Abstract
In vivo time lapse imaging of complete dendritic arbor structures in tectal neurons of Xenopus laevis tadpoles has served as a powerful in vivo model to study activity-dependent structural plasticity in the central nervous system during early development. In addition to quantitative analysis of gross arbor structure, dynamic analysis of the 4-dimentional data offers particularly valuable insights into the structural changes occurred in subcellular domains over experience/development-driven structural plasticity events, allowing not only quantifiable characterization of branch additions and retractions with high temporal resolution, but also identification of the loci of action. Such analysis provides critical information for a better understanding of the spatial-temporal association of structural changes to functional relevance. Here we describe a protocol for in vivo time lapse imaging of complete dendritic arbors from individual neurons in the brain of anesthetized tadpoles with 2-photon microscope and data analysis of the time series of 3D dendritic arbors. For data analysis, we focus on dynamic analysis of reconstructed neuronal filaments using a customized open source computer program we developed (4D SPA), which allows aligning and matching of 3-D neuronal structures across different time points with greatly improved speed and reliability. File converters are provided to convert reconstructed filament files from commercial reconstruction software to be used in 4D SPA. The program and user manual are publicly accessible and operate through a graphical user interface on Windows and Mac OSX.
Reagents
0.1X Steinberg’s solution: 58.0 mM NaCl, 0.67 mM KCl, 0.34 mM Ca(NO3)2, 0.83 mM MgSO4, 3.0 mM HEPES, pH 7.2
MS-222 solution: 0.01% MS222 (3- aminobenzoic acid ethyl ester, Sigma-Aldrich) in 0.1% Steinberg’s solution.
Fast-green solution: 0.1% Fast green (FCF, Sigma-Aldrich) in ddH2O.
DNA plasmid (1–3μg/μl, in ddH2O and mixed with fast-green): For time lapse structural imaging of complete dendritic arbor, GFP or other fluorescent protein expressing DNA constructs can be used. Amplification systems such as the Gal4-UAS binary system are recommended to ensure high expression for GFP. The Gal4 construct can be either subcloned into the same DNA construct or co-electroporated with the UAS-E1b-eGFP construct, which has a minimal e1b promoter under 14 repeats of the gal4 response element UAS.
Sylgard™ 184 (Electron Microscopy Sciences)
SeaPlaque Low-melting temperature agarose (Lonza)
Equipment
Two-photon microscope with a water-emersion objective (20x, NA 0.95).
We use a two-photon microscope custom modified from an Olympus (BX51) FV300 microscope (Bestman et al., 2008). GFP signal is two-photon excited by 910 nm laser generated by a Chameleon Ultra II laser (Coherent, Santa Clara, CA). Emitting fluorescence signal was detected by a R3896 photomultiplier tube (Hamamatsu, Shizuoka, Japan), mounted to the side of a filter cube equipped with a 700LP dichroic mirror and a T560LPXR beam splitter and ET510/80M-2P; ET605/70M-2P filters (Chroma Technology, Brattleboro, VT) optimized for the detection of GFP and RFP signals. The objective was a 20x water immersion objective (Olympus XLUMPlanFL 0.95NA).
Epifluorescence microscope for screening
Computer (PC or Mac)
Method
Animal care
Albino Xenopus laevis embryos are reared in 0.1X Steinberg’s solution at 22°C with 12 hrs dark/12 hrs light cycles. Animals should be fed with tadpole food from stage 47 (Nieuwkoop and Faber 1956), when the yolk completely disappears (McKeown and Cline 2019).
Starting from the first day of imaging, animals should be housed individually in 6-well plates to keep track of individual animals (and neurons) imaged.
Tectal Cell electroporation
-
1
Electroporate animals with GFP-expressing DNA constructs at stage 45–46. Two methods are commonly used to sparsely label individual tectal neurons with fluorescent protein for time-lapse imaging (Ruthazer et al. 2013): single cell electroporation (Bestman et al. 2006); or whole brain electroporation (Haas et al. 2002; Bestman et al. 2012). To achieve sparse labeling with whole brain electroporation, moderate concentration of DNA plasmids (1–3μg/μl), lower voltage (34V), and fewer (1–2) pulses are preferred.
-
2
Screen electroporated animals 1–7days post electroporation for well isolated tectal neurons with high GFP expression that fills the whole neuron including all dendritic processes. The best ones to be used for subsequent structural imaging are those with 1 brightly transfected GFP+ neuron in each tectal lobe.
In vivo time-lapse imaging of complete dendritic arbor
-
3
Anesthetize the animal by immersing it in 0.01% MS-222 solution for one minute. Move the anesthetized animal in 0.01% MS-222 to a Sylgard chamber that is shaped to snuggly fit the tadpole’s body. Seal the chamber tightly with a glass cover slip to prevent movement of the animal during imaging. Animals can stay healthy in the chamber for up to 10 minutes and should be removed from the chamber after each imaging session. For time-lapse imaging with shorter intervals, it is better to embed animals in 1% low melting temperature agarose and perfuse with rearing solution (Ruthazer et al. 2013).
-
4
Take z-stack image of the whole neuron under the two-photon microscope. Ensure that the complete dendritic arbor is encompassed. Collect time-lapse image stacks at regular intervals (can range from 10min to 24hours). Use the same zoom factor and Z-step size (we use 1um) throughout the time series. Choose the zoom factor for the first time point wisely by leaving room for potential neuronal growth. Maintain the laser intensity constant. Each time when fitting the animal in the Sylgard chamber for imaging, use care to position the animal the same way as in the previous time point to help with alignment of images in subsequent analysis.
Structural data reconstruction
-
5Reconstruction of the complete dendritic arbor from raw image stacks can be accomplished with multiple commercially available and open-source software.We use the semi-manual function in the Filament module of Imaris (Bitplane, US) to trace the dendritic arbor in the 3D image stack. Once the reconstruction is completed, the software generates measurements for basic filament characteristics, such as quantifications of arbor size and complexity (total branch tip number, total branch length, branch order, Sholl analysis, etc). An example of a 3D image of a GFP-labeled neuron and the reconstructed dendritic arbor is shown in online resource Video 1.
Dynamic Analysis of the 4D time-lapses structural data
We use a customized C++ software 4DSPA (Lee et al. 2013) to analyze the dynamic changes (growth and retraction of individual branches, branch tip dynamics, etc) of the reconstructed dendritic arbors across different time points.
-
6
Data preparation. Export the full filament data set as txt data file to be used as inputs for 4D SPA. In the txt filament file, the branches are represented as data points defined by their 3D location (coordinates) and is organized block-wise. Each block starts with a line defining the branch index (P #) and the number of nodes in that branch (N #). The first point is the starting (branching) point of the branch, and the last point in the block is the branch tip. The first point in the first block of the file represents the soma position. Converting tools (HOCtoBCF and ASCtoBCF) are available through supplemental online resources to convert reconstructed filament files generated from two of the most commonly used reconstruction software programs (.hoc file in Imaris and .asc file in Neurolucida) to txt files that meet the requirement for 4D SPA. The converting tools run in the terminal of any operation system. In cases where the soma location is not the first point in the first block of the filament file, the coordinates of the actual soma location can be specified as input parameters in the converting command to allow reorganizing of the filament file structure to meet the requirements for 4D SPA. Reconstructed filament data files in txt format generated by other reconstruction software should also be usable as long as the data structure abides by the above-mentioned rules as in BCF format.
-
7
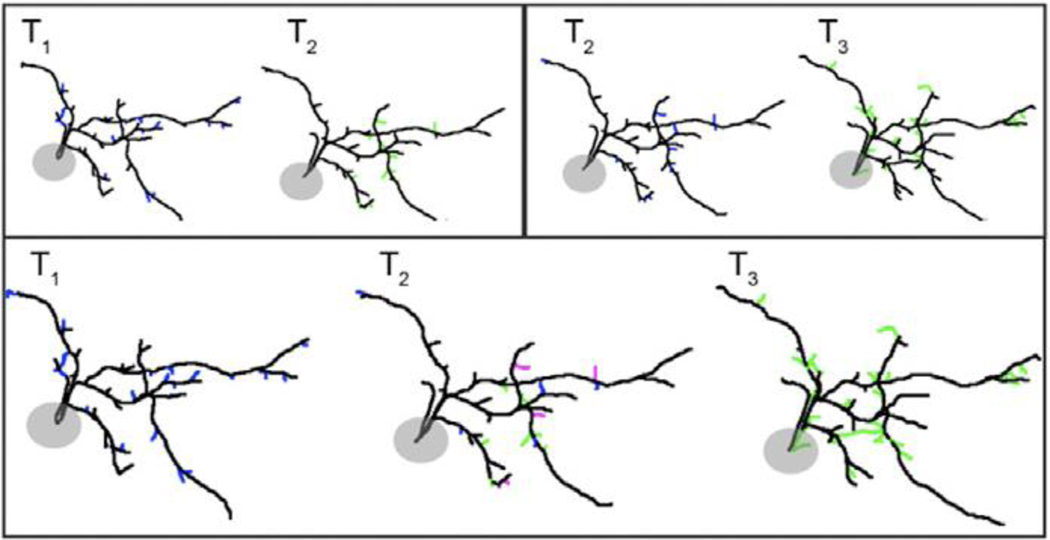
Pairwise matching analysis. Load the pair of reconstructed filament txt files of the same neuron into the 4DSPA software for alignment. Usually, these should be filaments traced from image stacks acquired at two consecutive time points (T1 and T2). The software will automatically highlight the branch to be aligned (matched) in T1 in the order of branch length (from longest to shortest). For each branch in T1, a list of candidate matching branches in the T2 filament will be provided, in the order of calculated matching score. User can choose the best matching branch and confirm the match by clicking the ‘add’ button. For branches in T1 that do not have a matching branch in T2, the software will still provide a candidate matching list, it is up to the user’s discretion to designate the branch as retracted by clicking the ‘retract’ button. After every branch in T1 is designated either as ‘matched’ or ‘retracted’, all of the un-matched branches in T2 will be automatically designated as ‘newly-added’. If any mistake was made during the matching process, the user can go back to the branch in question in T1 and reassign the proper matching branch (or designate as ‘retracted’). If the re-assigned matching branch had been assigned to another T1 branch, the new operation will overwrite the previous matching result and render the other T1 branch ‘unmatched’, which will be prompted for re-matching. For an example of the matching process, see online resource video 2. One tip to double check for matching mistakes is to examine the matching results with the color coding on. If any branch appears at the same locations in T1 and T2 but is labeled as ‘retracted’ in T1 and ‘newly-added’ in T2, it is likely a stable branch mis-categorized. Once the alignment is completed for the pair, save the results by clicking ‘SaveFiles’ – ‘Reports’. This will generate a separate ‘T2_matched.txt’ file for the T2 filament. Do this pairwise matching analysis for the next two time points (e.g. T2 and T3). Use the new ‘T2_matched.txt’ file instead of the original T2.txt file for T2. Examples of the pairwise matching analysis results are shown in Fig1 (top panel).
-
8
Serial dynamic analysis. Serial analysis of sequential images in a multi-time-point time-lapse data set requires that branches have unique identifiers that are maintained through the image series. This is achieved through serial pair wise matching analysis: T1 vs T2, T2 vs T3, and so on. Upon the completion of each pairwise analysis, the 4D SPA program adjusts the indices so that matching (stable) branches are assigned with the same identifier that is consistent with the starting time point indices. To combine the multiple pairwise alignment results in data sets with more than 2 time points, run ‘MultiplePairAnalysis’ in the terminal window with the report files generated by each pairwise matching analysis within the whole time series (*.rep.txt). A spreadsheet.txt file will be generated, in which branches are categorized as stable, newly added, retracted or transient. A stable branch is a branch that is present at all time-points. A retracted branch is a branch that is present at the first time-point but is lost at a later time-point. A ‘newly added’ branch is a branch that does not exist at the first time-point but appears later and remains in place until the end time-point. The ‘added’ branch category includes those that appear at the last time point. A transient branch is a branch that appears at a time-point after the first time-point and is then retracted by the last time-point (Fig 1, bottom panel). The output file is a .txt file (default ‘spreadsheet.txt’). An example of the data format is shown below. In addition to categorization of the branches, the dynamic changes (growth or retraction) of the stable branches can be analyzed subsequently. This spreadsheet file can also be loaded into 4DSPA together with the matched neuron files to generate filaments images with branches color coded by their categories (as shown in Fig 1, bottom panel). New neuron files (*_mat.txt) can then be saved with the branch category information to be used for further analysis in Matlab. An example of the format of the output spreadsheet file is shown below.
Total Time points <int>
BranchID BranchType BaseBranchLength ΔBranchLength@T0, …, Tn
Example:
3
index type base T0 T1 T2
0 Stable 131.658 0 −1.809 −8.333
1 Stable 130.953 0 −2.664 −8.099
…
27 Retracted 42.5477 0 −4.5266 #
28 Retracted 41.2327 0 −5.3916 −2.7764
…
65 Transient 39.9811 * 4.0722 −4.0722
…
94 Newly_Added 12.8521 * * 3.8426
Figure 1.
Example of dynamic analysis of reconstructed complete dendritic arbors of the same neuron in a time series. Top panels: pairwise matching analysis (T1 versus T2; T2 versus T3). Bottom panels: serial analysis. Dendritic branches are color-coded by their dynamic categories: black (stable branches), blue (retracted branches), green (newly added branches), magenta (transient branches). Adapted from (He et al. 2016)
Discussion
Quantitative analysis of time-lapse data requires particular care be taken in the initial 3D reconstructions of the neuronal structures, so errors in the identification of branch dynamic events do not arise from misidentification of branches throughout the imaging sequence. Recently, new reconstruction software has been made available that allows modification of the reconstructed filament file from the previous time point (Feng et al. 2015). This significantly facilitates the matching and alignment process by having the reconstructed filaments of the same neuron done in the same primary structure frame work. Thus, potentially eliminating the necessity of alignment when it comes to 4D analysis. This is especially useful when reconstructing time series collected with short time intervals, with the major arbor structure remaining stable across time points. On the other hand, 4D SPA can be used for dynamic analysis on data not suitable for such serial reconstruction, as well as data already collected and reconstructed and avoid the time-consuming steps of redoing the reconstruction in the newer software. Further analysis of the 3D location of the dynamic branches is also possible by combining the MultiplePairAnalysis result with the matched txt files from the pairwise alignment.
Supplementary Material
Video 1 – example of a 3D image of a GFP-labeled neuron and the reconstructed dendritic arbor.
Video 2 – an example of the matching process using 4D SPA.
Footnotes
Online resources:
Download link for 4D SPA and accessory programs: https://4dspa.web.nctu.edu.tw/
Contributor Information
Hai-yan He, Department of Biology, Georgetown University, Washington DC, USA 20057.
Chih-Yang Lin, Department of Optoelectronics and Materials Engineering, Chung Hua University, Hsinchu, Taiwan 30012.
Hollis T. Cline, Neuroscience Department, Dorris Neuroscience Center, Scripps Research Institute, La Jolla, CA USA 92037
References:
- Bestman JE, Ewald RC, Chiu SL, Cline HT. 2006. In vivo single-cell electroporation for transfer of DNA and macromolecules. Nat Protoc 1: 1267–1272. [DOI] [PubMed] [Google Scholar]
- Bestman JE, Lee-Osbourne J, Cline HT. 2012. In vivo time-lapse imaging of cell proliferation and differentiation in the optic tectum of Xenopus laevis tadpoles. J Comp Neurol 520: 401–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Zhao T, Kim J. 2015. neuTube 1.0: A New Design for Efficient Neuron Reconstruction Software Based on the SWC Format. eNeuro 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas K, Jensen K, Sin WC, Foa L, Cline HT. 2002. Targeted electroporation in Xenopus tadpoles in vivo--from single cells to the entire brain. Differentiation 70: 148–154. [DOI] [PubMed] [Google Scholar]
- He HY, Shen W, Hiramoto M, Cline HT. 2016. Experience-Dependent Bimodal Plasticity of Inhibitory Neurons in Early Development. Neuron 90: 1203–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PC, He HY, Lin CY, Ching YT, Cline HT. 2013. Computer aided alignment and quantitative 4D structural plasticity analysis of neurons. Neuroinformatics 11: 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown CR, Cline HT. 2019. Nutrient restriction causes reversible G2 arrest in Xenopus neural progenitors. Development 146: dev178871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. 1956. Normal table of Xenopus laevis (Daudin); a systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. North-Holland Pub. Co., Amsterdam,. [Google Scholar]
- Ruthazer ES, Schohl A, Schwartz N, Tavakoli A, Tremblay M, Cline HT. 2013. In vivo time-lapse imaging of neuronal development in Xenopus. Cold Spring Harb Protoc 2013: 804–809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1 – example of a 3D image of a GFP-labeled neuron and the reconstructed dendritic arbor.
Video 2 – an example of the matching process using 4D SPA.