Abstract
A survey of 22 farms confirmed the presence of paratuberculosis in wild rabbits in Scotland. Regional differences were apparent in the prevalence of the disease in rabbits, with a significantly higher incidence occurring in the Tayside region. Statistical analysis showed a significant relationship between a previous history or current problem of paratuberculosis in cattle and the presence of paratuberculosis in rabbits on the farms. Molecular genetic typing techniques could not discriminate between selected rabbit and cattle isolates from the same or different farms, suggesting that the same strain may infect and cause disease in both species and that interspecies transmission may occur. The possibility of interspecies transmission and the involvement of wildlife in the epidemiology of paratuberculosis have important implications for the control of the disease.
Paratuberculosis (Johne’s disease) is a chronic granulomatous enteritis caused by Mycobacterium avium subsp. paratuberculosis. In most species the disease is characterized by diarrhea, emaciation, and loss of body condition culminating in death. Paratuberculosis principally affects ruminants and is responsible for significant economic losses to the livestock industry worldwide (3, 36). Diagnosis is difficult, particularly of asymptomatic carriers. There is no single diagnostic test available that can diagnose the disease at every stage. Current control programs rely on culling or removing animals that test positive, usually as determined by bacteriological culture or serum antibody test such as the enzyme-linked immunosorbent assay.
Effective disease control programs depend on a clear understanding of the sources of infection and the routes of transmission. The most important mode of transmission of paratuberculosis is the fecal-oral route, although transmission in symptomatic animals is known to occur in utero and via infected semen, colostrum, and milk. What is less clear and of particular importance is whether paratuberculosis can be transmitted between species. Experimental infection of ruminants has been demonstrated with different strains of M. avium subsp. paratuberculosis isolated from different species and a variety of laboratory animals, including rabbits, have been experimentally infected with ruminant strains (22–24). In addition, there are a few reports of natural disease among sheep and goats that grazed with infected cattle (31, 32). Infected livestock is undoubtedly the principal source of infection, but there is still the question of the involvement of wildlife reservoirs. M. avium subsp. paratuberculosis has been reported previously in wildlife, including white-tailed deer (4, 18), red deer (35), roe deer (9), exotic deer (30), tule elk (13), bighorn sheep (39, 40) and, more recently, rabbits (7). The possibility of interspecies transmission, coupled with the data implicating wildlife in the epidemiology of paratuberculosis, have important implications for the control of the disease. If this occurs in areas where livestock interact with wildlife reservoirs, the current detection and cull policy will be inefficient in the long term, and vaccination may be a more suitable alternative.
The epidemiological study reported here was initiated to provide further information on the role of wild rabbits in the epidemiology of paratuberculosis. A pilot survey revealed that on four farms in the Tayside region of Scotland, 67% of wild rabbits were infected with M. avium subsp. paratuberculosis (7). This region also had a high prevalence of bovine paratuberculosis (38a). It was decided to extend the survey to cover 22 farms throughout Scotland to examine the prevalence of M. avium subsp. paratuberculosis infection in the wild-rabbit population in different regions. Farms with or without a history of paratuberculosis were selected to establish whether there was any association between the presence of paratuberculosis in livestock and rabbits. To identify whether interspecies transmission could have occurred, M. avium subsp. paratuberculosis strains isolated from rabbits and cattle present on two farms were compared by pulsed-field gel electrophoresis (PFGE), IS900 restriction fragment length polymorphism (RFLP), and chemotype profiles.
MATERIALS AND METHODS
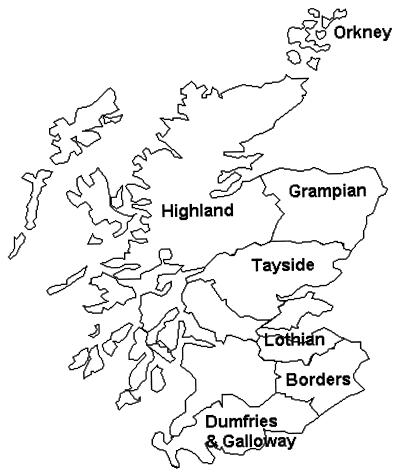
Two hundred and ten rabbits were killed between October and March 1996 on 22 farms located throughout Scotland. The regions sampled in the survey are shown in Fig. 1. Fourteen of the farms (A through N) had a history of paratuberculosis among livestock, and eight of the farms (O through V) had no observed or recorded cases of paratuberculosis. Paratuberculosis was diagnosed on the affected farms by microscopic examination of feces and serum antibody tests.
FIG. 1.
Map of Scotland showing regions sampled in the survey.
The rabbit carcasses were weighed, sexed, and examined at necropsy for visible lesions. Representative samples of the small and large intestine and mesenteric lymph nodes were removed aseptically and transferred to sterile containers. Separate instruments were used for each animal. Small portions of tissue were fixed in 10% formal saline for histopathological analysis.
Fecal samples were taken from cattle and sheep on farms A and C to culture M. avium subsp. paratuberculosis isolates from the livestock for typing. Samples were processed as outlined below.
Histopathology.
Fixed tissue samples were embedded in paraffin wax, and 4-μm sections were stained with hematoxylin and eosin and by the Ziehl-Neelsen procedure for acid-fast bacilli.
Primary culture.
Tissue and fecal homogenates were prepared as described previously (7). Briefly, 1 g of feces or 0.5 cm3 of finely chopped tissues were homogenized for 30 s in 10 ml of sterile distilled water with a Colworth Stomacher 80 (Seward Medical, London, United Kingdom). The homogenates were decontaminated by adding 10 ml of 1.5% hexadecyl pyridinium chloride and left overnight at room temperature to allow particulate materials to settle. The supernatants were centrifuged at 3,800 × g for 30 min at 4°C, and each pellet was resuspended in 10 ml of sterile distilled water. The centrifugation step was repeated, and each pellet was resuspended in 1 ml of sterile distilled water. The suspension was transferred to a microfuge tube and centrifuged at 6,500 × g for 5 min. The pellet was resuspended finally in 0.5 ml of sterile distilled water. Two slants of Middlebrook 7H11 agar supplemented with Selectatabs (amphotericin B, polymixin B, carbenicillin, and trimethoprim; Code MS 24; MAST Laboratories, Ltd., Merseyside, United Kingdom), 10% Middlebrook oleic acid-albumin-dextrose-catalase (OADC) enrichment medium (Difco, Surrey, United Kingdom) and 2 μg of mycobactin J (Allied Monitor, Fayette, Mo.) per ml were inoculated with 0.1 ml of the prepared suspension. The cultures were incubated at 37°C for up to 16 weeks and examined regularly for bacterial growth.
Analysis by PCR.
The identity of the mycobacterium isolates was confirmed by PCR. Two-hundred microliters of sterile distilled water was inoculated with a single bacterial colony from each positive culture. The mycobacteria were lysed by beating with 0.1-mm silica-zirconium beads (Biospec Products, Bartlesville, Okla.) in an ESPE capmix (Cottrell & Co.) twice for 40 s, with cooling on ice between each treatment. The DNA was extracted by using guanidine hydrochloride as described by Challans et al. (2). Five-microliter aliquots of DNA were analyzed by PCR assays directed against the IS900 sequence of M. avium subsp. paratuberculosis (33). PCR-amplified product was detected by continuous polyacrylamide gel electrophoresis followed by silver staining (2).
Statistical analysis.
The data were analyzed by using Genstat 5 software (release 3.2) with the generalized linear modelling facilities with a logit link function. Excel 97 was used to manage the dataset, to examine the residuals, and to carry out simple hypothesis testing on the models generated by Genstat.
Molecular typing of M. avium subsp. paratuberculosis isolates.
Four leporine isolates (R1, R7, R8, and R10) and four bovine isolates (F13, F14, F16, and F17) from farm A and seven leporine isolates (R186, R191, R193, R194, R195, R197, and R198) and four bovine isolates (JD1, JD2, JD3, and JD4) from farm C were selected for molecular typing. The primary cultures were subcultured on Middlebrook 7H11 agar supplemented with Selectatabs, OADC, and mycobactin J as described above and were sent to the various laboratories for typing. Due to a shortage of cultures at the time, it was not possible to analyze isolate F16 by IS900 RFLP or to chemotype isolate F17.
PFGE.
Ten milliliters of Middlebrook 7H9 broth supplemented with Tween 80 (4% [wt/vol]), OADC (10% [vol/vol]), glycerol (2.5% [vol/vol]), and 2 μg of mycobactin J per ml was inoculated with a single colony and incubated at 37°C until the cell density was at least McFarland standard 2, as assessed by using a densimat (BioMerieux, Lyon, France). Cultures were stirred during incubation with a magnetic stirrer bar to prevent clumping of the cells.
Cells were centrifuged at 3,500 × g for 20 min at 4°C. The cells were resuspended in spheroplasting buffer (citrate phosphate buffer, pH 5.6 [0.2 M citrate, 0.5 M phosphate]; 50 mM EDTA; 0.1% [wt/vol] Tween 80) to give 1.8 × 1010 cells/ml. One-half milliliter of the cell suspension was warmed to 45°C, mixed with 0.5 ml of prewarmed molten 1% (wt/vol) low-melting-point agarose (InCert Agarose; FMC Bioproducts, Flowgen, Staffordshire, United Kingdom), and poured into precooled moulds. Agarose plugs were allowed to set at 4°C for 15 min. Each plug was transferred to a plastic bijou bottle and incubated in 0.5 ml of lysis solution (10 mM Tris-HCl, pH 8.0; 1 mM EDTA, pH 8.0; 1 to 2 mg of lysozyme per ml) overnight at 37°C. After incubation, the lysis solution was discarded and replaced with 0.5 ml of ESP (0.5 M EDTA, pH 8.0; 1% [wt/vol] lauroyl sarcosine; 1 to 2 mg of proteinase K per ml) and then incubated with gentle agitation for at least 7 days at 55°C. The plugs were then transferred to fresh bijoux, and 5 ml of TE (10 mM Tris-HCl; 1 mM EDTA, pH 8.0) plus 1 mM phenylmethylsulfonyl fluoride was added; the mixture was then shaken gently for 30 min at room temperature. The samples were washed three times for 30 min in TE at room temperature with gentle agitation. Samples were stored in 0.5 M EDTA (pH 8.0) at 4°C until required for restriction analysis. Plugs were washed three times for 15 min with 5 ml of TE at room temperature, with shaking, prior to restriction analysis.
After equilibration in the appropriate restriction buffer for 1 h, the DNA was restricted with 20 U of HindIII or SpeI overnight at 37°C. Samples were electrophoresed on a 1% (wt/vol) pulsed-field certified agarose gel in 0.5× TBE (89 mM Tris-borate; 89 mM boric acid, pH 8.3) by using a CHEF Mapper (Bio-Rad Laboratories, Ltd., Hertfordshire, United Kingdom) with parameters designed to give optimum separation in the 20- to 110-kb size range (gradient, 6 V/cm; included angle, 120°; linear ramping with an initial switch time of 2.98 s and a final switch time of 9.39 s; overall time, 26.56 h). PFGE profiles were analyzed with Phoretix PC software (Pharmacia Biotech, Ltd., Hertfordshire, United Kingdom).
IS900 RFLP analysis.
DNA was prepared as previously described (17). Mycobacteria were treated with lysozyme, immobilized in low-melting-point agarose, and lysed in a solution containing EDTA, sodium dodecyl sulfate, and proteinase K. Agarose blocks, each containing approximately 4 μg of DNA, were digested with PstI and BstEII (New England Biolabs, Hertfordshire, United Kingdom) according to the manufacturer’s instructions. The resulting restriction fragments were separated by field inversion gel electrophoresis at 80 V (4 V/cm) with linear ramping from 0.3 to 10 s for 16 h. The restriction fragments were vacuum blotted onto Hybond-N nylon membrane (Amersham Life Science, Ltd.) and hybridized to an IS900 probe as described previously (27, 28). The IS900 probe was prepared by PCR amplification of a 453-bp fragment with the primers 5′-TGGACAATGACGGTTACGGAGGTGG-3′ and 5′-GATCGGAACGTCGGCTGGTCAGGCT-3′ according to the procedure described by Kunze et al. (14). The product was electrophoresed on a 2% agarose gel, purified by using a Wizard PCR purification kit (Promega), and labelled by using the ECL Direct Labelling Kit (Amersham Life Science, Ltd.) according to the manufacturer’s instructions. DNA fingerprints were scanned with a CCD camera and processed with the software Gel Manager (Biosystematika, Tavistock, United Kingdom). RFLP types were defined as previously described (27, 28). There are currently nine RFLP types generated by digestion with PstI which have been labelled A to K (27, 28). RFLP types generated by BstEII can be divided into three groups designated C, S, and I (27, 28). These three groups are further divided and designated with a new numerical series (C1–17, S1–3, and I1–2) (27, 28). Individual RFLP strain types were designated as an RFLP type after digestion with both restriction endonucleases PstI and BstEII (for example, B-C1).
Chemotyping.
Isolates of M. avium subsp. paratuberculosis were grown on Middlebrook 7H10 agar plates containing 100 μl of Middlebrook OADC enrichment medium per ml and 2 μg of mycobactin J per ml added at a pH of 7.9 for 13 weeks at 37°C and 99% humidity.
Matrix solid-phase dispersion (MSPD) was performed as previously described by Hines et al. (11) and as modified by Hines and Frazier (10). Briefly, 60 mg of mycobacteria was scraped from the plates and blended with 400 mg of C18 reversed-phase high-pressure liquid chromatography (HPLC) packing resin (Bakerbond; catalog no. 702500; J. T. Baker Co., Phillipsburg, N.J.). The blended material was transferred to a 10-ml syringe column, where it was eluted with 3 ml of each of the five solvents (hexane, methylene chloride, acetonitrile, methanol, and HPLC-grade water) in sequential order. An additional 7 ml of 100% methanol was mixed with the water-derived fraction to enhance evaporation. All five fractions from each bacterial sample were then evaporated to dryness under nitrogen gas at 40°C. Once dried, the samples were resolubilized in 60 μl of chloroform-methanol-water (5:4:1) and vortexed briefly. Thin-layer chromatography (TLC) was performed by prescribed methods (38), with the following modifications. Five microliters of each of the five MSPD fractions from each organism were spotted onto separate lanes of a silica gel TLC plate (Baker S1250-PA, 19C; catalog no. 7009-04; J. T. Baker Co.). This was repeated three times for a total of 15 μl of each fraction per lane. The plates were allowed to dry at room temperature and then developed in a TLC chamber containing 100 ml of mobile phase comprised of chloroform-methanol-water (5:4:1). Plates were removed when the mobile phase had risen approximately 10 cm. After being dried in a chemical fume hood, the plates were sprayed with Bial’s reagent (Sigma Chemical, St. Louis, Mo.), air dried, and then heated at 110 to 120°C for 15 to 20 min and photographed.
The Rx values were determined for each band by dividing the distance from the origin of each band by the distance from the origin of a common reference band in lane 4 (34). For standardization, band measurements were taken from the leading edge to prevent problems associated with thick bands. For crescent- or arrow-shaped bands, measurements were taken at the apex of the arc.
RESULTS
The results from the farm survey are detailed in Table 1 and summarized in Table 2. A total of 130 rabbits were collected from 14 farms with a history of Johne’s disease, and 80 rabbits were collected from 8 farms with no history of Johne’s disease. M. avium subsp. paratuberculosis was cultured from rabbits from 3 of the 14 paratuberculosis-affected farms which were located all in the Tayside region. On these farms, between 8 and 53% of wild rabbits investigated were found to be infected. M. avium subsp. paratuberculosis also was cultured from a rabbit on one farm in the Borders region with no known history of Johne’s disease. In addition, an acid-fast organism was cultured from a rabbit on a paratuberculosis-affected farm in Orkney, although it was not possible to confirm the identity of the isolate due to desiccation of the media and, for this reason, this result was excluded from the statistical analysis. The rabbits were generally in good condition, as judged by their fat reserves. No visible lesions were observed. Histopathological changes were apparent in 28 (22%) rabbits on nine farms with a history of Johne’s disease and in 6 (8%) rabbits on two farms with no known history of Johne’s disease. Acid-fast bacilli were associated with lesions in 10 (8%) of the rabbits on paratuberculosis-affected farms and 2 (3%) of the rabbits on the farms with no known history of Johne’s disease.
TABLE 1.
Rabbit survey dataa
| Farm | Location (region) | Livestock held and JD historyb | No. of rabbits investigated | No. of culture+ rabbits | Histopathology (no. of rabbits)
|
||
|---|---|---|---|---|---|---|---|
| Lesion+, AFB+ | Lesion+, AFB− | Culture+ and/or lesion+ | |||||
| A | Tayside | Cattle | 10 | 5 | 3 | 2 | 8c |
| B | Tayside | Sheep | 6 | 0 | 0 | 1 | 1 |
| C | Tayside | Cattle, sheep | 15 | 8 | 3 | 2 | 9 |
| D | Tayside | Cattle | 12 | 1 | 2 | 2 | 4 |
| E | Lothian | Sheep, cattle | 10 | 0 | 0 | 1 | 1 |
| F | Borders | Cattle, sheep | 5 | 0 | 0 | 0 | 0 |
| G | Borders | Cattle, sheep, goats | 4 | 0 | 0 | 0 | 0 |
| H | Dumfries and Galloway | Cattle | 9 | 0 | 0 | 3 | 3 |
| I | Dumfries and Galloway | Sheep | 10 | 0 | 0 | 0 | 0 |
| J | Dumfries and Galloway | Cattle | 9 | 0 | 0 | 0 | 0 |
| K | Grampian | Cattle | 10 | 0 | 1 | 4 | 5 |
| L | Highland | Cattle, sheep | 10 | 0 | 0 | 0 | 0 |
| M | Orkney | Cattle, sheep | 10 | 1d | 1 | 2 | 3d |
| N | Orkney | Cattle, sheep | 10 | 0 | 0 | 1 | 1 |
| O | Tayside | Cattle, sheep | 10 | 0 | 0 | 0 | 0 |
| P | Lothian | Cattle, sheep | 10 | 0 | 0 | 2 | 2 |
| Q | Borders | Cattle, sheep | 10 | 1 | 2 | 2 | 4 |
| R | Dumfries and Galloway | Cattle | 9 | 0 | 0 | 0 | 0 |
| S | Grampian | Cattle, sheep | 10 | 0 | 0 | 0 | 0 |
| T | Highland | Cattle, sheep | 10 | 0 | 0 | 0 | 0 |
| U | Highland | Cattle, sheep | 11 | 0 | 0 | 0 | 0 |
| V | Highland | Cattle, sheep | 10 | 0 | 0 | 0 | 0 |
| Total | 210 | 15 | 12 | 22 | 41 | ||
AFB, acid-fast bacilli; culture+, positive for M. avium subsp. paratuberculosis.
Livestock with a known history of Johne’s disease (JD) are indicated in boldface type.
Includes two rabbits which had lesions and from which a mycobacterium was cultured that was not IS900 positive or IS901 positive by PCR.
Cultured organism not identified due to dessication of media.
TABLE 2.
Relationship between farms with or without a known history of Johne’s disease and the presence of rabbits from which M. avium subsp. paratuberculosis was cultured or for which lesions were observed with or without acid-fast bacilli
| Rabbit typea | No. of farms with (+) or without (−) history of Johne’s disease
|
||
|---|---|---|---|
| + | − | Total | |
| Culture+ and/or lesion+ | 9 | 2 | 11 |
| Culture− and lesion− | 5 | 6 | 11 |
| Culture+ | 3 | 1 | 4 |
| Culture− | 11 | 7 | 18 |
| Lesion+ and AFB+ | 5 | 1 | 6 |
| Lesion+ and AFB− | 9 | 2 | 11 |
Culture+, positive for M. avium subsp. paratuberculosis; AFB, acid-fast bacilli.
Statistical analysis.
For the statistical evaluation, the farms were divided into two nonexclusive groups: farms with cattle and farms with sheep. The species of ruminant on the farm was considered important, since ovine M. avium subsp. paratuberculosis isolates are notoriously difficult to grow in vitro, a factor which could result in a lower number of isolates from sheep farms. Two sets of variates were examined: rabbits which were culture positive for M. avium subsp. paratuberculosis (a highly specific, minimalist definition of presence) and rabbits which were either culture positive or exhibited lesions on autopsy (a general, maximalist definition of presence, since the lesions could have been caused by other agents). Four situations were evaluated as follows.
(i) Cattle farms, culture-positive rabbits.
Initial analysis showed that the prevalence of M. avium subsp. paratuberculosis infection in rabbits was significantly higher in Tayside than elsewhere in Scotland. There were no discernible differences between samples from any of the other regions included in the study and, therefore, these regions were consolidated into one overall “baseline” region. The effects of region, history of Johne’s disease in cattle, presence of sheep, farm size (acreage), and number of cattle on the farm were evaluated relative to a constant which was deliberately chosen to be equivalent to a farm located outside Tayside that had no history of Johne’s disease in cattle and no sheep. The significance of each term in the analysis was evaluated by using a chi-squared approximation. The analysis indicated that the probability of finding M. avium subsp. paratuberculosis-infected rabbits was greater on farms in Tayside with a previous history of Johne’s disease (P < 0.001). Cattle farms with sheep were more likely to exhibit M. avium subsp. paratuberculosis-infected rabbits (P = 0.008), as were small acreage farms (P = 0.008). Farms with larger numbers of cattle were less likely to exhibit positive rabbits (P = 0.03).
(ii) Cattle farms, culture-positive and/or lesion-positive rabbits.
The same regional divisions and factors were used as for situation i. Once again, there was a significantly greater prevalence of M. avium subsp. paratuberculosis-infected rabbits on cattle farms in Tayside with a past history of Johne’s disease (P = 0.013). The acreage of farm was marginally significant (P = 0.05). The presence of sheep and the number of cattle were not statistically significant. There is the possibility that some of the observed lesions in the rabbits were caused by agents other than M. avium subsp. paratuberculosis and that these agents may be found on certain farms only. This results in clustering of the data and the statistical model having a large dispersion factor.
(iii) Sheep farms, culture-positive rabbits.
Initial examination of the data suggested that a different division of Scotland was appropriate in the case of sheep farms. Lothian, Dumfries, Highland, and Grampian remain grouped together as a control baseline, while Tayside and the Borders were grouped together as a “hot” region. The analysis indicated that sheep farms in the hot region that also maintained cattle had an enhanced probability of exhibiting M. avium subsp. paratuberculosis-infected rabbits, with a highly significant region by cattle interaction (P = 0.013). In the sample of sheep farms studied, it was impossible to identify whether positive rabbits were associated with the presence of cattle or with a history of Johne’s disease in sheep. However, the analysis described above, which assumed that the presence of cattle is the important factor, gave more consistent and more stable results and, therefore, was preferred.
(iv) Sheep farms, all culture-positive and/or lesion-positive rabbits.
With the same regional division and factors as in situation iii, the optimal analysis exhibits similar relationships. A clear regional effect was still apparent (P = 0.003), and rabbit infection was found to be associated with larger cattle numbers (P = 0.036).
As part of the survey, farmers were asked to estimate the size of the wild-rabbit population on their farms (high, medium, or low). The size of the rabbit population was not found to be a risk factor in any of the analyses evaluated.
Molecular typing.
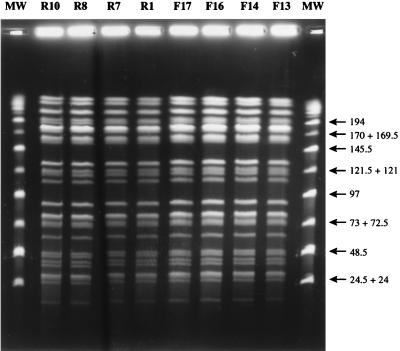
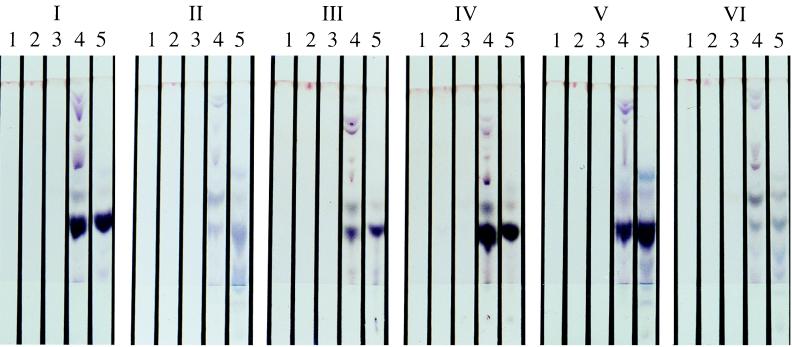
The molecular genetic typing technique PFGE did not detect any differences between the rabbit and cattle isolates from the same farm or between farms. A typical PFGE result with HindIII is shown in Fig. 2. There are currently no published PFGE profiles of M. avium subsp. paratuberculosis restricted with HindIII. The SpeI profiles obtained were identical to the S2 PFGE profile described previously (5). The results of the IS900 RFLP analysis are shown in Fig. 3. IS900 RFLP analysis identified all of the rabbit and cattle isolates, except R8, as RFLP type B-C17, which is the predominant RFLP type found in cattle and sheep strains in the United Kingdom (26). The isolate R8 was found to be RFLP type B-C16. The chemotype profiles of the rabbit and cattle strains revealed minor differences which allowed division of the isolates into six groups as follows (the farm identity is given in parentheses): group I, rabbit isolates R1(A) and R186(C); group II, rabbit isolate R196(C) and cattle isolate F16(A); group III, rabbit isolates R7(A), R8(A), R10(A), R191(C), and R194(C) and cattle isolates JD3(C) and JD4(C); group IV, rabbit isolates R193(C) and R198(C) and cattle isolates JD1(C), JD2(C), and F14(A); group V, cattle isolate F13(A); and group VI, rabbit isolate R197(C). Groups III and IV are so similar that they probably represent the same strain. These two chemotype profiles were represented by both cattle and rabbit isolates and were the most common, comprising 39% (group III) and 28% (group IV) of the isolates. Representative chemotype profiles for the six groups are shown in the composite photograph in Fig. 4. The distribution of chemotype profiles was different on the two farms, as shown in Table 3. There appeared to be no overlap between the chemotype profiles of cattle and rabbit isolates on farm A, whereas on farm C there is an overlap of chemotype groups 3 and 4.
FIG. 2.
Long restriction fragments of M. avium subsp. paratuberculosis isolates from rabbits (R prefix) and cattle (F prefix) from farm A. Chromosomal DNA was digested with HindIII and subjected to PFGE as described in Materials and Methods. MW, PFGE molecular weight markers (Mid-Range II; New England Biolabs); values are indicated in kilobases.
FIG. 3.
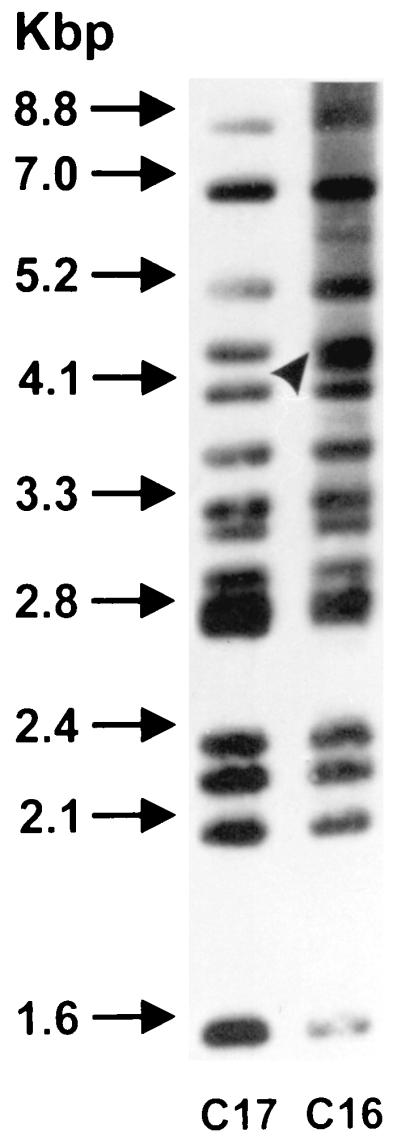
DNA fingerprinting of isolates from rabbits and cattle (DNA types C16 and C17).
FIG. 4.
Composite photograph showing the six representative chemotypes of M. avium subsp. paratuberculosis isolates from rabbits and cattle on farms A and C, as determined by the MSPD-TLC method. Lanes: 1, hexane extract; 2, methylene chloride extract; 3, acetonitrile extract; 4, methanol extract; and 5, H2O extract. Note the close similarity of groups III and IV.
TABLE 3.
Distribution of chemotypes of cattle and rabbit isolates from two positive farmsa
| Farm | Chemotype profile in (host):
|
|
|---|---|---|
| Cattle | Rabbits | |
| A | II, IV, V | I, III |
| C | III, IV | I, II, III, IV, VI |
Values represent the chemotype profile of isolates from each farm.
DISCUSSION
This study has corroborated the findings of the pilot study (7) and confirmed the presence of paratuberculosis in wild rabbits in Scotland. The survey revealed that the Tayside region was a definite hot spot for M. avium subsp. paratuberculosis infection of rabbits. Up to 53% of wild rabbits were found to be infected on farms in this area, where paratuberculosis was a problem in livestock. M. avium subsp. paratuberculosis also was cultured from a rabbit on a farm in the Borders region, which is geographically distant from Tayside.
Analysis of the data showed a statistically significant relationship between a past or current problem of paratuberculosis in cattle and in the wild rabbit population on the Tayside farms. This could be interpreted as evidence for interspecies transmission or it may be simply a further reflection of the regional prevalence of M. avium subsp. paratuberculosis in both host species. No evidence was found of a positive relationship between a history of Johne’s disease in sheep and the occurrence of paratuberculosis in rabbits. However, this should not be taken as evidence that no such relationship exists, since the result may be a consequence of having cultured fewer isolates from rabbits on sheep farms due to the difficulties in isolating and growing ovine M. avium subsp. paratuberculosis strains and the relatively small number of farms in the survey.
From the disease control perspective, the important question is whether M. avium subsp. paratuberculosis can be transmitted from rabbits to farm ruminants. Various typing techniques were employed to determine whether there were two distinct, noninteractive populations of M. avium subsp. paratuberculosis in the livestock and the rabbits or whether one population existed. The latter finding would support the concept of interspecies transmission. The rabbit and cattle isolates of M. avium subsp. paratuberculosis were morphologically indistinguishable and had comparable growth rates in vitro on primary isolation. Molecular genetic typing could not discriminate between rabbit and cattle isolates from the same or different farms, suggesting that a single strain may be responsible for the disease in either host and that interspecies transmission could have occurred. Unfortunately, it was not possible to compare sheep isolates with cattle and rabbit isolates from farm C due to difficulties encountered in culturing these isolates in vitro. IS900 RFLP analysis is currently the most successful technique for typing M. avium subsp. paratuberculosis and is considered to be the only technique with sufficient discriminatory power to be used in epidemiological studies (27, 28). However, there appears to be little variation in the genomic fingerprints of these isolates, which may indicate that M. avium subsp. paratuberculosis exhibits little diversity or simply that current techniques are inadequate for detecting genetic variation in populations of this organism.
It was possible to discriminate between isolates according to their chemotype profile. Six chemotype profiles were identified, and a difference in their distribution between cattle and rabbit isolates on the two farms was observed. On farm C there was overlap in the distribution of chemotypes III and IV between cattle and rabbit isolates, thus indicating that interspecies transmission could have occurred. On farm A, the chemotype profiles of the cattle and rabbit isolates were different, suggesting the existence of two independent populations of M. avium subsp. paratuberculosis associated with the two host species. However, if chemotype groups III and IV represent the same strain, interspecies transmission could have occurred. It will be necessary to phenotype a larger number of isolates to evaluate the epidemiological relationship between the cattle and rabbit isolates on these farms.
Since molecular typing per se cannot prove that interspecies transmission occurs, it will be necessary to demonstrate interspecies transmission experimentally under controlled conditions. Preliminary experiments have shown that lambs experimentally infected with a rabbit isolate develop lesions (unpublished observations). Further experiments are in progress.
This study highlights the fact that M. avium subsp. paratuberculosis has a broad host range. The organism principally affects ruminants, but the infection of lagomorphs may be more common than was first thought. There have been previous reports of the isolation of a mycobactin-dependent mycobacterium from the European hare (19) and lesions containing acid-fast bacilli in a wild rabbit in Scotland (1). More recently, culture and PCR analysis have confirmed the presence of M. avium subsp. paratuberculosis in a single wild rabbit in Spain (29). Other monogastrics known to have become infected include macaques (20) and humans (8). A variety of laboratory animals, such as mice (25, 37), rats (12), hamsters (6), gerbils (16), guinea pigs (21), and chickens (15), have been infected experimentally with M. avium subsp. paratuberculosis. However, although the mycobacterium replicates in some of these hosts, it does not produce the pathology and clinical signs characteristic of paratuberculosis in ruminants.
ACKNOWLEDGMENTS
We thank Kathleen Connor, Amanda Pirie, and Karen Rudge for maintaining the strain collection, Lenka Dvorska for the DNA fingerprinting, and Lisa Whittington for assistance with the chemotype profiles.
This work was funded by the Scottish Office Agriculture, Environment and Fisheries Department; the Animal Health Trust; the Ministry of Agriculture of the Czech Republic (grant number EP0960006087); and the Veterinary Medical Experiment Station, University of Georgia, Tifton, Ga.
REFERENCES
- 1.Angus K W. Intestinal lesions resembling paratuberculosis in a wild rabbit (Oryctolagus cuniculus) J Comp Pathol. 1990;103:101–105. doi: 10.1016/s0021-9975(08)80140-5. [DOI] [PubMed] [Google Scholar]
- 2.Challans J A, Stevenson K, Reid H W, Sharp J M. A rapid method for the extraction and amplification of Mycobacterium paratuberculosis DNA from clinical samples. Vet Rec. 1994;134:95–96. doi: 10.1136/vr.134.4.95. [DOI] [PubMed] [Google Scholar]
- 3.Chiodini R J, van Kruiningen H J, Merkal R S. Ruminant paratuberculosis (Johne’s disease): the current status and future prospects. Cornell Vet. 1984;74:218–262. [PubMed] [Google Scholar]
- 4.Chiodini R J, Van Kruiningen H J. Eastern white-tailed deer as a reservoir of ruminant paratuberculosis. J Am Vet Med Assoc. 1983;182:168–169. [PubMed] [Google Scholar]
- 5.Feizabadi M M, Robertson I D, Hope A, Cousins D V, Hampson D J. Differentiation of Australian isolates of Mycobacterium paratuberculosis using pulsed-field gel electrophoresis. Aust Vet J. 1997;75:887–889. doi: 10.1111/j.1751-0813.1997.tb11259.x. [DOI] [PubMed] [Google Scholar]
- 6.Gilmour N L, Campbell J, Brotherstone J G. The pathogenesis of Mycobacterium johnei in orally dosed hamsters. J Comp Pathol Ther. 1963;73:98–106. doi: 10.1016/s0368-1742(63)80013-2. [DOI] [PubMed] [Google Scholar]
- 7.Greig A, Stevenson K, Perez V, Pirie A A, Grant J M, Sharp J M. Paratuberculosis in wild rabbits (Oryctolagus cuniculus) Vet Rec. 1997;140:141–143. doi: 10.1136/vr.140.6.141. [DOI] [PubMed] [Google Scholar]
- 8.Hermon-Taylor J, Moss M, Tizard M, Malik Z, Sanderson J. Molecular biology of Crohn’s disease mycobacteria. Balliere’s Clin Gastroenterol. 1990;4:23–42. doi: 10.1016/0950-3528(90)90037-h. [DOI] [PubMed] [Google Scholar]
- 9.Hillermark K. A disease resembling paratuberculosis (Johne’s disease) in roe deer (Capreolus capreolus L.). An aetiological and pathological anatomical study. Acta Vet Scand. 1966;7:330–363. doi: 10.1186/BF03547122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hines M E, II, Frazier K S. Differentiation of mycobacteria on the basis of chemotype profiles by using matrix solid-phase dispersion and thin-layer chromatography. J Clin Microbiol. 1993;31:610–614. doi: 10.1128/jcm.31.3.610-614.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hines M E, II, Long A R, Snider III T G, Barker S A. Lysis and fractionation of Mycobacterium paratuberculosis and Escherichia coli by matrix solid-phase dispersion. Anal Biochem. 1991;195:197–206. doi: 10.1016/0003-2697(91)90317-m. [DOI] [PubMed] [Google Scholar]
- 12.Hole N H. Johne’s disease. Adv Vet Sci. 1958;4:341–387. [Google Scholar]
- 13.Jessup D A, Abbas B, Behymer D, Gogan P. Paratuberculosis in tule elk in California. J Am Vet Med Assoc. 1981;179:1252–1254. [PubMed] [Google Scholar]
- 14.Kunze Z M, Portaels F, McFadden J J. Biologically distinct subtypes of Mycobacterium avium differ in possession of insertion sequence IS901. J Clin Microbiol. 1992;30:2366–2372. doi: 10.1128/jcm.30.9.2366-2372.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen A B, Moon H W. Experimental Mycobacterium paratuberculosis infection in chickens. Am J Vet Res. 1972;33:1231–1235. [PubMed] [Google Scholar]
- 16.Larsen A B, Millar J M, Kopecky K E. Susceptibility of the Mongolian gerbil (Meriones unguiculatus) to Mycobacterium paratuberculosis. Am J Vet Res. 1976;37:1113–1114. [PubMed] [Google Scholar]
- 17.Levy-Frebault V V, Thorel M F, Varnerot A, Gicquel B. DNA polymorphism in Mycobacterium paratuberculosis, wood pigeon mycobacteria and related mycobacteria analyzed by field-inversion gel electrophoresis. J Clin Microbiol. 1989;27:2823–2826. doi: 10.1128/jcm.27.12.2823-2826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Libke K G, Walton A M. Presumptive paratuberculosis in a Virginia white-tailed deer. J Wildl Dis. 1975;11:552–553. doi: 10.7589/0090-3558-11.4.552. [DOI] [PubMed] [Google Scholar]
- 19.Matthews P R, Sargent A. The isolation of mycobacteria from the brown hare (Lepus europaeus) Br Vet J. 1977;133:399–404. doi: 10.1016/s0007-1935(17)34041-1. [DOI] [PubMed] [Google Scholar]
- 20.McClure H M, Chiodini R J, Anderson D C, Swenson R B, Thayer W R, Coutu J A. Mycobacterium paratuberculosis infection in a colony of stumptail macaques (Macaca arctoides) J Infect Dis. 1987;155:1011–1019. doi: 10.1093/infdis/155.5.1011. [DOI] [PubMed] [Google Scholar]
- 21.Mohler W M. Johne’s disease infection of laboratory animals with aid of paraffin oil. J Am Vet Med Assoc. 1939;94:590–594. [Google Scholar]
- 22.Mokresh A H, Butler D G. Granulomatous enteritis following oral inoculation of newborn rabbits with Mycobacterium paratuberculosis of bovine origin. Can J Vet Res. 1990;54:313–319. [PMC free article] [PubMed] [Google Scholar]
- 23.Mokresh A H, Czuprynski C J, Butler D G. A rabbit model for study of Mycobacterium paratuberculosis infection. Infect Immun. 1989;57:3798–3807. doi: 10.1128/iai.57.12.3798-3807.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mondal D, Sinha R P. Pathogenicity of caprine strain of Mycobacterium paratuberculosis in rabbit. Ind J Anim Sci. 1992;62:1117–1120. [Google Scholar]
- 25.Mutwiri G K, Butler D G, Rosendal S, Yager J. Experimental infection of severe combined immunodeficient beige mice with Mycobacterium paratuberculosis of bovine origin. Infect Immun. 1992;60:4074–4079. doi: 10.1128/iai.60.10.4074-4079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavlik, I., A. Horvathova, L. Dvorska, P. Svastova, R. du Maine, B. Fixa, and I. Rychlik. Unpublished results.
- 27.Pavlik, I., A. Horvathova, L. Dvorska, J. Bartl, P. Svastova, R. du Maine, and I. Rychlik. Restriction-fragment-length polymorphisms with the probe IS900 in Mycobacterium avium subspecies paratuberculosis. J. Microbiol. Methods, in press. [DOI] [PubMed]
- 28.Pavlik I, Bejckova L, Pavlas M, Rozsypalova Z, Koskova S. Characterisation by restriction endonuclease analysis and DNA hybridisation using IS900 of bovine, ovine, caprine and human mycobactin-dependent strains of Mycobacterium paratuberculosis isolated in various localities. Vet Microbiol. 1995;45:311–318. doi: 10.1016/0378-1135(94)00130-o. [DOI] [PubMed] [Google Scholar]
- 29.Perez-Perez, V., and F. Garcia-Marin. Personal communication.
- 30.Riemann H, Zaman M R, Ruppanner R, Aalund O, Jorgensen J B, Worsaae H, Behymer D. Paratuberculosis in cattle and free-living exotic deer. J Am Vet Med Assoc. 1979;174:841–843. [PubMed] [Google Scholar]
- 31.Ris D R, Hamel K L, Weaver A M. Natural transmission of Johne’s disease to feral goats. N Z Vet J. 1988;36:98–99. doi: 10.1080/00480169.1988.35496. [DOI] [PubMed] [Google Scholar]
- 32.Ris D R, Hamel K L, Ayling J M. Can sheep become infected by grazing pasture contaminated by cattle with Johne’s disease? N Z Vet J. 1987;35:137. doi: 10.1080/00480169.1987.35414. [DOI] [PubMed] [Google Scholar]
- 33.Sanderson J, Moss M T, Tizard M L V, Hermon-Taylor J. Mycobacterium paratuberculosis DNA in Crohn’s disease tissue. Gut. 1992;33:890–896. doi: 10.1136/gut.33.7.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer W B. Serologic identification and classification. Am Rev Respir Dis. 1965;92(Suppl.):85–93. doi: 10.1164/arrd.1965.92.6P2.85. [DOI] [PubMed] [Google Scholar]
- 35.Sharp J M, Stevenson K, Challans J A, Ramage C, Hitchcock D, Reid H W. Proceedings of the Fifth International Colloquium on Paratuberculosis. International Association for Paratuberculosis, Inc., East Providence, R.I. 1996. Mycobacterial infections of free-living deer in Scotland; pp. 180–182. [Google Scholar]
- 36.Sweeney, R. W. (ed.). 1996. Paratuberculosis (Johne’s disease). Vet. Clin. North Am. Food Anim. Pract. 12:No. 2. [DOI] [PubMed]
- 37.Tanaka S, Sato M, Taniguch T, Yokomizo Y. Histopathological and morphometrical comparison of granulomatous lesions in BALB/c and C3H/HeJ mice inoculated with Mycobacterium paratuberculosis. J Comp Pathol. 1994;110:381–388. doi: 10.1016/s0021-9975(08)80315-5. [DOI] [PubMed] [Google Scholar]
- 38.Touchstone J C, Dobbins M F. Practice of thin-layer chromatography. 2nd ed. New York, N.Y: Wiley Interscience; 1983. pp. 9–15. [Google Scholar]
- 38a.Veterinary Laboratories Agency. Veterinary Investigation Diagnostic Analysis report, 1991–1993. 1993. Veterinary Laboratories Agency, Addlestone, United Kingdom. [Google Scholar]
- 39.Williams E S, Spraker T R. Annual Proceedings of the AAZV. Denver, Colo: American Association of Zoo Veterinarians; 1979. Paratuberculosis in free-ranging bighorn sheep and a Rocky Mountain goat with a brief review of the disease in wild species; pp. 122–124. [Google Scholar]
- 40.Williams E S, Snyder S P, Martin K L. Pathology of spontaneous and experimental infection of North American wild ruminants with Mycobacterium paratuberculosis. Vet Pathol. 1983;20:274–290. doi: 10.1177/030098588302000304. [DOI] [PubMed] [Google Scholar]