low-cost protocol, performing SYBR Green-Based RT-PCR. In this work, we propose primer sets that can be applied as a second step to confirm the diagnosis in cases that were already detected as positive to SARS-CoV-2 when using the Applied Biosystems TaqPath RT-PCR COVID-19 kit (Thermo Fisher Scientific, Waltham, USA).
It is well known that confirmatory diagnosis based on specific diagnostic biomarkers remains a great challenge allowing the further control and eradication of infections including COVID-19 [11]. Therefore, this novel method may lead to specifically identify individuals carrying the Δ69/Δ70 and the Δ106/Δ107/Δ108 mutations.
Materials and methods
Clinical specimens
20 clinical samples for SARS-CoV-2 positive patients, with Ct (cycle threshold) value < 32, were selected for this study. All samples were previously tested positive by the Applied Biosystems™ TaqPath™ COVID-19 assay which targeted the RdRP, N and Spike genes in the Molecular Laboratories at Bahman Hospital and Beirut Cardiac Institute in the city of Beirut. 16 of these samples were S-negative and 4 were S-positive.
All patients provided written and signed informed consents.
RNA extraction and cDNA synthesis
RNA was extracted from the clinical samples using Qiamp viral RNA mini kit (Qiagen). Total RNA was converted to cDNA using iScript cDNA synthesis kit (BioRad), following the manufacturer’s recommended procedures.
Primer design
Primers were designed based on the recently available full sequence of the new variant VUI (202012/01) from the NCBI Reference Sequence Database (https://www.ncbi.nlm.nih.gov/nuccore/NC_045512). We utilized NCBI-Primer BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) to design specific primers (Fig. 1). The specificity of the primers was verified using in silico prediction analyses with the online Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi). None of our designed primers showed genomic cross-reactivity with other viruses, the human genome, or other probable interfering genomes in the BLAST database analysis. The primer sets were synthesized and delivered by Macrogen (Republic of Korea), they are listed in Table 1.
Fig. 1.
The Localization of the selected primers. Deleted nucleotides are written in bold
Table 1.
List of primer sets
| Target gene | Primer name | Forward sequence (5′–3′) | Reverse sequence (5′–3′) |
|---|---|---|---|
| Spike | Spike WT | GGTTCCATGCTATACATGTCTC | GGTCTTCGAATCTAAAGTAGTACCA |
| Spike | Spike del 69/70 | GTTCCATGCTATCTCTGG | GGGACTGGGTCTTCGAATCT |
| NSP6 | NSP6 WT | GGTTGATACTAGTTTGTCTGGTTTT | AACGAGTGTCAAGACATTCATAAG |
| N | WH-NIC | CGTTTGGTGGACCCTCAGAT | CCCCACTGCGTTCTCCATT |
Deleted nucleotides are in bold and underlined
Genome sequencing and lineage analysis
Genomic sequencing was performed for the 20 samples, collected between 03/01/2021 and 04/04/2021. Samples were sequenced using the ARTIC network methodology (https://artic.network/) with the V3 amplicon scheme, V3-LoCost library prep method (Quick 2020) and consensus sequences were generated using the bioinformatics SOP (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html) in Nano polish mode. 8,9 Consensus sequences were assigned a lineage using pangolin v2.0 (github.com/cov-lineages/pangolin).
SARS-CoV-2 rRT-PCR
Quantitative RT-PCR was carried out using iTaq universal SYBR Green super mix (Bio Rad). In brief, each reaction consisted of a total volume of 20 µL containing 2 µL of each primer [10 pM/µL], 5 µL of cDNA, 10 µL SYBR Green super mix and 1 µL of Rnase free Water.
Real-time PCR was performed using Bio Rad CFX96 Real-Time PCR Machine. The thermal cycling conditions used were as follows: 94 °C for 2 min, followed by 40 cycles of amplification at 94 °C for 10 s, and 60 °C for 1 min. The reaction was completed by determining the dissociation curve of all amplicons.
Results
In order to validate our method, we re-tested 20 samples that had been tested as positive for SARS-CoV-2 with Ct < 32, by the TaqPath kit. 16 samples with S-negative profile and four samples with S-positive profile (Table 2). N primer pairs were used as a control of cDNA synthesis and integrity of the sample, spike WT and NSP6 WT primer pairs were used to detect the variants not harboring the deletion mutations Δ69/Δ70 in the spike nor the Δ106/Δ107/Δ108 in the NSP6, respectively. Spike del 69/70 primer pairs were designed to detect the deletions Δ69/Δ70 in the spike protein (Fig. 1).
Table 2.
The Ct values from the TaqPath Kit and SYBER Green-Based assay
| Samples | Probe based (TaqPath Kit) Ct values | SYBER Green-based Ct values | ||||||
|---|---|---|---|---|---|---|---|---|
| ORF1 | N | Spike (S) | N (positive control) |
NSP6 (ORF1) |
Spike (S) | Spike del 69/70 | ||
| S-negative samples | 1 | 15 | 16 | ND | 15 | ND | ND | 16 |
| 2 | 25 | 25 | ND | 27 | ND | ND | 28 | |
| 3 | 17 | 17 | ND | 19 | ND | ND | 20 | |
| 4 | 15 | 16 | ND | 25 | ND | ND | 23 | |
| 5 | 15 | 17 | ND | 21 | ND | ND | 22 | |
| 6 | 23 | 22 | ND | 23 | ND | ND | 24 | |
| 7 | 15 | 16 | ND | 21 | ND | ND | 21 | |
| 8 | 22 | 23 | ND | 18 | ND | ND | 17 | |
| 9 | 20 | 19 | ND | 26 | ND | ND | 26 | |
| 10 | 19 | 21 | ND | 22 | ND | ND | 24 | |
| 11 | 13 | 14 | ND | 18 | ND | ND | 19 | |
| 12 | 14 | 14 | ND | 17 | ND | ND | 20 | |
| 13 | 32 | 31 | ND | 33 | ND | ND | 31 | |
| 14 | 20 | 21 | ND | 19 | ND | ND | 22 | |
| 15 | 20 | 22 | ND | 25 | ND | ND | 27 | |
| 16 | 17 | 17 | ND | 19 | ND | ND | 18 | |
| S-positive samples | 17 | 24 | 24 | 24 | 25 | 26 | 27 | ND |
| 18 | 19 | 19 | 18 | 21 | 19 | 19 | ND | |
| 19 | 23 | 23 | 25 | 26 | 27 | 28 | ND | |
| 20 | 20 | 20 | 21 | 19 | 21 | 22 | ND | |
Ct cycle threshold, ND not detected
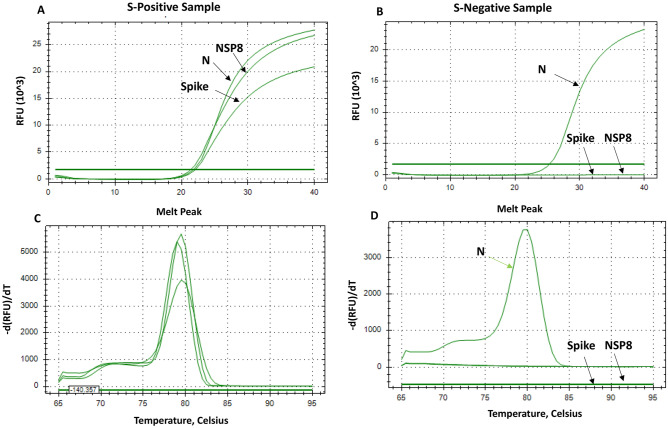
Our results showed that the N-gene amplicons were detected in both, S-negative and S-positive profiles (Table 2; Fig. 2), whereas the spike del 69/70-gene amplicons were detected only in the S-negative profile confirming the absence of the amino acids 69 and 70 in the spike protein of these samples (Fig. 2B). For the spike WT and NSP6 WT primers, amplicons of both, were detected only in the S-positive profile confirming the presence of the two amino acids 69 and 70 in the spike protein and the three amino acids 106, 107 and 108 in the NSP6 protein of S-positive samples (Fig. 2A).
Fig. 2.
Real time PCR results, from SYBR Green-Based assay, with primers targeting the non-mutated N, Spike and NSP6 genes in S-positive and S-negative samples (N-gene is used as a positive control). Panel A show the amplification curves of the targeted regions in N, S and NSP6 genes in the S-positive samples. Panel B show the amplification curves of the targeted region in N gene in the S-negative samples. Panel C and D show the melting curves of the targeted regions in S-positive and S-negative samples, respectively. In panel D only one melting peak in S-negative samples, corresponding to the N gene
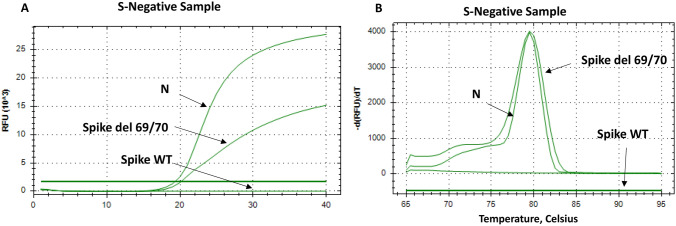
The results of spike del 69/70 primers were fully concordant with spike WT and NSP6 WT primers, 100% of the S-negative profile had the Spike deletions Δ69/Δ70 (Fig. 3A), while 100% of the S-positive profile did not contain the deletions Spike Δ69/Δ70 and NSP6 Δ106/Δ107/Δ108 (Fig. 2A).
Fig. 3.
SYBR Green-based PCR with primers targeting the mutated Spike del 69/70 genes in S-negative samples (N-gene is used as a positive control). Panel A show the amplification curves of the targeted regions in N, and Spike genes in the S-negative samples using three primer sets corresponding to N, Spike WT and Spike del 69/70. Panel B show the melting curves for the products amplified in S-negative samples by N and Spike del 69/70 primers only
To confirm the lineage present in these 20 patient samples, genomic sequencing was performed and the obtained sequences were deposited in Genbank under NCBI accession number MZ772870 to MZ772873, MZ772924, MZ772926, MZ772929, MZ772933, MZ772935, MZ772973 to MZ772975, MZ773014, MZ773900, MZ773902, MZ773903, MZ773924 and MZ773926 to MZ773928. Genomic sequencing revealed that all the 20 samples belonged to the B.1.1.7 lineage (Table 3).
Table 3.
Genome sequence recovered per sample and Pangolin defined lineage
| NCBI accession number | Percentage of genome recovered | Pangolin defined lineage | B1.1.7 defining SNPS confirmed |
|---|---|---|---|
| MZ772871 | 95.66 | B1.1.7 | 17/17 |
| MZ772872 | 98.11 | B1.1.7 | 17/17 |
| MZ772873 | 93.6 | B1.1.7 | 17/17 |
| MZ772924 | 94.27 | B1.1.7 | 17/17 |
| MZ772926 | 98.1 | B1.1.7 | 17/17 |
| MZ772929 | 97.93 | B1.1.7 | 17/17 |
| MZ772933 | 98.09 | B1.1.7 | 17/17 |
| MZ772935 | 96.74 | B1.1.7 | 17/17 |
| MZ772973 | 98.15 | B1.1.7 | 17/17 |
| MZ772974 | 98.15 | B1.1.7 | 17/17 |
| MZ772975 | 98.1 | B1.1.7 | 17/17 |
| MZ772975 | 98.08 | B1.1.7 | 17/17 |
| MZ773014 | 96.74 | B1.1.7 | 17/17 |
| MZ773900 | 94.4 | B1.1.7 | 17/17 |
| MZ773902 | 97.99 | B1.1.7 | 17/17 |
| MZ773903 | 98.11 | B1.1.7 | 17/17 |
| MZ773924 | 98.11 | B1.1.7 | 17/17 |
| MZ773926 | 98.12 | B1.1.7 | 17/17 |
| MZ773927 | 95.93 | B1.1.7 | 17/17 |
| MZ773928 | 94.3 | B1.1.7 | 17/17 |
Presence of the 17 B.1.1.7 lineage SNPS in each consensus sequencePresence of the 17 B.1.1.7 lineage SNPS in each consensus sequence
Analytical sensitivity and limit of detection
The analytical sensitivity of the assays was determined with 5 serials tenfold dilutions using an isolated sample having Ct values 25 by TaqMan RT-PCR (TaqPath kit) and 28 by our SYBR Green- based protocol. All diluted samples were tested in triplicate by gold standard TaqMan RT- qPCR (TaqPath kit) and by our SYBR Green- based protocol. The unamplified dilution by SYBR Green-based RT-PCR has Ct value 37.55 by the TaqMan RT-qPCR. Thus, the tested RT-PCRs could detect positive samples with maximal Ct of 34.45 by TaqMan RT-PCR or with Ct value of 37.67 by SYBR Green RT-PCR. All results generated by the serials tenfold dilution are shown in Table 4.
Table 4.
Determination of the sensitivity by 5 serials ten-fold dilutions
| Dilution | Probe based (TaqPath Kit) Ct values | SYBER Green-based Ct values |
|---|---|---|
| 1 | 25 | 27 |
| 1/10 | 27.92 | 30.64 |
| 1/100 | 31.34 | 34.04 |
| 1/1000 | 34.45 | 37.67 |
| 1/10,000 | 37.55 | ND |
| 1/100,000 | ND | ND |
The limit of detection of TaqPath kit is 250 copies/mL (according to the manufacturer). Starting with the same RNA concentration, the Ct values of both TaqPath kit and SYBR Green-based RT-PCR were similar to a difference in Ct of 2.1 on average (Table 2). This difference means a decrease of 4.2-folds in the limit of detection for the SYBR Green-based RT-PCR (~ 1050 copies/mL) compared to TaqPath kit.
One drawback of the SYBR Green assay is that the dye is nonspecific, and this lack of specificity can generate false-positive signals if nonspecific products or primer-dimers are present in the sample. However, including a melting curve analysis at the end of each PCR assay to determine the specificity and efficiency of each RT-PCR reaction will ensure the accuracy of the results when multiple peaks or primer-dimers are not observed. Our primer set “spike del 69/70” gave specific melt peak at 79.5 °C C in S-negative samples (Fig. 3B), while primers “spike” and “NSP8” gave melt peaks at 79 and 79.5 °C, respectively, in S-positive samples (Fig. 2C, D).
Discussion
Sequencing is the current gold standard diagnostic method to confirm the presence of any new variant, its efficient but a time consuming method and not accessible to all laboratories especially in developing countries based on the elevated costs and the present global demands for supplies and reagents. In this study, we have developed a rapid, low-cost, large-scale screening protocol for the detection of the deletions Δ69/Δ70 and Δ106/Δ107/Δ108. We adopted SYBR Green-based RT-PCR method over Taqman-based RT-PCR method. Taqman-based diagnosis is accepted in the field as more accurate method but also it’s not accessible to all laboratories especially in developing countries. In the present work we are dealing with samples already confirmed SARS-CoV-2 positive by TaqPath Kit (ThermoFisher), so even if SYBR Green-based RT-PCR method is less accurate it will be very useful since we need just to confirm the presence of the two mutations Δ69/Δ70 in the spike and Δ106/Δ107/Δ108 in the NSP6.
Conclusions
We hope that our efforts will be helpful and can contribute to the early detection of the new variant (VOC 202012/01) belonging to the B.1.1.7 lineage, for the prevention of transmission and early intervention. This protocol should not be limited to this variant, but also for any other future variant to come, just by designing the appropriate primers.
Acknowledgements
This work was supported by the Lebanese University and Fundación La Caixa (LCF/PR/PR13/11080005), Fundación Caja Navarra, Fundación Roviralta, Ubesol, Inversiones Garcilaso de la Vega, COST Actions CA18217 and CA18218, and EU Project uncover (Grant/Award Number: 101016216).
Abbreviations
- CDC
Centre for Disease Prevention and Control
- N
Nucleocapsid
- NSP6
Non-structural protein 6
- RdRP
RNA-dependent RNA polymerase
- RT-PCR
Reverse transcriptase polymerase chain reaction
- SARS CoV-2
Severe acute respiratory syndrome coronavirus 2
- S
Spike
- VUI
Variant under investigation
Author contributions
KH analysed and interpreted the data and wrote the draft of the manuscript. FAS analysed and interpreted the data and was involved in revising the manuscript critically. MY, HN and PG revised the manuscript critically. All the authors have given the final approval of the version to be published. All authors read and approved the final manuscript.
Funding
No funding was provided for this work.
Data availability
We declared that materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.
Declarations
Ethical approval and consent to participate
All presented cases or their legal guardian provided consent to data collection and use according to institutional guidelines.
Consent for publication
All presented cases or their legal guardian provided consent to publish according to institutional guidelines.
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
The original online version of this article was revised due to first author name correction.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/2/2022
A Correction to this paper has been published: 10.1007/s11033-021-07086-2
Contributor Information
Paul Nguewa, Email: panguewa@unav.es.
Kassem Hamze, Email: kassem.hamze@ul.edu.lb.
References
- 1.Lopez-Rincon A, Alberto T, Lucero MM, Eric C et al (2020) Design of specific primer set for detection of B. 1.1. 7 SARS-CoV-2 variant using deep learning. bioRxiv. 10.1101/2020.12.29.424715
- 2.Erik V, Swapnil M, Meera C, Jeffrey CB, Robert J, Lily G, et al (2020) Transmission of SARS-CoV-2 lineage B. 1.1. 7 in England: insights from linking epidemiological and genetic data. medRxiv. 10.1101/2020.12.30.20249034
- 3.Meera C, Susan H, Gavin D, Hester A, Theresa L, Obaghe E et al (2020) Investigation of novel SARS-COV-2 variant: variant of concern 202012/01. Public Health England. https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201
- 4.Andrew R, Nick L, Oliver P, Wendy B, Jeff B, Alesandro C et al (2020) Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations: COVID-19 genomics UK Consortium. https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-ukdefined-by-a-novel-set-of-spike-mutations/563
- 5.Afzal A. Molecular diagnostic technologies for COVID-19: limitations and challenges. J Adv Res. 2020;26:149–159. doi: 10.1016/j.jare.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonin B, Gregory D, Alexandre G, Hadrien R, Quentin S et al (2020) Two-step strategy for the identification of SARS-CoV-2 variants co-occurring with spike deletion H69-V70, Lyon, August to December. medRxiv. 10.1101/2020.11.10.20228528
- 7.Nicole LW, Simon W, Kelly MSB et al (2020) S gene dropout patterns in SARS-CoV-2 tests suggest spread of the H69del/V70del mutation in the US. medRxiv. 10.1101/2020.12.24.20248814.
- 8.SARS-CoV-2 variant: United Kingdom of Great Britain and Northern Ireland. https://www.who.int/csr/don/21-december-2020-sars-cov2-variant-united-kingdom/en/
- 9.Younes M, Hamze K, Nassar H, Makki M, Ghadar M, Nguewa P, Abdel Sater F (2021) Emergence and fast spread of B.1.1.7 lineage in Lebanon. medRxiv. 10.1101/2021.01.25.21249974
- 10.Younes M, Hamze K, Carter DP, Osman KL, Vipond R, Carroll M, Pullan ST, Nassar H, Mohamad N, Makki M, Ghadar M (2021) B. 1.1. 7 became the dominant variant in Lebanon. medRxiv. 10.1101/2021.03.17.21253782
- 11.Peña-Guerrero J, Nguewa PA, García-Sosa AT. Machine learning, artificial intelligence, and data science breaking into drug design and neglected diseases. WIREs Comput Mol Sci. 2021 doi: 10.1002/wcms.1513. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
We declared that materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.